Abstract
High-mobility group AT-hook 2 (HMGA2) is an architectural transcription factor that plays essential roles in embryonic development and cancer progression. However, the mechanism of HMGA2 regulation remains largely uncharacterized. Here, we demonstrate that HMGA2 can be modulated by hepatitis B X-interacting protein (HBXIP), an oncogenic transcriptional coactivator, in esophageal squamous cell carcinoma (ESCC). HMGA2 expression was positively associated with HBXIP expression in clinical ESCC tissues, and their high levels were associated with advanced tumor stage and reduced overall and disease-free survival. We found that oncogenic HBXIP could posttranslationally upregulate HMGA2 protein level in ESCC cells. HBXIP induced HMGA2 acetylation at the lysine 26 (K26), resulting in HMGA2 protein accumulation. In this process, HBXIP increased the acetyltransferase p300/CBP-associated factor (PCAF) phosphorylation and activation via the Akt pathway, then PCAF directly interacted with HMGA2, leading to HMGA2 acetylation in the cells. HMGA2 K26 acetylation enhanced its DNA binding capacity and blocked its ubiquitination and then inhibited proteasome-dependent degradation. Functionally, HBXIP-stabilized HMGA2 could promote ESCC cell growth in vitro and in vivo. Strikingly, aspirin suppressed ESCC growth by inhibiting HBXIP and HMGA2. Collectively, our findings disclose a new mechanism for the posttranslational regulation of HMGA2 mediated by HBXIP in ESCC.
INTRODUCTION
Esophageal cancer (EC) is an aggressive and lethal malignancy, ranking sixth in terms of mortality and eighth in terms of incidence among all cancer types (1). EC is primarily classified into two pathological subtypes: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (2). ESCC, which is the most severe pathological subtype of EC, accounts for ∼90% of all esophageal carcinomas at the time of diagnosis and has a high-incidence in China (2). Considering late-stage tumor detection and limited clinical therapeutic strategies, patients with ESCC show an extremely poor prognosis, with the overall 5-year survival rate of ∼17% (3). Although extensive efforts have been devoted to overcome this disease, information on its molecular drivers remains limited. Therefore, a better understanding of the molecular foundation of the occurrence and development of ESCC is urgently required for earlier diagnosis and more efficient treatment.
High-mobility group AT-hook 2 (HMGA2), an architectural transcription factor, constitutes ∼109 amino acid residues and three basic DNA-binding domains called ‘AT-hooks’, which bind to the AT-rich regions in DNA (4). HMGA2 modulates transcription by inducing structural changes in the chromatin, enabling the transcriptional machinery to access the target regions to regulate the expression of many mammalian genes in terms of both activation and repression (4). HMGA2 is highly expressed during tumorigenesis but rarely in normal adult tissues (4,5). Various studies indicate that high expression of HMGA2 is related to poor survival rates in breast cancer (6), colorectal cancer (7) and lung cancer patients (8). Furthermore, there is evidence that oncogenic HMGA2 participates in DNA damage repair (9), stem cell self-renewal (10), aggressive tumor growth (11) and tumor cell differentiation (12). Importantly, HMGA2 is considered to promote tumorigenesis in part through the modulation of a group of target genes. For instance, HMGA2 counteracts the repression activity of the transcription repressor p120E4F to induce cyclinA expression, which controls cell cycle progression (13). Additionally, HMGA2 stimulates human telomerase reverse transcriptase (hTERT) expression, preventing the gradual telomere shortening in cancer cells (14). Moreover, HMGA2 directly activates the transcription of pro-metastatic genes, including SNAIL, SLUG, and CXCR4 (15–17). Equally, much attention has been focused on the regulatory cascades of HMGA2 expression during cancer progression. HMGA2 can be positively regulated via the active Wnt/β-catenin pathway (18) and repressed via the ZBRK1/BRCA1/CtIP pathway (19). Interestingly, posttranslational modifications (PTMs) of HMGA2 confer a profound effect on its biological functions. For example, HMGA2 phosphorylation at the acidic C-terminal tail may affect its DNA-binding properties (20), and HMGA2 SUMOylation may promote promyelocytic leukemia (PML) protein degradation (21). However, whether PTM functions in the regulation of HMGA2 expression remains largely unknown.
Mammalian hepatitis B X-interacting protein (HBXIP), also known as LAMTOR5 (22), is a conserved 18-kDa protein, which was identified initially based on its binding to the C-terminus of hepatitis B virus X proteins (23). HBXIP is expressed in nearly all tissues (24). It can function as a cofactor of survivin to control cell apoptosis and regulate centrosome duplication and cytokinesis to mediate cell growth (24,25). Additionally, HBXIP can serve as a regulatory component required for the activation of mammalian target of rapamycin complex 1 via amino acids (22). Our group has reported that HBXIP is highly expressed in breast carcinoma and that it acts as an oncogenic transcriptional coactivator of multiple transcription factors, such as c-Myc, LXR, Sp1 and E2F1 to promote breast cancer growth and metastasis (26–29). Moreover, it supports the migration of breast cancer cells through GCN5-mediated modulation of microtubule acetylation (30). Our study has revealed that HBXIP as an important oncoprotein can regulate PTMs of some transcription factors. For instance, HBXIP can induce the acetylation of transcription factor HOXB13 to prevent HOXB13 degradation in the promotion of tamoxifen resistance of breast cancer (31). In addition, HBXIP can increase the phosphorylation levels of c-Fos through activating ERK1/2, which is a benefit for the nuclear localization of c-Fos in breast cancer (32). One study found that the abnormal expression of HBXIP was associated with poor prognosis in ESCC (33). Accordingly, in the present study we are interested in whether HBXIP is involved in HMGA2 PTM in ESCC development.
Aspirin (ASA), a nonsteroidal anti-inflammatory drug, displays anti-cancer effect and has been applied in colorectal cancer therapy (34). Substantial evidence indicates that regular aspirin use is useful for the reduction of incidence, mortality and distant metastasis of cancers including breast cancer, liver cancer, and colorectal cancer (35–37). Several epidemiologic studies have proven that the use of aspirin and other nonsteroidal anti-inflammatory drugs protects against the development of esophageal cancer (38,39). We have recently revealed that aspirin can target HBXIP to inhibit HBXIP/HOXB13 axis, overcoming tamoxifen resistance in breast cancer (31). Based on these previous findings, we focus on the investigation of the role of aspirin in HBXIP-associated ESCC.
In the present study, we explored the function and regulation of HMGA2 in the development of ESCC. HBXIP enhances HMGA2 acetylation at the lysine 26 residue (K26) through the Akt pathway-induced PCAF phosphorylation and activation in ESCC. HMGA2 K26 acetylation functionally enhances its DNA binding ability on the target genes and blocks its ubiquitination and proteasomal degradation, thus leading to HMGA2 accumulation and carcinogenesis. Intriguingly, aspirin can suppress ESCC growth through repressing HBXIP and HMGA2. Thus, our studies identify a novel regulatory mechanism of HMGA2 in ESCC growth, which probably provides an effective strategy for ESCC therapy.
MATERIALS AND METHODS
Tissue specimens
The ESCC tissue microarray containing 151 primary ESCC tissues and 43 normal esophageal tissues with information of patients' overall survival and disease-free survival was acquired from Shantou University Medical College between February 2011 and November 2016. The patient records are presented in Supplementary Table S1. The other two ESCC tissue microarrays (Catalog No.: Es-kx03c and Catalog No.: Es-kx14c) containing 124 cases of human ESCC tissues, two cases of human esophagus basal cell carcinoma tissues and 10 cases of normal esophagus tissues in total were purchased from Aomeibio Company (Xian, China). The clinical characteristics are presented in Supplementary Tables S5 and S6 respectively. All samples were approved by Ethics Committee of Hospital providing tissues. Written informed consent was obtained from patients before samples were collected. All specimens, including tumor tissues of ESCC patients and normal esophageal tissues, were obtained during surgery.
Cell culture and reagents
The ESCC cell lines KYSE2, KYSE180, KYSE450, KYSE510 and the human embryonic kidney cell line 293T (HEK293T) were obtained from the American Type Culture Collection (ATCC). ESCC cell lines were cultured in RPMI 1640 (Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco). HEK293T was maintained in Dulbecco's Modified Eagle's Medium (Gibco) supplemented with 10% FBS. All cells were cultured at 37°C in a humidified atmosphere with 5% CO2. Cells were collected and seeded in 6-, 24- or 96-well plates for 24 h and then transfected with plasmids or small interference RNAs (siRNAs) using Lipofectamine 2000 (Invitrogen, Life Technologies, Grand Island, NY, USA), according to the manufacturer's protocol. All experiments were conducted in cells with ∼80% convergence. The reagents used in this study were trichostatin A (TSA), cycloheximide (CHX), the inhibitors of Akt, ERK1/2 and p38, and aspirin (ASA). TSA and CHX were purchased from MedChem Express (USA). GSK690693 (an inhibitor of Akt), PD98059 (an inhibitor of the upstream kinase of ERK1/2), and SB202190 (an inhibitor of p38) were all purchased from MedChem Express (USA). ASA were purchased from Sigma-Aldrich (USA).
Plasmid construction and siRNAs
Plasmids, including pCMV-Tag2B, pCMV-HBXIP, pcDNA3.1(+), pcDNA3.1 (+)-HBXIP, pSilencer 3.1-neo and shRNA construct pSilencer-HBXIP, were kept in our laboratory. The complete human HMGA2 (NCBI Reference Sequence: NM_003483.4) cDNA sequence was amplified by polymerase chain reaction (PCR) and subcloned into the pEGFP-C2 or pCMV-Tag2B vector to generate GFP-HMGA2 or pCMV-HMGA2 (FLAG-HMGA2). All point or deletion mutants of HMGA2 were generated by PCR and subcloned into the pCMV-Tag2B vector and verified by sequencing. The vector expressing FLAG-tagged human full-length PCAF was kindly provided by Prof. Hongquan Zhang (Peking University Health Science Center, Beijing, China). The complete PCAF cDNA sequence was amplified by PCR and subcloned into the pEGFP-C2 to generate GFP-PCAF. All siRNAs and related primers are listed in Supplementary Table S2. All siRNAs were purchased from Riobio Co. (Guangzhou, China).
Western blotting analysis
Western blotting analysis was carried out with the standard protocol. Tissues or cells were lysed in RIPA buffer (Solarbio, Beijing, China). Equal amounts of total protein were loaded for western blotting. Following SDS-PAGE, resolved proteins were transferred onto PVDF membranes (Millipore, USA). The membranes were blocked in 5% skim milk for 2 h at room temperature, and then probed with primary antibodies (Supplementary Table S3) for 2 h at room temperature or overnight at 4°C. The rabbit polyclonal antibody recognizing the acetylated HMGA2 at lysine 26 residue was produced with a synthetic acetylated human HMGA2 peptide: APQ(AcK)RGRGRPR (Jia Xuan Zhi Rui, Beijing, China). After incubation with secondary antibody for 1 hour, the membrane was visualized by ECL (Millipore). The Image J software was used to quantify the intensity in western blotting analysis.
Immunohistochemistry staining
Immunohistochemical staining was carried out as described previously (40). The ESCC tissue samples were incubated with HBXIP and HMGA2 primary antibodies (Supplementary Table S3), or incubated with specific AcK26-HMGA2 antibody at 4°C for overnight, followed by incubation with horseradish peroxidase-conjugated secondary antibody at room temperature for 30 min. Immunostaining was developed by using a diaminobenzidine (DAB) substrate kit (Zhong Shan Jin Qiao, Beijing, China), and counterstained with hematoxylin. The staining levels of HBXIP, HMGA2 and AcK26-HMGA2 were classified into four groups using a modified scoring method based on the intensity of staining (0 = negative; 1 = low; 2 = moderate; 3 = high) and the percentage of stained cells (0 = 0% stained; 1 = 1–29% stained; 2 = 30–65% stained; 3 = 66–100% stained). A multiplied score (intensity score × percentage score) lower than 1 was considered to be negative staining (0), 1, 2 and 3 were considered to be weak staining (1), 4 and 6 were considered to be moderate staining (2) and 9 was considered to be intense staining (3). For Ki67 and AcK26-HMGA2 staining in tumor xenograft, tumor slides were fixed in 4% paraformaldehyde for 2 days. Then, sections were stained using a primary antibody against Ki67 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or specific AcK26-HMGA2 antibody. The positive Ki67 and AcK26-HMGA2 staining were identified by Image-Pro Plus software.
Co-immunoprecipitation (Co-IP) assay
Indicated cells were harvested and lysed in a lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 10% glycerine, 1 mM protease inhibitor PMSF). The lysates were incubated with Anti-FLAG M2 affinity gel (Sigma-Aldrich) at 4°C for 3 h or incubated with rabbit anti-HMGA2 antibody (Supplementary Table S3) at 4°C for 2 h before incubated with Protein G Magnetic beads (Pierce, Waltham, MA, USA) at 4°C for 1 h. After extensive washing (3 times), precipitated proteins were eluted from the gel or beads by 0.1 M glycine–HCl (pH 3.0) buffer and neutralized with 1 M Tris–HCl (pH 7.5) containing 1.5 M NaCl. Immunoprecipitated samples were resolved by SDS-PAGE followed by western blotting with appropriate antibodies (Supplementary Table S3).
In vivo ubiquitination assays
Cells were transfected with the indicated plasmids and treated with 40 μM MG-132 for 6 h before harvest. The cells were washed with cold PBS and then lysed in 200 μl denaturing buffer (150 mM Tris–HCl pH 7.4, 1% SDS). After incubation at 4°C for 10 min, the lysate was sonicated and boiling for 10 min. Lysates were added with lysis buffer to 1 ml and incubated with anti-FLAG M2 agarose beads (Sigma-Aldrich, St. Louis, MO, USA) for 3 h at 4°C. The immunoprecipitates were washed five times with 1× PBS before being resolved by SDS-PAGE and immunoblotted with the indicated antibodies (Supplementary Table S3).
Immunofluorescence staining and confocal microscopy
Indicated cells were cultured on acid-treated glass coverslips. The cells were fixed in 4% paraformaldehyde for 15 min at room temperature, washed in pre-cooled PBS three times and permeabilized with 0.1% Triton X-100 for 20 min. After blocking non-specific antibody-binding sites with PBS containing 3% BSA (w/v) for 1 h, the cells were stained with primary antibodies such as rabbit anti-HMGA2 (GeneTex, USA), mouse anti-PCAF (Santa Cruz), or rabbit anti-FLAG tag (Santa Cruz) (Supplementary Table S3) at room temperature for 60 min. Following three times washing with PBS, the secondary antibodies such as Alexa Fluor 488 goat anti-mouse IgG (Invitrogen), and Alexa Fluor 594 goat anti-rabbit IgG (Invitrogen) were added at room temperature for 30 min. Nuclei were counterstained with DAPI (Sigma-Aldrich, St. Louis, MO, USA). Fluorescent micrographs were obtained using laser scanning confocal microscopy (Leica, Germany).
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation (ChIP) assays were performed in KYSE180 cells transfected with the indicated plasmids using the EpiQuik™ chromatin immunoprecipitation kit from Epigentek Group Inc. (Brooklyn, NY, USA) as reported previously (41). The protein-DNA complexes were immunoprecipitated with anti-FLAG antibody (Supplementary Table S3), using normal mouse IgG as a negative control. DNA purified from these samples was analyzed by real-time RT-PCR. The primers used are listed in Supplementary Table S2.
Total RNA isolation, reverse transcription-PCR (RT-PCR) and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells using Trizol reagent (Invitrogen, USA) according to manufacturer's instructions. First-strand cDNA was synthesized with the Prime Script reverse transcriptase Kit (TaKaRa Bio, China). RT-PCR and qRT-PCR assays were carried out as previously described (40). All primers were listed in Supplementary Table S2.
GST pull-down assays
The sequences of the primers used to amplify PCAF cDNA are as follows: forward: 5′-ATGTCCGAGGCTGGCGGGGCCGG-3′, reverse: 5′-TCACTTGTCAATTAATCCAGCTT-3′. The sequences of the primers used to amplify HMGA2 cDNA are as follows: forward: 5′-GGAGGCAGGATGAGCGCA-3′, reverse: 5′-CTAGTCCTCTTCGGCAGACTC-3′. The PCAF cDNA was inserted into pGEX-4T-1 vector, and HMGA2 cDNA was cloned into pET-28a vector. Proteins were expressed in E. coli BL21 after induction with 0.5 mM IPTG at 16°C overnight (∼12 h). The GST-PCAF or His-HMGA2 fusion proteins expressed in bacteria were purified with glutathione Sepharose 4B (GE Healthcare, USA) or Ni2+-NTA agarose beads (Qiagen, USA) as described previously (42). In brief, the beads were washed, and purified His-HMGA2 was added. The binding reaction was performed in binding buffer (20 mM Tris, pH 7.5; 150 mM NaCl; 0.1% PMSF). After the incubation and washes, proteins were eluted by boiling in SDS-PAGE loading buffer and separated by SDS-PAGE. Precipitated HMGA2 was detected by western blotting analysis.
5-Ethynyl-2′-deoxyuridine (EdU) assay
The EdU assay was used to measure the cell proliferative capacity. The cells (5 × 103 cells/well) were seeded into 96-well plates. All operations were performed based on instructions of the Cell-Light TM EdU imaging detecting kit (RiboBio, Guangzhou, China). The images were acquired and analyzed under fluorescence microscope (Zeiss Axio Imager Z1, Carl Zeiss, Oberkochen, Germany).
MTT assays
Cell viability assays were carried out using 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide (MTT) reagent (Sigma) as described previously (43). Briefly, transfected cells were trypsinized, counted, and plated into 96-well plates at a density of 1000 cells per well. After forming a confluent monolayer, the cells were treated with aspirin (2.5 mM) for different time points. Then, MTT was added directly to each well. Four hours later, the supernatant was removed and dimethyl sulfoxide (DMSO) was added to stop the reaction. Absorbance at 490 nm was measured using a reader system (Labsystem, Multiskan Ascent).
Monolayer colony formation assay
The transfected cells were trypsinized and seeded in six-well plates. The medium added with corresponding reagents was replaced every 3 days. After growth for 2 weeks, the cells were washed with PBS three times and fixed in methanol for 20 min at room temperature. Fixed cells were then stained with crystal violet for 30 min at room temperature. After extensive washing and air drying, monoclonal colonies were photographed. The number of colonies was counted and the colony forming efficiency was determined with the formula: colony-forming efficiency = number of colonies counted / number of cells plated × 100%.
Xenograft
All experimental procedures involving animals were conducted in accordance with the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Briefly, 5-week-old female BALB/c athymic nude mice from Experimental Animal Center of Peking (Beijing, China) were fed and housed. The cells transfected with corresponding plasmids were harvested and suspended at a density of 5 × 107 cells/ml in phosphate saline and then subcutaneously injected into the right flank of each mouse (0.2 ml of cell suspension). Daily oral administration of saline or aspirin at 75 mg/kg was initiated after the tumor size exceeded 100 mm3 ∼10–14 days after injection. Tumor volume and body weight were monitored every 3 days. Tumor volume (V) was monitored by measuring the length (L) and width (W) with calipers and was calculated using the following formula: (L × W2) × 0.5. The mice were sacrificed when the tumor size reached ∼1000 mm3. The tumors were excised and assessed. The investigators who assessed the outcome data were blinded to the treatment groups.
Statistical analysis
Each experiment was repeated at least three times. The statistical significance of in vitro and in vivo data was assessed by comparing mean values (±SD) using Student's t-test, and significance was assumed at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and not significant (NS). The association between HBXIP and HMGA2 expression in ESCC tissue microarray was statistically analyzed by Pearson chi-square independence test using the SPSS software program (SPSS, Chicago, USA). Survival rates were calculated using the Kaplan–Meier method, and differences in survival curves were analyzed by log-rank tests using the GraphPad Prism 6.0.
RESULTS
HMGA2 is upregulated by HBXIP in human ESCC tissues and cells
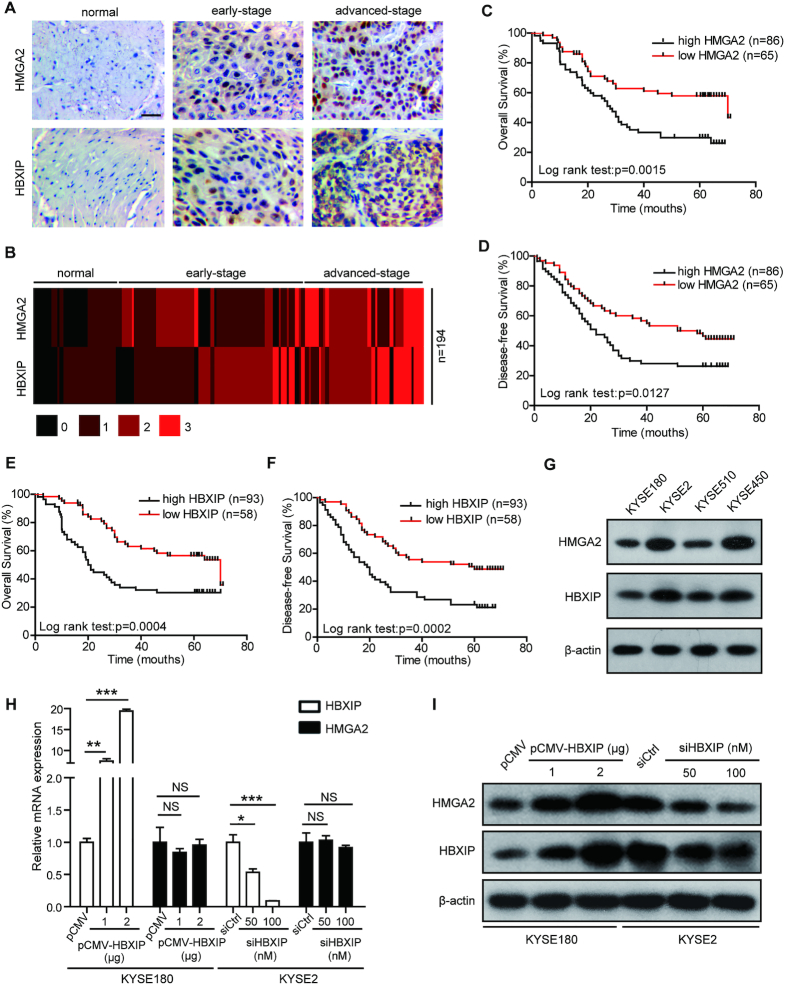
HMGA2, an architectural transcription factor, plays a pivotal role in carcinogenesis (13–15,44). HBXIP emerges as an oncogenic protein in many cancers (26–28,31). Either HMGA2 or HBXIP exhibits increased expression and is related to worse prognosis in ESCC (33,45). Therefore, we are wondering whether HMGA2 is associated with HBXIP in ESCC development. The immunohistochemical staining of HMGA2 and HBXIP in human tissue microarray (Supplementary Table S1) containing 43 normal esophageal tissues and 151 primary ESCC tissues revealed that HMGA2 and HBXIP were both overexpressed in clinical human ESCC tisses (Figure 1A and B). Furthermore, in 43 normal esophageal tissues and 151 primary ESCC tissues (Supplementary Table S1) a positive correlation was revealed between HMGA2 and HBXIP expression (Pearson chi-square independence test, χ2 = 25.05, P < 0.01. Supplementary Table S4). In addition, the Kaplan–Meier survival analysis of 151 ESCC patients (Supplementary Table S1) for 80 months demonstrated that high HMGA2 and HBXIP expressions were associated with advanced tumor stage (Supplementary Figure S1A and B) and reduced overall and disease-free survival (Figure 1C–F). To further explore the relevance of HBXIP and HMGA2, we evaluated HBXIP and HMGA2 expressions in four ESCC cell lines including KYSE180, KYSE2, KYSE510, and KYSE450. Western blotting analysis presented a close connection between HMGA2 and HBXIP in these four ESCC cell lines (Figure 1G). We then analyzed the effect of HBXIP on the protein levels of HMGA1, the other member of the HMGA family. The data showed that the protein levels of HMGA1 were not changed by HBXIP (Supplementary Figure S1C). Taken a step further, HBXIP overexpression dose-dependently enhanced HMGA2 protein level without altering HMGA2 mRNA level in both KYSE180 and KYSE510 cells (Figure 1H, I and Supplementary Figure S1D, E). Conversely, HBXIP knockdown by siRNA dose-dependently inhibited HMGA2 protein level, whereas had no effect on HMGA2 mRNA level in either KYSE2 or KYSE450 cells (Figure 1H, I and Supplementary Figure S1F, G), implying that HMGA2 could be modulated by oncogenic HBXIP through a posttranscriptional mechanism. We also tested the effect of HBXIP overexpression and HBXIP knockdown on the protein levels of HMGA2 in a liver cancer cell line Hep3B and a breast cancer cell line MCF-7. The protein levels of HMGA2 were obviously increased by HBXIP overexpression, while the knockdown of HBXIP significantly reduced the protein levels of HMGA2 in Hep3B cells (Supplementary Figure S1H). However, the HMGA2 protein levels were not changed by HBXIP in MCF-7 cells (data not shown). Collectively, HMGA2 and HBXIP expression exhibit a positive relationship in clinical ESCC tissues, and HMGA2 can be upregulated by HBXIP in ESCC cells.
Figure 1.
HMGA2 is upregulated by HBXIP in human ESCC tissues and cells. (A) HMGA2 and HBXIP expressions were assessed by immunohistochemical staining in an esophageal tissue microarray containing 43 normal esophageal tissues and 151 ESCC tissues from Shantou University Medical College between February 2011 and November 2016. Scale bars, 50 μm. (B) Heatmap view of HMGA2 and HBXIP expressions of the esophageal tissue microarray used in (A). Numbers 0, 1, 2 and 3 represent negative, week, moderate, and intense staining, respectively. (C and D) Kaplan–Meier plots of the overall survival (C) and disease-free survival (D) rates of 151 ESCC patients stratified according to HMGA2 expression from the esophageal tissue microarray containing 43 normal esophageal tissues and 151 ESCC tissues used in (A). (E and F) Kaplan–Meier plots of the overall survival (E) and disease-free survival (F) rates of 151 ESCC patients stratified according to HBXIP expression from the esophageal tissue microarray containing 43 normal esophageal tissues and 151 ESCC tissues used in (A). (G) HMGA2 and HBXIP expressions were measured by western blotting in four ESCC cell lines. (H) Real-time PCR analysis of HBXIP and HMGA2 mRNA levels in KYSE180 cells transfected with the pCMV or pCMV-HBXIP plasmids and in KYSE2 cells transfected with control siRNA (siCtrl) or HBXIP siRNA (siHBXIP). Each bar shows the means ± SD (n = 3). (I) The HBXIP and HMGA2 protein levels were measured by western blotting in KYSE180 cells transiently transfected with pCMV or pCMV-HBXIP plasmids and KYSE2 cells transfected with siRNA (siCtrl) or HBXIP siRNA (siHBXIP). Statistically significant differences are indicated: *P < 0.05, **P < 0.01 and ***P < 0.001; NS, not significant; Student's t-test.
HBXIP contributes to HMGA2 stabilization through inducing its K26 acetylation
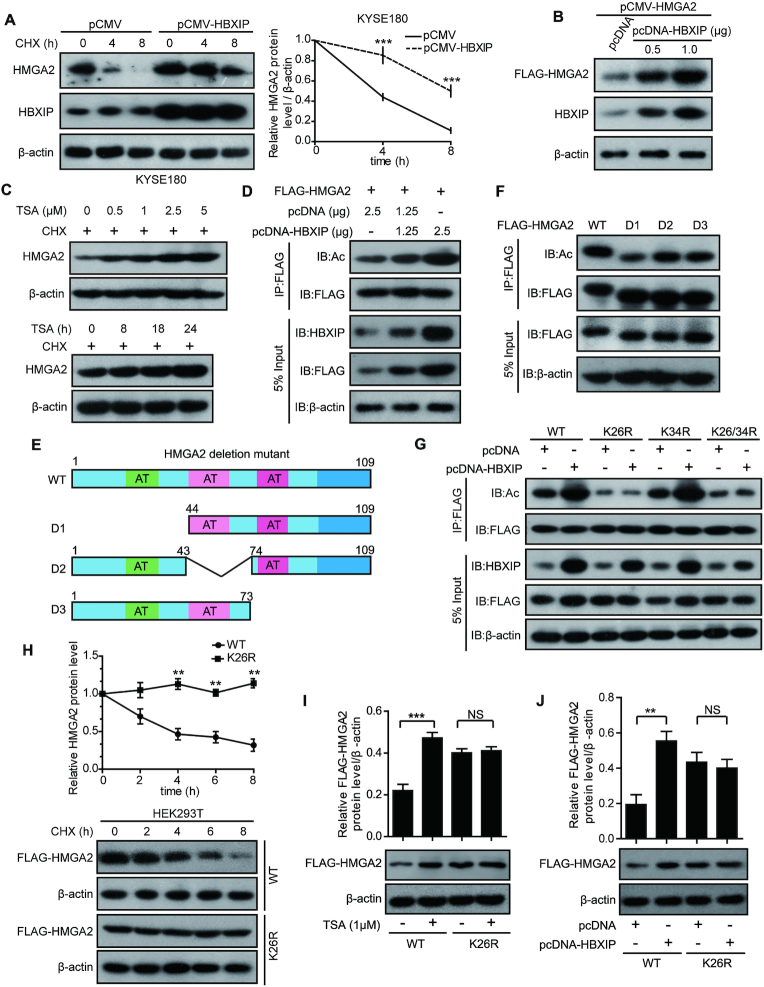
To explore how HBXIP upregulates HMGA2, KYSE180 and KYSE2 cells were treated with CHX (a protein synthesis inhibitor) for different periods to block protein translation, and the degradation rates of the existing HMGA2 protein were assessed by western blotting. Compared with the control groups, HBXIP overexpression in KYSE180 cells prolonged the half-life of endogenous HMGA2 protein (Figure 2A), whereas HBXIP knockdown in KYSE2 cells accelerated HMGA2 degradation (Supplementary Figure S2A). Moreover, the elevated expression of HBXIP led to dose-dependent increase of exogenous FLAG-HMGA2 at the protein level (Figure 2B), supporting that HBXIP could stabilize HMGA2 at the posttranslational level. Emerging studies demonstrated that lysine acetylation as a frequent posttranslational modification (PTM) is critical for modulating protein stability and function (46–48). Thus, we are wondering whether HBXIP could stabilize HMGA2 through modulating its acetylation. We first examined whether HMGA2 could be acetylated. The Co-IP assay showed that both endogenously and exogenously expressed HMGA2 could be acetylated in vivo, and its acetylation dramatically increased upon treatment with TSA, an inhibitor of histone deacetylases (Supplementary Figure S2B and C). To determine whether HMGA2 acetylation affects its stability, we treated KYSE180 cells with TSA and/or CHX. Then, we found an increase in endogenous HMGA2 protein levels (Figure 2C and Supplementary Figure S2D), but HMGA2 mRNA levels had no significant changes in the presence of TSA (Supplementary Figure S2E and F), suggesting that HMGA2 remains stable upon its acetylation.
Figure 2.
HBXIP contributes to HMGA2 stabilization through inducing its K26 acetylation. (A) Degradation of HMGA2 protein was measured in KYSE180 cells treated with 100 μg/ml CHX for the indicated periods after transient transfection with pCMV or pCMV-HBXIP vectors. The HMGA2 intensity normalized to β-actin was plotted. Each bar shows the means ± SD (n = 3). (B) Exogenous FLAG-HMGA2 was measured by western blotting in HEK293T cells transiently transfected with pCMV-HMGA2 co-transfected with pcDNA or pcDNA-HBXIP plasmids. The FLAG-HMGA2 protein level was detected using the anti-FLAG antibody. (C) Endogenous HMGA2 in KYSE180 cells were measured by western blotting after treatment with 100 μg/ml CHX along with different concentrations of TSA for 18 h or 1 μM TSA for the indicated periods. (D) Exogenous FLAG-HMGA2 was immunoprecipitated with FLAG beads in HEK293T cells, and then the acetylation of HMGA2 protein in precipitation was measured by western blotting with an anti-acetylated-lysine antibody. The cells were transiently transfected with the indicated plasmids. (E) A Schematic of FLAG-tagged full-length or serial deletion mutant HMGA2. (F) HEK293T were transfected with FLAG-tagged full-length HMGA2 expression vectors (WT) or serial deletion mutant HMGA2 expression vectors (D1, D2 or D3). Cell lysates were immunoprecipitated with FLAG beads, followed by western blotting with anti-FLAG and anti-acetylated-lysine antibodies. (G) FLAG-tagged WT, K26R, K34R, or K26/34R of HMGA2 along with pcDNA or pcDNA-HBXIP plasmids was transfected into HEK293T cells. Exogenous FLAG-tagged HMGA2 was immunoprecipitated with FLAG beads, and then acetylation levels of HMGA2 protein in precipitation were tested by western blotting with an anti-acetylated-lysine antibody. (H) The stabilities of WT and K26R mutant HMGA2 were measured by western blotting. HEK293T cells expressing FLAG-tagged WT or K26R mutant HMGA2 were treated with CHX for the indicated periods. Each bar shows the means ± SD (n = 3). (I) Ectopically expressed FLAG-tagged WT and K26R mutant HMGA2 levels were measured after the treatment of 1 μM TSA for 18 h in KYSE180 cells by western blotting with an anti-FLAG antibody. The upper panel is the quantification of the intensity relative to β-actin. Each bar shows the means ± SD (n = 3). (J) Ectopically expressed FLAG-tagged WT and K26R mutant HMGA2 levels were measured in KYSE180 cells transfected with pcDNA or pcDNA-HBXIP vectors by western blotting with an anti-FLAG antibody. The upper panel is the quantification of the intensity relative to β-actin. Each bar shows the means ± SD (n = 3). All experiments were repeated at least three times. Statistically significant differences are indicated: **P < 0.01 and ***P < 0.001; NS not significant; Student's t-test.
Next, we ascertained the role of HBXIP in acetylation modulation of HMGA2 protein. As expected, Co-IP assay showed that HBXIP dose-dependently enhanced the acetylation level of HMGA2 in HEK293T cells (Figure 2D), suggesting that HBXIP is involved in HMGA2 acetylation. To identify the acetylation sites of HMGA2, the acetylation levels of a series of deletion mutants of HMGA2 were evaluated (Figure 2E). Compared with other mutants including D2 and D3, the D1 mutant lacking 43 amino acids (1–43 aa) showed obviously decreased HMGA2 acetylation, indicating that the main acetylation residues of HMGA2 might be located in this region (Figure 2F). The fragment from 1 to 43 aa contains two lysine residues (K26 and K34) that might be modified by acetylation. To further confirm the acetylation site, the K26 and K34 residues were mutated to arginine (R, mimics of acetylation-deficient HMGA2) or glutamine (Q, mimics of hyperacetylated HMGA2), either alone (K26R and K34R or K26Q and K34Q) or together (K26/34R or K26/34Q). The acetylation level of HMGA2 were no longer elevated by HBXIP in the K26R, K26/34R, K26Q, and K26/34Q mutants (Figure 2G and Supplementary Figure S2G), suggesting that K26 could be the major acetylation site of HMGA2 mediated by HBXIP. Subsequently, we compared the amino acid sequence of HMGA2 aligned with the other five species. Interestingly, we found that HMGA2-K26 was highly conserved in multiple species during evolution (Supplementary Figure S2H). Based on these findings, we hypothesized that the acetylation of K26 in HMGA2 is important for its stability. Accordingly, we transfected HEK293T cells with different vectors of HMGA2 including wild-type (WT), K26R mutant and K26Q mutant. The K26R and K26Q mutant HMGA2 were more stable than WT HMGA2 (Figure 2H and Supplementary Figure S2I). Moreover, the K26R and K26Q mutations abolished the TSA-induced increase in HMGA2 and abrogated the HBXIP-induced stability of HMGA2 (Figure 2I, J and Supplementary Figure S2J, K). Thus, we conclude that HBXIP posttranslationally stabilizes HMGA2 via acetylation modification at K26 in ESCC cells.
The acetylase PCAF is responsible for HMGA2 acetylation at K26 by HBXIP
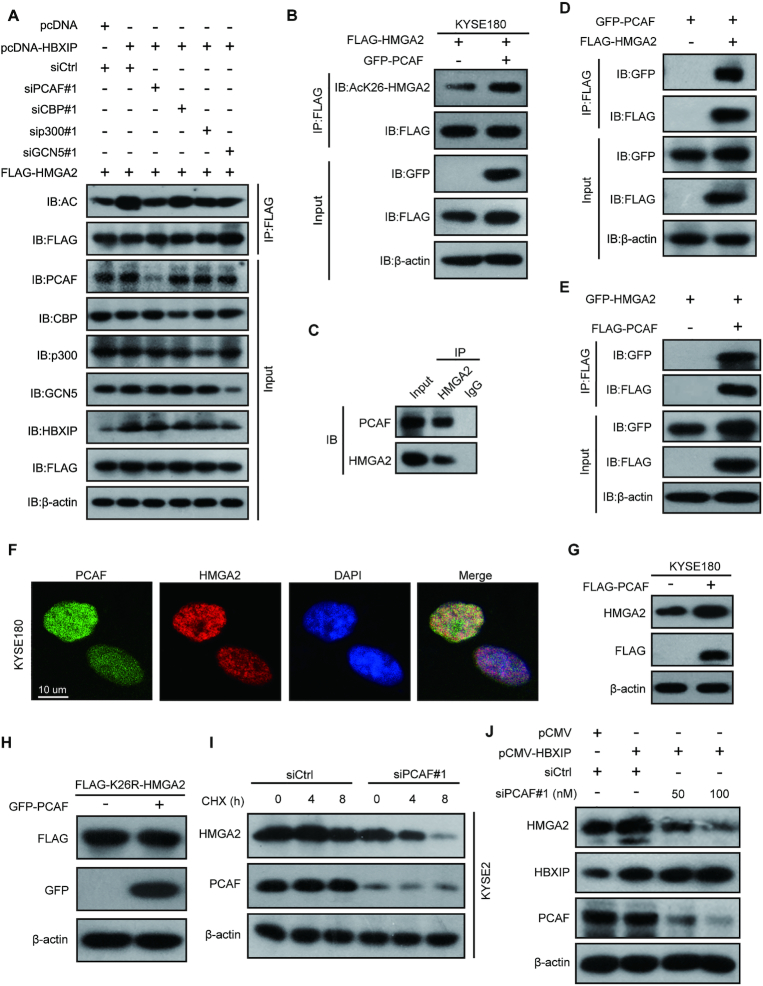
To investigate the acetyltransferase responsible for HMGA2 acetylation, we evaluated the effect of different acetyltransferases including PCAF, CBP, p300 and GCN5 on HMGA2 acetylation. Notably, the knockdown of PCAF effectively abolished the HBXIP-mediated increase in HMGA2 acetylation in KYSE2 cells, whereas the silencing of other acetyltransferases, including CBP, p300 and GCN5, showed little effect on HMGA2 acetylation (Figure 3A and Supplementary Figure S3A-D). Based on the same amount of FLAG-tagged HMGA2 in IP samples, we found that the acetylation levels of FLAG-HMGA2 were increased upon PCAF overexpression in IP samples immunoprecipitated with the same amount of anti-FLAG-coated agarose beads, demonstrating that PCAF can enhance the HMGA2 acetylation (Supplementary Figure S3E). To further confirm whether K26 of HMGA2 can be acetylated by PCAF, we entrusted a company (Jia Xuan Zhi Rui, Beijing, China) to generate a rabbit polyclonal antibody specifically recognizing the acetylated HMGA2-K26 (AcK26-HMGA2). The specificity of the AcK26-HMGA2 antibody was verified as it recognized the K26-acetylated peptide, but not the unacetylated HMGA2 peptide (Supplementary Figure S3F). Furthermore, the acetylation signal was blocked by preincubation of the antibody with antigen peptides (Supplementary Figure S3G), indicating the specificity of the AcK26-HMGA2 antibody for the recognition of HMGA2 acetylation at K26. Further data showed that the AcK26-HMGA2 antibody recognized acetylated HMGA2-WT at K26, but did not recognize K26Q or K26R mutations (Supplementary Figure S3H). Using this antibody, we found that the acetylation level of HMGA2 at K26 was clearly increased after PCAF was overexpressed in KYSE180 cells (Figure 3B). Given that PCAF might serve as the acetyltransferase of HMGA2, we wondered whether PCAF could interact with HMGA2. The data revealed endogenous interaction of HMGA2 with PCAF in KYSE180 cells (Figure 3C). Furthermore, exogenously expressed HMGA2 and PCAF could be co-immunoprecipitated by each other using different tag antibodies, implying that HMGA2 interacts with PCAF in the cells (Figure 3D and E). Moreover, GST pull-down assays in vitro revealed that GST-PCAF could bind to His-HMGA2 directly (Supplementary Figure S3I). Additionally, confocal microscopic analysis showed that endogenous HMGA2 and PCAF (or exogenously overexpressed EGFP-HMGA2 and FLAG-PCAF) were colocalized in the nucleus (Figure 3F and Supplementary Figure S3J). Intriguingly, PCAF overexpression increased endogenous HMGA2 protein levels (Figure 3G), whereas in K26R mutant-overexpressed cells PCAF lost the capacity of increasing HMGA2 protein levels (Figure 3H), suggesting that K26 in HMGA2 is required for PCAF-induced HMGA2 acetylation. Furthermore, PCAF knockdown destabilized HMGA2 (Figure 3I). HBXIP-mediated stabilization of HMGA2 was largely abrogated by PCAF knockdown in KYSE180 cells (Figure 3J and Supplementary Figure S3K). Altogether, these findings demonstrate that PCAF is required for HBXIP-induced HMGA2 acetylation at K26 in ESCC.
Figure 3.
The acetylase PCAF is responsible for HMGA2 acetylation at K26 by HBXIP. (A) FLAG-HMGA2 vectors accompanied by the indicated plasmids or siRNAs against a variety of acetyltransferases were transfected into KYSE2 cells. Exogenous FLAG-HMGA2 was immunoprecipitated with FLAG beads, and then the acetylation of HMGA2 protein in precipitation was tested by western blotting with an anti-acetylated-lysine antibody. (B) FLAG-HMGA2 vectors were cotransfected with or without GFP-PCAF plasmids into KYSE180 cells. Exogenous FLAG-HMGA2 was immunoprecipitated with FLAG beads, and then the acetylation of HMGA2 protein in precipitation was tested by western blotting with the anti-AcK26-HMGA2 antibody. (C) Co-IP assays were performed to detect the interaction of endogenous PCAF with HMGA2 in KYSE180 cells with anti-HMGA2 antibody or control IgG. (D) GFP-PCAF and FLAG-HMGA2 plasmids were co-transfected into HEK293T cells, immunoprecipitated with FLAG beads followed by western blotting with anti-GFP and anti-FLAG antibodies. (E) HEK293T cells were co-transfected with FLAG-PCAF and GFP-HMGA2 plasmids, immunoprecipitated with FLAG beads followed by western blotting with anti-GFP and anti-FLAG antibodies. (F) The co-localization of endogenous PCAF and HMGA2 in KYSE180 cells was examined by confocal microscopy. KYSE180 cells were stained with an anti-PCAF monoclonal antibody (green) and an anti-HMGA2 polyclonal antibody (red). Nuclei were stained with DAPI (blue), followed by visualization with confocal microscopy. Scale bars, 10 μm. (G) Endogenous HMGA2 was determined by western blotting in KYSE180 cells transfected with the indicated plasmids. (H) FLAG-K26R mutant HMGA2 vector was co-transfected with or without GFP-PCAF into KYSE180 cells. The levels of FLAG-K26R mutant HMGA2 were measured by western blotting with an anti-FLAG antibody. (I) Western blotting analysis of endogenous HMGA2 protein in KYSE2 cells. The cells were treated with CHX for the indicated periods after transfection with control siRNA (siCtrl) or PCAF siRNA#1 (siPCAF#1). (J) Endogenous HMGA2 protein levels were detected by western blotting in KYSE180 cells cotransfected with pCMV-HBXIP plasmids and/or PCAF siRNA#1. All experiments were repeated at least three times.
The Akt pathway activates the acetylase PCAF to promote HMGA2 acetylation mediated by HBXIP
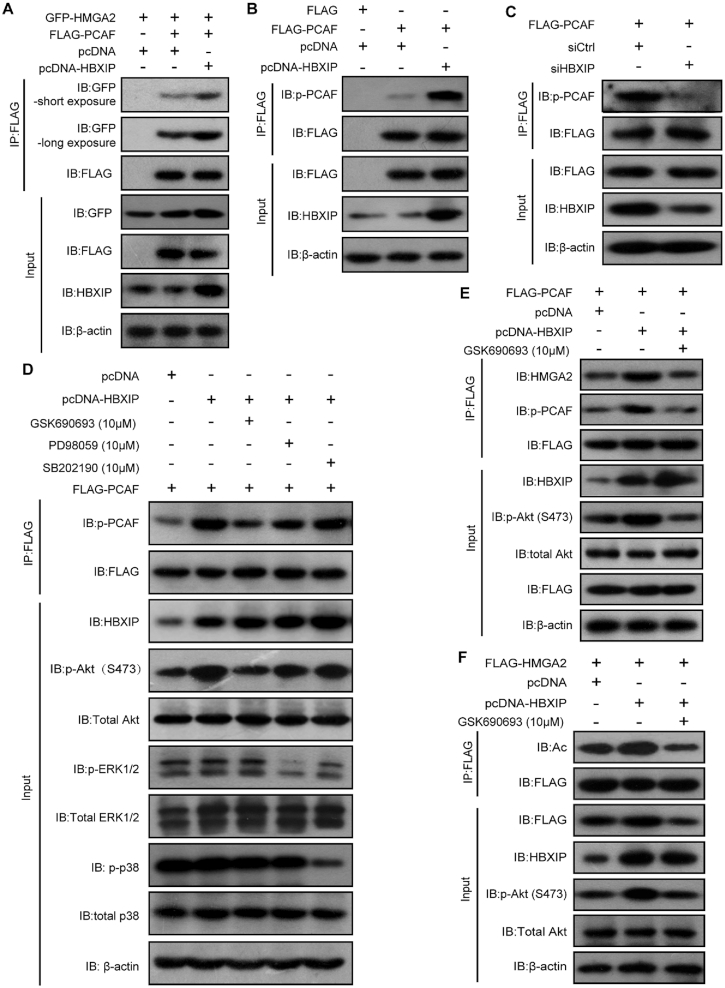
To clarify how HBXIP regulates the HMGA2 acetylation by PCAF, we assessed whether HBXIP modulated the binding affinity between HMGA2 and PCAF by Co-IP assays. HEK293T cells were transiently transfected with GFP-HMGA2 in the presence or absence of FLAG-PCAF and/or HBXIP, followed by Co-IP assays with an anti-FLAG antibody. Indeed, the levels of GFP-HMGA2 co-immunoprecipitated with the same amount of FLAG-PCAF were increased upon HBXIP overexpression, demonstrating that HBXIP can enhance the HMGA2-PCAF interaction (Figure 4A). It was shown previously that the histone acetyltransferase (HAT) activity of PCAF could be enhanced by its phosphorylation (49). Therefore, we subsequently examined whether PCAF activity was regulated in response to HBXIP overexpression. PCAF phosphorylation level was measured after HBXIP overexpression or silencing. As a result, HBXIP overexpression dramatically promoted PCAF phosphorylation, whereas HBXIP silencing efficiently prevented PCAF phosphorylation (Figure 4B and C). It has been reported that HBXIP can activate the Akt signaling in hepatocellular carcinoma, and activate ERK1/2 or p38 signaling pathways in breast cancer to promote tumor cell proliferation and migration (50–52). To screen the kinases responsible for PCAF phosphorylation, we treated KYSE180 cells with three kinase inhibitors, including GSK690693 (an inhibitor of Akt), PD98059 (an inhibitor of the upstream kinase of ERK1/2), and SB202190 (an inhibitor of p38). We found that the Akt inhibitor markedly reduced HBXIP-enhanced PCAF phosphorylation, whereas the ERK1/2 and p38 inhibitors showed little effects on PCAF phosphorylation (Figure 4D). Moreover, exclusive treatment with the Akt inhibitor reduced PCAF phosphorylation and suppressed the interactions between HMGA2 and PCAF, even upon HBXIP overexpression (Figure 4E). Furthermore, treatment with the Akt inhibitor dramatically inhibited HBXIP-induced HMGA2 acetylation (Figure 4F). These results were confirmed using siRNA-mediated Akt knockdown (Supplementary Figure S4A and B). Thus, we conclude that the Akt pathway is activated by HBXIP to increase PCAF phosphorylation and sequentially enhance HMGA2 acetylation.
Figure 4.
The Akt pathway activates the acetylase PCAF to promote HMGA2 acetylation mediated by HBXIP. (A) Co-IP assays were performed to examine the interaction of exogenous FLAG-PCAF with GFP-HMGA2 in HEK293T cells. GFP-HMGA2 and FLAG-PCAF along with pcDNA or pcDNA-HBXIP plasmids were coexpressed into HEK293T, immunoprecipitated with FLAG beads, and detected with indicated antibodies. (B) KYSE180 cells were transiently transfected with FLAG-PCAF along with pcDNA or pcDNA-HBXIP plasmids. Exogenous FLAG-PCAF was immunoprecipitated with FLAG beads from cell lysates, and then the phosphorylation of PCAF protein in precipitation was tested by western blotting with an anti-phospho-serine antibody. (C) KYSE2 cells were transiently transfected with FLAG-PCAF along with control siRNA (siCtrl) or HBXIP siRNA (siHBXIP). Exogenous FLAG-PCAF was immunoprecipitated with FLAG beads from cell lysates, and then the phosphorylation of PCAF protein in precipitation was examined by western blotting using an anti-phospho-serine antibody. (D) KYSE180 cells were transiently transfected with FLAG-PCAF along with pcDNA or pcDNA-HBXIP plasmids and treated with AKTi (GSK690693, 10 μM), ERKi (PD98059, 10 μM) or p38i (SB202190, 10 μM) for 6 h. Exogenous FLAG-PCAF was immunoprecipitated with FLAG beads from cell lysates, and then the phosphorylation of PCAF protein in precipitation was examined by western blotting with an anti-phospho-serine antibody. (E) KYSE180 cells were transiently transfected with FLAG-PCAF along with pcDNA or pcDNA-HBXIP plasmids and treated with or without AKTi (GSK690693, 10 μM) for 6 h. The interaction of HMGA2 with PCAF and the phosphorylation level of PCAF were detected by a Co-IP assay using FLAG beads followed by western blotting with anti-HMGA2 and anti-phospho-serine antibodies. (F) KYSE180 cells were transiently transfected with FLAG-HMGA2 along with the indicated plasmids and treated with or without AKTi (GSK690693, 10 μM) for 6 h. Exogenous FLAG-HMGA2 was immunoprecipitated with FLAG beads from cell lysates, and then the acetylation of HMGA2 protein in precipitation was tested by western blotting with an anti-acetylated-lysine antibody. All experiments were repeated at least three times.
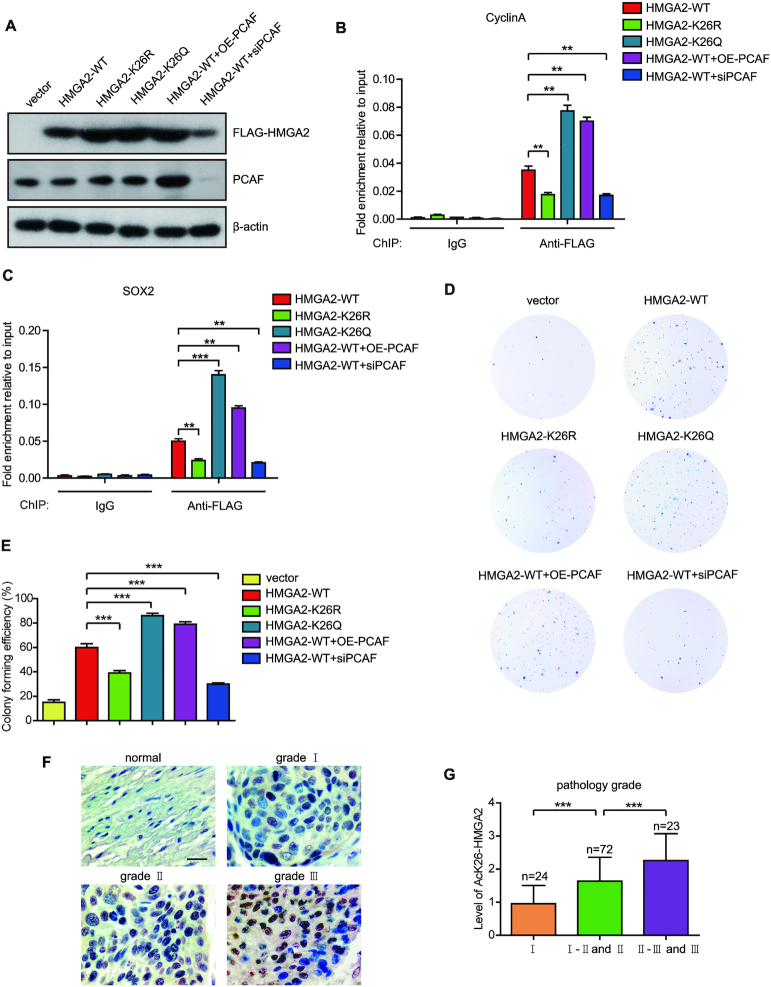
HMGA2 K26 acetylation enhances its DNA binding ability and predicts a poor prognosis of ESCC patients
To uncover the physiological function of HMGA2 acetylation at K26, we investigated the effect of HMGA2 K26 acetylation on its DNA binding ability. We firstly transfected KYSE180 cells with plasmids expressing FLAG-HMGA2-WT, FLAG-HMGA2-K26R, FLAG-HMGA2-K26Q, FLAG-HMGA2-WT with PCAF overexpression, and FLAG-HMGA2-WT with PCAF knockdown separately (Figure 5A). Two common HMGA2 target genes including cyclin A and SOX2 were selected to investigate the effect of HMGA2 acetylation at K26 on its DNA binding ability. It has been reported that HMGA2 binds directly to the cyclin A and SOX2 promoter (13,53). Results showed that HMGA2-K26Q occupied the promoter regions of the target genes to a high extent than the HMGA2-WT as determined by ChIP assays (Figure 5B and C). However, HMGA2-K26R decreased the DNA binding capacity of HMGA2 to the target gene promoters compared to HMGA2-WT in ChIP assays (Figure 5B and C). Notably, PCAF overexpression enhanced the HMGA2-WT occupancy to the target gene promoters by ChIP assays (Figure 5B and C). In contrast, knockdown of PCAF decreased the DNA binding capacity of HMGA2-WT to the target gene promoters (Figure 5B and C). These data suggest that HMGA2 K26 acetylation enhances its DNA binding ability to the target genes.
Figure 5.
HMGA2 K26 acetylation enhances its DNA binding ability and predicts a poor prognosis of ESCC patients. (A) KYSE180 cells were transiently transfected with FLAG-HMGA2-WT, FLAG-HMGA2-K26R, FLAG-HMGA2-K26Q, FLAG-HMGA2-WT along with FLAG-PCAF, or FLAG-HMGA2-WT along with PCAF small interfering RNA (siRNA). Cell lysates from these treatments were prepared and subjected to western blotting with indicated antibodies. (B) KYSE180 cells transfected with the indicated plasmids were subjected to ChIP assays with control IgG or an anti-FLAG antibody. The occupancy of HMGA2 in the promoters of cyclin A was examined by qRT-PCR. Each bar shows the means ± SD (n = 3). (C) KYSE180 cells transfected with the indicated plasmids were subjected to ChIP assays with control IgG or an anti-FLAG antibody. The occupancy of HMGA2 in the promoters of SOX2 was examined by qRT-PCR. Each bar shows the means ± SD (n = 3). (D) Monolayer colony-formation assay of KYSE180 cells transfected with the indicated plasmids. (E) Colony forming efficiency of KYSE180 cells expressing the indicated vectors. Each bar shows the means ± SD (n = 3). (F) HMGA2 K26 acetylation was assessed by immunohistochemical staining in 10 normal esophageal tissues and 124 ESCC tissues from human ESCC tissue microarrays. Scale bars, 50 μm. (G) The acetylation level of HMGA2-K26 was analyzed based on pathological grade. Statistically significant differences are indicated: **P < 0.01, and ***P < 0.001; Student's t-test.
We next investigated the effect of HMGA2 K26 acetylation on ESCC cell proliferation. The expression of HMGA2-K26Q obviously increased its ability in promoting the proliferation of KYSE180 cells compared to HMGA2-WT (Figure 5D and E). However, compared to the HMGA2-WT, the cell proliferation of HMGA2-K26R-expressed group was significantly decreased (Figure 5D and E). Moreover, cells ectopically expressing HMGA2-WT with PCAF overexpression in order to increase the acetylation of HMGA2 showed a similar effect to HMGA2-K26Q in promoting the cell proliferation. However, knockdown of PCAF decreased the ability of HMGA2-WT in promoting the proliferation of KYSE180 cells (Figure 5D and E). These results demonstrate that the acetylation at K26 enhances the ability of HMGA2 in promoting ESCC cell proliferation.
To further show the clinical relevance of HMGA2 K26 acetylation, we examined the levels of HMGA2 K26 acetylation in two human ESCC tissue microarrays (Supplementary Tables S5 and S6) containing 10 normal esophageal tissues and 124 ESCC tissues by immunohistochemistry staining analysis using the anti-AcK26-HMGA2 specific antibody. Importantly, we found that the levels of HMGA2 K26 acetylation were significantly higher in ESCC tissues than that in normal esophageal tissues and HMGA2 K26 acetylation was gradually increased along with the cancer pathological grade (Figure 5F and G). These findings indicate that elevated HMGA2 K26 acetylation may predict a poor outcome for ESCC patients.
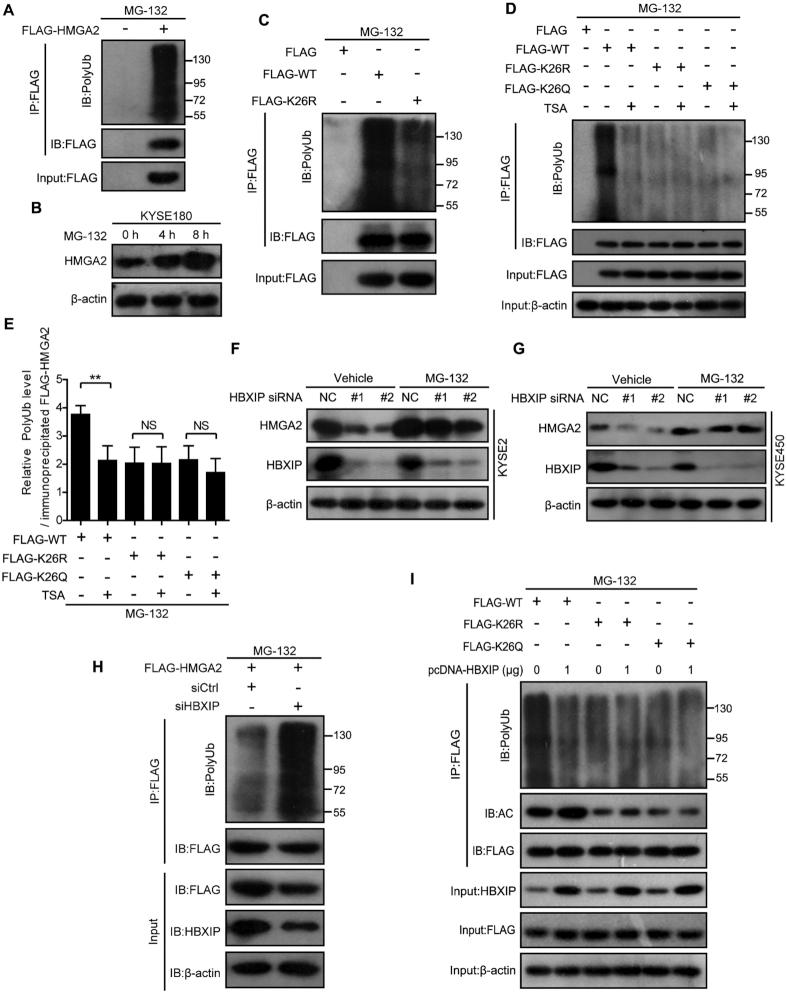
HMGA2 acetylation by HBXIP inhibits its ubiquitination to stabilize HMGA2
The acetylation of specific lysines can increase the stability of a protein through preventing ubiquitination of the same lysine residues (47,54,55). To probe the potential mechanism by which HBXIP-mediated acetylation regulates HMGA2 protein stability, we first examined HMGA2 ubiquitination and found that it was actively ubiquitinated in HEK293T cells (Figure 6A). Moreover, HMGA2 protein was accumulated in cells treated with the proteasome inhibitor MG-132 at different time points, indicating that HMGA2 stability could be regulated by the ubiquitin–proteasome pathway (Figure 6B). According to PhosphoSitePlus (http://www.phosphosite.org/homeAction.action), there existed an ubiquitination modification of HMGA2 at K26. Therefore, we assessed the ubiquitination level of the K26R HMGA2 mutant in cells. Interestingly, the K26R mutation dramatically reduced HMGA2 ubiquitination, indicating that K26 might also be the target residue for ubiquitination (Figure 6C). Importantly, the inhibition of deacetylases with TSA decreased the ubiquitination of WT but not of the K26R or K26Q HMGA2 mutants (Figure 6D and E). These results indicate a competitive interaction between the acetylation and ubiquitination of HMGA2 at K26. We subsequently investigated whether HMGA2 ubiquitination was regulated by HBXIP, which stimulates HMGA2 acetylation and stabilizes HMGA2. We found that the proteasome inhibitor MG132 blocked the HBXIP silencing-induced loss of endogenous HMGA2 (Figure 6F and G), and this was associated with the accumulation of polyubiquitinated HMGA2 (Figure 6H). Consistently, HBXIP overexpression upregulated the acetylation and downregulated the ubiquitination of WT HMGA2 but showed no effects on the K26R or K26Q HMGA2 mutants (Figure 6I). These results indicate that the acetylation and ubiquitination of HMGA2 at K26 occur mutually exclusively and that HBXIP-induced acetylation at K26 blocks HMGA2 ubiquitination and proteasomal degradation.
Figure 6.
HMGA2 acetylation by HBXIP inhibits its ubiquitination to stabilize HMGA2. (A) HEK293T cells were transfected with FLAG-HMGA2 and treated with 40 μM proteasome inhibitor MG-132 for 6 h. Exogenous FLAG-HMGA2 was immunoprecipitated with FLAG beads in HEK293T cells, and then the ubiquitylation level of HMGA2 protein in precipitation was tested by western blotting with an anti-ubiquitin antibody. (B) Endogenous HMGA2 level was detected by western blotting in KYSE180 cells treated with or without 40 μM proteasome inhibitor MG-132 for the indicated periods. (C) Exogenous FLAG-tagged WT and K26R mutant HMGA2 were immunoprecipitated with FLAG beads in HEK293T cells pretreated with 40 μM proteasome inhibitor MG-132 for 6 h, and then the ubiquitylation level of HMGA2 protein in precipitation was tested by western blotting with an anti-ubiquitin antibody. (D) Exogenous FLAG-tagged WT, K26R, or K26Q mutant HMGA2 was immunoprecipitated with FLAG beads in HEK293T cells, and then the ubiquitylation level of HMGA2 protein in precipitation was tested by Western blotting with an anti-ubiquitin antibody. The cells were treated with or without TSA for 12 h and then treated with 40 μM proteasome inhibitor MG-132 for additional 6 h. (E) The quantification of the polyubiquitination intensity relative to immunoprecipitated FLAG-HMGA2 in Figure 6D. Each bar shows the means ± SD (n = 3). (F and G) Endogenous HMGA2 level was detected by western blotting in KYSE2 cells (F) and KYSE450 cells (G) transfected with control siRNA (siCtrl) or HBXIP siRNA (siHBXIP) for 40 h and then treated with 40 μM proteasome inhibitor MG-132 for additional 6 h. (H) KYSE2 cells were transfected with control siRNA (siCtrl) or HBXIP siRNA (siHBXIP) for 40 h and then treated with 40 μM proteasome inhibitor MG-132 for additional 6 h. Exogenous FLAG-HMGA2 was immunoprecipitated with FLAG beads from cell lysates, and then the ubiquitylation level of HMGA2 protein in precipitation was tested by western blotting with an anti-ubiquitin antibody. (I) Exogenous FLAG-tagged WT, K26R or K26Q mutant HMGA2 were immunoprecipitated with FLAG beads in HEK293T cells transfected with or without pcDNA-HBXIP plasmids, and then the ubiquitylation level and acetylation level of HMGA2 protein in precipitation were tested by western blotting with anti-ubiquitin and anti-acetylated lysine antibodies, respectively. All experiments were repeated at least three times. Statistically significant differences are indicated: **P < 0.01; NS, not significant; Student's t-test.
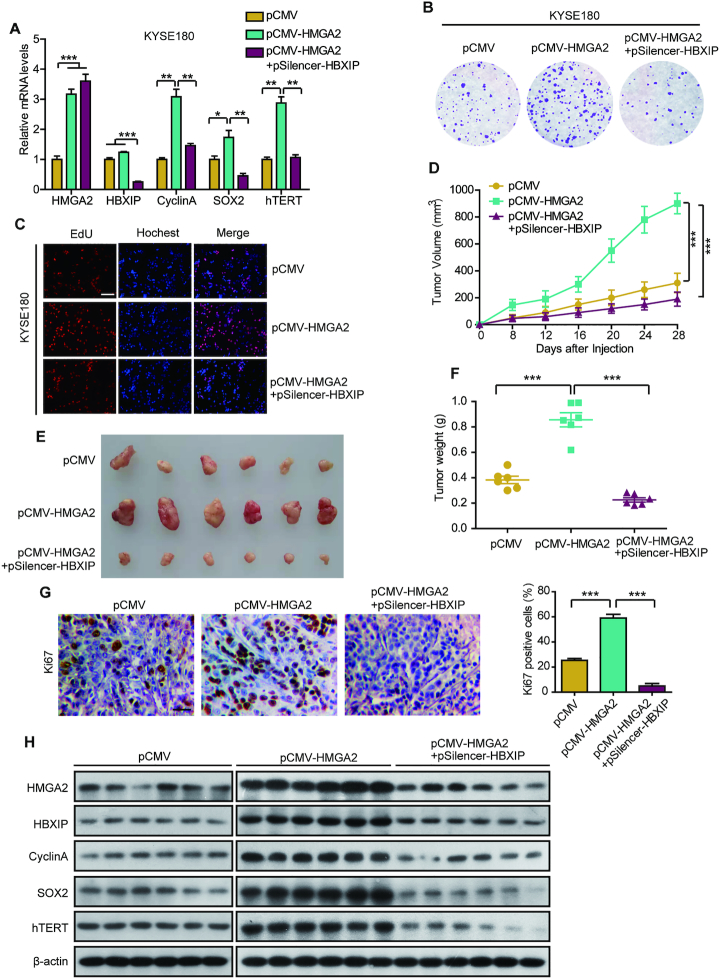
Stable accumulation of oncogenic HMGA2 mediated by HBXIP promotes its target gene expression and ESCC growth
Previous studies have shown that HMGA2 can modulate the transcription of cyclin A, SOX2, and hTERT to promote tumorigenesis (13,14,56). As expected, shRNA-mediated HBXIP silencing disrupted the HMGA2-enhanced cyclin A, SOX2 and hTERT expression at the mRNA level in KYSE180 and KYSE510 cells (Figure 7A and Supplementary Figure S5A), suggesting that HBXIP contributes to HMGA2-mediated transcription. We then investigated the effect of HMGA2 accumulation on ESCC growth in vitro and in vivo. EdU and colony formation assays revealed that ectopic HMGA2 expression significantly promoted KYSE180 and KYSE510 cell proliferation. However, this promotion was dramatically abrogated by HBXIP knockdown in the cells (Figure 7B, C and Supplementary Figure S5B–G). Moreover, the growth of KYSE180 xenografts in mice was markedly reinforced by HMGA2 overexpression, while this stimulation of tumor xenograft growth was largely abrogated by HBXIP knockdown (Figure 7D–F). Additionally, no difference was observed in the baseline weight of the mice, indicating that the mice remained healthy throughout the above treatment schedule (Supplementary Figure S5H). Immunohistochemical staining displayed that compared with the vector group, Ki67 as a marker of cell proliferation was more induced in the HMGA2 overexpressed group and then HBXIP knockdown reduced the HMGA2-elevated expression of Ki67, in the tumor tissues from mice (Figure 7G). Interestingly, the expression of HMGA2 target genes (cyclin A, SOX2 and hTERT) was significantly reduced upon HBXIP silencing in HMGA2-overexpressing tumor tissues from mice (Figure 7H). Collectively, our findings indicate that the HBXIP-induced stabilization of oncogenic HMGA2 activates target gene expression and therefore promotes ESCC growth in vitro and in vivo.
Figure 7.
Stable accumulation of oncogenic HMGA2 mediated by HBXIP promotes target gene expression and ESCC growth. (A) Real-time PCR analysis of HMGA2, HBXIP, cyclin A, SOX2, and hTERT mRNA levels in KYSE180 cells separately transfected with pCMV vectors, pCMV-HMGA2 vectors, or cotransfected with pCMV-HMGA2 and pSilencer-HBXIP vectors. Each bar shows the means ± SD (n = 3). (B) Monolayer colony-formation assay of KYSE180 cells transfected with the indicated plasmids. (C) EdU incorporation assays were used to assess EdU-positive cells among KYSE180 cells expressing the indicated vectors. Scale bars, 100 μm. (D) The curves of tumor growth in nude mice transplanted with KYSE180 cells is shown. Each bar shows the means ± SD (each group, n = 6). (E) Imaging of the tumors derived from nude mice transplanted with KYSE180 cells pretreated with indicated plasmids. (F) Weights of tumors in each group were shown. Each bar shows the means ± SD (each group, n = 6). (G) The expression levels of Ki67 and the statistics of Ki67-positive cells from above tumor tissues were examined by immunohistochemical assay. Scale bars, 50 μm. Each bar shows the means ± SD (n = 3). (H) The levels of HMGA2, HBXIP, CyclinA, SOX2 and hTERT from above tumor tissues were detected by western blotting. All experiments were repeated at least three times. Statistically significant differences are indicated: *P < 0.05, **P < 0.01 and ***P < 0.001; Student's t-test.
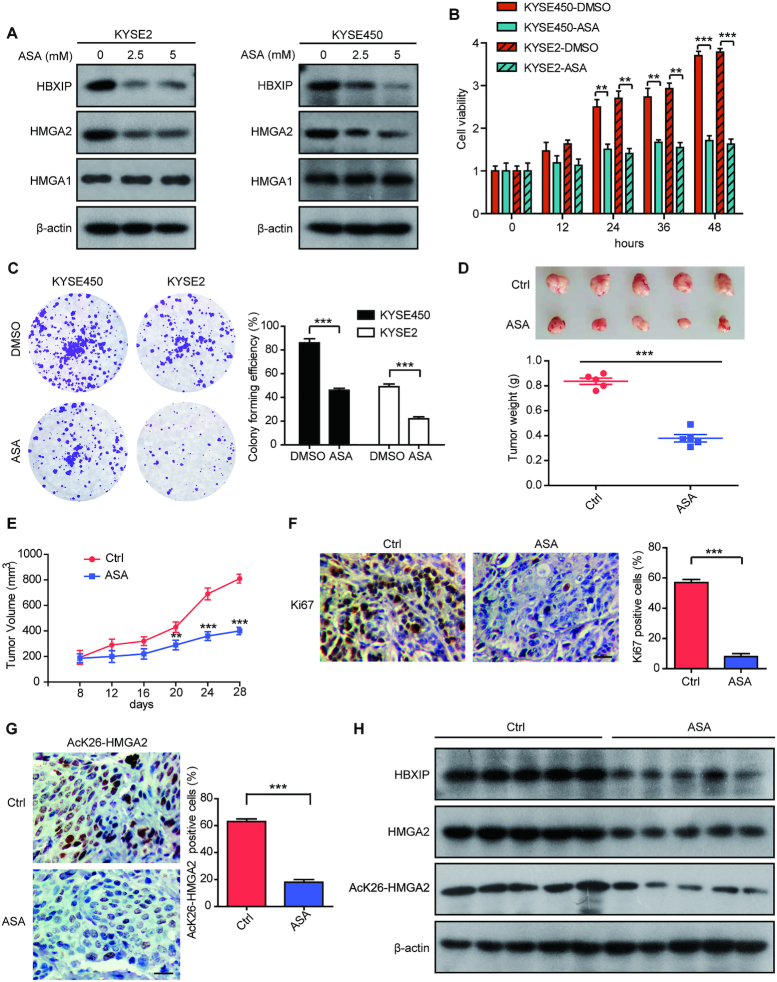
Aspirin attenuates ESCC growth by suppressing both HBXIP and HMGA2
Accumulative evidence reveals that the classic anti-inflammatory drug, aspirin can exert inhibitory effects on multiple cancers (35–37). We have recently reported that aspirin can suppress HBXIP expression to restraint tamoxifen resistance in breast cancer (31). Here, we are interested in whether aspirin can inhibit ESCC growth by targeting HBXIP or HMGA2. We treated KYSE2 and KYSE450 cells with aspirin and the results showed that aspirin markedly decreased HBXIP and HMGA2 levels in a dose-dependent manner (Figure 8A). We also evaluated the effect of aspirin on the protein levels of HMGA1, the other member of the HMGA family. The results showed that the protein levels of HMGA1 were not affected by aspirin (Figure 8A). Next, we explored the role of aspirin in ESCC growth in vitro and in vivo. MTT and colony formation assays revealed that aspirin treatment remarkably inhibited the proliferation of KYSE2 cells and KYSE450 cells (Figure 8B and C). Importantly, xenograft mice subcutaneously injected with KYSE2 cells demonstrated that administration with aspirin significantly restrained tumor growth in vivo (Figure 8D and E). Moreover, we found that the Ki67 staining dramatically weakened in aspirin-treated group (Figure 8F). Using specific AcK26-HMGA2 antibody, we tested the acetylation levels of HMGA2 at K26 in tumor samples from aspirin-treated or control mice via immunohistochemical staining and western blotting. Our data displayed that the acetylation levels of HMGA2 were dramatically decreased in the aspirin-treated group compared to controls (Figure 8G and H), indicating that aspirin treatment is able to suppress HBXIP-mediated HMGA2 acetylation at K26. Furthermore, western blotting analysis demonstrated that the expression of HBXIP and HMGA2 was dampened in mice tumor samples treated with aspirin (Figure 8H). Collectively, aspirin can decrease HBXIP and HMGA2 expression to restrict ESCC growth.
Figure 8.
Aspirin attenuates ESCC growth by suppressing both HBXIP and HMGA2. (A) KYSE2 and KYSE450 cells were treated with different concentrations of aspirin (ASA) for 24 h. Endogenous HBXIP, HMGA2 and HMGA1 levels were examined by western blotting. (B) MTT assays with KYSE2 and KYSE450 cells separately treated with DMSO or ASA (2.5 mM) for the indicated periods. Each bar shows the means ± SD (n = 3). (C) Colony photograph and colony forming efficiency of KYSE2 and KYSE450 cells separately treated with DMSO or ASA (2.5 mM). Each bar shows the means ± SD (n = 3). (D) The imaging and the weights of tumors from saline- or ASA-treated groups were shown. Each bar shows the means ± SD (each group, n = 5). (E) The nude mice were subcutaneously injected with KYSE2 cells. After the tumour size exceeded 100 mm3, the mice were daily oral administrated with saline as the control or ASA (75mg/kg/day). The xenograft tumors were monitored for 4 weeks. The tumor growth curve of mice was shown. Each bar shows the means ± SD (each group, n = 5). (F) The expression levels of Ki67 and the statistics of Ki67-positive cells from above tumor tissues were examined by immunohistochemical assay. Scale bars, 50 μm. Each bar shows the means ± SD (n = 3). (G) The levels of HMGA2 K26 acetylation and the statistics of AcK26-HMGA2-positive cells from above tumor tissues were examined by immunohistochemical assay. Scale bars, 50 μm. Each bar shows the means ± SD (n = 3). (H) The levels of HMGA2, HBXIP and HMGA2 K26 acetylation from above tumor tissues were detected by western blotting. All experiments were repeated at least three times. Statistically significant differences are indicated: **P < 0.01, and ***P < 0.001; Student's t-test.
DISCUSSION
HMGA2 is associated with aggressive tumor growth, early metastasis, and poor prognosis in various cancers through the modulation of the transcription of numerous genes (5–7,44). HMGA2 overexpression is common in many cancers, however, the molecular mechanisms of HMGA2 overexpression are yet to be determined. Thus, we aim to explore the function of HMGA2 and its regulatory mechanisms in ESCC. Multitude of evidence has indicated that the levels of HBXIP are elevated in several malignancies and that HBXIP acts as an oncogenic transcriptional coactivator in controlling cell growth, proliferation, gluconeogenesis or aberrant lipid metabolism (26,28,50). Our study has revealed that HBXIP as an important oncoprotein can regulate PTMs of some transcription factors. HBXIP can enhance the acetylation of transcription factor HOXB13 to prevent HOXB13 degradation in the promotion of tamoxifen resistance of breast cancer (31). In addition, HBXIP is able to induce c-Fos phosphorylation through p-ERK1/2 to facilitate the nuclear import of c-Fos in breast cancer (32). Meanwhile, increased HBXIP is correlated with an aggressive ESCC phenotype and a poor prognosis (33). Therefore, we first investigated whether HBXIP was involved in HMGA2 regulation to promote malignant phenotype of ESCC. Interestingly, we observed a positive correlation between HMGA2 and HBXIP expression in clinical ESCC tissues and ESCC cell lines. Moreover, high HBXIP and HMGA2 levels were associated with advanced tumor stage and poor overall and progression-free survival. Furthermore, HBXIP upregulated the protein levels of HMGA2 but not its mRNA levels in ESCC cell lines and restrained HMGA2 degradation in the presence of CHX, a protein synthesis inhibitor. These data suggest that HBXIP can enhance HMGA2 at the posttranslational level.
Apparently, lysine acetylation involving histone and non-histone acetylation regulates protein stability and function. Histone acetylation primarily regulates gene transcription, whereas non-histone acetylation regulates diverse biological functions, including DNA damage response, energy metabolism, and cytoskeleton dynamics (47,57). For instance, replication protein A acetylation mainly stabilizes XPA and NER downstream factors at ultraviolet damage sites to promote nucleotide excision repair (49). Acetylation modifications stimulated lactate dehydrogenase A (LDHA) to undergo degradation leading to weakened glycolysis and lactate production (48). Additionally, α-tubulin acetylation promotes microtubule stabilization, resulting in persistent directional movement (58). In the present study, we discovered that HBXIP stimulated HMGA2 acetylation modification and HBXIP-mediated acetylation was able to enhance the stability of HMGA2 protein in ESCC cell lines. Furthermore, using mutagenic analysis, the primary HMGA2 acetylation site was mapped to the conserved K26 residue. Thus, we expose that HBXIP augments HMGA2 protein levels by stimulating its acetylation at K26. Our data showed that HMGA2 stability became increasingly evident following treatment with the histone deacetylase inhibitor TSA. Then, we discovered that HMGA2 served as a novel substrate of acetylase PCAF, which acetylates various non-histone proteins, including replication protein A, p53, and transcription factor KLF13 (49,59,60). PCAF can acetylate HMGA2, leading to HMGA2 upregulation, and K26 is required for this event. Unexpectedly, HBXIP could activate PCAF through promoting its phosphorylation via the Akt pathway, sequentially enhancing the acetylation level of HMGA2. Future studies are warranted to determine how Akt regulates PCAF activation upon HBXIP stimulation.
Posttranslational modifications of HMGA2 have been shown to have a profound effect on its biological function. For example, the phosphorylation of the HMGA2 by Cdc2 kinase at Ser-43 and Ser-58 changes its conformation and weakens the strength of binding to the β-interferon promoter and phosphorylation of HMGA2 by Nek2 is essential for chromatin condensation in meiosis (4,61). In addition, SUMOylation of HMGA2 plays a critical role in stimulating HMGA2 function in decreasing PML protein level (21). As report here we found that HMGA2 K26 acetylation increased the binding capacity of HMGA2 to its target genes including cyclin A and SOX2. It is known that HMGA2 functions as a tumor promoting gene in cancer progression (5,44). Consistent with above observation, overexpression of HMGA2-WT increases proliferation of the human ESCC KYSE180 cells. Interestingly, HMGA2 K26 acetylation enhances KYSE180 cells proliferation compared to HMGA2-WT. Furthermore, the levels of HMGA2 acetylation at K26 are increased in ESCC tissues compared to normal esophageal tissues and HMGA2 K26 acetylation is gradually increased along with the cancer pathological grade, indicating the status of HMGA2 acetylation is of prognostic value for ESCC patients.
In the next investigation, we explored the underlying mechanism responsible for acetylation-mediated HMGA2 stability. Importantly, we observed that HMGA2 underwent ubiquitin–proteasome degradation pathway. Several studies have shown that prior lysine acetylation can affect subsequent protein ubiquitination and prevent proteasomal degradation (54). Interestingly, both acetylation and ubiquitination modify the ϵ-amino group of the substrate lysine residues. Moreover, several identified acetylation lysine residues are also potential ubiquitination sites (47). It has been reported that a Lys→Gln mutation can mimic the acetylated state of lysine (K) mainly because of the structure similarity between the Gln (Q) residue and the acetylated-lysine residue and a Lys→Arg mutation eliminates the ϵ-amino group of the lysine residues to mimic the acetylation-deficient state (55). In the present study, we found that the acetylation-mimetic K26Q and acetylation-deficient K26R mutants were more stable than WT HMGA2 following CHX treatment; a possible explanation is that there might be a competition between polyubiquitination and acetylation at the same lysine 26 residue in HMGA2 stability regulation. In terms of the K26Q mutants which mimic acetylated HMGA2 at K26, the ubiquitination of HMGA2 at K26 will never happen. The acetylation-deficient K26R mutants lost the ϵ-amino group of the lysine 26 residue for ubiquitination, therefore preventing ubiquitination and ubiquitination-dependent proteasomal degradation and then leading to HMGA2 protein stability. Accordingly, both HMGA2 K26Q and HMGA2 K26R are stable.
Consistently, our results reveal a previously unknown mechanism of HMGA2 stability that involves increased HMGA2 acetylation mediated via HBXIP, which competitively inhibits its ubiquitination at the same lysine residue and proteasomal degradation, resulting in HMGA2 accumulation and augmented HMGA2 target gene expression. Future research will be focused on the polyubiquitination machinery of HMGA2 at the K26 residue. Functionally, HBXIP-mediated HMGA2 stability promoted ESCC cell growth both in vitro and in vivo. More importantly, our findings revealed a crosstalk between acetylation and ubiquitination competing for the same lysine residue in the regulation of HMGA2 stability and tumor growth in response to HBXIP. Therapeutically, the status of HMGA2 acetylation is of prognostic significance for ESCC patients, and the potential drug-induced inhibition of HMGA2 acetylation merits its exploration as a target in ESCC.
In recent years, the classic anti-inflammation drug aspirin has reportedly acted as an anti-tumour agent in various cancers, including colorectal cancer, breast cancer and other cancers (34,36,37). Some epidemiologic studies have proven that the use of aspirin and other nonsteroidal anti-inflammatory drugs protects against the development of esophageal cancer (38,39). We recently have revealed that aspirin can target HBXIP to inhibit HBXIP/HOXB13 axis, overcoming tamoxifen resistance in breast cancer (31). Accordingly, in further study we attempted to evaluate the function of aspirin in the development of ESCC. We assessed HBXIP and HMGA2 expression in aspirin-treated ESCC cells. Interestingly, we found that the expression of HBXIP and HMGA2 were dramatically impaired by aspirin treatment. Moreover, aspirin could retard the growth of ESCC in vitro and in vivo. Western blotting and IHC assays revealed decreases in HBXIP and HMGA2 expression in the mouse tumor with aspirin treatment, which supported that aspirin could sabotage ESCC growth by targeting HBXIP and HMGA2. However, the mechanism underlying how aspirin limits HBXIP and HMGA2 expression remains to be addressed in future studies. Our results uncovered the therapeutic potential of aspirin for ESCC with high HBXIP and HMGA2 expression.
In conclusion, we identified a novel mechanism underlying HBXIP-mediated regulation of HMGA2 protein stability in ESCC and a novel HMGA2 modification (acetylation) (Supplementary Figure S6). HBXIP can prevent ubiquitination-dependent degradation of HMGA2 by increasing its acetylation. HMGA2 is acetylated at the K26 residue. The oncoprotein HBXIP activates PCAF via the Akt pathway, resulting in HMGA2 acetylation and HMGA2 protein accumulation in ESCC cells. Moreover, HMGA2 K26 acetylation increased the binding capacity of HMGA2 to its target genes. Furthermore, HMGA2 acetylation at K26 prevented subsequent ubiquitination at the same residue, thereby blocking its proteasomal degradation; this finding indicates that the competition between acetylation and ubiquitination controls HMGA2 stability. Finally, the stability of oncogenic HMGA2 facilitates ESCC cell growth. Aspirin ameliorates ESCC tumor growth through blocking HBXIP and HMGA2. Our findings provide new insights into the mechanism of HMGA2 regulation in ESCC. Therapeutically, aspirin can act as a promising tool for ESCC therapy via inhibiting HBXIP and HMGA2.
Supplementary Material
ACKNOWLEDGEMENTS
Authors’ contributions: Y. Wu, W. Zhang and L. Ye conceived and designed the study. Y. Wu designed the research methods. Y. Wu and X. Wang conducted the experiments, collected and analyzed the data. F. Xu, L. Zhang, T. Wang, X. Fu, and T. Jin participated in the experiments. Y. Wu wrote the manuscript. Y. Wu, W. Zhang and L. Ye designed and revised the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China (973 Program) [2015CB553905]; National Natural Scientific Foundation of China [31670771, 31870752]; Fundamental Research Funds for the Central Universities; Ph.D. Candidate Research Innovation Fund of Nankai University; Project of Prevention and Control of Key Chronic Non-Infectious Diseases [2016YFC1303401]. Funding for open access charge: National Natural Scientific Foundation of China [31670771, 31870752].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F.. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015; 136:E359–E386. [DOI] [PubMed] [Google Scholar]
- 2. Smyth E.C., Lagergren J., Fitzgerald R.C., Lordick F., Shah M.A., Lagergren P., Cunningham D.. Oesophageal cancer. Nat. Rev. Dis. Primers. 2017; 3:17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Napier K.J., Scheerer M., Misra S.. Esophageal cancer: a review of epidemiology, pathogenesis, staging workup and treatment modalities. World. J. Gastrointest. Oncol. 2014; 6:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sgarra R., Rustighi A., Tessari M.A., Di Bernardo J., Altamura S., Fusco A., Manfioletti G., Giancotti V.. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett. 2004; 574:1–8. [DOI] [PubMed] [Google Scholar]
- 5. Fedele M., Visone R., De Martino I., Troncone G., Palmieri D., Battista S., Ciarmiello A., Pallante P., Arra C., Melillo R.M. et al.. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006; 9:459–471. [DOI] [PubMed] [Google Scholar]
- 6. Langelotz C., Schmid P., Jakob C., Heider U., Wernecke K.D., Possinger K., Sezer O.. Expression of high-mobility-group-protein HMGI-C mRNA in the peripheral blood is an independent poor prognostic indicator for survival in metastatic breast cancer. Br. J. Cancer. 2003; 88:1406–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang X., Liu X., Li A.Y., Chen L., Lai L., Lin H.H., Hu S., Yao L., Peng J., Loera S. et al.. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin. Cancer Res. 2011; 17:2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sarhadi V.K., Wikman H., Salmenkivi K., Kuosma E., Sioris T., Salo J., Karjalainen A., Knuutila S., Anttila S.. Increased expression of high mobility group A proteins in lung cancer. J. Pathol. 2006; 209:206–212. [DOI] [PubMed] [Google Scholar]
- 9. Summer H., Li O., Bao Q., Zhan L., Peter S., Sathiyanathan P., Henderson D., Klonisch T., Goodman S.D., Droge P.. HMGA2 exhibits dRP/AP site cleavage activity and protects cancer cells from DNA-damage-induced cytotoxicity during chemotherapy. Nucleic Acids Res. 2009; 37:4371–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nishino J., Kim I., Chada K., Morrison S.J.. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008; 135:227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng Y., Laser J., Shi G., Mittal K., Melamed J., Lee P., Wei J.J.. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol. Cancer Res. 2008; 6:663–673. [DOI] [PubMed] [Google Scholar]
- 12. Shell S., Park S.M., Radjabi A.R., Schickel R., Kistner E.O., Jewell D.A., Feig C., Lengyel E., Peter M.E.. Let-7 expression defines two differentiation stages of cancer. PNAS. 2007; 104:11400–11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tessari M.A., Gostissa M., Altamura S., Sgarra R., Rustighi A., Salvagno C., Caretti G., Imbriano C., Mantovani R., Del Sal G. et al.. Transcriptional activation of the cyclin A gene by the architectural transcription factor HMGA2. Mol. Cell. Biol. 2003; 23:9104–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li A.Y., Lin H.H., Kuo C.Y., Shih H.M., Wang C.C., Yen Y., Ann D.K.. High-mobility group A2 protein modulates hTERT transcription to promote tumorigenesis. Mol. Cell. Biol. 2011; 31:2605–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y., Zhao Z., Xu C., Zhou Z., Zhu Z., You T.. HMGA2 induces transcription factor Slug expression to promote epithelial-to-mesenchymal transition and contributes to colon cancer progression. Cancer Lett. 2014; 355:130–140. [DOI] [PubMed] [Google Scholar]
- 16. Thuault S., Tan E.J., Peinado H., Cano A., Heldin C.H., Moustakas A.. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J. Biol. Chem. 2008; 283:33437–33446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yun J., Frankenberger C.A., Kuo W.L., Boelens M.C., Eves E.M., Cheng N., Liang H., Li W.H., Ishwaran H., Minn A.J. et al.. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011; 30:4500–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wend P., Runke S., Wend K., Anchondo B., Yesayan M., Jardon M., Hardie N., Loddenkemper C., Ulasov I., Lesniak M.S. et al.. WNT10B/beta-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol. Med. 2013; 5:264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahmed K.M., Tsai C.Y., Lee W.H.. Derepression of HMGA2 via removal of ZBRK1/BRCA1/CtIP complex enhances mammary tumorigenesis. J. Biol. Chem. 2010; 285:4464–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sgarra R., Maurizio E., Zammitti S., Lo Sardo A., Giancotti V., Manfioletti G.. Macroscopic differences in HMGA oncoproteins post-translational modifications: C-terminal phosphorylation of HMGA2 affects its DNA binding properties. J. Proteome Res. 2009; 8:2978–2989. [DOI] [PubMed] [Google Scholar]
- 21. Cao X., Clavijo C., Li X., Lin H.H., Chen Y., Shih H.M., Ann D.K.. SUMOylation of HMGA2: selective destabilization of promyelocytic leukemia protein via proteasome. Mol. Cancer Ther. 2008; 7:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M.. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012; 150:1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Melegari M., Scaglioni P.P., Wands J.R.. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J. Virol. 1998; 72:1737–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fujii R., Zhu C., Wen Y., Marusawa H., Bailly-Maitre B., Matsuzawa S., Zhang H., Kim Y., Bennett C.F., Jiang W. et al.. HBXIP, cellular target of hepatitis B virus oncoprotein, is a regulator of centrosome dynamics and cytokinesis. Cancer Res. 2006; 66:9099–9107. [DOI] [PubMed] [Google Scholar]
- 25. Marusawa H., Matsuzawa S., Welsh K., Zou H., Armstrong R., Tamm I., Reed J.C.. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003; 22:2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yue L., Li L., Liu F., Hu N., Zhang W., Bai X., Li Y., Zhang Y., Fu L., Zhang X. et al.. The oncoprotein HBXIP activates transcriptional coregulatory protein LMO4 via Sp1 to promote proliferation of breast cancer cells. Carcinogenesis. 2013; 34:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y., Wang Z., Shi H., Li H., Li L., Fang R., Cai X., Liu B., Zhang X., Ye L.. HBXIP and LSD1 Scaffolded by lncRNA Hotair Mediate Transcriptional Activation by c-Myc. Cancer Res. 2016; 76:293–304. [DOI] [PubMed] [Google Scholar]
- 28. Zhao Y., Li H., Zhang Y., Li L., Fang R., Li Y., Liu Q., Zhang W., Qiu L., Liu F. et al.. Oncoprotein HBXIP modulates abnormal lipid metabolism and growth of breast cancer cells by activating the LXRs/SREBP-1c/FAS signaling cascade. Cancer Res. 2016; 76:4696–4707. [DOI] [PubMed] [Google Scholar]
- 29. Wang Y., Cui M., Cai X., Sun B., Liu F., Zhang X., Ye L.. The oncoprotein HBXIP up-regulates SCG3 through modulating E2F1 and miR-509-3p in hepatoma cells. Cancer Lett. 2014; 352:169–178. [DOI] [PubMed] [Google Scholar]
- 30. Li L., Liu B., Zhang X., Ye L.. The oncoprotein HBXIP promotes migration of breast cancer cells via GCN5-mediated microtubule acetylation. Biochem. Biophys. Res. Commun. 2015; 458:720–725. [DOI] [PubMed] [Google Scholar]
- 31. Liu B., Wang T., Wang H., Zhang L., Xu F., Fang R., Li L., Cai X., Wu Y., Zhang W. et al.. Oncoprotein HBXIP enhances HOXB13 acetylation and co-activates HOXB13 to confer tamoxifen resistance in breast cancer. J. Hematol. Oncol. 2018; 11:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Zhao Y., Li H., Li Y., Cai X., Shen Y., Shi H., Li L., Liu Q., Zhang X. et al.. The nuclear import of oncoprotein hepatitis B X-interacting protein depends on interacting with c-Fos and phosphorylation of both proteins in breast cancer cells. J. Biol. Chem. 2013; 288:18961–18974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xia H., Ma L., Li J., Bai H., Wang D.. Elevated HBXIP expression is associated with aggressive phenotype and poor prognosis in esophageal squamous cell carcinoma. Am. J. Cancer Res. 2017; 7:2190–2198. [PMC free article] [PubMed] [Google Scholar]
- 34. Drew D.A., Cao Y., Chan A.T.. Aspirin and colorectal cancer: the promise of precision chemoprevention. Nat. Rev. Cancer. 2016; 16:173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thun M.J., Jacobs E.J., Patrono C.. The role of aspirin in cancer prevention. Nat. Rev. Clin. Oncol. 2012; 9:259–267. [DOI] [PubMed] [Google Scholar]
- 36. Chen W.Y., Holmes M.D.. Role of aspirin in breast cancer survival. Curr. Oncol. Rep. 2017; 19:48. [DOI] [PubMed] [Google Scholar]
- 37. Sitia G., Aiolfi R., Di Lucia P., Mainetti M., Fiocchi A., Mingozzi F., Esposito A., Ruggeri Z.M., Chisari F.V., Iannacone M. et al.. Antiplatelet therapy prevents hepatocellular carcinoma and improves survival in a mouse model of chronic hepatitis B. PNAS. 2012; 109:E2165–E2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Corley D.A., Kerlikowske K., Verma R., Buffler P.. Protective association of aspirin/NSAIDs and esophageal cancer: a systematic review and meta-analysis. Gastroenterology. 2003; 124:47–56. [DOI] [PubMed] [Google Scholar]
- 39. Liao L.M., Vaughan T.L., Corley D.A., Cook M.B., Casson A.G., Kamangar F., Abnet C.C., Risch H.A., Giffen C., Freedman N.D. et al.. Nonsteroidal anti-inflammatory drug use reduces risk of adenocarcinomas of the esophagus and esophagogastric junction in a pooled analysis. Gastroenterology. 2012; 142:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu N., Zhang J., Cui W., Kong G., Zhang S., Yue L., Bai X., Zhang Z., Zhang W., Zhang X. et al.. miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J. Biol. Chem. 2011; 286:13714–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shen W., Wang C., Xia L., Fan C., Dong H., Deckelbaum R.J., Qi K.. Epigenetic modification of the leptin promoter in diet-induced obese mice and the effects of N-3 polyunsaturated fatty acids. Sci. Rep. 2014; 4:5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johansson L., Bavner A., Thomsen J.S., Farnegardh M., Gustafsson J.A., Treuter E.. The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol. Cell. Biol. 2000; 20:1124–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shan C., Xu F., Zhang S., You J., You X., Qiu L., Zheng J., Ye L., Zhang X.. Hepatitis B virus X protein promotes liver cell proliferation via a positive cascade loop involving arachidonic acid metabolism and p-ERK1/2. Cell Res. 2010; 20:563–575. [DOI] [PubMed] [Google Scholar]
- 44. Fusco A., Fedele M.. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007; 7:899–910. [DOI] [PubMed] [Google Scholar]
- 45. Palumbo A. Jr, Da Costa N.M., Esposito F., De Martino M., D’Angelo D., de Sousa V.P., Martins I., Nasciutti L.E., Fusco A., Ribeiro Pinto L.F.. HMGA2 overexpression plays a critical role in the progression of esophageal squamous carcinoma. Oncotarget. 2016; 7:25872–25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang C., Zhang Z., Chen L., Lee H.W., Ayrapetov M.K., Zhao T.C., Hao Y., Gao J., Yang C., Mehta G.U. et al.. Acetylation within the N- and C-Terminal Domains of Src Regulates Distinct Roles of STAT3-Mediated Tumorigenesis. Cancer Res. 2018; 78:2825–2838. [DOI] [PubMed] [Google Scholar]
- 47. Yang X.J., Seto E.. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. 2008; 31:449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao D., Zou S.W., Liu Y., Zhou X., Mo Y., Wang P., Xu Y.H., Dong B., Xiong Y., Lei Q.Y. et al.. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell. 2013; 23:464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao M., Geng R., Guo X., Yuan R., Zhou X., Zhong Y., Huo Y., Zhou M., Shen Q., Li Y. et al.. PCAF/GCN5-Mediated acetylation of RPA1 promotes nucleotide excision repair. Cell Rep. 2017; 20:1997–2009. [DOI] [PubMed] [Google Scholar]
- 50. Shi H., Fang R., Li Y., Li L., Zhang W., Wang H., Chen F., Zhang S., Zhang X., Ye L.. The oncoprotein HBXIP suppresses gluconeogenesis through modulating PCK1 to enhance the growth of hepatoma cells. Cancer Lett. 2016; 382:147–156. [DOI] [PubMed] [Google Scholar]
- 51. Li Y., Zhang Z., Zhou X., Li L., Liu Q., Wang Z., Bai X., Zhao Y., Shi H., Zhang X. et al.. The oncoprotein HBXIP enhances migration of breast cancer cells through increasing filopodia formation involving MEKK2/ERK1/2/Capn4 signaling. Cancer Lett. 2014; 355:288–296. [DOI] [PubMed] [Google Scholar]
- 52. Cai X., Cao C., Li J., Chen F., Zhang S., Liu B., Zhang W., Zhang X., Ye L.. Inflammatory factor TNF-alpha promotes the growth of breast cancer via the positive feedback loop of TNFR1/NF-kappaB (and/or p38)/p-STAT3/HBXIP/TNFR1. Oncotarget. 2017; 8:58338–58352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chiou G.Y., Chien C.S., Wang M.L., Chen M.T., Yang Y.P., Yu Y.L., Chien Y., Chang Y.C., Shen C.C., Chio C.C. et al.. Epigenetic regulation of the miR142-3p/interleukin-6 circuit in glioblastoma. Mol. Cell. 2013; 52:693–706. [DOI] [PubMed] [Google Scholar]
- 54. Caron C., Boyault C., Khochbin S.. Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays. 2005; 27:408–415. [DOI] [PubMed] [Google Scholar]
- 55. Li M., Luo J., Brooks C.L., Gu W.. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 2002; 277:50607–50611. [DOI] [PubMed] [Google Scholar]
- 56. Chien C.S., Wang M.L., Chu P.Y., Chang Y.L., Liu W.H., Yu C.C., Lan Y.T., Huang P.I., Lee Y.Y., Chen Y.W. et al.. Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a Stem-like State. Cancer Res. 2015; 75:2553–2565. [DOI] [PubMed] [Google Scholar]
- 57. Latham J.A., Dent S.Y.. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 2007; 14:1017–1024. [DOI] [PubMed] [Google Scholar]
- 58. Liu X., Xiao W., Wang X.D., Li Y.F., Han J., Li Y.. The p38-interacting protein (p38IP) regulates G2/M progression by promoting alpha-tubulin acetylation via inhibiting ubiquitination-induced degradation of the acetyltransferase GCN5. J. Biol. Chem. 2013; 288:36648–36661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu L., Scolnick D.M., Trievel R.C., Zhang H.B., Marmorstein R., Halazonetis T.D., Berger S.L.. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999; 19:1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Song C.Z., Keller K., Chen Y., Stamatoyannopoulos G.. Functional interplay between CBP and PCAF in acetylation and regulation of transcription factor KLF13 activity. J. Mol. Biol. 2003; 329:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwanbeck R., Manfioletti G., Wisniewski J.R.. Architecture of high mobility group protein I-C.DNA complex and its perturbation upon phosphorylation by Cdc2 kinase. J. Biol. Chem. 2000; 275:1793–1801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.