Recent studies have found that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) could be detected in fecal samples and that the live virus could be successfully isolated from stool and used for successful infection in cell cultures, indicating a possible fecal-oral transmission.1, 2, 3 Although important for disease management, information on the time sequences of oral and fecal viral shedding in patients with coronavirus disease 2019 (COVID-19) needs more investigation.1 , 4 In this study, we tested in 401 patients with COVID-19 for viral RNA in both respiratory and rectal specimens and tracked them for more than 7 weeks to present a clear elucidation of the respiratory and rectal viral shedding profile, such as the duration, viral load, and relationship to patient symptoms.
Methods
A retrospective cohort of 401 patients with laboratory-confirmed COVID-19 admitted to Shenzhen Third People’s Hospital in China were included in this study. Viral RNA of SARS-CoV-2 was tested by quantitative reverse-transcription polymerase chain reaction in both the respiratory and rectal specimens. The results were further analyzed in combination with clinical data. Patients were divided into severe and nonsevere groups based on the disease severity defined according to China National Health Commission’s “Guidelines for Diagnosis and Treatment of SARS-CoV-2 Infection,” seventh version.5 The study was approved by the Third People’s Hospital of Shenzhen Ethics Committee (2020-105). Further details are provided in the Supplementary Methods.
Results
Positive Detection of Severe Acute Respiratory Syndrome Coronavirus 2 RNA in Rectal Swabs Lasted for 7 Weeks
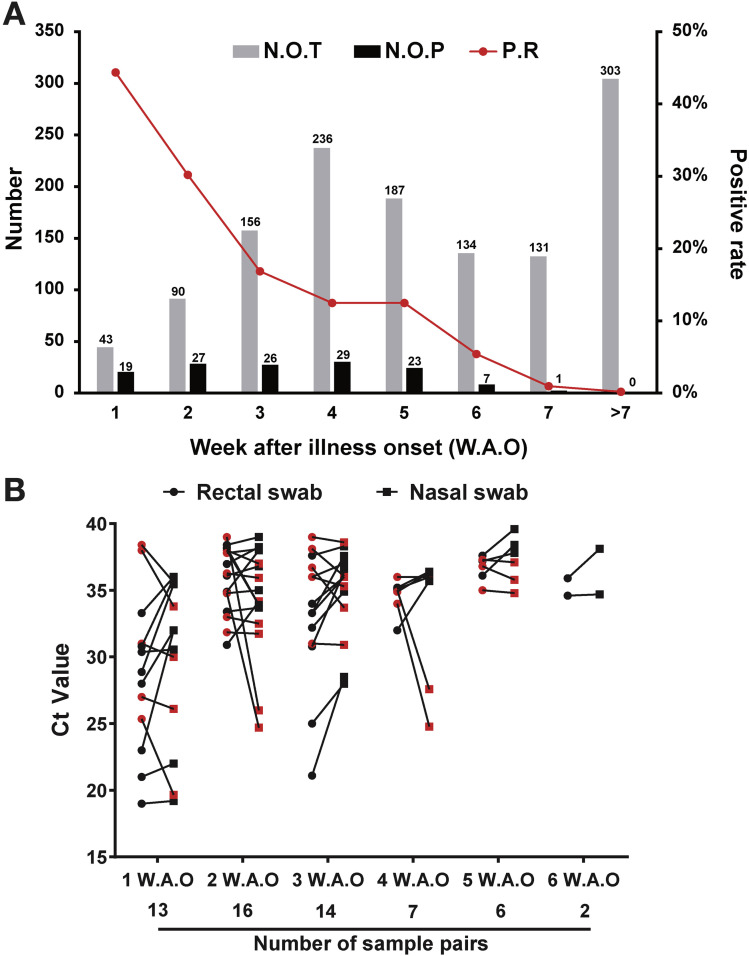
A total of 1758 rectal swabs from 401 patients with COVID-19 were collected during 0 to 98 days after illness onset (median, 33 d; interquartile range, 23–52). Grouping the rectal samples according to the collection timepoint, quantitative reverse-transcription polymerase chain reaction assay for SARS-CoV-2 RNA showed 44.19%, 30%, 16.67%, 12.29%, 12.3%, 5.22%, 0.76%, and 0% positive for 1–7 weeks after illness onset (WAO) and >7 WAO, respectively (Figure 1 A). Associating the sample testing results to the individual patients, altogether, 80 out of 401 (19.95%) patients with COVID-19, including 32 (40%) male and 48 (60%) female patients, tested positive for SARS-CoV-2 in the rectal swabs (Supplementary Table 1), suggesting that the collection time (WAO) of the rectal swabs affects the testing result. Pediatric (<18 y) patients had a positive rate of 56.67% compared to 16.98% in adults (≥18 y) patients. The clinical characteristics were similar between patients with rectal samples that were negative for viral RNA (n = 321) and those with rectal samples that were positive (n = 80) (Supplementary Table 1).
Figure 1.
Time sequences of the respiratory and rectal viral shedding of patients with COVID-19. (A) Detection of viral RNA in rectal samples collected at different timepoints after illness onset. The sample collection times were stratified into 8 groups according to WAO, as indicated in the figure. (B) Comparison of the viral loads between rectal and nasal swabs. Paired samples collected on the same day with double positive results were used for the comparison, and higher Ct values indicated lower viral loads. The pairs with higher viral loads in the nasal swabs are marked in red. Ct, cycle threshold; N.O.T, number of tested patients; N.O.P, number of patients with positive results; P.R, positivity rate.
Paired Rectal and Respiratory Samples Showed More Positive Cases and Higher Viral Load in Rectal Swabs Than Respiratory Samples
Respiratory and rectal samples collected at the same day were defined as a pair, and 517 pairs were collected from the 80 patients who tested positive for SARS-CoV-2 RNA on rectal swabs. Overall, 58 (11.8%) pairs were double positive, 112 (21.7%) pairs were positive in the rectal swab but negative in the respiratory swab, and 40 (7.7%) pairs were positive in the respiratory swab but negative in the rectal swab. Most of the pairs with positive results in only the rectal swab were found within 5 WAO (Supplementary Table 2). The overall coincidence rate of the respiratory and rectal samples was 70.6%, and this gradually increased during the disease progression (Supplementary Table 2). At weeks 1–6 WAO, 11 (61.54%), 9 (56.25%), 9 (64.29%), 4 (51.14%), 3 (50%), and 2 (100%) pairs with higher viral loads in the rectal than respiratory samples were found, respectively (Figure 1 B).
Factors Associated With the Duration of Rectal Viral Shedding
Patients positive for rectal viral RNA were divided into 2 groups according to the durations of rectal viral shedding, including the >4 weeks group and ≤4 weeks group. Using a multivariate logistic regression model, we found that both the neutrophil levels and the intervals between antiviral treatment and illness onset were independently associated with the duration of rectal viral shedding. The odds ratios for the neutrophil levels and the intervals between antiviral treatment and illness onset were 1.549 (95% confidence interval, 1.055–2.405; P = .034) and 1.168 (95% confidence interval: 1.011–2.369; P = .042), respectively.
Discussion
Transmission routine varies in different viral infections, and elucidation of the viral shedding profile is crucial for the diagnosis, treatment, and control of COVID-19. The positive rate of rectal SARS-CoV-2 RNA in children (<18 years) was significantly higher than in adults. SARS-CoV-2 has a strong transmission capacity in children, but the clinical presentation of COVID-19 was generally milder in children than adults.6 , 7 Therefore, viral diagnosis in rectal specimens is crucial in the diagnosis and management of COVID-19 in children. Patients with COVID-19, whether with positive or negative rectal viral RNA, showed no difference in gastrointestinal symptoms, suggesting that SARS-CoV-2 may use the intestine as a reservoir without altering the intestine functions. Histologic and immunofluorescent imaging of a the gastrointestinal tissues of a patient with COVID-19 supports this hypothesis,8 and the underlying mechanisms merit further investigation.
Notably, we found that SARS-CoV-2 RNA in rectal samples remained for an unexpectedly long period, with a higher positive rate and higher viral load than the paired respiratory samples. The longest duration observed was 43 days, much longer than the usual 3–5 weeks from symptom onset to discharge for most patients,4 suggesting that SARS-CoV-2 test of rectal swabs is crucial to minimize false negatives for the laboratory diagnosis of COVID-19 and to reduce the potential fecal-oral transmission from asymptomatic patients with COVID-19. An obvious difference in the interval from illness onset to antiviral therapy was observed, raising the possibility that early antiviral treatment might reduce SARS-CoV-2 persistence within the intestinal tract.
Our study provides a comprehensive respiratory and rectal viral shedding profile of a large cohort of patients with COVID-19, with the longest tracking duration to our knowledge. The gastrointestinal viral reservoir is potentially a long-lasting fomite for SARS-CoV-2 transmission even for asymptomatic patients.
Acknowledgments
The authors appreciate the support from Dr Chuming Chen, Ling Qing, and Xiaohe Li (Shenzhen Third People’s Hospital) on patient sample collection and Dr Quanying Liu, Zhichao Liang, and Haihui Wang (Southern University of Science and Technology) on data analysis.
CRediT Authorship Contributions
Fang Zhao, PhD (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Writing – original draft: Equal); Yang, MD (Formal analysis: Equal; Validation: Equal; Writing – review & editing: Equal); Zhaoqin Wang, MD (Project administration: Lead; Resources: Lead; Supervision: Lead; Validation: Equal); Liang Li, PhD (Conceptualization: Lead; Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Lead); Lei Liu, MD (Funding acquisition: Equal; Supervision: Lead); Yingxia Liu, MD (Funding acquisition: Lead; Project administration: Lead; Supervision: Lead; Validation: Lead; Writing – review & editing: Equal).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the Ministry of Science and Technology (2020YFC0846300), the National Natural Science Foundation of China for Major Project (2018ZX10711001, 2017ZX10103011, 2017ZX10204401), Shenzhen Science and Technology Innovation Commission for Research and Development Project (Grants JCYJ20180228162201541, 202002073000001), Sanming Project of Medicine in Shenzhen (SZSM201412003, SZSM201512005) and China Postdoctoral Science Foundation (2019T120147, 2019M660836).
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2020.05.035.
Supplementary Methods
Patients
A total of 401 patients with laboratory-confirmed COVID-19 hospitalized in our hospital were included in this study. Each patient had at least 1 rectal sample tested for viral RNA of SARS-CoV-2. Clinical information was collected retrospectively through the hospital informational system. A total of 1758 rectal swabs from 401 patients with COVID-19 were collected during 0– 98 days after illness onset. Paired respiratory and rectal samples were collected on the same day from the same patient for patients confirmed positive for SARS-CoV-2 RNA in their rectal swabs. A total of 517 such pairs of samples were collected.
Quantitative Reverse-Transcription Polymerase Chain Reaction
Nasal and rectal swabs were collected from patients with laboratory-confirmed COVID-19 upon admission and thereafter. The sample collection time was further divided into 8 groups based on the disease progression, including 1, 2, 3, 4, 5, 6, and 7 WAO and later (>7 WAO). Viral RNAs were extracted from clinical specimens using the QIAamp RNA Viral Kit (Qiagen, Hilden, Germany). The China Food and Drug Administration approved a commercially kit (GeneoDX Co, Shanghai, China) used for the detection of SARS-CoV-2–specific RNA. Samples with a cycle threshold (Ct) value of ≤37.0 were considered putatively positive. Samples with a Ct of >37 were retested and considered positive if Ct was ≥37 but ≤40 and negative if viral RNA was undetectable on the second test.
Statistical Analyses
Categorical variables are reported as frequency rates and percentages. Continuous variables are reported as mean and standard deviation or median and interquartile range values. Means for continuous variables were compared using independent-group t tests when the data were normally distributed; otherwise, the Mann-Whitney U test was performed. Comparisons of categorical variables were done with the chi-squared test, whereas Fisher’s exact test was used when the data were limited. The pairwise comparison between groups was performed, and Bonferroni correction was applied to correct for multiple comparisons. To explore factors associated with the positive detection of viral RNA in rectal specimens and the clearance time of rectal virus RNA >4 weeks, we used a multivariable logistic regression model. A 2-sided α of >.05 was considered statistically significant. All statistical analysis was done using R, version 3.6.2 (R http://www.R-project.org/).
Supplementary Table 1.
Epidemiologic and Clinical Features of the Enrolled Patients With COVID-19 in This Study
| Characteristics | Total (n = 401) | Viral RNA in rectal samples |
||
|---|---|---|---|---|
| Positive (n = 80) | Negative (n = 321) | P value | ||
| Age, y, median (IQR) | 47 (33–60) | 38 (20–54) | 49 (3–61) | .0001 |
| Age subgroups, n (%) | ||||
| Children: 0–17 y | 30 (7.5) | 17 (56.67) | 13 (43.33) | <.0001 |
| Adults: 18 and >18 y | 371 (92.5) | 63 (16.98) | 308 (83.02) | <.0001 |
| Male, n (%) | 190 (47.4) | 32 (40.0) | 158 (49.2) | .176 |
| BMI, median (IQR) | 23.1 (20.9–25.3) | 21.3 (18.5–23.9) | 23.4 (21.4–25.5) | <.0001 |
| Disease severity, n (%) | ||||
| Mild/moderate | 316 (78.8) | 69 (86.3) | 247 (76.9) | .037 |
| Severe/critical | 85 (21.2) | 11 (13.7) | 74 (23.1) | .037 |
| Initial symptoms, n (%) | ||||
| Fever | 268 (66.8) | 50 (62.5) | 218 (67.9) | .431 |
| Cough | 139 (34.7) | 26 (32.5) | 113 (35.2) | .747 |
| Expectoration | 11 (2.7) | 0 (0) | 11(3.4) | .131 |
| Headache | 12 (3.0) | 1 (1.3) | 11(3.4) | .473 |
| Myalgia | 13 (3.2) | 2 (2.5) | 11(3.4) | 1.00 |
| Chills | 3 (0.7) | 1 (1.3) | 2 (0.6) | .488 |
| Nausea or vomiting | 1 (0.2) | 1 (0.3) | 0 (0) | 1 |
| Diarrhea | 25 (6.2) | 6 (7.5) | 19 (5.9) | .61 |
| Coexisting chronic medical conditions, n (%) | ||||
| Chronic heart disease | 8 (2.0) | 1 (1.3) | 7 (2.2) | 1.00 |
| Chronic lung disease | 3 (0.7) | 0 (0) | 3 (0.9) | 1.00 |
| Chronic renal disease | 6 (1.5) | 0 (0) | 6 (1.9) | .604 |
| Chronic liver disease | 23 (5.7) | 1 (1.3) | 22 (6.9) | .06 |
| Diabetes | 14 (3.5) | 0 (0) | 14 (4.4) | .082 |
| Hypertension | 22 (5.5) | 1 (1.3) | 21 (6.5) | .095 |
| Cancer | 1 (0.2) | 0 (0) | 1 (0.3) | 1.00 |
| Exposure history, n (%) | ||||
| Wuhan/Hubei | 297 (74.1) | 49 (61.3) | 248 (77.3) | .005 |
| Interval, d, median (IQR) | ||||
| Onset to admission | 3 (1–6) | 3 (1–6) | 3 (1–6) | .50 |
| Onset to laboratory confirmation | 2 (0–4) | 1 (0–4) | 2 (0–5) | .31 |
| Onset to antiviral treatment | 3 (1–6) | 5 (2–7) | 2 (1–5) | .028 |
BMI, body mass index; IQR, interquartile range.
Supplementary Table 2.
The Coincidence Rate of the SARS-CoV-2 RNA Detection From Paired Rectal Swabs and Nasal Swabs
| Collection date, WAO | All, n | R+/N+ n (%) | R+/N– n (%) | R–/N+ n (%) | R–/N– n (%) | Coincidence rate, % |
|---|---|---|---|---|---|---|
| 1 | 26 | 13 (50) | 7 (26.9) | 3 (11.5) | 3 (11.5) | 61.54 |
| 2 | 62 | 16 (25.8) | 20 (32.3) | 10 (16.1) | 16 (25.8) | 51.61 |
| 3 | 74 | 14 (18.9) | 23 (31.1) | 5 (6.8) | 32 (43.2) | 62.16 |
| 4 | 80 | 7 (8.8) | 28 (35) | 2 (2.5) | 43 (53.8) | 62.50 |
| 5 | 69 | 6 (8.7) | 24 (34.8) | 6 (8.7) | 33 (47.8) | 56.52 |
| 6 | 61 | 2 (3.3) | 9 (14.8) | 3 (4.9) | 47 (77) | 80.33 |
| 7 | 44 | 0 (0) | 1 (2.3) | 1 (2.3) | 42 (95.5) | 95.45 |
| >7 | 101 | 0 (0) | 0 (0) | 10 (9.9) | 91 (90.1) | 90.10 |
| All | 517 | 58 (11.8) | 112 (21.7) | 40 (7.7) | 307 (59.4) | 70.60 |
N, nasal swab; R, rectal swab; WAO, weeks after illness onset.
References
- 1.Wang W. JAMA. 2020 [Google Scholar]
- 2.Zhang Y., Chen C. China CDC Wkly. 2020;2:123–124. [PMC free article] [PubMed] [Google Scholar]
- 3.Yao H., Lu X., Chen Q. MedRχiv. 2020 [Google Scholar]
- 4.Cai Q., Huang D., Ou P. Allergy. 2020 [Google Scholar]
- 5.National Health Commission of China http://www.chinacdc.cn/jkzt/crb/zl/szkb_11803/jszl_11815/202003/W020200305456621460977.pdf
- 6.Cao Q. J Formos Med Assoc. 2020;119:670–673. doi: 10.1016/j.jfma.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Q. Lu. J Med Virol. 2020;92:564–567. doi: 10.1002/jmv.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao F., Tang M., Zheng X. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]