Abstract
Cellular stress causes multifaceted reactions to trigger adaptive responses to environmental cues at all levels of the gene expression pathway. RNA-binding proteins (RBP) are key contributors to stress-induced regulation of RNA fate and function. Here, we uncover the plasticity of the RNA interactome in stressed cells, differentiating between responses in the nucleus and in the cytoplasm. We applied enhanced RNA interactome capture (eRIC) analysis preceded by nucleo-cytoplasmic fractionation following arsenite-induced oxidative stress. The data reveal unexpectedly compartmentalized RNA interactomes and their responses to stress, including differential responses of RBPs in the nucleus versus the cytoplasm, which would have been missed by whole cell analyses.
INTRODUCTION
RNA-binding proteins (RBP) are key effectors of biological responses and tightly interconnected with RNAs throughout their life cycles. The collective of RNA-binding proteins of a cell, the RNA interactome, thus plays a central role in RNA function at all levels from transcription, processing, transport, translation, and turnover, and defines a central regulatory layer of cell biology. The interactions of RBPs with their target RNAs are highly dynamic and enable cell responses to changing environmental conditions such as stress. Stress responses may involve translational silencing or activation by regulated inclusion/exclusion of RBPs and ribonucleoproteins (RNPs) into stress granules (1,2), regulated shuttling of RBPs between subcellular compartments (3,4), or modulation of RNA processing and post-transcriptional modifications (5,6). Many stress-induced changes in protein-RNA interactions and their molecular mechanisms have been characterized in earlier studies. Among others, a recent study characterized the stress-induced changes in the total RNA interactome from whole cell lysates (7), while others have focused on specific protein–RNA complexes (8,9). The development of techniques that enable the analysis of global mRNA interactomes in vivo and in cellulo has advanced the field by opening the view into layers of cellular regulation that have previously been challenging to explore. Specifically, hundreds of RBPs with unexpected RNA-binding activity have been identified (10–12), and functional analyses of previously unknown RBPs have revealed the process of riboregulation, i.e. the regulation of protein function by RNA (13,14).
Here, we have used enhanced RNA interactome capture (eRIC) (15) to explore RBP responses to stress, distinguishing between changes within the nuclear and the cytoplasmic cell compartments, and focusing on RBPs targeting polyadenylated mRNAs.
MATERIALS AND METHODS
Experimental model and subject details
HuH7 human hepatocellular carcinoma cells (RRID:CVCL_0336) were grown in low glucose (1 g/l) Dulbeco's modified Eagle's medium (DMEM) (#21885108, Thermo Fisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin and streptomycin. Forty eight hours prior to stress experiments, the cells were seeded on culture plates at a density of 1.1 × 104 cells/cm2. For the stress-experiments the cells were changed into a culture media containing 100 μM arsenite (AsNaO2, #S7400-100G, Sigma Aldrich) for 1 h. For non-stressed controls, normal fresh culture media was used.
Cell fractionation and lysate preparation for interactome capture
The cell fractionation protocol was adapted from (16). In short, cells grown on 500 cm2 Nunc™ Square BioAssay Dishes (#166508, Thermo Fisher Scientific) were washed twice with ice-cold PBS and placed on a cold plate. The cells were subjected to UV-crosslinking at 254 nm (150 mJ/cm2) in a UV Stratalinker 2400 (Stratagene) or left non-crosslinked as controls. For fractionation the cells were scraped on ice directly into 4 ml of ice cold fractionation buffer (20 mM Tris pH 7.5, 10 mM KCl, 0.2% Nonidet P-40) supplemented with cOmplete protease inhibitor cocktail (#04693132001, Sigma Aldrich) per plate, and the lysates from cells subjected to the same conditions were pooled into 50 ml DNA LoBind Tubes (#0030122232, Eppendorf). The crude nuclei were pelleted by centrifugation with 1000 × g for 3 min at +4°C. The supernatant containing the cytoplasmic fraction was transferred to a new tube and snap frozen in liquid nitrogen. The nuclear pellet was subsequently transferred to a 15 ml DNA LoBind Tube (#0030122208, Eppendorf) and washed twice with 12 ml of the fractionation buffer combined with gentle resuspension and low speed centrifugation at 300 × g for 2 min at +4°C. After the last wash, 10 μl of nuclei was taken for analysis of nuclear integrity and efficient removal of ER. The washed nuclear pellet was snap frozen and stored at –80°C until processed by eRIC.
For LNA captures, 20 mg of cytoplasmic fractions and 3.5 mg of nuclear fractions were used. The protein concentrations were measured using the Protein DC Assay (# 5000116, Bio Rad) according to manufacturer's instructions.
The cytoplasmic fractions were adjusted with 5× interactome buffer (20 mM Tris pH 7.5, 2.5M LiCl (#3739.2, Carl Roth GmbH), 5 mM EDTA, 2.5% LiDs (#L9781-5G, Sigma Aldrich), 25 mM DTT, cOmplete) to obtain optimal conditions for LNA-capture.
The nuclear pellets were thawed on ice and resuspended in 3.5 ml DNase buffer (20 mM Tris pH 7.5, 2.5 mM MgCl2, 0.5 mM CaCl2). The suspensions were loaded into tubes containing 500 μl of 0.5 mm zirconia/silica beads (# N034.1, Carl Roth GmbH). The nuclei were disrupted and genomic DNA fragmented with a PreCellys tissue homogenizer combined with Cryolys cooling unit (Bertin Instruments) in 2 × 20 s bursts with a 5 s interval at +4°C. The bottom of the tubes were punctured with a hot needle and the lysates were collected into new tubes by centrifugation (450 × g, +4°C, 5 min).
The nuclear lysates were treated with 1 kU DNase I (#M0303L, New England BioLabs) each for 30 min at +37°C in a rotator mixer. After the treatment, 1 ml of 5× DNase stopping buffer (20 mM Tris pH 7.5, 25 mM EDTA) was added to the lysates. The protein concentration was measured and the lysates were supplemented with 5× interactome buffer as above before proceeding to the LNA-capture.
Analysis of isolated nuclei for nuclear integrity and ER removal
The analysis was performed as described in (16). 10 μl of washed nuclei were mixed with 90 μl PBS, and centrifuged at 500 × g for 1 min. The nuclear pellet was gently resuspended in 10 μl PBS with 1 μM ER Tracker Red (#E34250, Thermo Fisher Scientific) and incubated on ice for 20 min. The sample was mixed with 90 μl 4% (w/v) paraformaldehyde (PFA) in PBS and 5 μl was spotted on a glass slide. The partially air-dried spot was covered with Vectashield HardSet with DAPI mounting medium (#H-1500, Vector Laboratories) and sealed with a coverslip. The samples were imaged with an inverted fluorescent microscope (Olympus iX50 with ColorView 12 CCD-camera). The pseudocolored images were processed and combined in ImageJ (17).
Coupling of LNA probes to carboxylated beads
LNA oligos (/5AmMC6/+TT+TT+TT+TT+TT+TT+TT +TT+TT+TT, #339415, Qiagen) resuspended in nuclease-free water to a final concentration of 100 μM were coupled to carboxylated M-PVA C11 magnetic beads (#CMG-203, PerkinElmer chemagen Technology). All the steps were performed using 2 ml and 15 ml DNA LoBind Tubes (#0030108078 / #0030122208, Eppendorf). The uncoupled beads were washed 3× with 5 volumes of 50 mM MES pH 6 (#M3671-50, Sigma Aldrich). The beads were aliquoted into 2 ml tubes with 200 μl of original bead slurry per tube. The beads were magnetized and the washing buffer was removed. Per sample, 200 μl of 100 μM LNA oligo was mixed with 1 ml of freshly prepared solution of 20 mg/ml N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) (#E7750-1G, Sigma Aldrich) in MES buffer. For coupling, 200 μl of washed and magnetized beads were resuspended in 1200 μl of LNA-EDC-MES solution. The coupling solution was incubated for 5 h in a thermomixer at +50°C and 800 rpm. To remove condensation from the lids, the tubes were occasionally shortly centrifuged and the beads resuspended by vortexing. The coupled beads were washed 2× with 1.5 ml phosphate buffered saline (PBS) and resuspended by soft vortexing. To inactivate any uncoupled carboxyl residues, the beads were resuspended in 1.2 ml of 200 mM ethanolamine pH 8.5 (E9508-500ML, Sigma Aldrich) and incubated for 1 h at +37°C and 800 rpm. The beads were finally washed 3× with 1.5 ml of 1 M NaCl and resuspended in 200 μl of 0.1% PBS–Tween. The coupled beads were stored at +4°C.
Enhanced RNA interactome capture
The interactome capture was performed essentially as described in (15). All steps were performed in low binding tubes (DNA LoBind Tubes, Eppendorf). The samples were incubated for 15 min at +60°C, shortly cooled down on ice, and fresh dithiotreitol (DTT) was added to a final concentration of 5 mM. The samples were centrifuged at 1700 × g for 10 min at +4°C, and transferred into fresh 15 or 50 ml tubes. Aliquots to be used as input samples for full proteome analysis and for quality controls were collected in fresh 1.5 ml tubes (Eppendorf, #0030108501) and snap-frozen in liquid nitrogen. The rest of the samples were subjected to interactome capture. The LNA-coupled beads were washed 3× with 3 volumes of eRIC lysis buffer (20 mM Tris–HCl (pH 7.5), 500 mM LiCl, 1 mM EDTA, 5 mM DTT, 0.5% (w/v) LiDs). To capture protein-RNA complexes, the samples were incubated with 300 μl of washed LNA-coupled beads each for 1 h in a +37°C incubator with gentle rotation. The beads were captured with a magnet and the supernatant was transferred to a fresh tube and stored at +4°C for a second round of capture. The beads were subjected to successive rounds of washes of 5 min each at +37°C with gentle rotation with 8 ml of pre-warmed (+37°C) buffers. The buffers and washes used were as follows: one wash with eRIC lysis buffer, two washes with buffer 1 (20 mM Tris–HCl (pH 7.5), 500 mM LiCl, 1mM EDTA, 5 mM DTT, 0.1% (w/v) LiDs), two washes with buffer 2 (20 mM Tris–HCl (pH 7.5), 500 mM LiCl, 1mM EDTA, 5 mM DTT, 0.02% (v/v) Nonidet P-40), and two washes with buffer 3 (20 mM Tris–HCl (pH 7.5), 200 mM LiCl, 1mM EDTA, 5 mM DTT, 0.02% (v/v) Nonidet P-40). After the last wash, the beads were resuspended in 1 ml of buffer 3 and transferred to a 1.5 ml tube. For pre-elution of unspecific binders, the beads were resuspended in 170 μl of nuclease free water and incubated in a thermomixer for 5 min at +40°C and 800 rpm. 20 μl of the beads were transferred to a fresh tube for heat elution of the captured RNA. The rest (150 μl) were used for RNase elution of captured proteins. For heat elution, the beads were resuspended in fresh nuclease free water and incubated for 10 min at +90°C and 800 rpm. The tubes were centrifuged shortly and the eluate transferred to a fresh tube and snap-frozen in liquid nitrogen. For RNase elution, the beads were resuspended in 150 μl of RNase buffer (10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM DTT, 0.01% Nonidet P-40, 1 μl/ml RNase A (#R4642-10MG, Sigma Aldrich), 1 μl/ml RNase T1 (#R1003-100KU, Sigma Aldrich)) and incubated for 45 min at +37°C and 800 rpm. The eluate was transferred to a new tube and placed again on magnet to remove any remaining beads. Finally, the eluate was again transferred to a fresh tube and snap frozen in liquid nitrogen. The beads from RNase and heat eluates were resuspended in nuclease-free water and combined (beads from different samples were not combined). The beads were resuspended in 300 μl nuclease-free water and incubated for 10 min at +90°C and 800 rpm. Subsequently, the beads were washed 3× with 1 ml of lysis buffer and finally resuspended in the lysates for a second round of capture. Before any downstream analysis, the eluates from the two capture rounds were pooled, supplemented with 2 μl of 10% SDS and concentrated to 100 μl using a SpeedVac. After the captures, the beads were resuspended in 300 μl of nuclease-free water and incubated for 10 min at +90°C and 800 rpm. The beads were subsequently washed 3× with 1ml nuclease-free water, 3× with 1 ml eRIC lysis buffer, and stored in 0.1% PBS×Tween at +4°C.
Immunofluorescence staining
In addition to observing increased EIF2S1 (also known as eIF2α) phosphorylation by western blot, stress-induction was confirmed by observing stress granule formation as indicated by cytoplasmic ELAVL1 (also known as HuR) aggregation. Sub-confluent HuH7 cells grown on glass coverslips were changed to fresh growth media or to growth media containing 100 μM arsenite for 1 h. After the treatment, the media was removed and the cells were washed twice with phosphate buffered saline (PBS). Cells were fixed with 4% PFA in PBS for 15 min at room temperature (RT) and washed twice with PBS. The cells were permeabilized for 15 min with 0.2% Triton X-100 in PBS on a low-speed orbital shaker and washed 3× with PBS. The coverslips were blocked with 1% BSA in PBST (0.1% Tween 20 in PBS) for 1h at RT. Next, the coverslips were incubated with antibody against ELAVL1 (#11910-1-AP, Proteintech) (1:250 in 1% BSA-PBST) for 1h at RT or overnight at +4°C in a humidified chamber and washed 3× with PBST. The coverslips were incubated with secondary antibody (Alexa 488 Goat-anti-rabbit, #4412, Cell Signaling) (1:500 in 1% BSA-PBST) for 1 h at RT in a humidified chamber, washed 3× with PBST, once with PBS, rinsed with ddH2O, and mounted on glass slides using Vectashield Hardset Mounting Media with DAPI for nuclear counterstain (#H-1500, Vector Laboratories). The samples were imaged using an inverted fluorescent microscope (Olympus iX50 with ColorView 12 CCD-camera) and the pseudocolored images were processed and combined in ImageJ.
Western blot and immunodetection
High purity cell fractionation and LNA capture were analyzed by western blot and immunodetection. For cell fractionation equal volumes of cytoplasmic or nuclear lysates were separated in 10% SDS-PAGE gels under reducing conditions. For interactome captures 0.05% of inputs and 10% of eluates were separated as mentioned above. The proteins were subsequently transferred to PVDC membranes by western blotting and the membranes were probed using antibodies against the proteins of interest. Primary antibodies at indicated dilutions against proteins of interest were used for fractionation samples: α-tubulin (1:30 000; #T5168, Sigma Aldrich), calnexin (1:1000; #2433, Cell Signaling Technology), heat-shock protein 60 (HSP60) (1:1000; #4870, Cell Signaling Technology), Lamin A/C (1:1000; #2032, Cell Signaling Technology), Histone deacetylase 1 (HDAC1) (1:1000; #5356, Cell Signaling Technology), eukaryotic initiation factor 2 α (EIF2S1) (1:1000; #9722, Cell Signaling Technology), phosphorylated eIF2α (pEIF2S1) (1:1000; #3597, Cell Signaling Technology), T-intracellular antigen 1 (TIA-1) (1:1000; #86050, Cell Signaling Technology), Hu-antigen R (HuR / ELAVL1) (1:1000; #11910-1-AP, Proteintech). For the interactome captures, antibodies against TIA-1, ELAVL1, and α-tubulin was used as described above. In addition, the membranes were probed with an antibody against histone H4 (H4) (1:1000; #ab31830, Abcam). As secondary antibodies, 1:10 000 dilutions of horseradish peroxidase (HRP) conjugated goat-anti-rabbit (#A0545, Sigma Aldrich) or rabbit-anti-mouse antibodies (#A9044, Sigma Aldrich) were used. The plots were developed with enhanced chemiluminescence (ECL) (Western lightning Plus-ECL, #NEL104001EA, PerkinElmer) and visualized using a Fusion FX7 imaging system (Vilber Lourmat).
Silver staining
For evaluating successful protein-RNA UV-crosslinking and interactome capture, aliquots of the inputs and RNase eluates were separated by SDS-PAGE and the proteins visualized by silver staining. 250 ng of inputs and 10% of eluates were loaded into the wells. Silver staining was performed according to manufacturer's instructions (SilverQuest™ Staining Kit, #LC6070, Thermo Fisher Scientific).
RNA extraction, bioanalyzer, cDNA synthesis and real-time qPCR
Total RNA from lysates was extracted using RNA Clean & Concentrator-5 (#R1016, Zymo Research) according to manufacturer's instructions. RNA concentration of total RNA extractions and heat-eluted aliquots from interactome captures were measured using NanoDrop 2000 (Thermo Fisher Scientific). For analyzing the profiles of captured RNA, 5 ng of non-crosslinked heat eluates were analyzed in an Agilent 2100 Bioanalyzer Sytem with RNA 6000 Pico Kit (#5067-1513 Agilent) according to the manufacturer's instructions. 500 ng of total RNA or RNA from non-crosslinked heat eluates were reverse transcribed into cDNA using random hexamers and SuperScript III (#18080044, Thermo Fisher Scientific) according to manufacturer's instructions. For evaluating genomic DNA (gDNA) contamination, control reactions without reverse transcriptase were used. Real-time qPCR was performed using PrimaQuant SYBRGreen Master Mix with ROX (#SL-9912-50mL, Steinbrenner Laborsysteme GmbH) in a StepOnePlus system (Thermo Fisher Scientific). Previously published primer pairs against following targets were used for the qPCR (in 5′-3′ direction): RPS6 (forward: TGAAGTGGACGATGAACGCA, reverse: CCATTCTTCACCCAGAGCGT), 18S rRNA (forward: GAAACTGCGAATGGCTCATTAAA, reverse: CACAGTTATCCAAGTGGGAGAGG), 28S rRNA (forward: TTACCCTACTGATGATGTGTTGTTG, reverse: CCTGCGGTTCCTCTCGTA), β-actin (forward: CGCGAGAAGATGACCCAGAT, reverse: TCACCGGAGTCCATCACGAT) (15), HPRT mRNA (forward: GACCAGTCAACAGGGGACAT, reverse: AACACTTCGTGGGGTCCTTTTC), HPRT pre-mRNA (forward: ACGTCAGTCTTCTCTTTTGTAAT, reverse: ACACTTCGTGGGGTCCTTTT) (5).
Sample preparation and TMT labeling
Reduction of disulfide bonds in cysteine-containing proteins was performed with dithiothreitol (56°C, 30 min, 10 mM in 50 mM HEPES, pH 8.5). Reduced cysteines were alkylated with 2-chloroacetamide (room temperature, in the dark, 30 min, 20 mM in 50 mM HEPES, pH 8.5). Samples were prepared using the SP3 protocol (18,19) and trypsin (sequencing grade, Promega) was added in an enzyme to protein ratio of 1:50 for overnight digestion at 37°C. Next day, peptides were recovered in HEPES buffer by collecting the supernatants on a magnet and combining with second elution wash of beads with HEPES buffer. Peptides were labeled with TMT10plex (20) Isobaric Label Reagent (Thermo Fisher Scientific) according the manufacturer's instructions. For further sample clean up an OASIS® HLB μElution Plate (Waters) was used. Offline high pH reverse phase fractionation was carried out on an Agilent 1200 Infinity high-performance liquid chromatography system, equipped with a Gemini C18 column (3 μm, 110 Å, 100 × 1.0 mm, Phenomenex) (21). For the interactomes, eight fractions were pooled and for input samples 12 fractions, each fraction was subjected individually to mass spectrometry.
Mass spectrometry (MS) data acquisition
An UltiMate 3000 RSLC nano LC system (Dionex) fitted with a trapping cartridge (μ-Precolumn C18 PepMap 100, 5μm, 300 μm i.d. × 5 mm, 100 Å) and an analytical column (nanoEase™ M/Z HSS T3 column 75 μm × 250 mm C18, 1.8 μm, 100 Å, Waters). Trapping was carried out with a constant flow of solvent A (0.1% formic acid in water) at 30 μl/min onto the trapping column for 6 minutes. Subsequently, peptides were eluted via the analytical column with a constant flow of 0.3 μL/min with increasing percentage of solvent B (0.1% formic acid in acetonitrile) from 2% to 4% in 6 min, from 4% to 8% in 1 min, then 8% to 25% for a further 71 min, and finally from 25% to 40% in another 5 min. The outlet of the analytical column was coupled directly to a Fusion Lumos (Thermo Fisher Scientific) mass spectrometer using the proxeon nanoflow source in positive ion mode.
The peptides were introduced into the Fusion Lumos via a Pico-Tip Emitter 360 μm OD ×× 20 μm ID; 10 μm tip (New Objective) and an applied spray voltage of 2.4 kV. The capillary temperature was set at 275°C. Full mass scan was acquired with mass range 375–1500 m/z in profile mode in the orbitrap with resolution of 120,000. The filling time was set at maximum of 50 ms with a limitation of 4 × 105 ions. Data dependent acquisition (DDA) was performed with the resolution of the Orbitrap set to 30 000, isolation window of 0.7 m/z, with a fill time of 64 ms and a limitation of 1 × 105 ions. A normalized collision energy of 38 was applied. MS2 data was acquired in profile mode.
MS data analysis
IsobarQuant (22) and Mascot (v2.2.07) were used to process the acquired data, which was searched against a Uniprot Homo sapiens proteome database (UP000005640) containing common contaminants and reversed sequences. The following modifications were included into the search parameters: Carbamidomethyl (C) and TMT10 (K) (fixed modification), Acetyl (N-term), Oxidation (M) and TMT10 (N-term) (variable modifications). For the full scan (MS1) a mass error tolerance of 10 ppm and for MS/MS (MS2) spectra of 0.02 Da was set. Further parameters were set: Trypsin as protease with an allowance of maximum two missed cleavages: a minimum peptide length of seven amino acids; at least two unique peptides were required for a protein identification. The false discovery rate on peptide and protein level was set to 0.01.
The protein.txt output files of IsobarQuant were processed using the R programming language (ISBN 3-900051-07-0). To ensure good data quality, only proteins which were quantified with at least two unique peptides were considered for the analysis. Furthermore, proteins had to be quantified in at least two out of three replicates. The ‘signal_sum’ column of the protein.txt output files were first cleaned for batch-effects using the ‘removeBatchEffect’ function of the limma package (23) and second normalized with a variance stabilization normalization (vsn) method (24). During the normalization process all cross-link (CL) conditions and all non-cross-link (NoCL) conditions were normalized separately in order to keep the abundance difference between the two groups. Missing values were imputed using the ‘impute’ function of the Msnbase package (25) and the k-nearest neighborhood method (knn). In order to test for differential abundance, limma was employed again. Imputed values were given a weight of 5%. For comparisons of CL against NoCL conditions, adjusted P-values directly from limma were used. For comparison of CL against another CL conditions, t-values from limma were used to recalculate false discovery rates (q-values) using the fdrtool package (26). A protein was annotated as ‘hit’ with an FDR smaller than 5% and a fold-change of at least 100% and annotated as ‘candidate’ with an FDR smaller than 20% and a fold-change of at least 50%.
Comparison of interactomes and RBP functions
The identified RBPs were annotated with the curated RIC datasets (13) and combined with recent RIC studies (7,15,27–30). The functional annotation was used according to Hentze 2018 (13): The IntEnz database (release May 2017) was used to classify enzymes, enzymes which are members of ‘metabolism’ in Reactome, plus the subunits of the ATP synthase and the respiratory chain complexes where called metabolic enzymes. Hits were compared to a) mRNA RIC/eRIC approaches (10–12,15,31,32) and (b) to total RNA interactome captures (7,27–30,33). RBPs which were not identified in (a), did not have known RNA-binding domains (12,34), and were not found in a set of known RBPs (35) were classified as novel mRNA RBPs. RBPs that were also not identified in b) were classified as novel RBPs.
Quantification and statistical analysis
For qPCR experiments the data is displayed as mean + s.d. scatter dot plots. Unpaired 2-tailed t-test was used. P<0.05 was considered as significant. Fisher's exact tests with multiple hypothesis correction were used to calculate enrichment of protein domains and GO terms. Plots were created with GraphPad Prism 8.
RESULTS
Validation and quality control of the experimental system
Enhanced RNA interactome capture (eRIC) using locked nucleic acid (LNA)-coupled magnetic beads enables more specific and sensitive capture of mRNA interactomes than previous RNA interactome capture (RIC) methods employing oligo-dT probes (15). We reasoned that these refinements should facilitate an exploration of how mRNA–protein interactions differ between the nuclear and the cytoplasmic compartments of the cell and how environmental cues such as stress affect the composition of the mRNA interactomes in both compartments.
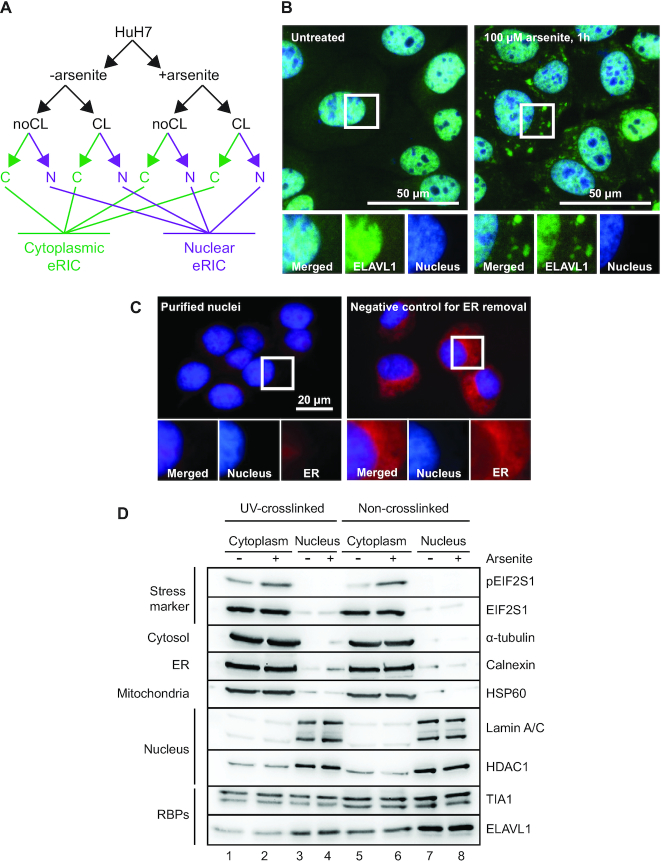
HuH7 cells were grown either as controls or under conditions of arsenite-induced stress. Arsenite is a well-established and potent stressor known to affect protein-RNA interactions (7,36). The cells were then either subjected to UV254 to crosslink RBPs to RNA (CL), or left unexposed (noCL) as negative controls. Subsequently nuclear and cytoplasmic fractions were carefully separated (see Materials and Methods for details) and polyadenylated RNA was captured using eRIC (Figure 1A). To avoid secondary effects resulting from prolonged stress, we limited the treatment with 100 μM sodium arsenite (NaAsO2) to 1h. This treatment caused the expected induction of cytoplasmic stress granules, indicated by ELAVL1 (also known as HuR) aggregation (Figure 1B), and increased phosphorylation of EIF2S1 (also known as eIF2α) in the cytoplasmic fraction (Figure 1D).
Figure 1.
Experimental setup and cell fractionation for compartment-specific eRIC from steady state and stressed cells. (A) A schematic representation of the experimental strategy. HuH7 cells under steady state conditions or following treatment with arsenite were subjected to UV-crosslinking (CL) or not (noCL), and processed to generate clean cytoplasmic (C) and nuclear (N) fractions prior to compartment-specific eRIC. (B) HuH7 cells grown under steady state conditions or exposed to 100 μM Na-arsenite were fixed and immuno-stained against ELAVL1 (green). Cytoplasmic aggregation of ELAVL1 positively controls for stress induction. Nuclear counterstain is shown in blue. (C) Nuclear (blue) and ER (red) staining showing nuclear integrity and removal of ER by cell fractionation (left panel). For a negative control of ER removal, NP-40 in the lysis buffer was replaced with the less effective Tween-20 (right panel). (D) Analysis of cell fractionation. Equal aliquots of cell fractions were separated by SDS-PAGE and analyzed by immunoblotting against the indicated marker proteins.
We confirmed the purity of cell fractionation with nuclear fractions containing intact nuclei devoid of endoplasmic reticulum (ER) remnants. The nuclear integrity and ER removal was analyzed by fluorescent staining of the nuclei and the ER (Figure 1C). We also conducted immunoblotting for several marker proteins. The cytoplasmic fractions were enriched in α-tubulin, calnexin and HSP60, representing primarily cytosolic, ER, and mitochondrial marker proteins, respectively. These proteins were highly underrepresented in the nuclear fractions that instead were substantially enriched for the nuclear marker proteins lamin A/C and HDAC1. As expected, the known shuttling nucleo-cytoplasmic RBPs TIA-1 and ELAVL1 were observed in both fractions (Figure 1D).
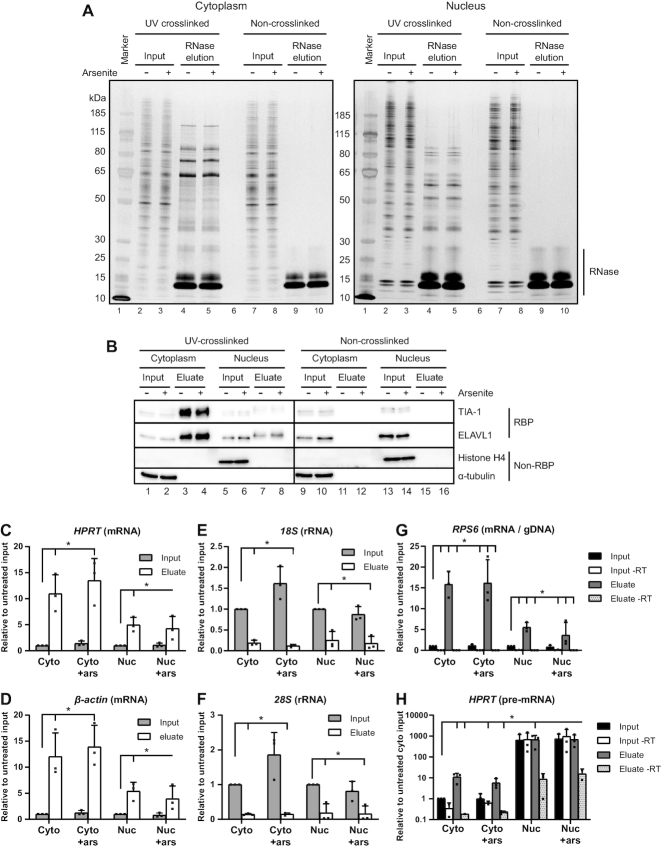
Having established robust experimental conditions, we performed eRIC on the nuclear and the cytoplasmic fractions (15). Sample quality was controlled by SDS-PAGE followed by silver-staining or immunoblotting of aliquots from the inputs and RNase-eluted samples from UV-treated and control cells. The eluates from the UV-crosslinked samples reveal a distinctive pattern of proteins that profoundly differs between cytoplasmic and nuclear fractions (Figure 2A, lanes 4 and 5) as well as from the inputs (lanes 2 and 3 and 7 and 8). The eluates of the non-crosslinked samples are clean apart from the added RNase (lanes 9 and 10) and with only traces of stained cellular proteins, reflecting the specificity and low background of the eRIC procedure. Arsenite treatment did not change the protein profiles of inputs nor eluates, as assessed by silver staining (compare uneven with even numbered lanes in Figure 2A). The clear differences between the cytoplasmic and nuclear samples suggest that the RNA-binding proteomes of the nucleus and the cytoplasm substantially differ.
Figure 2.
Compartment-specific RBP capture by eRIC is highly specific and sensitive. (A, B) Inputs and 10% of RNase eluates from eRIC captures were separated by SDS-PAGE and the proteins were visualized by silver staining (A) and western blotting (B). 250 ng and 0.05% of inputs were loaded onto the gels for silver staining and western blotting, respectively. (B) The western blots were immunoblotted against the known RBPs TIA-1 and ELAVL1, or against the non-RNA-binding proteins Histone H4 and α-tubulin as negative controls. The observed upwards shift in gel mobility of TIA-1 and ELAVL1 between the inputs and eluates of the UV-crosslinked samples is likely due to the presence of small cross-linked RNA fragments in the eluates. (C–H) captured RNA species were analyzed by RT-qPCR. The relative abundance of mRNA (C, D), rRNA (E, F), gDNA (G), and pre-mRNA (H) in cytoplasmic (Cyto) and nuclear (Nuc) inputs and eluates, from untreated and arsenite-treated (+ars) cells were quantified. The data represent the mean with standard deviation from three biological replicates. For analysis of captured pre-mRNA and gDNA samples where reverse transcriptase (-RT) was omitted from the cDNA synthesis was included to evaluate the presence of a gDNA-derived template. Note that mRNA is effectively captured by eRIC while rRNA is strongly selected against compared to inputs. Also gDNA contamination is low as observed by minor amplification in –RT samples. Further, the pre-mRNA capture is restricted to the nuclear fractions and eRIC efficiently discriminates against gDNA capture as observed by only minor amplification in –RT eluates (note the log10 scale in the Y-axis). The horizontal lines marked (*) above the columns indicate the compared groups. Long vertical ticks indicate the control-samples and short ticks indicate P< 0.05 (unpaired two-tailed t-test).
Western blotting was used to further validate the specificity of the interactome captures (Figure 2B). The UV-crosslinked interactome captures are expected to contain known RBPs while lacking non-RNA-binding proteins. As expected, the RBPs ELAVL1 and TIA-1 are present in the eluates of the UV-crosslinked cytoplasmic or nuclear samples whereas these RBPs are present only in the inputs but not in the eluates of the non-crosslinked samples. Further, α-tubulin and histone H4, chosen as negative controls for non-RNA-binding cytoplasmic and nuclear proteins, respectively, were detected exclusively in the inputs. These results show that UV-exposure specifically crosslinks RBPs to RNAs both, in the cytoplasmic and in the nuclear fractions. A comparison of arsenite-treated and untreated cells shows that treatment does not negatively affect the quality the cell fractionation or the eRIC selection (compare uneven with even numbered lanes in Figure 2B).
Next, we assessed the effective and specific capture of polyadenylated RNAs by bioanalyzer size separation and by qPCR from aliquots eluted by heat instead of RNase treatment (Figures 2C–H and Supplementary Figure S1). The bioanalyzer profiles show a typical length distribution for mRNAs and strongly reduced 18S and 28S rRNAs, which otherwise dominate total RNA profiles (Supplementary Figure S1). This general enrichment pattern was confirmed by qPCR of specific RNAs (Figures 2C–H). The qPCR analyses show that the LNA oligo-mediated capture is highly selective for the tested polyadenylated RNAs (mRNAs and pre-mRNA): using equal amounts of RNA from inputs and eluates for cDNA synthesis and normalizing the qPCR results to untreated inputs shows the expected strong enrichment of β-actin and HPRT1 mRNAs in the eluates both, in nuclear and in cytoplasmic fractions regardless of arsenite treatment (Figures 2C, D). By contrast, eRIC selects against ribosomal RNAs in the eluates compared to inputs (Figures 2E, F), which is not affected by arsenite treatment. We also controlled for contaminating genomic DNA (gDNA) with primer pairs targeting an intra-exonic sequence of RPS6. There were only traces of signal when reverse transcriptase (RT) was omitted from the cDNA synthesis, reflecting minimal gDNA contamination. By contrast, there were strong signals when the RT was used, reflecting the presence of RPS6 mRNA (Figure 2G).
To evaluate the capture of pre-mRNA from the cytoplasmic and nuclear fractions, the qPCR quantifications were normalized to cytoplasmic inputs derived from RT-assisted cDNA synthesis. The results show that the amplification of HPRT pre-mRNA by intron-exon targeting primers is limited to the nuclear fractions further confirming successful cell fractionation (Figure 2H). In the input of the nuclear fractions some gDNA is still present after DNase treatment and RNA extraction as observed by amplification of the target without RT. However, the signals are greatly reduced in the eluates of interactome captures without the use of RT. These data confirm that the capture of gDNA is strongly discriminated against by the LNA-probes. In sum, these results document (i) the specificity and efficiency of eRIC for capturing polyadenylated RNAs, (ii) the quality of the nuclear-cytoplasmic fractionation and (iii) that arsenite treatment does not negatively affect the quality of the fractionation or the eRIC procedure.
Nuclear and cytoplasmic interactomes show profound differences in their RBP composition
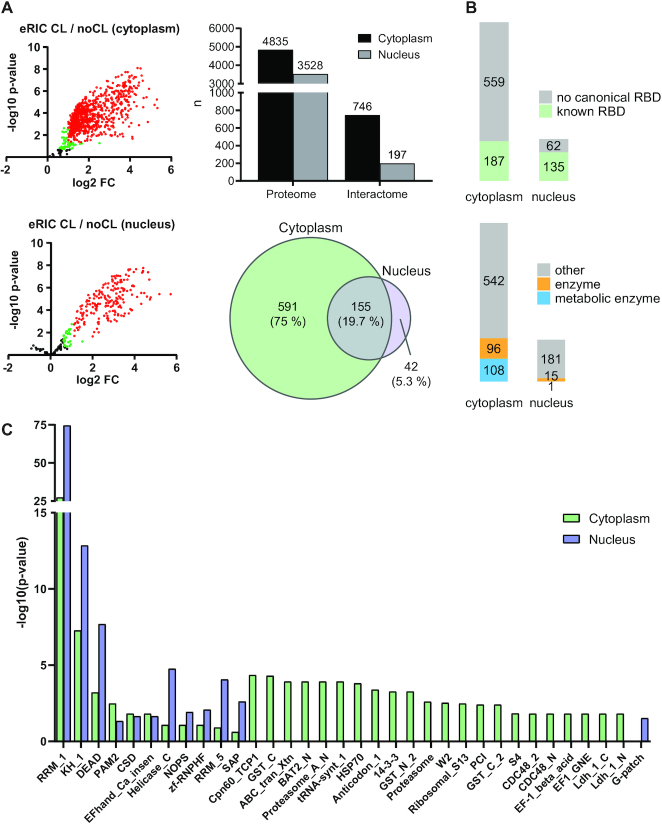
For the eRIC analyses, three biological replicates were used for each condition. Total proteomes and RBPs were identified by quantitative mass spectrometry (MS) from eRIC inputs and RNase eluates, respectively (37). A total of 4835 cytoplasmic and 3528 nuclear proteins were identified from the full proteomes of untreated cells (Figure 3A, Supplementary Table S1).
Figure 3.
Identification of nuclear and cytoplasmic RBPs. (A) Left panel: volcano plots showing the results of a differential abundance analysis by limma (23). Log2-fold change (FC) of UV-crosslinked over non-crosslinked (x-axis) and log10-fold changes of P-values (y-axis) of RBPs (red: FDR < 0.05, FC > 100%), candidate RBPs (green: FDR < 20%, FC > 50%), and ‘unspecific’ non-RBPs (black) captured by eRIC. The top plot shows the cytoplasmic and the bottom plot the nuclear RBPs. Top right panel: cytoplasmic and nuclear proteins identified in the full proteomes (proteome) and the RBPs identified by the interactome captures (interactome). Bottom right panel: overlap of common RBPs identified in the cytoplasmic and nuclear interactomes. (B) RBPs with known RNA-binding domains (top) and RBPs that are known (metabolic) enzymes (bottom). (C) Analysis of significantly enriched (P.adj < 0.05) protein domains within the identified RBPs.
UV-irradiation effectively crosslinks directly-bound RBPs to RNAs, and the stringent hybridization and washing conditions enabled by the LNA-modified antisense probes to poly(A) tails effectively remove non-crosslinked proteins (including indirectly-bound proteins), DNA and rRNA from the interactome (15). The eRIC eluates harbor 746 cytoplasmic and 197 nuclear RBPs with >2-fold enrichment and an FDR <0.05 in UV-crosslinked compared to non-crosslinked samples. Of these, 155 were common to both fractions while 591 and 42 were found exclusively in the cytoplasmic and in the nuclear fractions, respectively (Figure 3A, Supplementary Table S1). This makes a total of 788 unique RBPs from both fractions combined. Note that eRIC captures polyadenylated RNAs, thus excluding nuclear RBPs binding exclusively to RNAs that are not (yet) polyadenylated.
The fractionation-assisted eRIC revealed 245 cytoplasmic and six nuclear RBPs (total of 247 unique RBPs combined) that have been missed by earlier mRNA interactome captures from unfractionated human cells, lack any known RNA-binding domains, or are not included in curated lists of human RBPs. While 185 of these have been found in some total cellular RNA interactomes, the mRNA binding activity of these RBPs has not been known before (Supplementary Figure S2 and Supplementary Table S2) (7,10–13,15,27–35). These 247 novel mRNA binding RBPs are enriched in proteasomal proteins, some of which have been reported to possess RNase activity (Supplementary Figure S3) (38,39).
Previous interactome capture experiments revealed that many of the recently discovered RBPs do not contain well-defined RNA-binding domains (reviewed in (13)). In line with these observations, only approximately one quarter of the cytoplasmic and two thirds of the nuclear RBPs contain classical RNA-binding domains (RBD) (Figure 3B, Supplementary Table S2). This is in line with a report that nuclear RBPs mostly harbor canonical RBDs (32). RRMs and KH-domains represent the most prominent RBDs of both cytoplasmic and nuclear RBPs (Figure 3C). Enzymes represent 27% of the cytoplasmic, but only 8% of the nuclear RBPs (Figure 3B, Supplementary Table S2). Interestingly, the novel mRNA RBPs account for 45% (93) of the total of 207 RBPs with known enzymatic functions (Supplementary Table S2). Overall, we find numerous differences in the RBP function and domain composition of the nucleus and the cytoplasm.
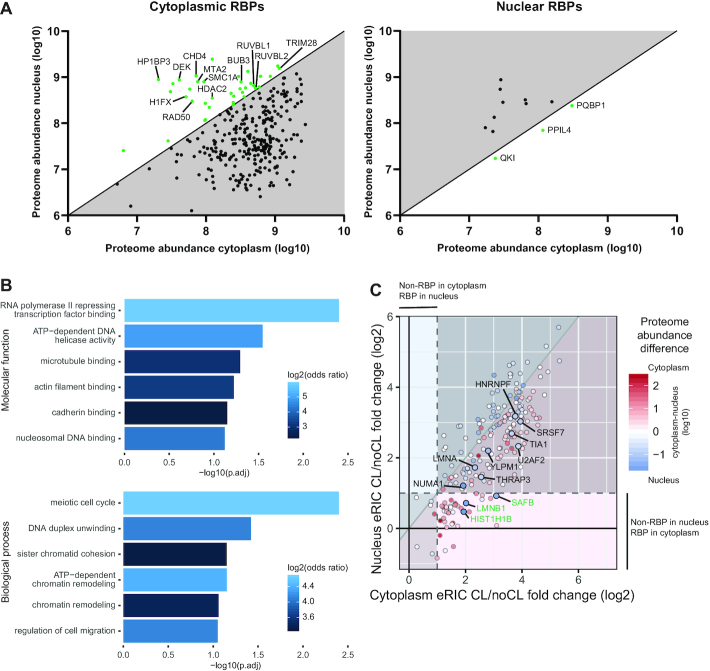
We next focused on a group of RBPs, which are expressed in both the nucleus and the cytoplasm (identified in total proteome of both cytoplasm and nucleus), but that reproducibly displayed RNA-binding activity in only one compartment. We were particularly interested in a remarkable subgroup of such RBPs, whose abundance in the total proteome was even lower in the compartment where they display RBP activity. We identified 37 proteins that bind RNA only in the cytoplasm although being more abundant in the proteome of the nucleus (green dots on white background) (Figure 4A, Supplementary Tables S1 and S3). GO-term enrichment analysis revealed that these proteins are enriched for chromatin- and DNA-associated proteins including a linker histone (H1FX), various histone modifiers, and proteins involved in chromosome/chromatin organization (Figures 4A, B). These findings reveal that these mostly nuclear proteins with well-established chromatin-associated functions bind RNA in the cytoplasm; the functional rationale for this will be interesting to define.
Figure 4.
A subset of RBPs exhibit compartment-controlled RNA-binding activity. (A) Scatter plots of RBPs identified in all full proteomes but only in either cytoplasmic (left) or nuclear (right) interactomes. The axes indicate the cytoplasmic (x-axis) and nuclear (y-axis) proteome abundances based on their top3 values. The top3 value represents the average ms1 intensity of the three most abundant peptides for a protein in the mass spectrometry in log10 scale. Proteins identified as RBPs only in the compartment where they are relatively less abundant are shown in green on white background. Of these, selected cytoplasmic RBPs that are otherwise annotated as nuclear histone and chromatin-associated proteins are named. (B) GO-term enrichment analysis of the cytoplasmic RBPs with higher protein abundance in the nucleus. (C) Correlation of RNA binding activity between cytoplasm and nucleus. The RNA-binding activity is reflected by CL/noCL fold change ratios (log2) of the RBPs in cytoplasmic (x-axis) and nuclear (y-axis) interactomes. Color of dots indicate their abundance difference in full proteomes. Dots on red or blue background show more pronounced RNA binding in the cytoplasm or in the nucleus, respectively. Red or blue dots indicates proteins with higher abundance in the cytoplasm or in the nucleus, respectively. Note that the color intensity of the dots indicates the difference in abundance on a log10 scale. The shaded bottom left area shows proteins identified in both nuclear and cytoplasmic interactomes but not showing significant RBP-activity in either. The upper right shaded are shows bona fide RBPs in both compartments. The light areas show bona fide RBPs only in one compartment as indicated. The highlighted and named proteins are at least 5-fold more abundant in the nucleus while showing an at least 50% higher CL/noCL ratio in the cytoplasm. Bona fide RBPs only in the cytoplasm while being significantly more abundant in the nucleus are named in green. Note that no proteins filled these criteria the other way around. The dashed lines represent the 2-fold enrichment threshold in CL/noCL ratios used for defining bona fide RBPs.
The twelve proteins with exclusively nuclear RBP activity largely represent RNA processing- and splicing-associated proteins. Three of these are more abundant in the cytoplasmic proteome (green dots on white background) (Figure 4A, Supplementary Table S3). These 3 RBPs include the nuclear mRNA processing factors PQBP1 and QKI, and PPIL4, an enzymatic RBP with a propyl isomerase domain involved in protein folding (40) (Supplementary Table S2). While QKI and PPIL4 bear KH and RRM motifs, respectively, PQBP1 is an intrinsically disordered protein, a feature of many non-canonical RBPs (13,41) (Supplementary Table S2).
While the analysis described above focused on proteins displaying RBP activity exclusively in either the cytoplasmic or the nuclear compartment, we next extended the analysis to proteins identified in the interactomes of both compartments, but showing more distinguishable RNA binding (CL/noCL fold change) in the compartment where they were found to be less abundant in the full proteome (blue dots on red background and vice versa) (Figure 4C). None of the RBPs that are most abundantly found in the full cytoplasmic proteome (red dots) show considerably more recognizable RNA binding (≥50% difference) in the nucleus (red dots on blue background). However, we identified 11 RBPs that are substantially more abundant (≥ 5-fold) in the nucleus, while exhibiting considerably more apparent RNA binding in the cytoplasm (Figure 4C, highlighted blue dots). Three of these proteins (SAFB, LMNB1, HIST1H1B) were particularly highly abundant in the nuclear proteome but only reproducibly identified as RBPs in the cytoplasm. Although all of these proteins are previously known RBPs, the unexpected subcellular bias in RNA-binding had not been recognized. In particular, it is noteworthy to find another member of the histone H1 family, HIST1H1B, as a noticeable cytoplasmic RBP. Histone H1 family members are known to localize to cytoplasm but their cytoplasmic functions have remained largely unknown (42–44). Overall, the nucleo-cytoplasmic fractionation combined with eRIC reveal a number of primarily nuclear proteins that exhibit more pronounced or exclusively cytoplasmic mRNA-binding.
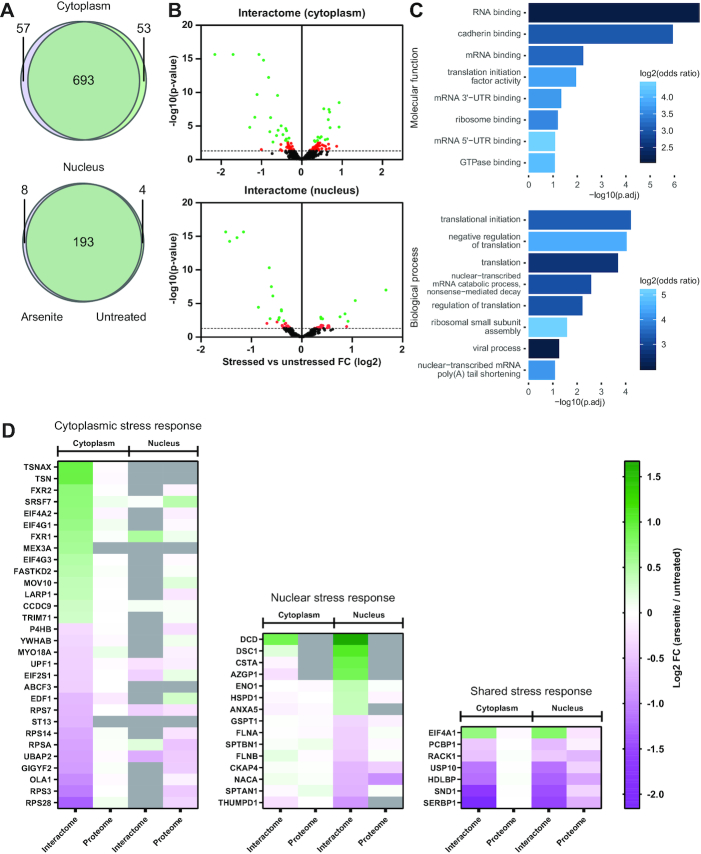
Compartment-restricted, stress-induced changes in RNA binding
We next analyzed how arsenite-induced oxidative stress affects the nuclear and cytoplasmic interactomes (Figures 5A, B). RBPs identified under the different conditions show high degree of overlap in both cytoplasmic and nuclear compartments (Figure 5A). It should be noted that the condition-specific RBPs were identified in the eRIC eluates of both conditions but considered as RBPs only in one. Integrating the interactomes of treated and untreated cells, we identified a total of 803 cytoplasmic and 205 nuclear RBPs representing 845 unique RBPs in total (Supplementary Table S1). As expected, most proteins in the full proteome did not respond to stress while known arsenite-induced effects such as the nuclear accumulation of TP53 and reduced oxidoreductase TXNL1 expression could be clearly observed (Supplementary Figure S4 and Supplementary Table S1) (45,46).
Figure 5.
Stress-induces specific nuclear and cytoplasmic changes of RBP activity. (A) Venn diagrams showing the overlap of RBPs identified in cytoplasmic (top) and nuclear (bottom) interactome captures from both untreated and arsenite-treated cells. (B) Volcano plots showing the arsenite-induced effects in cytoplasmic (top) and nuclear (bottom) RBPs. Limma was used to test for differential abundance between cross-linked fractions of arsenite-treated versus untreated. X-axis: fold change (log2) comparison of treated against untreated CL samples. Y-axis: P-values of the comparisons. Proteins showing arsenite-induced fold changes with FDR <0.05 are shown in green. Proteins with P-value <0.05 but FDR >0.05 are shown in red. The dashed line represents the 0.05 P-value threshold. (C) GO-term enrichment analysis of the stress-affected RBPs. (D) Heat maps of changes of RNA-binding activity induced by stress of RBPs with an FDR <0.05 (interactome) and the corresponding changes in the full proteome (proteome). The color-scale represents the log2 FC of the treated against untreated CL samples. The stress-affected RBPs are divided into three groups, RBPs showing a significant change only in the cytoplasm (left), in the nucleus (middle), or RBPs significantly affected in both compartments (right). Proteins not identified in a specific group are shown in grey.
When analyzing the stress-responsive RBPs by comparing the CL-samples of the interactome eluates from untreated and arsenite exposed cells, we found that a total of 52 displayed high confidence (FDR < 0.05) arsenite-induced changes (Figure 5B, Supplementary Tables S1 and S3). A GO-term enrichment analysis shows that RNA-binding and translation-associated functions lead the lists of molecular functions and biological processes, respectively (Figure 5C), in line with the known stress-induced control of translation (reviewed in (47)).
Interestingly, most of the RBPs that showed a stress-response in the cytoplasm were identified in the cytoplasmic interactome only (Figure 5D, Supplementary Table S4). Similarly, the small group of RBPs that reacted both, in the cytoplasm and in the nucleus, showed similarly significant quantitative differences in RNA binding in both compartments. By contrast, all stress responsive RBPs in the nucleus were also identified in the cytoplasmic interactomes, but showed less pronounced stress-responses with FDRs > 0.05.
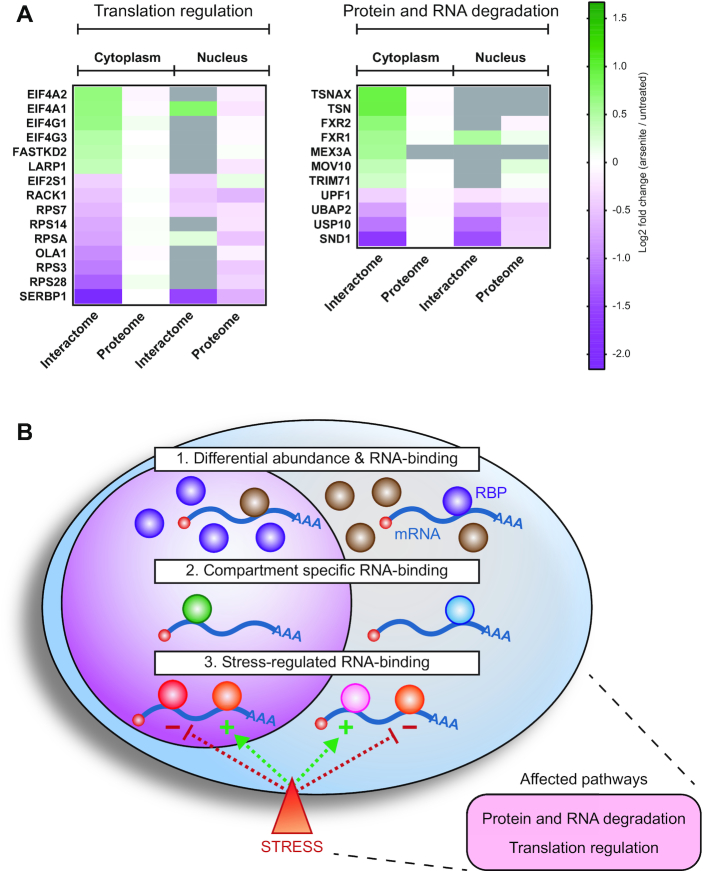
Stress remodels the RNA interactome to regulate translation and RNA degradation
In line with the interactomes of unfractionated cells (7), many of the stress-regulated RBPs identified here are cytoplasmic or mitochondrial ribosome-associated proteins, cap-dependent or IRES-dependent translation initiation factors (47–50), or proteins involved in the translational regulation of mRNAs containing a 5′ terminal oligopyrimidine (5′TOP) motif (51) (Figure 6A, Supplementary Table S5). These data strongly substantiate the importance of stress-regulated translation by altered RNA binding of RBPs.
Figure 6.
Stress-regulated RBPs are associated with translation regulation and degradation pathways. (A) Selected stress-affected RBPs from Figure 5D grouped based on common pathway associations. (B) A graphical conclusion of the results indicating the observed compartmentalized differences in RNA-binding and the associated pathways of stress-affected RBPs. 1: Nucleo-cytoplasmic proteins that moonlight as RBPs exclusively in the compartment where they are less abundant. 2: RBPs that bind RNA only in the cytoplasm or in the nucleus. 3: RBPs whose RNA-binding is stress-regulated. The dashed arrows indicate positive (+) or negative (−) effect on RNA-binding. The bottom right panel indicates the major stress-regulated and RBP-associated pathways identified in the current study.
Unlike earlier studies, the introduction of cell fractionation before interactome capture allowed the identification of components of the ubiquitination and protein degradation machinery as a previously unknown group of low-abundance RBPs whose RNA binding is regulated by stress (Figure 6A, Supplementary Table S5). This group includes MEX3A, an RBP with ubiquitin ligase RING finger domains and known to localize to p-bodies (52). Like its family member MEX3C, MEX3A could potentially regulate mRNA stability via proteasome-independent ubiquitination of the deadenylation machinery (53). Similarly, UBAP2, TRIM71 (also known as LIN41), and USP10 are proteins involved in protein ubiquitination/de-ubiquitination pathways (54–56). Several ubiquitin ligases have been identified as RBPs linking the ubiquitin-system to RNA-metabolism (57,58). While the role of these ubiquitination-associated proteins in RNA metabolism is poorly understood, our results unexpectedly indicate that some of them are stress-sensitive. Based on our data, it will be interesting to explore whether this group of RBPs is also linked to stress-regulated RNA degradation.
A functional link of low-abundance cytoplasmic RBPs to RNA degradation pathways is further highlighted by the identification of RBPs that are known components of the RNA-induced silencing complex (RISC) and of the nonsense-mediated decay (NMD) pathway (Figure 6A, Supplementary Table S5). RISC-targeted mRNAs can be subjected to both translational repression and degradation (reviewed in (59)). Specifically, RISC-associated proteins that have been identified here in the stress-regulated interactome include the endonucleases translin (TSN), translin associated factor X (TSNAX) and SND1 (also known as Tudor-SN) (60,61). Interestingly, these endonucleases show opposite stress responses in their RNA binding, suggesting that their functions are differentially regulated (Figure 6A, Supplementary Table S5). The role of RBPs associated with RISC is further highlighted by the stress-induced differential RNA-binding of the fragile-X mental retardation proteins FXR1 and FXR2 (Figure 6A, Supplementary Table S5) that are also known to interact with AGO2 (62). FXR1 and FXR2 are autosomal paralogs of the fragile-X mental retardation protein FMR1 that is also a component of RISC (62,63) and showed a trend toward stress-induced increased RNA-binding (Supplementary Table S1).
We also found the RNA-binding of UPF1 and MOV10 to be stress-regulated (Figure 6A, Supplementary Table S5). UPF1 is one of the key effectors of the nonsense-mediated decay (NMD) pathway, which is known to be down-modulated by cellular stress via a still unknown molecular mechanism (64–66). Further, the RNA helicase MOV10 has been shown to participate in UPF1 associated RNA degradation (67). The regulated binding of UPF1 and MOV10 to RNA under conditions of stress suggests that these effects may contribute to the stress-mediated control of NMD.
Taken together, we discovered unexpected stress-mediated changes in RBPs regulating ribosome function, translation, as well as RNA- and protein stability.
DISCUSSION
In vivo analyses of RNA interactomes have resulted in the identification of a large number of previously unknown RNA-binding proteins lacking canonical RBDs. Some of these novel RBPs have cell biological functions that may be controlled by RNA binding (13). One of the hypotheses motivating the current study has been that cellular stress would affect RNA binding not only of canonical RBPs but also of proteins with other known functions. We were also curious whether stress may elicit different responses in the cytoplasm and in the nucleus. The main results of this study are summarized in the graphical conclusion (Figure 6B). To analyze the nuclear and the cytoplasmic interactomes separately and sensitively from the same cells, we benefitted from the use of eRIC which employs LNAs in the capture reaction allowing highly stringent hybridization and washing, and hence the identification of low abundance RBPs against a low background (15). Therefore, we could confidently determine the relatively small nuclear RNA interactome. It chiefly consists of canonical RBPs, as had been indicated by earlier studies (32,33). We also could identify a substantial number of low abundance novel mRNA-binding RBPs in the cytoplasm, many of which without canonical RBDs and with enzymatic functions, extending the scope of proteins that could potentially be subject to riboregulation (14).
Most interestingly, many of the proteins found in both the nuclear and the cytoplasmic proteomes only function as RBPs in the cellular compartment where they are less abundant (Figure 4). Thus, they act as RBPs at low abundance while exerting their previously known, annotated function at higher abundance in the other compartment. Might RNA influence their nucleo-cytoplasmic transport? We also show here that a substantial number of RBPs, both in the nucleus and in the cytoplasm, differentially alter their RNA-binding activity in response to stress, expanding our knowledge of how stress affects RNA-protein interactions in a compartment specific fashion. Unexpectedly, the analysis of fractionated interactomes demonstrated that the stress-induced changes of RNA binding may show highly quantitative differences in their stress responses between the cytoplasm and the nucleus. SRSF7 and THUMPD1 are examples of this (Figure 5). Furthermore, the responses may even go into opposite directions indicating fundamentally differential and compartment-specific functions of these RBPs. AZGP1, CSTA and FLNB are illustrative examples of this surprising response pattern. Both of these compartmentalized differences would have been missed by whole cell analyses.
Many of the stress-responsive RBPs exert stress-related functions on the ribosome in general and in translational regulation in particular (Figure 6). These ribosome-associated stress-responses are largely in-line with a previous interactome study from arsenite-stressed cells focusing on total RNA with a predominant fraction of ribosomal RNAs from whole cell lysates (7). However, we identified another unexpected, mostly cytoplasmic group of RBPs with differential stress-induced binding with known functions in RNA- and protein degradation, thus suggesting that these pathways could also be subject to riboregulation. Alternatively, and as suggested previously (38,39,57,58), proteins known to play a role in proteasomal function and ubiquitination may have additional functions regulating RNA stability.
In conclusion, the results from this study further our understanding on RBP biology by charting the nucleo-cytoplasmic landscape and compartmentalized stress-responses of human RBPs and their mRNA interactomes. This adds further value to the existing datasets of human RBPs from other interactome studies. We believe the data reported here will serve as a valuable resource for the scientific community for further research.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (68) partner repository with the dataset identifier PXD016735. Scripts are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Rastislav Horos (EMBL) for HuH7 cells and advice, and to Beate Amthor, Daria Lavysh, Jonas Becker, Claudia Gruber, Gabriele Tolle and Margit Happich for their support.
Author contributions: M.B., M.W.H. and A.E.K. designed the project. M.B. performed most of the experiments and drafted the manuscript. M.B. and A.B. performed coupling of the LNA-beads. M.R. performed the MS sample preparation and MS runs. F.S. performed the MS data analysis. T.S. performed the domain enrichment, GO-enrichment, and novel RBP identification analyses. J.I.P.-P. contributed to the experimental design and training of the eRIC protocol. J.I.P.-P., G.N.Y., A.B. and Y.Z. contributed to the conceptualization of the study and interpretation of results. M.W.H. and A.E.K. coordinated the study and finalized the manuscript with input from all authors.
Notes
Annika Brosig: Candidate for joint PhD degree from EMBL and Faculty of Biosciences, Heidelberg University.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SFB1036 TP5 to A.E.K. and M.W.H]. Funding for open access charge: Deutsche Forschungsgemeinschaft [SFB1036 TP5].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kedersha N.L., Gupta M., Li W., Miller I., Anderson P.. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999; 147:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ivanov P., Kedersha N., Anderson P.. Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 2019; 11:a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Higa M., Oka M., Fujihara Y., Masuda K., Yoneda Y., Kishimoto T.. Regulation of inflammatory responses by dynamic subcellular localization of RNA-binding protein Arid5a. PNAS. 2018; 115:E1214–E1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Courteau L., Crasto J., Hassanzadeh G., Baird S.D., Hodgins J., Liwak-Muir U., Fung G., Luo H., Stojdl D.F., Screaton R.A. et al.. Hexokinase 2 controls cellular stress response through localization of an RNA-binding protein. Cell Death Dis. 2015; 6:e1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hollerer I., Curk T., Haase B., Benes V., Hauer C., Neu-Yilik G., Bhuvanagiri M., Hentze M.W., Kulozik A.E.. The differential expression of alternatively polyadenylated transcripts is a common stress-induced response mechanism that modulates mammalian mRNA expression in a quantitative and qualitative fashion. RNA. 2016; 22:1441–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Danckwardt S., Gantzert A.S., Macher-Goeppinger S., Probst H.C., Gentzel M., Wilm M., Grone H.J., Schirmacher P., Hentze M.W., Kulozik A.E.. p38 MAPK controls prothrombin expression by regulated RNA 3′ end processing. Mol. Cell. 2011; 41:298–310. [DOI] [PubMed] [Google Scholar]
- 7. Trendel J., Schwarzl T., Horos R., Prakash A., Bateman A., Hentze M.W., Krijgsveld J.. The human RNA-Binding proteome and its dynamics during translational arrest. Cell. 2019; 176:391–403. [DOI] [PubMed] [Google Scholar]
- 8. Backlund M., Paukku K., Kontula K.K., Lehtonen J.Y.. Endoplasmic reticulum stress increases AT1R mRNA expression via TIA-1-dependent mechanism. Nucleic Acids Res. 2016; 44:3095–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawai T., Lal A., Yang X., Galban S., Mazan-Mamczarz K., Gorospe M.. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 2006; 26:3295–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al.. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 11. Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. et al.. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012; 46:674–690. [DOI] [PubMed] [Google Scholar]
- 12. Castello A., Fischer B., Frese C.K., Horos R., Alleaume A.M., Foehr S., Curk T., Krijgsveld J., Hentze M.W.. Comprehensive identification of RNA-Binding domains in human cells. Mol. Cell. 2016; 63:696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hentze M.W., Castello A., Schwarzl T., Preiss T.. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018; 19:327–341. [DOI] [PubMed] [Google Scholar]
- 14. Horos R., Buscher M., Kleinendorst R., Alleaume A.M., Tarafder A.K., Schwarzl T., Dziuba D., Tischer C., Zielonka E.M., Adak A. et al.. The small Non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell. 2019; 176:1054–1067. [DOI] [PubMed] [Google Scholar]
- 15. Perez-Perri J.I., Rogell B., Schwarzl T., Stein F., Zhou Y., Rettel M., Brosig A., Hentze M.W.. Discovery of RNA-binding proteins and characterization of their dynamic responses by enhanced RNA interactome capture. Nat. Commun. 2018; 9:4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gagnon K.T., Li L., Janowski B.A., Corey D.R.. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 2014; 9:2045–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hughes C.S., Foehr S., Garfield D.A., Furlong E.E., Steinmetz L.M., Krijgsveld J.. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014; 10:757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hughes C.S., Moggridge S., Muller T., Sorensen P.H., Morin G.B., Krijgsveld J.. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019; 14:68–85. [DOI] [PubMed] [Google Scholar]
- 20. Werner T., Sweetman G., Savitski M.F., Mathieson T., Bantscheff M., Savitski M.M.. Ion coalescence of neutron encoded TMT 10-plex reporter ions. Anal. Chem. 2014; 86:3594–3601. [DOI] [PubMed] [Google Scholar]
- 21. Reichel M., Liao Y., Rettel M., Ragan C., Evers M., Alleaume A.M., Horos R., Hentze M.W., Preiss T., Millar A.A.. In planta determination of the mRNA-binding proteome of arabidopsis etiolated seedlings. Plant Cell. 2016; 28:2435–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Franken H., Mathieson T., Childs D., Sweetman G.M., Werner T., Togel I., Doce C., Gade S., Bantscheff M., Drewes G. et al.. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015; 10:1567–1593. [DOI] [PubMed] [Google Scholar]
- 23. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M.. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002; 18(Suppl. 1):S96–S104. [DOI] [PubMed] [Google Scholar]
- 25. Gatto L., Lilley K.S.. MSnbase-an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics. 2012; 28:288–289. [DOI] [PubMed] [Google Scholar]
- 26. Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008; 24:1461–1462. [DOI] [PubMed] [Google Scholar]
- 27. Bao X., Guo X., Yin M., Tariq M., Lai Y., Kanwal S., Zhou J., Li N., Lv Y., Pulido-Quetglas C. et al.. Capturing the interactome of newly transcribed RNA. Nat. Methods. 2018; 15:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang R., Han M., Meng L., Chen X.. Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. PNAS. 2018; 115:E3879–E3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Queiroz R.M.L., Smith T., Villanueva E., Marti-Solano M., Monti M., Pizzinga M., Mirea D.M., Ramakrishna M., Harvey R.F., Dezi V. et al.. Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat. Biotechnol. 2019; 37:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Urdaneta E.C., Vieira-Vieira C.H., Hick T., Wessels H.H., Figini D., Moschall R., Medenbach J., Ohler U., Granneman S., Selbach M. et al.. Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat. Commun. 2019; 10:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beckmann B.M., Horos R., Fischer B., Castello A., Eichelbaum K., Alleaume A.M., Schwarzl T., Curk T., Foehr S., Huber W. et al.. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015; 6:10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Conrad T., Albrecht A.S., de Melo Costa V.R., Sauer S., Meierhofer D., Orom U.A.. Serial interactome capture of the human cell nucleus. Nat. Commun. 2016; 7:11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kramer K., Sachsenberg T., Beckmann B.M., Qamar S., Boon K.L., Hentze M.W., Kohlbacher O., Urlaub H.. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods. 2014; 11:1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brannan K.W., Jin W., Huelga S.C., Banks C.A., Gilmore J.M., Florens L., Washburn M.P., Van Nostrand E.L., Pratt G.A., Schwinn M.K. et al.. SONAR discovers RNA-binding proteins from analysis of large-scale protein-protein interactomes. Mol. Cell. 2016; 64:282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerstberger S., Hafner M., Tuschl T.. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014; 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buchan J.R., Parker R.. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009; 36:932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bantscheff M., Lemeer S., Savitski M.M., Kuster B.. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 2012; 404:939–965. [DOI] [PubMed] [Google Scholar]
- 38. Brooks S.A. Functional interactions between mRNA turnover and surveillance and the ubiquitin proteasome system. Wiley Interdiscipl. Rev. RNA. 2010; 1:240–252. [DOI] [PubMed] [Google Scholar]
- 39. Kulichkova V.A., Tsimokha A.S., Fedorova O.A., Moiseeva T.N., Bottril A., Lezina L., Gauze L.N., Konstantinova I.M., Mittenberg A.G., Barlev N.A.. 26S proteasome exhibits endoribonuclease activity controlled by extra-cellular stimuli. Cell Cycle. 2010; 9:840–849. [DOI] [PubMed] [Google Scholar]
- 40. Zeng L., Zhou Z., Xu J., Zhao W., Wang W., Huang Y., Cheng C., Xu M., Xie Y., Mao Y.. Molecular cloning, structure and expression of a novel nuclear RNA-binding cyclophilin-like gene (PPIL4) from human fetal brain. Cytogenet. Cell Genet. 2001; 95:43–47. [DOI] [PubMed] [Google Scholar]
- 41. Takahashi M., Mizuguchi M., Shinoda H., Aizawa T., Demura M., Okazawa H., Kawano K.. Polyglutamine tract binding protein-1 is an intrinsically unstructured protein. Biochim. Biophys. Acta. 2009; 1794:936–943. [DOI] [PubMed] [Google Scholar]
- 42. Harshman S.W., Chen M.M., Branson O.E., Jacob N.K., Johnson A.J., Byrd J.C., Freitas M.A.. Isolation and analysis of linker histones across cellular compartments. J. Proteomics. 2013; 91:595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hsu L.W., Chen C.L., Nakano T., Lai C.Y., Chiang K.C., Lin Y.C., Kao Y.H., Chen S.H., Goto T., Sung W.C. et al.. The role of a nuclear protein, histone H1, on signalling pathways for the maturation of dendritic cells. Clin. Exp. Immunol. 2008; 152:576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Talati M., Seeley E., Ihida-Stansbury K., Delisser H., McDonald H., Ye F., Zhang X., Shyr Y., Caprioli R., Meyrick B.. Altered expression of nuclear and cytoplasmic histone H1 in pulmonary artery and pulmonary artery smooth muscle cells in patients with IPAH. Pulmonary Circ. 2012; 2:340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wiseman R.L., Chin K.T., Haynes C.M., Stanhill A., Xu C.F., Roguev A., Krogan N.J., Neubert T.A., Ron D.. Thioredoxin-related Protein 32 is an arsenite-regulated Thiol Reductase of the proteasome 19 S particle. J. Biol. Chem. 2009; 284:15233–15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yih L.H., Lee T.C.. Arsenite induces p53 accumulation through an ATM-dependent pathway in human fibroblasts. Cancer Res. 2000; 60:6346–6352. [PubMed] [Google Scholar]
- 47. Holcik M., Sonenberg N.. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005; 6:318–327. [DOI] [PubMed] [Google Scholar]
- 48. Popow J., Alleaume A.M., Curk T., Schwarzl T., Sauer S., Hentze M.W.. FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA. 2015; 21:1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Breyne S., Yu Y., Unbehaun A., Pestova T.V., Hellen C.U.. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. PNAS. 2009; 106:9197–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen H., Song R., Wang G., Ding Z., Yang C., Zhang J., Zeng Z., Rubio V., Wang L., Zu N. et al.. OLA1 regulates protein synthesis and integrated stress response by inhibiting eIF2 ternary complex formation. Sci. Rep. 2015; 5:13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Philippe L., Vasseur J.J., Debart F., Thoreen C.C.. La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 2018; 46:1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Buchet-Poyau K., Courchet J., Le Hir H., Seraphin B., Scoazec J.Y., Duret L., Domon-Dell C., Freund J.N., Billaud M.. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 2007; 35:1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cano F., Rapiteanu R., Sebastiaan Winkler G., Lehner P.J.. A non-proteolytic role for ubiquitin in deadenylation of MHC-I mRNA by the RNA-binding E3-ligase MEX-3C. Nat. Commun. 2015; 6:8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yuan J., Luo K., Zhang L., Cheville J.C., Lou Z.. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010; 140:384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nguyen D.T.T., Richter D., Michel G., Mitschka S., Kolanus W., Cuevas E., Wulczyn F.G.. The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death Differ. 2017; 24:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bai D.S., Wu C., Yang L.X., Zhang C., Zhang P.F., He Y.Z., Cai J.B., Song Z.J., Dong Z.R., Huang X.Y. et al.. UBAP2 negatively regulates the invasion of hepatocellular carcinoma cell by ubiquitinating and degradating Annexin A2. Oncotarget. 2016; 7:32946–32955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cano F., Miranda-Saavedra D., Lehner P.J.. RNA-binding E3 ubiquitin ligases: novel players in nucleic acid regulation. Biochem. Soc. Trans. 2010; 38:1621–1626. [DOI] [PubMed] [Google Scholar]
- 58. Hildebrandt A., Alanis-Lobato G., Voigt A., Zarnack K., Andrade-Navarro M.A., Beli P., Konig J.. Interaction profiling of RNA-binding ubiquitin ligases reveals a link between posttranscriptional regulation and the ubiquitin system. Sci. Rep. 2017; 7:16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van den Berg A., Mols J., Han J.. RISC-target interaction: cleavage and translational suppression. Biochim. Biophys. Acta. 2008; 1779:668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Caudy A.A., Ketting R.F., Hammond S.M., Denli A.M., Bathoorn A.M., Tops B.B., Silva J.M., Myers M.M., Hannon G.J., Plasterk R.H.. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003; 425:411–414. [DOI] [PubMed] [Google Scholar]
- 61. Liu Y., Ye X., Jiang F., Liang C., Chen D., Peng J., Kinch L.N., Grishin N.V., Liu Q.. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science (New York, N.Y.). 2009; 325:750–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jin P., Zarnescu D.C., Ceman S., Nakamoto M., Mowrey J., Jongens T.A., Nelson D.L., Moses K., Warren S.T.. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004; 7:113–117. [DOI] [PubMed] [Google Scholar]
- 63. Caudy A.A., Myers M., Hannon G.J., Hammond S.M.. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002; 16:2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Usuki F., Yamashita A., Fujimura M.. Environmental stresses suppress nonsense-mediated mRNA decay (NMD) and affect cells by stabilizing NMD-targeted gene expression. Sci. Rep. 2019; 9:1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Martin L., Gardner L.B.. Stress-induced inhibition of nonsense-mediated RNA decay regulates intracellular cystine transport and intracellular glutathione through regulation of the cystine/glutamate exchanger SLC7A11. Oncogene. 2015; 34:4211–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sieber J., Hauer C., Bhuvanagiri M., Leicht S., Krijgsveld J., Neu-Yilik G., Hentze M.W., Kulozik A.E.. Proteomic analysis reveals Branch-specific regulation of the unfolded protein response by Nonsense-mediated mRNA decay. Mol. Cell. Proteomics: MCP. 2016; 15:1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gregersen L.H., Schueler M., Munschauer M., Mastrobuoni G., Chen W., Kempa S., Dieterich C., Landthaler M.. MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol. Cell. 2014; 54:573–585. [DOI] [PubMed] [Google Scholar]
- 68. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (68) partner repository with the dataset identifier PXD016735. Scripts are available upon request.