Abstract
Post-translational histone modifications and chromatin remodelling play a critical role controlling the integrity of the genome. Here, we identify histone lysine demethylase PHF2 as a novel regulator of the DNA damage response by regulating DNA damage-induced focus formation of 53BP1 and BRCA1, critical factors in the pathway choice for DNA double strand break repair. PHF2 knockdown leads to impaired BRCA1 focus formation and delays the resolution of 53BP1 foci. Moreover, irradiation-induced RPA phosphorylation and focus formation, as well as localization of CtIP, required for DNA end resection, to sites of DNA lesions are affected by depletion of PHF2. These results are indicative of a defective resection of double strand breaks and thereby an impaired homologous recombination upon PHF2 depletion. In accordance with these data, Rad51 focus formation and homology-directed double strand break repair is inhibited in cells depleted for PHF2. Importantly, we demonstrate that PHF2 knockdown decreases CtIP and BRCA1 protein and mRNA levels, an effect that is dependent on the demethylase activity of PHF2. Furthermore, PHF2-depleted cells display genome instability and are mildly sensitive to the inhibition of PARP. Together these results demonstrate that PHF2 promotes DNA repair by homologous recombination by controlling CtIP-dependent resection of double strand breaks.
INTRODUCTION
The DNA damage response (DDR), which detects, signals and repairs DNA lesions, is essential in the maintenance of genome integrity and functions as a first defence in the early stages of cancer development (1). Among the different types of DNA lesions, DNA double strand breaks (DSBs) are particularly hazardous to the cell as, if not repaired adequately, they can lead to chromosomal rearrangements (2).
Mammalian cells have developed different pathways to repair DSBs. Non-homologous end joining (NHEJ) is a fast, and efficient, but also error-prone pathway in which the broken DNA-ends are directly ligated. On the other hand, during homologous recombination (HR), the sister chromatid is used as a repair template and thereby results in more accurate repair. Finally, the less efficient alternative non-homologous end joining (Alt-NHEJ), also called microhomology-mediated end joining (MMEJ), uses short microhomologous sequences during the alignment of broken ends (3). Several factors are important in DSB repair pathway choice, in which the availability of homologous sequence, and therefore the cell cycle stage, plays a critical role. Two DDR proteins that have an important influence on this decision are 53BP1 and BRCA1, that together control DNA end resection, the degradation of the DNA end in the 5′ to 3′ direction, resulting in singe-stranded (ss) DNA that is critical for DSB repair by HR (4). Whereas 53BP1, together with its partner RIF1 and the Shieldin complex (5,6), blocks DNA end-resection and thus stimulates repair through NHEJ, BRCA1 promotes DNA end-resection and the removal of 53BP1 from sites of DNA damage, thereby switching repair from NHEJ to HR (7,8). DNA end resection is initiated by endonuclease CtIP, in cooperation with the Mre11/Rad50/Nbs1 (MRN) complex, and thereafter extended by the EXO1 and DNA2 nucleases (9). The resulting ssDNA is protected by immediate coating with the RPA1-3 complex, which is replaced by the Rad51 protein that then mediates strand invasion (10).
Efficient DNA repair requires the correct and timely coordination of a multitude of signalling events, in which posttranslational modifications play a critical role. As DNA lesions occur and are repaired in the context of chromatin, it is not surprising that chromatin modifications impact on this process (11). One of the earliest modifications upon the induction of a DSB is the phosphorylation of histone H2AX at serine 139 (named γH2AX) by the central DDR kinase ATM, on either side of the lesion (12). H2AX phosphorylation triggers the initiation of protein ubiquitination by the RNF8 E3 ubiquitin ligase, which is recruited to DSBs via binding to MDC1, a direct reader of γH2AX (13). Ubiquitination of histones and other proteins by RNF8 and RNF168, another E3 ligase that is subsequently bound, serves to recruit additional proteins, among which are 53BP1 and BRCA1 (14,15). Also, the modification of histone and non-histone proteins by methylation and acetylation are involved in the regulation of DNA repair. For example, the recruitment of 53BP1 depends on the methylation of H4K20, the RNF8-dependent degradation of competing H4K20me readers, deacetylation of H4K16 and RNF168-mediated H2AK15 ubiquitination (16). In addition to promoting the direct recruitment of repair proteins, chromatin modifications can physically facilitate the accessibility of regulatory proteins to the lesion. An example is lysine acetylation, which opens up the chromatin (17). Finally, histone methylation regulates gene expression, by recruiting proteins involved in this process or by inhibiting the binding of transcription factors to DNA. Lysine methylation of histone H3 and H4 is associated to both transcriptional activation and repression, depending on the methylation site (18).
Interestingly, defects in the regulation of chromatin modifying enzymes were described to be linked to genome instability and tumour development (19). Here, we identified PHD Finger Protein 2 (PHF2), also known as KDM7C or JHDM1E, member of the KDM7 family of histone demethylases, in a search for additional chromatin modifications and regulators involved in DNA repair. PHF2 was described as a transcriptional activator by demethylating H3K9me2, although PHF2 was also shown to demethylate H4K20me3, another transcriptional repression mark, and suggested to demethylate non-histone proteins (20–23). Our results show that PHF2 regulates the resection of DSBs by controlling the levels of CtIP and BRCA1. By demonstrating that PHF2 regulates DNA DSB repair in this way, we identify a novel function for this lysine-specific demethylase.
MATERIALS AND METHODS
Cell culture and treatments
293T, U2OS and HeLa cells were grown in DMEM supplemented with 10% FBS and penicillin–streptomycin. U2OS DR-GFP, U2OS EJ5-GFP and U2OS SA-GFP reporter stable cell lines were maintained in medium supplemented with 1 μg/ml puromycin. Rap80 knock out U2OS cells were kindly provided by Dr. Daniel Durocher (Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Canada) and have been validated previously (24).
Cells were treated with camptothecin (CPT, 2 μM), hydroxyurea (HU, 10 mM), ultra violet light (UV, 40 J/m2), etoposide (ETP, 20 μM) or ionizing radiation using a CellRad (Faxitron) or a WOMed superficial X-Ray Therapy Unit and harvested 1 h later, unless stated otherwise.
siRNA oligos, plasmids and transfection
siRNA oligonucleotides (Sigma, Microsynth AG) were transfected into cells using Lipofectamine RNAiMax (Invitrogen). Sequences of oligonucleotides were as follows:
| Luc | CGUACGCGGAAUACUUCGA |
| Non-Target | UGGUUUACAUGUCGACUAA |
| PHF2#1 | GCUGGAAAUUCGAGAGCAA |
| PHF2#2 | GCUAGAGAAGUCGCCUCUA |
| PHF2#3 | CCACUUUAAGGACAGCCUU |
| CtIP | GCUAAAACAGGAACGAAUC |
| BRCA1 | AAGGAACCUGUCUCCACAAAG |
Plasmid DNA was transfected into cells using Polyethylenimine (Sigma Aldrich) or Lipofectamine 2000 (Invitrogen).
p3FLAG murine PHF2 was kindly provided by Dr Xiaobing Shi (Department of Epigenetics and Molecular Carcinogenesis, The University of Texas M.D. Anderson Cancer Center, Houston, USA). Three silent mutations (in capital, gctggaGatCAgGgagcaa) were introduced in the Flag-PHF2 plasmid) to make it resistant to siRNA oligonucleotide PHF2#1 (Flag-PHF2*). A demethylase inactive version of PHF2 containing the mutations H249A/D251A (catalytic inactive, CI) and PHF2 PHD domain mutant with W29E/H31A/C34A (PHDm) were generated in Flag-PHF2*. For mutagenesis, the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) was used and mutants were verified by Sanger-Sequencing. GFP-CtIP and mCherry-NBS1 expressing plasmids have been previously described (25,26).
Antibodies and western blot
Antibodies obtained from commercial sources were as following: β-actin and Histone H3 from Genscript, Ku86 (C-20) and p53 (DO-1) from Santa Cruz Biotechnology, 53BP1 (Ab172580) and NBS1 (Ab175800) from Abcam, pSer139-H2AX (clone: JWB301), BRCA1 (clone MS110) from Merck-Millipore, pSer345-CHK1 and PHF2 from Cell Signalling, PHF2 and pSer4/8-RPA2 from Bethyl, RPA2 from Novus Biologicals, Rad51 by Invitrogen and CtIP from Active Motif.
Also, antibodies against CtIP and Mre11 were generated by injecting rabbits with a His-tagged antigen (amino acids 150–500 and 182–480, respectively) that were obtained by expression in bacteria and purification with a Ni-NTA resin (Qiagen) following manufacturers recommendations.
Cell were lysed in Laemmli sample buffer. Lysates containing equal amounts of protein, measured by the BCA method (Thermo Scientific), were subjected to SDS-PAGE. The chemiluminescent images were obtained using the ImageQuant LAS 4000 mini and quantified using the ImageQuant TL software (GE Healthcare).
Comet assay
Single Cell Gel Electrophoresis (SCGE) was carried out using the CometAssay® ES II kit (Trevigen) according to the manufacturer's instructions. Images were taken using a Zeiss Cell Observer fluorescent microscope and the tail moment of at least 50 cells per experiment was analysed with the TriTek CometScore software.
RT-PCR
RNA was isolated using the RiboZol Extraction Reagent (VWR) according to the manufacturer's instructions. cDNA synthesis and PCR amplification were carried out in the same tube using the qScript One-Step SYBR Green RT-qPCR Kit (Quantabio) in a LightCycler480 II (Roche). All reactions were performed in triplicate. Transcript levels were normalized in parallel with test genes GAPDH. The primers used were the following:
BRCA1-F: ACCTTGGAACTGTGAGAACTCT
BRCA1-R: TCTTGATCTCCCACACTGCAATA
CtIP-F: CAGGAACGAATCTTAGATGCACA
CtIP-R: GCCTGCTCTTAACCGATCTTCT
GAPDH-F: GGAGCGAGATCCCTCCAAAAT
GAPDH-R: GGCTGTTGTCATACTTCTCATGG
Mre11-F: ATGCAGTCAGAGGAAATGATACG
Mre11-R: CAGGCCGATCACCCATACAAT
RPA2-F: GCACCTTCTCAAGCCGAAAAG
RPA2-R: CCCCACAATAGTGACCTGTGAAA
DR-GFP, SA-GFP and EJ5-GFP assays
U2OS cells bearing a single copy integration of the reporters DR-GFP (HR (27)), SA-GFP (SSA (28)) or EJ5-GFP (NHEJ (28)) were used to analyse the different DSB repair pathways. Cells were seeded in 6-well plates in duplicate and transfected with the indicated siRNA oligonucleotide. The next day, cells were infected with lentiviral particles containing I-SceI–BFP expression construct at MOI 10 using 8 μg/ml polybrene. Forty eight hours later, cells were collected by trypsinization and fixed with 4% paraformaldehyde for 20 min. Samples were analysed with a BD FACSAria with the BD FACSDiva Software.
The number of GFP-positive cells from at least 10 000 events positive for blue fluorescence (infected with the I-SceI–BFP construct) was scored. The average of both duplicates was calculated for each sample and normalized to siRNA control. At least three independent experiments were carried out for each condition and the mean and standard deviation of the three experiments represented.
Chromatin fractionation
Biochemical fractionation of cells was performed as previously described (29,30). Soluble cytoplasmic and soluble nuclear fractions were pooled to one soluble fraction.
Immunofluorescence
For immunostaining, cells were fixed in 4% paraformaldehyde for 15 min and then permeabilized with PBS + 0.2% Triton X-100 for 10 min at RT. Samples were blocked in PBS + 0.5% FCS, immunostained with antibodies as indicated and mounted with DAPI. For RPA2, BRCA1 and Rad51 focus formation, cells were pre-extracted (20 mM HEPES pH 8, 20 mM NaCl, 5 mM MgCl2, 0.5% NP40) for 5 min at 4°C before fixation.
Images of cells were taken using a Zeiss Cell Observer fluorescent microscope equipped with a 63× NA 1.3 objective and ZEN imaging software. The number of foci was evaluated using the ImageJ software. In all instances, >100 cells were analysed for each point and error bars on graphs represent the standard deviation of three independent experiments. Cells with >10 foci were scored as positive.
Laser micro-irradiation
Multiphoton laser micro-irradiation was essentially performed as described previously (31). Cells, grown on coverslips, were placed in a Chamlide CMB magnetic chamber and the regular culture medium was replaced by CO2-independent Leibovitz's L15 supplemented with 10% FBS and penicillin-streptomycin. Laser micro-irradiation was carried out on a Leica SP5 confocal microscope equipped with an environmental chamber set to 37°C. DSB-containing tracks (1.5 μm width) were generated with a Mira mode-locked titanium-sapphire (Ti:Sapphire) laser (λ = 800 nm, pulse length = 200 fs, repetition rate = 76 MHz, output power = 80 mW) using a UV-transmitting 63× 1.4 NA oil immersion objective (HCX PL APO; Leica). Confocal images were recorded before and after laser irradiation at 5 or 10 s time intervals over a period of 5–10 min.
To examine accumulation of Flag-PHF2, a different field was irradiated every minute for 20 min, after which the cells were fixed for immunofluorescence.
FokI assays
U2OS 2-6-3 cells expressing inducible FokI-mCherry-LacR (32) were treated with 300 nM 4-OHT and 1 μM Shield-I for 5 h for inducing stabilization and nuclear localization of the expressed product. Subsequently, cells were fixed with PFA and immunostained with the indicated antibodies as described above.
Cell cycle analysis
For cell cycle analysis, cells were fixed in 70% ethanol at 4°C o/n. After fixation, cells were washed with PBS, and the DNA was stained with propidium iodide (PI). Cells were analysed using a Macsquant Analyzer with Macsquantify software (Miltenyi).
Clonogenic survival
To determine cellular sensitivity to DNA damaging agents, HeLa cells were transfected with the indicated siRNA oligonucleotides. Forty eight hours later, 500 cells were seeded in six-well dishes and treated with the indicated concentrations of Olaparib (Cayman Chemical) or doses of IR. Following 7–10 days in culture, cells were fixed, stained and colonies were counted. Triplicate cultures were scored for each treatment. Shown is the relative survival as compared to the undamaged control and the error bars present the standard error of three independent experiments.
Gene expression correlation analysis
Gene expression correlation analysis was performed using the Gene Expression Profiling Interactive Analysis (GEPIA), a web-based tool containing high-throughput RNA sequencing data (TCGA and GTEx databases) (33). A pairwise gene correlation analysis for given tumour data sets of TGCA was performed, with PHF2 as ‘gene A’ and a correlation against ‘gene B’: RBBP8 (CtIP), BRCA1 or GAPDH. All genes were normalized to TUBA1A (Tubulin alpha-1A). The correlation coefficient was calculated using Spearman rank correlation test against the tumour dataset, which included a range different tumours of the female reproductive tract (cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC, n = 306), ovarian serous cystadenocarcinoma (OV, n = 426), uterine corpus endometrial carcinoma (UCEC, n = 174) and uterine carcinosarcoma (UCS, n = 57)), and breast invasive carcinoma (BRCA, n = 1085).
Statistical analysis and reproducibility
Gaussian (normal) distribution of the data was assessed using a D' Agostino–Pearson omnibus normality test. In case of normal distribution, an unpaired Student's t-test was used to determine whether the difference between the means of two sets of values was significant. In case the distribution was not normal, a Mann–Whitney test was used. *P < 0.1 **P < 0.01, ***P < 0.001, ****P < 0.0001. P < 0.05 was considered statistically significant. NS = not significant. Unless stated otherwise, representative experiments are shown out of at least two independent ones and depicted is the mean ± standard deviation. Additional details are listed in the individual figure legends.
RESULTS
The demethylase PHF2 controls the DNA damage response
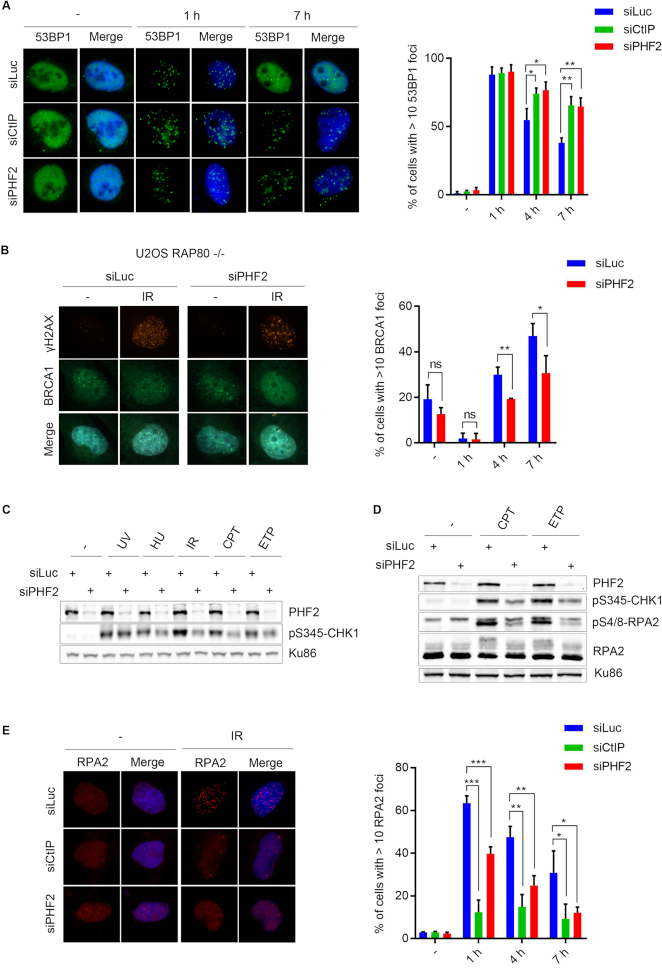
To identify novel enzymes that regulate DSB repair by NHEJ and HR, we analysed ionizing radiation (IR)-induced focus formation of 53BP1 and BRCA1, that together control the choice between these two DSB repair pathways, in cells depleted for individual enzymes involved in post-translation modifications of histones by siRNA. Modulation of the expression level of PHF2/KDM7C/JHDM1E, a lysine-specific histone demethylase hereafter called PHF2, changed the dynamics of 53BP1 focus formation in response to IR. Whereas irradiating U2OS cells triggered efficient 53BP1 focus formation, the number of cells with 53BP1 foci and the number of 53BP1 foci per cell stayed high at later time points (4–7 h) in cells depleted for PHF2 by siRNA whereas at these time points 53BP1 foci decreased again in control transfected populations (Figure 1A and Supplementary Figure S1A). The effect of PHF2 depletion on 53BP1 focus resolution was the same as that of downregulation of CtIP, which served as a control for negatively regulating HR by preventing the initiation of DNA resection (Figure 1A). In contrast, overexpression of Flag-PHF2 prevented IR-induced focus formation of 53BP1 (Supplementary Figure S1B). To examine the effect of modulating PHF2 levels on HR, we monitored the recruitment of BRCA1 in Rap80 knock out U2OS cells, in which Rap80-mediated recruitment of BRCA1, subsequent BRCA1-A complex formation and suppression of HR are prevented (31,34). Whereas treating control siRNA transfected cells with IR led to efficient focus formation of BRCA1 that increased in time, depletion of PHF2 by siRNA impaired IR-induced BRCA1 foci (Figure 1B). The observed altered dynamics of BRCA1 and 53BP1 focus formation upon modulating the levels of PHF2 suggests that this demethylase regulates DSB repair. As a consequence, depletion of PHF2 led to increased focus formation of phosphorylated H2AX (γH2AX), used as marker for DNA damage, as compared to control-depleted cells (Supplementary Figure S1C).
Figure 1.
PHF2 knockdown alters 53BP1 and BRCA1 focus dynamics in response to IR. (A) U2OS cells were depleted for Luciferase (Luc), CtIP or PHF2 by siRNA. After 48 h, cells were treated with IR (10 Gy) and fixed after 1, 4 or 7 h. 53BP1 focus formation was analysed by immunofluorescence. Left panel: representative images. Right panel: quantification of three independent experiments with each at least 100 cells. (B) U2OS Rap80 knockout cells were depleted for Luc or PHF2. Cells were treated with IR (10 Gy), fixed at the indicated time points and γH2AX (positive control for DNA damage induction) and BRCA1 focus formation was analysed as in (A). Left panel: representative images (IR, 4 h). (C) U2OS cells depleted for Luc or PHF2 were subjected to different DNA damaging agents and lysed after 1 h. Extracts were analysed by western blot using the indicated antibodies. (D) U2OS cells were transfected as in (C), treated with CPT or ETP and analysed by western blot with the indicated antibodies. (E) U2OS cells were depleted for CtIP or PHF2 by siRNA. Cells were treated with IR (3 Gy), fixed at the indicated time points and RPA2 focus formation was analysed as in (A). Left panel: representative images (IR, 1 h).
Depletion of PHF2 affects DSB resection and DNA repair
We continued to study what process in the DNA damage response is affected by PHF2. To this end, we first examined DNA damage-induced checkpoint activation by exposing U2OS cells to different types of DNA damage and monitoring the phosphorylation of CHK1, a critical effector kinase of this response (35). Although DNA damaging agents triggered efficient CHK1 phosphorylation in control depleted cells, PHF2 depletion reduced the DNA damage-induced phosphorylation of CHK1, especially after IR, camptothecin and etoposide (Figure 1C). These treatments result in a more efficient DSB formation compared to UV and HU, which only resulted in a moderate effect, substantiating a possible role for PHF2 in the DSB response. In addition, these treatments need DNA end resection to generate ssDNA, subsequently covered by the RPA1–3 complex, that triggers the activation of ATR and phosphorylation of its substrates such as CHK1 (36,37). We therefore examined if PHF2 depletion affected DNA end resection, by studying RPA2 phosphorylation using western blot analysis (38). Downregulation of PHF2 led to lower levels of RPA2 phosphorylation on Ser4/8 and reduced levels of total RPA2 phosphorylation, as demonstrated by a lower mobility shift using an antibody against total RPA2, in response to camptothecin and etoposide, as compared to control transfected cells (Figure 1D). To corroborate these findings, we also measured IR-induced focus formation of RPA2 by immunofluorescence. As previously published, depletion of CtIP completely inhibited focus formation of RPA2 (Figure 1E). Notably, although less pronounced when compared to that after CtIP knockdown, the downregulation of PHF2 also significantly reduced IR-induced focus formation of RPA (Figure 1E).
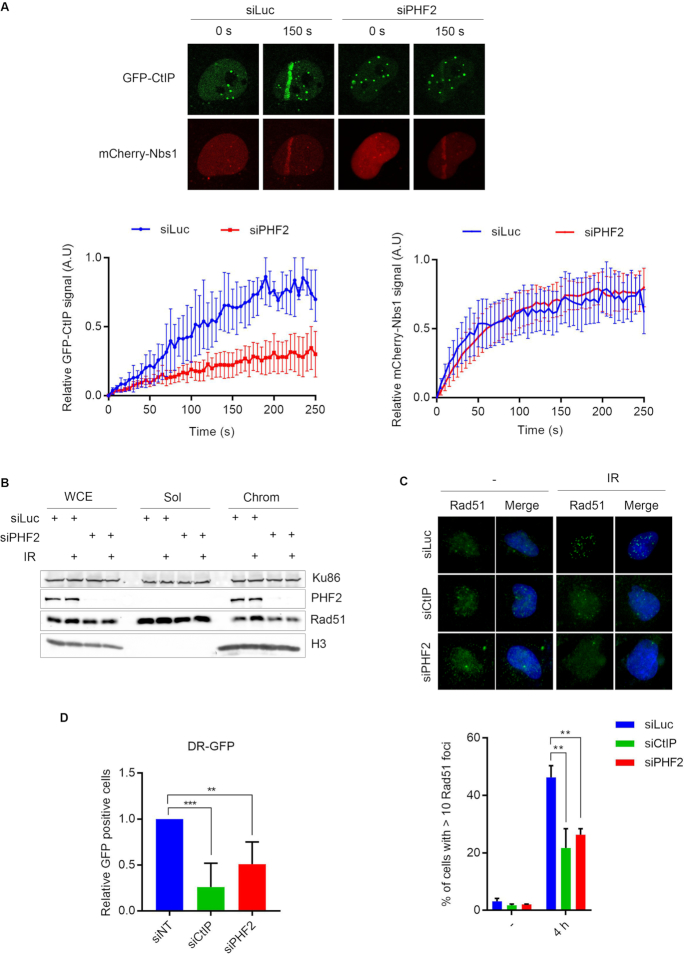
DSB resection is initiated by CtIP, together with the MRN complex. Given the effect of PHF2 on DNA end resection, we wondered if PHF2 functions at the level of CtIP. We consequently monitored the recruitment of GFP-CtIP to DSB-containing tracks in U2OS cells by laser micro-irradiation (25). An inhibition in the accumulation of GFP-CtIP to the laser tracks was observed upon depletion of PHF2, whereas the recruitment of mCherry-Nbs1, a component of the MRN complex, was unaffected in these conditions (Figure 2A). To investigate the consequences of the effect of PHF2 on DNA end resection, we examined the IR-induced accumulation of HR protein Rad51 on the chromatin. Downregulation of PHF2 prevented this accumulation (Figure 2B). In accordance, when analysing Rad51 focus formation in response to IR by immunofluorescence, we observed that knockdown of PHF2, as well as depletion of CtIP, inhibited focus formation by Rad51 (Figure 2C). Together these results indicate that PHF2 promotes the DSB response by regulating DNA resection and strongly suggest that the subsequent DSB repair is affected by PHF2 depletion.
Figure 2.
Depletion of PHF2 impairs DSB repair by homologous recombination. (A) U2OS cells, transfected with Luc or PHF2 siRNA oligos, and 24 h later transfected with GFP-CtIP and mCherry-Nbs1, were laser-irradiated and analysed by time-lapse imaging. Upper panel: representative images of the indicated time points. Lower panel: relative fluorescence (left: GFP-CtIP, right: mCherry-Nbs1) at laser stripes of at least 50 cells per experiment. (B) U2OS cells were depleted for Luc or PHF2 by siRNA, treated with IR (10 Gy) and subjected to chromatin fractionation (WCE: whole cell extracts, Sol: soluble and Chrom: chromatin). Samples were analysed by western blot using the indicated antibodies. (C) U2OS cells were depleted for Luc, CtIP or PHF2 by siRNA. Cells were treated with IR (3 Gy), fixed after 4 h and Rad51 focus formation was analysed by immunofluorescence. Top panel: representative images. Bottom panel: quantification from three independent experiments with each at least 100 cells. (D) U2OS cells stably expressing a single copy of the DR-GFP reporter construct were depleted of CtIP, PHF2 or control (non-target, NT). After 48 h, GFP fluorescence was analysed by flow cytometry. Presented is the relative fluorescence as compared to the control cells, of three independent experiments.
DNA end resection is a critical step in homology-directed DSB repair (4). To address whether PHF2 impacts on DSB repair by homologous recombination, we used a DR-GFP reporter assay in U2OS. In these cells, depletion of PHF2 caused a significant decrease in HR efficiency (Figure 2D). The efficiency of Single Strand Annealing (SSA), another form of homology-directed repair that also depends on CtIP-mediated DNA end resection but is Rad51-independent (39), measured using an SA-GFP reporter assay, was also dramatically affected by PHF2 knockdown (Supplementary Figure S1D). Surprisingly, depletion of PHF2 also slightly inhibited NHEJ, not dependent on DNA end resection, as measured by the EJ5-GFP reporter (Supplementary Figure S1E).
Cell cycle is a major determinant of DSB repair pathway choice. As HR depends on the availability of a sister chromatid as repair template, this type of repair is restricted to the S and G2 phases of the cell cycle (40). Importantly, PHF2 depletion only very slightly decreased the percentage of cells in S phase, making it unlikely that the decrease in DSB repair efficiency upon PHF2 knockdown is due to an indirect effect on the cell cycle (Supplementary Figure S1F).
PHF2 controls CtIP and BRCA1 mRNA levels by means of its demethylase activity
We next set out to address if PHF2 affects DSB repair in a direct manner by acting at sites of DNA damage. To this end, the accumulation of Flag-PHF2 at sites of DNA damages generated by laser micro-irradiation was examined. Although mCherry-Nbs1 and γH2AX were detected at damaged regions upon laser micro-irradiation, we did not observe detectable Flag-PHF2 accumulation to such laser-tracks (Supplementary Figure S2A). In addition, also no accumulation of Flag-PHF2 was observed to a DSB created in a single genomic locus containing an array of LacO repeats following expression and tethering of an mCherry-LacI-FokI nuclease fusion (Supplementary Figure S2B). Together these results could be an indication that PHF2 regulates DSB repair in an indirect and more global manner.
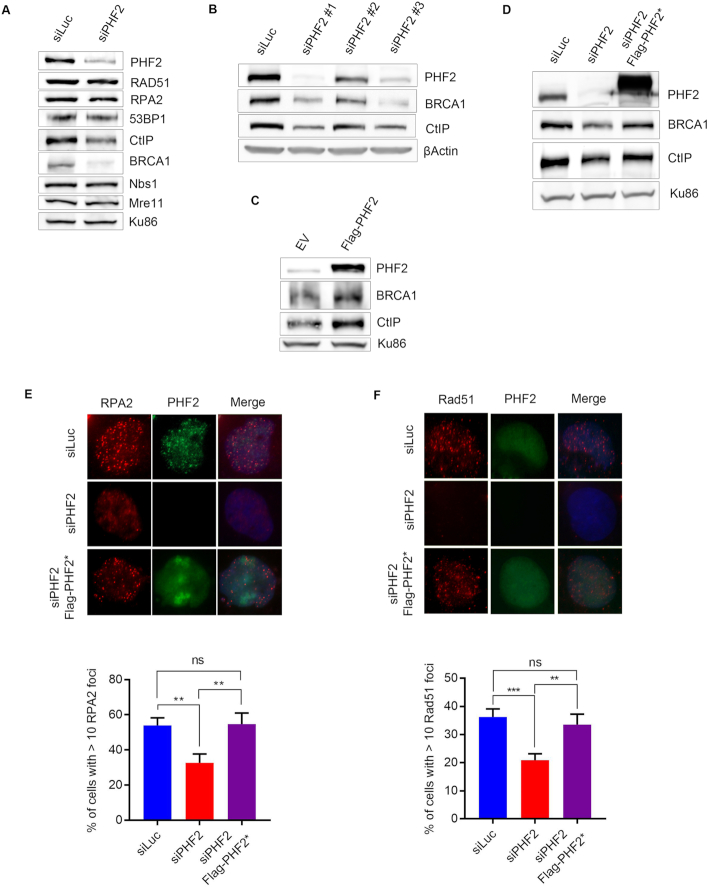
Interestingly, PHF2 contains a zinc finger-like PHD (plant homeodomain) finger, a motif found in proteins that are involved in transcriptional regulation, possibly by recognizing chromatin modifications (41), which led to the suggestion that PHF2 might regulate DNA repair in a transcriptional manner. Western blot analysis of the levels of proteins involved in DSB repair demonstrated that downregulation of PHF2 led to diminished protein levels of CtIP as well as BRCA1, while leaving Rad51, RPA2, 53BP1, Nbs1 and Mre11 unaffected (Figure 3A). Downregulating PHF2 by two additional siRNA oligonucleotides resulted in the same phenotype as seen before, namely a diminished abundance of CtIP and BRCA1 protein (Figure 3B). In contrast, overexpression of Flag-PHF2 had the opposite effect: both BRCA1 and CtIP protein levels increased under these conditions (Figure 3C). Furthermore, the lower protein levels of BRCA1 and CtIP upon depletion of PHF2 were partially rescued by expressing an siRNA-resistant version of Flag-PHF2 (Figure 3D). In addition, we could complement the effects of PHF2 depletion on focus formation of RPA2, Rad51 and 53BP1 by expressing siRNA-resistant Flag-PHF2 (Figure 3E, F and Supplementary Figure S3A, respectively). Together these data demonstrate that the effects of modulating PHF2 levels are genuinely due to the depletion of PHF2 instead of an off-target effect of the siRNA oligonucleotides used.
Figure 3.
PHF2 regulates HR by modulating CtIP and BRCA1 levels. (A) U2OS cells were depleted for Luc or PHF2 by siRNA. Forty eight hours later, the cells were lysed and extracts were analysed by western blot with the indicated antibodies. (B) U2OS cells were depleted for PHF2 using three different siRNA oligonucleotides and subsequently analysed by western blot using the indicated antibodies. (C) U2OS cells were transfected with empty vector (EV) or a Flag-PHF2 expression vector, followed by western blot analysis with the indicated antibodies. (D) U2OS cells were depleted for Luc or PHF2 by siRNA and 24 h later transfected with EV or siRNA-resistant Flag-PHF2 (Flag-PHF2*). The day after, extracts were prepared and analysed by western blot with the indicated antibodies. (E) U2OS cells were depleted for PHF2 and transfected with Flag-PHF2* the day after. One day later, cells were treated with IR (3 Gy) and fixed for IF after 1 h. RPA2 focus formation of Flag-positive cells was analysed by immunofluorescence. Top panel: representative images. Bottom panel: quantification of three independent experiments, counting at least 50 cells each. (F) U2OS cells were depleted for PHF2 and transfected with Flag-PHF2* the day after. One day later, cells were treated with IR (3 Gy) and fixed for IF after 4 h. Rad51 focus formation of Flag-positive cells was analysed as in (E).
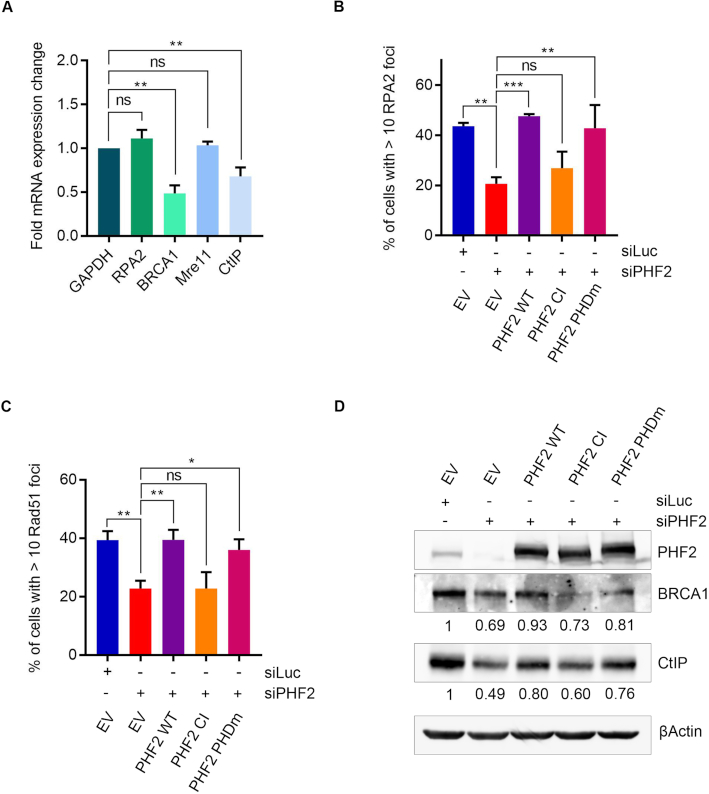
We next examined if PHF2 regulates CtIP and BRCA1 at the transcriptional level by investigating the effect of PHF2 depletion on CtIP and BRCA1 mRNA in U2OS by quantitative PCR. The mRNA levels of CtIP and BRCA1, but not those of Mre11 and RPA2, were decreased under conditions of PHF2 depletion when compared to control transfected cells (Figure 4A), indicating that PHF2 controls homology-directed DSB repair by regulating the mRNA levels of CtIP and BRCA1, two proteins critical for DNA end-resection and thereby initiation of HR.
Figure 4.
PHF2 controls the expression of CtIP and BRCA1 through demethylase activity. (A) U2OS were downregulated with Luc or PHF2 siRNA oligos. 72 h later, RNA was isolated and mRNA levels of GAPDH, RPA2, BRCA1, Mre11 and CtIP were determined by RT-qPCR. Shown is the fold mRNA change of PHF2-depleted samples as compared to the Luc control. (B) U2OS cells were depleted for PHF2 and transfected with EV, siRNA resistant Flag-PHF2 wild type (WT), demethylase inactive Flag-PHF2 (CI) or PHD domain mutant Flag-PHF2 (PHDm) the day after. One day later, cells were treated with IR (5 Gy) and fixed for IF after 1 h. RPA2 focus formation of Flag-positive cells was analysed by immunofluorescence. Quantification of three independent experiments with at least 50 cells each is shown. (C) U2OS cells were transfected as in (B). One day later, cells were treated with IR (10 Gy) and fixed for IF after 4 h. Rad51 focus formation of Flag-positive cells was analysed by immunofluorescence. Quantification of three independent experiments with at least 50 cells each. (D) U2OS cells were transfected as in (B) and the day after, extracts were prepared and analysed by western blot with the indicated antibodies. Quantifications of BRCA1 and CtIP levels, compared to the loading control, are shown below each panel.
As mentioned before, PHF2 harbours an N-terminal PHD domain, a chromatin reader module, in addition to the enzymatically active Jumonji-C (JmjC) domain (41). To gain more insight into the mechanism of how PHF2 controls DSB repair, we investigated if the effect of PHF2 on DNA end resection depends on the lysine demethylase activity and/or on the presence of a functional PHD domain. To do so, mutants of these domains were generated by introducing changes in critical residues within these regions in siRNA resistant Flag-PHF2 (42,43). These mutants were subsequently expressed in PHF2-depleted cells and IR-induced focus formation of RPA2 and Rad51 were studied. The defect in RPA2 and Rad51 focus formation caused by knockdown of PHF2 could be rescued by the wild type (WT) and PHD mutant (PHDm) version of PHF2 but not with catalytic inactive (CI) PHF2 (Figure 4B and C). In accordance with these data, whereas WT and PHDm PHF2 could complement the decrease in BRCA1 and CtIP protein levels caused by PHF2 depletion, expressing PHF2 CI did not rescue the reduced BRCA1 and CtIP levels (Figure 4D). Together these results demonstrate that the lysine demethylase activity is required for the effect of PHF2 in resection. In contrast, the PHD chromatin reader domain seems to be dispensable.
PHF2 downregulation causes genome instability
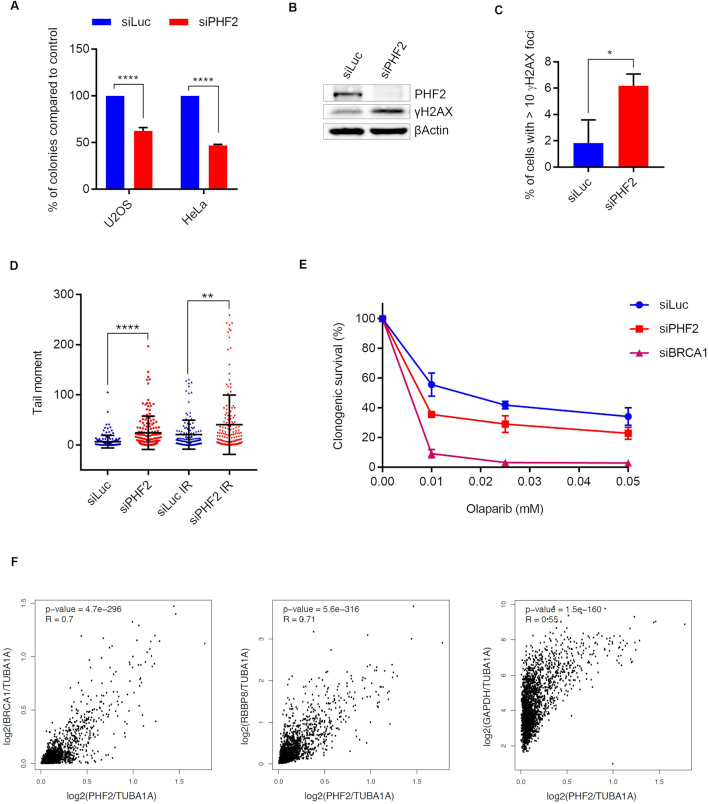
As DNA repair is also important in unperturbed cells, defective DSB repair caused by downregulation of PHF2 is likely to affect genome stability and cell survival in these conditions. Consistent with such a scenario, the colony forming capacity of both U2OS and HeLa cells decreased dramatically after depletion of PHF2 as compared to Luciferase knockdown control cells (Figure 5A). Interestingly, although downregulation of CtIP also affected clonogenic survival, the effect was less severe than after PHF2 depletion, suggesting that the effect of PHF2 on cell survival is not solely due to its function in controlling HR (Supplementary Figure S3B). Western blot analysis demonstrated an increase in γH2AX in PHF2-depleted U2OS cells in the absence of exogenous damage (Figure 5B), and this was confirmed by an elevated percentage of cells with γH2AX foci upon PHF2 knockdown as compared to Luc control cells by immunofluorescence (Figure 5C). To directly asses the appearance of DNA breaks resulting from downregulation of PHF2 in individual cells, we employed the alkaline comet assay. Compared to control depleted cells, decreasing PHF2 by siRNA led to a significant tail moment increase in U2OS and Hela cells, in the absence of exogenous DNA damage as well as after IR (Figure 5D and Supplementary Figure S3C). Together these data demonstrate that regulated levels of PHF2 are important for the maintenance of genome stability.
Figure 5.
PHF2 controls genome stability. (A) U2OS and HeLa cells were depleted for PHF2 by siRNA. Equal numbers of cells were seeded and incubated for 10 days for colonies to form. Bar graph shows the number of colonies compared to control depleted cells from three independent experiments. (B) U2OS cells were depleted for Luc or PHF2 by siRNA. 48 h later the cells were lysed and analysed by western blot with the indicated antibodies. (C) As in (B), but now γH2AX focus formation was analysed by IF. Quantification of three independent experiments with each at least 100 cells. (D) U2OS cells, depleted for PHF2 by siRNA, were left untreated or irradiated with 5 Gy and processed after 1 h for comet assay analysis. Depicted is the tail moment of three independent experiments (at least 50 cells each). (E) Clonogenic survival assays of HeLa cells that were depleted for Luc, PHF2 or BRCA1 by siRNA and incubated with the indicated concentrations of Olaparib. Shown is the relative survival as compared to the undamaged control. Error bars represent the SEM of three individual experiments. (F) Correlation analysis between expression of PHF2 and expression of BRCA1, RBBP8 and GAPDH by GEPIA. Scatter plots represent the correlation between gene expression of PHF2 and BRCA1 (left), RBBP8 (middle) or GAPDH (right) in each tumour (n = 2048), with R as the correlation coefficient.
Notably, HR deficiency is exploited in the treatment of tumours with BRCA1/BRCA2 mutations as cells in such conditions are sensitive to inhibition of poly(ADP-ribose) polymerase (PARP) (44). As PHF2 depletion affects HR, downregulation of this protein is likewise expected to result in sensitivity to PARP inhibition. Indeed, depletion of PHF2 mildly sensitized HeLa cells to inhibition of PARP1/2 by Olaparib (Figure 5E), which is in accordance to its effect on BRCA1 levels. Co-depletion of PHF2 and BRCA1 did not increase the sensitivity to PARP1/2 inhibition as compared to BRCA1 knockdown alone, suggesting that the Olaparib sensitivity after PHF2 depletion is due to its effect on BRCA1 (Supplementary Figure S3D). In addition, downregulation of PHF2 also caused sensitivity to IR (Supplementary Figure S3E). Together these data underscore the importance of PHF2 in HR.
Finally, we analysed the gene expression of PHF2, BRCA1 and RBBP8, the gene encoding CtIP, in a broad range of tumours of breast and the female reproductive tract by GEPIA (33). As shown in Figure 5F, the expression of PHF2 correlated with that of BRCA1 and RBBP8 in a high proportion of the tumours (R ≥ 0.7), whereas the correlation with GAPDH was lower (R = 0.55). These data are in accordance with our previous experiments and strongly suggest that the mechanism of regulation of BRCA1 and CtIP expression by PHF2 also occurs in tumours.
DISCUSSION
In this study, we show that the demethylase PHF2 contributes to the maintenance of genome integrity by controlling DNA repair, predominantly through homology-directed DSB repair. Specifically, we showed that downregulation of PHF2 affects the DNA damage-induced focus formation by BRCA1 and 53BP1, which together determine the choice between DSB repair by homologous recombination or non-homologous end joining. Concomitant with a decrease in BRCA1 focus formation, PHF2 depletion also affected the accumulation of CtIP to sites of DNA lesions. CtIP is known to initiate DNA end resection to generate ssDNA ends that are a prerequisite for HR (10,25). BRCA1, that forms a complex with CtIP, collaborates in ssDNA-end formation (39,45,46). Indeed, impaired DSB resection was observed after PHF2 downregulation, as demonstrated by decreased RPA phosphorylation and focus formation. Consequently, PHF2 depletion compromised IR-induced focus formation of Rad51 and resulted in a diminished efficiency of HR. Impaired DSB repair by HR in response to PHF2 depletion is explained by our data showing that PHF2 controls the DSB response by regulating CtIP and BRCA1 mRNA and protein levels. That PHF2 regulates expression of CtIP and BRCA1 simultaneously is in accordance to indications in the literature that BRCA1 supports the control of CtIP. CtIP was reported to control its own transcription, possibly via interaction with BRCA1 through its BRCT domains (47,48). BRCA1-dependent ubiquitination of CtIP is additionally required for CtIP chromatin binding and damage-induced focus formation (49), which likely explains the reported defect in the accumulation of GFP-CtIP at interstrand crosslinks induced by micro-irradiation (50). Our observation that depletion of PHF2 inhibits the accumulation of GFP-CtIP to the laser damage (Figure 2A) is therefore likely to be an indirect effect of the diminished BRCA1 levels in these conditions. However, we cannot exclude the possibility that PHF2 regulates CtIP on several levels, like the recently reported splicing complex SF3B, that controls CtIP both at the level of mRNA abundance and the recruitment of CtIP to the chromatin in response to DNA damage (51).
Interestingly, we also observed a modest decrease in the efficiency of NHEJ after knockdown of PHF2. A possible explanation for this result could be the decreased BRCA1 levels in these conditions, as depletion of BRCA1 has been reported to affect NHEJ as well as HR (52,53). Depletion of PHF2 also affected cell growth and led to genome instability in unperturbed cells, as demonstrated by H2AX phosphorylation and comet assay analysis, an effect also recently reported by others in mouse neural stem cells (54). Although these results could be due to DNA repair defects, these observations might also reflect a separate phenotype of PHF2. Indeed, PHF2 has been reported to be involved in the control of cell cycle regulation, cell differentiation and the inflammatory response (21,54,55).
The KDM7 family of histone demethylases, to which PHF2 belongs, can remove the methylation from H3K9, H3K27 or H4K20, which are responsible for transcriptional repression, presumably by the concerted action of their PHD methyl reader domain and the enzymatically active JmjC-domain (41,56). Indeed, PHF2 was described to regulate transcription by removing the dimethylated H3K9 and, to a lesser extent, trimethylated H4K20 (20–23). We hypothesize that PHF2 controls CtIP and BRCA1 gene transcription by erasing the transcription repression mark from the respective promoters (23). PHF2 was shown to stimulate the expression of genes driven by the transcription factors HNF4, CEBPα, p53 and NF-κB (20–23). Interestingly, NF-κB was shown to regulate HR by controlling BRCA1-CtIP complexes, although this effect is mediated by protein stabilization of BRCA1 rather than by transcriptional regulation of BRCA1 and CtIP (57).
The fact that a catalytic inactive version of PHF2 cannot rescue the defect in DNA end resection and the lower BRCA1/CtIP levels caused by PHF2 depletion suggests that PHF2 controls the transcription of CtIP and BRCA1 mRNA by direct demethylation of H3K9me2 at the respective promotors. Interestingly however, although biochemical data indicate that PHF2 demethylates H3K9me2 upon interaction with methylated H3K4 through its PHD domain (58), our experiments show that the PHD domain of PHF2 is dispensable for its function on DSB resection and control of BRCA1/CtIP levels. It is therefore possible that PHF2 controls BRCA1/CtIP mRNA levels in another, possibly indirect way. Moreover, as dimethylation of H4K20 is critical in the recruitment of 53BP1 to sites of DNA lesions (59,60), interfering with this methyl mark and/or competition for binding to methylated H4K20 by PHF2 could also contribute to the phenotype in disturbing homology-directed DSB repair observed in this study. We consider this possibility less likely though, since at this moment, we have no indications that PHF2 might function directly at the sites of DNA lesions.
By controlling DNA DSB repair, PHF2 emerges as a putative important regulator in maintaining genomic integrity. Therefore, our data might have an importance in pathologies in which PHF2 levels are changed or PHF2 is mutated. For example, high PHF2 levels were found in oesophageal carcinoma and renal cell carcinoma (61,62), whereas the PHF2 gene is deleted or hypermethylated in its promoter region in breast cancer (63). In addition, PHF2 mutants were reported in gastric and colon cancer (64). Together these observations suggest that PHF2 plays a role in the development and/or progression of cancer. Interestingly, the use of histone demethylases as therapeutic targets by pharmacological inhibitors is currently being investigated and might open new strategies for tumour therapy (65,66). Our results demonstrating that depletion of PHF2 affects the sensitivity to inhibition of PARP, together with the observed correlation between PHF2 and CtIP/BRCA1 levels in tumours, might suggest that targeting PHF2 could be particularly effective in breast and ovarian cancers without mutations in BRCA1/2 or other known HR proteins.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Daniel Durocher and Xiaobing Shi for sharing reagents, Esperanza Hernández for valuable assistance and Antonio Catalán and Luciano Benítez (Servicio de Física Médica) from the Hospital Universitario de Canarias for use of the WOmed X-Ray Therapy Unit.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia e Innovación [SAF2016-80626-R to V.A.J.S./R.F., SAF2016-74855-P to P.H. and BFU2017-90889-REDT to V.A.J.S./P.H.], co-funded by EU-ERDF; Fundación Canaria Instituto de Investigación Sanitaria de Canarias (FIISC) [PIFUN16/18 to V.A.J.S.]; I.A.V. is supported by a predoctoral fellowship from the Gobierno de Canarias; J.R.H.F. by the Asociación Española Contra el Cáncer; H.v.A. by an ERC Consolidator grant from the European Research Council and a VICI grant from the Netherlands Organisation for Scientific Research. Funding for open access charge: FIISC [PIFUN16/18]; Ministerio de Ciencia e Innovación [SAF2016-80626-R].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bartek J., Bartkova J., Lukas J.. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007; 26:7773–7779. [DOI] [PubMed] [Google Scholar]
- 2. Aguilera A., Gómez-González B.. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008; 9:204–217. [DOI] [PubMed] [Google Scholar]
- 3. Hiom K. Coping with DNA double strand breaks. DNA Repair (Amst.). 2010; 9:1256–1263. [DOI] [PubMed] [Google Scholar]
- 4. Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 2010; 17:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noordermeer S.M., Adam S., Setiaputra D., Barazas M., Pettitt S.J., Ling A.K., Olivieri M., Álvarez-Quilón A., Moatti N., Zimmermann M. et al.. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018; 560:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mirman Z., Lottersberger F., Takai H., Kibe T., Gong Y., Takai K., Bianchi A., Zimmermann M., Durocher D., de Lange T.. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature. 2018; 560:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bunting S.F., Callén E., Wong N., Chen H.-T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L. et al.. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010; 141:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chapman J.R., Taylor M.R.G., Boulton S.J.. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012; 47:497–510. [DOI] [PubMed] [Google Scholar]
- 9. Symington L.S. Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2016; 51:195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jasin M., Rothstein R.. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect Biol. 2013; 5:a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sulli G., Di Micco R., d’Adda di Fagagna F.. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat. Rev. Cancer. 2012; 12:709–720. [DOI] [PubMed] [Google Scholar]
- 12. Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J.. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001; 276:42462–42467. [DOI] [PubMed] [Google Scholar]
- 13. Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M. et al.. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007; 318:1637–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J. et al.. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009; 136:435–446. [DOI] [PubMed] [Google Scholar]
- 15. Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M. et al.. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009; 136:420–434. [DOI] [PubMed] [Google Scholar]
- 16. Zhao Y., Brickner J.R., Majid M.C., Mosammaparast N.. Crosstalk between ubiquitin and other post-translational modifications on chromatin during double-strand break repair. Trends Cell Biol. 2014; 24:426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation. EMBO J. 2000; 19:1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greer E.L., Shi Y.. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012; 13:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luijsterburg M.S., van Attikum H.. Chromatin and the DNA damage response: the cancer connection. Mol. Oncol. 2011; 5:349–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baba A., Ohtake F., Okuno Y., Yokota K., Okada M., Imai Y., Ni M., Meyer C.A., Igarashi K., Kanno J. et al.. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat. Cell Biol. 2011; 13:668–675. [DOI] [PubMed] [Google Scholar]
- 21. Stender J.D., Pascual G., Liu W., Kaikkonen M.U., Do K., Spann N.J., Boutros M., Perrimon N., Rosenfeld M.G., Glass C.K.. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol. Cell. 2012; 48:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Okuno Y., Ohtake F., Igarashi K., Kanno J., Matsumoto T., Takada I., Kato S., Imai Y.. Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes. 2013; 62:1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee K.-H., Park J.-W., Sung H.-S., Choi Y.-J., Kim W.H., Lee H.S., Chung H.-J., Shin H.-W., Cho C.-H., Kim T.-Y. et al.. PHF2 histone demethylase acts as a tumor suppressor in association with p53 in cancer. Oncogene. 2015; 34:2897–2909. [DOI] [PubMed] [Google Scholar]
- 24. Rabl J., Bunker R.D., Schenk A.D., Cavadini S., Gill M.E., Abdulrahman W., Andrés-Pons A., Luijsterburg M.S., Ibrahim A.F.M., Branigan E. et al.. Structural basis of BRCC36 function in DNA repair and immune regulation. Mol. Cell. 2019; 75:483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P.. Human CtIP promotes DNA end resection. Nature. 2007; 450:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luijsterburg M.S., de Krijger I., Wiegant W.W., Shah R.G., Smeenk G., de Groot A.J.L., Pines A., Vertegaal A.C.O., Jacobs J.J.L., Shah G.M. et al.. PARP1 links CHD2-Mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell. 2016; 61:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bennardo N., Cheng A., Huang N., Stark J.M.. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008; 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Méndez J., Stillman B.. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000; 20:8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smits V.A.J., Reaper P.M., Jackson S.P.. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr. Biol. 2006; 16:150–159. [DOI] [PubMed] [Google Scholar]
- 31. Typas D., Luijsterburg M.S., Wiegant W.W., Diakatou M., Helfricht A., Thijssen P.E., van den Broek B., van de Broek B., Mullenders L.H., van Attikum H.. The de-ubiquitylating enzymes USP26 and USP37 regulate homologous recombination by counteracting RAP80. Nucleic Acids Res. 2015; 43:6919–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., Greenberg R.A.. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013; 20:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z.. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017; 45:W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu Y., Scully R., Sobhian B., Xie A., Shestakova E., Livingston D.M.. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011; 25:685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smits V.A.J., Gillespie D.A.. DNA damage control: regulation and functions of checkpoint kinase 1. FEBS J. 2015; 282:3681–3692. [DOI] [PubMed] [Google Scholar]
- 36. Zou L., Elledge S.J.. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 37. Shiotani B., Zou L.. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 2009; 33:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maréchal A., Zou L.. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015; 25:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yun M.H., Hiom K.. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009; 459:460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huertas P., Jackson S.P.. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009; 284:9558–9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fortschegger K., Shiekhattar R.. Plant homeodomain fingers form a helping hand for transcription. Epigenetics. 2011; 6:4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Horton J.R., Upadhyay A.K., Hashimoto H., Zhang X., Cheng X.. Structural basis for human PHF2 Jumonji domain interaction with metal ions. J. Mol. Biol. 2011; 406:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fortschegger K., de Graaf P., Outchkourov N.S., van Schaik F.M.A., Timmers H.T.M., Shiekhattar R.. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol. Cell. Biol. 2010; 30:3286–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jackson S.P., Helleday T.. DNA REPAIR. Drugging DNA repair. Science. 2016; 352:1178–1179. [DOI] [PubMed] [Google Scholar]
- 45. Chen L., Nievera C.J., Lee A.Y.-L., Wu X.. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008; 283:7713–7720. [DOI] [PubMed] [Google Scholar]
- 46. Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E. et al.. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010; 6:e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu X., Wu L.C., Bowcock A.M., Aronheim A., Baer R.. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 1998; 273:25388–25392. [DOI] [PubMed] [Google Scholar]
- 48. Liu F., Lee W.-H.. CtIP activates its own and cyclin D1 promoters via the E2F/RB pathway during G1/S progression. Mol. Cell. Biol. 2006; 26:3124–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu X., Fu S., Lai M., Baer R., Chen J.. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006; 20:1721–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Duquette M.L., Zhu Q., Taylor E.R., Tsay A.J., Shi L.Z., Berns M.W., McGowan C.H.. CtIP is required to initiate replication-dependent interstrand crosslink repair. PLoS Genet. 2012; 8:e1003050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Prados-Carvajal R., López-Saavedra A., Cepeda-García C., Jimeno S., Huertas P.. Multiple roles of the splicing complex SF3B in DNA end resection and homologous recombination. DNA Repair (Amst.). 2018; 66-67:11–23. [DOI] [PubMed] [Google Scholar]
- 52. Fouquin A., Guirouilh-Barbat J., Lopez B., Hall J., Amor-Guéret M., Pennaneach V.. PARP2 controls double-strand break repair pathway choice by limiting 53BP1 accumulation at DNA damage sites and promoting end-resection. Nucleic Acids Res. 2017; 45:12325–12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu Y., Wang C., Huang K., Xia F., Parvin J.D., Mondal N.. Regulation of 53BP1 protein stability by RNF8 and RNF168 is important for efficient DNA double-strand break repair. PLoS ONE. 2014; 9:e110522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pappa S., Padilla N., Iacobucci S., Vicioso M., Álvarez de la Campa E., Navarro C., Marcos E., la Cruz X., Martínez-Balbás M.A.. PHF2 histone demethylase prevents DNA damage and genome instability by controlling cell cycle progression of neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:19464–19473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang J., Ma J., Xiong Y., Wang Y., Jin K., Xia W., Chen Q., Huang J., Zhang J., Jiang N. et al.. Epigenetic regulation of megakaryocytic and erythroid differentiation by PHF2 histone demethylase. J. Cell. Physiol. 2018; 233:6841–6852. [DOI] [PubMed] [Google Scholar]
- 56. Jaskelioff M., Peterson C.L.. Chromatin and transcription: histones continue to make their marks. Nat. Cell Biol. 2003; 5:395–399. [DOI] [PubMed] [Google Scholar]
- 57. Volcic M., Karl S., Baumann B., Salles D., Daniel P., Fulda S., Wiesmüller L.. NF-κB regulates DNA double-strand break repair in conjunction with BRCA1-CtIP complexes. Nucleic Acids Res. 2012; 40:181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wen H., Li J., Song T., Lu M., Kan P.-Y., Lee M.G., Sha B., Shi X.. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J. Biol. Chem. 2010; 285:9322–9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Botuyan M.V., Lee J., Ward I.M., Kim J.-E., Thompson J.R., Chen J., Mer G.. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006; 127:1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pei H., Zhang L., Luo K., Qin Y., Chesi M., Fei F., Bergsagel P.L., Wang L., You Z., Lou Z.. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011; 469:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sun L.-L., Sun X.-X., Xu X.-E., Zhu M.-X., Wu Z.-Y., Shen J.-H., Wu J.-Y., Huang Q., Li E.-M., Xu L.-Y.. Overexpression of Jumonji AT-rich interactive domain 1B and PHD finger protein 2 is involved in the progression of esophageal squamous cell carcinoma. Acta Histochem. 2013; 115:56–62. [DOI] [PubMed] [Google Scholar]
- 62. Lee C., Kim B., Song B., Moon K.C.. Implication of PHF2 expression in clear cell renal cell carcinoma. J. Pathol. Transl. Med. 2017; 51:359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sinha S., Singh R.K., Alam N., Roy A., Roychoudhury S., Panda C.K.. Alterations in candidate genes PHF2, FANCC, PTCH1 and XPA at chromosomal 9q22.3 region: pathological significance in early- and late-onset breast carcinoma. Mol. Cancer. 2008; 7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee J.H., Yoo N.J., Kim M.S., Lee S.H.. Histone demethylase gene PHF2 is mutated in gastric and colorectal cancers. Pathol. Oncol. Res. 2017; 23:471–476. [DOI] [PubMed] [Google Scholar]
- 65. Højfeldt J.W., Agger K., Helin K.. Histone lysine demethylases as targets for anticancer therapy. Nat. Rev. Drug Discov. 2013; 12:917–930. [DOI] [PubMed] [Google Scholar]
- 66. Morera L., Lübbert M., Jung M.. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenetics. 2016; 8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.