Abstract
Previously, we have shown that human sperm Prohibitin (PHB) expression is significantly negatively correlated with mitochondrial ROS levels but positively correlated with mitochondrial membrane potential and motility. However, the possible role of PHB in mammalian spermatogenesis has not been investigated. Here we document the presence of PHB in spermatocytes and its functional roles in meiosis by generating the first male germ cell-specific Phb-cKO mouse. Loss of PHB in spermatocytes resulted in complete male infertility, associated with not only meiotic pachytene arrest with accompanying apoptosis, but also apoptosis resulting from mitochondrial morphology and function impairment. Our mechanistic studies show that PHB in spermatocytes regulates the expression of STAG3, a key component of the meiotic cohesin complex, via a non-canonical JAK/STAT pathway, and consequently promotes meiotic DSB repair and homologous recombination. Furthermore, the PHB/JAK2 axis was found as a novel mechanism in the maintenance of stabilization of meiotic STAG3 cohesin complex and the modulation of heterochromatin formation in spermatocytes during meiosis. The observed JAK2-mediated epigenetic changes in histone modifications, reflected in a reduction of histone 3 tyrosine 41 phosphorylation (H3Y41ph) and a retention of H3K9me3 at the Stag3 locus, could be responsible for Stag3 dysregulation in spermatocytes with the loss of PHB.
INTRODUCTION
Prohibitin (PHB or PHB1) is an evolutionarily conserved mitochondrial inner membrane protein. Although the encoding gene has been shown to be pleiotropic, with PHB known to regulate a number of non-mitochondrial functions (1,2), the protein is highly expressed in cells that have a demand for a strong mitochondrial function. It has been shown that PHB is associated with mitochondrial respiratory chain subunits assembly, mitochondrial biogenesis, and mitophagy (degradation of mitochondria via autophagy) (3). Silencing of Phb in endothelial cells reduces mitochondrial membrane potential (MMP) and complex I activity (4). Our previous studies in sperm from patients with poor sperm motility and/or low sperm concentrations have shown that PHB expression has a significantly negative correlation with mitochondrial ROS level (mROS), but a positive one with MMP and sperm motility (5,6). These findings suggest that PHB expression levels could be used as an indicator of human sperm quality.
PHB expression in sperm is not limited to humans. PHB has been reported in spermatids and spermatozoa in bulls (7,8) and rhesus monkeys (8), where it was one of the ubiquitinated substrates prone to rapid degradation of sperm mitochondria after fertilization (8). However, in the rat model PHB has been shown not to be expressed in spermatids and sperm, but to be present in spermatogonia and to be highly expressed in spermatocytes (9). These findings reveal that PHB expression in mammalian species occurs in a variety of cell types at different stages of the sperm formation process, suggesting that it may perform multiple functional roles in the testis. Sperm formation or spermatogenesis is a complex and dynamic process that involves germ cell division and differentiation in the seminiferous tubules of the testes. The process of spermatogenesis includes mitosis and self-renewal of spermatogonia, meiosis of spermatocytes, and differentiation of haploid spermatids into sperm (spermiogenesis). Of these, the most complex is meiosis in spermatocytes which involves two divisions and is the key process that ensures the precise reduction of chromosome number in gametes and their genetic uniqueness due to recombination (10,11). Thus PHB expression during meiosis is likely to play a consequential role(s). However, these potential roles have not yet been investigated. The goal of the present study therefore was to determine if PHB is expressed during mouse spermatogenesis and its potential role in the process. In this study, we have investigated PHB expression in spermatocytes both in vitro and in vivo, and in the latter we have used germ-cell-specific Phb-cKO mice, generated for the first time, to determine PHB’s functional role.
Our results demonstrate that loss of PHB results in complete male infertility, associated with not only meiotic pachytene arrest due to a failure of double strand break (DSB) repair and homologous recombination with accompanying apoptosis, but also apoptosis resulting from an impairment of mitochondrial morphology and function of spermatocytes. Furthermore, we have identified that in spermatocytes, both in vivo and in vitro, PHB regulates the expression of STAG3, a key component of the meiotic cohesin complex, via a non-canonical JAK/STAT signaling pathway, and consequently promotes meiotic chromosome structure, meiotic DSB repair and homologous recombination. An axis of PHB-JAK2-H3Y41ph was found as a novel mechanism in both the maintenance of the stabilization of meiotic STAG3 cohesin complex and the modulation of the heterochromatin formation in spermatocytes. The latter was evidenced by JAK2-mediated reduction of H3Y41ph and a retention of H3K9me3 surrounding the promoter region of Stag3 in Phb−/− spermatocytes. These findings could be responsible for Stag3 dysregulation in spermatocytes with the loss of PHB.
MATERIALS AND METHODS
Cell culture, siRNA transfection and viral transduction
A mouse spermatocyte-derived GC-2spd(ts) cell line (12) was used as a cell-culture model that has been authenticated to be functional for biochemical studies (13,14). The GC-2spd(ts) cell line (abbreviated GC2 cells) was obtained from the American Type Culture Collection (ATCC) and grown in Dulbecco's modified Eagle's medium (DMEM, Corning) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% Penicillin–Streptomycin at 37°C under 5% CO2.
Cell transfection was performed using X-tremeGENE siRNA Transfection Reagent (Roche) according to the manufacturer's instructions. In general, 50 pmol of the siRNA oligonucleotide was used for each transfection in six-well plate. High-titer lentivirus for PHB shRNA was purchased from Genechem (Shanghai, China). GC2 cells were infected by incubation with lentivirus containing media (supplemented with 8 μg/ml polybrene) for 24 h. Puromycin (1 μg/ml) was added 24 h after transduction.
Generation of Phb conditional knockout mice and genotyping identification
Phb-floxed mice were designed via targeting exon 2 of Phb gene for flanking with loxP sequences (Biocytogen, China), as shown in Supplementary Figure S3A. The Stra8-GFPCre (officially named as Stra8<em1(GFP/cre)Smoc>) mouse was a generous gift from Tong Lab (University of Chinese Academy of Sciences) (15). To generate PhbFlox/−; Stra8-GFPCre (Phb-cKO) mice, the PhbFlox/Flox mice were crossed with PhbFlox/+; Stra8-GFPCre mice to introduce the germ cell-specific Cre reporter system. The resulting offspring were maintained on C57BL/6 genetic background. The design and conduct of all animal experiments were approved by the Institutional Animal Care and Use Committee at Fudan University. The primers for PCR genotyping were provided in Supplementary Table S1.
Sperm motility assay
Mouse caudal epididymal sperm from sexually mature males were harvested in 37°C pre-warmed Enriched Krebs-Ringer Bicarbonate Medium (EKRB medium; 120.1 mM NaCl, 4.8 mM KCl, 25.2 mM NaHCO3, 1.2 mM KH2PO4, 1.2 mM MgSO4, 1.3 mM CaCl2, supplemented with 11.1 mM glucose, 2 mM glutamine, 10 ml/l essential amino acids, 10 ml/l nonessential amino acids, 100 μg/ml streptomycin and 100 U/ml penicillin). Sperm concentration and motility parameters were assessed by using a computer-assisted sperm analysis (CASA) system (HTM-TOX IVOS, Hamilton-Thorne) with standard instrument settings (16).
Histological analysis, immunofluorescence and TUNEL assay
For histological analysis, the testes and epididymides were collected, fixed in Bouin's fixative solution for 24 h, processed, and embedded in paraffin using routine methods. Then, slices of 5 μm thickness were stained with Hematoxylin and Eosin.
For immunofluorescence, testes were collected and fixed in 4% paraformaldehyde (PFA) in PBS for 24 h while the cultured mouse spermatocytes fixed for 5 min. After routine preparation of paraffin section, the dewaxed sections were processed for antigen retrieval with citrate buffer while the fixed culture cells permeabilized in cold acetone for 5 min. After blocking with 5% BSA in PBS for 2 h at room temperature (RT), sections or cells were incubated with primary antibody for 2 h at RT and then treated with Alexa Fluor 488-, Alexa Fluor 647- or Cy3-conjugated secondary antibodies for 2 h at 4°C. Nuclei were counterstained with DAPI (Vector Laboratories). Images were captured with a LSM 880 confocal microscope (Zeiss). The images of Tomm-20-labeled mitochondria were acquired along the Z-axis, followed by three-dimensional (3D) reconstruction and measurement of mitochondrial length using Imaris software (Bitplane). The antibodies used with their dilutions are listed in Supplementary Table S2.
Apoptotic cells were analyzed by Fluorometric TUNEL System (Promega) as previously described (17). Paraffin-embedded testis sections were incubated with TUNEL reaction buffer under humidified atmosphere for 60 min at 37°C, then rinsed with 2× SSC for three times. The nuclei were stained with DAPI. TUNEL-positive cells were observed by the emission of green fluorescence.
Meiotic chromosome spreading and immunofluorescence
Chromosomes spreads were prepared as previously reported with slight modifications (18). In brief, testicular tubules were washed in PBS and then incubated with a hypotonic extraction buffer (30 mM Tris, 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 0.5 mM DTT, and 0.5 mM PMSF, pH 8.2) on ice for 45 min. Following this, the tubules were minced in 100 mM sucrose (pH 8.2) and allowed to form a cell suspension. The cell suspension (80 μl) was spread on slides with an equal volume of fixative buffer (1% PFA and 0.15% Triton X-100 in PBS, pH 9.2). After incubation in a humid chamber for 3 h at RT, the slides were washed twice with 0.4% Photoflo in PBS and air-dried for the following immunofluorescence staining mentioned above. The antibodies used with their dilutions are listed in Supplementary Table S2.
Intra-testicular injection
Intra-testicular injection was performed as previously described (14). In brief, control mice (8 weeks old) were anesthetized with sodium pentobarbital (0.06 mg/g, intraperitoneal injection) and intra-testicular efferent ductules were identified. Approximately 15 μl of DMSO (vehicle) or inhibitor TG101209 (5 mg/ml) were introduced slowly into seminiferous tubules through microinjection in the efferent ductules. Seven days later, the testes were then dissected for western blotting analysis and chromosomes spread preparation.
Fluorescence activated cell sorting (FACS) of mouse spermatogenic cells
Isolation of spermatogenic cells from mouse testes was performed using a previously reported method with slight modifications (19). In brief, seminiferous tubules were incubated in Gey's Balanced Salt Solution (GBSS) supplemented with 200 U/ml collagenase and 5 μg/ml DNase I for 10 min at 35°C. The dispersed tubules were re-suspended and incubated in 5 ml GBSS containing 200 U/ml collagenase and 0.025% trypsin for 20 min at 37°C with agitation. After that, FBS (Gibco) was added at a final concentration of 5% to inactivate the trypsin. The cell suspension was then filtered through 40 μm diameter strainers to remove small clumps and subsequently stained with Hoechst 33342 (5 μg/ml) for 40 min at 37°C. Germ cell populations were enriched by BD Aria II (BD Biosciences) with a 355 nm laser. Each isolated population (spermatogonia, spermatocytes, or round spermatids) was collected in PBS and subjected to immunofluorescence staining to evaluate the enrichment.
Electron microscopy
FACS-sorted spermatocytes were centrifuged at 1500 × g for 5 min. The cell pellets were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer for 2 h at 4°C. After treatment with 2% OsO4, the pellets were dehydrated in graded ethanol and embedded in epoxy resin (Poly/Bed 812, polysciences). Ultrathin sections (70 nm) were obtained and doubly stained with uranyl acetate and lead citrate before imaging with a 120 kV FEI Tecnai G2 Spirit transmission electron microscope.
Mitochondrial (Mt) DNA quantification
Total DNA was extracted from GC2 cells or FACS-isolated spermatocytes using the DNeasy Blood & Tissue Kit (QIAGEN) according to the manufacturer's instructions. mtDNA levels were assessed by qRT-PCR using primers specific to mitochondrial gene 16S, with nuclear apoB serving as a reference. All the primer sequences are provided in Supplementary Table S1.
Assessment of MMP by JC-1 assay
GC2 cells or FACS-isolated spermatocytes were incubated with JC-1 working solution (5 μg/ml) for 30 min at 37°C. The cells were harvested by centrifugation at 600 × g, then gently washed in HBSS three times and fixed in 4% PFA on a coverslip. JC-1 exhibits a MMP-dependent accumulation in living cells, in which JC-1 aggregates in mitochondria emits red fluorescence (590 nm) whereas JC-1 monomer in cytoplasm emits green fluorescence (525 nm). Images for each channel were captured with a LSM 880 confocal microscope (Zeiss). The ratio of red to green fluorescence was calculated as the indicator of MMP of spermatocytes.
Transcriptome-seq and bioinformatic analyses
Total RNA was isolated from GC2 cells and spermatocytes that were sorted from 14 dpp testes using RNeasy Mini Kit (Qiagen). The concentration and integrity of RNA were assessed using Qubit Fluorometer (Invitrogen) and Bioanalyzer 2100 system (Agilent), respectively. cDNA libraries were generated using NEB Next Ultra Directional RNA Library Prep Kit (NEB) following the manufacturer's instructions, and then sequenced using the Illumina sequencing technology on an Illumina Hiseq2500 at LC Bio (Zhejiang, China) according to the recommended protocols. Paired-end clean reads were mapped to the mouse reference genome GRCm38/mm10 using TopHat software (20). The genome-matching reads were used to measure mRNA abundance using Cufflinks software (21). The mRNAs at a cut-off of 10 reads were compared and considered as differentially expressed if the fold change >2 and P value <0.05. Gene set enrichment analysis (GSEA) was performed by inputting a list in which differentially expressed genes were ranked according to their fold change, into GSEA application (22). KEGG pathway analysis was performed using Cluster Profiler R package.
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay was performed as we described previously with a few modifications (23). Briefly, FACS-sorted spermatocytes were fixed in 1% formaldehyde and lysed in chromatin prep buffer supplemented with proteinase inhibitor cocktail (Roche). Extracted-nuclei were digested with micrococcal nuclease (NEB) and sonicated to achieve DNA fragments of 200–1,000 bp in length. Sheared chromatin was immunoprecipitated using ChIP-grade H3K9me3 or H3Y41ph antibodies. DNA was purified by AMPure XP beads (Beckman Coulter) and quantified by qRT-PCR analysis with the primers specific to the promoter region of Stag3 gene. All the primer sequences are provided in Supplementary Table S1. The antibodies used with their dilutions are listed in Supplementary Table S2.
Western blot
Testes or spermatocytes, cultured or FACS-sorted, were homogenized in lysis buffer [150 mM NaCl, 50 mM Tris–HCl (pH 7.4), 5 mM EDTA, 1% Triton X-100, 1× proteinase inhibitor cocktail (Roche)], and the protein concentration was measured using a BCA protein assay kit (Pierce). Cell/tissue extracts were subjected to standard SDS-PAGE and immunoblot procedures. The band intensities were quantified with a Tanon 5200 Image Analyzer (Tanon, China). The primary and secondary antibodies used with their dilutions are listed in Supplementary Table S2.
RNA extraction and RT-PCR
Total RNA was extracted from testes or spermatocytes, cultured or FACS-sorted, using TRIzol reagent (Thermo). The isolated RNA (0.5 μg) was reverse transcribed into cDNA using a PrimeScript Kit with gDNA Eraser (TaKaRa). The mRNA levels were detected by qRT-PCR with a SYBR premix reagent (TaKaRa) on an ABI QuantStudio 3 (Applied Biosystems). The results were calculated by the ΔΔCt quantification method using the Quant Studio Design & Analysis software, with β-actin mRNA serving as an internal reference. The validity of the qRT-PCR data was assured by following the MIQE guidelines (24). All the primers sequences are provided in Supplementary Table S1.
Statistical analysis
Results are presented as the mean ± S.D. Each experiment is repeated at least three times. Experimental groups are compared using the two-tailed unpaired Student's t-tests, except for the results of foci numbers that are analyzed using the non-parametric two-tailed Mann-Whitney test. P-values of <0.05 are considered statistically significant. ‘n.s.’ indicates non-significant.
RESULTS
PHB expression in mouse germ cells during spermatogenesis
In order to explore the function of PHB during mouse spermatogenesis, we first examined the expression pattern of PHB in mouse germ cells using immunostaining methods. We found that PHB is expressed in both germ cells and somatic cells of mouse testes, and is especially highly expressed in spermatocytes (Supplementary Figure S1A). To confirm this pattern of PHB expression in spermatocytes, the fluorescence-activated cell sorting (FACS) method was used to isolate specific germ cells from mouse testes, specifically spermatogonia, spermatocytes, and round spermatids (Supplementary Figure S1B, C). Compared to that of FACS-sorted spermatogonia and round spermatids, FACS-sorted spermatocytes showed higher levels of Phb mRNA and PHB protein expression after qRT-PCR (Supplementary Figure S1E) and immunostaining (Supplementary Figure S1C, D), respectively. These data indicate that PHB could be potentially involved in meiotic progression.
PHB depletion in mouse spermatocytes in vitro blocks the JAK-STAT signaling pathway
It has been reported that PHB could modulate various signaling pathways in different types of cells (25). In order to explore a potential signaling pathway mediated via PHB in spermatocytes, we thus analyzed the transcriptomic differences of GC-2spd(ts) cells (GC2 cells) stably transfected with PHB-targeting shRNA (briefly, PHB KD GC2) from those with a control shRNA (briefly, Ctrl GC2) (Supplementary Figure S2A, B). As shown in Figure 1A, the RNA sequencing data revealed a total of 519 differentially expressed genes according to the criteria of FC > 2 and P < 0.05. Gene set enrichment analysis (GSEA) then indicated that both JAK-STAT and PI3K-AKT pathways were the most enriched KEGG pathways for the differentially expressed genes (Figure 1B, Supplementary Figure S2C). The top-ranking gene candidates (fold change >2 or <0.5, P<0.05) involved in these pathways (Figure 1C), were validated, using qRT-PCR assay. This involved 7–8 candidate genes with the largest difference (Figure 1D).
Figure 1.
RNA-seq analysis reveals PHB-regulated genes in mouse spermatocyte-derived GC2 cells. (A) Scatterplot showing differential gene expression between PHB-knockdown GC2 cells and the controls. Values are presented as normalized log2(FPKM). Up- and down-regulated genes are colored in red and blue, respectively. (B) Gene set enrichment analysis (GSEA) for genes associated with the JAK-STAT (left) or PI3K-AKT (right) pathway. (C) Hierarchical clustering with heat map showing the mRNA levels of all differentially expressed genes associated with the JAK-STAT (top) or PI3K-AKT (bottom) pathway in PHB-knockdown GC2 cells and the controls. Genes are ranked based on their fold-change. (D) qRT-PCR validation of PHB-regulated genes (fold change >2 or <0.5, P<0.05). The results are normalized to the β-actin mRNA level. n = 3, Error bars, S.D. *P< 0.05; **P< 0.01 by two-tailed Student's t-test. (E). Quantitation of the protein expression levels of selected differential genes by Western blotting, with β-actin as a loading control.
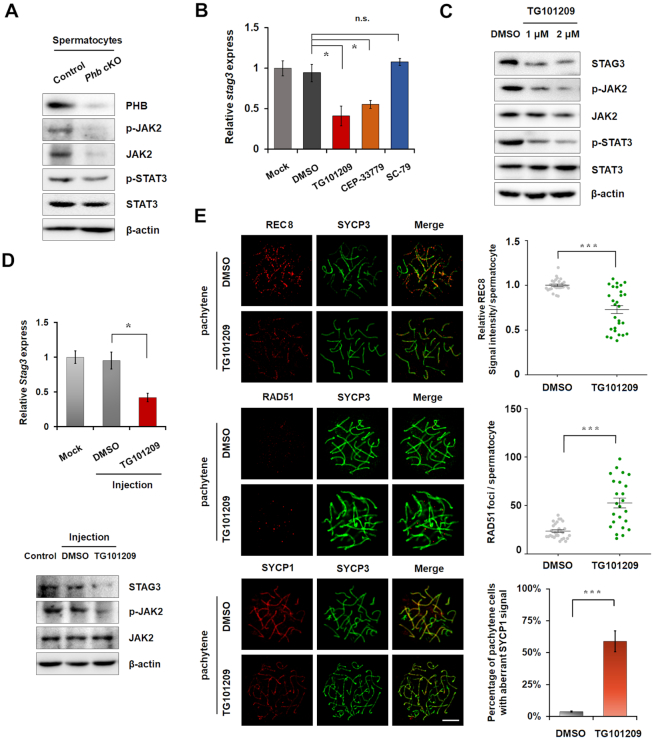
The findings in this study that both p-Akt and its downstream targets are up-regulated in PHB-knockdown GC-2spd(ts) cells (GC2 cells) (Figure 1E), are consistent with the previous report that the absence of PHB could highly activate the PI3K-AKT pathway by promoting ROS production in the vascular system (4). Additionally, PHB-knockdown GC-2spd(ts) cells (GC2 cells) showed a substantial reduction in the expression of JAK2 and p-STAT3 (Figure 1E), indicating that JAK-STAT signaling pathway was repressed therein. This repression of the JAK-STAT pathway is consistent with JAK2 and STAT3 phosphorylation levels which are positively correlated with the activation status of the JAK-STAT pathway (26,27). Based on our findings in PHB-knockdown GC2 cells (Figure 1E), it seems clear that PHB deficiency promotes the activation of PI3K-AKT pathway, while it represses the JAK2-STAT signaling. Moreover, the results in GC2 cells have been further validated in FACS-isolated spermatocytes from Phb-cKO and control testes (Figure 6A, and Supplementary Figure S10A). Accordingly, these data suggest that PHB could modulate JAK-STAT and PI3K-AKT pathways in mouse spermatocytes in vitro.
Figure 6.
JAK2 is involved in the down-regulation of STAG3 in Phb−/− spermatocytes. (A) Expression levels of p-JAK2, JAK2, p-STAT3 and STAT3 proteins in spermatocytes from control and Phb-cKO mice, with β-actin as a reference. (B) Stag3 mRNA levels in GC2 cells treated with either DMSO or JAK2 inhibitor (TG101209 or CEP-33779) or a PI3K-AKT activator (SC-79). β-actin is used as a reference. n = 3, error bars, S.D. *P< 0.05 by two-tailed Student's t-test. n.s., non-significant. (C) Expression levels of STAG3, p-JAK2, JAK2, p-STAT3 and STAT3 proteins in GC2 cells treated with DMSO or inhibitor TG101209, with β-actin as a loading control. (D) Left: Stag3 mRNA levels in testis injected with DMSO or inhibitor TG101209. Right: expression of STAG3 and p-JAK2 proteins in TG101209-treated testis lysates or the controls, with β-actin as a reference. n = 3, Error bars, S.D. *P< 0.05 by two-tailed Student's t-test. (E) Left: immunostaining of REC8 (red, top), RAD51 (red, middle) and SYCP1 (red, bottom) in meiotic spreads of pachytene spermatocytes prepared from DMSO- or inhibitor TG101209-treated testes, with SYCP3 (green) as a staining control. Right: quantification of RAD51 foci number, REC8 and SYCP1 signals between two groups. Error bars, S.D. Scale bar = 5 μm. ***P< 0.001 by Mann–Whitney test.
Mice lacking PHB in spermatocyte are infertile
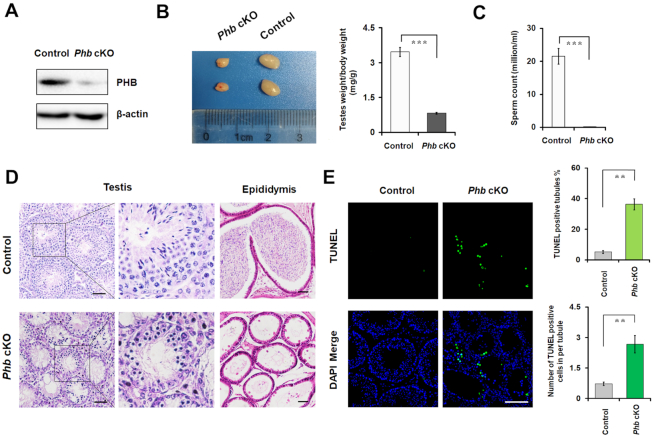
To further examine the function of PHB in spermatocytes in vivo, under the limit of the embryonic lethality in Phb knockout mouse, we generated and identified the first male germ cell-specific Phb knockout mice (PhbFlox/−; Stra8-GFPCre, called Phb-cKO) (Supplementary Figure S3). These Phb-cKO mice were obtained after crossing PhbFlox/Flox mice made using CRISPR/Cas9 system (Supplementary Figure S3A) with Stra8-GFPCre transgenic mice whose male germ cells contained active recombinase prior to the initiation of meiosis (Supplementary Figure S3B). The lack of PHB expression was subsequently detected in both testes from 14 day postpartum (dpp) mice (Figure 2A, Supplementary Figure S3C) and the spermatocytes of testes from 8-week-old mice (Supplementary Figure S3D), using Western blotting, qRT-PCR, and immunostaining assays. Compared to control mice with a normal genotype, the Phb-cKO mice showed a significant reduction in testis weight (Figure 2B) at 8 weeks of age, despite of a similar body stature (Supplementary Figure S3E). Furthermore, these Phb-cKO mice, after mating with WT females of proven fertility, were completely infertile (Supplementary Figure S3F). These Phb-cKO males had seminiferous tubules that were devoid of secondary spermatocytes and post-meiotic cell populations (spermatids and spermatozoa) and epididymides in which sperm were absent (Figure 2C, D, Supplementary Figure S3G). Also, a higher frequency of apoptotic germ cells, demonstrated by TUNEL assay (Figure 2E), was found in the seminiferous tubules of the Phb-cKO mice compared to that of control mice. Interestingly, spermatogonia, as detected by the positive PLZF marker, were found in both control and Phb-cKO mice; whereas spermatocytes, as detected by the positive SYCP3 marker, were predominantly found in control mice (Supplementary Figure S4A). These data suggest that the absence of PHB in male germ cells results in spermatogenesis failure and ultimately male infertility.
Figure 2.
Depletion of PHB in germ cells results in male infertility. (A) Expression of PHB protein in testes extracts prepared from Phb-cKO mice and the controls at 14 dpp, with β-actin as a loading control. (B) Left: testes of Phb-cKO mice (photo) are smaller than those of the controls at 8 weeks of age. Right: quantification of testis/body weight ratio in Phb-cKO mice and the controls. n = 3, Error bars, S.D. ***P<0.001 by two-tailed Student's t-test. (C) Determination of sperm counts collected from the caudal epididymis of Phb-cKO mice and the controls at 8 weeks old, using CASA assays. n = 3, Error bars, S.D. ***P< 0.001 by two-tailed Student's t-test. (D) H&E staining of histological sections of the testes (left) and caudal epididymis (right) prepared from Phb-cKO mice and the controls at 8 weeks old. Scale bar = 50 μm. (E) Left: TUNEL assays of testes sections prepared from Phb-cKO mice and the controls at 8 weeks of age. TUNEL-positive cells are shown in green. Right: Quantification of the numbers of TUNEL-positive seminiferous tubules (top) and TUNEL-positive cells per tubule (bottom), respectively. Scale bar = 50 μm. n = 3, error bars, S.D. **P< 0.01 by two-tailed Student's t-test.
Phb-cKO male mice have impaired meiosis due to a pachytene arrest
In order to test if PHB loss could affect spermatocytes as early as in the first wave of spermatogenesis, we next examined the first wave of spermatogenesis in Phb-cKO mice at the age of 10 dpp, which is just at the beginning of meiosis (28). Compared to the control testes at 10 dpp, some of the seminiferous tubules in Phb-cKO mice had already shown a decrease in the amount of spermatocytes (Supplementary Figure S5C). This is due to an increased incidence in apoptotic germ cells in the testes of Phb-cKO mice (Supplementary Figure S5D, E), despite a lack of significant difference in the testis weight compared to the wild-type control at 10 dpp (Supplementary Figure S5A, B). Furthermore, at 14 dpp when pachytene spermatocytes are present in seminiferous tubules during the first wave of spermatogenesis (29), Phb-cKO mice exhibited a significant decrease in both spermatocyte number and testis weight (Supplementary Figure S5A–C) as well as an increased frequency of apoptotic germ cells (Supplementary Figure S5D–G), compared to controls. Notably, spermatocytes with split SYCP3 signals were observed in the seminiferous tubules of Phb-cKO mice at ages of 10 dpp, 14 dpp and 8 weeks (Supplementary Figures S4B and S6), suggesting that meiotic defects could have occurred as early as in the first wave of spermatogenesis.
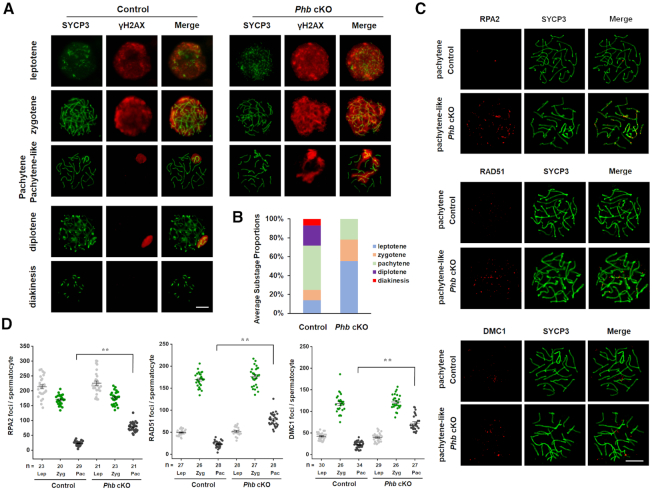
It is well-known that the chromosomal localization of γH2AX, a histone variant that detects DNA double strand breaks (DSBs), and the monitoring of foci formation of recombination sites can reveal whether or not DNA DSBs and their repair are normally initiated and completed (30,31). Using co-immunostaining of SYCP3 with γH2AX (Figure 3A), we show that meiosis in Phb−/− spermatocytes is impaired with a pachytene stage arrest. This impairment is also evidenced by an increase in the proportion of leptotene/zygotene spermatocytes in the testes of Phb-cKO mice (Figure 3B). Furthermore, in contrast to the controls where the γH2AX signal is restricted to the late-replicating XY body in pachytene spermatocytes, pachytene-like Phb−/− spermatocytes showed the persistent localization of γH2AX in both the autosomes and XY bodies (Figure 3A), despite the normal initiation of the programmed DSB in leptotene. This finding suggests that the DSB repair is severely impaired in the pachytene-like Phb−/− spermatocytes. Additionally, when the foci of three key recombination markers (RPA2, RAD51 and DMC1) were monitored, we were able to corroborate our findings for the localization of γH2AX in chromosome spreads of pachytene-like Phb−/− spermatocytes. Compared to the controls which had a reduction in the number of foci, marking the completion of DSB repair, there was a significant accumulation of all three foci in the autosomes of pachytene-like Phb−/− spermatocytes (Figure 3C, D), despite similar numbers of foci in leptotene/zygotene of Phb−/− and control spermatocytes (Figure 3D, Supplementary Figure S7). These findings allow us to propose that PHB is indispensable in meiosis and programmed DSB repair during meiosis/spermatogenesis.
Figure 3.
PHB is essential for meiotic recombination in spermatocytes. (A) Immunostaining of γH2AX (red) and SYCP3 (green) in chromosome spreads of spermatocytes prepared from the testes of Phb-cKO mice (right) and the controls (left). Scale bar = 5 μm. (B) Comparison of frequencies of meiotic stages in the testes of Phb-cKO mice and the controls. (C) Immunostaining of RPA2 (red, top), RAD51 (red, middle), and DMC1 (red, bottom) in meiotic spreads of pachytene spermatocytes prepared from the testes of Phb-cKO mice and the controls. SYCP3 (green) was used as a staining control. Scale bar = 5 μm. (D) Comparison of foci numbers of RPA2, RAD51, and DMC1 in leptotene, zygotene, and pachytene spermatocytes prepared from the testes of Phb-cKO mice and the controls, respectively. n = 3, Error bars, S.D. **P< 0.01 by one-tailed Mann–Whitney test.
PHB maintains mitochondrial function in spermatocytes
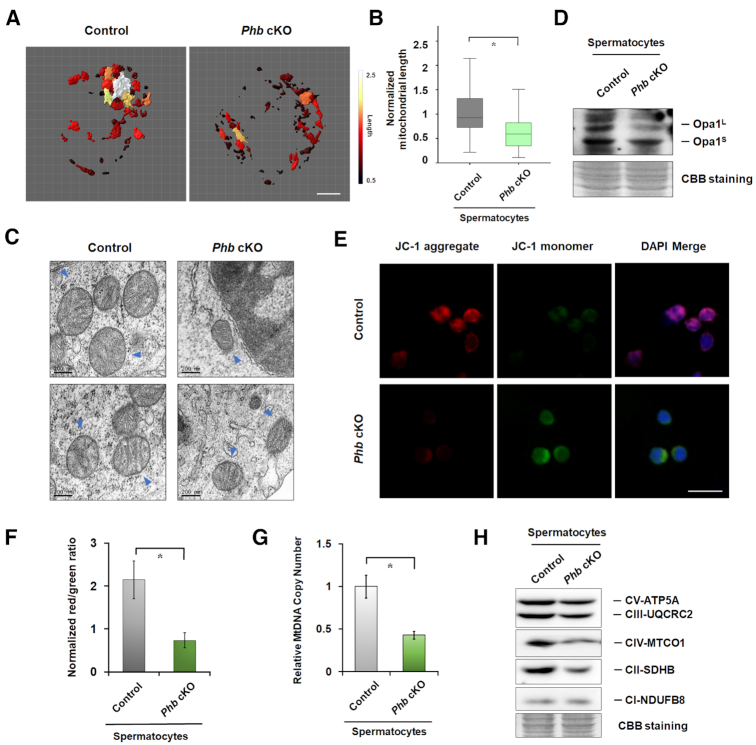
Our previous studies have shown that PHB expression is correlated with mitochondrial function in human sperm (5,6). We thus investigated whether PHB could also modulate the mitochondrial morphology and function in spermatocytes. Tomm-20, an outer mitochondrial membrane (OMM) protein, was used to evaluate the morphology and length of mitochondria based on its immune-positive signal (32). The results of both immunostaining of Tomm-20 (Figure 4A, B, Supplementary Figure S8A, B) and transmission electron microscopy (Figure 4C) confirmed that PHB deficiency could lead to a significant accumulation of short and fragmented mitochondria in spermatocytes in vivo and in vitro. This finding is also accompanied by the reduction of the long isoform of OPA1 (Figure 4D), a marker of mitochondrial fusion. Compared to the controls, PHB KD GC2 cells and Phb−/− spermatocytes exhibited a decrease in both mitochondrial membrane potential (Figure 4E, F, Supplementary Figure S8C, D) and copy numbers of mtDNA (Figure 4G, Supplementary Figure S8E), the latter of which could lead to the impairment of the respiratory complex. Indeed, the levels of respiratory complex subunits CII and CIV were significantly reduced in mitochondria isolated from the testes of Phb-cKO mice (Figure 4H). These data suggest that PHB also plays a key role in the maintenance of mitochondrial function in spermatocytes.
Figure 4.
PHB regulates mitochondrial morphology and function in spermatocytes. (A) Reconstruction of Tomm-20 (red) signals on the mitochondrial membranes of FACS-isolated control and Phb−/− spermatocytes. The lengths of the mitochondria were measured and color-coded. Scale bar = 5 μm. (B) Quantification analysis of mitochondrial length between two groups. n = 50, Error bars, S.D. *P<0.05 by two-tailed Student's t-test. (C). The structures of mitochondria in control and Phb−/− spermatocytes, as characterized by transmission electron microscopy, show a short mitochondria in Phb−/− spermatocytes (arrowed head). Scale bar = 200 nm. (D) Western blot analysis showing the proteolysis of long-isoform OPA1, a marker of mitochondrial fusion, with β-actin as a loading control. (E) Representative images showing aggregated (red) or monomeric (green) JC-1 in Phb−/− spermatocytes and the controls. Scale bar = 50 μm. (F) The fluorescence red/green intensity for JC-1 staining in control and Phb−/− spermatocytes. n = 3, Error bars, S.D. *P<0.05 by two-tailed Student's t-test. (G) The mtDNA copy number ratio to nuclear DNA in control and Phb−/− spermatocytes. n = 3, Error bars, S.D. *P< 0.05 by two-tailed Student's t-test. (H) Western blot analysis of respiratory complex subunits. Coomassie blue staining of the membrane was used as a control.
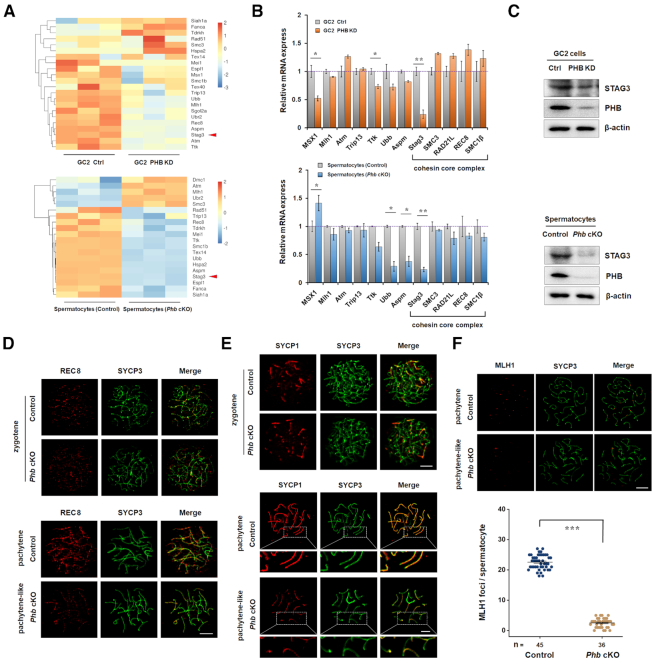
Impairment of the synapsis in Phb−/− spermatocytes is accompanied by loss of STAG3
To gain insight into the mechanism of meiotic arrest with PHB depletion, we analyzed the spermatocyte transcriptomes in the Phb-cKO mice and control (Figure 5). Based on a combined analysis of RNA-seq data from Phb−/− spermatocytes and PHB knockdown GC2 cells, we identified a set of meiosis-associated genes whose expression levels were substantially altered by the absence of PHB (Figure 5A). Among them, qRT-PCR validation indicated that Stag3 was the most repressed gene in both PHB KD GC2 cells and Phb−/− spermatocytes (Figure 5B). Western blotting also showed a significant reduction of STAG3 protein level in vitro and in vivo (Figure 5C). STAG3, a component of meiosis-specific cohesin (33), is required for chromosome pairing and formation of the synaptonemal complex (SC), by stabilizing the cohesin complexes and maintaining the protein level of REC8, a key component responsible for proper synapsis. As expected, we observed a substantial decrease of REC8 signals localized on the axial (or lateral) elements of synapsis complex (SC) in Phb−/− spermatocytes (Figure 5D). In germ cells, loss of STAG3 or other cohesin subunits routinely results in meiotic arrest with impaired DSB repair and/or aberrant synapsis (34–37). This is also consistent with our observation of incomplete SC formation in pachytene-like Phb−/− spermatocytes, evidenced by the discontinuous signal of SYCP1 (Figure 5E), a marker for transverse elements of SC. More importantly, the Phb−/− spermatocytes exhibited a reduced number of MLH1 foci per cell (control: 22.56 ± 2.36, Phb-cKO: 2.5 ± 1.52; Figure 5F), suggesting impaired crossover progression. These results suggest that STAG3 deficiency might play a role in inducing defects in synapsis and DSB repair in Phb−/− spermatocytes.
Figure 5.
Phb−/− spermatocytes have decreased STAG3 and impaired synapsis. (A) Hierarchical clustering with heat maps showing differentially expressed genes involved in the meiotic progress between PHB-knockdown and control GC2 cells (top) or between Phb−/− and control spermatocytes (bottom). Genes are ranked based on their fold change. (B) Confirmation of differential expression of meiosis related genes by qRT-PCR in PHB-knockdown GC2 cells (top) and Phb−/− spermatocytes (bottom), with the mRNA levels in control cells set to 1. β-actin was used as a reference. n = 3, error bars, S.D. *P< 0.05; **P< 0.01 by two-tailed Student's t-test. (C) Western blot showing the downregulated level of STAG3 in PHB-knockdown GC2 cells (top) and Phb−/−spermatocytes (bottom) compared with their respective controls, with β-actin serving as a loading control. n = 3. (D, E) Immunostaining of REC8 (red, D) and SYCP1 (red, E) in chromosome spreads of pachytene spermatocytes from Phb-cKO testes and the controls, with SYCP3 (green) serving as a staining control. Scale bar = 5 μm. (F) Top: Immunostaining of MLH1 (red) in chromosome spreads of control and Phb−/− pachytene spermatocytes, with SYCP3 (green) serving as a staining control. Scale bar = 5 μm. Bottom: Quantification analysis of MLH1 foci number per nucleus in control (22.56 ± 2.36) and Phb−/− (2.5 ± 1.52) pachytene spermatocytes. n = 3, error bars, S.D. ***P< 0.001 by Mann–Whitney test.
STAG3 is regulated by the PHB/JAK2 axis in spermatocytes
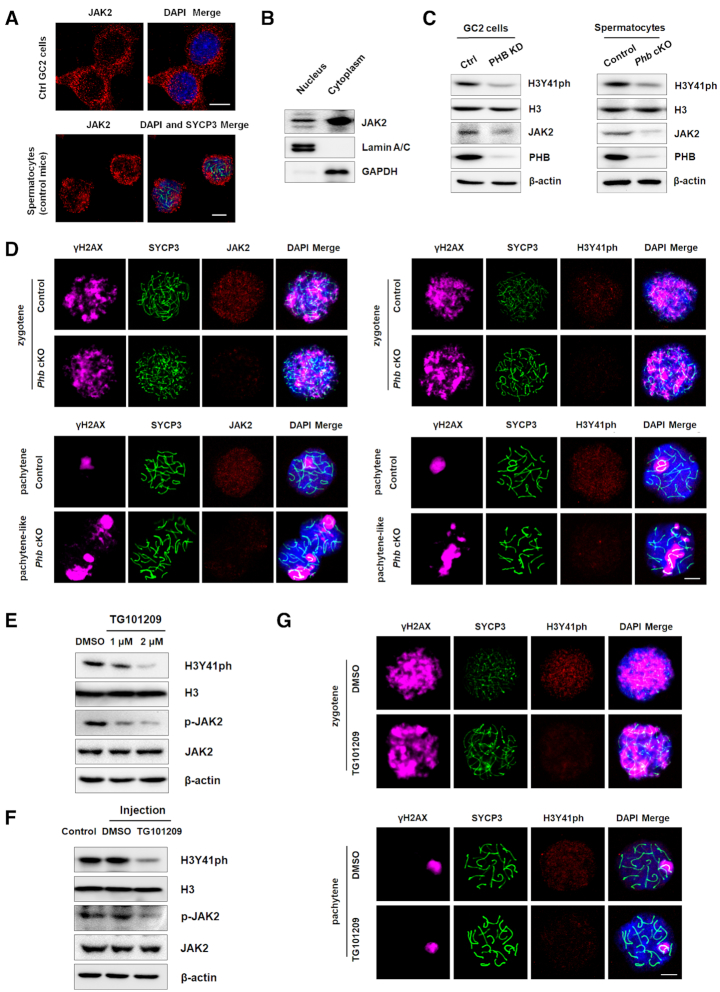
The finding that Stag3 mRNA expression decreased in both PHB knockdown GC2 cells and Phb−/− spermatocytes suggests that there could be a PHB-mediated pathway regulated at the transcriptional level. To test this hypothesis, we determined that PHB deficiency in FACS-sorted spermatocytes from Phb-cKO mice has the ability to affect JAK-STAT and PI3K-AKT pathways (Supplementary Figure S9, Figure 6A, Supplementary Figure S10A), similar to that seen in PHB knockdown GC2 cells (Figure 1E). Further investigations showed that in GC2 cells treatment with two specific and chemically distinct JAK2 inhibitors, TG101209 and CEP-33779, could block Stag3 expression at both transcriptional and protein levels in a dose-dependent manner (Figure 6B, C), whereas activating the PI3K-AKT pathway with SC-79 (an activator of PI3K-AKT pathway) showed no appreciable effect (Figure 6B, Supplementary Figure S10B). To address whether STAG3 was also altered by JAK-STAT pathway in vivo, we injected 15 μl of TG101209 into control testes and then isolated germ cells after 7 days. The results of Western blot confirmed that blocking JAK-STAT pathway indeed repressed STAG3 levels in mouse testes (Figure 6D). Furthermore, immunostaining results confirmed that both programmed DSB repair and cohesin complex formation were impaired in mouse spermatocytes after injection with TG101209 (Figure 6E). These phenotypes were similar to what we observed in Phb-cKO mice, which lend credence to our above-mentioned hypothesis that inhibition of the JAK–STAT pathway is associated with spermatogenic defects in Phb-cKO spermatocytes. Importantly, siRNA-induced suppression of JAK2, but not its downstream effectors (STAT3, STAT5 or both), was shown to be able to down-regulate the transcription of Stag3 in GC2 cells (Supplementary Figure S10C, D). Thus, we propose that STAG3 could be regulated by PHB in spermatocytes through a non-canonical JAK-STAT signaling pathway or in a STAT-independent manner.
Histone H3Y41 phosphorylation is repressed in Phb−/− spermatocytes
Previous studies indicate that JAK2 could act as a transcriptional activator in the nucleus by phosphorylating tyrosine 41 of the histone H3 tail (38). This prompted us to examine whether the crosstalk between PHB/JAK2 axis and H3Y41 phosphorylation (H3Y41ph) is implicated in meiosis. JAK2 was observed in both cytoplasm and nuclei of GC2 cells and spermatocytes from normal testes (Figure 7A, B). On the other hand, knockdown of PHB significantly suppressed the expression of H3Y41ph in spermatocytes (Figure 7C). These results were further corroborated by immunostaining of chromosomal spreads of zygotene/pachytene Phb-null spermatocytes (Figure 7D). Importantly, a similar inhibitory effect on H3Y41ph could be observed in TG101209-treated GC2 cells (Figure 7E) or spermatocytes isolated from TG101209-injected mouse testes (Figure 7F, G). Taken together, these data provide convincing evidence that the PHB/JAK2 axis is responsible for maintaining H3Y41ph in spermatocytes.
Figure 7.
Histone H3Y41 phosphorylation is down-regulated in Phb−/−spermatocytes. (A) Confocal immunofluorescence images confirming the nuclear localization of JAK2 (red) in GC2 cells and normal mouse spermatocytes. The nuclei are stained with DAPI (blue). Scale bar = 5 μm. (B) Expression levels of JAK2 protein in the cytoplasmic and nuclear fractions of FACS-isolated normal mouse spermatocytes. Lamin A/C and GAPDH are used as nuclear and cytoplasmic markers, respectively. (C) Western blot showing the down-regulated H3Y41ph in PHB-deficient spermatocytes (right) or GC2 cells (left), with H3 serving as a loading control. (D) Immunostaining of JAK2 (red, left) and H3Y41ph (red, right) in chromosome spreads from control and Phb-cKO testes, with γH2AX (magenta) and SYCP3 (green) serving as staining controls. The nuclei are stained with DAPI (blue). Scale bar = 5 μm. (E, F) Expression levels of H3Y41ph in inhibitor TG101209-treated GC2 cells (E) and mouse spermatocytes isolated from inhibitor TG101209-injected testes (F), with H3 serving as a loading control. (G) Immunostaining of H3Y41ph (red) in mouse spermatocytes isolated from DMSO- or inhibitor TG101209-injected testes, with γH2AX (magenta) and SYCP3 (green) serving as staining controls. The nuclei are stained with DAPI (blue). Scale bar = 5 μm.
PHB modulates the epigenome of spermatocytes
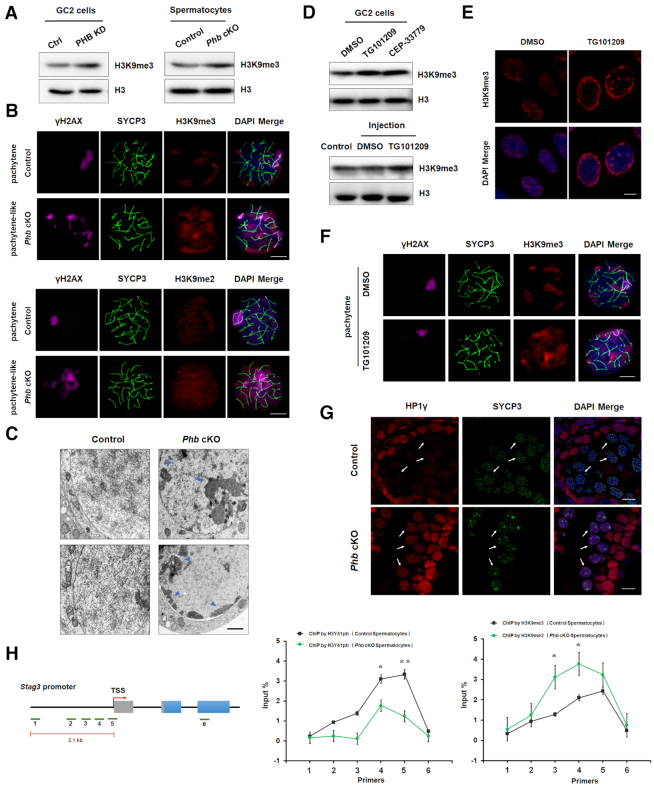
JAK2-H3Y41ph axis has been known to be involved in gene expression by modulating heterochromatin formation (38–40). Consistent with this report, the deletion of PHB in this study was shown to up-regulate H3K9me3, a transcriptionally repressive histone modification accumulated at constitutive heterochromatin, in both GC2 cells and spermatocytes (Figure 8A, Supplementary Figure S11A), especially in meiotic spreads of pachytene spermatocytes (Figure 8B). These spreads also exhibited elevated H3K9me2 levels (Figure 8B), suggesting the propagation of facultative heterochromatin. Transmission electron microscopic (TEM) imaging provided clear evidence for the accumulation of condensed chromatin at the nuclear periphery in Phb−/− spermatocytes (Figure 8C). Importantly, treatment with two specific and chemically distinct JAK2 inhibitors, TG101209 and CEP-33779, exhibited the same stimulatory effect as the PHB deficiency on H3K9me3 expression in both GC2 cells and spermatocytes (Figure 8D-F), suggesting that JAK2 repression might be responsible for the increase of histone H3 methylation in Phb−/− spermatocytes. H3K9me2/3 is known to modulate heterochromatin assembly by recruiting heterochromatin protein 1 (HP1) (41). Among the three members of the mammalian HP1 family, the levels of HP1α and HP1β were relatively weak in spermatocytes from both genotypes (Supplementary Figure S11C). Only HP1γ was abnormally accumulated in Phb−/− spermatocytes at pachytene stage (Figure 8G), which coincided with its pivotal role in facilitating H3K9 epigenetic modifications in male germ cells (42). Furthermore, ChIP assays revealed that Phb−/− spermatocytes harbored a retention of H3K9me3 surrounding the Stag3 promoter region, accompanied by a reduction of H3Y41ph levels (Figure 8H). These data suggest that JAK2-mediated epigenetic modifications could be responsible for Stag3 dysregulation in Phb−/− spermatocytes.
Figure 8.
Loss of PHB promotes heterochromatin formation in spermatocytes. (A) Western blot analysis of H3K9me3 in control and Phb−/− spermatocytes, with H3 serving as a loading control. (B) Immunostaining of H3K9me3 (top, red) and H3K9me2 (bottom, red) in control and Phb−/− spermatocytes at the pachytene stage, with γH2AX (magenta) and SYCP3 (green) serving as a staining control. Scale bar = 5 μm. (C) Representative TEM images of control and Phb−/− spermatocytes, showing increased levels of heterochromatin in the latter (arrowed head). Scale bar = 200 nm. (D). Western blot showing H3K9me3 levels in GC2 cells treated with JAK2 inhibitor (TG101209 or CEP-33779) and mouse spermatocytes isolated from inhibitor TG101209-injected testes, with H3 serving as a loading control. (E) Immunostaining of H3K9me3 (red) in DMSO- or inhibitor TG101209-treated GC2 cells. Nuclei were stained with DAPI (blue). Scale bar = 5 μm. (F) Immunostaining of H3K9me3 (red) in mouse spermatocytes isolated from DMSO- or inhibitor TG101209-injected testes, with SYCP3 (green) serving as a staining control. Scale bar = 5 μm. (G) Immunostaining of HP1γ (red) in testes sections prepared from Phb-cKO mice and the controls. SYCP3 (green) is used as a spermatocyte marker. The nuclei are stained with DAPI (blue). These arrows point to SYCP3-positive spermatocytes. Scale bar = 10 μm. (H) ChIP analyses of H3Y41ph (middle) and H3K9me3 (right) at Stag3 locus in control and Phb−/−spermatocytes. Left: Schematic view of the six regions investigated from Stag3 locus. The median value of signal was normalized to that of input DNA. n = 3, error bars, S.D. *P< 0.05; **P< 0.01 by two-tailed Student's t-test.
DISCUSSION
PHB performs important functional roles in spermatocytes
The present study shows that PHB, which is known to be expressed in human sperm where it plays a role in their motility (5,6), is also expressed throughout the murine spermatogenic process where its expression levels are high in spermatocytes. This expression pattern is different from that in the rat where PHB is not seen in spermatids or sperm, but is highly expressed in leptotene and pachytene spermatocytes (9). The similarity of high expression in spermatocytes in mice and rats suggests that PHB might play an important role in mammalian spermatocytes during meiosis. As we expected, a critical role for PHB in spermatocytes was seen when Phb was deleted in these cells via a conditional knockout using the CRISPR/Cas9 system. Our results show that male mice lacking PHB are sterile with significantly reduced testis weight and the absence of sperm in the epididymis. This is accompanied by the loss of post-meiotic germ cells due to meiotic arrest and apoptotic spermatocytes, as early as in the first wave of spermatogenesis.
It is known that male germ cell apoptosis is mainly induced by two major apoptotic pathways: the extrinsic or death receptor pathway and the intrinsic or mitochondrial pathway (43). In the present study, as shown in Figure 4 and Supplementary Figure S8, severely mitochondrial dysfunctions were found in PHB deficient spermatocytes, for example, accumulation of short and fragmented mitochondria, reduction of the long isoform of mitochondrial fusion marker (OPA1), a decrease in mitochondrial membrane potential and copy numbers of mtDNA, and reduced levels of respiratory complex subunits CII and CIV. These severe mitochondrial dysfunctions may result in the prevalence of TUNEL-positive spermatocytes in 10 dpp and 14 dpp Phb-cKO testis, based on their highly activated mitochondrial apoptotic effectors, for example, released Cytochrome c, increased pro-apoptotic protein BAX expression and caspase-9 activity (Supplementary Figure S12A-C). Collectively, our finding of spermatocyte apoptosis induced by an intrinsic pathway suggest that PHB plays a key role in the maintenance of mitochondrial function in spermatocytes and could be responsible for the apoptosis of spermatocytes observed in the seminiferous tubules of Phb-cKO mice, as early as in the first wave of spermatogenesis.
However, the lack of maintenance of mitochondrial function in Phb-deficient mice might only be partly responsible for the loss of spermatocytes, since we detected that the loss is also associated with meiotic defects, namely, pachytene arrest and impaired double strand break (DSB) repair during prophase I (Figure 3 and Supplementary Figure S7). Moreover, these recombination defects could trigger pachytene apoptosis as reported (44). This is because in Phb-cKO spermatocytes at 14 dpp, but not 10 dpp, we detected increased p53 signals (Supplementary Figure S12D), which is an effector responsible for activating recombination-dependent arrest (45). Accordingly, our above findings further suggest that spermatocyte apoptosis at early/mid-pachytene stage is recombination-dependent and could be promoted by p53-induced pro-apoptotic genes.
Meiotic cohesin, a conserved ring-shaped protein structure, has been shown to be necessary for the assembly of the synaptonemal complex (SC) during prophase I. This protein which is also known as multi-subunit complex binds the sister chromatids together and then loads them onto the axial elements which comprise a distinct protein substructure of the SC (46,47). It should be noted that meiotic cohesin is crucial for sister chromatid binding and the subsequent homolog pairing and synapsis (48). Our finding of pachytene arrest in Phb-cKO mice therefore suggests that PHB deficiency might in some way be obstructing the function of meiotic cohesin.
Recently, it was reported that mice with the loss of a core subunit protein of meiotic cohesin, for example, SMC1β, RAD21L, REC8 or STAG3, exhibit male infertility (37,48–50). Common to the knockout mice for the relevant gene for each of the above-mentioned proteins is the finding that meiotic arrest occurs prior to the late pachytene stage (37,48,49). Interestingly, STAG3 which has been reported to stabilize meiotic cohesin onto the axial elements of the SC (49) was significantly downregulated in Phb−/− spermatocytes both in vivo and in vitro, as shown in Figure 5. These results suggest that STAG3 deficiency might play a role in inducing defects in synapsis and DSB repair in Phb−/− spermatocytes. Thus PHB appears to play an essential role in maintaining the integrity of meiotic cohesin in spermatocytes. It is worth noting that these defects seen in Phb-KO mice are similar in nature to, but less severe than, what was reported in mice with a Stag3 mutation (34). This is supported by the finding that Phb−/− spermatocytes, shown in Figure 5D, have a higher residual level of REC8 loading onto the chromosome axes, when compared with the report that lower REC8 expression was found with a mutated Stag3 (34). Also, these Phb−/− spermatocytes, shown in Figure 5C, have a higher level of STAG3 than those of the reported Stag3 mutants (34). Collectively, the above results support the notion that PHB/STAG3 plays a key role in regulating the stabilization of meiotic STAG3/REC8 cohesin complex in a dose-dependent manner.
Involvement of spermatocyte PHB in a non-canonical JAK/STAT signaling pathway
The possible signal pathway mediated by PHB in regulating the function of spermatocytes during prophase I has not been investigated. Following a genome-wide analysis, we found that the deletion of PHB in spermatocytes predominantly affected JAK/STAT and PI3K/AKT pathways, in the absence of additional adverse effects (Supplementary Figure S13). The latter, PI3K/AKT signaling pathway, is the same target mediated by PHB as that in cancer cells and endothelial cells, as reported recently (4,51). On the other hand, for the JAK/STAT signaling pathway, PHB has been reported to be up-regulated by IL-6-dependent STAT3 phosphorylation in somatic cells (52). However, the crosstalk between the JAK/STAT pathway and PHB-mediated signaling network is still poorly understood in germ cells. Surprisingly, as shown in Figures 1, 6 and 7, in our present study we discovered that JAK2 was repressed in both FACS-sorted Phb−/− spermatocytes and PHB-knockdown GC2 cells, indicating that PHB is a critical regulator of the JAK/STAT pathway in germ cells. Furthermore, as shown in Figures 5D–F and 6E, inactivation of JAK2 via the deletion of PHB or the use of a chemical inhibitor disrupted the structure of the meiotic cohesin complex and thus caused defects in homologous synapsis. This clearly suggests a novel role for JAK2 in modulating sister chromatid cohesion, meiotic chromosome axis structure and synaptonemal complex, and recombination. These novel findings could help provide a better understanding of the function of JAK2 in spermatocytes.
To gain an insight into the downstream target mediated by the PHB/JAK2 axis in spermatocytes, further investigation was conducted and revealed that the PHB/JAK2 axis exerts its regulatory effects on meiotic cells via a non-canonical pathway, likely involving dynamic epigenetic changes in histone modifications. As shown in Figure 7, JAK2 and its induced partner, H3Y41ph, were detected at high levels in the nuclei of spermatocytes. This is consistent with the report that JAK2 localizes in the nuclei of somatic and embryonic stem (ES) cells where it acts as a transcriptional modulator by promoting the phosphorylation of H3Y41 (26,38,53). The above findings are consistent with the observation that treatment with JAK2 inhibitors could reverse the stimulatory effects of JAK2 on H3Y41ph levels in spermatocytes both in vitro and in vivo.
On the other hand, given that the inactivation of JAK2 and the dephosphorylation of STAT have been shown to promote heterochromatin formation in Drosophila and mammalian somatic cells (54–56), we have proposed that the loss of PHB could down-regulate the mRNA levels of its downstream targets in part through heterochromatin-mediated transcriptional silencing. Indeed, compared to controls, abnormally higher levels of H3K9me3 was observed in Phb-null spermatocytes at the zygotene and pachytene stages in prophase I of meiosis (Figure 8B and Supplementary Figure S11A-B). This was confirmed after JAK2 inhibitors treatment of spermatocytes both in vivo and in vitro (Figure 8D–F). Also, ChIP assays revealed that Phb−/− spermatocytes harbored a retention of H3K9me3 at the Stag3 promoter region, accompanied by a reduction of the level of H3Y41ph. Additionally, accumulation of heterochromatin in Phb−/− spermatocytes was corroborated directly by TEM imaging and indirectly by immunostaining of an increase in nuclear HP1γ, which has the ability of recognizing H3K9 methylation and promoting heterochromatin packaging in male germ cells (42). It should be noted that the regulatory roles of JAK2 seemed to be target-dependent in different types of cells or tissues. For example, JAK2 could mediate the self-renewal of ES cells by modulating the expression of Nanog (26), and support the proliferation of lymphoma cells by remodeling the epigenetic modification at the Myc locus (39). Our present study shows that JAK2 mediates meiotic recombination in spermatocytes by remodeling the epigenetic modifications at the Stag3 promoter region. This finding suggests that identification of a specific target may contribute to understanding the unique biological function of JAK2-mediated transcriptional regulation in different tissues.
Since the majority of mitochondrial proteins are encoded in the nuclear genome, the altered chromatin structure and aberrant patterns of gene transcription in Phb−/− spermatocytes may, in turn, affect the expression levels of mitochondrial proteins, which could drive further mitochondrial dysfunction and oxidative damage. In somatic cells, this feedback loop was previously proven to be mediated by metabolism-epigenome-genome axis, in which disturbances in mitochondrial dynamics influence the histone modification via modulating the metabolite levels (57). We therefore hypothesize that the PHB/JAK2 axis may act as a novel second messenger in the communication between the nucleus and mitochondria in spermatocytes, which could be essential for promoting meiotic recombination and maintaining proper mitochondrial function. This scenario underscores the pleiotropic nature of the Phb gene.
In summary, the results of our present study indicate a novel function of PHB in regulating the stabilization of meiotic STAG3 cohesin in spermatocytes during meiotic prophase I. The related mechanism occurs most probably through transcriptional silencing of Stag3 via a non-canonical JAK/STAT pathway. This pathway likely involves dynamic epigenetic changes in histone modifications mediated via the PHB/JAK2 axis. Further studies are needed to identify PHB-regulated genes at different stages of spermatogenesis or oogenesis. Such studies would offer new insights into the biological functions of mitochondrial proteins in mammalian gametogenesis.
DATA AVAILABILITY
RNA-seq data have been deposited in the NCBI Gene Expression Omnibus database. The GEO accession number is GSE140346.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Ming-Han Tong at the CAS Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences for kindly gifting the Stra8-GFPCre mice and advices on data analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81873855, 81270738]; Major State Basic Research Development Program of China (973 Program) [2014CB943104]. Funding for open access charges: National Natural Science Foundation of China [81873855]; Major State Basic Research Development Program of China (973 Program) [2014CB943104].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ande S.R., Nguyen K.H., Nyomba B.L.G., Mishra S.. Prohibitin in adipose and immune functions. Trends Endocrinol. Metab. 2016; 27:531–541. [DOI] [PubMed] [Google Scholar]
- 2. Ko K.S., Tomasi M.L., Iglesias-Ara A., French B.A., French S.W., Ramani K., Lozano J.J., Oh P., He L., Stiles B.L. et al.. Liver-specific deletion of prohibitin 1 results in spontaneous liver injury, fibrosis, and hepatocellular carcinoma in mice. Hepatology. 2010; 52:2096–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hernando-Rodriguez B., Artal-Sanz M.. Mitochondrial quality control mechanisms and the PHB (Prohibitin) complex. Cells. 2018; 7:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schleicher M., Shepherd B.R., Suarez Y., Fernandez-Hernando C., Yu J., Pan Y., Acevedo L.M., Shadel G.S., Sessa W.C.. Prohibitin-1 maintains the angiogenic capacity of endothelial cells by regulating mitochondrial function and senescence. J. Cell Biol. 2008; 180:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang M.J., Ou J.X., Chen G.W., Wu J.P., Shi H.J., O W.S., Martin-DeLeon P.A., Chen H.. Does prohibitin expression regulate sperm mitochondrial membrane potential, sperm motility, and male fertility. Antioxid. Redox Signal. 2012; 17:513–519. [DOI] [PubMed] [Google Scholar]
- 6. Chai R.R., Chen G.W., Shi H.J., O W.S., Martin-DeLeon P.A., Chen H.. Prohibitin involvement in the generation of mitochondrial superoxide at complex I in human sperm. J. Cell. Mol. Med. 2017; 21:121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sutovsky P., Moreno R.D., Ramalho-Santos J., Dominko T., Simerly C., Schatten G.. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 2000; 63:582–590. [DOI] [PubMed] [Google Scholar]
- 8. Thompson W.E., Ramalho-Santos J., Sutovsky P.. Ubiquitination of prohibitin in mammalian sperm mitochondria: possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biol. Reprod. 2003; 69:254–260. [DOI] [PubMed] [Google Scholar]
- 9. Choongkittaworn N.M., Kim K.H., Danner D.B., Griswold M.D.. Expression of prohibitin in rat seminiferous epithelium. Biol. Reprod. 1993; 49:300–310. [DOI] [PubMed] [Google Scholar]
- 10. Jan S.Z., Hamer G., Repping S., de Rooij D.G., van Pelt A.M., Vormer T.L.. Molecular control of rodent spermatogenesis. Biochim. Biophys. Acta. 2012; 1822:1838–1850. [DOI] [PubMed] [Google Scholar]
- 11. Handel M.A., Schimenti J.C.. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2010; 11:124–136. [DOI] [PubMed] [Google Scholar]
- 12. Hofmann M.C., Abramian D., Millan J.L.. A haploid and a diploid cell coexist in an in vitro immortalized spermatogenic cell line. Dev. Genet. 1995; 16:119–127. [DOI] [PubMed] [Google Scholar]
- 13. Dai P., Wang X., Gou L.T., Li Z.T., Wen Z., Chen Z.G., Hua M.M., Zhong A., Wang L., Su H. et al.. A Translation-Activating function of MIWI/piRNA during mouse spermiogenesis. Cell. 2019; 179:1566–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gou L.T., Dai P., Yang J.H., Xue Y., Hu Y.P., Zhou Y., Kang J.Y., Wang X., Li H., Hua M.M. et al.. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014; 24:680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin Z., Hsu P.J., Xing X., Fang J., Lu Z., Zou Q., Zhang K.J., Zhang X., Zhou Y., Zhang T. et al.. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 2017; 27:1216–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Y., Wu F., Zhang M., Xiong Z., Yin Q., Ru Y., Shi H., Li J., Mao S., Li Y. et al.. EMC10 governs male fertility via maintaining sperm ion balance. J. Mol. Cell Biol. 2018; 10:503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L.F., Lou J.T., Lu M.H., Gao C., Zhao S., Li B., Liang S., Li Y., Li D., Liu M.F.. Suppression of miR-199a maturation by HuR is crucial for hypoxia-induced glycolytic switch in hepatocellular carcinoma. EMBO J. 2015; 34:2671–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bellani M.A., Romanienko P.J., Cairatti D.A., Camerini-Otero R.D.. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm-/- spermatocytes. J. Cell Sci. 2005; 118:3233–3245. [DOI] [PubMed] [Google Scholar]
- 19. Gaysinskaya V., Soh I.Y., van der Heijden G.W., Bortvin A.. Optimized flow cytometry isolation of murine spermatocytes. Cytometry A. 2014; 85:556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J.P.. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007; 23:3251–3253. [DOI] [PubMed] [Google Scholar]
- 23. Jiang S., Zhang L.F., Zhang H.W., Hu S., Lu M.H., Liang S., Li B., Li Y., Li D., Wang E.D. et al.. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012; 31:1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al.. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 25. Mishra S., Ande S.R., Nyomba B.L.. The role of prohibitin in cell signaling. FEBS J. 2010; 277:3937–3946. [DOI] [PubMed] [Google Scholar]
- 26. Griffiths D.S., Li J., Dawson M.A., Trotter M.W., Cheng Y.H., Smith A.M., Mansfield W., Liu P., Kouzarides T., Nichols J. et al.. LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat. Cell Biol. 2011; 13:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He J., Zhang Y.. Janus kinase 2: an epigenetic ‘writer’ that activates leukemogenic genes. J. Mol. Cell Biol. 2010; 2:231–233. [DOI] [PubMed] [Google Scholar]
- 28. Bellve A.R., Cavicchia J.C., Millette C.F., O’Brien D.A., Bhatnagar Y.M., Dym M.. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 1977; 74:68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ellis P.J., Furlong R.A., Wilson A., Morris S., Carter D., Oliver G., Print C., Burgoyne P.S., Loveland K.L., Affara N.A.. Modulation of the mouse testis transcriptome during postnatal development and in selected models of male infertility. Mol. Hum. Reprod. 2004; 10:271–281. [DOI] [PubMed] [Google Scholar]
- 30. Ribeiro J., Abby E., Livera G., Martini E.. RPA homologs and ssDNA processing during meiotic recombination. Chromosoma. 2016; 125:265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gerton J.L., Hawley R.S.. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 2005; 6:477–487. [DOI] [PubMed] [Google Scholar]
- 32. Ward J.M., Stoyas C.A., Switonski P.M., Ichou F., Fan W., Collins B., Wall C.E., Adanyeguh I., Niu C., Sopher B.L. et al.. Metabolic and organelle morphology defects in mice and human patients define spinocerebellar ataxia type 7 as a mitochondrial disease. Cell Rep. 2019; 26:1189–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bayes M., Prieto I., Noguchi J., Barbero J.L., Perez Jurado L.A.. Evaluation of the Stag3 gene and the synaptonemal complex in a rat model (as/as) for male infertility. Mol. Reprod. Dev. 2001; 60:414–417. [DOI] [PubMed] [Google Scholar]
- 34. Fukuda T., Fukuda N., Agostinho A., Hernandez-Hernandez A., Kouznetsova A., Hoog C.. STAG3-mediated stabilization of REC8 cohesin complexes promotes chromosome synapsis during meiosis. EMBO J. 2014; 33:1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Llano E., Gomez H.L., Garcia-Tunon I., Sanchez-Martin M., Caburet S., Barbero J.L., Schimenti J.C., Veitia R.A., Pendas A.M.. STAG3 is a strong candidate gene for male infertility. Hum. Mol. Genet. 2014; 23:3421–3431. [DOI] [PubMed] [Google Scholar]
- 36. Winters T., McNicoll F., Jessberger R.. Meiotic cohesin STAG3 is required for chromosome axis formation and sister chromatid cohesion. EMBO J. 2014; 33:1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hopkins J., Hwang G., Jacob J., Sapp N., Bedigian R., Oka K., Overbeek P., Murray S., Jordan P.W.. Meiosis-specific cohesin component, Stag3 is essential for maintaining centromere chromatid cohesion, and required for DNA repair and synapsis between homologous chromosomes. PLoS Genet. 2014; 10:e1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dawson M.A., Bannister A.J., Gottgens B., Foster S.D., Bartke T., Green A.R., Kouzarides T.. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009; 461:819–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rui L., Emre N.C., Kruhlak M.J., Chung H.J., Steidl C., Slack G., Wright G.W., Lenz G., Ngo V.N., Shaffer A.L. et al.. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010; 18:590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsurumi A., Zhao C., Li W.X.. Canonical and non-canonical JAK/STAT transcriptional targets may be involved in distinct and overlapping cellular processes. BMC Genomics. 2017; 18:718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nestorov P., Tardat M., Peters A.H.. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr. Top. Dev. Biol. 2013; 104:243–291. [DOI] [PubMed] [Google Scholar]
- 42. Takada Y., Naruse C., Costa Y., Shirakawa T., Tachibana M., Sharif J., Kezuka-Shiotani F., Kakiuchi D., Masumoto H., Shinkai Y. et al.. HP1gamma links histone methylation marks to meiotic synapsis in mice. Development. 2011; 138:4207–4217. [DOI] [PubMed] [Google Scholar]
- 43. Shaha C., Tripathi R., Mishra D.P.. Male germ cell apoptosis: regulation and biology. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 2010; 365:1501–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barchi M., Mahadevaiah S., Di Giacomo M., Baudat F., de Rooij D.G., Burgoyne P.S., Jasin M., Keeney S.. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol. Cell. Biol. 2005; 25:7203–7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marcet-Ortega M., Pacheco S., Martinez-Marchal A., Castillo H., Flores E., Jasin M., Keeney S., Roig I.. p53 and TAp63 participate in the recombination-dependent pachytene arrest in mouse spermatocytes. PLoS Genet. 2017; 13:e1006845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klein F., Mahr P., Galova M., Buonomo S.B., Michaelis C., Nairz K., Nasmyth K.. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999; 98:91–103. [DOI] [PubMed] [Google Scholar]
- 47. Llano E., Herran Y., Garcia-Tunon I., Gutierrez-Caballero C., de Alava E., Barbero J.L., Schimenti J., de Rooij D.G., Sanchez-Martin M., Pendas A.M.. Meiotic cohesin complexes are essential for the formation of the axial element in mice. J. Cell Biol. 2012; 197:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ishiguro K.I. The cohesin complex in mammalian meiosis. Genes Cells. 2019; 24:6–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ward A., Hopkins J., McKay M., Murray S., Jordan P.W.. Genetic interactions between the meiosis-specific cohesin components, STAG3, REC8, and RAD21L. G3 (Bethesda, Md.). 2016; 6:1713–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Biswas U., Hempel K., Llano E., Pendas A., Jessberger R.. Distinct roles of meiosis-specific cohesin complexes in mammalian spermatogenesis. PLoS Genet. 2016; 12:e1006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chiu C.F., Ho M.Y., Peng J.M., Hung S.W., Lee W.H., Liang C.M., Liang S.M.. Raf activation by Ras and promotion of cellular metastasis require phosphorylation of prohibitin in the raft domain of the plasma membrane. Oncogene. 2013; 32:777–787. [DOI] [PubMed] [Google Scholar]
- 52. Theiss A.L., Obertone T.S., Merlin D., Sitaraman S.V.. Interleukin-6 transcriptionally regulates prohibitin expression in intestinal epithelial cells. J. Biol. Chem. 2007; 282:12804–12812. [DOI] [PubMed] [Google Scholar]
- 53. Kamakura S., Oishi K., Yoshimatsu T., Nakafuku M., Masuyama N., Gotoh Y.. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat. Cell Biol. 2004; 6:547–554. [DOI] [PubMed] [Google Scholar]
- 54. Shi S., Calhoun H.C., Xia F., Li J., Le L., Li W.X.. JAK signaling globally counteracts heterochromatic gene silencing. Nat. Genet. 2006; 38:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Silver-Morse L., Li W.X.. JAK-STAT in heterochromatin and genome stability. Jak-stat. 2013; 2:e26090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shi S., Larson K., Guo D., Lim S.J., Dutta P., Yan S.J., Li W.X.. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat. Cell Biol. 2008; 10:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aon M.A., Cortassa S., Juhaszova M., Sollott S.J.. Mitochondrial health, the epigenome and healthspan. Clin. Sci. (Lond.). 2016; 130:1285–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the NCBI Gene Expression Omnibus database. The GEO accession number is GSE140346.