Abstract
Public health practitioners and researchers have used traditional medical databases to study and understand public health for a long time. Recently, social media data, particularly Twitter, has seen some use for public health purposes. Every large technological development in history has had an impact on the behaviour of society. The advent of the internet and social media is no different. Social media creates public streams of communication, and scientists are starting to understand that such data can provide some level of access into the people's opinions and situations. As such, this paper aims to review and synthesize the literature on Twitter applications for public health, highlighting current research and products in practice. A scoping review methodology was employed and four leading health, computer science and cross-disciplinary databases were searched. A total of 755 articles were retreived, 92 of which met the criteria for review. From the reviewed literature, six domains for the application of Twitter to public health were identified: (i) Surveillance; (ii) Event Detection; (iii) Pharmacovigilance; (iv) Forecasting; (v) Disease Tracking; and (vi) Geographic Identification. From our review, we were able to obtain a clear picture of the use of Twitter for public health. We gained insights into interesting observations such as how the popularity of different domains changed with time, the diseases and conditions studied and the different approaches to understanding each disease, which algorithms and techniques were popular with each domain, and more.
Keywords: Public health, Syndromic surveillance, Pharmacovigilance, Event forecasting, Disease tracking
Highlights
-
•
Twitter is a very popular microblogging platform with over 300 million active users.
-
•
We analyse the literature to understand Twitter's capability to provide a useful tool for public health.
-
•
We found that Twitter can be used for surveillance, event detection, pharmacovigilance, disease tracking and forecasting.
-
•
Twitter is mostly used in the context of flu, drug abuse, depression and dengue.
-
•
Our analysis presents the modern landscape of public health using social media data in combination with Machine Learning.
1. Introduction
Surveillance, described by the World Health Organisation (WHO) as ‘‘the cornerstone of public health security’’ [1], is aimed at the detection of elevated disease and death rates, implementation of control measures and reporting to the WHO of any event that may constitute a public health emergency or international concern. Syndromic surveillance can be described as the real-time (or near real-time) collection, analysis, interpretation, and dissemination of health-related data, to enable the early identification of the impact (or absence of impact) of potential human or veterinary public health threats that require effective public health action [2]. The task of syndromic surveillance is an undertaking motivated by the notion of public health. Public health has been defined as the science and art of preventing disease, prolonging life and promoting human health through organized efforts and informed choices of society, organizations, public and private, communities and individuals [3]. In this sense, the concept of health encompasses the physical, emotional and social well-being. Historically, public health practitioners have used data from multiple sources for measuring the burden of diseases and other health outcomes, preventing and controlling diseases and guiding healthcare activities. Emergency department attendances or general practitioner (GP, family doctor) consultations are some of the sources traditionally used to track specific syndromes such as influenza-like illnesses (ILI). With the proliferation of the internet and the advent of modern technology, potential new data sources present themselves. In recent years, researchers have recognized that social media platforms, such as Twitter and Facebook, could also provide data about national-level health and behaviour [4]. Among these social media platforms, Twitter offers a unique and potentially powerful data source due to its ease of access, real-time nature and richness in detail. In this paper, we look towards Twitter with the aim of investigating and assessing its utility as a public health tool by performing a scoping review on the subject. While we seek to review the literature of Public health research making use of Twitter, our interest in such literature is limited to research concerning the monitoring, detection and forecasting of public health conditions. We are not interested in social science research investigating the use of Twitter for recruitment or public awareness and dissemination of public health information. We are similarly not interested in research concerned with opinion mining to understand public opinion on public health issues.A scoping review such as ours is pertinent as there exist no broad and recent evidence-reviews on the use of Twitter data for health research purposes. Wargon et al. [5] performed a systematic review on syndromic surveillance models used in forecasting emergency department visits, however, only 9 studies were found and none of them made use of Twitter or any social media. Subsequently, Charles-Smithe et al. [6] carried out a systematic review of the use of social media (not limited to Twitter) specifically for disease surveillance and outbreak management. Sinnenberg et al. performed another systematic review looking at Twitter as a tool for health research [7]. Their systematic review encompassed research in both the sciences and social sciences. We seek to carry out a scoping review in order to map the broad area of Twitter for public health research as well as to produce an updated review containing more recent studies carried out since the above reviews were published. Hence, our research question is: ‘‘What is known from the existing literature about the use of Twitter data in the context of monitoring, detection and forecasting of public health conditions?’‘. We are particularly interested in the type of conditions/illnesses being studied; in the sources of data being used; in the data analysis techniques being applied; and in the geographical and time trends of such studies. (see Table 3, Table 4, Table 5, Table 6, Table 7)
Table 3.
Summary of statistical and machine learning methods and data sources for event detection using Twitter data.
| Public Health Issue | Method | Complementary Data |
|---|---|---|
| Cancer | Support Vector Machine [61] | CDC |
| Smoking | Bayesian Logistic Regression [62] | |
| Suicide | ARIMA (Autoregressive Integrated Moving Average [26] | |
| Harmful Algal Blooms (HABS) | Deep Learning (CNN) [59] | |
| HIV | Decision Tree [63], Support Vector Machine [63], Graph Modelling [27], Multilayer Perceptron [63] | |
| Allergies | [64], Bayesian Inference [64] | pollen.com, National Climatic Data Center Climate Data Online (CDO) |
| Drug Abuse | Biterm Topic Model [55], Decision Tree [65], Support Vector Machine [58], Topic Model [66] | |
| HPV | Decision Tree [67], Linear Classifier [67] | |
| Infectious Intestinal Diseases (IID) | Word2Vec [68], Gaussian Process [68] | Public Health England |
| Adverse Drug Events (ADE) | Multi-Instance Logistic Regression [69] | |
| Depression | Non-Negative Matrix Factorization [70], ARIMA (Autoregressive Integrated Moving Average) [26], Simple Statistical Analysis [71], Stepwise Regression [60] | National Climatic Data Center, National Oceanic and Atmospheric Administration (NOAA) |
| Ebola | Lexicon Analysis [56], Support Vector Machine [56] | |
| Back Pain | Logistic Regression [72] | |
| Vomiting | TSVM [22], ARIMA (Autoregressive Integrated Moving Average) [22] | Public Health England |
| Gastroenteritis | TSVM [22], ARIMA (Autoregressive Integrated Moving Average) [22] | Public Health England |
| Asthma | Support Vector Machine [61] | CDC |
| Food Borne Illness | K-Nearest Neighbour [73], Support Vector Machine [32] | Southern Nevada Health District (SNHD), CDC |
| Earthquake | Clustering [19], Bayesian Inference [19] | |
| Diabetes | Support Vector Machine [61] | CDC |
| Dental Pain | Simple Statistical Analysis [74] | |
| Influenze-like Illnesses (ILIs) | Clustering [75], Lexicon Analysis [35,57,76], Deep Learning (RNN) [36], Logistic Regression [77], Gaussian Process [78], Deep Learning (CNN) [36], Outlier Detection [46], Bayesian Inference [35,57], Fasttext [36], ARIMA (Autoregressive Integrated Moving Average) [22], GloVe [36], FP-Growth [37], Trap Model [79], Support Vector Machine [37,77,80], Shallow MLP [81], TSVM [22], Word2Vec [75], Regression [80] | Penn State's Health Services, Infectious Disease Surveillance Center, Royal College of General Practitioners (RCGP), Public Health England, CDC |
| General Healtha | Support Vector Machine [82], Lexicon Analysis [82] | |
| Diarrhoea | TSVM [22], ARIMA (Autoregressive Integrated Moving Average) [22] | Public Health England |
| Obesity | Dbscan (Clustering) [54] | |
| Middle East Respiratory Syndrome (Mers) | Lexicon Analysis [56], Support Vector Machine [56] |
Generic feelings of unwellness and non-specific illness.
Table 4.
Summary of statistical and machine learning methods and data sources for pharmacovigilance using Twitter data.
| Public Health Issue | Method | Complementary Data |
|---|---|---|
| Smoking | Bayesian Logistic Regression [62] | |
| HIV | Support Vector Machine [63], Word2Vec [28], Doc2Vec [28], Multilayer Perceptron [63], Decision Tree [63], Dynamic Topic Modeling [28] | |
| Vaccination | Semantic Network Analysis [87] | |
| Drug Abuse | Decision Tree [65], Support Vector Machine [58], Topic Model [66], Simple Statistical Analysis [88] | National Surveys on Drug Usage and Health (NSDUH) |
| Adverse Drug Reactions (ADRs) | Conditional Random Field [85,89], Lexicon Analysis [89,90], Deep Learning (RNN) [86], Word Embeddings (Glove) [86], Word2Vec [85] | ADRMine |
| Adverse Drug Events (ADEs) | Multi-Instance Logistic Regression (Milr) [69], Semi-Supervised Multi-Instance (Nssm) [91], Bayesian Inference [83], Support Vector Machine [83,92], Lexicon Analysis [92] | |
| Alcoholism | Simple Statistical Analysis [84] | |
| Miscellaneous | Decision Tree [93], Support Vector Machine [94], Latent Dirichlet Allocation [94] |
Table 5.
Summary of statistical and machine learning methods and data sources for forecasting using Twitter data.
| Public Health Issue | Method | Complementary Data |
|---|---|---|
| Cancer | Simple Statistical Analysis [23], Linear Regression [99] | CDC |
| E Coli | Latent Dirichlet Allocation [31], Lexicon Analysis [31] | Robert Koch Institute |
| Vomiting | TSVM [22], ARIMA (Autoregressive Integrated Moving Average) [22] | Public Health England |
| Gastroenteritis | TSVM [22], Latent Dirichlet Allocation [31], Lexicon Analysis [31], ARIMA (Autoregressive Integrated Moving Average) [22] | Public Health England, Robert Koch Institute |
| Asthma | Decision Tree [95], Shallow MLP [95] | Children's Medical Center (CMC) |
| Influenze-like Illnesses (H1N1) | Support Vector Regression [100] | CDC |
| Influenze-like Illnesses | Deep Learning (RNN) [36], Deep Learning (MLP) [40], Fasttext [36], Deep Learning (CNN) [36], ARIMA (Autoregressive Integrated Moving Average) [22,97], GloVe [36], Temporal Topic Model [14], Dynamic Regression [96], TSVM [22], Partial Differential Equation [44], Simple Statistical Analysis [23], Autoregressive Moving Average (ARMA) [45] | Boston Public Health Commission, Public Health England, Pan American Health Organization (PAHO), Chinese CDC, CDC |
| General Healtha | Temporal Ailment Topic Aspect Model (TM-ATAM) [51] | CDC |
| Dengue | Simple Statistical Analysis [53] | Brazilian Official Dengue case data |
| Diarrhoea | TSVM [22], ARIMA (Autoregressive Integrated Moving Average) [22] | Public Health England |
Generic feelings of unwellness and non-specific illness.
Table 6.
Summary of statistical and machine learning methods and data sources for disease tracking using Twitter data.
| Public Health Issue | Method | Complementary Data |
|---|---|---|
| Measles | Semantic Network Analysis [101] | CDC |
| Influenze-like Illnesses (Hemophilus) | Bayesian Inference [15] | Genbank |
| Influenze-like Illnesses (H1N1) | Semi-Superviseddeep Learning (MLP) [104], Support Vector Regression [100] | CDC |
| Influenze-like Illnesses | Bayesian Inference [39], Bayesian Inference [41], Dynamic Regression [96], Maximum Entropy [41] | FluWatch, Boston Public Health Commission, Chinese CDC |
| General Healtha | Temporal Ailment Topic Aspect Model (TM-ATAM) [51] | CDC |
| Dengue | Time-Series Susceptible-Infected-Recovered Model [103], Simple Statistical Analysis [53] | Brazilian Official Dengue case data |
| Miscellaneous | Gaussian Mixture Regression (Gmr) [102] | Map data |
Generic feelings of unwellness and non-specific illness.
Table 7.
Summary of statistical and machine learning methods and data sources for geographic identification using Twitter data.
| Public Health Issue | Method | Complementary Data |
|---|---|---|
| Depression | Non-Negative Matrix Factorization (Nmf) [70] | |
| Dengue | Time-Series Susceptible-Infected-Recovered Model [103], Dbscan (Clustering) [21] | Brazilian Health Ministry |
| Obesity | Dbscan (Clustering) [54] | |
| Miscellaneous | Latent Dirichlet Allocation [105], Support Vector Machine [105,106], Bayesian Inference [105], Random Forest [105], Multilayer Perceptron [105], Gaussian Mixture Regression (GMR) [102], HDBSCAN (Clustering) [106] | Map data |
We deliver a summary of what has been done so far, which will enable researchers to quickly and efficiently understand this field in terms of the volume, nature and characteristics of the primary research undertaken and any gaps in research that may need prompt attention. Such evidence is particularly necessary in new but fast moving areas of research such as analysis of Twitter data for health applications.
2. Method
A scoping review methodology was chosen to achieve our goal of investigating the state of Twitter applications in the field of public health research, our research question. The scoping review is defined by Arskey and O'Malley [8] as a study that aims ‘‘to map rapidly the key concepts underpinning a research area and the main sources and types of evidence available, and can be undertaken as stand-alone projects in their own right, especially where an area is complex’‘. For our scoping review, we made use of the Arksey and O'Malley framework which adopts a rigorous process of transparency, enabling replication of the search strategy and increasing the reliability of the study findings. As Arksey and O'Malley [8] explain, the method consists of a number of stages such as: identifying the research question; identifying relevant studies; study selection; charting the data and collating, summarizing and reporting the results (i.e. analysis). We elaborate on specific application of the method to our scenario next.
2.1. Search strategy to identify relevant studies
To gain a broad coverage of the available literature, the general terms ‘‘Twitter’’ and ‘‘Public Health’’ were used as search keywords. We chose these two keywords as ‘‘Twitter’’ covers every discussion of the Twitter platform, and used together with ‘‘Public Health’’ covers all mention of Twitter in a health context. As our work is multidisciplinary in that it spans multiple fields, we conducted our search in both health and Information Technology (IT) databases. First, we performed a literature search in the health/medical database PubMed. Next, we searched the IT databases IEEE Xplore and the ACM Digital Library. Finally, we searched a general database that indexed both fields, Scopus. Our searches were refined such that we only included research articles which were peer-reviewed and in English. We also limited our search to only return results within the date range of January 2009 and March 2019, which was when the search was carried out. We started our search from 2009 because of the highly influential Google Flu Trends paper published that year which inspired and kickstarted the use of social media as a data source for public health research [9].
2.2. Study selection
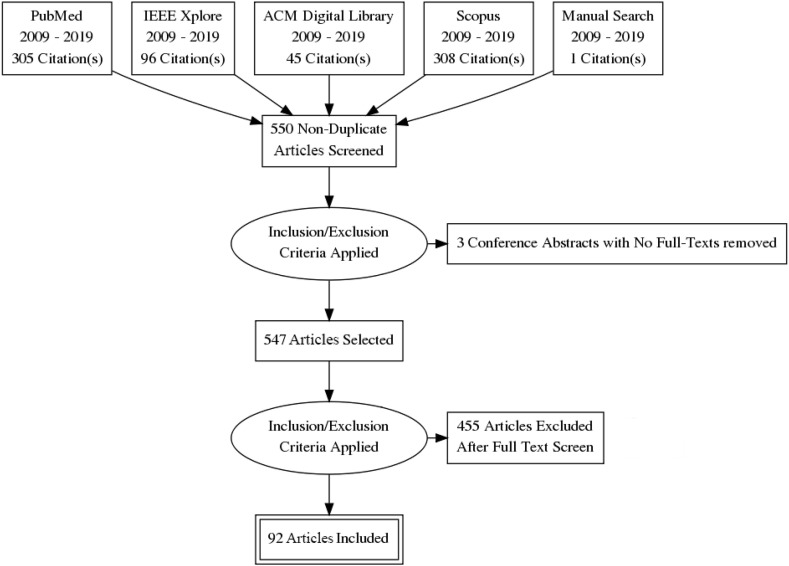
In accordance to best practice for systematic reviews and meta-analysis, we applied the guidelines for Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) [10] to select studies for inclusion in the analysis. The flowchart for PRISMA that corresponds to our review is shown in Fig. 1 .
Fig. 1.
PRISMA flow diagram for the identification and selection of studies.
754 research articles were returned by our search and 1 paper was added from the bibliographic listings of relevant retrieved papers. Of these 755 articles, we found 550 to be unique. We then drew up a list of criteria for inclusion and exclusion of articles in our review similar to those used by Shatte et al. [11]. These criteria are shown in Table 1 . In short, articles were included if all the following criteria were met: (i) the article reported on a method or application of Twitter data to address a public health issue; (ii) the article evaluated the performance of the statistical or machine learning technique used in drawing utility from the Twitter data; (iii) the article was published in a peer-reviewed publication and (iv) the article was available in English. Articles were excluded if any of the following criteria were met: (i) the article did not report an original contribution (e.g. review papers or articles commenting or speculating on the state or future of such research); (ii) the article was focused on the use of Twitter for public health in the context of recruitment and outreach, public awareness and communication, information dissemination or opinion mining; (iii) the article did not make known the statistical or machine learning technique being used; (iv) the full text of the article was not available (e.g. conference abstracts). Guided by our inclusion and exclusion criteria, we identified and selected 92 articles to be included for the review (see Table 2).
Table 1.
Inclusion and exclusion criteria.
| Criterion | Inclusion | Exclusion |
|---|---|---|
| Time period | 2009–2019 | Studies outside these dates |
| Language | English | Non-english articles |
| Article Type | Original peer-reviewed research | Research that was not peer-reviewed |
| Literature focus |
|
|
Table 2.
Summary of statistical and machine learning methods and data sources for surveillance using Twitter data.
| Public Health Issue | Method | Comparative Data Source |
|---|---|---|
| Cancer | Simple Statistical Analysis [23] | CDC |
| Hepatitis A | Support Vector Machine [24] | |
| Gastrointestinal Illnesses | Correlation Analysis [25] | Government of ontario, Kingston, Frontenac and Lennox & Addington Public Health |
| Suicide | ARIMA (Autoregressive Integrated Moving Average [26] | |
| HIV | Graph Modelling [27], Word2Vec [28], Doc2Vec [28], Dynamic Topic Modeling [28] | |
| Allergies | K-Nearest Neighbour [20], Bayesian Inference [20], Support Vector Machine [20] | |
| Heat Wave | Near Regression [18], ARIMA (Autoregressive Integrated Moving Average) [18] | The US National Oceanic and Atmospheric Administration (NOAA) National Centers for Environmental Information (NCEI) |
| Heat Related Illnesses | Correlation Analysis [25] | Government of ontario, Kingston, Frontenac and Lennox & Addington Public Health |
| Depression | ARIMA (Autoregressive Integrated Moving Average [26] | |
| Syphilis | Binomial Regressions [29] | CDC |
| Ebola | Bayesian Inference [30], Lexicon Analysis [30] | |
| Respiratory Illness | Correlation Analysis [25] | Government of ontario, Kingston, Frontenac and Lennox & Addington Public Health |
| E Coli | Latent Dirichlet Allocation [31], Lexicon Analysis [31] | Robert Koch Institute |
| Measles | Support Vector Machine [24] | |
| Influenze-like Illnesses (Hemophilus) | Bayesian Inference [15] | Genbank |
| Vomiting | TSVM [22], ARIMA (Autoregressive Integrated Moving Average) [22] | Public Health England |
| Gastroenteritis | TSVM [22], Latent Dirichlet Allocation [31], Lexicon Analysis [31], ARIMA (Autoregressive Integrated Moving Average) [22] | Public Health England, Robert Koch Institute |
| Salmonella | Support Vector Machine [24] | |
| Food Borne Illness | Support Vector Machine [32] | Southern Nevada Health District (SNHD) |
| Earthquake | Clustering [19], Bayesian Inference [19] | |
| Stress | Ordinal Regression [33] | |
| Air Pollution | Self-Organizing Map (Clustering) [34], Cross-Correlation [17] | The European Centre for Medium-Range Weather Forecasts (ECMWF), London Air Quality Network |
| Influenze-like Illnesses (ILI) | Lexicon Analysis [35], Deep Learning (CNN) [36], Fp-Growth [37], Bayesian Inference [38,39], Correlation Analysis [25], Deep Learning (RNN) [36], Deep Learning (MLP) [40], Fasttext [36], Bayesian Inference [35,41], ARIMA (Autoregressive Integrated Moving Average) [22,42], Simple Statistical Analysis [23], Support Vector Machine [37,43], Glove [36], Maximum Entropy [41], TSVM [22], Partial Differential Equation [44], Autoregressive Moving Average (Arma) [45], Outlier Detection [46], Topic Model [47], Temporal Topic Model [14], Logistic Regression [42], Count Correlation [16] | Public Health England, Frontenac and Lennox & Addington Public Health, Chinese CDC, Pan American Health Organization (PAHO), CDC, HHS data, Kingston, FluWatch, Government of ontario, The Pan American Health Organi-zation (PAHO) |
| General Healtha | Topic Model (Ailment Topic Aspect Model (Atam)) [48], Lexicon Analysis [49], Regression [49], Simple Statistical Analysis [50], Temporal Ailment Topic Aspect Model (TM-ATAM) [51] | CDC, U.S. Census' State-Based Counties Gazetteer |
| Dengue | Dbscan (Clustering) [21], Deep Learning (RNN) [52], Word Embeddings (Glove) [52], Simple Statistical Analysis [53] | Brazilian Health Ministry, Philippine's Department of Health, Brazilian Official Dengue case data |
| Diarrhoea | TSVM [22], ARIMA (Autoregressive Integrated Moving Average) [22] | Public Health England |
| Obesity | Dbscan (Clustering) [54] |
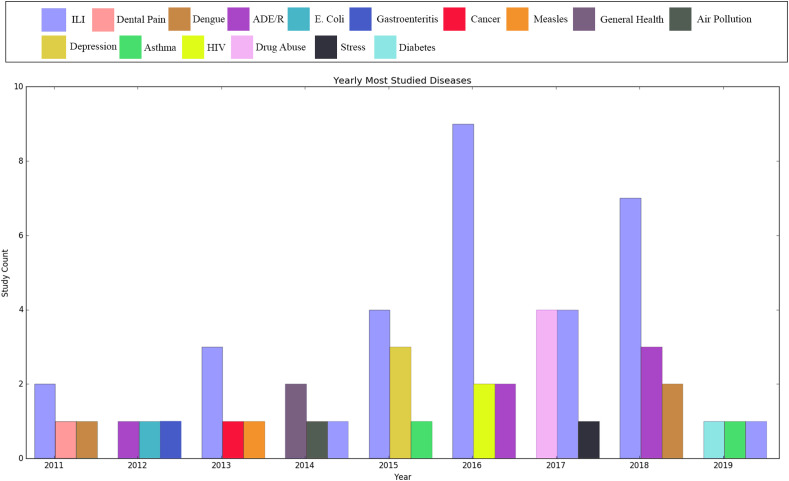
Note that the information shown for 2019 is not comparable to that for other years due to the fact that, at the time of plotting the graph, 2019 had not elapsed.
2.3. Information extraction and analysis plan
The focus of our review was to get an exploratory map of the key problems and concepts being tackled in the public health space through the use of Twitter and the techniques being used. To this effect, for each article in our review, data was collected on (i) the aim of the research (ii) the disease or illness of focus (iii) sources of data for the study (iv) statistical or machine learning algorithms and methods used (v) the country for which the study was carried out (vi) the year in which the study was carried out. To analyse the collected information, we used a narrative review synthesis to capture the broad range of research studying Twitter for public health in our scoping review.
3. Results
3.1. Study characteristics
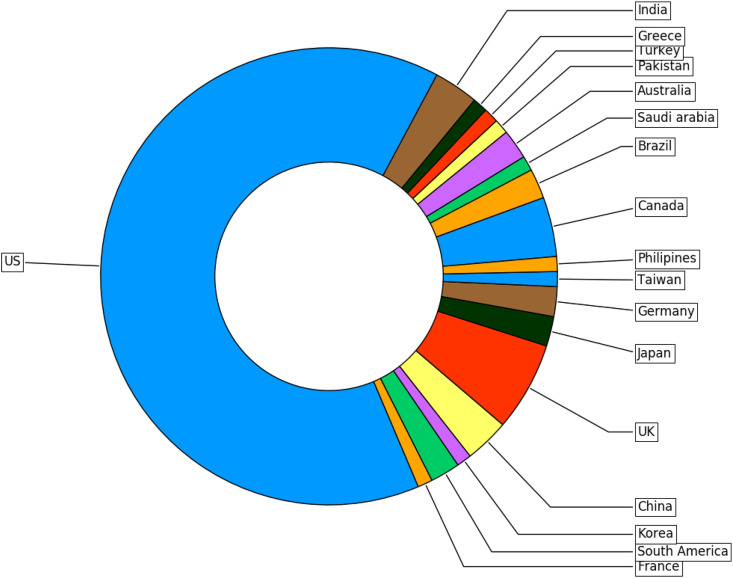
As explained in section 2.2, the search strategies identified 755 articles, with 92 of these articles meeting the criteria for inclusion in this review. The mode publication year for articles was 2017 with a range of 2011–2019.19 countries were represented in the studies, with the top 5 countries being the United States of America (US), United Kingdom (UK), Canada, India and China. See Fig. 3 for a breakdown of study activity by country.
Fig. 3.
Breakdown of studies by country.
The use of Twitter data was evident for a varied number of different diseases and health conditions. We observed a range of applications dealing with physical health and illnesses (n = 82) [e.g. influenza-like illnesses (ILIs), adverse drug events and reactions, sexually transmitted diseases, food-borne illneses], mental health (n = 6) [e.g. suicide and depression], natural disasters and environmental issues (n = 5) [e.g. earthquakes, heat waves, air pollution] and social issues (n = 8) [e.g. drug abuse, smoking, alcoholism]. We examined the subjects of the studies for trends in Twitter applications. We analysed and plotted the three most studied diseases for each year. Fig. 4 shows the result of this analysis. Taking a closer look at the diseases, conditions and public health phenomena studied using Twitter data, we observed ILIs to be the most common. The next most common subject of public health research using Twitter were drug abuse and adverse drug events and/or reactions (ADE/R). Furthermore, we observed a general rise in the quantity of research into the use of Twitter for public health. Research activity appears to have peaked in 2016 but seems to be on the rise from 2018. As this scoping review looks at studies up until March 2019, the data for 2019 is incomplete. This limitation is due to the fact that this review can only investigate studies until the time of its writing, which happened to be early in the year.
Fig. 4.
Most studied diseases each year.
Generic feelings of unwellness and non-specific illness.
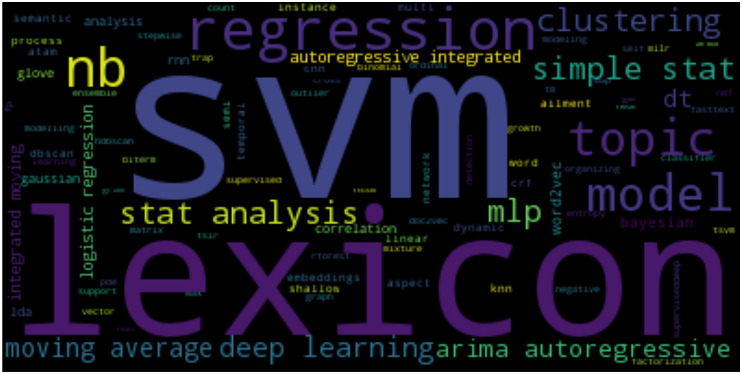
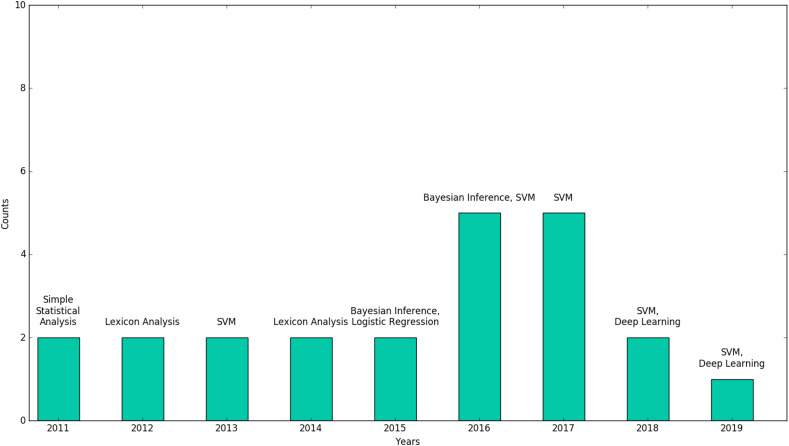
A myriad of statistical and machine learning techniques were used in the analysis of Twitter data for public health (see Fig. 2) . Most studies implemented just one technique (n = 54) but some others made use of a mix of methods and techniques (n = 38). The articles made use of a range of statistical and machine learning techniques including supervised learning (n = 70) [e.g.Support Vector Machine (SVM), naive bayes, decision trees, logistic regression], unsupervised learning (n = 18) [e.g. clustering, association rule mining], semi-supervised learning (n = 4) [e.g. graph learning, transductive support vector machine (t-SVM)], text analysis and natural language processing (n = 23) [e.g. latent Dirichlet allocation (LDA), biterm topic modelling, lexicon analysis], deep learning (n = 16) [e.g. Recurrent Neural Networks (RNNs), Convolutional Neural Networks (CNNs), word and document embeddings], statistical modelling and analysis (n = 12) [e.g. correclation analysis, partial differntial equation (PDE), TRAP] and time series analysis (n = 7) [e.g. Autoregressive Integrated Moving Average (ARIMA), time-series Susceptible-Infected-Recovered (TSIR) model]. The average number of Tweets used in the reviewed studies was roughly twenty thousand. A closer look at the research towards Twitter use for public health revealed that the SVM was a popular tool in this research field. We hypothesize that this is due to the SVM's popularity and strength in text classification problems [12]. We also analysed the surveyed studies to find out which statistical or machine learning algorithms were popular, as well as if and how this might have shifted over time. Fig. 5 shows a plot of the most used algorithms for each year covered in this review. Lexicon-based analysis proved popular between 2012 until 2014. After this, Bayesian learning seemed to be the method of choice, followed by the SVM. From 2018, the widespread popularity of deep learning appears to have made its way into public health research with Twitter data, as it is becoming the dominant method used since then.
Fig. 2.
Word cloud of statistical and machine learning methods discovered in review.
Fig. 5.
Most applied algorithms each year.
3.2. Application domains of Twitter in public health
Through the synthesis of the data obtained from the reviewed articles, we broadly identified 6 different ways in which Twitter data is used for public health research. The identified domains were: (i) surveillance (n = 41); (ii) event detection (n = 38); (iii) pharmacovigilance (n = 19); (iv) forecasting (n = 15); (v) disease tracking (n = 12) and (vi) geographic identification (n = 7). Note that these domains where not always mutually exclusive. Surveillance includes articles aiming to monitor some status over a period of time. Event detection includes articles that aim to discover and/or identify a health-related event from Twitter data. Pharmocovigilance includes articles which were concerned with public drug consumption and reactions to said drugs. Forecasting includes articles which aim to predict the trends for health-related events. Disease tracking includes articles attempting to observe or predict the spread of diseases in the public through Twitter. Geographic identification includes articles whose aim is to geolocate Twitter users, usually in order to facilitate or improve the application of one of the other domains.
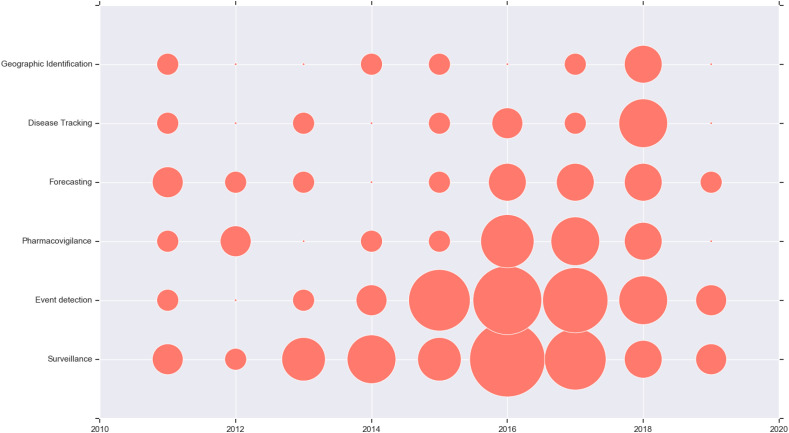
We were interested in examining the trends, if any, in the public health application domains studied over the years. We constructed a bubble trend chart from the reviewed papers. This chart, included in Fig. 6 , illustrates the research activity in each domain for each year with the size of the bubble representing the number of articles for a given year and public health domain. It shows that there appears to indeed be a trend in activity for different public health domains. In 2011, there is little to moderate activity across the board. In the years following that, we see research in some domains drop off and on the map, and some growing steadily in size. Event detection, surveillance and pharmacovigilance appear to have seen steady increases in activity, leading the other domains. However, since 2016, research in those three domains has reduced slightly, with some focus switching to the other domains. The data for the year 2019 is not particularly informative, as the scoping review was only carried out in the first quarter of 2019.
Fig. 6.
Bubble chart showing the trends of research activity in public health application domains with time. The size of the bubble represents the number of articles in each category and year.
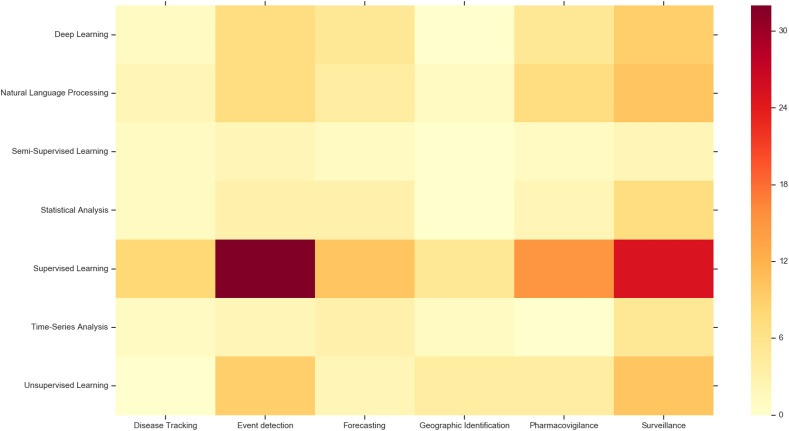
We were also interested in the different techniques applied across different public health research domains. We computed a matrix of the application domains against the techniques applied and visualised it as a heatmap. This heatmap is shown in Fig. 7 . Darker colours in the heatmap indicate higher activity for that cell. Supervised learning appears to see a lot of utility across the board. Deep learning and natural language processing also see a fair amount of utility, particularly in event detection, pharmacovigilance and surveillance. Unsupervised learning seems to see some utility use in surveillance and event detection. On the other hand, semi-supervised learning appears to see the least use across the board.
Fig. 7.
Most applied algorithms each year.
The reviewed articles were found to exist within one or more of these domains. These domains are discussed in more detail below.
3.2.1. Surveillance
Surveillance was the most popular research domain with around 43% of the reviewed articles represented. Research on surveillance focused on employing machine learning in order to utilize Twitter as an alternative or augmentative resource to traditional health surveillance systems. Naturally, the surveillance domain encompasses the field of syndromic surveillance [[13], [14], [15]]. However, it is broad and also includes additional applications such as the tracking of vaccination efforts [16] and monitoring of environmental conditions [17,18], as well as for natural disaster reporting and alarming [19]. That being said, the most common application was the syndromic surveillance of influenza-like illnesses (ILIs). Besides ILIs, other diseases and conditions that were studied include dengue, HIV, gastroenteritis, ebola, diarrhoea and allergies. Due to the extensive research carried out in this area, a wide range of techniques were used. For example, supervised learning applied in the form of k-Nearest Neighbours (kNN) was used to monitor allergy trends and occurences [20]. Unsupervised learning was used in the form of Density-based Spatial Clustering of Applications with Noise (DBSCAN) clustering in order to exploit the spatial and temporal properties of the Twitter stream for dengue surveillance [21]. Semi-supervised learning was used in the form of transductive SVMs for the surveillance of ILIs, gastroenteritis, diarrhoea and vomiting [22].
3.2.2. Event detection
Detection was another popular domain which saw around 40% of the reviewed articles represented. Research in this domain sought to automatically detect events and describe the magnitude and trend of disease, as well as the impact of control measures. Examples of applications in this domain are automatically detecting drug abuse within the population [55], depression and suicide [26], ebola [56] and most common of all, ILI [57]. Such research tends to be fairly recent with the mode publication year being 2016. The statistical and machine learning techniques used were typically supervised, with most studies employing either classification or regression to make the predictions necessary for detection. For example, SVMs were used to detect mention of ‘‘dabbing’‘, a method of marijuana consumption that involves inhaling vapors from heating marijuana concentrates [58]. CNNs were used to detect harmful algal blooms from pictures posted on Twitter [59]. Additionally, stepwise regression was used to detect depression from Tweets in order to explore the effect of climate and seasonality on mood [60].
3.2.3. Pharmacovigilance
Research in pharmocovigilance focused mainly on adverse drug reactions and events, but also investigated with recreational drug use and abuse. Usually, when studying the use of Twitter to detect adverse drug reactions and events, articles searched for a range of names obtained from a thesaurus of drugs and events, such as the Medline Plus Drug Information [83]. However, other such studies focused on a drug for a particular disease such as HIV [63]. In addition, studies also investigated drug habits and their effects on the population. For example, one article studied the use of e-cigarettes and their utility for smoking cessation [62]. Another article studied the variability of alcoholism with time [84]. A number of the pharmacovigilance studies utilized sentiment analysis, usually a form of supervised text classification, to aid in their efforts [28,63,83]. In fact, most of the studies make use of supervised learning in the form of text classification using mostly SVMs and decision trees. Of the 19 articles in this domain, three made use of deep learning [28,85,86], one employed a semi-supervised multi-instance learning approach [86] and three used unsupervised natural language processing [28,66,87].
3.2.4. Forecasting
Forecasting research studies the prediction of public health trends, as well as means of nowcasting which is the prediction of the present state of public health. It can be seen as a part of the syndromic surveillance effort, aimed at predicting epidemics in order to improve crisis response. Research in this domain is focused predominantly on ILIs. Around 67% of the reviewed literature studied ILI. However, other diseases such as dengue, gastroenteritis, cancer and asthma were also studied [22,23,53,95]. While a mix of statistics and machine learning is used in this domain, there is a heavier focus on statistics. In fact most studies made use of statistical techniques like regression and time series analysis. For example, dynamic regression was used to predict infuenza trends in Boston, USA [96]. AutoRegressive Integrated Moving Average (ARIMA) was used to forecast influenza cases on a city level in Chongqing, China, as well as for predicting gatroenteritis in the UK [22,97]. Partial differential equations were used to forecast influenza cases on a regional level across the USA [44]. Deep learning was also used to aid in the forecasting problem of predicting influenza cases [40] and in the creation of SENTINEL, a software system capable of nowcasting diseases being monitored by the US Centre for Disease Control (CDC) [98]. Unsupervised learning was used in the form of topic modelling in a study aiming to predict health transition trends without any a priori diseases [51].
3.2.5. Disease tracking
Disease tracking is a domain that seeks to support epidemiology by offering insight into the spread of infectious diseases. Research in this domain is primarily interested in understanding the way in which diseases spread through a population. It looks toward not only gaining a better understanding of the spread of diseases, but also to keep track of the public health state during recognized outbreaks and mass gatherings which could be a breeding ground for disease. For example, one study investigated and proposed a means of traking flu transmission in China using Twitter [39]. Another study retrospectively tracked the spread of measles during the 2015 outbreak [101]. Additionally, there was a study to detect the occurrence and spread of disease symptoms which could signify a potential outbreak at a number of British music festivals and a religious event in Mecca, Saudi Arabia [50]. Most studies in this domain made use of machine learning methods, leaning towards supervised learning. In particular, regression learning proved popular, as two studies utilized dynamic regression and support vector regression to track the spread of influenza [96,100]. Another study proposed a gaussian mixture regression approach to estimating the geographic origin of a tweet for use during an outbreak [102]. There were also some studies which used statistical analysis to obtain impressive results. One of such studies made use of the TSIR (time-series Susceptible-Infected-Recovered) model to understand human mobility and the spread of the dengue virus in Lahore, Pakistan [103]. While it was rare, one study made use of semi-supervised learning and deep learning to simulate influenza epidemics.
3.2.6. Geographic identification
Geographic identification is a small domain which involves the extraction of geographical information from Twitter data and typically sees little use alone. Rather, it is used in conjunction with other domains to improve the efficacy of solutions or provide added benefit. It is most often used with surveillance and disease tracking. Methods used in geograhic identification are typically based on unsupervised learning. For example, DBSCAN clustering was used to monitor and track obesity levels within the population [54], as well as track the spread of the dengue virus [21]. Another study utilized hot spot analysis to examine spatial patterns of depression on Twitter. Some supervised learning, typically in the form of classification is also used in geographic identification. Here, a classifier is used to predict the location of a tweet based on some features of the tweet, usually its word collocations. As an example, one study in the review made use of a random forest classifier to predict which city and province a tweet determined to be from Canada (according to the Twitter API), was from Ref. [105]. While geographic identification in itself is not of major use to the field of public health, when combined with other identified public health research domains, it offers improvements on the specificity and granularity of their results.
4. Discussion
This review has compiled and analysed the published literature on the use of Twitter data for public health, highlighting popular and current research and applications. In terms of research undertaken so far, three findings were produced from the review. First, we identified the key application domains being studied: (i) surveillance; (ii) event detection; (iii) pharmacovigilance; (iv) forecasting; (v) disease tracking and (vi) geographic identification. Studies were found to predominantly be concerned with surveillance, event detection and pharmacovigilance. Next, the conditions and diseases being tackled using Twitter data were identified. We discovered a wide range of illnesses to which Twitter data is being applied to including infectious diseases, mental health problems, environmental issues and social issues. Finally, we mapped out the statistical and machine learning algorithms and approaches being used to process and analyse Twitter data for public health purposes. In doing so, we observed trends in these approaches. Bayesian learning and SVMs appear to be popular algorithms of choice, however, in the past two years the focus seems to have shifted towards deep learning.
So far our findings will enable researchers working in health data to identify relevant studies in different application areas, tackling different diseases or conditions and will also provide evidence of analysis techniques that have been applied in each context. This will enable faster development of new applications, which is an important contribution of our research with the growth on the user of Twitter around the world, and particularly in Low and Middle Income Countries (LMIC). The use of Twitter in a health context can present new practical and affordable solutions for implementing disease monitoring and surveillance in countries with weak health systems.
While research toward using Twitter for public health has been extensive, our study has also identified some gaps for future researchers to fill. The identification of gaps is an important deliverable of a scoping review and hence a contribution of our work.
In terms of diseases tackled so far, understandably, studies are focused on infectious diseases because of their global importance. In particular, the reviewed research focused heavily on the surveillance and detection of influenza. However, we have identified significant scope to explore the use of Twitter data in other infectious diseases. Some such studies are beginning to take place (e.g. dengue or ebola) but much more work is expected in the light of recent outbreaks. Often outbreaks are fast moving situations and research needs to progress very quickly so our findings will facilitate such endevours. Whereas we may not expect Twitter data to be of use for the study of sexually transmitted diseases (STDs) as such a study would rely on Twitter user-reporting what may be quite sensitive information, other infectious diseases such as cholera could be studied. Furthermore, we have also identified the potential utility of Twitter and social media for public health in the context of non-infectious diseases, such as asthma or celiac disease as little work has so far been reported in the literature, yet those diseases can represent a large health burden. An additional area of application may be the occurrence of positive health states/outcomes. Our review did not identify any articles that used Twitter for this, although it might be a result of the limitations of our scoping methodology.
In terms of analysis techniques employed so far, there was wide application of supervised learning techniques. This is somewhat understandable as the most popular application domains were surveillance and detection, which are related to the supervised learning tasks of classification and prediction. The average number of Tweets used in the reviewed studies was roughly twenty thousand. This suggests that most of the reviewed articles had large amounts of labelled Twitter data available to them which leads to supervised learning tasks. Unfortunately, such labeling could constitute a sizeable effort so we have identified the use of unsupervised learning, and particularly semi-supervised learning, as another potential area for new exploration. Such approaches would reduce the amount of labelled Twitter data required by also taking advantage of the unlabeled data. Some articles are already starting to emerge [91,104] but mostly only focused on ILI so far.
Furthermore, in terms of application areas despite the rich potential for success from using Twitter data for public health which was identified in the literature, there were few articles describing active Twitter-based systems and/or their evaluation in an operational context for routine public health practice. This may suggest that it is somewhat difficult to translate research using Twitter for public health into practice. We believe the bulk of this challenge might come from the ethical issues involved and the lack of an ethical framework for the integration of social media into surveillance systems. Hence the development of robust ethical frameworks could be an important area for future work. That being said, public health institutions around the world may already be using Twitter as such a tool, and just not reporting their efforts.
It is also important to note that this review had some limitations. Constraints in the search methodology such as the use of broad search terms and the exclusion of works-in-progress may have resulted in some relevant studies being missed. However, this is a common limitation of scoping reviews as they are intended to broadly map topics, achieving a good balance of breadth and depth in a relatively quick time-frame [107].
5. Conclusion
This review makes an important contribution by successfully giving an overview of the use of Twitter data in the context of monitoring, detection and forecasting of public health conditions. We providing insightful analysis of the existing literature in the field, including the type of conditions being monitored; the data analysis techniques being used and the application areas most commonly found. We also analysed time trends to understand how research in this area is evolving over time. Such information will be useful in aiding researchers, clinicians and policy makers in understanding the modern landscape of public health applications for social media.
To conclude, research into the application of Twitter data for public health has uncovered interesting and inspiring advances, especially in recent years, and identified gaps in the knowledge thus allowing targeted research in the future. Overall, we see that Twitter data has been used to aid in pubic health efforts concerned with surveillance, event detection, pharmacovigilance, forecasting, disease tracking and geographic identification, demonstrating positive results. We have uncovered the need to evaluate the use of Twitter in less studied epidemiological diseases and other non-epidemiological conditions. We also uncovered scope to apply semi-supervised algorithms to the task in hand to reduce labelling efforts. Furthermore, we have identified the need for a robust framework including ethics to translate research into an operational context and produce working systems.
With the richness of Twitter as a data source, is semi-real time nature, the take up of mobile devices in LMIC that give access to such platforms and with the development of machine learning tools and their increasing accessibility, we expect to see more interesting ideas and applications of Twitter to public health.
Declaration of competing interest
None Declared.
Acknowledgements
We acknowledge support from Grant Number ES/L011859/1, from The Business and Local Government Data Research Centre, funded by the Economic and Social Research Council to provide economic, scientific and social researchers and business analysts with secure data services.
References
- 1.World Health Organisation Who The world health report 2007 - a safer future: global public health security in the 21st century. 2007. http://www.who.int/whr/2007/en/
- 2.Triple S. Assessment of syndromic surveillance in europe. Lancet. 2011;378(9806):1833. doi: 10.1016/S0140-6736(11)60834-9. [DOI] [PubMed] [Google Scholar]
- 3.Winslow C.-E. The untilled fields of public health. Science. 1920:23–33. doi: 10.1126/science.51.1306.23. [DOI] [PubMed] [Google Scholar]
- 4.Neiger B.L., Thackeray R., Burton S.H., Giraud-Carrier C.G., Fagen M.C. Evaluating social media's capacity to develop engaged audiences in health promotion settings: use of twitter metrics as a case study. Health Promot. Pract. 2013;14(2):157–162. doi: 10.1177/1524839912469378. [DOI] [PubMed] [Google Scholar]
- 5.Wargon M., Guidet B., Hoang T., Hejblum G. A systematic review of models for forecasting the number of emergency department visits. Emerg. Med. J. 2009;26(6):395–399. doi: 10.1136/emj.2008.062380. [DOI] [PubMed] [Google Scholar]
- 6.Charles-Smith L.E., Reynolds T.L., Cameron M.A., Conway M., Lau E.H., Olsen J.M., Pavlin J.A., Shigematsu M., Streichert L.C., Suda K.J. Using social media for actionable disease surveillance and outbreak management: a systematic literature review. PloS One. 2015;10(10) doi: 10.1371/journal.pone.0139701. e0139701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinnenberg L., Buttenheim A.M., Padrez K., Mancheno C., Ungar L., Merchant R.M. Twitter as a tool for health research: a systematic review. Am. J. Publ. Health. 2017;107(1) doi: 10.2105/AJPH.2016.303512. e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arksey H., O'Malley L. Scoping studies: towards a methodological framework. Int. J. Soc. Res. Methodol. 2005;8(1):19–32. [Google Scholar]
- 9.Ginsberg J., Mohebbi M.H., Patel R.S., Brammer L., Smolinski M.S., Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457(7232):1012. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 10.Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. 2010. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the Prisma Statement. [PMC free article] [PubMed] [Google Scholar]
- 11.Shatte A.B., Hutchinson D.M., Teague S.J. Machine learning in mental health: a scoping review of methods and applications. Psychol. Med. 2019:1–23. doi: 10.1017/S0033291719000151. [DOI] [PubMed] [Google Scholar]
- 12.Joachims T. European Conference on Machine Learning, Springer. 1998. Text categorization with support vector machines: learning with many relevant features; pp. 137–142. [Google Scholar]
- 13.C. Hankin, O. Serban, N. Thapen, B. Maginnis, V. Foot, Real-time processing of social media with sentinel: a syndromic surveillance system incorporating deep learning for health classification.
- 14.Chen L., Hossain K.S.M.T., Butler P., Ramakrishnan N., Prakash B.A. Syndromic surveillance of flu on twitter using weakly supervised temporal topic models. Data Min. Knowl. Discov. 2015;30(3):681–710. doi: 10.1007/s10618-015-0434-x. doi:10.1007/s10618-015-0434-x. [DOI] [Google Scholar]
- 15.Janies D., Witter Z., Gibson C., Kraft T., Senturk I.F., Çatalyürek Ü. Syndromic surveillance of infectious diseases meets molecular epidemiology in a workflow and phylogeographic application. Stud. Health Technol. Inf. 2015;216:766–770. [PubMed] [Google Scholar]
- 16.Song S., Miled Z.B. 2017 IEEE 14th International Conference on Mobile Ad Hoc and Sensor Systems (MASS) IEEE; 2017. Digital immunization surveillance: monitoring flu vaccination rates using online social networks. doi:10.1109/mass.2017.96. [DOI] [Google Scholar]
- 17.Hswen Y., Qin Q., Brownstein J.S., Hawkins J.B. Feasibility of using social media to monitor outdoor air pollution in london, england. Prev. Med. 2019;121:86–93. doi: 10.1016/j.ypmed.2019.02.005. doi:10.1016/j.ypmed.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung J., Uejio C.K. Social media responses to heat waves. Int. J. Biometeorol. 2017;61(7):1247–1260. doi: 10.1007/s00484-016-1302-0. doi:10.1007/s00484-016-1302-0. [DOI] [PubMed] [Google Scholar]
- 19.Auxilia R., Gandhi M. Earthquake reporting system development by tweet analysis with approach earthquake alarm systems. Res. J. Pharmaceut. Biol. Chem. Sci. 2016;7(3):501–506. [Google Scholar]
- 20.Nargund K., Natarajan S. 2016 International Conference on Advances in Computing, Communications and Informatics (ICACCI) IEEE; 2016. Public health allergy surveillance using micro-blogs. doi:10.1109/icacci.2016.7732248. [DOI] [Google Scholar]
- 21.Gomide J., Veloso A., Meira W., Almeida V., Benevenuto F., Ferraz F., Teixeira M. vol. 11. ACM Press; 2011. Dengue surveillance based on a computational model of spatio-temporal locality of twitter. (Proceedings of the 3rd International Web Science Conference on - WebSci). doi:10.1145/2527031.2527049. [DOI] [Google Scholar]
- 22.Thapen N., Simmie D., Hankin C., Gillard J., Defender Detecting and forecasting epidemics using novel data-analytics for enhanced response. PloS One. 2016;11(5) doi: 10.1371/journal.pone.0155417. e0155417. doi:10.1371/journal.pone.0155417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K., Agrawal A., Choudhary A. vol. 13. ACM Press; 2013. Real-time disease surveillance using twitter data. (Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD). doi:10.1145/2487575.2487709. [DOI] [Google Scholar]
- 24.Krieck M., Otrusina L., Smrz P., Dolog P., Nejdl W., Velasco E., Denecke K. How to exploit twitter for public health monitoring? Methods Inf. Med. 2013;52(4):326–339. doi: 10.3414/me12-02-0010. doi:10.3414/me12-02-0010. [DOI] [PubMed] [Google Scholar]
- 25.Khan Y., Leung G.J., Belanger P., Gournis E., Buckeridge D.L., Liu L., Li Y., Johnson I.L. Comparing twitter data to routine data sources in public health surveillance for the 2015 pan/parapan american games: an ecological study. Can. J. Public Health. 2018;109(3):419–426. doi: 10.17269/s41997-018-0059-0. doi:10.17269/s41997-018-0059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClellan C., Ali M.M., Mutter R., Kroutil L., Landwehr J. Using social media to monitor mental health discussions - evidence from twitter. J. Am. Med. Inf. Assoc. 2016 doi: 10.1093/jamia/ocw133. ocw133. doi:10.1093/jamia/ocw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thangarajan N., Green N., Gupta A., Little S., Weibel N. vol. 15. ACM Press; 2015. Analyzing social media to characterize local HIV at-risk populations. (Proceedings of the Conference on Wireless Health - WH). doi:10.1145/2811780.2811923. [DOI] [Google Scholar]
- 28.Breen P., Kelly J., Heckman T., Quinn S. IEEE International Conference on Data Science and Advanced Analytics (DSAA) IEEE; 2016. Mining pre-exposure prophylaxis trends in social media. 2016. doi:10.1109/dsaa.2016.29. [DOI] [Google Scholar]
- 29.Young S.D., Mercer N., Weiss R.E., Torrone E.A., Aral S.O. Using social media as a tool to predict syphilis. Prev. Med. 2018;109:58–61. doi: 10.1016/j.ypmed.2017.12.016\. doi:10.1016/j.ypmed.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ofoghi B., Mann M., Verspoor K. Biocomputing 2016: Proceedings of the Pacific Symposium. World Scientific; 2016. Towards early discovery of salient health threats: a social media emotion classification technique; pp. 504–515. [PubMed] [Google Scholar]
- 31.Diaz-Aviles E., Stewart A. Proceedings of the 3rd Annual ACM Web Science Conference on - WebSci '12. ACM Press; 2012. Tracking twitter for epidemic intelligence. doi:10.1145/2380718.2380730. [DOI] [Google Scholar]
- 32.Sadilek A., Kautz H., DiPrete L., Labus B., Portman E., Teitel J., Silenzio V. Deploying nemesis: preventing foodborne illness by data mining social media. AI Mag. 2017;38(1):37–48. [Google Scholar]
- 33.Liu S., Zhu M., Yu D.J., Rasin A., Young S.D. Using real-time social media technologies to monitor levels of perceived stress and emotional state in college students: a web-based questionnaire study. JMIR Mental Health. 2017;4(1) doi: 10.2196/mental.5626. e2. doi:10.2196/mental.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riga M., Karatzas K. Proceedings of the 4th International Conference on Web Intelligence, Mining and Semantics (WIMS14) - WIMS '14. ACM Press; 2014. Investigating the relationship between social media content and real-time observations for urban air quality and public health. doi:10.1145/2611040.2611093. [DOI] [Google Scholar]
- 35.Lin G., Zaeem R.N., Sun H., Barber K.S. Intelligent Systems Conference (IntelliSys) IEEE; 2017. Trust filter for disease surveillance: Identity. 2017. doi:10.1109/intellisys.2017.8324259. [DOI] [Google Scholar]
- 36.Şerban O., Thapen N., Maginnis B., Hankin C., Foot V. Real-time processing of social media with SENTINEL: a syndromic surveillance system incorporating deep learning for health classification. Inf. Process. Manag. 2019;56(3):1166–1184. doi: 10.1016/j.ipm.2018.04.011. doi:10.1016/j.ipm.2018.04.011. [DOI] [Google Scholar]
- 37.Parker J., Yates A., Goharian N., Frieder O. Health-related hypothesis generation using social media data. Social Network Analysis and Mining. 2015;5(1) doi: 10.1007/s13278-014-0239-8. doi:10.1007/s13278-014-0239-8. [DOI] [Google Scholar]
- 38.J. D. Sharpe, R. S. Hopkins, R. L. Cook, C. W. Striley, Evaluating google, twitter, and wikipedia as tools for influenza surveillance using bayesian change point analysis: a comparative analysis, JMIR public health and surveillance 2 (2). [DOI] [PMC free article] [PubMed]
- 39.Huang J., Zhao H., Zhang J. IEEE International Conference on Green Computing and Communications and IEEE Internet of Things and IEEE Cyber, Physical and Social Computing. IEEE; 2013. Detecting flu transmission by social sensor in China. 2013. doi:10.1109/greencom-ithings-cpscom.2013.216. [DOI] [Google Scholar]
- 40.Lee K., Agrawal A., Choudhary A. IEEE International Conference on Healthcare Informatics (ICHI) IEEE; 2017. Forecasting influenza levels using real-time social media streams. 2017. doi:10.1109/ichi.2017.68. [DOI] [Google Scholar]
- 41.Byrd K., Mansurov A., Baysal O. Proceedings of the International Workshop on Software Engineering in Healthcare Systems. ACM; 2016. Mining twitter data for influenza detection and surveillance; pp. 43–49. [Google Scholar]
- 42.Broniatowski D.A., Dredze M., Paul M.J., Dugas A. Using social media to perform local influenza surveillance in an inner-city hospital: a retrospective observational study. JMIR public health and surveillance. 2015;1(1) doi: 10.2196/publichealth.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen C., Tsou M.-H., Aslam A., Nagel A., Gawron J.-M. Applying GIS and machine learning methods to twitter data for multiscale surveillance of influenza. PloS One. 2016;11(7) doi: 10.1371/journal.pone.0157734. e0157734. doi:10.1371/journal.pone.0157734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang F., Wang H., Xu K., Raymond R., Chon J., Fuller S., Debruyn A. Regional level influenza study with geo-tagged twitter data. J. Med. Syst. 2016;40(8) doi: 10.1007/s10916-016-0545-y. doi:10.1007/s10916-016-0545-y. [DOI] [PubMed] [Google Scholar]
- 45.Achrekar H., Gandhe A., Lazarus R., Yu S.-H., Liu B. IEEE Conference on Computer Communications Workshops (INFOCOM WKSHPS) IEEE; 2011. Predicting flu trends using twitter data. 2011. doi:10.1109/infcomw.2011.5928903. [DOI] [Google Scholar]
- 46.Dai X., Bikdash M. SoutheastCon 2016. IEEE; 2016. Distance-based outliers method for detecting disease outbreaks using social media. doi:10.1109/secon.2016.7506752. [DOI] [Google Scholar]
- 47.Chen L., Hossain K.T., Butler P., Ramakrishnan N., Prakash B.A. IEEE International Conference on Data Mining. IEEE; 2014. Flu gone viral: syndromic surveillance of flu on twitter using temporal topic models. 2014. doi:10.1109/icdm.2014.137. [DOI] [Google Scholar]
- 48.Parker J., Wei Y., Yates A., Frieder O., Goharian N. vol. 13. ACM Press; 2013. A framework for detecting public health trends with twitter. (Proceedings of the 2013 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining - ASONAM). doi:10.1145/2492517.2492544. [DOI] [Google Scholar]
- 49.Culotta A. vol. 14. ACM Press; 2014. Estimating county health statistics with twitter. (Proceedings of the 32nd Annual ACM Conference on Human Factors in Computing Systems - CHI). doi:10.1145/2556288.2557139. [DOI] [Google Scholar]
- 50.Yom-Tov E., Borsa D., Cox I.J., McKendry R.A. Detecting disease outbreaks in mass gatherings using internet data. J. Med. Internet Res. 2014;16(6) doi: 10.2196/jmir.3156. e154. doi:10.2196/jmir.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sidana S., Amer-Yahia S., Clausel M., Rebai M., Mai S.T., Amini M.-R. Health monitoring on social media over time. IEEE Trans. Knowl. Data Eng. 2018;30(8):1467–1480. doi: 10.1109/tkde.2018.2795606. doi:10.1109/tkde.2018.2795606. [DOI] [Google Scholar]
- 52.Livelo E.D., Cheng C. IEEE International Conference on Agents (ICA) IEEE; 2018. Intelligent dengue infoveillance using gated recurrent neural learning and cross-label frequencies. 2018. doi:10.1109/agents.2018.8459963. [DOI] [Google Scholar]
- 53.de Almeida Marques-Toledo C., Degener C.M., Vinhal L., Coelho G., Meira W., Codeço C.T., Teixeira M.M. Dengue prediction by the web: tweets are a useful tool for estimating and forecasting dengue at country and city level. PLoS Neglected Trop. Dis. 2017;11(7) doi: 10.1371/journal.pntd.0005729. e0005729. doi:10.1371/journal.pntd.0005729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khanaferov D., Luc C., Wang T. IEEE International Conference on Semantic Computing. IEEE; 2014. Social network data mining using natural language processing and density based clustering. 2014. doi:10.1109/icsc.2014.48. [DOI] [Google Scholar]
- 55.Mackey T.K., Kalyanam J. Detection of illicit online sales of fentanyls via twitter. F1000Research. 2017;6:1937. doi: 10.12688/f1000research.12914.1. doi:10.12688/f1000research.12914.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudra K., Sharma A., Ganguly N., Imran M. vol. 17. ACM Press; 2017. Classifying information from microblogs during epidemics. (Proceedings of the 2017 International Conference on Digital Health - DH). doi:10.1145/3079452.3079491. [DOI] [Google Scholar]
- 57.Dai X., Bikdash M. SoutheastCon 2015. IEEE; 2015. Hybrid classification for tweets related to infection with influenza. doi:10.1109/secon.2015.7133015. [DOI] [Google Scholar]
- 58.Ginart A.A., Das S., Harris J.K., Wong R., Yan H., Krauss M., Cavazos-Rehg P.A. 2016 IEEE International Conference on Healthcare Informatics (ICHI) IEEE; 2016. Drugs or dancing? using real-time machine learning to classify streamed “dabbing” homograph tweets. doi:10.1109/ichi.2016.97. [DOI] [Google Scholar]
- 59.Kumar A.C., Bhandarkar S.M. IEEE Winter Conference on Applications of Computer Vision (WACV) IEEE; 2017. A deep learning paradigm for detection of harmful algal blooms. 2017. doi:10.1109/wacv.2017.88. [DOI] [Google Scholar]
- 60.Yang W., Mu L., Shen Y. Effect of climate and seasonality on depressed mood among twitter users. Appl. Geogr. 2015;63:184–191. doi: 10.1016/j.apgeog.2015.06.017. doi:10.1016/j.apgeog.2015.06.017. [DOI] [Google Scholar]
- 61.Esperanca A., Miled Z.B., Mahoui M. 2019 IEEE 9th Annual Computing and Communication Workshop and Conference (CCWC) IEEE; 2019. Social media sensing framework for population health. doi:10.1109/ccwc.2019.8666534. [DOI] [Google Scholar]
- 62.Aphinyanaphongs Y., Lulejian A., Brown D.P., Bonneau R., Krebs P. Biocomputing 2016: Proceedings of the Pacific Symposium. World Scientific; 2016. Text classification for automatic detection of e-cigarette use and use for smoking cessation from twitter: a feasibility pilot; pp. 480–491. [PMC free article] [PubMed] [Google Scholar]
- 63.Adrover C., Bodnar T., Huang Z., Telenti A., Salathé M. Identifying adverse effects of HIV drug treatment and associated sentiments using twitter. JMIR Public Health and Surveillance. 2015;1(2) doi: 10.2196/publichealth.4488. doi:10.2196/publichealth.4488 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee K., Agrawal A., Choudhary A. vol. 15. ACM Press; 2015. Mining social media streams to improve public health allergy surveillance. (Proceedings of the 2015 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining 2015 - ASONAM). doi:10.1145/2808797.2808896. [DOI] [Google Scholar]
- 65.Phan N., Chun S.A., Bhole M., Geller J. IEEE 33rd International Conference on Data Engineering (ICDE) IEEE; 2017. Enabling real-time drug abuse detection in tweets. 2017. doi:10.1109/icde.2017.221. [DOI] [Google Scholar]
- 66.Mackey T.K., Kalyanam J., Katsuki T., Lanckriet G. Twitter-based detection of illegal online sale of prescription opioid. Am. J. Publ. Health. 2017;107(12):1910–1915. doi: 10.2105/ajph.2017.303994. doi:10.2105/ajph.2017.303994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.P. M. Massey, A. Leader, E. Yom-Tov, A. Budenz, K. Fisher, A. C. Klassen, Applying multiple data collection tools to quantify human papillomavirus vaccine communication on twitter, J. Med. Internet Res. 18 (12). [DOI] [PMC free article] [PubMed]
- 68.Zou B., Lampos V., Gorton R., Cox I.J. vol. 16. ACM Press; 2016. On infectious intestinal disease surveillance using social media content. (Proceedings of the 6th International Conference on Digital Health Conference - DH). doi:10.1145/2896338.2896372. [DOI] [Google Scholar]
- 69.Wang J., Zhao L., Ye Y., Zhang Y. Adverse event detection by integrating twitter data and VAERS. J. Biomed. Semant. 2018;9(1) doi: 10.1186/s13326-018-0184-y. doi:10.1186/s13326-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang W., Mu L. GIS analysis of depression among twitter users. Appl. Geogr. 2015;60:217–223. doi: 10.1016/j.apgeog.2014.10.016. doi:10.1016/j.apgeog.2014.10.016. [DOI] [Google Scholar]
- 71.Nambisan P., Luo Z., Kapoor A., Patrick T.B., Cisler R.A. 2015 48th Hawaii International Conference on System Sciences. IEEE; 2015. Social media, big data, and public health informatics: ruminating behavior of depression revealed through twitter. doi:10.1109/hicss.2015.351. [DOI] [Google Scholar]
- 72.Lee H., McAuley J.H., Hübscher M., Allen H.G., Kamper S.J., Moseley G.L. Tweeting back: predicting new cases of back pain with mass social media data. J. Am. Med. Inf. Assoc. 2015;23(3):644–648. doi: 10.1093/jamia/ocv168. doi:10.1093/jamia/ocv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harris J.K., Mansour R., Choucair B., Olson J., Nissen C., Bhatt J. vol. 63. 2014. p. 681. (Health Department Use of Social Media to Identify Foodborne Illness––chicago, illinois, 2013–2014, MMWR. Morbidity and mortality weekly report). 32. [PMC free article] [PubMed] [Google Scholar]
- 74.Heaivilin N., Gerbert B., Page J., Gibbs J. Public health surveillance of dental pain via twitter. J. Dent. Res. 2011;90(9):1047–1051. doi: 10.1177/0022034511415273. doi:10.1177/0022034511415273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dai X., Bikdash M., Meyer B. SoutheastCon. IEEE; 2017. From social media to public health surveillance: word embedding based clustering method for twitter classification. 2017. doi:10.1109/secon.2017.7925400. [DOI] [Google Scholar]
- 76.Lim S., Tucker C.S., Kumara S. An unsupervised machine learning model for discovering latent infectious diseases using social media data. J. Biomed. Inf. 2017;66:82–94. doi: 10.1016/j.jbi.2016.12.007. doi:10.1016/j.jbi.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 77.Broniatowski D.A., Paul M.J., Dredze M. National and local influenza surveillance through twitter: an analysis of the 2012-2013 influenza epidemic. PloS One. 2013;8(12) doi: 10.1371/journal.pone.0083672. e83672. doi:10.1371/journal.pone.0083672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wagner M., Lampos V., Cox I.J., Pebody R. The added value of online user-generated content in traditional methods for influenza surveillance. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-32029-6. doi:10.1038/s41598-018-32029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wakamiya S., Kawai Y., Aramaki E. Twitter-based influenza detection after flu peak via tweets with indirect information: text mining study. JMIR Public Health and Surveillance. 2018;4(3) doi: 10.2196/publichealth.8627. e65. doi:10.2196/publichealth.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woo H., Cho H.S., Shim E., Lee J.K., Lee K., Song G., Cho Y. Identification of keywords from twitter and web blog posts to detect influenza epidemics in korea. Disaster Med. Public Health Prep. 2017;12(3):352–359. doi: 10.1017/dmp.2017.84. doi:10.1017/dmp.2017.84. [DOI] [PubMed] [Google Scholar]
- 81.Hu H., Wang H., Wang F., Langley D., Avram A., Liu M. Prediction of influenza-like illness based on the improved artificial tree algorithm and artificial neural network. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-23075-1. doi:10.1038/s41598-018-23075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Küçük E.E., Yapar K., Küçük D., Küçük D. Ontology-based automatic identification of public health-related Turkish tweets. Comput. Biol. Med. 2017;83:1–9. doi: 10.1016/j.compbiomed.2017.02.001. doi:10.1016/j.compbiomed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 83.Peng Y., Moh M., Moh T.-S. 2016 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM) IEEE; 2016. Efficient adverse drug event extraction using twitter sentiment analysis. doi:10.1109/asonam.2016.7752365. [DOI] [Google Scholar]
- 84.West J.H., Hall P.C., Hanson C.L., Prier K., Giraud-Carrier C., Neeley E.S., Barnes M.D. Temporal variability of problem drinking on twitter. Open J. Prev. Med. 2012;2:43. 01. [Google Scholar]
- 85.Lin W.-S., Dai H.-J., Jonnagaddala J., Chang N.-W., Jue T.R., Iqbal U., Shao J.Y.-H., Chiang I.-J., Li Y.-C. 2015 Conference on Technologies and Applications of Artificial Intelligence (TAAI) IEEE; 2015. Utilizing different word representation methods for twitter data in adverse drug reactions extraction. doi:10.1109/taai.2015.7407070. [DOI] [Google Scholar]
- 86.Gupta S., Pawar S., Ramrakhiyani N., Palshikar G.K., Varma V. Semi-supervised recurrent neural network for adverse drug reaction mention extraction. BMC Bioinf. 2018;19(S8) doi: 10.1186/s12859-018-2192-4. doi:10.1186/s12859-018-2192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kang G.J., Ewing-Nelson S.R., Mackey L., Schlitt J.T., Marathe A., Abbas K.M., Swarup S. Semantic network analysis of vaccine sentiment in online social media. Vaccine. 2017;35(29):3621–3638. doi: 10.1016/j.vaccine.2017.05.052. doi:10.1016/j.vaccine.2017.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chary M., Genes N., Giraud-Carrier C., Hanson C., Nelson L.S., Manini A.F. Epidemiology from tweets: estimating misuse of prescription opioids in the USA from social media. J. Med. Toxicol. 2017;13(4):278–286. doi: 10.1007/s13181-017-0625-5. doi:10.1007/s13181-017-0625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korkontzelos I., Nikfarjam A., Shardlow M., Sarker A., Ananiadou S., Gonzalez G.H. Analysis of the effect of sentiment analysis on extracting adverse drug reactions from tweets and forum posts. J. Biomed. Inf. 2016;62:148–158. doi: 10.1016/j.jbi.2016.06.007. doi:10.1016/j.jbi.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O'Connor K., Pimpalkhute P., Nikfarjam A., Ginn R., Smith K.L., Gonzalez G. vol. 2014. American Medical Informatics Association; 2014. Pharmacovigilance on twitter? mining tweets for adverse drug reactions; p. 924. (AMIA Annual Symposium Proceedings). [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J., Zhao L., Ye Y. 2018 IEEE International Conference on Big Data (Big Data) IEEE; 2018. Semi-supervised multi-instance interpretable models for flu shot adverse event detection. doi:10.1109/bigdata.2018.8622434. [DOI] [Google Scholar]
- 92.Bian J., Topaloglu U., Yu F. Proceedings of the 2012 International Workshop on Smart Health and Wellbeing - SHB '12. ACM Press; 2012. Towards large-scale twitter mining for drug-related adverse events. doi:10.1145/2389707.2389713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hamed A.A., Roose R., Branicki M., Rubin A. 2012 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining. IEEE; 2012. T-recs: time-aware twitter-based drug recommender system. doi:10.1109/asonam.2012.178. [DOI] [Google Scholar]
- 94.Kagashe I., Yan Z., Suheryani I. Enhancing seasonal influenza surveillance: topic analysis of widely used medicinal drugs using twitter data. J. Med. Internet Res. 2017;19(9) doi: 10.2196/jmir.7393. e315. doi:10.2196/jmir.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ram S., Zhang W., Williams M., Pengetnze Y. Predicting asthma-related emergency department visits using big data. IEEE Journal of Biomedical and Health Informatics. 2015;19(4):1216–1223. doi: 10.1109/jbhi.2015.2404829. doi:10.1109/jbhi.2015.2404829. [DOI] [PubMed] [Google Scholar]
- 96.Lu F.S., Hou S., Baltrusaitis K., Shah M., Leskovec J., Sosic R., Hawkins J., Brownstein J., Conidi G., Gunn J., Gray J., Zink A., Santillana M. Accurate influenza monitoring and forecasting using novel internet data streams: a case study in the boston metropolis. JMIR Public Health and Surveillance. 2018;4(1) doi: 10.2196/publichealth.8950. e4. doi:10.2196/publichealth.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Su K., Xiong Y., Qi L., Xia Y., Li B., Yang L., Li Q., Tang W., Li X., Ruan X., Lu S., Chen X., Shen C., Xu J., Xu L., Han M., Xiao J. 2018 IEEE International Conference on Big Data (Big Data) IEEE; 2018. City-wide influenza forecasting based on multi-source data. doi:10.1109/bigdata.2018.8622413. [DOI] [Google Scholar]
- 98.Serban O., Thapen N., Maginnis B., Hankin C., Foot V. Real-time processing of social media with sentinel: a syndromic surveillance system incorporating deep learning for health classification. Inf. Process. Manag. 2019;56(3):1166–1184. [Google Scholar]
- 99.Valli P.A., Uma M., Sasikala T. International Conference on Energy, Communication, Data Analytics and Soft Computing. ICECDS; IEEE: 2017. Tracing out various diseases by analyzing twitter data applying data mining techniques. 2017. doi:10.1109/icecds.2017.8389714. [DOI] [Google Scholar]
- 100.Signorini A., Segre A.M., Polgreen P.M. The use of twitter to track levels of disease activity and public concern in the u.s. during the influenza a h1n1 pandemic. PloS One. 2011;6(5) doi: 10.1371/journal.pone.0019467. e19467. doi:10.1371/journal.pone.0019467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tang L., Bie B., Zhi D. Tweeting about measles during stages of an outbreak: a semantic network approach to the framing of an emerging infectious disease. Am. J. Infect. Contr. 2018;46(12):1375–1380. doi: 10.1016/j.ajic.2018.05.019. doi:10.1016/j.ajic.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iso H., Wakamiya S., Aramaki E. vol. 245. IOS Press; 2018. Conditional density estimation of tweet location: a feature-dependent approach; p. 408. (MEDINFO 2017: Precision Healthcare through Informatics: Proceedings of the 16th World Congress on Medical and Health Informatics). [PubMed] [Google Scholar]
- 103.Kraemer M.U.G., Bisanzio D., Reiner R.C., Zakar R., Hawkins J.B., Freifeld C.C., Smith D.L., Hay S.I., Brownstein J.S., Perkins T.A. Inferences about spatiotemporal variation in dengue virus transmission are sensitive to assumptions about human mobility: a case study using geolocated tweets from lahore, Pakistan. EPJ Data Science. 2018;7(1) doi: 10.1140/epjds/s13688-018-0144-x. doi:10.1140/epjds/s13688-018-0144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao L., Chen J., Chen F., Wang W., Lu C.-T., Ramakrishnan N. 2015 IEEE International Conference on Data Mining. IEEE; 2015. Simnest: social media nested epidemic simulation via online semi-supervised deep learning; pp. 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Samuel H., Noori B., Farazi S., Zaiane O. 2018 IEEE/WIC/ACM International Conference on Web Intelligence (WI) IEEE; 2018. Context prediction in the social web using applied machine learning: a study of canadian tweeters. doi:10.1109/wi.2018.00-85. [DOI] [Google Scholar]
- 106.Jenson S., Reeves M., Tomasini M., Menezes R. 2017 IEEE SmartWorld, Ubiquitous Intelligence & Computing, Advanced & Trusted Computed, Scalable Computing & Communications, Cloud & Big Data Computing, Internet of People and Smart City Innovation (SmartWorld/SCALCOM/UIC/ATC/CBDCom/IOP/SCI) IEEE; 2017. Mining location information from users' spatio-temporal data. doi:10.1109/uic-atc.2017.8397519. [DOI] [Google Scholar]
- 107.Pham M.T., Rajić A., Greig J.D., Sargeant J.M., Papadopoulos A., McEwen S.A. A scoping review of scoping reviews: advancing the approach and enhancing the consistency. Res. Synth. Methods. 2014;5(4):371–385. doi: 10.1002/jrsm.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]