Abstract
Background
Carbapenem-resistant Enterobacteriaceae (CRE) is a major concern leading to morbidity and mortality in the world. CRE often is becoming a cause of therapeutic failure in both hospital and community-acquired infections.
Aim
This study aimed to investigate the resistance mechanisms of CRE by phenotypic and molecular methods.
Materials and Methods
Sixty CRE (50 Klebsiella pneumoniae, 6 Escherichia coli, and 4 Enterobacter spp.) were isolated from October 2018 to June 2019. Antimicrobial susceptibility testing was carried out using phenotypic methods. The carbapenem resistance mechanisms including efflux pump hyperexpression, AmpC overproduction, carbapenemase genes, and deficiency in OmpK35 and OmpK36 were determined by phenotypic and molecular methods, respectively.
Results
Sixty CRE (50 Klebsiella pneumoniae, 6 Escherichia coli, and 4 Enterobacter spp.) were isolated from October 2018 to June 2019. Amikacin was found to be the most effective drug against CRE isolates. All isolates were resistant to imipenem and meropenem by the micro-broth dilution. AmpC overproduction was observed in all Enterobacter spp. and three K. pneumoniae isolates. No efflux pump activity was found. Carba NP test and Modified Hodge Test could find carbapenemase in 59 (98%) isolates and 57 (95%) isolates, respectively. The most common carbapenemase gene was blaOXA-48-like (72.8%) followed by blaNDM (50.8%), blaIMP (18.6%), blaVIM (11.8%), and blaKPC (6.7%). The ompK35 and ompK36 genes were not detected in 10 and 7 K. pneumoniae isolates, respectively.
Conclusion
The amikacin is considered as a very efficient antibiotic for the treatment of CRE isolates in our region. Carbapenemase production and overproduction of AmpC are the main carbapenem resistance mechanisms in CRE isolates. Finally, Carba NP test is a rapid and reliable test for early detection of carbapenemase-producing isolates.
Keywords: amikacin, carbapenemase genes, carbapenem-resistant Enterobacteriaceae, Carba NP test
Introduction
Enterobacteriaceae as a Gram-negative bacteria is the cause of various acquired infections including urinary tract, bloodstream, and lower respiratory tract infections.1 Carbapenems are often considered as a last resort of therapeutic option for infections due to Enterobacteriaceae.2 Carbapenem resistance refers to the ability of bacteria to survive and grow in the presence of clinically relevant concentrations of carbapenems.3 Emergence and dissemination of carbapenem resistance have been increasingly reported across the world which limits the usage of carbapenems.4 Carbapenem-resistant Enterobacteriaceae (CRE) is a significant clinical and public health concern.5 Considering their transmissibility and limited treatment options against CRE, the Centers for Disease Control and Prevention (CDC) considers CRE as specially dangerous.6 CRE is often resistant to all β-lactam drugs and simultaneously they often carry mechanisms conferring resistance to other antimicrobial classes. The infections with these bacteria are associated with increased mortality rates and higher healthcare costs.7,8 Currently, treatment options for CRE infections are limited to few antibiotics including polymyxin, tigecycline, fosfomycin, and aminoglycosides, or in combination with other antibiotics.9 To initiate/optimize antibiotic therapy and to control CRE infections, early detection of carbapenemase production and other mechanisms is necessary.10 There are three primary mechanisms by which Enterobacteriaceae employ resistance to carbapenems:3 (a) enzymatic hydrolysis of carbapenems by carbapenemases, enzymes that break down carbapenems (such as carbapenemase encoding genes, NDM, KPC, VIM, OXA, and IMP), (b) expression of efflux pumps, actively extruding carbapenems from the bacterial cell, and (c) reduction of outer membrane permeability via production of beta-lactamases (AmpC) in combination with alterations in the bacterial cell membrane (porin mutations in OmpK35 and OmpK36).1,3,11 Recent breakpoints for CRE often detect all clinically important resistance mechanisms (especially carbapenemase genes).12
Several Iranian studies during the last decade are clarified CRE, but most of these studies focus on carbapenemase genes detection.13–15 According to the data on the prevalence of CRE, the molecular characteristics and carbapenem resistance mechanisms in CRE are poorly known in our region. The present study aimed to assess the prevalence rate of CRE and to monitor the antimicrobial susceptibility, and to perform phenotypic and genotypic evaluation of resistance mechanisms of CRE in Tabriz, Iran.
Materials and Methods
Bacterial Strains
Enterobacteriaceae isolates were recovered from clinical samples of Tabriz hospitals, Iran. The isolates were identified using conventional biochemical tests in the Department of Microbiology, Tabriz University of Medical Sciences, Iran. The inclusion criteria were reduced susceptibility to at least one carbapenem (imipenem, meropenem, and ertapenem) according to the Clinical and Laboratory Standards Institute (CLSI) (2019, 29th) guideline. A total of 60 non-duplicate CRE isolates were collected during a 9-month period from October 2018 to June 2019.
Antimicrobial Susceptibility Testing
We performed the susceptibility testing by the Kirby–Bauer method in Mueller–Hinton agar according to CLSI (2019, 29th) guideline. The antibiotic disks of imipenem (10 µg), meropenem (10 µg), ertapenem (10 µg), aztreonam (30 µg), cefixime (5 µg), ceftazidime (30 µg), cefotaxime (30 µg), cefepime (30 µg), cefazolin (30 µg), tobramycin (10 µg), amikacin (30 µg), gentamicin (10 µg), tetracycline (30 µg), and Piperacillin-tazobactam, Trimethoprim-sulfamethoxazole (1.25/23.75 μg) were tested for the antimicrobial susceptibility.16 The results of susceptibility testing were validated using the American Type Culture Collection (ATCC) quality control strain P. aeruginosa ATCC 27853 and Escherichia coli ATCC 25922. The minimum inhibitory concentrations (MICs) for the imipenem, meropenem, and colistin were determined using the micro-broth dilution method. The MICs were defined as the lowest concentration inhibiting an evident growth of the bacteria and interpreted according to the guidelines of the CLSI (2019, 29th).16
Detection of AmpC Overproduction
AmpC overproduction was confirmed according to the method described by Martínez et al. The isolates were considered as AmpC overproducer when there was at least a twofold dilution difference between the MICs of imipenem and those of imipenem plus cloxacillin.17
Efflux Pump Inhibitor Tests
MICs of meropenem were determined alone and in the presence of phenylalanine arginine-naphthylamide dihydrochloride (PaβN) as an inhibitor of RND pumps of Enterobacteriaceae. A two-fold decline in MICs after addition of PAN was considered significant.18
Detection of Carbapenemase Enzymes by the Phenotypic Tests
All the carbapenem-resistant isolates were subjected to further evaluation by two methods. Modified Hodge Test (MHT) was performed according to the protocol recommended by CLSI instructions.19 The Carba NP test was performed as described previously.20 Briefly, one inoculation loop (10 μL) of the isolate, recovered from MHA, was suspended in 200 μL of 0.02% cetyl trimethyl ammonium bromide (CTAB); then 100 μL of the bacterial suspension was added to 100 μL of diluted phenol red solution containing 0.1 mM ZnSO 4 (pH = 7.5) supplemented with 6 mg/mL of imipenem. The phenol red solution, with no antibiotic, was used as a control tube for each isolate. Both tubes were vortexed and incubated at 37°C for a maximum of two hours. The color of the test tube changed to full yellow or orange, indicating carbapenemase-producing isolate, while the control tube remained red.20
PCR Amplification of Carbapenem Resistance Genes and Porin Coding Genes
All isolates that were phenotypically resistant to carbapenems were screened for the carbapenemase and porin encoding genes. The DNA was extracted by the CTAB method as described previously.21 PCR assays were performed using primers specific for the blaKPC, blaVIM, blaIMP, blaNDM, and blaOXA-48-like carbapenemase encoding genes, as well as OmpK35 and OmpK36 as porin coding genes (Table 1).22,23 PCR products underwent electrophoresis in 1.5% agarose gel, and after staining with 0.5 µg/mL safe stain, they were visualized under ultraviolet (UV) light. P. aeruginosa (blaVIM and blaIMP,) and Klebsiella pneumoniae (blaKPC, blaNDM, and blaOXA-48-like) were used as controls in this study.
Table 1.
List of Primers and Expected Amplicon Size in This Study
| Gene | Forward Sequence [5–3] | Reverse Sequence [5–3] | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| blaKPC | CGTCTAGTTCTGCTGTCTTG | CTTGTCATCCTTGTTAGGCG | 798 | [22] |
| blaNDM | GGTTTGGCGATCTGGTTTTC | CGGAATGGCTCATCACGATC | 621 | [22] |
| blaOXA-48-like | GCGTGGTTAAGGATGAACAC | CATCAAGTTCAACCCAACCG | 438 | [22] |
| blaIMP | GGAATAGAGTGGCTTAAYTCTC | GGTTTAAYAAAACAACCACC | 232 | [22] |
| blaVIM | GATGGTGTTTGGTCGCATA | CGAATGCGCAGCACCAG | 390 | [22] |
| OmpK35 | CTCCAGCTCTAACCGTAGCG | GGTCTGTACGTAGCCGATGG | 241 | [23] |
| OmpK36 | GAAATTTATAACAAAGACGGC | GACGTTACGTCGTATACTACG | 305 | [23] |
Statistical Method
The results were analyzed using the descriptive statistics in SPSS software for Windows (version 23 SPSS Inc., Chicago, IL, USA). In this study, p≤ 0.05 was considered statistically significant.
Results
Sixty isolates were resistant to at least one of the tested carbapenem (imipenem, meropenem, and ertapenem) antibiotics. Based on conventional microbiologic tests, 50 K. pneumoniae isolates, 6 E. coli isolates, and 4 Enterobacter spp. isolates were identified.
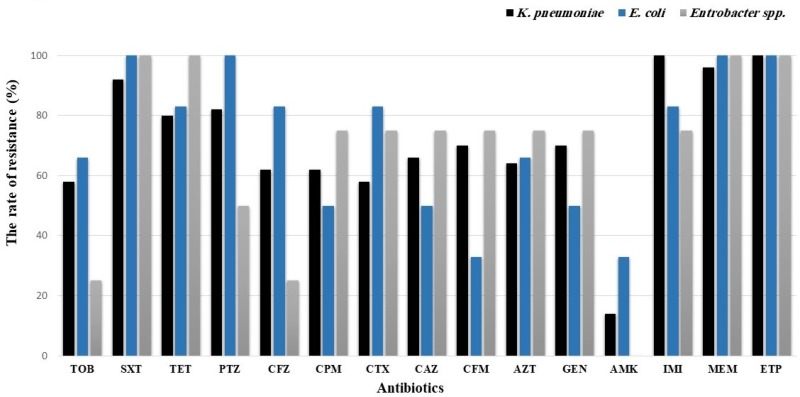
The resistance rates to imipenem, meropenem, and ertapenem were 96%, 96%, and 100%, respectively. Amikacin had the highest susceptibility rate (85%) followed by tobramycin (43%) (Figure 1). Amikacin was found to be the most effective drug against KPC-positive and VIM-positive isolates while other tested carbapenemase coding genes had variable susceptibilities to amikacin.
Figure 1.
Antimicrobial susceptibility patterns of K. pneumoniae, E. coli, and Enterobacter spp. isolates.
The micro-broth dilution results indicated that all isolates were resistant to imipenem and meropenem (MIC ≥4 µg/mL). Further, 10 isolates were highly resistant (MIC ≥ 64 mg/mL) to imipenem and meropenem, respectively (Figure 2). The micro-broth dilution results showed that all isolates were susceptible to colistin with MIC≤0.5 mg/mL. MIC50 for imipenem and meropenem was 8 µg/mL and 16 µg/mL, while the MIC90 of agents was 32 µg/mL and 64 µg/mL, respectively. MIC90 and MIC50 values were defined as the lowest concentration of the antibiotic at which 90% and 50% of the isolates were inhibited, respectively. Note that the MIC90 of isolates observed to be within the non-susceptible range.
Figure 2.
Distribution of imipenem and meropenem MICs in K. pneumoniae, E. coli, and Enterobacter spp. isolates.
Reduced MICs of meropenem and imipenem in AmpC overproduction isolates in CRE with/without inhibitor were found in 7 (11.6%) isolates. These isolates had carbapenem activity with a decline of MIC up to twofold by overproduction of AmpC. AmpC overproduction was observed in all of Enterobacter spp. and three K. pneumoniae isolates.
In this study, the isolates showed less than a threefold reduction in the MIC of meropenem in the presence of efflux pump inhibitor (PAbN) among 60 CRE isolates.
Carba NP test detected carbapenemase in 59 (98%) isolates, except one isolate (Enterobacter spp.) which was negative for carbapenemase genes. In addition, carbapenemase activity was detected by MHT in 57 (95%) isolates. Three carbapenemase producer isolates were not detected by MHT assay, where two isolates were K. pneumoniae (NDM-positive), and one isolate was Enterobacter spp. (overproduction of AmpC). The sensitivity and specificity of Carba NP test and MHT were 98% and 95%, and 100% and 100%, respectively.
Fifty-nine of 60 isolates (98.3%) were positive for carbapenemase genes. The most common carbapenemase genes were blaOXA-48-like (72.8%) followed by blaNDM (50.8%), blaIMP (18.6%), blaVIM (11.8%), and blaKPC (6.7%) (Table 2). Among the isolates, the percentage of carbapenemase genes in K. pneumoniae was blaOXA-48-like (78%), blaNDM (48%), blaIMP (22%), blaVIM (12%), blaKPC (8%); in E. coli was blaNDM (100%), blaVIM (16.7%), and in Enterobacter spp. was blaOXA-48-like (75%). In one isolate of Enterobacter spp., none of the tested carbapenemase genes was detected. Among the 59 isolates that were positive to at least one of the tested genes, 29 isolates had one carbapenemase gene, 25 isolates had two genes, and 5 isolates had three genes. The blaIMP and blaKPC were detected only in K. pneumoniae isolates (Table 3). As reported in Table 2, 12 distinct combination patterns were recognized among 59 carbapenemase genes PCR-positive isolates.
Table 2.
The Susceptibility Testing Results in Carbapenemase-Producing Isolates
| Genotype | Species N (%) | Antibiotics | MIC (µg/mL) | ||||
|---|---|---|---|---|---|---|---|
| 4 | 8 | 16 | 32 | 64 | |||
| blaOXA-48-like |
K. pneumoniae 15 (25.4) Enterobacter spp. 3 (5.1) |
IMI MEM |
5 2 |
7 1 |
4 10 |
2 5 |
0 0 |
| blaNDM |
K. pneumoniae 6 (10.1) E. coli 5 (8.8) |
IMI MEM |
2 1 |
4 3 |
4 5 |
0 1 |
1 1 |
| blaOXA-48-like + blaNDM | K. pneumoniae 8 (13.5) | IMI MEM |
3 1 |
1 3 |
2 3 |
2 1 |
0 0 |
| blaNDM + blaVIM |
K. pneumoniae (1.6) E. coli 1 (1.6) |
IMI MEM |
0 0 |
1 1 |
0 0 |
0 0 |
1 1 |
| blaNDM + blaIMP | K. pneumoniae 3 (3.3) | IMI MEM |
0 1 |
2 1 |
1 0 |
0 1 |
0 0 |
| blaOXA-48-like + blaIMP | K. pneumoniae 6 (10.1) | IMI MEM |
0 0 |
4 3 |
1 1 |
1 2 |
0 0 |
| blaOXA-48-like+ blaVIM | K. pneumoniae 4 (6.7) | IMI MEM |
1 0 |
1 1 |
1 1 |
1 2 |
0 0 |
| blaOXA-48-like+ blaKPC | K. pneumoniae 1 (1.6) | IMI MEM |
0 0 |
0 1 |
1 0 |
0 0 |
0 0 |
| blaNDM+ blaKPC | K. pneumoniae 1 (1.6) | IMI MEM |
0 0 |
0 0 |
1 1 |
0 0 |
0 0 |
| blaOXA-48-like+ blaNDM + blaIMP | K. pneumoniae 2 (3.3) | IMI MEM |
0 0 |
1 1 |
0 0 |
0 0 |
1 1 |
| blaOXA-48-like+ blaNDM + blaVIM | K. pneumoniae 1 (1.6) | IMI MEM |
0 0 |
0 0 |
0 0 |
0 0 |
1 1 |
| blaOXA-48-like+ blaNDM + blaKPC | K. pneumoniae 2 (3.3) | IMI MEM |
0 0 |
0 0 |
0 0 |
2 0 |
0 2 |
Abbreviations: MIC, minimum inhibitory concentration; IMI, imipenem; MEM, meropenem.
Table 3.
The Frequency of Carbapenemase Genes and AmpC Overproduction in CRE Isolates
| Genotype | K. pneumoniae | E. coli | Enterobacter spp. |
|---|---|---|---|
| blaOXA-48-like | 39 | 0 | 3 |
| blaNDM | 24 | 6 | 0 |
| blaVIM | 6 | 1 | 0 |
| blaIMP | 11 | 0 | 0 |
| blaKPC | 4 | 0 | 0 |
| AmpC | 3 | 0 | 4 |
Of 50 isolates of K. pneumoniae, the genes coding for porin protein (ompK35 and ompK36) were not detected in 10 and 7 K. pneumoniae isolates, respectively. Genes coding for the porins (ompK35 and ompK36) were detected in all K. pneumoniae recovered from body fluid specimens in blood, wound, and urine samples.
Discussion
Resistance to carbapenems as the last-resort antibiotics for the treatment of infections caused by the CRE is a major health concern. The increase in CRE is a serious matter due to high case-fatality rates (>30%).24–26 Studying the mechanisms of CRE is an essential step in planning a national public health strategy for this emerging pathogen.27 Several studies have been studied the mechanisms of resistance to carbapenem in CRE isolates from Iran. However, most of these studies focus on the carbapenemase associated mechanisms.15,28 In the present study, carbapenemase-associated mechanisms and also other mechanisms of CRE such as efflux pump, AmpC overproduction, and porin related mechanism were studied. Because of the impact geographical characteristics on susceptibility pattern CRE, the results of such a study can be applied in the treatment of CRE isolates from Tabriz, Iran.
In this research, K. pneumoniae was the most common CRE which was isolated from 50 specimens followed by E. coli and Enterobacter spp. This agrees with the previous studies reporting that K. pneumoniae and E. coli were predominant CRE among Enterobacteriaceae.4,29-31 We found a high rate of antimicrobial resistance. In addition, these CRE pose the greatest risk to the public health because of their high prevalence, high potential for causing a wide range of clinical infections, co-resistance to Beta-lactam as well as other antimicrobial agents (such as aminoglycosides and fluoroquinolones).4,29
Understanding the antibiotic susceptibility patterns of CRE is important for antibiotic therapy of infections. In our study, amikacin showed the highest susceptibility rate (85%). Similar reports previously described by Gao et al,31 Park et al,32 Freire et al,33 and Chen et al34 showed that amikacin had high in vitro activity against CRE isolates. In contrast, some of the studies reported high rates of resistance to aminoglycosides chiefly amikacin.29,35 These findings could be attributed to the differences in the characteristics of the various carbapenemase types, geographical area, and usage of antibiotics.30 Overall, this result and previous reports suggested that some active agents such as amikacin might be appropriate options for the treatment of CRE.34 Our study revealed that all isolates had MIC ≥4 µg/mL and resistance to imipenem plus meropenem. The MIC50 and MIC90 of carbapenems were 8–16 µg/mL and 32–64 µg/mL, respectively. Other studies confirmed that the most frequent non-susceptible carbapenems were meropenem and imipenem.4,31 Therefore, the CLSI and EUCAST recommend the MIC test for routine clinical isolates.16,36
The carbapenem-hydrolyzing activity of class C-beta lactamases has been reported.18 In this study, the overproduction of AmpC was confirmed in all isolates of Enterobacter spp. and 6% K. pneumoniae isolates. Several studies have reported that AmpC producer in K. pneumoniae ranged from 7.8% to 43%.37–39 Similar to a previous study, resistance to carbapenem among Enterobacter spp. was high.40 Class C-beta lactamase enzymes appeared as an important contribution to carbapenem resistance among Enterobacter spp. in our region.
Antibiotic efflux pumps are not considered as an important mechanism of antimicrobial resistance in CRE.41 Similar to other studies carried by Kim et al,42 Dupont et al,18 and Osei Sekyere et al,43 in the present study, we did not find any efflux pump activity by the phenotypic method. Previous studies found the efflux pump by the molecular technique such as PCR and observed that AcrAB efflux pump and mdtK efflux pump existed in CRE isolates.23,41
In the current study, carbapenemase enzymes were detected by Carba NP test and MHT in 98% and 95%, respectively, which was in line with another study carried by Elawady et al44 and Garg et al45 who reported good sensitivity and specificity of Carba NP test.
The presence of carbapenemase genes was found in 42 isolates for blaOXA-48-like. Also, 30 isolates were positive for blaNDM, 11 isolates were positive for blaIMP, 7 isolates were positive for blaVIM, and 4 isolates were for blaKPC. blaOXA-48-like was the most common gens in K. pneumoniae, while blaNDM was the most common in E. coli. These findings are also congruent with other results obtained by the previous studies.46,47 In our study, blaVIM and blaKPC were the least frequent, which is in agreement with other results reported by Shibl et al48 and Cakirlar et al.49 This similarity can be due to the population size studied, the proximity of geographic regions, and the similarity of antibiotic usage.
Porins such as OmpK35 and OmpK36 have a critical role in the penetration of antibiotics into cells and susceptibility to cephalosporins and carbapenems. It has also been reported that loss of OmpK35 and OmpK36 plays a key role in K. pneumoniae virulence and infection.23 Deletion of OmpK36 and OmpK35 can lead to a reduction in virulence and infection. In the current study, K. pneumoniae isolates harbored high distribution of OmpK35 and OmpK36 genes coding for porin protein which was in accordance with studies carried by Wasfi et al23 and Ranjbar et al41 who reported a high prevalence of these genes.23 The authors suggest the detection of porin coding genes and efflux pump inhibitors by Real-time PCR. Furthermore, doing molecular typing methods could help to identify the origin of resistant strains and designing the program to control their spread. The resistance rate to carbapenems is high. We found that amikacin is an effective antibiotic for the treatment of CRE isolates. The increasing resistance to amikacin in CRE isolates is an alarming signal for monotherapy with amikacin. This study revealed that carbapenemase production and overproduction of AmpC are the main resistance mechanisms to carbapenem among CRE isolates in the West Azerbaijan, Iran. The most common carbapenemase gene was blaOXA-48-like. Based on the results, early detection of carbapenemase-producing isolates with rapid and reliable tests such as Carba NP test may be useful for the treatment of infections induced by CRE isolates.
Funding Statement
This article was financially supported by the Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
Ethical Approval
This study was approved by the research ethics committee (IR.TBZMED.VCR.REC.1397.035) at Tabriz University of Medical Sciences, Tabriz, Iran.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. Study concept and design: Naser Alizadeh, Mohammad Ahangarzadeh Rezaee, Reza Ghotaslou. Acquisition of data and sampling: Aidin Lalehzadeh, Fatemeh Yeganeh Sefidan, Mohammad Hossein Soroush Barhaghi. Analysis and interpretation of data: Naser Alizadeh, Mohammad Yousef Memar, Hossein Samadi Kafil, Alka Hasani. Original draft preparation: Naser Alizadeh, Reza Ghotaslou, Mohammad Ahangarzadeh Rezaee. Review and editing: Morteza Milani, Reza Ghotaslou. Study supervision: Reza Ghotaslou, Mohammad Ahangarzadeh Rezaee.
Disclosure
The authors have reported no conflict of interest.
References
- 1.Alizadeh N, Rezaee MA, Kafil HS, et al. Detection of carbapenem-resistant Enterobacteriaceae by chromogenic screening media. J Microbiol Methods. 2018;153:40–44. doi: 10.1016/j.mimet.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 2.Lutgring JD, Limbago BM. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol. 2016;54(3):529–534. doi: 10.1128/JCM.02771-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durante-Mangoni E, Andini R, Zampino R. Management of carbapenem-resistant Enterobacteriaceae infections. J Clin Microbiol. 2019;25:943–950. [DOI] [PubMed] [Google Scholar]
- 4.Sekar R, Srivani S, Kalyanaraman N, et al. NDM and other mechanisms of carbapenemases among Enterobacteriaceae in rural South India. J Glob Antimicrob Resist. 2019;18:207–214. doi: 10.1016/j.jgar.2019.05.028 [DOI] [PubMed] [Google Scholar]
- 5.Guh AY, Limbago BM, Kallen AJ. Epidemiology and prevention of carbapenem-resistant Enterobacteriaceae in the United States. Expert Rev Anti-Infect Ther. 2014;12(5):565–580. doi: 10.1586/14787210.2014.902306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley N, Lee Y. Practical implications of new antibiotic agents for the treatment of carbapenem-resistant Enterobacteriaceae. Microbiol Insights. 2019;12:1178636119840367. doi: 10.1177/1178636119840367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53(1):60–67. doi: 10.1093/cid/cir202 [DOI] [PubMed] [Google Scholar]
- 8.Eichenberger EM, Thaden JT. Epidemiology and mechanisms of resistance of extensively drug resistant Gram-negative bacteria. Antibiotics. 2019;8(2):37. doi: 10.3390/antibiotics8020037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheu -C-C, Chang Y-T, Lin S-Y, Chen Y-H, Hsueh P-R. Infections caused by carbapenem-resistant enterobacteriaceae: an update on therapeutic options. Front Microbiol. 2019;10. doi: 10.3389/fmicb.2019.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greissl C, Saleh A, Hamprecht A. Rapid detection of OXA-48-like, KPC, NDM, and VIM carbapenemases in Enterobacterales by a new multiplex immunochromatographic test. Eur J Clin Microbiol Infect Dis. 2019;38(2):331–335. doi: 10.1007/s10096-018-3432-2 [DOI] [PubMed] [Google Scholar]
- 11.Girlich D, Grosperrin V, Naas T, Dortet L. CHROMagar™ ESBL/mSuperCARBA bi-plate medium for detection of ESBL-and carbapenemase-producing Enterobacteriaceae from spiked stools. Diagn Microbiol Infect Dis. 2019;95(2):107–112. doi: 10.1016/j.diagmicrobio.2019.05.002 [DOI] [PubMed] [Google Scholar]
- 12.Codjoe FS, Donkor ES. Carbapenem resistance: a review. Med Sci. 2018;6(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadeghi MR, Ghotaslou R, Akhi MT, Asgharzadeh M, Hasani A. Molecular characterization of extended-spectrum β-lactamase, plasmid-mediated AmpC cephalosporinase and carbapenemase genes among Enterobacteriaceae isolates in five medical centres of East and West Azerbaijan, Iran. J Med Microbiol. 2016;65(11):1322–1331. doi: 10.1099/jmm.0.000356 [DOI] [PubMed] [Google Scholar]
- 14.Shahcheraghi F, Aslani MM, Mahmoudi H, et al. Molecular study of carbapenemase genes in clinical isolates of Enterobacteriaceae resistant to carbapenems and determining their clonal relationship using pulsed-field gel electrophoresis. J Med Microbiol. 2017;66(5):570–576. doi: 10.1099/jmm.0.000467 [DOI] [PubMed] [Google Scholar]
- 15.Solgi H, Badmasti F, Aminzadeh Z, et al. Molecular characterization of intestinal carriage of carbapenem-resistant Enterobacteriaceae among inpatients at two Iranian university hospitals: first report of co-production of bla NDM-7 and bla OXA-48. Eur J Clin Microbiol Infect Dis. 2017;36(11):2127–2135. doi: 10.1007/s10096-017-3035-3 [DOI] [PubMed] [Google Scholar]
- 16.Wayne P; Clinical and Laboratory Standards Institute. Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing; 2019:21 [Google Scholar]
- 17.Rodríguez-Martínez J-M, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(11):4783–4788. doi: 10.1128/AAC.00574-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupont H, Gaillot O, Goetgheluck A-S, et al. Molecular characterization of carbapenem-nonsusceptible enterobacterial isolates collected during a prospective interregional survey in France and susceptibility to the novel ceftazidime-avibactam and aztreonam-avibactam combinations. Antimicrob Agents Chemother. 2016;60(1):215–221. doi: 10.1128/AAC.01559-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CLSI C. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. M100-S24 January; 2014.
- 20.Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18(9):1503. doi: 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akhi MT, Ghotaslou R, Alizadeh N, Beheshtirouy S, Memar MY. High frequency of MRSA in surgical site infections and elevated vancomycin MIC. Wound Med. 2017;17:7–10. doi: 10.1016/j.wndm.2017.01.002 [DOI] [Google Scholar]
- 22.Candan ED, Aksöz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochimica Polonica. 2015;62:4. doi: 10.18388/abp.2015_1148 [DOI] [PubMed] [Google Scholar]
- 23.Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6:38929. doi: 10.1038/srep38929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teethaisong Y, Nakouti I, Evans K, Eumkeb G, Hobbs G. Nitro-Carba test, a novel and simple chromogenic phenotypic method for rapid screening of carbapenemase-producing Enterobacteriaceae. J Glob Antimicrob Resist. 2019;18:22–25. doi: 10.1016/j.jgar.2019.01.012 [DOI] [PubMed] [Google Scholar]
- 25.Correa AAF, Fortaleza CMCB. Incidence and predictors of health care–associated infections among patients colonized with carbapenem-resistant Enterobacteriaceae. Am J Infect Control. 2019;47(2):213–216. doi: 10.1016/j.ajic.2018.08.007 [DOI] [PubMed] [Google Scholar]
- 26.Hsueh P-R, Jean -S-S, Lee N-Y, Tang H-J, Lu M-C. Carbapenem-resistant Enterobacteriaceae infections: Taiwan aspects. Front Microbiol. 2018;9:2888. doi: 10.3389/fmicb.2018.02888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livorsi DJ, Chorazy ML, Schweizer ML, et al. A systematic review of the epidemiology of carbapenem-resistant Enterobacteriaceae in the United States. Antimicrob Resist Infect Control. 2018;7(1):55. doi: 10.1186/s13756-018-0346-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosavian M, Koraei D. Molecular detection of IMP carbapenemase-producing gram-negative bacteria isolated from clinical specimens in Ahvaz, Iran. JJHR. 2016;7(6). [Google Scholar]
- 29.Saeed NK, Alkhawaja S, Azam NFAEM, Alaradi K, Al-Biltagi M. Epidemiology of carbapenem-resistant Enterobacteriaceae in a Tertiary Care Center in the Kingdom of Bahrain. J Lab Physicians. 2019;11(2):111. doi: 10.4103/JLP.JLP_101_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makhoul IR, Sujov P, Smolkin T, Lusky A, Reichman B. Epidemiological, clinical, and microbiological characteristics of late-onset sepsis among very low birth weight infants in Israel: a national survey. Pediatrics. 2002;109(1):34–39. doi: 10.1542/peds.109.1.34 [DOI] [PubMed] [Google Scholar]
- 31.Gao B, Li X, Yang F, et al. Molecular epidemiology and risk factors of ventilator-associated pneumonia infection caused by carbapenem-resistant enterobacteriaceae. Front Pharmacol. 2019;10:262. doi: 10.3389/fphar.2019.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park JW, Lee H, Park SY, Kim TH. Epidemiological, clinical, and microbiological characteristics of carbapenemase-producing Enterobacteriaceae bloodstream infection in the Republic of Korea. Antimicrob Resist Infect Control. 2019;8:48. doi: 10.1186/s13756-019-0497-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freire MP, de Oliveira Garcia D, Cury AP, et al. The role of therapy with aminoglycoside in the outcomes of kidney transplant recipients infected with polymyxin-and carbapenem-resistant Enterobacteriaceae. Eur J Clin Microbiol Infect Dis. 2019;38(4):755–765. doi: 10.1007/s10096-019-03468-4 [DOI] [PubMed] [Google Scholar]
- 34.Chen C-W, Tang H-J, Chen -C-C, et al. The microbiological characteristics of carbapenem-resistant enterobacteriaceae carrying the mcr-1 gene. J Clin Med. 2019;8(2):261. doi: 10.3390/jcm8020261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu F, Chen S, Xu X, et al. Emergence of carbapenem-resistant clinical Enterobacteriaceae isolates from a teaching hospital in Shanghai, China. J Med Microbiol. 2012;61(1):132–136. doi: 10.1099/jmm.0.036483-0 [DOI] [PubMed] [Google Scholar]
- 36.Leclercq R, Cantón R, Brown DF, et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect. 2013;19(2):141–160. doi: 10.1111/j.1469-0691.2011.03703.x [DOI] [PubMed] [Google Scholar]
- 37.Bhaskar BH, Mulki SS, Joshi S, Adhikary R, Venkatesh BM. Molecular characterization of extended spectrum β-lactamase and carbapenemase producing klebsiella pneumoniae from a tertiary care hospital. Indian J Crit Care Med. 2019;23(2):61–66. doi: 10.5005/jp-journals-10071-23118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazemian H, Heidari H, Ghanavati R, et al. Phenotypic and genotypic characterization of ESBL-, AmpC-, and carbapenemase-producing Klebsiella pneumoniae and Escherichia coli isolates. Med Princ Pract. 2019;28(6):547–551. doi: 10.1159/000500311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hertz FB, Jansaker F, Okon KO, et al. ESBL-production in Escherichia coli and Klebsiella pneumoniae isolates from Nigeria. Microbiol Open. 2019;8:e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majewski P, Wieczorek P, Ojdana D, et al. Altered outer membrane transcriptome balance with AmpC overexpression in carbapenem-resistant enterobacter cloacae. Front Microbiol. 2016;7:2054. doi: 10.3389/fmicb.2016.02054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranjbar R, Kelishadrokhi AF, Chehelgerdi M. Molecular characterization, serotypes and phenotypic and genotypic evaluation of antibiotic resistance of the Klebsiella pneumoniae strains isolated from different types of hospital-acquired infections. Infect Drug Resist. 2019;12:603. doi: 10.2147/IDR.S199639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim SY, Shin J, Shin SY, Ko KS. Characteristics of carbapenem-resistant Enterobacteriaceae isolates from Korea. Diagn Microbiol Infect Dis. 2013;76(4):486–490. doi: 10.1016/j.diagmicrobio.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 43.Osei Sekyere J, Amoako DG. Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front Microbiol. 2017;8:228. doi: 10.3389/fmicb.2017.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elawady B, Ghobashy M, Balbaa A. Rapidec Carba NP for detection of carbapenemase-producing enterobacteriaceae in clinical isolates: a cross-sectional study. Surg Infect. 2019;20(8):672–676. doi: 10.1089/sur.2019.084 [DOI] [PubMed] [Google Scholar]
- 45.Garg A, Garg J, Kumar S, Bhattacharya A, Agarwal S, Upadhyay GC. Molecular epidemiology & therapeutic options of carbapenem-resistant Gram-negative bacteria. Indian J Med Res. 2019;149(2):285–289. doi: 10.4103/ijmr.IJMR_36_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Girlich D, Bouihat N, Poirel L, Benouda A, Nordmann P. High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin Microbiol Infect. 2014;20(4):350–354. doi: 10.1111/1469-0691.12325 [DOI] [PubMed] [Google Scholar]
- 47.Sonnevend A, Ghazawi AA, Hashmey R, et al. Characterization of carbapenem-resistant Enterobacteriaceae with high rate of autochthonous transmission in the Arabian Peninsula. PLoS One. 2015;10(6):e0131372. doi: 10.1371/journal.pone.0131372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibl A, Al-Agamy M, Memish Z, Senok A, Khader SA, Assiri A. The emergence of OXA-48-and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int J Infect Dis. 2013;17(12):e1130–e1133. doi: 10.1016/j.ijid.2013.06.016 [DOI] [PubMed] [Google Scholar]
- 49.Cakirlar FK, Gonullu N, Kalayci F, Kiraz N. Detection of carbapenemase genes OXA-48, VIM, IMP, KPC and NDM in carbapenemase-producing klebsiella pneumoniae isolates from blood cultures of hospitalized patients in Istanbul, Turkey. Int J Infect Dis. 2016;45:99. doi: 10.1016/j.ijid.2016.02.259 [DOI] [Google Scholar]