Significance
The corpus callosum connects left and right cerebral cortices, integrating sensory-motor and associative functions, and is the largest connection in the human brain. While all mammals have a cerebral cortex, only eutherians evolved a corpus callosum. However, how this occurred remains largely unknown. We compared transcription factors that control subcerebral versus callosal neuron projection fates in eutherians and marsupials and found remarkably high similarity of their gene sequences and functions. However, expression of the callosal gene SATB2 was delayed in mice relative to dunnarts, and premature overexpression was sufficient for reversion to an ancestral-like brain phenotype. Our results suggest that transcriptional heterochrony might have influenced callosal evolution, and that complex traits can originate by differential deployment of existing regulatory genes.
Keywords: Bcl11b/Ctip2, corpus callosum, cortical evolution, evolutionary innovations, heterochrony
Abstract
A unique combination of transcription factor expression and projection neuron identity demarcates each layer of the cerebral cortex. During mouse and human cortical development, the transcription factor CTIP2 specifies neurons that project subcerebrally, while SATB2 specifies neuronal projections via the corpus callosum, a large axon tract connecting the two neocortical hemispheres that emerged exclusively in eutherian mammals. Marsupials comprise the sister taxon of eutherians but do not have a corpus callosum; their intercortical commissural neurons instead project via the anterior commissure, similar to egg-laying monotreme mammals. It remains unknown whether divergent transcriptional networks underlie these cortical wiring differences. Here, we combine birth-dating analysis, retrograde tracing, gene overexpression and knockdown, and axonal quantification to compare the functions of CTIP2 and SATB2 in neocortical development, between the eutherian mouse and the marsupial fat-tailed dunnart. We demonstrate a striking degree of structural and functional homology, whereby CTIP2 or SATB2 of either species is sufficient to promote a subcerebral or commissural fate, respectively. Remarkably, we reveal a substantial delay in the onset of developmental SATB2 expression in mice as compared to the equivalent stage in dunnarts, with premature SATB2 overexpression in mice to match that of dunnarts resulting in a marsupial-like projection fate via the anterior commissure. Our results suggest that small alterations in the timing of regulatory gene expression may underlie interspecies differences in neuronal projection fate specification.
A unique feature of the mammalian brain is the presence of a six-layered cerebral cortex, also known as the neocortex, that is remarkably different from the nuclear arrangement of telencephalic neurons in other vertebrates (1). Moreover, whereas in egg-laying monotremes (e.g., platypus, echidna) and marsupials the neocortical hemispheres are interconnected via the anterior commissure, eutherian mammals (e.g., rodents, humans) evolved the corpus callosum as a novel axonal route over the fused septum (2–4). We recently showed that adult monotremes and marsupials share a similar interhemispheric map of cortical connections (i.e., commissural connectome) to that of eutherians via the corpus callosum, suggesting the ancient conservation of a developmental program of cortical wiring, despite the different routes taken by their axons (5). However, the conservation or divergence of the molecular networks that regulate the development of long-range neocortical projections throughout mammalian evolution remains largely unknown.
Among the wide range of molecular regulators known to direct the fate of neocortical long-range projection neurons in eutherians (6–8), the transcription factors CTIP2 and SATB2 have been well established to specify subcerebral (layer [L]5) versus callosal (L2/3 and L5) neurons, respectively, via a complex balance of expression during development (9). Subcerebrally projecting neurons express CTIP2 and extend axons that descend through the internal capsule and cerebral peduncle on their way to the midbrain, hindbrain, and spinal cord (10, 11). Ctip2 knockout mice display a loss of subcerebrally projecting axons, and the ectopic overexpression of CTIP2 in mouse upper layer “callosal” neurons induces subcerebral rerouting (10, 11). In contrast, SATB2 is expressed by intracortical projection neurons, most notably callosal neurons, where it acts by directly repressing CTIP2 expression (9, 12–15) in conjunction with the protooncogene SKI (16). Satb2 knockout mice display agenesis (absence) of the corpus callosum, and their neuronal projections instead reroute to subcerebral targets, as well as through the anterior commissure, which in wild types predominantly carries connections between the olfactorecipient areas (12, 13, 17). A mechanism by which CTIP2 and SATB2 specify these distinct fates is thought to be by regulating the initial lateral versus medial axonal extension of migrating neurons, respectively (18, 19).
A putative role of CTIP2 and SATB2 as transcriptional regulators of lateral (subcerebral) versus medial (callosal) projection fates is also suggested by certain cases of neuroplasticity in humans. For example, in line with the result of Satb2 knockout mouse displaying rerouting through the anterior commissure, in some cases of human callosal agenesis interhemispheric connections are retained through the anterior commissure, resembling the preeutherian connectome (20, 21). This potentially atavistic phenomenon of long-range axonal plasticity may underlie the improved functional outcomes of individuals with developmental callosal agenesis compared to those who have had their corpus callosum surgically severed in adulthood to treat drug-resistant epilepsy.
In order to gain insight into the evolution and development of the molecular and cellular mechanisms regulating cortical projection wiring and plasticity, we applied a comparative approach that integrates both the similarities and the differences (22, 23) of neocortical development between two extant mammalian species: mouse (Eutheria: Muridae) and the Australian fat-tailed dunnart (Metatheria: Dasyuridae). Surprisingly, despite the striking differences in the medial- versus lateral-turning and subsequent commissural routes taken by the neocortical axons of each species, we report a remarkable degree of conservation in their molecular regulation, with CTIP2 specifying subcerebral projections, and SATB2 specifying intracortical connections likely via CTIP2 repression. We also demonstrate that subtle changes in the developmental timing of expression of SATB2 can dramatically alter axon turning and commissural route. These results highlight transcriptional heterochronies as potential mechanisms for the origin of variation in neural projection strategies relevant for the evolution, development, and malformation of cortical circuits.
Results
CTIP2 and SATB2 Predicted Proteins Share Remarkable Conservation of Functional Domains between Eutherians and Marsupials.
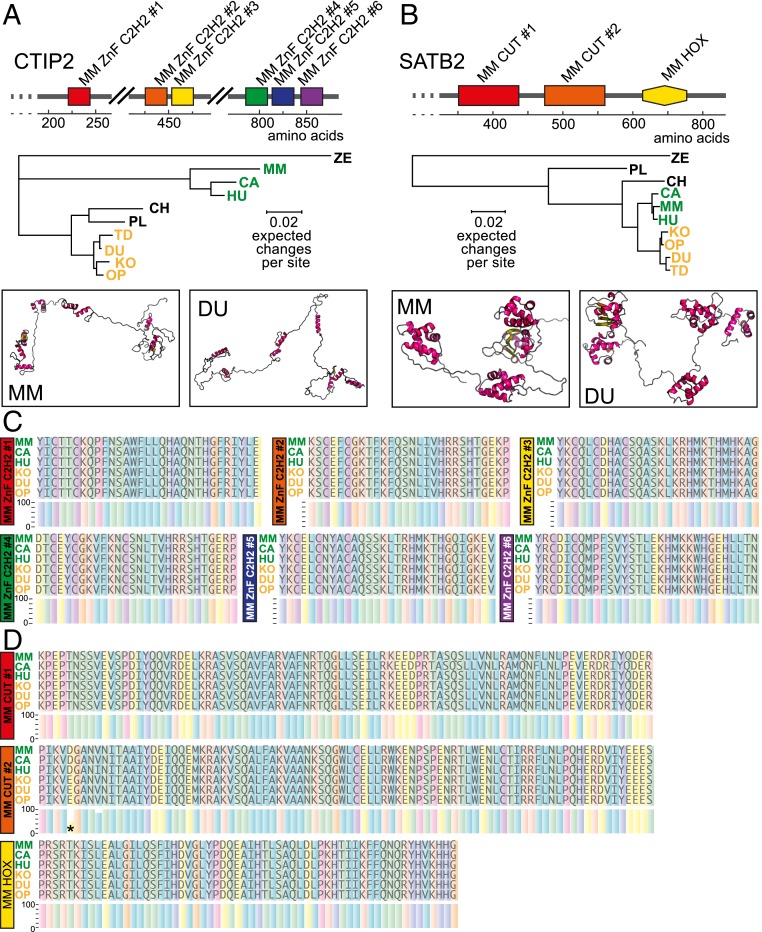
CTIP2 and SATB2 regulate the development of neocortical connectivity in eutherian mammals, but the expression, distribution, or function of these proteins has never been examined in the marsupial brain. We sequenced mRNA extracted from the developing dunnart cortex and isolated sequences homologous to eutherian Satb2 and Ctip2 genes to compare with other species. The sequences of Satb2 and Ctip2 transcripts were predicted in some marsupial species based on genomic data, such as in the South American gray short-tailed opossum (Monodelphis domestica, order Didelphimorphia; National Center for Biotechnology Information [NCBI] reference sequences: XM_001379413.3 and XM_007473605.1), the Tasmanian devil (Sarcophilus harrisii, order Dasyuromorphia; NCBI reference sequences: XM_012544916.1 and XM_012550451.1), and more recently the koala (Phascolarctos cinereus, order Diprotodontia; NCBI reference sequences: XM_020993639.1 and XM_021007322.1). We used Bayesian inference and model choice (MrBayes; http://nbisweden.github.io/MrBayes/) to construct phylogenetic trees based on these full-protein sequences in relation to those of other Therian mammals, using zebrafish as an outgroup root, and found that, as expected, dunnart CTIP2 and SATB2 amino acid sequences clustered within those of other marsupials, whereas those of eutherians tended to be more closely related to each other (Fig. 1 A and B).
Fig. 1.
CTIP2 and SATB2 predicted proteins share remarkable conservation of functional domains between eutherians and marsupials. (A and B) Functional domain representations, lineage analyses, and three-dimensional (3D) structure predictions of CTIP2 (A; six functional domains) and SATB2 (B; three functional domains) between mouse (MM) and dunnart (DU). (C and D) Sequences of the predicted functional domains for three eutherian and three marsupial species for CTIP2 (C) and SATB2 (D). Asterisk indicates predicted amino acid substitution. CA, dog; CH, chicken; HU, human; KO, koala; OP, opossum; PL, platypus; ZE, zebrafish.
We next used RaptorX (http://raptorx.uchicago.edu) to predict CTIP2 and SATB2 protein structures from mRNA transcripts and compared these with those of other marsupials and eutherians. Mouse CTIP2 comprises 884 amino acids, and comparisons with the predicted dunnart CTIP2 revealed 111 amino acids located outside the functional domains that are marsupial specific (12.5%) and 40 amino acids showing inconsistency between marsupials and eutherians (4.5%) (SI Appendix, Fig. S1A). Similarly, using mouse SATB2 as a reference (733 amino acids), we found that 11 out of 733 amino acids (1.5% of the whole protein) were consistently different in marsupials compared to eutherians, but conserved among marsupial species, suggesting that they might be lineage specific. One amino acid was different in marsupials compared to eutherians and also differed among marsupial species (SI Appendix, Fig. S1B). Strikingly, when only the predicted functional DNA-binding domains of each protein were compared across species, CTIP2 showed 100% sequence homology across all six domains, while only one amino acid differed across the three functional domains of SATB2 (Fig. 1 C and D). The machine learning trained classifier SNAP2 (screening for nonacceptable polymorphisms, version 2; https://www.predictprotein.org/) (24) was employed to predict the effects of this single variant in the SATB2 mouse protein; a substitution of aspartic acid for glutamic acid was identified in position 477, which produced a low score (−24; from a −100 to 100 scale of none-to-strong effect) that predicted a very weak, if any, effect on SATB2 protein function. Collectively these results suggest that there is a high degree of homology among Therian mammals in CTIP2 and SATB2 protein sequences.
Cells Expressing SATB2 and CTIP2 mRNA and Protein Populate Similar Areas and Layers in the Brain of Adult Mice and Dunnarts.
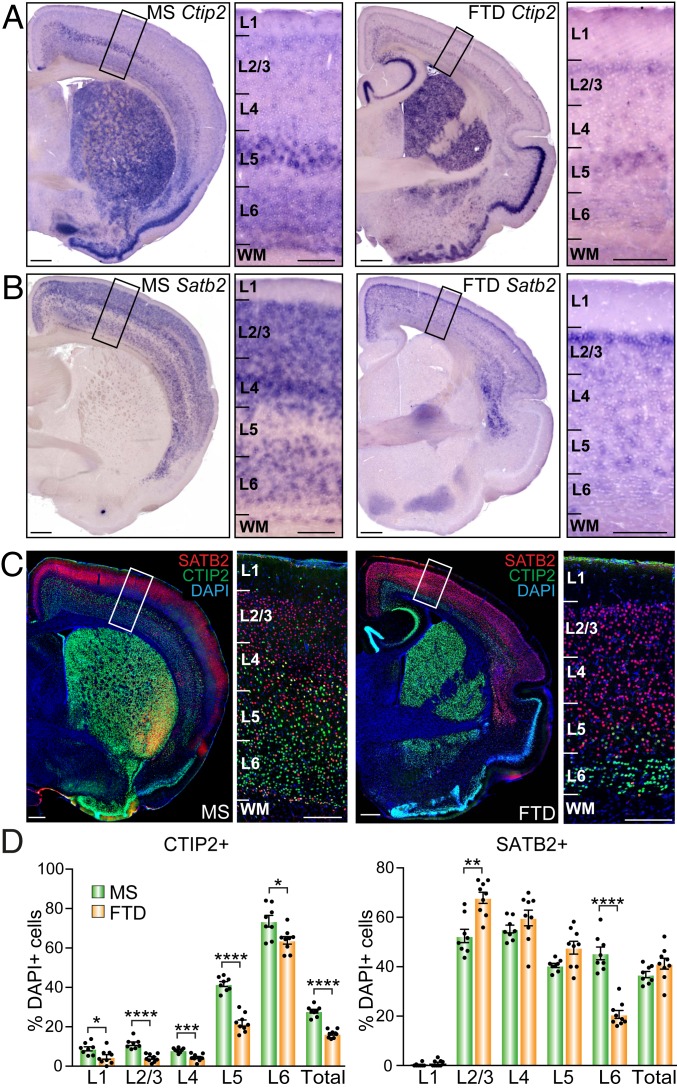
Although genes homologous to eutherian Ctip2 and Satb2 have been predicted in the genomes of a variety of marsupials, the presence and spatial distribution of their mRNA and protein in brain tissue has never been studied. We first performed in situ hybridizations using probes specifically designed against mouse or dunnart Ctip2 or Satb2 sequences. We detected both mRNAs in the adult brain of both species, with broadly comparable expression patterns. Ctip2 was expressed strongly in the piriform cortex and striatum, and detectable in the neocortex, where it formed a strong band of expression in the deeper cortical layers (L5/6) (Fig. 2A). Satb2 was predominantly expressed in the neocortex of both species, showing strong bands of expression in the upper cortical layers (Fig. 2B).
Fig. 2.
Cells expressing SATB2 and CTIP2 mRNA and protein populate similar areas and layers in the brain of adult mice and dunnarts. (A and B) In situ hybridization with species-specific probes against Ctip2 or Satb2 mRNA in adult brains of each species. (C) Immunohistochemistry against SATB2 (red) and CTIP2 (green) in adult brain sections of mouse (Left) and dunnart (Right). (D) Cell counts of CTIP2- or SATB2-positive neurons per neocortical layer reveal similarities and differences in the distribution and proportion of cells expressing the proteins between mouse and dunnart. Data are presented as mean ± SEM, n ≥ 8 animals per condition; each dot represents a single animal average. Mann–Whitney U tests, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. FTD, fat-tailed dunnart; L, layer; Ms, mouse; WM, white matter. (Scale bars, 400 µm in A–C for hemibrains, 200 µm for Insets.)
Similarly, immunohistochemistry against CTIP2 and SATB2 revealed that both proteins were detectable in the dunnart neocortex, which displayed gross banding patterns of SATB2 expression in the upper cortical layers and CTIP2 in the deeper cortical layers, very similar to mice (Fig. 2C). Cell counts of SATB2- and/or CTIP2-positive neurons within comparable regions of the primary somatosensory cortex (S1), expressed as a percentage of 4′,6-diamidino-2-phenylindole (DAPI)-positive cells throughout the layers, confirmed this general trend, while also revealing small but intriguing differences between species (Fig. 2D). For instance, the proportion of CTIP2-positive neurons was significantly lower in all layers of the dunnart neocortex as compared to mice, possibly reflecting a global cortical difference, such as in the number of interneurons, which are known to express CTIP2 and populate all layers of the cortex (25). The proportions of SATB2-positive neurons were more similar between species, with only an increase in L2/3 and a decrease in L6 in dunnarts as compared to mice. We also found a higher proportion of cells coexpressing both SATB2 and CTIP2 in mice than in dunnarts (SI Appendix, Fig. S2A). It has previously been shown that the proportion of SATB2/CTIP2 coexpression changes throughout mouse ontogeny, and that these dynamics may affect the functional properties of these transcription factors across development (26). To examine whether these patterns of expression are specific to S1, or instead are representative of whole-cortex trends, we performed averaged cell counts in a more anterior region (frontal association cortex; SI Appendix, Fig. S2B), and a more posterior region (primary auditory cortex; SI Appendix, Fig. S2C) in both species. We found similar distribution patterns in these areas to those observed in S1, including nonsignificant trends of increased proportions of CTIP2+ neurons in the mouse, increased proportions of SATB2+ neurons in the dunnart, and slightly lower proportions of coexpressing neurons in the dunnart, indicating that the species-specific patterns of expression are likely generalizable to the whole cortex. To further investigate potential interspecies differences in CTIP2 and SATB2, we subsequently explored the neurogenic dynamics of the cells that express each protein.
Birth-Dating Analysis Reveals Overlapping Peaks in the Neurogenic Timing of Mature SATB2- and CTIP2-Positive Cells between Mice and Dunnarts.
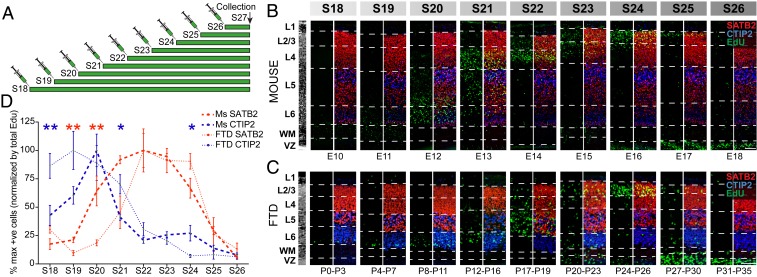
We next performed 5-ethynyl-2′-deoxyuridine (EdU) injections across developmental stages in both dunnarts and mice and examined the incidence of colocalization within CTIP2- or SATB2-positive neurons in the mature cortex (Fig. 3A). We employed a staging system for dunnarts that is equivalent to human (Carnegie) and mouse (Thieler) embryonic developmental stages (27). We confirmed previous reports that the marsupial cortex develops in an inside-out pattern (28–30), and found that comparable populations of neurons are born at equivalent stages of mouse and dunnart ontogeny, further consolidating this staging system for interspecies developmental comparisons (Fig. 3 B and C). By counting the total number of EdU-positive cells throughout the mature cortical layers for each age of injection with CTIP2 or SATB2 expression, we were also able to outline the neurogenic dynamics of neurons that ultimately express either transcription factor in mature animals. In both species, we found that the peak (100% maximum) of the proportion of EdU-positive cells expressing CTIP2 preceded the peak of SATB2-positive neuron generation (Fig. 3D). In accordance with gene electroporation studies in mouse (31) and dunnart (30), these peaks approximately coincided with the birth of deeper layer neurons for CTIP2 (stages 19 to 20: embryonic day [E]11 to E13 in mice, and postnatal day [P]4 to P11 in dunnarts) and upper layer neurons for SATB2 (stages 22 to 23, E13.5 to E15.5 in mice, and P17 to P23 in dunnarts). We therefore selected stages 20 and 23 in both species for subsequent electroporation-based transfections of deeper layer CTIP2-expressing neurons and upper layer SATB2-expressing neurons, respectively. Despite broadly similar peaks of neurogenesis for CTIP2- and SATB2-expressing neurons, we found small yet significant differences in their dynamics, specifically a relative delay in reaching the peak of cell birth for dunnart SATB2-expressing neurons, as well as a relative delay to reach the peak of cell birth for mouse CTIP2-expressing neurons (Fig. 3D).
Fig. 3.
Birth-dating analysis reveals overlapping peaks in the neurogenic timing of mature SATB2- and CTIP2-positive cells between mice and dunnarts. (A) Schematic of EdU injection protocol, where animals of both species were injected at various stages (S) of development and collected at the same stage of mature cortical development (late stage 27). (B and C) Neocortical Insets showing qualitative dynamics of EdU localization (green) between mouse (B) and dunnart (C) injected with EdU at different stages of development. (D) Counts of cells labeled with EdU and costained for either SATB2 (red in B and C) or CTIP2 (blue in B and C), normalized by the average number of EdU-labeled cells (green in B and C) at each stage per species, and presented as a percentage of the peak value for each cell type (% max). Data are presented as mean ± SEM, n ≥ 3 animals per condition. Mann–Whitney U tests, *P < 0.05; **P < 0.01. E, embryonic day; FTD, fat-tailed dunnart; L, layer; Ms, mouse; P, postnatal day; S, stage; VZ, ventricular zone; WM, white matter. (Scale bars, 100 µm.)
Subcerebral and Commissural Long-Range Projection Neurons Predominantly Express CTIP2 and SATB2, Respectively, in Both Mouse and Dunnart.
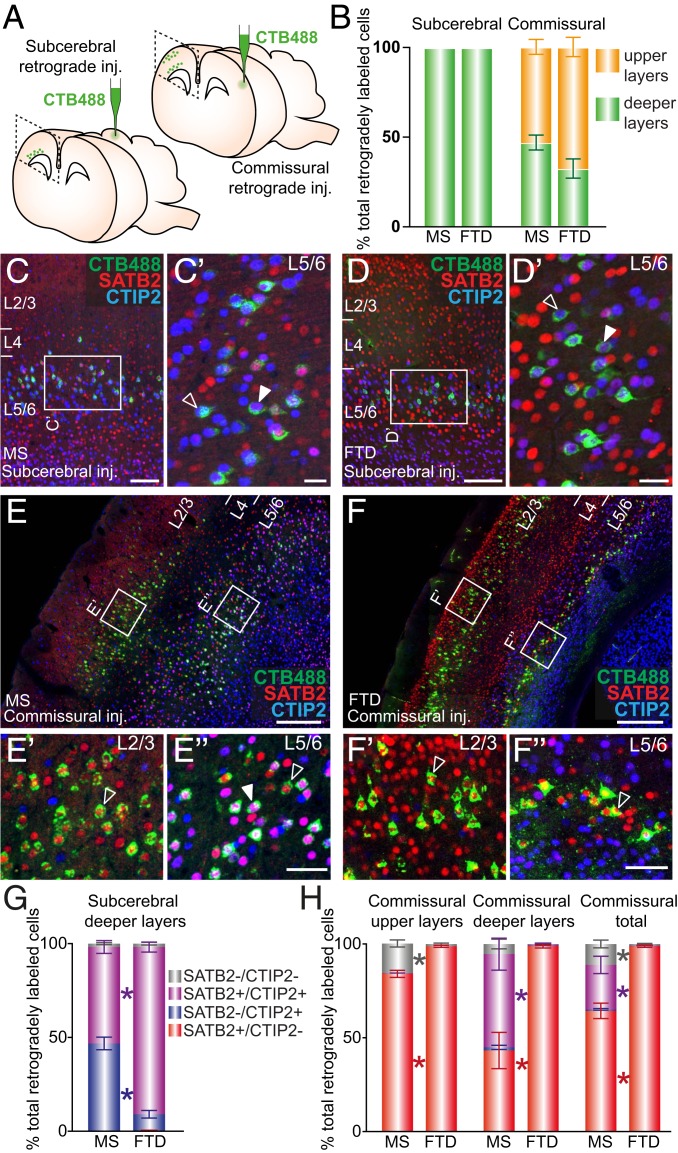
To investigate whether the broadly conserved expression patterns and neurogenic timing of CTIP2- and SATB2-positive cells among Therians reflect conserved specification of long-range projection neuron identities, we injected the nontoxic tracer cholera toxin B subunit (CTB) fluorophore-conjugated dyes into the ipsilateral superior colliculus or into the contralateral primary somatosensory cortex (S1) of adult mice and dunnarts, and examined retrogradely labeled cell bodies (Fig. 4A). We found an interspecies conservation of the laminar distribution of subcerebrally projecting neurons (to the ipsilateral superior colliculus) and commissurally projecting neurons (to contralateral S1), with subcerebral projection neurons located exclusively in L5/6, and interhemispheric projection neurons located predominantly in L2/3, as well as L5/6 (Fig. 4B). We next performed immunohistochemistry against CTIP2 and SATB2 on sections of CTB-injected brains and counted the proportions of retrogradely labeled cells positive for CTIP2, SATB2, both, or neither (Fig. 4 C–H). Almost all subcerebrally projecting neurons in both mice and dunnarts expressed CTIP2; however, although around half of these also coexpressed SATB2 in mice (in accordance with previous reports) (26), a significantly larger proportion coexpressed SATB2 in dunnarts (Fig. 4G). We examined the upper (L2/3) and deeper (L5/6) layer neurons with commissural fates separately, as well as in combination, finding that the vast majority of these neurons expressed SATB2 in both species. However, a small population in L2/3 of mice expressed neither transcription factor, with this population being almost indiscernible in dunnarts, and significantly more L5/6 commissural neurons also coexpressed CTIP2 in mice than in dunnarts (Fig. 4H).
Fig. 4.
Subcerebral and commissural long-range projection neurons predominantly express CTIP2 and SATB2, respectively, in both mouse and dunnart. (A) Schematics depict experimental protocol. (B) Quantification of the distribution of retrogradely labeled cell bodies in the primary somatosensory cortex in each species upon each CTB injection reveals conservation of the laminar distributions. (C and D) Example of a mouse (C) and dunnart (D) primary somatosensory cortex after CTB (green) injection into the tectum, followed by immunostaining for SATB2 (red) and CTIP2 (blue). Filled arrowheads in C′ and D′ show corticotectal cells coexpressing both SATB2 and CTIP2, whereas empty arrowheads indicate corticotectal cells expressing CTIP2 alone. (E and F) Example of a mouse (E) and dunnart (F) primary somatosensory cortex after CTB (green) injection into the contralateral homotopic cortex, and immunostaining for SATB2 (red) and CTIP2 (blue). Filled arrowheads show cells coexpressing SATB2 and Ctip2 and empty arrowheads indicate cells expressing SATB2 alone. (G and H) Quantification of the percentage of retrogradely labeled cells expressing SATB2 and/or CTIP2 or neither following subcerebral injections (G) or contralateral homotopic injections (H). Data are presented as mean ± SEM, n ≥ 4 animals per condition. Mann–Whitney U tests, *P < 0.05. Inj., Injection; CTB, cholera-toxin B; Ms, mouse; FTD, fat-tailed dunnart; L, layer. (Scale bars: [C–D] 125 µm and 25 µm for Insets; [E and F] 250 µm and 50 µm for Insets.)
Given that complete Satb2 knockout in mouse has been reported to result in rerouting of axons laterally toward subcerebral targets through the internal capsule, as well as through the anterior commissure, instead of medially to form a corpus callosum (12, 13), we investigated whether medially versus laterally projecting populations in the dunnart differed in their expression of SATB2. To label medially projecting neurons in the adult dunnart, we injected CTB into the cingulate cortex, which we have previously described receives dense cortical projections (5), and examined the labeled cell bodies in the ipsilateral S1 for SATB2 and CTIP2 expression (SI Appendix, Fig. S2D). We found that these ipsilaterally projecting neurons were predominantly located in layer 2/3, similar to dunnart commissurally projecting neurons, with a minority situated in L5/6 (SI Appendix, Fig. S2E). These neurons were also predominantly SATB2-positive and CTIP2-negative, similar to dunnart commissurally projecting neurons, although there was a large population that was negative for both transcription factors, in contrast with the commissural population (SI Appendix, Fig. S2 F and G).
These data collectively suggest that adult SATB2 expression, rather than marking callosal and/or medially projecting populations alone, may be an evolutionarily conserved marker of corticocortical projecting neurons of many projection fates.
The Ectopic Overexpression of Either Mouse- or Dunnart-Specific Ctip2 Constructs Is Sufficient to Specify a Subcerebral Projection Fate in Upper Layer Neurons in Both Species.
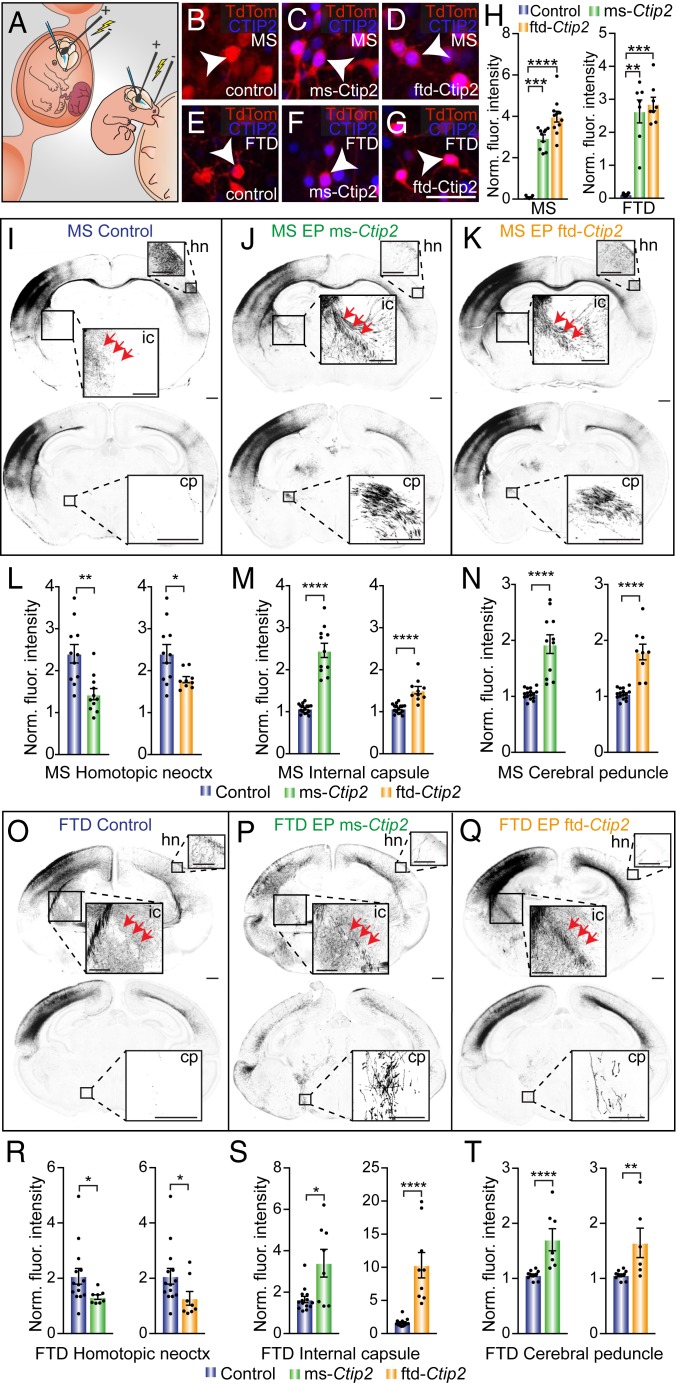
To better understand whether the conservation of CTIP2/SATB2 expression patterns between species is indicative of conserved cell-type specification properties, we next manipulated their expression during development in vivo. To elucidate whether mouse and dunnart CTIP2 proteins have differing functions, we used overexpression constructs for each species. We then ectopically overexpressed each of these constructs via in utero or in pouch electroporation in mice or dunnarts, respectively, at stage 23 (E15 mouse, P20 dunnart; Fig. 5A). This stage was previously identified for both species as a comparable timepoint of relatively high neurogenesis for SATB2-expressing neurons and low neurogenesis for CTIP2-expressing neurons (Fig. 3) and is the peak of upper layer (L2/3) neurogenesis (30). All animals for this and subsequent in vivo manipulations were collected at stage 28 (P10 mouse, P50 dunnart), an age at which long-range projection patterns are well established in the brain of both species (30, 32). We confirmed that overexpression of either construct resulted in an increase of CTIP2 immunofluorescence in electroporated cells, as compared with control electroporated cells (Fig. 5 B–H), as well as an increase in the overall number of cells expressing CTIP2 compared to control (SI Appendix, Fig. S3 A–D). Ectopic overexpression of mouse Ctip2 in L2/3 neurons resulted in a significant decrease in commissural projections to the contralateral homotopic neocortex (Fig. 5 I, J, and L), and a significant increase in axonal projections coursing subcerebrally through the internal capsule and cerebral peduncle (Fig. 5 M and N) in the P10 mouse brain, corroborating previous reports of Ctip2 overexpression in mouse (11). Interestingly, we found the same phenotype following ectopic overexpression of the dunnart-specific Ctip2 construct in the mouse brain, suggesting that any differences in the sequence of these proteins may not affect their subcerebral-fate specification capacity in mouse (Fig. 5 K and L–N). The decrease in contralateral innervation concomitant with increased subcerebral projections indicates that the overexpression of Ctip2 from either species in L2/3 neurons causes a switch in axon projection fate from contralateral to subcerebral in mouse.
Fig. 5.
The ectopic overexpression of either mouse- or dunnart-specific Ctip2 construct is sufficient to specify a subcerebral projection fate in upper layer neurons in both species. (A) Schematic of in utero and in pouch electroporation in mouse (Left) and dunnart (Right), respectively. (B–G) Example images of upper layer neurons electroporated at stage (S)23 (E15 for mice, P20 for dunnarts) and examined at S28 (P10 in mice, P50 in dunnarts) in control (TdTomato alone; Left), ms-Ctip2 (Middle), and ftd-Ctip2 (Right) for both mouse (Top row) and dunnart (Bottom row) conditions, immunostained against CTIP2 (blue). Arrowheads indicate example electroporated cells. (H) Quantification of fluorescence intensity of CTIP2 immunostaining normalized to the maximum value of nonelectroporated CTIP2-positive cells (norm. fluor. intensity) for each electroporated construct in both mouse and dunnart. (I–K) Example images of the brains of mice electroporated at S23 (E15) with either control (TdTomato alone), ms-Ctip2, or ftd-Ctip2 constructs and collected at S28 (P10). Red arrows trace the course of the internal capsule. (L–N) Quantification of the fluorescence intensity of labeled axons normalized to the background intensity value for the homotopic contralateral neocortex, the internal capsule, and the cerebral peduncle. (O–Q) Example images of the brains of dunnarts electroporated at S23 (P20) with either control (TdTomato alone), ms-Ctip2, or ftd-Ctip2 constructs and collected at S28 (P50). Red arrows trace the course of the internal capsule. (R–T) Quantification of the fluorescence intensity of labeled axons normalized to the background intensity value for the homotopic contralateral neocortex, the internal capsule, and the cerebral peduncle in dunnarts. Data are presented as mean ± SEM, n ≥ 6 animals per condition; each scatterdot represents a single animal. Mann–Whitney U tests, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Cp, cerebral peduncle; FTD, fat-tailed dunnart; hn, homotopic neocortex; ic, internal capsule; MS, mouse; neoctx, neocortex. (Scale bars: [B–G] 50 µm; [I–K, O–Q, and internal capsule Insets] 500 µm; [cerebral peduncle and homotopic neocortex Insets] 250 µm.)
To understand whether or not the developmental context of the marsupial brain results in a differing function of CTIP2, we transfected both mouse- and dunnart-specific Ctip2 constructs ectopically in the comparable age of developing dunnarts and examined the resulting axonal phenotypes at stage 28 (P50). We found that comparable ectopic projections resulted from overexpression of the protein from each species. Similar to the ectopic projections found in the mouse, transfected dunnart neurons showed decreased contralateral homotopic projections and increased subcerebral projection fates detectable in both the internal capsule and cerebral peduncle (Fig. 5 O–T). Interestingly, although they produced the similar effects compared to control, some significant differences between constructs were found that tended toward a pattern of greater effectiveness of each species-specific construct when transfected into its own host species (SI Appendix, Fig. S3F); however, the reasons that might underlie this effect remain unclear. Taken together, these results demonstrate the interchangeability of CTIP2 between mice and dunnarts in the ectopic specification of a subcerebral fate in upper layer neurons of both species. As the developmental stage in which a specific transcription factor is expressed can affect its function, to consolidate the conserved role of CTIP2, we performed immunohistochemical staining of wild-type mouse and dunnart brains across pertinent ages. Our data revealed a broadly comparable distribution of CTIP2-positive neurons at similar stages of development, further supporting the conserved dynamics of CTIP2 across Therian evolution (SI Appendix, Fig. S3E).
CRISPR-Cas9 Knockdown of Endogenous Satb2 in Upper Layer Neurons Induces an Ectopic Subcerebral Projection Fate.
Previous studies have shown that SATB2 is responsible for repressing Ctip2, thereby suppressing subcerebral fates in favor of corticocortical projections during neocortical development (12, 13, 17). Given the different routes taken by commissural axons between eutherians and marsupials, we next investigated the effect of reducing SATB2 expression in both species using CRISPR-Cas9 electroporatable constructs, followed by examination of projection phenotypes in each species.
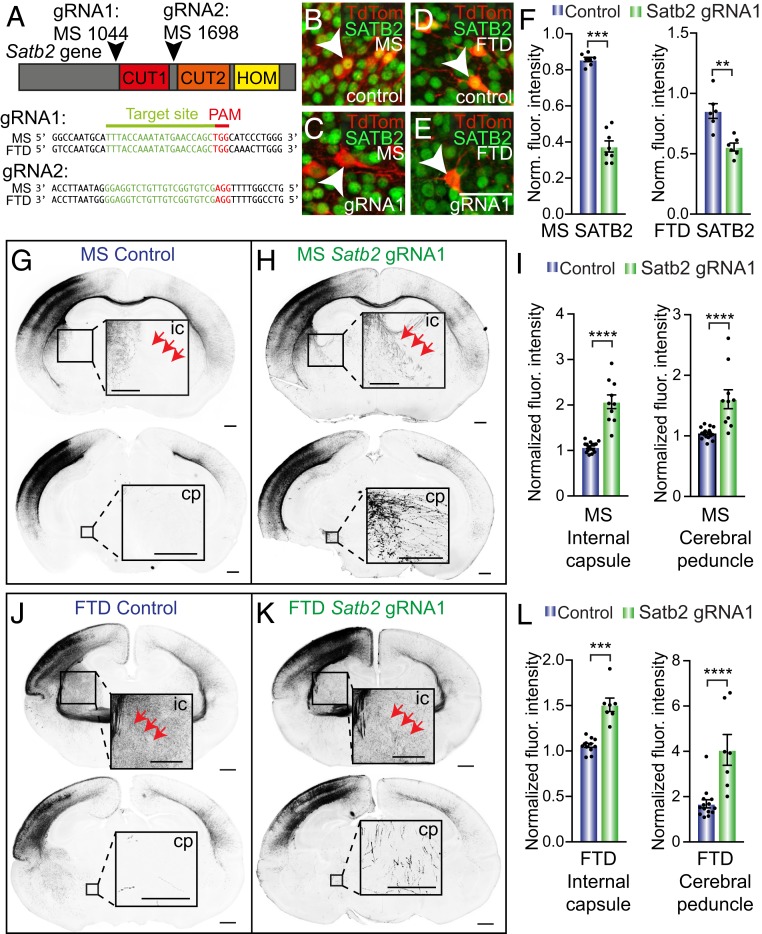
As a complete dunnart genome was not available at the time, we were not able to confidently predict potential off-target effects of CRISPR-Cas9 gRNAs in this species. We addressed this by designing two distinct gRNAs, each targeted to flanking sequences of the CUT1 domain of the Satb2 gene that share identical sequence between species (and therefore likely eliciting the most comparable effects and efficiencies possible; Fig. 6A). We performed all in vivo experiments in duplicate using each of these gRNAs, reasoning that any phenotypes that could be attributable to off-target effects instead of Satb2 knockdown would be very unlikely to be duplicated, thereby increasing our confidence in the specificity of our results.
Fig. 6.
CRISPR-Cas9 knockdown of endogenous Satb2 in upper layer neurons induces an ectopic subcerebral projection fate. (A) Schematic of sites of two gRNAs targeting Satb2 gene knockdown at the whole-gene level (Upper), as well as a comparison of the homology of these sites between mouse and dunnart (Lower). (B–E) Cells electroporated at stage (S)23 with either TdTomato (TdTom; red) alone (control; B and D), or together with Satb2 gRNA1 (C and E) in mouse (B and C) or dunnart (D and E), and immunostained for SATB2 (green). (F) Quantification of the fluorescence (fluor.) intensity of SATB2 immunostaining for control and gRNA1 electroporated cells of mouse and dunnart. (G and H) Example images at a more rostral (Upper) and caudal (Lower) level for mice electroporated with control (G) or gRNA1 (H) at S23 (embryonic day 15). (I) Quantification of the normalized fluorescence intensity of fluorescently labeled axons in the internal capsule or cerebral peduncle upon control or gRNA1 electroporation in mice. (J and K) Example images at a more rostral (Upper) and caudal (Lower) level for dunnarts electroporated with control (J) or gRNA1 (K) at S23 (postnatal day 20). (L) Quantification of the normalized fluorescence intensity of fluorescently labeled axons in the internal capsule or cerebral peduncle upon control or gRNA1 electroporation in the dunnart. Data are presented as mean ± SEM, n ≥ 6 animals per condition; each scatterdot represents a single animal. Mann–Whitney U tests, **P < 0.01; ***P < 0.001; ****P < 0.0001. Cp, cerebral peduncle; FTD, fat-tailed dunnart; gRNA, guide RNA; ic, internal capsule; Ms, mouse; PAM, protospacer adjacent motif. (Scale bars: [B–E] 50 µm; [G, H, J, and K] 500 µm for whole brains and internal capsule Insets; 250 µm for cerebral peduncle Insets.)
We first performed immunohistochemical staining on brains that had been transfected with the Satb2 CRISPR-Cas9 constructs at stage 23, aiming to verify a decrease in the expression of SATB2 in the targeted L2/3 neurons of each species. By performing a fluorescence intensity analysis on transfected cell bodies, we confirmed that both of our gRNA constructs caused a significant reduction, albeit not a complete knockout, of SATB2 protein in both mice and dunnarts (Fig. 6 B–F and SI Appendix, Fig. S4 A–F), similar to a previous study which used electroporatable CRISPR-Cas9 constructs targeting Satb2 in mice (15).
Analyses of axon projection phenotypes following these manipulations revealed similar patterns of ectopic projections to those found in the CTIP2 overexpression experiments, with a significant increase in L2/3 neuron projections to the internal capsule and cerebral peduncle of both species, as previously reported in Satb2 complete knockout mice (12, 13) and electroporated CRISPR-Cas9 knockdown (15) (Fig. 6 G–L). However, unlike our CTIP2 overexpression manipulations, this was not accompanied by a decrease in commissural axons in the contralateral homotopic cortex, perhaps due to the incomplete reduction of SATB2 expression (Fig. 6H and SI Appendix, Fig. S4 A–F). Given the previous reports that Satb2 knockout mice display rerouting of cortical projections through the anterior commissure (12, 13), we also measured the fluorescence of the mouse anterior commissure, finding that it was not significantly different from the control (SI Appendix, Fig. S4I). This disparity may again be due to the incomplete knockdown of Satb2 resulting from CRISPR-Cas9 manipulations and/or noncell-autonomous effects of Satb2 knockout on the entire brain versus a small population of transfected neurons. All of the above results using our Satb2 gRNA1 construct were replicated with the gRNA2 construct targeting a different locus of the Satb2 gene (SI Appendix, Fig. S4 J–T) and we found no significant differences in the effects of the two gRNAs for any measurement (SI Appendix, Fig. S4U), strongly suggesting that these effects result from Satb2 knockdown, and not off-target effects, in both species. These data collectively suggest that SATB2 has a conserved role in suppressing a subcerebral projection fate during development in both eutherians and marsupials.
The Overexpression of Either a Mouse- or Dunnart-Specific Satb2 Construct at Stage 20 Produces Similar Phenotypes within Species, but Different Projection Routes between Species.
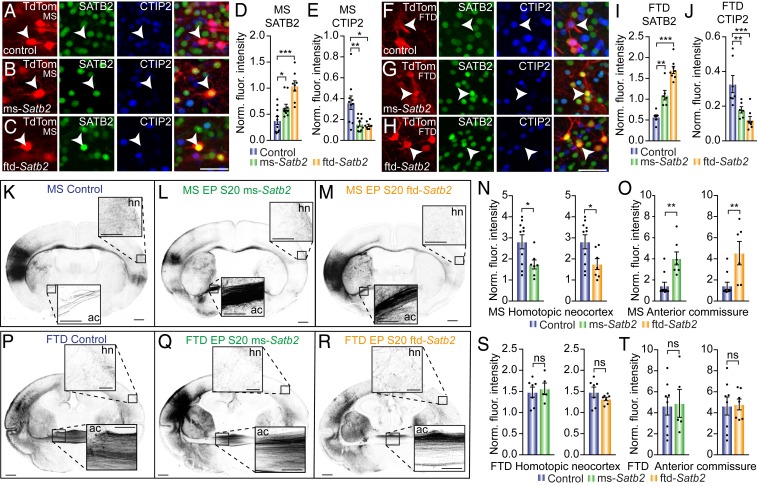
To better understand the projection fate that SATB2 promotes in both species, we next ectopically overexpressed the protein in vivo. We transfected deeper layer neurons at stage 20 (mouse E12, dunnart P10) with either mouse- or dunnart-specific Satb2 cDNA, together with species-specific overexpression constructs of the cofactor SKI, which has previously been shown to be necessary for SATB2 function (16). Both mouse and dunnart constructs resulted in increased SATB2 expression in transfected cells of both species (Fig. 7 A–D and F–I). Similarly, cell counts for SATB2-positive or -negative electroporated cells revealed significantly more SATB2-positive cells in both Satb2 overexpression conditions for both species (SI Appendix, Fig. S5 A–D). Moreover, given that SATB2 is known to suppress CTIP2 in mice (12, 13), we analyzed the fluorescence intensity of CTIP2 following this manipulation, finding a significant decrease in CTIP2 protein staining as a result of transfection of either construct in both species, as compared with controls (Fig. 7 E and J). This suggests that the suppressive effects of SATB2 on CTIP2 are conserved throughout Therian evolution.
Fig. 7.
The overexpression of either a mouse- or dunnart-specific Satb2 construct at stage 20 produces similar phenotypes within species, but different projection routes between species. (A–C) Mouse cells electroporated at S20 (E12) with TdTomato (red; TdTom) either alone (A; control), or with a Satb2 overexpression construct cloned from mouse DNA (B; ms-Satb2) or dunnart DNA (C; ftd-Satb2), and immunostained for SATB2 (green) and CTIP2 (green). Arrowheads indicate example electroporated neurons. (D and E) Quantification of the fluorescence intensity of SATB2 (D) or CTIP2 (E) immunostaining for each electroporated construct in mouse. (F–H) Dunnart cells electroporated at S20 (P10) with TdTomato (red; TdTom) either alone (F; control) or with a Satb2 overexpression construct cloned from mouse DNA (G; ms-Satb2) or dunnart DNA (H; ftd-Satb2), and immunostained for SATB2 (green) and CTIP2 (green). Arrowheads indicate example electroporated neurons. (I and J) Quantification of the fluorescence intensity of SATB2 (I) or CTIP2 (J) immunostaining for each electroporated construct in dunnart. (K–M) Example images of mouse brains electroporated at S20 (E12) with control (K), ms-Satb2 (L), or ftd-Satb2 (M) constructs and collected at S28 (P10). (N–O) Quantification of the normalized fluorescence intensity of electroporated axons in the contralateral homotopic neocortex and anterior commissure following electroporation with each construct. (P–R) Example images of dunnart brains electroporated at S20 (P10) with control (P), ms-Satb2 (Q), or ftd-Satb2 (R) constructs and collected at S28 (P50). (S and T) Quantification of the normalized fluorescence intensity of electroporated axons in the contralateral homotopic neocortex and anterior commissure upon electroporation with each construct in dunnarts. Data are presented as mean ± SEM, n ≥ 5 animals per condition; each scatterdot represents a single animal. Mann–Whitney U tests, *P < 0.05; **P < 0.01; ***P < 0.001. Ac, anterior commissure; FTD, fat-tailed dunnart; hn, homotopic neocortex; Ms, mouse; ns, not significant; S, stage. (Scale bars: [A–C and F–H] 50 µm, [K–M and P–R], 500 µm for whole brains, 250 µm for anterior commissure and homotopic neocortex Insets.)
Surprisingly, despite SATB2 previously being suggested to act as a transcription factor that predominantly specifies callosal fate (12, 13), mice ectopically overexpressing the mouse SATB2 protein at stage 20 (E12) showed a significant decrease in axonal innervation into the contralateral homotopic neocortex (Fig. 7 K–O). This was accompanied by a remarkable increase in axonal projections through the anterior commissure (Fig. 7 K–O), with no change in subcerebral projections (cerebral peduncle; SI Appendix, Fig. S5E) nor in the innervation of the contralateral piriform cortex (SI Appendix, Fig. S5F). These findings were replicated with no significant differences between the two constructs (SI Appendix, Fig. S5M) when the dunnart-specific Satb2 construct was transfected into mouse (Fig. 7 K–O), suggesting that species differences in the Satb2 gene sequence do not affect its functionality in mice.
We then performed the same experiment in dunnarts and found that, surprisingly, overexpression of either mouse or dunnart Satb2 constructs at stage 20 produced no significant difference in axonal projections to the contralateral homotopic neocortex (Fig. 7 P–S), or through the anterior commissure (Fig. 7T) or subcerebrally (cerebral peduncle; SI Appendix, Fig. S5G). Despite the fact that overexpression of Satb2 in mouse at stage 20 (E12) caused more projections to turn laterally and course through the anterior commissure, previous studies have suggested that SATB2 is involved in axonal medial turning during cortical development (18). For this reason, we also tested whether or not this manipulation caused axons from L5/6 dunnart neurons to switch to a more medial projection fate by quantifying their presence in the ipsilateral cingulate cortex. However, this value was also unchanged between conditions (SI Appendix, Fig. S5H), demonstrating that overexpressing Satb2 at this stage is not sufficient to elicit a more medial/callosal-like projection pattern in dunnarts. Together, these data indicate the seemingly counterintuitive fact that Satb2 overexpression at stage 20 (E12) in mouse elicits a more “marsupial-like” phenotype of increased neocortical axons coursing through the anterior commissure, whereas the same manipulation at an equivalent stage in dunnarts does not alter the axonal phenotypes.
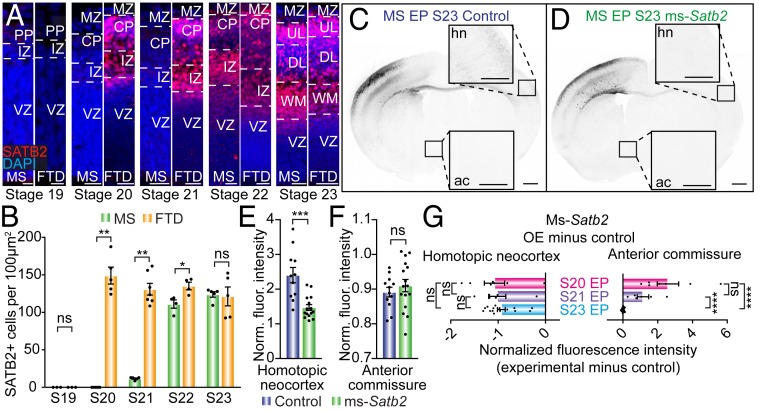
Heterochronic Expression of SATB2 May Underlie the Difference in Commissural Routes between Eutherians and Marsupials.
To elucidate the potential mechanisms underlying this disparity between species, we investigated the timing of developmental SATB2 expression in mice and dunnarts, which has previously been suggested to be an important factor in fate determination (18) and axonal pathfinding of neurons from different cortical layers (17). Unlike the broadly comparable dynamics that we observed for CTIP2 (SI Appendix, Fig. S3E), we found a striking difference in the onset of SATB2 expression between species, with detectable expression starting at stage 20 in dunnarts, but not until stage 22 (E14) in the mouse dorsal neocortex (Fig. 8A). When we quantified the density of SATB2-positive cells in dunnarts and mice of equivalent stages, we found that although both species show negligible SATB2 expression by stage 19, the developing cortex has significantly more SATB2-positive cells in dunnarts than mice between stages 20 and 22, with both species reaching similar values by stage 23 (Fig. 8B). Notably, the period of heterochronic SATB2 expression includes the stage at which we found differing results of Satb2 overexpression between species (stage 20; E12 in mice Figs. 7 and 8 A and B). Therefore, we reasoned that the interspecies differences in axon rewiring upon ectopic Satb2 overexpression may be due to the fact that SATB2 is already highly expressed in dunnarts but not mice at this timepoint. This raised the intriguing possibility that, although SATB2 promotes a corticocortical/commissural projection fate in both eutherians and marsupials, the precise timing of its onset of expression influences the route that neocortical axons take, with an earlier onset leading to a lateral/anterior commissural fate, and later onset leading to a medial/callosal fate. To test this hypothesis, we ectopically overexpressed Satb2 at a later stage in mice (stage 23; E15), at which point endogenous SATB2 expression is equivalent to that of dunnarts (Fig. 8 A and B). We found that, whereas the contralateral homotopic neocortical innervation was decreased relative to control, as in the stage 20 (E12) mouse electroporations, there was no effect on axons entering the anterior commissure (Fig. 8 C–F). Furthermore, Satb2 overexpression at stage 21 (E13), i.e., when SATB2 is detectable although still at very low levels in the mouse dorsal neocortex as compared to dunnarts, had a similar effect to stage 20 (E12) overexpression, including decreased contralateral homotopic projections and increased ectopic projections through the anterior commissure, resembling the marsupial commissural route (SI Appendix, Fig. S5 and Fig. 8G). Notably, the degree of change in anterior commissure axonal routing between experimental and control manipulations at each stage revealed an inverse trend to the pattern of endogenous SATB2 expression (compare Fig. 8G with Fig. 8B). Taken together, these findings point toward the conservation of SATB2 function as a specifier of corticocortical/commissural projections among Therian mammals, and suggest that differences in the developmental timing of expression of SATB2 might determine the interhemispheric routes taken by neocortical axons.
Fig. 8.
Heterochronic expression of SATB2 may underlie the difference in commissural routes between eutherians and marsupials. (A) Time course of mouse (Left) or dunnart (Right) sections, immunostained for SATB2 (red) and counterstained with DAPI (blue). (B) Quantification of the number of SATB2-positive cells averaged across the cortical plate and intermediate zone for mouse and dunnart at each developmental stage, revealing a significant delay in the onset of SATB2 expression in mouse compared to dunnart. (C and D) Example images of mice electroporated at stage 23 (E15) with TdTomato, either alone (control; c) or with ms-Satb2 (D). (E–F) Quantification of the normalized fluorescence intensity of electroporated axons in control or ms-Satb2 overexpression conditions in the contralateral homotopic neocortex or anterior commissure. (G) Quantification of the degree change between experimental (ms-Satb2 overexpression) and control conditions for three ages of Satb2 electroporation, all collected at S28/P10 (S20/E12 raw values from Fig. 7; S21/E13 raw values in SI Appendix, Fig. S5) for the homotopic neocortex and anterior commissure. Data are presented as mean ± SEM, n ≥ 3 animals per condition; each scatterdot represents a single animal. Mann–Whitney U tests, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Ac, anterior commissure; CP, cortical plate; DL, deeper layers; FTD, fat-tailed dunnart; hn, homotopic neocortex; IZ, intermediate zone; MS, mouse; MZ, marginal zone; ns, not significant; OE, overexpression; PP, preplate; S, stage; UL, upper layers; VZ, ventricular zone; WM, white matter. (Scale bars: [A] 10 µm for stage 19, 25 µm for stages 20 to 22, 50 µm for stage 23; [C and D] 500 µm for whole brains, 250 µm for homotopic neocortex and anterior commissure Insets.)
Discussion
Our findings advance our understanding of the function and evolution of CTIP2 and SATB2 in specifying long-range projection neurons of the cortex. We reveal a striking conservation of CTIP2 as a subcerebral-specifying protein across mammalian evolution. We similarly report conservation in the ability of SATB2 to repress CTIP2 between species, as evidenced by the decrease in CTIP2 immunostaining upon ectopic Satb2 expression, as well as an increase in subcerebral fate of cortical axons following CRISPR-Cas9-induced Satb2 knockdown. The role of SATB2 itself is more complex, with retrograde tracing revealing that it is not only a primary marker of intracortically projecting neurons in both species, but also coexpressed with CTIP2 in many subcerebrally projecting neurons. There have been previous reports that the degree to which CTIP2 and SATB2 are coexpressed changes throughout development (26), making the developmental and functional differences between these populations intriguing areas for future study. Retrograde tracing from the ipsilateral cingulate cortex in dunnarts revealed that medially projecting neurons also predominantly express SATB2, consolidating that this transcription factor likely does not specify medial versus lateral projection fates, but rather corticocortical ipsi- and contralateral identities. Finally, by overexpressing Satb2 ectopically in mouse before its endogenous onset of expression, we were surprised to find that axons follow a striking ectopic route through the anterior commissure, resembling the marsupial commissural projection. A study of the developmental dynamics of SATB2 expression revealed a significantly delayed initiation in the mouse cortex compared to that of dunnarts, thereby highlighting a potential mechanism by which the timing of SATB2 expression relative to other developmental milestones may influence the projection fate between species. In favor of this hypothesis, later ectopic overexpression in mice, when SATB2 expression has reached equivalent levels to those in the dunnart, did not alter the degree of anterior commissure innervation.
One notable difference between our data and previous work on Satb2 mutants is that we did not observe a change in the number of axons (measured via fluorescence intensity) in the anterior commissure or contralateral callosal projection in response to knockdown of Satb2 with CRISPR-Cas9 in the mouse. In contrast, earlier studies of whole-animal knockout of Satb2 reported callosal agenesis, as well as increased axonal routing through the anterior commissure (12, 13). This difference could be explained by the fact that our fluorescence intensity of SATB2 protein measurements reveal that the CRISPR-Cas9 manipulation reduced the levels of SATB2, but did not eliminate it completely. Similarly, our manipulation only targeted a subpopulation of cells. Previous work in Satb2 knockout mice has demonstrated that SATB2 is a repressor of Ctip2, and that upper layer neurons that lack SATB2 ectopically upregulate CTIP2, likely underlying their subcortical projection identity (12). Our analyses in SATB2 overexpressing cells indicate that this repressive effect of SATB2 on Ctip2 is conserved between mouse and fat-tailed dunnarts, and therefore this relationship may underlie the similar subcortical projection fates upon Satb2 knockdown in both species. Future experiments are required to dissect the phenotypes that result from differing levels of SATB2 expression, or indeed whether some phenotypes can only be elicited from whole-brain deletion of the protein in a noncell-autonomous manner (33).
An interesting comparison can be made between our EdU neurogenesis data and the developmental dynamics of SATB2 expression (Figs. 3D and 8 A and B). The EdU injections at different stages of development revealed broadly overlapping dynamics of neurogenesis for mature SATB2-expressing neurons, with mice showing a slight shift toward earlier birth dates of these cells compared to dunnarts. This is strikingly contrasted by our cross-species developmental time course of SATB2 expression, which revealed a significant delay in the onset of SATB2 expression developmentally in the mouse compared to dunnart, investigated with immunohistochemistry. Given that it has been reported that SATB2 is only expressed in postmitotic cells (13), these data may indicate differences in progenitor cell cycle dynamics between species. This is particularly interesting given the recent descriptions of fate-restricted delayed progenitors that give rise to callosal neurons, some of which have been shown to be absent in the chicken, which has no corpus callosum (34, 35). Similarly, protracted neuronal development has been linked to increased production of upper layer neurons and associated genes (36). However, the precise mechanisms of heterochronic molecular and cellular processes, including their role in the evolution of brain processes, are largely unknown and remain intriguing areas for future study.
Our Satb2 overexpression experiments in stages 20 (E12) and 21 (E13) mouse revealed a striking difference in the ectopic projections into the anterior commissure compared to controls electroporated at the same stages. However, this did not occur following Satb2 overexpression at stage 23 (E15), and is in contrast to the contralateral homotopic neocortex projections, which were significantly reduced in all three stages (Fig. 8G). This could indicate that, whereas the anterior commissure projection fate is sensitive to the timing of SATB2 expression, the reduced contralateral innervation is due to other factors. For example, overexpression of Satb2 has previously been shown to alter the migration of neurons (12, 13, 37), and this or other factors (such as cytotoxicity of excessive Satb2 expression) may therefore have decreased the total number of axons exiting the cortex in an independent mechanism from the projection fate. This issue also relates to an open question regarding the contralateral fate of the ectopic anterior commissure projection following Satb2 overexpression at stage 20 (E12) in mice, as we also found no significant difference in axonal innervation in the piriform cortex, the major target of axons projecting through the anterior commissure in control mice (SI Appendix, Fig. S5F). It therefore remains to be determined whether these ectopic axons terminate diffusely across the entire cortex, or specifically target the contralateral homotopic neocortex to a degree that does not reach control levels of innervation, and so is still registered as a deficit by our intensity analyses.
In summary, this study describes and functionally manipulates key molecular regulators of eutherian and metatherian neocortical development. Our results reveal a surprisingly high degree of homology of transcriptional regulators, in terms of protein sequence, cell-type localization, and functional consequence of knockdown or overexpression. We also provide compelling evidence to suggest that a potential mechanism for the origin of phenotypic diversity in complex brain traits includes changes in the developmental timing (i.e., heterochrony) of preexisting regulatory genes, rather than variations in the genes themselves. Future work will be required to elucidate candidates for upstream mechanisms coordinating the heterochronic timing of SATB2 expression (e.g., via whole cortex RNA sequencing at multiple developmental stages to screen for genes or gene networks expressed in similarly heterochronic patterns), as well as potential interspecies differences in the downstream targets of SATB2 and CTIP2 (e.g., via ChIP-seq with SATB2 and CTIP2 antibodies and/or RNA-sequencing of cells overexpressing or downregulating SATB2 or CTIP2 in each species).
Materials and Methods
Animal Ethics.
All animal procedures, including laboratory breeding, were approved by The University of Queensland Animal Ethics Committee and the Queensland Government Department of Environment and Science, and were performed according to the current Australian Code for the Care and Use of Animals for Scientific Purposes (38), as well as international guidelines on animal welfare.
In Utero and in Pouch Electroporation.
Time-mated CD1 pregnant dams were used at stage 20 (E12, deeper layer neurogenesis in the neocortex) or stage 23 (E15, upper layer neurogenesis) for all experiments. In pouch electroporation of dunnart joeys was performed at stage 20 (P8–P11, deeper layer neurogenesis) or stage 23 (P20–P23, upper layer neurogenesis), as described before (30). Briefly, 0.5 μL of a 1-mL/mg cDNA solution was injected into the lateral ventricles with glass-pulled micropipettes and a picospritzer, and five 100-ms square pulses of 30 to 35 V delivered at 1 Hz with 1- to 3-mm paddles (ECM 803, BTX). Further details can be found in SI Appendix.
Histology and Analysis.
The brain atlas of the stripe-faced dunnart (Sminthopsis macroura) was used as an indicative reference for stereotaxic injections in fat-tailed dunnarts (39). The targeted regions were: primary somatosensory cortex (S1), cingulate/motor cortex, and the superior colliculus. The tracer used for the experiments was 0.5 to 1 μL of 0.5 to 1.0% cholera toxin subunit B conjugated to a florescent dye (Alexa Fluor 488, Alexa Fluor 647, and Alexa Fluor 555). Injected animals were then recovered and collected via transcardial perfusion after 7 d. Digoxygenin-labeled antisense riboprobes for in situ hybridization were transcribed from cDNA plasmids. Primers for dunnart probes were designed based on the RNA-seq dataset generated in the laboratory and can be found in SI Appendix, while mouse probe primer designs were obtained from the website of the Allen Brain Atlas. For dunnart joeys and mouse pups collected after stage 20 (joeys older than P13 and pups older than E12.5), the brains were dissected out of the skull and sectioned in 50-µm coronal slices. Fluorescence images were acquired with confocal microscopy and analyzed with Imaris and FIJI. Experimental details, including antibodies for fluorescent immunohistochemistry, primers, reagents, analysis, and statistics, can be found in SI Appendix.
Data Availability.
All relevant data are included in the main text and SI Appendix.
Supplementary Material
Acknowledgments
We thank Qiongyi Zhao of the Queensland Brain Institute (QBI) bioinformatics core for assembling the fat-tailed dunnart transcriptome that produced the fat-tailed dunnart Ctip2 and Satb2 transcripts. The Queensland Brain Institute’s Histology Facility and Advanced Microscopy Facility assisted with histology preparation and imaging. We thank The University of Queensland (UQ) Biological Resources and the Native Wildlife Teaching and Research Facility for help with all animal husbandry. Thanks to Rowan Tweedale, Tobias Bluett, Laura R. Morcom, Caitlin Bridges, Timothy J. Edwards, Jens Bunt, Ilan Gobius, and Ching Moey for productive discussions and experimental support. We also thank Victor Tarabykin for interesting discussions and for providing the Satb2 construct, Nenad Sestan for the mouse Ctip2 construct, and Suzana Atanasoski for the mouse Ski construct. This work was supported by the Australian Research Council (Discovery Early Career Researcher Award DE160101394 to R.S., Discovery Project Grant DP160103958 to L.J.R. and R.S., and Discovery Project Grant DP200103093 to R.S. and L.R.F.), a UQ-QBI Doctoral Scholarship (A.P.), a UQ Development Fellowship (L.R.F.), a UQ Amplify Fellowship (R.S.), and a National Health and Medical Research Council Principal Research Fellowship (GNT1120615 to L.J.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922422117/-/DCSupplemental.
References
- 1.Briscoe S. D., Ragsdale C. W., Homology, neocortex, and the evolution of developmental mechanisms. Science 362, 190–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suárez R., Gobius I., Richards L. J., Evolution and development of interhemispheric connections in the vertebrate forebrain. Front. Hum. Neurosci. 8, 497 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gobius I., et al. , Astroglial-mediated remodeling of the interhemispheric midline during telencephalic development is exclusive to eutherian mammals. Neural Dev. 12, 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suárez R., “Evolution of telencephalic commissures: Conservation and change of developmental systems in the origin of brain wiring novelties” in Evolution of Nervous Systems, Kaas J. H., Ed. (Academic Press, Oxford, UK, 2nd Ed., 2017), pp. 205–223. [Google Scholar]
- 5.Suárez R., et al. , A pan-mammalian map of interhemispheric brain connections predates the evolution of the corpus callosum. Proc. Natl. Acad. Sci. U.S.A. 115, 9622–9627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paolino A., Fenlon L. R., Suárez R., Richards L. J., Transcriptional control of long-range cortical projections. Curr. Opin. Neurobiol. 53, 57–65 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Rakic P., The radial edifice of cortical architecture: From neuronal silhouettes to genetic engineering. Brain Res. Brain Res. Rev. 55, 204–219 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molyneaux B. J., et al. , DeCoN: Genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan K., et al. , A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc. Natl. Acad. Sci. U.S.A. 109, 19071–19078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arlotta P., et al. , Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Chen B., et al. , The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 105, 11382–11387 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcamo E. A., et al. , Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364–377 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Britanova O., et al. , Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57, 378–392 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Srivatsa S., et al. , Unc5C and DCC act downstream of Ctip2 and Satb2 and contribute to corpus callosum formation. Nat. Commun. 5, 3708 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shinmyo Y., et al. , CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation. Sci. Rep. 6, 20611 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranek C., et al. , Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proc. Natl. Acad. Sci. U.S.A. 109, 3546–3551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leone D. P., et al. , Satb2 regulates the differentiation of both callosal and subcerebral projection neurons in the developing cerebral cortex. Cereb. Cortex 25, 3406–3419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lickiss T., Cheung A. F. P., Hutchinson C. E., Taylor J. S. H., Molnár Z., Examining the relationship between early axon growth and transcription factor expression in the developing cerebral cortex. J. Anat. 220, 201–211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatanaka Y., Namikawa T., Yamauchi K., Kawaguchi Y., Cortical divergent projections in mice originate from two sequentially generated, distinct populations of excitatory cortical neurons with different initial axonal outgrowth characteristics. Cereb. Cortex 26, 2257–2270 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Tovar-Moll F., et al. , Structural and functional brain rewiring clarifies preserved interhemispheric transfer in humans born without the corpus callosum. Proc. Natl. Acad. Sci. U.S.A. 111, 7843–7848 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Meer N., et al. , Interhemispheric connections between the primary visual cortical areas via the anterior commissure in human callosal agenesis. Front. Syst. Neurosci. 10, 101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullock T. H., Comparative neuroscience holds promise for quiet revolutions. Science 225, 473–478 (1984). [DOI] [PubMed] [Google Scholar]
- 23.Krubitzer L., The magnificent compromise: Cortical field evolution in mammals. Neuron 56, 201–208 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Hecht M., Bromberg Y., Rost B., Better prediction of functional effects for sequence variants. BMC Genomics 16 (suppl. 8), S1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikouei K., Muñoz-Manchado A. B., Hjerling-Leffler J., BCL11B/CTIP2 is highly expressed in GABAergic interneurons of the mouse somatosensory cortex. J. Chem. Neuroanat. 71, 1–5 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Harb K., et al. , Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. eLife 5, e09531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suárez R., et al. , Development of body, head and brain features in the Australian fat-tailed dunnart (Sminthopsis crassicaudata; Marsupialia: Dasyuridae); A postnatal model of forebrain formation. PLoS One 12, e0184450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanderson K. J., Weller W. L., Gradients of neurogenesis in possum neocortex. Brain Res. Dev. Brain Res. 55, 269–274 (1990). [DOI] [PubMed] [Google Scholar]
- 29.Puzzolo E., Mallamaci A., Cortico-cerebral histogenesis in the opossum Monodelphis domestica: Generation of a hexalaminar neocortex in the absence of a basal proliferative compartment. Neural Dev. 5, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paolino A., Fenlon L. R., Kozulin P., Richards L. J., Suárez R., Multiple events of gene manipulation via in pouch electroporation in a marsupial model of mammalian forebrain development. J. Neurosci. Methods 293, 45–52 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Kozulin P., Almarza G., Gobius I., Richards L. J., “Investigating early formation of the cerebral cortex by in utero electroporation: Methods and protocols” in Prenatal and Postnatal Determinants of Development, Walker D. W., Ed. (Humana Press, New York, NY, 2016), pp. 3–20. [Google Scholar]
- 32.Fenlon L. R., Suárez R., Richards L. J., The anatomy, organisation and development of contralateral callosal projections of the mouse somatosensory cortex. Brain Neurosci. Adv. 1, 2398212817694888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q., Huang Y., Zhang L., Ding Y.-Q., Song N.-N., Loss of Satb2 in the cortex and Hippocampus leads to abnormal behaviors in mice. Front. Mol. Neurosci. 12, 33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.García-Moreno F., Molnár Z., Subset of early radial glial progenitors that contribute to the development of callosal neurons is absent from avian brain. Proc. Natl. Acad. Sci. U.S.A. 112, E5058–E5067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franco S. J., et al. , Fate-restricted neural progenitors in the mammalian cerebral cortex. Science 337, 746–749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendy J. P., Takahashi E., van der Kouwe A. J., Charvet C. J., Brain wiring and supragranular-enriched genes linked to protracted human frontal cortex development. bioRxiv:10.1101/746248 (24 August 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., et al. , Satb2 is required for dendritic arborization and soma spacing in mouse cerebral cortex. Cereb. Cortex 22, 1510–1519 (2012). [DOI] [PubMed] [Google Scholar]
- 38.National Health and Medical Research Council, Australian Code for the Care and Use of Animals for Scientific Purposes, (National Health and Medical Research Council, Canberra, Australia, 8th Ed., 2013). [Google Scholar]
- 39.Ashwell K., The Neurobiology of Australian Marsupials: Brain Evolution in the Other Mammalian Radiation (Cambridge University Press, 2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the main text and SI Appendix.