Abstract
The N6-methyladenosine modification at position 43 (m6A43) of U6 snRNA is catalyzed by METTL16, and is important for the 5′-splice site recognition by U6 snRNA during pre-mRNA splicing. Human METTL16 consists of the N-terminal methyltransferase domain (MTD) and the C-terminal vertebrate conserved region (VCR). While the MTD has an intrinsic property to recognize a specific sequence in the distinct structural context of RNA, the VCR functions have remained uncharacterized. Here, we present structural and functional analyses of the human METTL16 VCR. The VCR increases the affinity of METTL16 toward U6 snRNA, and the conserved basic region in VCR is important for the METTL16–U6 snRNA interaction. The VCR structure is topologically homologous to the C-terminal RNA binding domain, KA1, in U6 snRNA-specific terminal uridylyl transferase 1 (TUT1). A chimera of the N-terminal MTD of METTL16 and the C-terminal KA1 of TUT1 methylated U6 snRNA more efficiently than the MTD, indicating the functional conservation of the VCR and KA1 for U6 snRNA biogenesis. The VCR interacts with the internal stem-loop (ISL) within U6 snRNA, and this interaction would induce the conformational rearrangement of the A43-containing region of U6 snRNA, thereby modifying the RNA structure to become suitable for productive catalysis by the MTD. Therefore, the MTD and VCR in METTL16 cooperatively facilitate the m6A43 U6 snRNA modification.
INTRODUCTION
N6-Methyladenosine (m6A) is an abundant modification observed in mRNA (1–4) and plays pivotal roles in various cellular processes, encompassing stem cell differentiation, development, and cell signaling such as circadian rhythms (5–9). The m6A modification in RNAs affects splicing, translation, localization, decay and stability (10–15).
Most m6A modifications in mRNA are produced by the heteromeric methyltransferase-like protein 3 (METTL3) and methyltransferase-like protein 14 (METTL14) complex, and the METTL3/METTL14 complex methylates mRNA at the RRACH (R = A or G, H = A, C or U) motif close to the stop codon or 3′-UTR of mRNAs (2,3,16–18). METTL3 is the catalytic active subunit and interacts with S-adenosylmethionine (SAM), while METTL14 is catalytically inactive and contributes to RNA binding (19–21).
Another methyltransferase-like protein, METTL16, was recently shown to have distinct target RNAs for m6A modification. METTL16 methylates the 3′ UTR of the SAM synthetase (MAT2A) mRNA and U6 snRNA (22–24). METTL16 methylates the adenosine in the UACAGAGAA (methylated adenosine is underlined) conserved sequence in a specific structural context (22–26).
The METTL16-mediated methylation of specific adenosines in UACAGAGAA (methylated adenosine is underlined) in the MAT2A 3′ UTR hairpins is involved in the regulation of the stability and splicing of pre-MAT2A mRNA, thereby maintaining the homeostasis of the intracellular SAM concentration (22,23). METTL16 also methylates a specific adenosine (A43 in human) of U6 snRNA in the conserved motif sequence, as observed in MAT2A 3′ UTR hairpins (23–25). A43 lies within the highly conserved ACAGAGA (methylated adenosine is underlined) box of U6 snRNA, which base pairs with the 5′ splice site of mRNA during pre-mRNA splicing (27–31). In yeast, mutations in this sequence are lethal (32), implying that the m6A modification at this position is essential for pre-mRNA splicing regulation, although the precise functions of this modification remain elusive (24).
Human METTL16 consists of the N-terminal methyltransferase domain (MTD) and C-terminal vertebrate conserved region (VCR). Recent biochemical and structural studies of METTL16 showed that the MTD itself is capable of recognizing the 5′-UACAGAGAA motif in the specific structural context of RNA (25,26,33). However, the function of the VCR in the methylation of the MAT2A hairpin and U6 snRNA has remained elusive.
In this study, we characterized the structure and functions of the VCR of METTL16 in U6 snRNA methylation. Biochemical and structural analyses of the METTL16 VCR provided mechanistic insights into the efficient modification of A43 in the U6 snRNA by METTL16, using the N-terminal MTD and C-terminal VCR. The VCR interacts with the ISL (internal stem-loop) within the U6 snRNA, and this interaction would induce the conformational rearrangement required for productive catalysis by the MTD. Thereby, the MTD and VCR in METTL16 cooperatively facilitate the m6A43 modification of U6 snRNA.
MATERIALS AND METHODS
Expression and purification of human METTL16 and its variants
The DNA fragment encoding human METTL16 was synthesized by Eurofins Genomics Japan, and cloned into the NdeI and XhoI sites of the pET22b vector (Merck Millipore), yielding pET22_METTL16. The nucleotide sequence of the synthetic METTL16 gene is shown in Table S1. The DNA fragments encoding the N-terminal methyltransferase domain (MTD) and the C-terminal vertebrate conserved region (VCR) of METTL16 were PCR amplified from the pET22_METTL16 plasmid template and cloned into the NdeI and XhoI sites of the pET22b vector. The chimeric gene encoding the N-terminal MTD of METTL16 and the C-terminal KA-1 of TUT1 (34) was generated by overlap PCR methods and cloned into the NdeI and XhoI sites of pET22b. Mutations were introduced by the overlap PCR methods. The oligonucleotide sequences used for the plasmid constructions and mutations are listed in Table S2.
For expression of the proteins, Escherichia coli BL21(DE3) cells (Merck Millipore) were transformed with the plasmids and grown in LB medium containing 50 μg/ml ampicillin at 37°C, until the A600 reached 1.0. The expression of METTL16 and its variant proteins was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 0.1 mM, and the cultures were continued for 16 hours at 18°C. The cells were harvested and lysed in buffer, containing 20 mM Tris–HCl, pH 7.0, 500 mM NaCl, 10 mM β-mercaptoethanol, 20 mM imidazole and 5% (v/v) glycerol. The proteins were first purified using a Ni-NTA agarose column (Qiagen, Japan), and then further purified on a HiTrap Heparin column (GE Healthcare, Japan) as described (35). Finally, the proteins were applied to a HiLoad 16/60 Superdex 200 or 75 column (GE Healthcare, Japan), equilibrated with buffer containing 20 mM Tris–HCl, pH 7.0, 200 mM NaCl and 10 mM β-mercaptoethanol. The purified proteins were concentrated and stored at −80°C until use.
Crystallization and structural determination of VCR
VCR_ΔL (amino acid residues 310–410 and 509–562), VCR lacking the linker between VCR1 and VCR2 (Figure 1A), was crystallized by the sitting drop vapor diffusion method at 20°C. The concentration of VCR_ΔL was adjusted to 5 mg/ml, with 10 mM DTT (dithiothreitol) and 5 mM dextran sulfate sodium salt supplementation. A 200 nl portion of the protein solution was mixed with 200 nl of reservoir solution, containing 100 mM Tris–HCl, pH 7.4, 20% (w/v) PEG3350, 200 mM sodium citrate, and 4% (w/v) formamide. For crystallization of the selenomethionine derivative VCR_ΔL, the protein concentration was adjusted to 5 mg/ml, with 10 mM DTT and 3 mM dextran sulfate sodium salt supplementation. A 300 nl portion of the protein solution was mixed with 200 nl of reservoir solution, containing 100 mM Tris–HCl, pH 7.0, 20% (w/v) PEG3350, 200 mM sodium citrate, and 4% (w/v) formamide. To facilitate the crystallization, the mixture was supplemented with 100 nl of crystal seed solution, prepared with Seed Bead (Hampton Research).
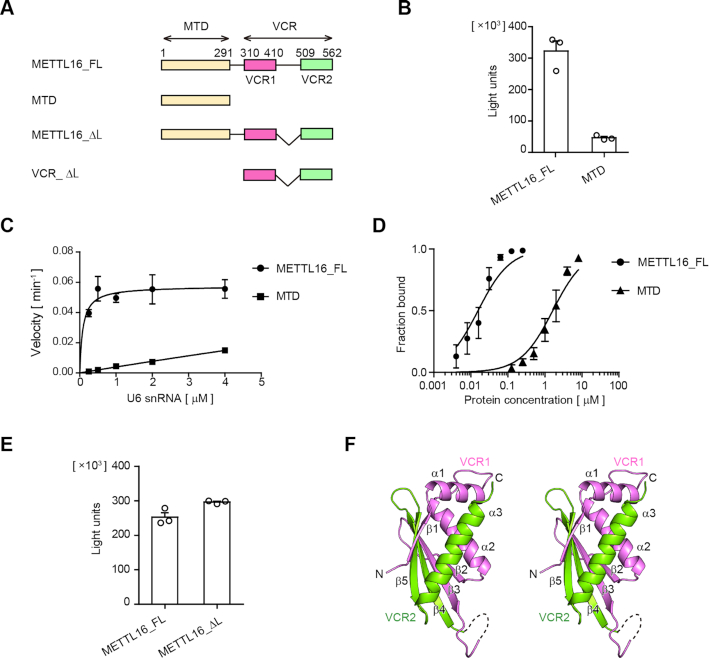
Figure 1.
The VCR of METTL16 enhances U6 snRNA methylation. (A) Schematic diagrams of human METTL16 and its variants. METTL16_FL: full-length METTL16, MTD: methyltransferase domain, and VCR: vertebrate conserved region. VCR consists of VCR1 (pink) and VCR2 (green) connected by a linker. VCR_ΔL is VCR with the deletion of the linker and was used for crystallization and structural determination. (B) Methylation of U6 snRNA by METTL16_FL and MTD under standard conditions (1 μM of U6 snRNA). (C) Steady-state kinetics of m6A modification of U6 snRNA by METTL16_FL and MTD (Supplementary Figure S1A). (D) Gel-shift of U6 snRNA by METTL16_FL (0–0.125 μM) and MTD (0–8 μM) (Supplementary Figure S1B). (E) Methylation of U6 snRNA by METTL16_FL and METTL16_ΔL under standard conditions, as in (B). (F) A stereo view of the structure of VCR_ΔL. Amino acid residues 310–381 and 513–562 are modeled in the structure. VCR1 and VCR2 are colored purple and green, respectively. Bars in the graphs in (B)–(E) are standard deviations (SD) of three independent experiments.
Data sets were collected at beamline 17A at the Photon Factory at KEK, Japan. The crystal was cryoprotected in the reservoir solution supplemented with 25% (v/v) ethylene glycol and flash-cooled in a 100 K nitrogen stream. The data were indexed, integrated, and scaled with XDS (36). The initial phase was determined by the single-wavelength anomalous dispersion (SAD) method, using the selenomethionine derivative crystals. Selenium sites were located by SHELX (37) and initial phase was calculated by Phaser (38), and then the phase was extended to high resolution using native data sets. The model structure was built and refined with phenix.refine (39) and manually modified with Coot (40).
In vitro methylation assay
In vitro methylation of RNA was performed using an MTase-GloTM system (Promega, Japan), according to the manufacturer's instructions with modifications (41). A reaction mixture (12.5 μl volume), containing 50 mM HEPES–KCl, pH 8.0, 120 mM NaCl, 2 mM MgCl2, 1 mM DTT, 1 mM SAM (S-adenosyl-l-methionine, Cayman Chemical, USA), 1 × MTase-Glo® Reagent, 0.4 (or 0.1) μM METTL16 (or its variants or mutants), and various amounts of RNA substrate (0.05–4.0 μM), was incubated at 37°C for 4 min. MTase-Glo® Reagent converts SAH (S-adenosyl-l-homocysteine), the byproduct of methylation, to ADP. Subsequently, the reaction mixture was mixed with 12.5 μl MTase-Glo™ Detection Solution and incubated at room temperature for 30 min to convert ADP to ATP, which was assessed using a luciferase/luciferin reaction. The luminescence was measured with a GloMax-Multi Detection System (Promega, Japan). The luminescence value of the reaction in the absence of RNA substrate was taken as the background and subtracted from the measured luminescence of the reactions in the presence of RNA substrate. Under the reaction conditions, the methylation reaction proceeded in a linear range up to 10 min. Thus, for the steady state kinetic analyses, the methylation reactions were stopped at 4 min to calculate the initial velocities of the reactions. The SAH standard curve was prepared using serial concentrations of SAH (Sigma Aldrich, Japan) to assess the linearity of the reactions, and the amounts of SAH produced from methylation reactions were calculated. One picomole of SAH corresponded to 190 000 LU (light units).
RNA substrates (human U6 snRNA and its variants and MAT2A hairpin RNA) were synthesized by T7 RNA polymerase in vitro, using plasmids encoding the respective DNA sequences downstream of the T7 promoter as templates. The synthesized RNAs were purified by 10% (w/v) polyacrylamide gel electrophoresis under denaturing conditions, eluted from the gel and precipitated by ethanol. The nucleotide sequences of the RNAs used in this study are listed in Table S2.
Gel-shift assay
For the gel-shift assay, increasing amounts of recombinant METTL16 or its variants were incubated with 1 nM of 5′-32P-labeled RNA, in buffer containing 50 mM Tris–HCl, pH 8.0, 120 mM NaCl, 10 mM MgCl2 and 10% (v/v) glycerol, for 15 min at room temperature. The samples were separated by 6% (w/v) polyacrylamide electrophoresis under native conditions in TBE buffer. The intensities of the 32P-labeled RNAs were quantified by a BAS-5000 or FLA-3000 imager (Fuji Film, Japan). The affinities between METTL16 (or its variants) and RNAs were quantified using all the shifted bonds in the gel-shift assays.
RESULTS
The C-terminal VCR of METTL16 enhances U6 snRNA methylation
To evaluate the effect of the C-terminal VCR of human METTL16 on the m6A43 modification of U6 snRNA, the recombinant full-length human METTL16 (METTL16_FL) and its N-terminal methyltransferase domain (MTD: residues 1–291) proteins were expressed in Escherichia coli. The enzymatic activities of the purified proteins were analyzed using the U6 snRNA transcript as the substrate (Figure 1A).
Under the standard conditions (1 μM of U6 snRNA), METTL16_FL methylated U6 snRNA more efficiently than the MTD (Figure 1B). Detailed steady state kinetics showed that METTL16_FL methylates U6 snRNA more efficiently than the MTD by over two to three orders of magnitude (Figure 1C). The Km value of U6 snRNA toward METTL16_FL was estimated to be 0.025 ± 0.01 μM (Supplementary Figure S1A), while the Km value of U6 snRNA toward MTD was much greater than 4 μM (>>4 μM), suggesting that the affinity between METTL16_FL and U6 snRNA is higher than that between MTD and U6 snRNA, by more than two orders of magnitude (Figure 1C). To further compare the affinities between METTL16_FL and U6 snRNA and the MTD and U6 snRNA, gel-shift assays were performed. The results showed that the interaction between METTL16_FL and U6 snRNA is much stronger than that between the MTD and U6 snRNA, and the Kd values of U6 snRNA toward METTL16_FL and the MTD were estimated to be 0.016 ± 0.002 and 1.6 ± 0.2 μM, respectively (Figure 1D, Supplementary Figure S1B).
These results indicate that the C-terminal VCR of METTL16 enhances the methylation of U6 snRNA by increasing the affinity of METTL16 for U6 snRNA.
Structure determination of the human METTL16 VCR
To address the structural basis of the enhancement of U6 snRNA methylation by the C-terminal VCR of METTL16, the VCR (amino acid residues 310–562) and its variant, VCR_ΔL (amino acid residues 310–410 and 509–562), lacking the putative disordered linker region between VCR1 and VCR2 (amino acid residues: 411–508) in VCR (Figure 1A, Supplementary Figure S3) were expressed in Escherichia coli, purified, and subjected to crystallization screenings. Note that METTL16_ΔL (Figure 1A) can methylate U6 snRNA as efficiently as METTL16_FL (Figure 1E) under the standard conditions, as shown in Figure 1B.
Although our attempt to obtain the crystal of the VCR was unsuccessful, we crystallized VCR_ΔL and solved its structure. The initial phase was determined by the single-wavelength anomalous dispersion (SAD) method, using the selenomethionine derivative crystal of VCR_ΔL (Supplementary Figure S2). The crystal belongs to the space group R32 and contains one VCR_ΔL molecule in the asymmetric unit. The structure was modeled and refined to an R factor of 23.4% (Rfree of 26.3%), using reflection data up to 2.8 Å resolution. Data collection and refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| SeMet-METTL16_VCRΔL | Native-METTL16_VCRΔL | |
|---|---|---|
| Data collection | ||
| Space group | R32 | R32 |
| Cell dimensions | ||
| a, b, c (Å) | 139.59 | 139.61 |
| 139.59 | 139.61 | |
| 53.31 | 53.13 | |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Wavelength (Å) | 0.97800 | 0.98000 |
| Resolution (Å)* | 50–3.0 (3.11–3.00) | 50–2.8 (2.89–2.79) |
| R sym* | 0.332 (2.973) | 0.305 (4.265) |
| I/σI * | 14.8 (1.9) | 14.9 (1.1) |
| CC1/2* | 0.998 (0.684) | 0.999 (0.523) |
| Completeness (%)* | 100.0 (100.0) | 99.8 (98.4) |
| Redundancy* | 40.6 (42.2) | 40.9 (41.6) |
| Phasing | ||
| Se sites | 3 | |
| Mean FOM | 0.378 | |
| Refinement | ||
| Resolution (Å) | 50-2.8 | |
| No. reflections | 5013 | |
| R work/Rfree (%) | 23.36/26.31 | |
| No. atoms | ||
| Protein | 122 | |
| B-factors (Å2) | ||
| Protein | 85.4 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.012 | |
| Bond angles (°) | 1.44 | |
| Ramachandran | ||
| Preferred (%) | 92.4 | |
| Allowed (%) | 7.6 | |
| Outliers (%) | 0 |
*Values in parentheses are for the highest-resolution shell.
The VCR_ΔL consists of a five-stranded anti-parallel β-sheet (β1–β5) and three α-helices (α1–α3) (Figure 1F). The flexible linker, removed for crystallization, resides between the β3- and β4-strands, and thus VCR1 (β1, α1, α2, β2 and β3, pink) and VCR2 (β4, β5 and α3, green) together compose a single domain, and five β-strands compose the anti-parallel β-sheet. In the present structure, several N- and C- terminal residues and residues 382–410 could not be model-built due to the absence of the corresponding clear electron densities.
VCR is structurally and functionally equivalent to KA-1 of TUT1
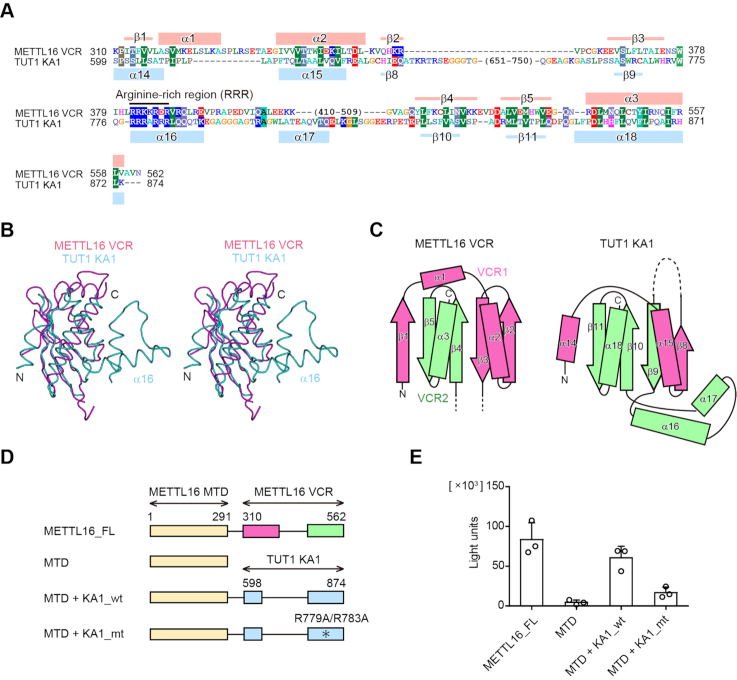
The primary amino acid sequence of the VCR of human METTL16 is not homologous to other proteins with known function, except for the C-terminal region of other putative U6 snRNA-specific m6A methyltransferase. However, the structure-based homologous search using the Dali server (42) revealed that the VCR_ ΔL structure is topologically homologous to the kinase-associated 1 (KA1) domain structure of various proteins (43), including that of the U6 snRNA-specific terminal uridylyltransferase 1 (TUT1) (34,44–46) [Z-score of 8.2] (Figure 2A–C). The KA1 domain resides at the C-terminal half of TUT1. The C-terminal KA1 domain of TUT1 reportedly functions as an RNA-binding domain to facilitate the oligo-uridylylation of the 3′-end of U6 snRNA (34,47), which is required for the maturation and recycling of U6 snRNA (48–50). The structures of VCR of METTL16 and KA1 of TUT1 superimposed well on each other, with an RMSD of 2.4 Å for 69 structurally equivalent Cα atoms (Figure 2B, C).
Figure 2.
The VCR is structurally and functionally equivalent to KA1 of TUT1. (A) Sequence alignments of the VCR of human METTL16 and the KA1 domain of human TUT1. The secondary structural elements of the VCR and KA1 domain are indicated above and below the alignments, respectively. (B) A stereo view of the superimposed structures of the VCR of METTL16 (purple) and the KA1 domain of TUT1 (cyan). (C) Topology diagrams of the VCR of METTL16 (left) and the KA1 domain of TUT1 (right). The N- and C-terminal halves are colored pink and green, respectively. (D) Schematic diagrams of METTL16 and its variants used for assays in (E). METTL16_FL: full-length METTL16, MTD: methyltransferase domain, MTD +KA1_wt: a chimeric protein of the N-terminal METTL16 MTD and the C-terminal TUT1 KA1, MTD+KA1_mt: mutant protein of MTD+KA1_wt. The asterisk indicates the R779A/R783A mutation. VCR1 and VCR2 of METTL16 are colored pink and green, respectively. (E) Methylation of U6 snRNA by METTL16_FL and its variants shown in (D), under the conditions in which 50 nM of U6 snRNA was used for the assays. Bars in the graphs are standard deviations (SD) of three independent experiments.
We examined the compatibility of the functions of the KA1 domain of TUT1 and the VCR of METTL16. A chimeric enzyme consisting of the N-terminal MTD of METTL16 and the C-terminal KA1 of TUT1 (MTD+KA1_wt) (Figure 2D) was expressed in E. coli and purified, and the enzymatic activity toward the U6 snRNA transcript was analyzed. As expected, the chimeric protein, MTD+KA1_wt, could methylate the U6 snRNA transcript more efficiently than the MTD (Figure 2E). Furthermore, a chimeric protein with the R779A/R783A mutations in KA1 (MTD+KA1_mt) was prepared and the enzymatic activity was analyzed. The R779A/R783A mutation in TUT1 reduced the oligo-uridylylation activity of TUT1 toward the U6 snRNA, due to the decreased RNA binding activity of KA1 (34). Consistent with this, MTD+KA1_mt could not enhance the methylation to the same extent as MTD+KA1_wt (Figure 2E). These results imply that the VCR of METTL16 and the KA1 of TUT1 have similar functions in the specific recognition of the U6 snRNA, as described below, and they are functionally compatible.
RNA binding activity of VCR facilitates U6 snRNA methylation by METTL16
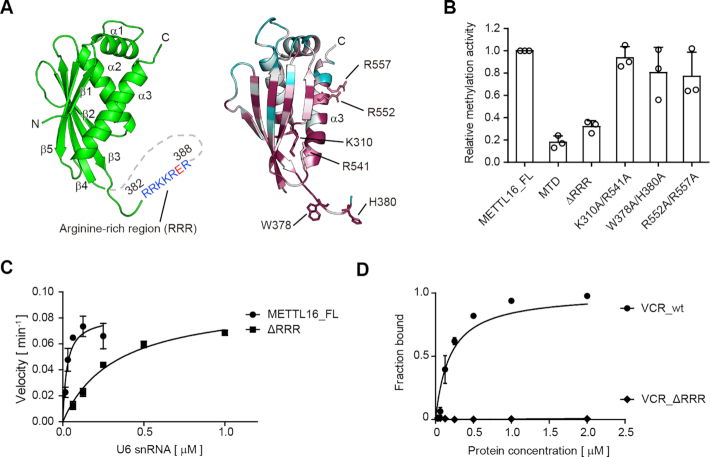
The C-terminal KA1 domain of TUT1 facilitates the oligo-uridylylation of U6 snRNA, and KA1 itself has RNA-binding activity (34). An arginine rich α-helix (α16) in KA1 is important for the RNA binding. The amino acid sequence alignment indicated that the VCR also possesses the arginine-rich region (amino acid residues 382–388), termed the ‘RRR’ hereafter, at the corresponding position in KA1 of TUT1 (Figure 2A). Although the RRR between β3 and β4 of VCR is disordered in the present structure (Figures 1F, 2B, 3A), the RNA-binding ability of the VCR of METTL16 is likely to be similar to that of the KA1 domain of TUT1, to facilitate U6 snRNA methylation.
Figure 3.
RNA binding activity of VCR facilitates U6 snRNA methylation. (A) The arginine-rich region (RRR: amino acid residues 382–388) resides between β3 and β4 (left). Conservation analysis of the VCR of METTL16 (right). Conserved and non-conserved residues are colored purple and cyan, respectively (Supplementary Figure S3). (B) Methylation of U6 snRNA by METTL16 variants. Relative activities of METTL16 variants. The activity of METTL_FL under the conditions described in Figure 2E was taken as 1.0. ΔRRR: METTL16 lacking RRR in the VCR. (C) Steady-state kinetics of m6A modification of U6 snRNA by METTL16_FL and ΔRRR. (D) Gel shift of U6 snRNA by wild-type VCR (VCR_wt) and its variant lacking RRR, VCR_ΔRRR (0–2.0 μM) (Supplementary Figure S1C). Bars in the graphs in (B)–(D) are standard deviations (SD) of three independent experiments.
To evaluate the relationship between the RNA binding property of VCR and the methylation activity of METTL16 toward U6 snRNA, the conserved residues in the VCR of METTL16 were mutated or deleted (Figure 3A, Supplementary Figure S3) and the enzymatic activities of the mutants were analyzed. The mutations, K310A/R541A, W378A/H380A or R552A/R557A, in the VCR did not significantly reduce the methylation activity towards U6 snRNA (Figure 3B). In contrast, the RRR deletion (amino acid residues 382–388: METTL16_ΔRRR) reduced the methylation activity of METTL16 toward U6 snRNA to the extent of the methylation by the MTD (Figure 3B). Detailed steady state kinetics analyses of the methylation of U6 snRNA by METTL16_ΔRRR showed that the decreased activity of METTL16_ΔRRR resulted from the increased Km value toward U6 snRNA. The Km value toward U6 snRNA was estimated to be 0.32 ± 0.05 μM, which is elevated by about twelve-fold by the RRR deletion in the VCR (Figure 3C). Furthermore, to confirm the reduced RNA binding activity by the RRR deletion, gel-shift assays were performed using wild-type VCR and a mutant VCR with the RRR deletion (VCR_ΔRRR). As expected, VCR_ΔRRR reduced the RNA binding activity (Figure 3D, Supplementary Figure S1C). Altogether, these results suggest that the RNA binding activity of VCR facilitates U6 snRNA methylation by METTL16, using the conserved RRR in the VCR.
Secondary structure of U6 snRNA for methylation by METTL16
METTL16 was reportedly to methylate RNAs containing 5′-UACAGAGAA-3′ sequence (methylated adenosine is underlined), such as U6 snRNA and MAT2A 3′ UTR hairpin RNAs (22–24). Recently, the crystal structures of the MTD of METTL16 and its complex with the MAT2A short hairpin 1 RNA were reported (25,26,33). The complex structure with RNA revealed the molecular basis for the specific recognition of the structured hairpin RNA by the MTD and identified the RNA conformation necessary for productive catalysis by the MTD.
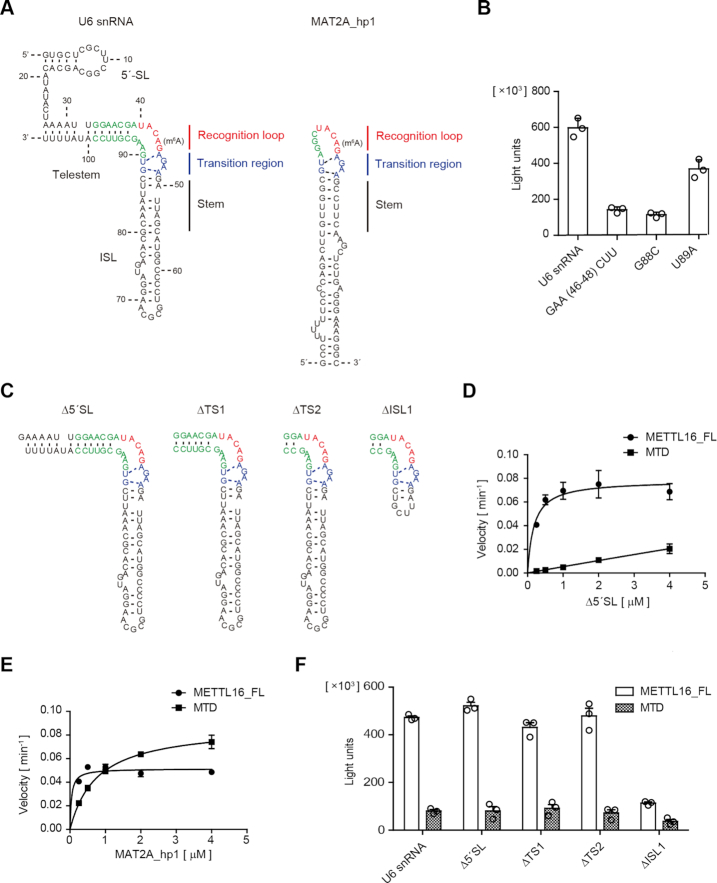
The MAT2A 3′ UTR hairpin RNA structure in the complex with MTD consists of the recognition loop containing the adenosine to be methylated, transition region, and stem (Figure 4A, right). The 5′-UACAG-3′ (methylated adenosine is underlined) in the recognition loop is specifically recognized by the MTD. The transition region also interacts with the MTD, and the AGAA and GU motifs form three unusual base pairs, while the stem does not interact with the MTD (33). By referring to the structure of the MAT2A hairpin complexed with the MTD, the secondary structure of U6 snRNA necessary for methylation by the MTD was modeled (Figure 4A, left).
Figure 4.
Secondary structure of U6 snRNA for productive catalysis. (A) The secondary structural model of U6 snRNA for methylation by METTL16 (left) is similar to the structure of the MAT2A hairpin (MAT2A_hp1) in the complex with the MTD (right). The MAT2A_hp1 consists of the recognition loop, transition region, and stem required for productive catalysis after complex formation with the MTD. (B) The methylation of U6 snRNA variants with a mutation in the putative transition region, under the standard conditions (1 μM of RNAs). (C) Secondary structures of the U6 snRNA variants used for the assays in (D) and (F). (D) Steady-state kinetics of m6A modification of U6 snRNA_Δ5′SL by METTL16_FL and MTD. (E) Steady-state kinetics of m6A modification of MAT2A_hp1 by METTL16_FL and MTD (Supplementary Figure S1D). (F) The methylation of U6 snRNA variants by METTL16_FL and MTD under the standard conditions (1 μM of RNAs). Bars in the graphs in (B) and (D)–(F) are standard deviations (SD) of three independent experiments.
Unlike the conventional straight telestem-bulge-ISL structure of U6 snRNA (51), the modeled secondary structure has a bent telestem-bulge-ISL structure, with a bend in the joint between the telestem and ISL (Figure 4A, Supplementary Figure S4). In the model, the U6 snRNA also forms the same transition region as the MAT2A 3′ UTR hairpin (Figure 4A, blue), and the region (nucleotides at positions 33–39) upstream of the 5′-UACA43G-3′ (Figure 4A, red) constitutes the telestem (Figure 4A, left, green), and the 5′-UACA43G-3′ is bulged-out at the junction between the telestem and ISL. As a result, the secondary structure of the region around the methylation site of U6 snRNA forms a quasi-loop similar to the MAT2A 3′ UTR hairpin recognition loop, and the telestem is extended from the recognition loop. Thus, the bent-form of the telestem-bulge-ISL with the transition region would be the productive conformation of the U6 snRNA necessary for methylation by METTL16.
To evaluate the secondary structural model, we examined the methylation of U6 snRNA mutants by METTL16_FL. The mutations in the putative transition region, GAA(46–48)CUU, G88C or U89A, of the U6 snRNA all reduced the methylation efficiency by METTL16 under the standard conditions (1 μM of RNA) (Figure 4B), as the corresponding mutations in the MAT2A 3′ UTR reduced the methylation efficiencies by METTL16 (23,25,33). Thus, the U6 snRNA would adopt the modeled secondary structure shown in Figure 4A after the complex formation, and the bent form of U6 snRNA with the transition region would be a productive conformation for methylation by METTL16.
VCR interacts with ISL within U6 snRNA
To identify the VCR binding region within the U6 snRNA, we first deleted the 5′ stem-loop (5′SL) (U6 snRNA_Δ5′SL) and analyzed the methylation by METTL16_FL and MTD (Figure 4C). Detailed steady state kinetics revealed that the VCR enhances the methylation of U6 snRNA_Δ5′SL by a similar order of magnitude as the full-length U6 snRNA (Figures 1C, 4D). Thus, the VCR would not interact with the 5′SL of U6 snRNA.
METTL16 methylates the MAT2A 3′ UTR hairpin as well as U6 snRNA. We next analyzed the methylation efficiencies of the MAT2A 3′ UTR hairpin 1 (MAT2A_hp1) by METTL16_FL and MTD (Figures 1A, 4E). The methylation efficiencies of MAT2A_hp1 by METTL16_FL and MTD were almost the same under the standard conditions (1 μM of MAT2A_hp1 RNA). However, detailed steady state kinetics analyses of the methylation of MAT2A_hp1 yielded estimated Km values for MAT2A_hp1 toward METTL16_FL and MTD of 0.027 ± 0.05 μM and 0.76 ± 0.1 μM, respectively (Figure 4E, Supplementary Figure S1D). Thus, VCR also facilitates the methylation of MAT2A_hp1 by increasing the affinity between METTL16 and the RNA substrate, although the impact of the VCR on MAT2A_hp1 methylation is smaller than that on the U6 snRNA methylation by one order of magnitude (Figures 1C, 4E, Supplementary Figure S1E), as discussed below.
Since the VCR of METTL16 could facilitate the methylation of both U6 snRNA_Δ5′ SL and MAT2A_hp1 through increasing the affinities between METTL16 and the RNA substrates, the VCR would interact with the stem region of ISL, rather than the telestem of U6 snRNA (Figure 4A). Consistent with this notion, U6 snRNA_Δ5' SL variants with shorter telestems (U6 snRNA_ΔTS1 and ΔTS2, Figure 4C) could be methylated by METTL16_FL more efficiently than by the MTD (Figure 4F) under the standard conditions. However, further shortening of the ISL of U6 snRNA_ΔTS2 reduced the methylation efficiency by METTL16_FL to the extent of the MTD alone (U6 snRNA_ΔISL1, Figure 4C, F). Therefore, the VCR is likely to interact with the stem region of ISL, rather than the telestem of U6 snRNA.
Relaxed telestem structure increases the methylation of U6 snRNA by the MTD
As described above, the impact of VCR on the methylation of U6 snRNA is larger than that on the methylation of MAT2A_hp1, by one order of magnitude (Figures 1C, 4E, Figures S1A, D). This implies that the difference of the VCR impacts on the methylation between the two RNAs would arise from the structural and/or conformational differences in the vicinity of the recognition sites of the RNAs. At the reaction stage, U6 snRNA should adopt a bent conformation at the junction between the telestem and ISL and form a productive conformation for efficient methylation by the MTD (Figure 4A). Thus, the flexibility at the junction between the telestem and ISL of U6 snRNA could affect the methylation of A43 by the MTD.
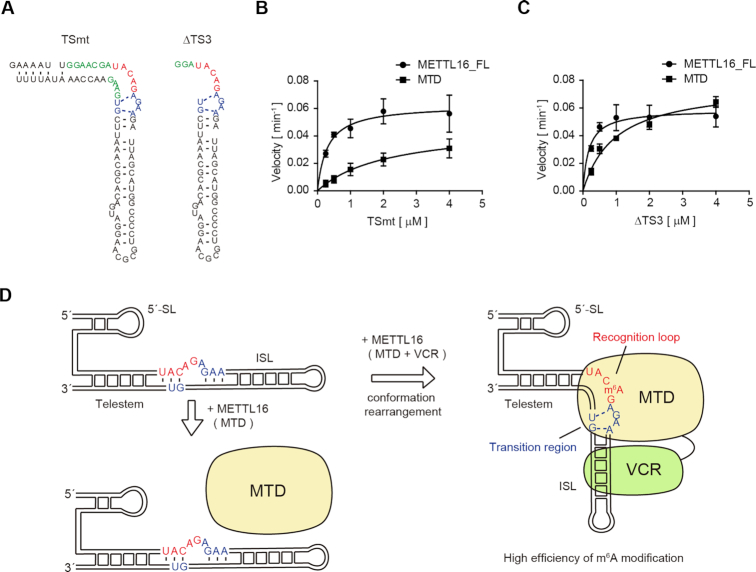
Consistent with this notion, the mutant U6 snRNA with mis-pairs in the telestem region (U6 snRNA_TSmt, Figure 5A, left), in which the junction between the telestem and ISL would be more flexible than in the wild-type U6 snRNA, was more efficiently methylated by the MTD than U6 snRNA_Δ5′ SL (Figure 5B). The steady state kinetics showed that the estimated Km value of U6 snRNA_TSmt toward the MTD is 2.1 ± 1.4 μM and smaller than the Km value of U6 snRNA_Δ5′ SL (larger than 4 μM, >>4 μM) (Figure 5B). Furthermore, a mutant U6 snRNA with the telestem deletion (U6 snRNA_ΔTS3, Figure 5A, right) could be methylated as efficiently by the MTD as MAT2A_hp1 (Figure 5C). The Km value of U6 snRNA_ΔTS3 was estimated to be 0.98 ± 0.19 μM, which is comparable to that of MAT2A_hp1 toward the MTD (0.76 ± 0.10 μM) (Figures 4E, 5C, Supplementary Figure S1D).
Figure 5.
A model of cooperative U6 snRNA methylation by MTD and VCR. (A) Secondary structures of U6 snRNA variants used for assays in (B) and (C). (B) Steady-state kinetics of m6A modification of TSmt by METTL16_FL and MTD. (C) Steady-state kinetics of m6A modification of ΔTS3 by METTL16_FL and MTD. Bars in the graphs in (B) and (C) are standard deviations (SD) of three independent experiments. (D) A model of the cooperative methylation of U6 snRNA by the MTD and VCR of METTL16. VCR binding to ISL would promote the transition of the structural configurations of telestem-bulge-ISL in U6 snRNA to the bent form and enable the transition structure to be formed in the junction between the telestem and ISL for productive catalysis by the MTD.
These results suggest that, by relaxing the telestem structure or destabilizing the bulge junction between the telestem and ISL, the MTD can methylate A43 of U6 snRNA efficiently. Thus, the different impacts of the VCR on the methylation of U6 snRNA and MAT2A_hp1 could arise from the structural differences around the methylation sites between these RNAs.
DISCUSSION
In this study, we analyzed the structure and functions of the C-terminal VCR of human METTL16 in the m6A43 modification of U6 snRNA. The m6A43 lies in the 5′-ACAGAG-3′ box (modified A is underlined) of the U6 snRNA, which base pairs with the 5′ splice site of pre-mRNA during splicing (27–29), and the modification is presumably involved in the regulation of pre-mRNA splicing (24,30–32).
Our biochemical studies revealed that the C-terminal VCR of METTL16 facilitates the methylation of U6 snRNA in the N-terminal methyltransferase domain (MTD) more efficiently than the methylation by MTD alone, by two to three orders of magnitude (Figure 1C). The structure of VCR is topologically homologous to that of the recently identified C-terminal KA1 domain in the U6 snRNA-specific terminal uridylyltransferase, TUT1 (34) (Figure 2B, C), and the VCR of METTL16 could be functionally replaced with the KA1 of TUT1 (Figure 2E). Furthermore, the conserved arginine-rich regions between KA1 and VCR are involved in the U6 snRNA–VCR interactions (Figure 3C, D). Thus, KA1 of TUT1 and VCR of METTL16 share common functions and are attached to the catalytic domains of TUT1 and METTL16, respectively, to promote the different steps of U6 snRNA biogenesis.
The previous protection analysis of U6 snRNA by TUT1 and its variant lacking KA1 showed that KA1 would interact with the ISL in U6 snRNA (34). It was also shown that, upon the binding of KA1 of TUT1 to U6 snRNA, a structural and/or conformational alteration in the vicinity of the bulge between the telestem and ISL would be induced. KA1 promotes the oligo-uridylylation of U6 snRNA by TUT1, by preventing the U6 snRNA from being dislodged from the enzyme surface during the reaction. Considering the structural and functional similarities between KA1 of TUT1 and VCR of METTL16, they would interact with the same region within the U6 snRNA. Consistent with this notion, a U6 snRNA variant with a shorter ISL could no longer be methylated efficiently by the full-length METTL16 (Figure 4C, F). Thus, the VCR interacts with the ISL of the U6 snRNA in a similar manner to the KA1 of TUT1. In addition, upon VCR binding to the ISL of the U6 snRNA, the structure and/or conformation change of the region in the vicinity of the bulge between the telestem and ISL of U6 snRNA would be probably be induced, as in the interaction with KA1.
The recent crystallographic study of MTD complexed with a short MAT2A hairpin identified the intrinsic structural features of RNA that affect the methylation efficiencies by the MTD of METTL16 (26) (Figure 4A). The MTD itself is capable of recognizing the specific sequence in the context of the distinct structural features. For efficient methylation, the U6 snRNA should adopt the bent structure at the junction between the telestem and ISL, rather than the conventional straight telestem-bulge-ISL structure (Figure 4A, Supplementary Figure S4), and the region in the vicinity of the methylation site of U6 snRNA should adopt similar configurations to those of the MAT2A hairpin for productive catalysis (Figure 4A). The bent telestem-bulge-ISL structure is observed in the structure of U6 snRNA - Prp24 complex (52). Thus, a bent telestem-bulge-ISL could be introduced into U6 snRNA by METTL16 with the help from VCR. The present results suggest that the VCR would shift the structure of the U6 snRNA to that required for productive catalysis by the MTD (Figure 5D).
The MTD could methylate U6 snRNA variants with mismatches in the telestem more efficiently than the wild-type U6 snRNA (Figure 5A, B). Furthermore, the MTD methylates the U6 snRNA variant without a telestem as efficiently as the full-length METTL16 (Figure 5A, C). These results imply that the relaxation of the telestem-region would allow the structural configuration of the region containing the methylation site to resemble that observed in the MAT2A hairpin structure. The interaction of the VCR with the ISL would have the same effects on the structural configuration changes. VCR binding to the ISL and MTD binding to the bulge between the telestem and ISL are likely to compress the RNA structure (Figure 5D), and the telestem-region might become relaxed and/or warped. This would facilitate the rearrangement of the structural configurations of the U6 snRNA, making them similar to those observed in the MAT2A hairpin for productive catalysis (Figures 4A, 5D). In the gel-shift of U6 snRNA by METTL16, the multiple shifted bands were observed (Supplementary Figure S1B). These shifted bands probably correspond to the sequential and cooperative binding of VCR and MTD to ISL and the bulge between telestem and ISL, respectively, and reflect the rearrangement of the structural configurations of U6 snRNA, as the METTL16 concentration is increased. In these ways, the MTD and VCR of METTL16 cooperatively methylate A43 of the U6 snRNA.
The METTL16 binding and methylation of the MAT2A 3′ UTR hairpin regulate the MAT2A mRNA splicing and stability, in response to changes in the intracellular SAM levels (22,23). The recent crystal structure of the MTD complex with a short MAT2A hairpin showed that the MTD manifests the full activity without the VCR (25,26), and the VCR itself recruits splicing factors to mRNA to promote splicing (23). The present detailed biochemical analysis of MAT2A hairpin methylation by the full-length METTL16 and MTD showed that the VCR increases the affinity of METTL16 toward the MAT2A hairpin and facilitates the methylation (Figures 4E, Supplementary Figure S1D). Thus, the VCR binding to the stem would stabilize the transition region of the MAT2A hairpin required for productive catalysis, and the VCR complements the methylation of the MAT2A 3′ UTR by the MTD to maintain intracellular SAM homeostasis.
The VCR would interact with double-stranded RNA, but it is presently not clear whether the interactions are sequence-specific. The addition of the VCR protein in trans does not enhance the methylation of U6 snRNA by the MTD alone (Supplementary Figure S1F). Thus, the VCR binding site in the U6 snRNA would be partly governed by the MTD in METTL16. METTL16 also reportedly interacts with various cellular non-coding RNAs (24), including the 3′-terminal RNA triple helix of MALAT1 (metastasis-associated lung adenocarcinoma transcript-1) (53,54). Full-length METTL16 binds the MALAT1 RNA triple helix, while the MTD (amino acid residues 1 – 291) does not (33). This implies that the C-terminal VCR of METTL16 interacts with the MALAT1 RNA triple helix. At present, the detailed recognition mechanism of the MALAT1 RNA triple helix by METTL16 remains poorly understood. The detailed mechanisms of the RNA recognition by VCR, the RNA–VCR interactions, and the cooperative methylation of specific RNAs by the MTD and VCR of METTL16 await further structural determinations.
DATA AVAILABILITY
Coordinates and structure factors for the crystal structure of VCR of human METTL16 have been deposited in the Protein Data Bank, under the accession code 6M1U.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the beamline staff of BL-17A (KEK, Tsukuba) for technical assistance during data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Scientific Research (A) [18H03980 to K.T.]; Grant-in-Aid for JSPS Fellows [18J23142 to T.A.] from JSPS; Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [26113002 to K.T.]; Takeda Science Foundation, Uehara Memorial Foundation, Terumo Foundation for Life Science and Art, Takahashi Industrial and Economic Research Foundation and Princess Takamatsu Cancer Research Fund. Funding for open access charge: Grant-in-Aid for Scientific Research (A) from JSPS [to K.T.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 2013; 38:204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dominissini D. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 3. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yue Y., Liu J., He C.. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015; 29:1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fustin J.-M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., Isagawa T., Morioka Masaki S., Kakeya H., Manabe I. et al.. RNA-Methylation-Dependent RNA processing controls the speed of the circadian clock. Cell. 2013; 155:793–806. [DOI] [PubMed] [Google Scholar]
- 6. Batista Pedro J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley Donna M., Lujan E., Haddad B., Daneshvar K. et al.. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014; 15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Batista P.J. The RNA Modification N6-methyladenosine and Its Implications in Human Disease. Genomics Proteomics Bioinformatics. 2017; 15:154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geula S. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science (New York, N.Y.). 2015; 347:1002–1006. [DOI] [PubMed] [Google Scholar]
- 9. Meyer K.D., Jaffrey S.R.. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017; 33:319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang X. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiao W., Adhikari S., Dahal U., Chen Y.-S., Hao Y.-J., Sun B.-F., Sun H.-Y., Li A., Ping X.-L., Lai W.-Y. et al.. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- 13. Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu P.J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., Qi M., Lu Z., Shi H., Wang J. et al.. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017; 27:1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meyer K.D. 5′ UTR m6A promotes cap-independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ke S., Alemu E.A., Mertens C., Gantman E.C., Fak J.J., Mele A., Haripal B., Zucker-Scharff I., Moore M.J., Park C.Y. et al.. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015; 29:2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang P., Doxtader Katelyn A., Nam Y.. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016; 63:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C. et al.. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature. 2016; 534:575–578. [DOI] [PubMed] [Google Scholar]
- 21. Sledz P., Jinek M.. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife. 2016; 5:e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shima H., Matsumoto M., Ishigami Y., Ebina M., Muto A., Sato Y., Kumagai S., Ochiai K., Suzuki T., Igarashi K.. S-Adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 2017; 21:3354–3363. [DOI] [PubMed] [Google Scholar]
- 23. Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K.. The U6 snRNA m(6)A methyltransferase METTL16 Regulates SAM synthetase intron retention. Cell. 2017; 169:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Hobartner C., Sloan K.E., Bohnsack M.T.. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017; 18:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mendel M., Chen K.M., Homolka D., Gos P., Pandey R.R., McCarthy A.A., Pillai R.S.. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell. 2018; 71:986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doxtader K.A., Wang P., Scarborough A.M., Seo D., Conrad N.K., Nam Y.. Structural basis for regulation of METTL16, an S-Adenosylmethionine homeostasis factor. Mol. Cell. 2018; 71:1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sawa H., Abelson J.. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5′ splice site during the splicing reaction in yeast. Proc. Natl. Acad. Sci. 1992; 89:11269–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sawa H., Shimura Y.. Association of U6 snRNA with the 5′-splice site region of pre-mRNA in the spliceosome. Genes Dev. 1992; 6:244–254. [DOI] [PubMed] [Google Scholar]
- 29. Wassarman D., Steitz J.. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science (New York, N.Y.). 1992; 257:1918–1925. [DOI] [PubMed] [Google Scholar]
- 30. Gu J., Patton J.R., Shimba S., Reddy R.. Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA. 1996; 2:909–918. [PMC free article] [PubMed] [Google Scholar]
- 31. Brow D.A., Guthrie C.. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988; 334:213–218. [DOI] [PubMed] [Google Scholar]
- 32. Madhani H.D., Bordonne R., Guthrie C.. Multiple roles for U6 snRNA in the splicing pathway. Genes Dev. 1990; 4:2264–2277. [DOI] [PubMed] [Google Scholar]
- 33. Ruszkowska A., Ruszkowski M., Dauter Z., Brown J.A.. Structural insights into the RNA methyltransferase domain of METTL16. Sci. Rep. 2018; 8:5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamashita S., Takagi Y., Nagaike T., Tomita K.. Crystal structures of U6 snRNA-specific terminal uridylyltransferase. Nat. Commun. 2017; 8:15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martinez A., Yamashita S., Nagaike T., Sakaguchi Y., Suzuki T., Tomita K.. Human BCDIN3D monomethylates cytoplasmic histidine transfer RNA. Nucleic Acids Res. 2017; 45:5423–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kabsch W. XDS. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sheldrick G.M. A short history of SHELX. Acta Crystallogr. A, Found. Crystallogr. 2008; 64:112–122. [DOI] [PubMed] [Google Scholar]
- 38. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D, Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsiao K., Zegzouti H., Goueli S.A.. Methyltransferase-Glo: a universal, bioluminescent and homogenous assay for monitoring all classes of methyltransferases. Epigenomics. 2016; 8:321–339. [DOI] [PubMed] [Google Scholar]
- 42. Holm L., Rosenström P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moravcevic K., Mendrola J.M., Schmitz K.R., Wang Y.H., Slochower D., Janmey P.A., Lemmon M.A.. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010; 143:966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trippe R., Guschina E., Hossbach M., Urlaub H., Luhrmann R., Benecke B.J.. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006; 12:1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trippe R., Richly H., Benecke B.J.. Biochemical characterization of a U6 small nuclear RNA-specific terminal uridylyltransferase. Eur. J. Biochem. 2003; 270:971–980. [DOI] [PubMed] [Google Scholar]
- 46. Trippe R., Sandrock B., Benecke B.J.. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998; 26:3119–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yashiro Y., Tomita K.. Function and regulation of human terminal uridylyltransferases. Front. Genet. 2018; 9:538–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Achsel T., Brahms H., Kastner B., Bachi A., Wilm M., Lührmann R.. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999; 18:5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vidal V.P., Verdone L., Mayes A.E., Beggs J.D.. Characterization of U6 snRNA-protein interactions. RNA. 1999; 5:1470–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bell M., Schreiner S., Damianov A., Reddy R., Bindereif A.. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002; 21:2724–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mougin A., Gottschalk A., Fabrizio P., Luhrmann R., Branlant C.. Direct probing of RNA structure and RNA-protein interactions in purified HeLa cell's and yeast spliceosomal U4/U6.U5 tri-snRNP particles. J. Mol. Biol. 2002; 317:631–649. [DOI] [PubMed] [Google Scholar]
- 52. Montemayor E.J., Curran E.C., Liao H.H., Andrews K.L., Treba C.N., Butcher S.E., Brow D.A.. Core structure of the U6 small nuclear ribonucleoprotein at 1.7-A resolution. Nat. Struct. Mol. Biol. 2014; 21:544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ji P., Diederichs S., Wang W., Böing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E. et al.. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003; 22:8031–8041. [DOI] [PubMed] [Google Scholar]
- 54. Brown J.A., Kinzig C.G., DeGregorio S.J., Steitz J.A.. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. PNAS. 2016; 113:14013–14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the crystal structure of VCR of human METTL16 have been deposited in the Protein Data Bank, under the accession code 6M1U.