Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused more than 200,000 reported COVID-19 cases in Spain resulting in more than 20,800 deaths as of April 21, 2020. Faecal shedding of SARS-CoV-2 RNA from COVID-19 patients has extensively been reported. Therefore, we investigated the occurrence of SARS-CoV-2 RNA in six wastewater treatments plants (WWTPs) serving the major municipalities within the Region of Murcia (Spain), the area with the lowest COVID-19 prevalence within Iberian Peninsula. Firstly, an aluminum hydroxide adsorption-precipitation concentration method was validated using a porcine coronavirus (Porcine Epidemic Diarrhea Virus, PEDV) and mengovirus (MgV). The procedure resulted in average recoveries of 10 ± 3.5% and 10 ± 2.1% in influent water (n = 2) and 3.3 ± 1.6% and 6.2 ± 1.0% in effluent water (n = 2) samples for PEDV and MgV, respectively. Then, the method was used to monitor the occurrence of SARS-CoV-2 from March 12 to April 14, 2020 in influent, secondary and tertiary effluent water samples. By using the real-time RT-PCR (RT-qPCR) Diagnostic Panel validated by US CDC that targets three regions of the virus nucleocapsid (N) gene, we estimated quantification of SARS-CoV-2 RNA titers in untreated wastewater samples of 5.4 ± 0.2 log10 genomic copies/L on average. Two secondary water samples resulted positive (2 out of 18) and all tertiary water samples tested as negative (0 out 12). This environmental surveillance data were compared to declared COVID-19 cases at municipality level, revealing that members of the community were shedding SARS-CoV-2 RNA in their stool even before the first cases were reported by local or national authorities in many of the cities where wastewaters have been sampled. The detection of SARS-CoV-2 in wastewater in early stages of the spread of COVID-19 highlights the relevance of this strategy as an early indicator of the infection within a specific population. At this point, this environmental surveillance could be implemented by municipalities right away as a tool, designed to help authorities to coordinate the exit strategy to gradually lift its coronavirus lockdown.
Keywords: Environmental surveillance, Influent water, Reclaimed water, Concentration protocol, RNA virus, Coronavirus
Graphical abstract
Highlights
-
•
An adsorption-precipitation concentration method was validated using a porcine coronavirus.
-
•
First detection of SARS-CoV-2 RNA in untreated wastewater in Spain.
-
•
11% secondary treated water samples tested positive for at least one SARS-CoV-2 RT-qPCR target.
-
•
None of the tertiary effluent samples (n = 12) tested positive for SARS-CoV-2.
-
•
SARS-CoV-2 RNA was detected in wastewater before the first COVID-19 cases were declared by local authorities.
1. Introduction
Coronaviruses (CoVs) are a family of viruses pathogenic for humans and animals associated to respiratory and gastro-intestinal infections. CoVs used to be considered as minor human pathogens as they were responsible of common cold or mild respiratory infections in immunocompetent people (Channappanavar and Perlman, 2017). Nonetheless, the emergence of novel and highly pathogenic zoonotic diseases caused by CoVs such as Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS) and most recently SARS-CoV-2 brings to light questions to be addressed to guide public health response.
CoVs are mainly transmitted through respiratory droplets (Meselson, 2020). However, as for SARS and MERS, SARS-CoV-2 RNA has been detected in stool samples from patients exhibiting symptoms of COVID-19 and from asymptomatic carriers (He et al., 2020; Pan et al., 2020; Wölfel et al., 2020; Young et al., 2020; Zhang et al., 2020). The duration of viral shedding has been observed to vary among patients with means of 14–21 days (Y. Wu et al., 2020b; Xu et al., 2020). As well as the magnitude of shedding varies from 102 up to 108 RNA copies per gram (Lescure et al., 2020; Pan et al., 2020; Wölfel et al., 2020).
Infectious viruses deriving from fecal and urine specimen have reportedly been cultured in Vero E6 cells (Sun et al., 2020; W. Wang et al., 2020b). In addition, gastric, duodenal, and rectal epithelial cells are infected by SARS-CoV-2 and the release of the infectious virions to the gastrointestinal tract supports the possible fecal-oral transmission route (Xiao et al., 2020). Even though the possibility of faecal-oral transmission has been hypothesized, the role of secretions in the spreading of the disease is not clarified yet (W. Wang et al., 2020b; Y. Wu et al., 2020b; Xu et al., 2020; Yeo et al., 2020).
Wastewater monitoring has been a successful strategy pursued to track chemical and biological markers of human activity including illicit drugs consumption, pharmaceuticals use/abuse, water pollution, and occurrence of antimicrobial resistance genes (Choi et al., 2018; de Oliveira et al., 2020; Lorenzo and Picó, 2019; Mercan et al., 2019). Viral diseases have been also surveilled by the detection of genetic material into wastewater as for enteric viruses (Hellmer et al., 2014; Prevost et al., 2015; Santiso-Bellón et al., 2020), re-emerging zoonotic hepatitis E virus (Cuevas-Ferrando et al., 2020; Miura et al., 2016), and poliovirus during the global eradication programme (Asghar et al., 2014).
Currently, various studies detected SARS-CoV-2 RNA in wastewater worldwide (Ahmed et al., 2020; La Rosa et al., 2020; Lodder and de Roda Husman, 2020; Medema et al., 2020; Rimoldi et al., 2020; F. Wu et al., 2020a; Wurtzer et al., 2020), and wastewater testing has been suggested as a non-invasive early-warning tool for monitoring the status and trend of COVID-19 infection and as an instrument for tuning public health response (Daughton, 2020; Mallapaty, 2020; Naddeo and Liu, 2020). Under current circumstance, this environmental surveillance could be implemented in wastewater treatment plants as a tool designed to help authorities to coordinate the exit strategy to gradually lift its coronavirus lockdown.
Here, we report the first detection of SARS-CoV-2 RNA in untreated wastewater samples in Spain collected from six different wastewater treatment plants (WWTPs) in Murcia, the lowest prevalence area in Iberian Peninsula. Additionally, the efficacy of the tertiary treatments implemented in the WWPTs against SARS-CoV-2 has been confirmed. The outcomes of the environmental surveillance reflect the epidemiological data in a low COVID-19 diagnosed cases setting, thus supporting the need of developing and implementing advanced models for wastewater-based epidemiology (WBE).
2. Material and methods
2.1. Sampling sites and samples collection
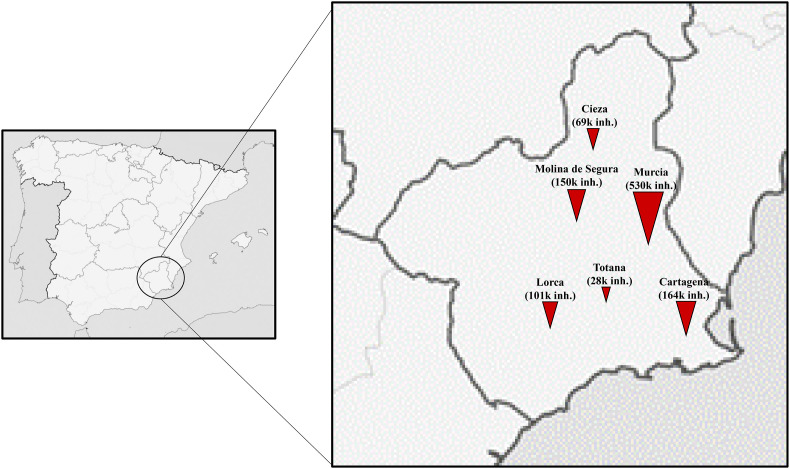
Influent, secondary and tertiary treated effluent water samples were collected from six WWTPs located in the main cities of the Region of Murcia, Spain (Fig. 1 ). Technical data on WWTPs are provided in Table 1 .
Fig. 1.
Maps of the sampling location. Symbols represents WWTPs and are sized according to the number of equivalent inhabitants (inh.).
Table 1.
Data on population and operating characteristics of WWTPs in the area of study.
| Served Populationa | Population Equivalentb | Capacity (m3/y)c |
Reclamation processes | Reuse | ||
|---|---|---|---|---|---|---|
| Designed | Currentd | |||||
| Murcia | 370,893 | 530,499 | 36,500,000 | 36,952,999 | Activated sludge (A2O process), Disinfection, NaClO | Public domain |
| Cartagena | 175,870 | 163,969 | 12,775,000 | 8,625,103 | Activated sludge, Disinfection | Irrigation |
| Molina de Segura | 67,455 | 150,545 | 9,125,000 | 5,699,930 | Activated sludge, Decantation, Coagulation, Flocculation, Sand filtration, Disinfection, UV, NaClO | Irrigation |
| Lorca | 73,057 | 101,161 | 7,300,000 | 3,366,919 | Activated sludge, Coagulation, Flocculation, Sand filtration, Disinfection, UV, NaClO | Irrigation |
| Cieza | 33,744 | 69,502 | 3,650,000 | 2,338,673 | Activated sludge (Extended aeration), Disinfection, Coagulation, Flocculation, Sand filtration, Disinfection, UV | Irrigation |
| Totana | 29,113 | 28,289 | 2,190,000 | 1,440,463 | Activated sludge (Extended aeration), Disinfection, UV | Irrigation |
Population connected to the wastewater treatment facility.
Calculated based on the organic biodegradable load having a five-day biochemical oxygen demand (BOD5) of 60 g of oxygen per day.
m3/y, water flow expressed as volume per year.
Average water flow observed during the period of study.
A total of 42 influent, and 18 secondary and 12 tertiary treated effluent water samples were collected from 12 March to 14 April 2020 and investigated for the occurrence of SARS-CoV-2 RNA. All samples were grabbed early in the morning (7–12am) by collecting 500–1000 mL of water in sterile HDPE plastic containers (Labbox Labware, Spain). Collected samples were transferred on ice to the laboratory, kept refrigerated at 4 °C and concentrated within 24 h. To this end, subsamples of 200 mL were processed as detailed hereafter.
2.2. Wastewater and effluent water concentration
The porcine epidemic diarrhea virus (PEDV) strain CV777, an enveloped virus member of the Coronaviridae family, genus Alphacoronavirus, and etiological agent of porcine epidemic diarrhea (PED), was preliminary used to evaluate the water concentration protocol together with the mengovirus (MgV) vMC0 (CECT 100000), a non-enveloped member of the Picornaviridae designated in the ISO 15216-1, 2017 standard method as process control.
The concentration method consisted in an aluminum hydroxide adsorption-precipitation protocol previously described for concentrating enteric viruses from wastewater and effluent water (AAVV, 2011; Cuevas-Ferrando et al., 2020; Randazzo et al., 2019). The validation was carried out by using biobanked influent (n = 2) and effluent water samples (n = 2) collected in July and October 2019 and stored at −80 °C until processed. In brief, 200 mL of sample was transferred in 250 mL PPCO centrifuge bottles (Thermo Fisher Scientific, Rochester, US) and artificially inoculated with PEDV and MgV. Then pH was adjusted to 6.0 and Al(OH)3 precipitate formed by adding 1 part 0.9N AlCl3 (Acros organics, Geel, Belgium) solution to 100 parts of sample. The pH was readjusted to 6.0 and sample mixed using an orbital shaker at 150 rpm for 15 min at room temperature. Then, viruses were concentrated by centrifugation at 1,700×g for 20 min in a RC-5B Sorvall centrifuge with SS-34 rotor. The pellet was resuspended in 10 mL of 3% beef extract pH 7.4, transferred in 50 mL PPCO centrifuge tubes and shaken for 10 min at 150 rpm. Concentrate was recovered by centrifugation at 1,900×g for 30 min in a RC-5B Sorvall centrifuge with F14S rotor and pellet resuspended in 1 mL of PBS. Alternatively, ST16R Sorvall centrifuge (Thermo Fisher Scientific, Rochester, US) with a TX-1000 ROTOR for 225 mL PPCO centrifuge bottles was used for the two concentration steps following the conditions previously indicated.
All wastewater and effluent water samples included in this study were processed as described and MgV (5 log10 PCR units, PCRU) was spiked as process control.
2.3. Viral extraction, detection and quantification
Viral RNA was extracted from concentrates using the NucleoSpin RNA virus kit (Macherey-Nagel GmbH & Co., Düren, Germany) according to the manufacturer’s instructions with some modifications. Briefly, 150 μL of the concentrated sample was mixed with 25 μL of Plant RNA Isolation Aid (Thermo Fisher Scientific, Vilnius, Lithuania) and 600 μL of lysis buffer from the NucleoSpin virus kit and subjected to pulse-vortexing for 1 min. Then, the homogenate was centrifuged for 5 min at 10,000×g to remove the debris. The supernatant was subsequently processed according to the manufacturer’s instructions and eluted in 100 μL of RNAse free dH2O.
Viral RNA was detected by TaqMan real-time RT-PCR (RT-qPCR) on LightCycler 480 instrument (Roche Diagnostics, Germany) for all reactions. MgV RNA was quantified by using UltraSense One-Step kit (Invitrogen, SA, US) and the RT-qPCR assay as in ISO 15216–1:2017 (Costafreda et al., 2006; ISO 15216-1, 2017). Reaction mix (10 μL) consisted of 2.00 μL 5X Reaction Mix, 0.50 μL 20X Bovine Serum Albumin, 0.20 μL ROX Reference Dye, 0.50 μL Enzyme Mix, 0.90 pmol/μL Mengo 209 REV primer, 0.5 pmol/μL Mengo 110 FW primer and 0.25 pmol/μL Mengo FAM probe. The cycling parameters were as RT at 55 °C for 1 h, preheating at 95 °C for 5 min and 45 cycles of amplification at 95 °C for 15 s, 60 °C for 1 min and 65 °C for 1 min. Undiluted and ten-fold diluted MgV RNA was tested to check for RT-qPCR inhibitors.
PEDV RNA was detected by using One Step PrimeScript™ RT-PCR Kit (Perfect Real Time) (Takara Bio, USA) and the TaqMan RT-qPCR assay described by (Zhou et al., 2017). Reaction mix (10 μL) consisted of 5.00 μL 2X One Step RT-PCR Buffer III, 0.20 μL PrimeScript RT enzyme Mix II, 0.20 μL TaKaRa Ex Taq HS, 0.20 μL ROX, 0.50 μL REV primer (10 μM), 0.50 μL FW primer (10 μM), 0.50 μL FAM labelled TaqMan probe (10 μM). The thermal cycling conditions were as RT at 45 °C for 15 min, preheating at 95 °C for 5 min and 45 cycles of amplification at 95 °C for 15 s and 60 °C for 1 min. SARS-CoV-2 RNA was detected by using One Step PrimeScript™ RT-PCR Kit (Perfect Real Time) and the RT-qPCR diagnostic panel assays validated by the US Centers for Disease Control and Prevention (CDC, 2020). The first version of the kit with three sets of oligonucleotide primers and probes was used to target three different SARS-CoV-2 regions of the nucleocapsid (N) gene. The sets of primers and probe (2019-nCoV RUO Kit) as well as the positive control (2019-nCoV_N_Positive Control, 2 × 105 genome copies (gc)/μL) were provided by IDT (Integrated DNA Technologies, Leuven, Belgium). Reaction mix (10 μL) consisted of 5.00 μL 2X One Step RT-PCR Buffer III, 0.20 μL PrimeScript RT enzyme Mix II, 0.20 μL TaKaRa Ex Taq HS, 0.75 μL for each sets of primers and probe. The thermal cycling conditions were as RT at 50 °C for 10 min, preheating at 95 °C for 3 min and 45 cycles of amplification at 95 °C for 3 s and 55 °C for 30 s. Each RNA was analyzed in duplicate and every RT-qPCR assay included negative (nuclease-free water) and positive controls.
Biobanked samples (n = 4) collected in October 2019, before the first COVID-19 case was documented, were used as relevant negative control to exclude false positive reactions.
SARS-CoV-2 RNA was quantified as gc by plotting the quantification cycles (Ct) to an external standard curve built with 10-fold serial dilution of a quantified plasmid control (IDT). Calibration curves for N1 (y = −3.3774x + 41.515, R2 = 0.95), N2 (y = −3.7752x + 43.951, R2 = 0.989), and N3 (y = -3.6006x + 43.142, R2 = 0.99) showed a linear dynamic range between 5 × 10 and 5 × 104. The limit of detection (LOD) resulted as 50 gc per reaction with Ct values of 37.05 ± 0.77, 38.12 ± 0.24 and 37.29 ± 1.48 for N1, N2 and N3, respectively. The theoretical limits of quantification of the overall method resulted as 4.45, 4.91, and 4.75 log10 gc/L for N1, N2 and N3, respectively.
MgV and PEDV RNA were quantified by plotting the Cts to external standard curves generated by serial end-point dilution method using RNA extracted from purified cell culture suspensions. Quantification were referred as PCRU. Standard curve showed a linear dynamic range between 10 and 107 and between 10 and 105 for MgV (y = −3.603x + 38.02, R2 = 0.99) and PEDV (y = −3.8281x+36.81, R2 = 0.98), respectively.
MgV recovery rates were calculated and used as quality assurance parameters according to ISO 15216–1:2017 (ISO 15216-1, 2017).
3. Results and discussion
3.1. Performance of the concentration method
The aluminum hydroxide adsorption-precipitation method was tested by spiking influent and effluent samples with MgV and PEDV. On average, MgV was recovered at ranges of 11 ± 2.1% in influent and 6.2 ± 1.0% in effluent water. PEDV was recovered at ranges of 11 ± 3.5% in influent and 3.3 ± 1.6% in effluent water. Notably, not significant differences (p > 0.05) were detected between recovery rates in influent waters. This finding implies that a non-enveloped virus may be used as process control for coronavirus detection in influent waters upon method validation. In contrast, significant differences (p < 0.05) were reported between PEDV and MgV recoveries in effluent waters.
These results are in line with the MgV recoveries reported for enteric viruses concentration in water samples by the same aluminum-based method (Cuevas-Ferrando et al., 2020; Randazzo et al., 2019) and higher than the 1% as the quality assurance parameter indicated for bottled water into ISO 15216–1:2017 (ISO 15216-1, 2017).
Similarly, MgV was successfully used as recovery control for hepatitis E virus concentration from influent and effluent water samples (5–13%) by applying a polyethylene glycol (PEG) precipitation method (Miura et al., 2016). A similar PEG-based protocol was recently used to recover SARS-CoV-2 from wastewater, although recovery control was not included in the study (F. Wu et al., 2020a).
Moreover, filtration through 10 kDa Centricon® Plus-70 centrifugal device successfully recovered SARS-CoV-2 in wastewater with recovery efficiencies of F-specific RNA phages of 73% (Medema et al., 2020). However, concentration by electropositive membrane should be further evaluated given a SARS-CoV recovery from wastewater of 1% (Wang et al., 2005).
Rigorous limits of detection should be established by spiking SARS-CoV-2 cell-culture adapted strain or positive COVID-19 fecal samples in influent and effluent wastewater samples to be concentrated following the aluminum hydroxide adsorption-precipitation method. Nonetheless, the need of a BSL3 laboratory facility to handle SARS-CoV-2 represents the main limitation of this experiment.
3.2. SARS-CoV-2 titers in wastewater and effluent water
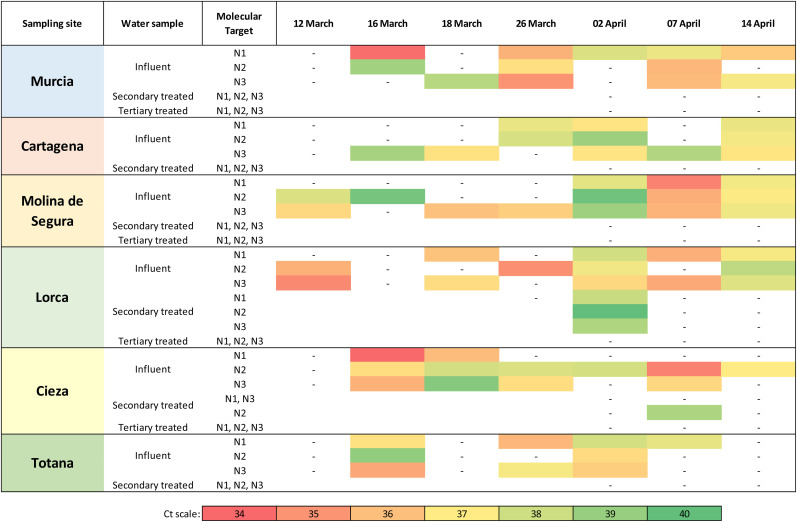
A total of 42 influent, and 18 secondary and 12 tertiary treated effluent water samples were collected from 12 March to 14 April 2020 and investigated for the occurrence of SARS-CoV-2 RNA. Samples were considered positive for Ct below 40 (as in Medema et al., 2020 and F. Wu et al., 2020a) and titrated by using the quantified plasmid control for each of the RT-qPCR targets. As expected, biobanked samples collected in October 2019, before the first COVID-19 case was documented, tested negative for all the three RT-qPCR assays thus excluding false positive reactions. The 83% (35 positive samples out of 42) influent samples and the 11% (2 out of 18) secondary treated water samples were tested positive for at least one SARS-CoV-2 RT-qPCR target. None of the tertiary effluent samples (0 out of 12) tested positive for any of the SARS-CoV-2 RT-qPCR target (Fig. 2 ). A relevant number of influent water samples (12%) showed Ct ranging between 37 and 40, even though lower Ct of 34–37 were observed (29%).
Fig. 2.
Mean amplification cycles of SARS-CoV-2 RNA in influent, secondary and tertiary effluent waters in monitored WWTPs within Murcia Region (Spain). Results are reported for each of the three regions of the virus nucleocapsid (N) gene according to the first version of the Real-Time RT-PCR Diagnostic Panel by US CDC. Abbreviations: , negative; white boxes, not tested.
In influent samples, a poor positive correlation among RT-qPCR assays was detected, being 0.5, 0.3, and 0.6 the resulting coefficients between N1 and N2, N1 and N3, N2 and N3, respectively. The total number of RT-qPCR determinations was 84 for each target. For N1, 23 results showed Ct below 37 out of 33 positive samples (70%), for N2 18 out of 31 (58%), and for N3 28 out of 36 (78%). In all samples, MgV recoveries were above 1% (11 ± 15%). MgV recovery for each sample and Ct values for each SARS-CoV-2 target are reported in Table S1 in Supplementary Material.
On average, SARS-CoV-2 RNA titers of 5.1 ± 0.3, 5.5 ± 0.2, and 5.5 ± 0.3 log10 gc/L were quantified in wastewater by using N1, N2 and N3 primer/probe mixes, respectively. Titers of 4 and 5 to more than 6 log10 gc/L have been reported in Massachusetts and France, respectively (F. Wu et al., 2020a; Wurtzer et al., 2020).
A secondary effluent sample resulted positive for N2 and quantified as 5.4 log10 gc/L. An additional secondary effluent sample was positive for the three molecular targets and below the limit of quantification.
Detection of SARS-CoV-2 RNA in influent water has been reported worldwide (Ahmed et al., 2020; La Rosa et al., 2020; Lodder and de Roda Husman, 2020; Medema et al., 2020; Wu et al., 2020a), and only one study tested treated wastewater that resulted positive (Paris) (Wurtzer et al., 2020). We observed discrepancies among RT-qPCR N1, N2 and N3 assays for several water samples in agreement to a previous report (Medema et al., 2020). This could be due to the different analytical sensitivity among the assays as well as the detection of possible false positive samples by RT-qPCR N3 in low concentrated clinical samples (Jung et al., 2020; Vogels et al., 2020). The latter possibility has been solved by excluding the N3 primers/probe set from the US CDC 2019-nCoV RT-qPCR diagnostic panel in its last revision (March, 30) (CDC a, CDC b, n.d.). In addition, a partial inhibitory effect of the matrix is not to be completely excluded despite the controls included in the assays. A more sensitive estimation of SARS-CoV-2 loads in wastewater should be studied by digital RT-qPCR (dRT-qPCR). dRT-qPCR could be used to quantify samples with low viral loads as reported for norovirus in wastewater (Monteiro and Santos, 2017) and SARS-CoV-2 in clinical samples (Dong et al., 2020; Suo et al., 2020), even though it may not be the best practical and economically sustainable option for environmental surveillance (Abachin et al., 2018).
Even though the SARS-CoV-2 RNA detection in wastewater is functional for WBE purposes, the risk for human health associated to the water cycle is still under debate as infectivity of viral particles in sewage and faeces remain to be confirmed as well as its potential fecal-oral transmission. A pre-print report suggests that the risk of infection from wastewater and river is negligible given the failure in cell culturing SARS-CoV-2 from water samples despite the high number of RNA copies (Rimoldi et al., 2020).
In spite of the high concentration of viral RNA in specimen and the evidence of gastrointestinal infection (Xiao et al., 2020), infectious viruses from stools have been isolated in one study (W. Wang et al., 2020b) while another attempt resulted without success (Wölfel et al., 2020).
The potential transmission of SARS-CoV-2 via wastewater has not been proven (CDC a, CDC b, n.d.; WHO, 2020) and it seems unlikely given the poor stability of viable SARS-CoV-2 in wastewater (Rimoldi et al., 2020; J. Wang et al., 2020a) that resembles some previous studies made with representative coronaviruses (Gundy et al., 2008) and enveloped surrogates (Casanova and Weaver, 2015). As well, the elevated sensitivity of human pathogenic coronaviruses to environmental conditions (Chin et al., 2020; Darnell et al., 2004; Darnell and Taylor, 2006) and disinfectants (Chin et al., 2020; J. Wang et al., 2020a) suggests a poor risk of transmission via wastewater, even though formal risk analysis needs to be performed (Haas, 2020).
3.3. Environmental surveillance
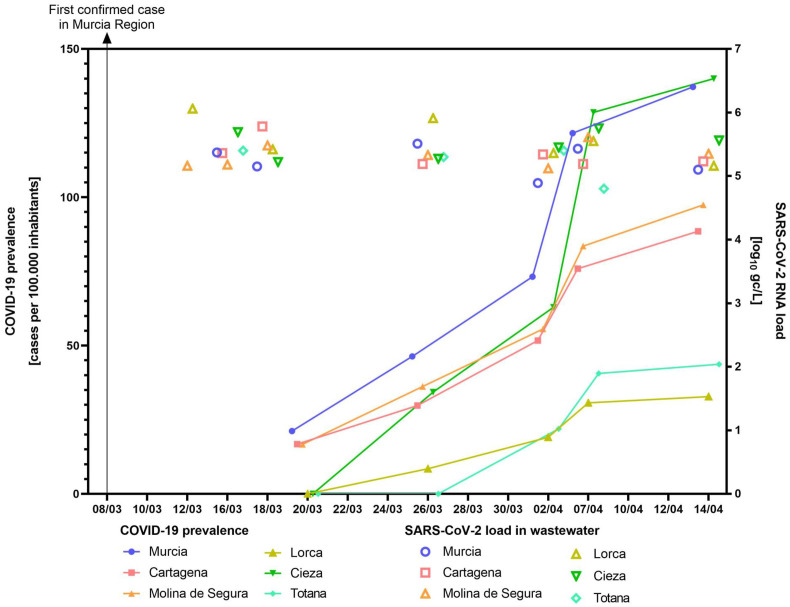
Epidemiological data on COVID-19 in the Murcia Region have been retrieved from the publically available repository of the “Servicio de epidemiologia” of the “Consejería de Salud de la Región de Murcia” (available at http://www.murciasalud.es/principal.php) (Table 2 ) and plotted to the SARS-CoV-2 RNA mean loads as detected by three RT-qPCR assays (Fig. 3 ).
Table 2.
Epidemiological dataa summary of COVID-19 cases in the area of study.
| 20/03/2020 |
25/03/2020 |
30/03/2020 |
08/04/2020 |
15/04/2020 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Prevalenceb | Cases | Prevalence | Cases | Prevalence | Cases | Prevalence | Cases | Prevalence | |
| Murcia | 96 | 21.18 | 210 | 46.33 | 332 | 73.2 | 551 | 121.6 | 622 | 137.2 |
| Cartagena | 36 | 16.76 | 64 | 29.79 | 111 | 51.7 | 163 | 75.9 | 190 | 88.5 |
| Molina de Segura | 12 | 16.69 | 26 | 36.17 | 40 | 55.6 | 60 | 83.5 | 70 | 97.4 |
| Lorca | – | – | 8 | 8.47 | 18 | 19.1 | 29 | 30.7 | 31 | 32.8 |
| Cieza | – | – | 12 | 34.30 | 22 | 62.9 | 45 | 128.6 | 49 | 140.0 |
| Totana | – | – | – | – | 7 | 21.9 | 13 | 40.6 | 14 | 43.7 |
Data retrieved from the public repository of the “Servicio de epidemiologia” of the “Consejería de Salud de la Región de Murcia” (available at http://www.murciasalud.es/principal.php).
Prevalence, percentage of diagnosed cases per 100.000 inhabitants.
Fig. 3.
Epidemiological surveillance of COVID-19 by wastewater SARS-CoV-2 RT-qPCR in six municipalities.
In general, RT-qPCR amplification signals have been detected in wastewaters when cases were diagnosed within the municipality. Positive wastewater samples have been detected with at least two out of three RT-qPCR assays in low prevalence municipalities as in Murcia (96 cases, 21.18 cases per 100,000 inhabitants), Cartagena (36 cases, 16.76) and Molina de Segura (12 cases, 16.69). Of note, positive wastewater samples were detected 12–16 days before COVID-19 cases were declared in Lorca, Cieza and Totana municipalities.
A similar study conducted in Paris (France) demonstrated the detection of viral genome before the exponential phase of the epidemic (Wurtzer et al., 2020). However, our results indicate that SARS-CoV-2 can be detected weeks before the first confirmed case. The early detection of SARS-CoV-2 RNA in wastewater could have alerted about the imminent danger, giving a valuate time to the managers to coordinate and implement actions to slow the spread of the disease. Therefore, our outcomes support that WBE could be used as an early warning tool to monitor the status of COVID-19 infection within a community.
On the other hand, we believe that this environmental surveillance could be used as an instrument to drive the right decisions to reduce the risk of lifting restrictions too early. For instance, a key question is how to reduce the risk of a “second wave” and/or recurring local outbreaks. Massive population tests are the first choice, but in their absence, wastewater monitorization of SARS-CoV-2 RNA can give a reliable picture of the current situation. Our wastewater data do not quantitatively resemble the prevalence of COVID-19 confirmed cases. To this end, a quantitative model that includes and corrects all the variables affecting these wastewater surveillance data would be useful for a better interpretation. For instance, not all COVID-19 positive patients excrete SARS-CoV-2 RNA in faeces, and when it occurs, the titers and the duration of shedding vary among individuals and across time (He et al., 2020; Pan et al., 2020; Wölfel et al., 2020; Xu et al., 2020). On the other hand, the real number of positive cases within the Murcia Region remains unknown because of the large number of mild or asymptomatic carriers that have not been included in epidemiological statistics.
These aspects together with environmental variables (e.g., rainfall events, temperature, hydraulic retention time in sewers) increase the uncertainties linked to the correlation between SARS-CoV-2 RNA detection in wastewater samples and the prevalence of COVID-19 that could be explored by using complex models.
4. Conclusion
Overall, wastewater surveillance and WBE may represent a complementary approach to estimate the presence and even the prevalence of COVID-19 in communities. This represents an effective tool that needs to be further explored in order to direct public health response, especially in cases of limited capacity for clinical testing.
Funding
The study was funded by the projects 20180705 of ESAMUR, 202070E101 of CSIC and “VIRIDIANA” AGL2017-82909 (AEI/FEDER, UE) of MICIU. WR is supported by APOSTD/2018/150 postdoctoral fellowship of Generalitat Valenciana. EC-F is recipient of a predoctoral contract from the MICINN, Call 2018. PT is holder of the RYC2018- 025510-I Ramón y Cajal contract from the MICIU.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge the “Entidad Regional de Saneamiento y Depuración de Aguas Residuales (ESAMUR)” for authorizing the sampling and Prof. Ana Carvajal (Faculty of Veterinary Medicine, University of Leon, Spain) for kindly providing PEDV.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.watres.2020.115942.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- AAVV . Detection of Enteric Viruses. American Public Health Association; 2011. Standard methods for the examination of water and wastewater (9510) [Google Scholar]
- Abachin E., Convers S., Falque S., Esson R., Mallet L., Nougarede N. Comparison of reverse-transcriptase qPCR and droplet digital PCR for the quantification of dengue virus nucleic acid. Biologicals. 2018;52:49–54. doi: 10.1016/j.biologicals.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., El Bassioni L., Akande A.O., Al Maamoun E., Zaidi S., Adeniji A.J., Burns C.C., Deshpande J., Oberste M.S., Lowther S.A. Environmental surveillance for polioviruses in the global polio eradication initiative. J. Infect. Dis. 2014;210(Suppl. l):S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Weaver S.R. Inactivation of an enveloped surrogate virus in human sewage. Environ. Sci. Technol. Lett. 2015;2:76–78. doi: 10.1021/acs.estlett.5b00029. [DOI] [Google Scholar]
- CDC CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. https://www.fda.gov/media/134922/download n.d.[WWW Document]. URL. [DOI] [PMC free article] [PubMed]
- CDC Water transmission and COVID-19: questions and answers. https://www.cdc.gov/coronavirus/2019-ncov/php/water.html n.d.[WWW Document]. URL. (accessed 4.21.20b)
- CDC CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. 2020. https://www.fda.gov/media/134922/download [WWW Document]. URL. [DOI] [PMC free article] [PubMed]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. The Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends Anal. Chem. (Reference Ed.) 2018 doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- Costafreda M.I., Bosch A., Pintó R.M. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 2006;72:3846–3855. doi: 10.1128/AEM.02660-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ferrando E., Randazzo W., Pérez-Cataluña A., Sánchez G. HEV occurrence in waste and drinking water treatment plants. Front. Microbiol. 2020;10:2937. doi: 10.3389/fmicb.2019.02937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M.E.R., Taylor D.R. Evaluation of inactivation methods for severe acute respiratory syndrome coronavirus in noncellular blood products. Transfusion. 2006;46:1770–1777. doi: 10.1111/j.1537-2995.2006.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M.E.R., Subbarao K., Feinstone S.M., Taylor D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome. SARS-CoV. J. Virol. Methods. 2004;121:85–91. doi: 10.1016/j.jviromet.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138149. 138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira M., Frihling B.E.F., Velasques J., Filho F.J.C.M., Cavalheri P.S., Migliolo L. Pharmaceuticals residues and xenobiotics contaminants: occurrence, analytical techniques and sustainable alternatives for wastewater treatment. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2019.135568. [DOI] [PubMed] [Google Scholar]
- Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S., Wang X., Zhang Y., Yang J., Liu M., Zhao Yang, Zhang X., Zhu T., Peng T., Xie J., Gao Y., Wang D., Zhao Yun, Dai X., Fang X. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. medRxiv. 2020 doi: 10.1101/2020.03.14.20036129. 2020.03.14.20036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2008;1:10. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Haas C. Coronavirus and risk analysis. Risk Anal. 2020;40:660–661. doi: 10.1111/risa.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hellmer M., Paxeus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergstrom T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISO 15216-1 . 2017. Microbiology of the Food Chain – Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR – Part 1: Method for Quantification. ISO 15216-1:2017. [Google Scholar]
- Jung Y.J., Park G.-S., Moon J.H., Ku K., Beak S.-H., Kim Seil, Park E.C., Park D., Lee J.-H., Byeon C.W., Lee J.J., Maeng J.-S., Kim S.J., Kim Seung Il, Kim B.-T., Lee M.J., Kim H.G. bioRxiv; 2020. Comparative Analysis of Primer-Probe Sets for the Laboratory Confirmation of SARS-CoV-2; p. 964775. 2020.02.25. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection OF SARS-COV-2 IN untreated wastewaters IN Italy. medRxiv. 2020 doi: 10.1101/2020.04.25.20079830. 2020.04.25.20079830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J.-C., Mentre F., Duval X., Descamps D., Malvy D., Timsit J.-F., Lina B., van-der-Werf S., Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020 doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo M., Picó Y. Wastewater-based epidemiology: current status and future prospects. Curr. Opin. Environ. Sci. Heal. 2019 doi: 10.1016/j.coesh.2019.05.007. [DOI] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020 doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. medRxiv. 2020 doi: 10.1101/2020.03.29.20045880. 2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- Mercan S., Kuloglu M., Asicioglu F. Monitoring of illicit drug consumption via wastewater: development, challenges, and future aspects. Curr. Opin. Environ. Sci. Heal. 2019 doi: 10.1016/j.coesh.2019.05.002. [DOI] [Google Scholar]
- Meselson M. Droplets and aerosols in the transmission of SARS-CoV-2. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T., Lhomme S., Le Saux J.-C., Le Mehaute P., Guillois Y., Couturier E., Izopet J., Abranavel F., Le Guyader F.S. Detection of hepatitis E virus in sewage after an outbreak on a French island. Food Environ. Virol. 2016;8:194–199. doi: 10.1007/s12560-016-9241-9. [DOI] [PubMed] [Google Scholar]
- Monteiro S., Santos R. Nanofluidic digital PCR for the quantification of Norovirus for water quality assessment. PloS One. 2017;12 doi: 10.1371/journal.pone.0179985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial Perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 2020 doi: 10.1039/D0EW90015J. [DOI] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost B., Lucas F.S., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Piqueras J., Evtoski Z., Sastre G., Sancho R., Gonzalez C., Sanchez G. Interlaboratory comparative study to detect potentially infectious human enteric viruses in influent and effluent waters. Food Environ. Virol. 2019 doi: 10.1007/s12560-019-09392-2. [DOI] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Moja L., Gismondo M.R., Salerno F. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. medRxiv. 2020 doi: 10.1101/2020.05.01.20086009. 2020.05.01.20086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiso-Bellón C., Randazzo W., Pérez-Cataluña A., Vila-Vicent S., Gozalbo-Rovira R., Muñoz C., Buesa J., Sanchez G., Díaz J.R. Epidemiological surveillance of norovirus and rotavirus in sewage (2016–2017) in Valencia (Spain) Microorganisms. 2020 doi: 10.3390/microorganisms8030458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhu A., Li H., Zheng K., Zhuang Z., Chen Z., Shi Y., Zhang Z., Chen S., Liu X., Dai J., Li X., Huang S., Huang X., Luo L., Wen L., Zhuo J., Li Yuming, Wang Y., Zhang L., Zhang Y., Li F., Feng L., Chen X., Zhong N., Yang Z., Huang J., Zhao J., Li Yi-min. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microb. Infect. 2020:1–8. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Qiuhan, Wang X., Sajid M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Yuan, Zhang Qi, Liu Yingle, Xiong Y., Chen G., Lan K., Chen Y. ddPCR: a more sensitive and accurate tool for SARS-CoV-2 detection in low viral load specimens. medRxiv. 2020:20029439. doi: 10.1101/2020.02.29.20029439. 2020.02.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Muenker M.C., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takehashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O.-E., Wong P., Yang Y., Cheemarla N.R., White E., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C., Iwasaki A., Ko A.I., Landry M.-L., Foxman E.F., Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays. medRxiv. 2020 doi: 10.1101/2020.03.30.20048108. 2020.03.30.20048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Jin M., Zhen B., Kong Q.-X., Song N., Xiao W.-J., Yin J., Wei W., Wang G.-J., Si B.-Y., Guo B.-Z., Liu C., Ou G.-R., Wang M.-N., Fang T.-Y., Chao F.-H., Li J.-W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO W.H.O. Water, sanitation, hygiene and waste management for COVID-19. 2020. https://www.who.int/publications-detail/water-sanitation-hygiene-and-waste-management-for-covid-19 [WWW Document]. URL. (accessed 4.21.20)
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.05.20051540. 2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. 2020.04.12.20062679. [DOI] [Google Scholar]
- Xiao F., Tang M., Zheng X., Li C., He J., Hong Z., Huang S., Zhang Z., Lin X., Fang Z., Lai R., Chen S., Liu J., Huang J., Xia J., Li Z., Jiang G., Liu Y., Li X., Shan H. 2020. Evidence for Gastrointestinal Infection of SARS-CoV-2. medRxiv 2020.02.17.20023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatology. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.-T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K., Anderson D.E., Chan K.S., Tan T.Y., Ng T.Y., Cui L., Said Z., Kurupatham L., Chen M.I.-C., Chan M., Vasoo S., Wang L.-F., Tan B.H., Lin R.T.P., Lee V.J.M., Leo Y.-S., Lye D.C., Team, for the S. 2019 N.C.O.R. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zhang T., Song D., Huang T., Peng Q., Chen Y., Li A., Zhang F., Wu Q., Ye Y., Tang Y. Comparison and evaluation of conventional RT-PCR, SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Mol. Cell. Probes. 2017;33:36–41. doi: 10.1016/j.mcp.2017.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.