Abstract
G-quadruplexes represent unique roadblocks to DNA replication, which tends to stall at these secondary structures. Although G-quadruplexes can be found throughout the genome, telomeres, due to their G-richness, are particularly predisposed to forming these structures and thus represent difficult-to-replicate regions. Here, we demonstrate that exonuclease 1 (EXO1) plays a key role in the resolution of, and replication through, telomeric G-quadruplexes. When replication forks encounter G-quadruplexes, EXO1 resects the nascent DNA proximal to these structures to facilitate fork progression and faithful replication. In the absence of EXO1, forks accumulate at stabilized G-quadruplexes and ultimately collapse. These collapsed forks are preferentially repaired via error-prone end joining as depletion of EXO1 diverts repair away from error-free homology-dependent repair. Such aberrant repair leads to increased genomic instability, which is exacerbated at chromosome termini in the form of dysfunction and telomere loss.
INTRODUCTION
Mammalian telomeres are comprised of a unique DNA:protein architecture, which protectively caps the ends of chromosomes (1–3). The DNA portion of a human telomere consists of ∼2–12 kb of a six-nucleotide (TTAGGG) repeat. The extreme terminus of the telomere ends with a 3′ G-rich, single-stranded overhang that loops back into the double-stranded telomere to form a t-loop (4). The protein portion of a telomere is provided by a six-protein complex known as Shelterin that binds to the bulk of the telomere (5). Shelterin provides at least two critical activities to telomere maintenance: (i) it facilitates telomere looping and (ii) it masks the ends of telomeres from the DNA repair machinery (1). Importantly, this proper sheltering of chromosome ends is essential to protect them from undue erosion and fusion events, which would otherwise contribute to genomic instability and cellular death.
Integral to faithful telomere maintenance is the error-free replication of telomeric DNA. Telomere replication is inherently difficult due to (i) the repetitive nature of the telomeric DNA sequence; (ii) the secondary structures it is capable of forming; and (iii) the heterochromatic nature of this genomic region (6,7). Because of these features, telomeres are formally defined as ‘fragile sites’—genomic locations that have an intrinsic tendency to induce replication fork stalling and fork collapse (8,9). One of the most potentially pathological features of a telomere is the frequent presence of G-quadruplexes (G4s), which act as a roadblock to telomere replication (7). Classical G4s consist of four tracts of guanine trios bonded together in a square-planar orientation (10). However, G4s can also arise wherever four sets of three guanine bases or more are separated by several base pairs of any sequence (11). These quadruplexes have been proposed to form in both an inter- and intrastranded manner as well as in both parallel and antiparallel formations. Computational estimates for the number of sequences capable of forming a G4 in the genome vary, but most agree that there are likely hundreds of thousands of such putative sequences with at least 10,000 existing at any given time in any given cell (12,13). Due to the triplicated run of guanines in the telomere tandem repeat (TTAGGG), telomeric DNA has a high propensity to form these structures. While positive, regulatory roles have been proposed for these arrangements (14,15), G4s are unequivocally impediments to the replication machinery. Consistent with this belief, somatic copy-number alteration breakpoints are enriched at sequences with the potential to form G4s (16). A parsimonious interpretation of these data is that it is likely that replication forks stall when they encounter these secondary structures, which leads to improper replication and repair resulting in copy-number alterations. A corollary of this interpretation is that the proper resolution and replication of these regions is likely essential for telomere integrity and genomic stability.
To overcome their inherent replication difficulties, telomeres rely heavily on a number of specialized proteins, specifically helicases and nucleases, to combat fork stalling and to resolve secondary structures (17,18). Arguably, two of the most important helicases for faithful telomere replication are Werner (WRN) and regulator of telomere elongation 1 (RTEL1). WRN is essential for faithful lagging-strand replication at the telomere and for prevention of telomere loss (19). In contrast, the RTEL1 helicase is allowed transient entry at the t-loop site to both unwind it and permit access of the replication machinery (20). In the absence of RTEL1, rapid telomere attrition is observed due to the loss of the t-loop, presumably after replication fork collapse (21). In a complementary fashion, nucleases such as exonuclease 1 (EXO1), C-terminal interacting protein (CtIP), DNA exonuclease 2 (DNA2) and meiotic recombination defective 11 (MRE11) have been proposed to have multiple roles in telomeric replication ranging from overhang formation (22) to rescue, restart and reversal of stalled forks (18).
EXO1 is 5′ → 3′ exonuclease whose canonical roles are in DNA mismatch repair and resection at the sites of DNA double-stranded breaks (DSBs) in the preliminary stages of homology-dependent repair (HDR) (23). EXO1 possesses both an exonuclease domain and a structure-specific endonuclease domain, which are indispensable for its functions (23). EXO1 has a well-documented role in processing telomere ends to form the 3′ overhang necessary for t-loop generation (24). In addition, EXO1 has also been implicated in roles in replication dynamics, especially at difficult-to-replicate-through secondary structures. To this end, EXO1 has been suggested to participate in replication fork reversal, HDR-mediated rescue and nascent strand processing of stalled forks (25,26). For these functions, EXO1 is thought to be at least partially redundant with DNA2 (27,28). Relevantly, nucleolytic processing by DNA2 has also been implicated in G4 resolution and cleavage in vitro (29,30).
In this study, we show that EXO1-knockout human cells are hypersensitive to G4-stabilizing agents. Additionally, we demonstrate that telomere defects are elevated in the absence of EXO1 and are exacerbated by combining this absence with a G4 stabilizer. Mechanistically, we find that replication forks are more likely to colocalize with G4s in the absence of EXO1, consistent with increased fork stalling. Moreover, less resection takes place proximal to the G4s in these mutants compared to the parental cells. In toto, we demonstrate that resection by EXO1 is required for both faithful replication through G4s and the HDR-mediated rescue of stalled forks at these structures. Specifically, we propose that EXO1 plays an important role in the processing of replication forks that have stalled at G4s. Thus, the enrichment or stabilization of telomeric G4s and/or the removal of EXO1 result in deleterious effects on telomere integrity and impede cellular survival. Since EXO1 is often under- or overexpressed in cancer cells, these observations may have clinical significance.
MATERIALS AND METHODS
Cell lines
Human HeLa cells were obtained from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with l-glutamine, penicillin–streptomycin and 10% fetal bovine serum. Cells were maintained at 37°C in a humidified incubator with 5% CO2. U-2 OS, ARPE-19 and HCT116 cells were similarly obtained from ATCC and cultured as described for the HeLa cells with the exception that McCoy's medium, DMEM–F12 and McCoy's medium, respectively, were utilized. EXO1 was functionally inactivated in these cell lines using the CRISPR/Cas9 system. Briefly, a single-guide RNA (sgRNA) targeting the second coding exon of EXO1 was delivered in tandem with a Cas9 expression cassette (hSpCas9-2A-GFP/px459). Candidate monoclonal populations were screened and tentatively identified by polymerase chain reaction and then verified to contain EXO1 frameshift mutations via Sanger sequencing. These candidate clones were confirmed to lack EXO1 expression by western blot analysis and one confirmed clone from each background was stably complemented with an EXO1 cDNA expressed from a pcDNA3.1 plasmid obtained from Addgene.
Proximity ligation assay
Cells were plated on chamber slides and fixed with 4% paraformaldehyde for 10 min at room temperature. Samples were then permeabilized with 0.5% NP-40 for 10 min at room temperature and blocked with blocking solution (0.5% bovine serum albumin and 0.2% fish gelatin). The proximity ligation assays (PLAs) were subsequently carried out according to the manufacturer's guidelines for the Duolink anti-Mouse MINUS and Rabbit PLUS red kit (Sigma) with the exception of an additional 20 min of amplification. Images were acquired with a Leica DM LB2 and processed with the FIJI software.
G-overhang assays
Genomic DNA was purified, and the negative control samples were pre-digested with bacterial Exo1. Subsequently, all of the samples were digested with RsaI and HinfI restriction enzymes. The samples were then subjected to gel electrophoresis for 2 h at 35 V. Following electrophoresis, the gels were dried with a Bio-Rad Model 583 gel dryer for 3 h. Dried gels were hybridized with a 32P-end-labeled oligonucleotide probe (CCCTAA)4 overnight. Following hybridization and washing, the gels were exposed to a PhosphoImager screen and scanned using a Typhoon FLA 9500 imager. After native imaging, the gels were denatured, neutralized, re-probed and imaged in the denatured condition. Images were analyzed and quantified with FIJI software.
Telomere combing
Actively dividing cells were labeled with 25 μM iodo-deoxyuridine (IdU) for 1 h followed by a 2-h period without IdU; this regimen was repeated four times. Cells were then embedded into low melting point agarose plugs and digested with proteinase K for 48 h and subsequently digested with the restriction enzymes HinfI and RsaI for 24 h at 37°C. DNA was then combed onto vinylsilane-coated coverslips using the Genomic Vision™ combing system. Combed coverslips were then incubated in a 60°C chamber for 2 h. Combed DNA was subsequently fixed and denatured to allow for antibody/probe binding. IdU was detected with a primary antibody (BD, BD347580) and an AF555-conjugated secondary antibody (Invitrogen, A21127). Telomeres were identified with an AF488-conjugated PNA probe (C3TA2)3 (PNA Bio, F1004). Slides were mounted and cured overnight prior to imaging with a Zeiss total internal reflection fluorescence (TIRF) microscope, processed with Image J/FIJI and quantitated in a blinded fashion.
Single molecule analysis of resection tracks
Actively dividing cells were labeled with CldU for 24 h. Cells were then harvested and embedded in low melting point agarose plugs and digested with proteinase K for 48 h at 37°C. Plugs were then melted and the DNA was combed onto coverslips as described earlier for telomere combing. Combed DNA was fixed and processed in the native state, to allow for the detection of ssDNA and G4s. CldU was detected with a primary antibody (Abcam, ab6326) and AF555-conjugated secondary antibody (Invitrogen, A21434). G4s were detected with a primary antibody (Millipore, MABE1126 and MABE917) and AF488-conjugated secondary antibody (Invitrogen, A11001). Images were acquired with a Zeiss TIRF microscope and processed with Image J/FIJI software. ssDNA fibers were imaged, scored and measured in a double-blind manner. Relative resection was measured in pixels and then normalized.
G4 combing
Actively diving cells were labeled with EdU for 30 min. Cells were harvested, prepped and combed as described for single molecule analysis of resection tracks (SMART). After fixation, EdU was detected using a Click-iT™ reaction as described with AF488 (31). G4s were detected with a primary antibody (Millipore, MABE1126) and an AF555-conjugated secondary antibody (Invitrogen, A21127). DNA fibers were processed with a Zeiss TIRF microscope and the images were then analyzed with Image J/FIJI software and scored in a blinded fashion.
Telomere restriction fragment analysis
Genomic DNA was collected, digested with the restriction enzymes HinfI and RsaI, and separated via gel electrophoresis overnight at 35 V for ∼16 h. Gels were then depurinated with 0.25 M HCl treatment and subsequently denatured and neutralized. The DNA was then transferred to a nucleotide-optimized membrane overnight by capillary transfer for ∼18 h. DNA was then cross-linked to the membrane via ultraviolet (UV) light exposure and hybridized with a 32P-end-labeled oligonucleotide probe (CCCTAA)4 overnight at 55°C. Membranes were exposed to phosphoimaging screens for a minimum of 24 h and imaged with a Typhoon FLA 9500 imager. Images were then analyzed with FIJI software.
Telomere dysfunction-induced foci assay
Cells were cultured on chamber slides and fixed with 4% paraformaldehyde at the time of harvest at ∼70% confluency. Cells were subsequently permeabilized with 0.5% NP-40 and blocked with 0.2% fish gelatin and 0.5% bovine serum albumin in phosphate buffered saline (PBS). Primary antibodies to gamma-H2AX (Bethyl, A300-081) and telomere recognition factor 2 (TRF2; Santa Cruz, sc-271710) were used to detect DNA damage and telomeres, respectively. Primary antibodies were incubated with slides overnight at 4°C and secondary antibodies (Invitrogen, A21127 and A11008) were incubated for 45 min at room temperature. Cells were counterstained with diamidino-phenylindole (DAPI) and imaged on a Nikon TiE deconvolution microscope in z-stacks. Z-stack depth was taken with recommended optimization. Images were analyzed with MATLAB and processed with the FIJI software. Colocalization events were quantified as a minimum of 10% gamma-H2AX individual foci overlap with an individual TRF2 foci.
Immunofluorescence
Cells were cultured on chamber slides and fixed with 4% paraformaldehyde when harvested. Cells were permeabilized and blocked as described earlier for TIFs. The primary antibodies used included gamma-H2AX (Bethyl, A300-081), pRPA32 (Bethyl, A300-245) and 53BP1 (Abcam, ab175933). Secondary antibodies conjugated to an appropriate fluorophore were used to detect the proteins of interest (Invitrogen, A21127, A11001, A11008 and A31572). Cells were counterstained with DAPI to determine the nuclear localization of the protein of interest. Images were acquired with a Zeiss TIRF microscope in a blinded fashion and analyzed with FIJI software.
Immunoblotting
Cells were collected and lysed in radioimmunoprecipitation assay (RIPA) buffer plus protease inhibitors. Lysates were incubated in RIPA for 15 min at 4°C and centrifuged at 4°C for 15 min. Protein concentrations were assessed using a Bradford protein assay. Samples were then resuspended in 4× lithium dodecyl sulfate sample buffer and denatured using heat. Protein samples were electrophoresed on 4–20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes in transfer buffer (20% methanol, 11% glycine and 3.4% Tris) at 100 V. Membranes were blocked with 5% milk in Tris-buffered saline–0.1% Tween (TBST) at room temperature for 45 min. The membranes were then incubated overnight in 5% milk in TBST at 4°C with an EXO1 (Thermo Fisher, 266) or actin (Novus, NB600-503) antibody. Membranes were subsequently washed in TBST and exposed to horseradish peroxidase-conjugated secondary antibodies in 5% milk in TBST at room temperature for 1–2 h. Membranes were subsequently developed and enhanced using a commercial enhanced chemiluminescent kit.
Telomere fluorescent in situ hybridization
Cells were pre-treated with colcemid at 10 mg/ml for 3 h prior to collection. Cells were then fixed with methanol and acetic acid at a ratio of 3:1, dropped over a wet slide at a 45° angle and then washed and dried overnight in a fume hood at room temperature. Metaphase-containing slides were then rehydrated with PBS, fixed with 4% paraformaldehyde, dehydrated in a cold ethanol series and heat denatured to allow for hybridization of a Cy3-conjugated PNA telomere probe (C3TA2)3 (PNA Bio, F1002). Slides were then washed and counterstained with DAPI and dehydrated in a cold ethanol series. Images were acquired by a Zeiss TIRF microscope and analyzed with FIJI software. Images were scored in a blind fashion.
Chromosome orientation fluorescent in situ hybridization
Cells were grown in 7.5 mM BrdU and 2.5 mM BrdC for 14 h prior to treatment with colcemid at 10 mg/ml for 3 h prior to collection. The cells were then fixed, dropped over a wet slide at a 45° angle and then washed and dried overnight as described for telomere fluorescent in situ hybridization (T-FISH). Metaphase cell-containing slides were then rehydrated in PBS, stained with Hoechst 33258 and exposed to 365 nm UV light. Slides were then digested with ExoIII, washed in PBS and dehydrated in a cold ethanol series. Slides were then individually hybridized with a G- or C-strand-specific PNA probe conjugated to either Cy3 or AF488 (PNA Bio, F1008 and F1002). The slides were then washed, counterstained with DAPI and dehydrated in a cold ethanol series. Images were acquired on a Zeiss TIRF microscope and analyzed with FIJI software. Images were scored in a blind manner.
MTS assay
Actively dividing cells were plated in triplicate in 96-well plates. On the following day, cells were treated with a designated drug. After an appropriate time period, plates were incubated with CellTiter 96 Aqueous reagent for 3 h to assess viability. Optical densities were then acquired and normalized to untreated control samples for each cell line.
Cancer expression assessment
Fragments per kilobase of transcript per million mapped reads (FPKM) expression values across 17 cancer subtypes were obtained from the TCGA (The Cancer Genome Atlas) dataset. Data were obtained through The Human Protein Atlas website using a custom web scraping R script (https://github.umn.edu/linx1048/exo1). FPKM expression distributions were plotted from these data using a ‘ggplot2’ R package (Figure 7).
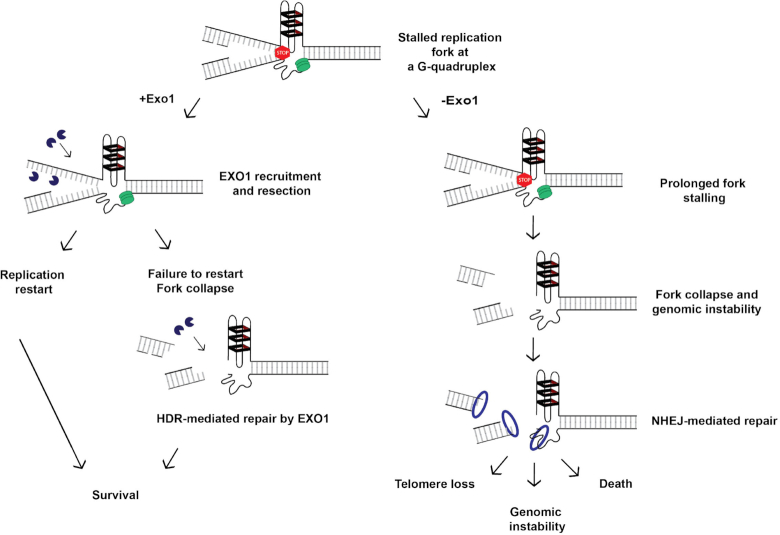
Figure 7.
Model for EXO1’s role at G4s. A schematic of a replication fork is presented. The green ovals represent the replicative helicase and associated replication machinery. The red stop sign indicates that DNA replication may stall if the lagging-strand template forms a G4 structure (hairpins with black rectangles). If the fork does stall in a cell where EXO1 is present (+Exo1; left panel), then the EXO1 (PacMan™ symbols) can facilitate either resection of the Okazaki fragments to yield an ssDNA strand with an asymmetrically located G4 (as shown in Figure 4A) or regressed fork processing (not shown). In either case, these events will lead to cellular survival. Alternatively, the fork may collapse. In this event, EXO1 will now be recruited to assist HDR to repair/restart the fork. Again, the successful completion of this process promotes cellular survival. In contrast, if the replication fork should stall in a cell lacking EXO1 (−Exo1; right panel), the fork cannot be restarted and it will ultimately break. This fork is now a substrate for NHEJ factors (generically indicated as the blue ring) such as 53BP1 (as shown in Figure 5B and D) that lead to pathological outcomes such as telomere loss, genomic instability and death due to the error-prone nature of NHEJ.
Survival data for 17 cancer subtypes were also obtained from The Human Protein Atlas website (original source: TCGA). A custom web scraping R script (see above) was used to obtain the following data for each sample: ‘living years’, ‘FPKM’ and ‘Dead’ status, and the recommended FPKM expression cutoff values for ‘high’ versus ‘low’ expression. Kaplan–Meier survival curves were generated using the ‘survival’, ‘survminer’ and ‘dplyr’ R packages. The four cancer subtypes showing significant association after multiple test correction (false discovery rate <0.05, Benjamini–Hochberg) are shown in Figure 7.
Statistics
Statistical analyses were carried out using the Prism software. Asterisks represent the degree of significance determined by the stated statistical test: *, **, *** and **** denote <0.05, <0.01, <0.001 and <0.0001, respectively. Error bars indicate the standard deviation unless otherwise noted.
RESULTS
Generation of viable EXO1-null human cells
To study the function of EXO1 at telomeres and G4s, EXO1-null cell lines were generated in multiple human cell line backgrounds. To achieve this scientific end, a clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 (CRISPR/Cas9) approach was utilized (32). sgRNAs were designed to target the second coding exon of the EXO1 gene (Supplementary Figure S1A) and frameshift mutations were successfully introduced and confirmed by Sanger sequencing. Two independent EXO1-null clones were isolated in HeLa (where the gene is triploid; Figure 1A), ARPE-19, U-2 OS (where the gene is also triploid) and HCT116 (Supplementary Figure S1B–D, respectively) backgrounds and confirmed by western blot. These cell lines represent hTERT (human telomerase)-positive cancerous (HeLa and HCT116), and ALT (alternative lengthening of telomeres)-positive cancerous (U-2 OS) and non-cancerous (ARPE-19) genetic backgrounds. Additionally, select clones from these different backgrounds were stably complemented with an EXO1-expressing cDNA to restore nuclease expression (Figure 1A; Supplementary Figure S1B–D). These complemented EXO1-mutant lines were used to analyze functional rescue of observed phenotypes in subsequent experiments. Of note, the EXO1-null HeLa cells, which were utilized most frequently, had no notable growth defect (Supplementary Figure S1E). Altogether, these data indicate that EXO1 is non-essential in human cells and that its absence does not, on its own, significantly limit proliferation.
Figure 1.
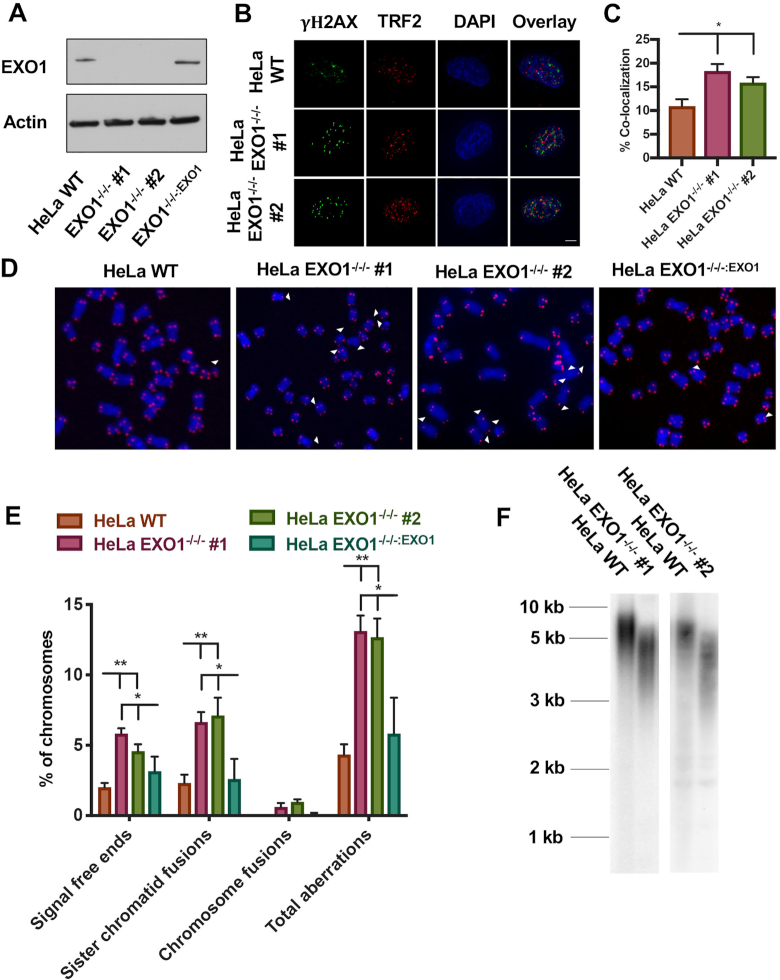
Generation and characterization of telomere defects in EXO1-null HeLa cells. (A) Western blot confirmation of EXO1 expression in the parental wild-type (WT) HeLa cells, as well as two EXO1-null clones (#1 and #2) and EXO1−/−/−:EXO1, an EXO1-null clone complemented with an EXO1 cDNA. Actin was used as a loading control. (B) DNA damage assessment at telomeres with a TIF analysis. TRF2 was used as a telomere marker and gamma-H2AX as a marker of DNA damage. Scale bar = 10 μm. (C) Quantification of the TIF analysis from (B). A paired t-test was used to evaluate statistical significance; n = 150 from three independent experiments. (D) Representative images of T-FISH analyses of EXO1-HeLa cell lines. Chromosomes were DAPI stained (blue) and telomeres imaged with a PNA probe (red). Abnormalities are marked with white arrowheads. (E) Quantification of T-FISH from (D). Sister chromatid fusions, chromosome:chromosome fusions and signal-free ends were scored. A one-way ANOVA analysis followed by Tukey's post-hoc test was used to determine statistical significance; n = 6000 from three independent experiments. (F) Telomere restriction fragment (TRF) assessment of telomere length in HeLa EXO1-null cell lines.
EXO1-null human cells have telomeric defects
EXO1 plays a role in 3′ overhang generation at telomeric ends in the mouse (24) and in yeast (28). To see whether this phenotype could be extended to human cells, we carried out a conventional non-denaturing, in-gel hybridization analysis [i.e. a G-overhang analysis (33)]. Genomic DNA was prepared from the indicated cell lines and digested to completion with frequent-cutting restriction enzymes that will not cleave the telomeric TTAGGG sequence. The samples were then pre-treated either without (−Exo1) or with (+Exo1) purified bacterial Exo1 (a 3′ → 5′ exonuclease) as a control to demonstrate that the ssDNA that hybridized in this assay corresponded to a G-overhang. Following gel electrophoresis under native conditions, the samples were hybridized with a telomere-specific probe and the signal quantitated. Finally, the gel was denatured and rehybridized with a telomere-specific probe. The ratio of the native to denatured telomeric signal constitutes an arbitrary measurement for G-overhangs (33). G-overhangs were shortened significantly in both EXO1-null cell lines (Supplementary Figure S2A and B). Thus, the loss of EXO1 in human somatic cells manifests itself in demonstrable telomere defects, although these deficits are not reflected in the overall cellular fitness (Supplementary Figure S1E).
EXO1 has also been implicated in a bevy of replication dynamics including HDR-mediated rescue of stalled forks (25,26) that are relevant for replication fork progression through a telomere. Thus, we posited that the removal of EXO1 may have deleterious effects on overall telomere integrity in human somatic cells. To determine whether EXO1-null cells have elevated levels of telomere damage, a TIF analysis was performed. Colocalization of gamma-H2AX, a marker for DNA damage, with TRF2, a Shelterin complex subunit that binds the telomeric sequence (1), was quantified (Figure 1B and C). In the EXO1-deficient HeLa cells, DNA damage at telomeres was significantly, but not dramatically, elevated compared to their WT counterparts. To expand on this, we assessed for telomere abnormalities by T-FISH (Supplementary Figure S3A). Specifically, EXO1-deficient HeLa cells displayed at least a 2-fold increase in telomere aberrations including signal-free chromosome ends and sister chromatid fusions and a modest elevation in telomere fragility (Figure 1D and E; Supplementary Figure S3D). Importantly, in every instance, the re-expression of EXO1 protein in the null cell lines suppressed these phenotypes (Figure 1D and E; Supplementary Figure S3D). To evaluate whether this increase in telomeric aberrations and damage had a negative effect on total telomere length, we performed TRF analyses. EXO1-null cells possessed significantly shorter telomeres than passage-matched WT cells (Figure 1F). ARPE-19 and HCT116 cell lines lacking EXO1 also exhibited this telomere shortening profile (Supplementary Figure S3B and C) demonstrating the generality of this defect. (Note that the extremely long telomeres in U-2 OS cells preclude their characterization by a traditional TRF analysis.) In conclusion, EXO1-null human cells exhibit spontaneous telomere defects including shortening and increased instability, although, again, these deficits are not reflected in the overall cellular fitness (Supplementary Figure S1E).
EXO1-null cells are sensitive to G4 stabilizers
To determine whether the telomeric defects observed in untreated EXO1 nuclease-deficient cells could be enhanced, the HeLa EXO1-null, complemented and WT cell lines were treated with the G4 stabilizer, pyridostatin (Supplementary Figure S4A), and toxicity assays were carried out. The EXO1-deficient HeLa cells were quite sensitive to increasing doses of pyridostatin, whereas the WT and complemented cell lines displayed only slight sensitivities at high dosages (Supplementary Figure S4B). To confirm that this sensitivity was not exclusive to HeLa cells, we performed toxicity analyses in the three additional EXO1-null human cell lines: ARPE-19, U-2 OS and HCT116. In these backgrounds, to varying degrees, an EXO1 deficiency equated to significant pyridostatin sensitivity, at least at high doses (Supplementary Figure S4C–E). To verify that this sensitivity was not due to unique non-specific effects of pyridostatin, we repeated the toxicity analyses with the small molecule, CX-5461 (Supplementary Figure S4A). CX-5461 also selectively stabilizes G4 structures and has been validated as a potential therapeutic for certain DNA repair-deficient cancers (34). Mirroring the effects of pyridostatin, EXO1-deficient HeLa cells were hypersensitive to CX-5461 treatment (Supplementary Figure S4F). These data demonstrated that an EXO1 deficiency leads to a sensitivity toward G4-stabilizing compounds in both cancerous and non-cancerous backgrounds.
EXO1 deficiency leads to telomere replication defects under G4-stabilizing conditions
To determine the source of G4 stabilizer hypersensitivity in the absence of EXO1, we explored the possibility of a replication impairment in the mutant cells. Telomeres represent a large genomic region that is highly enriched in G4s and thus a suitable region to explore the replication dynamics of these structures. Thus, to specifically gauge EXO1’s role in telomere replication, we performed telomere-specific DNA combing using modifications of an existing protocol (35). Cells were pulsed with IdU, genomic DNA was collected, a telomere enrichment was performed by restriction enzyme digestion removal of non-telomeric sequences, individual DNA fibers were combed and both telomeres and IdU incorporation were quantitated. This procedure yielded a high percentage of telomere-containing DNA fibers (Supplementary Figure S5A). Telomere fibers were then analyzed for replication tracts that coincided with the beginning of the telomere sequence, were partially replicated through the telomere or were completely replicated to the end of the telomere (left to right, respectively, Figure 2A). In unchallenged conditions, there was no significant difference between the mutant and EXO1-proficient cells (Figure 2B). However, after treatment with pyridostatin, telomere replication defects increased preferentially in the EXO1-null cells (Figure 2C). Specifically, replication tracts that ended at the beginning of the telomere, presumably in the subtelomere region, increased 2-fold in the EXO1-null mutants compared to the parental cells. Additionally, forks that had only partially replicated through the telomere were also increased. As a result, faithful, complete replication of telomeres was decreased in the absence of EXO1 (Figure 2C). Again, restoration of EXO1 expression in the mutant cell lines complemented all of these phenotypes. Importantly, there was no overall change in the frequency of unreplicated versus attempted replication events in any genetic background (Supplementary Figure S5B and C) demonstrating that the differences were not simply due to more or less replication in the mutant cell lines.
Figure 2.
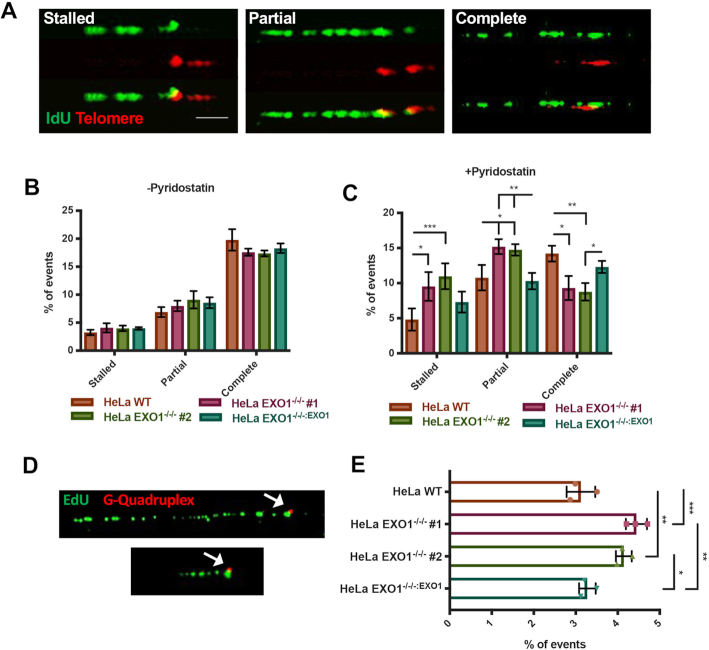
EXO1 deficiency leads to telomere replication defects under G4-stabilizing conditions. (A) Representative images of telomere replication events. Replication tracts that extend to the end of the telomere were scored as complete, tracts that are partially replicated were scored as partial and tracts at the beginning of the telomere sequence were denoted as stalled. Scale bar = 10 kb. (B) Quantification of telomere replication events in EXO1 WT, EXO1−/−/− #1 and #2, and EXO1−/−/−:EXO1; n = 200 from three independent experiments. (C) Quantification of telomere replication events in EXO1 WT, EXO1−/−/− #1 and #2, and EXO1−/−/−:EXO1 with 7.5 μM pyridostatin for 5 h prior to and through labeling. Statistical significance was determined by one-way ANOVA analysis followed by Tukey's post-hoc test; n = 200. (D) Representative images of native replication tracts (green) stalled at a G4 (red). (E) Quantification of replication forks stalled at G4s with 7.5 μM pyridostatin for 5 h prior to and through labeling. Significance was determined by one-way ANOVA analysis followed by Tukey's post-hoc test; n = 600 from three independent experiments.
To corroborate that replication fork progression into telomeres after G4 stabilization was specifically due to fork arrest at individual G4 structures, we performed a novel G4-specific DNA combing using a technique that we developed based on modifications of an existing protocol (36). As a control, native and denatured DNA fibers were stained with a G4-specific antibody (1H6) and puncta were observed only in the native, but not denatured, state indicating that the antibody was in fact detecting G4 secondary structures (Supplementary Figure S5D) (37). To detect G4 structures in combed DNA fibers, we utilized this G4 antibody and click-it chemistry and probed for replication tracts stalled at G4s (Figure 2D). Under G4-stabilized conditions, a small, but significant, increase in replication forks stalled at these structures in the absence of EXO1 was observed (Figure 2E). The above results indicate that replication through telomeric G4s is impaired under these conditions in EXO1-null cells.
Finally, to address whether the increase in replication forks accumulating at G4 structures in EXO1-null cells was possibly due to dysregulation in G4 processing factors such as RTEL1, we carried out PLAs using antibodies directed against G4s, RTEL1 [a known G4-interacting protein (21)] and EXO1. An interaction between RTEL1 and G4s was readily observed in the parental cell line and, importantly, the frequency of this interaction did not increase in the EXO1-null cells (Supplementary Figure S6A and B). Moreover, an interaction between RTEL1 and EXO1 proteins was observed in the parental HeLa cell line and completely abolished in the EXO1−/− cell line. These experiments suggest that EXO1 is, like RTEL1, a legitimate G4-interacting (and likely processing) factor. Moreover, and more importantly, they suggest that there is not a defect in RTEL1 helicase processing of G4s in the EXO1-null cells.
Resection occurs proximal to G4s as part of the repair/resolution process
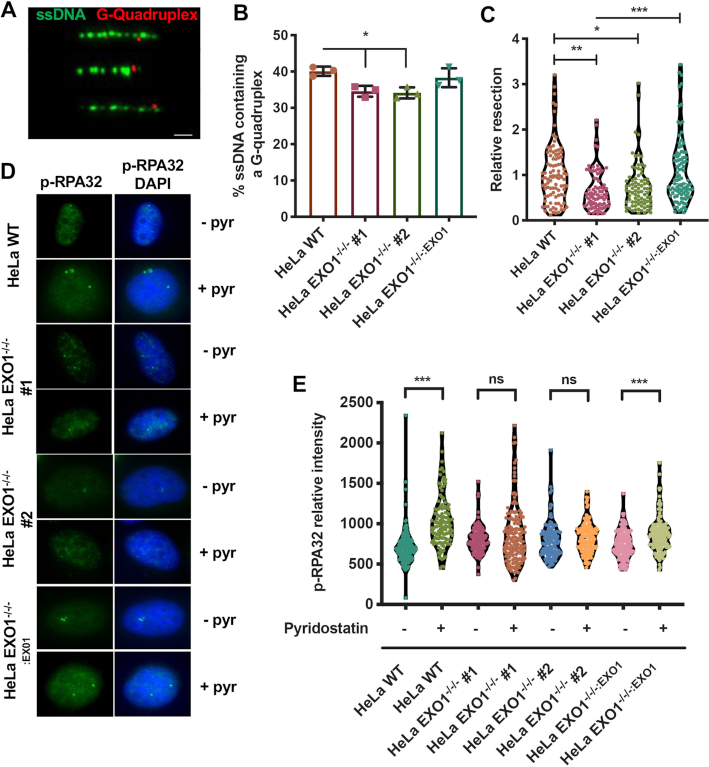
EXO1 is an exonuclease responsible for the 5′ → 3′ resection of DNA as well as cleavage of 5′-flap structures (23). Thus, we explored the possibility that EXO1’s role in G4 resolution was due to either resection of nascent DNA at stalled/collapsed replication tracts or cleavage of the quadruplex itself. To determine whether resection takes place around G4 structures, we performed a modified SMART assay (38) under G4-stabilized conditions. G4 structures were detected lying on the extremities of ssDNA tracts, indicative of resection taking place predominately on one side of the quadruplex (Figure 3A). To assess whether EXO1-deficient cells are defective in resection around these structures, we quantified the total percentage of ssDNA fibers that contained a G4 structure under G4-stabilized conditions. In the absence of EXO1, fewer ssDNA fibers contained a G4 (Figure 3B). Although the difference was modest, it was statistically significant. Further, we measured the length of the ssDNA tracts that contained G4s and observed that the magnitude of resection flanking G4s was negatively impacted by the absence of EXO1 (Figure 3C). We validated these findings with a second G4 antibody (BG4) and achieved similar results (Supplementary Figure S5E and F). Finally, to further confirm that in the absence of EXO1 there is a lack of resection globally, including at non-telomeric G4s, we probed for the presence of phosphorylated replication protein A (p-RPA32)—RPA32 phosphorylated at serines 4 and 8 is a marker of DNA resection (39)—during pyridostatin stabilization (Figure 3D). While EXO1-proficient cells exhibited increased levels of p-RPA32 in response to pyridostatin treatment, EXO1-deficient cells did not (Figure 3E). These data imply that resection around stabilized G4s is part of normal resolution and repair. In the absence of EXO1, resection is limited at these sites, presumably forcing the cell to engage in alternative pathway(s) to repair the compromised replication forks stalled at these secondary structures.
Figure 3.
Resection occurs proximal to G4s as part of the repair/restart process. (A) Representative images of SMART combing for G4s. ssDNA is displayed in green and G4s in red; scale bar = 1.5 kb. (B) Quantification of the percentage of ssDNA fibers from the SMART assay containing a G4 after 24 h of pyridostatin treatment. A paired t-test was used to determine statistical significance; n = 250. (C) Measurement of G4-containing ssDNA fibers as a readout of resection. Significance of relative resection was evaluated by one-way ANOVA analysis and a Tukey's post-hoc test. For EXO1 WT, EXO1−/−/− #1 and #2, and EXO1−/−/−:EXO1, n values are 94, 80, 83 and 95, respectively. (D) Representative images of p-RPA32 staining in HeLa WT, EXO1−/−/− #1 and #2, and EXO1−/−/−:EXO1 with (+pyr) and without (−pyr) pyridostatin treatment for 24 h. (E) Mean fluorescence quantification of (D). One-way ANOVA analysis followed by Tukey's post-hoc test was utilized to evaluate statistical significance; ns is not significant. For EXO1 WT −/+ pyridostatin, EXO1−/−/− #1 and #2 −/+ pyridostatin, and EXO1−/−/−:EXO1 −/+ pyridostatin, n values are 124, 170, 151, 162, 133, 133, 139 and 147, respectively.
G4 stabilization results in increased non-homologous end joining when EXO1 is absent
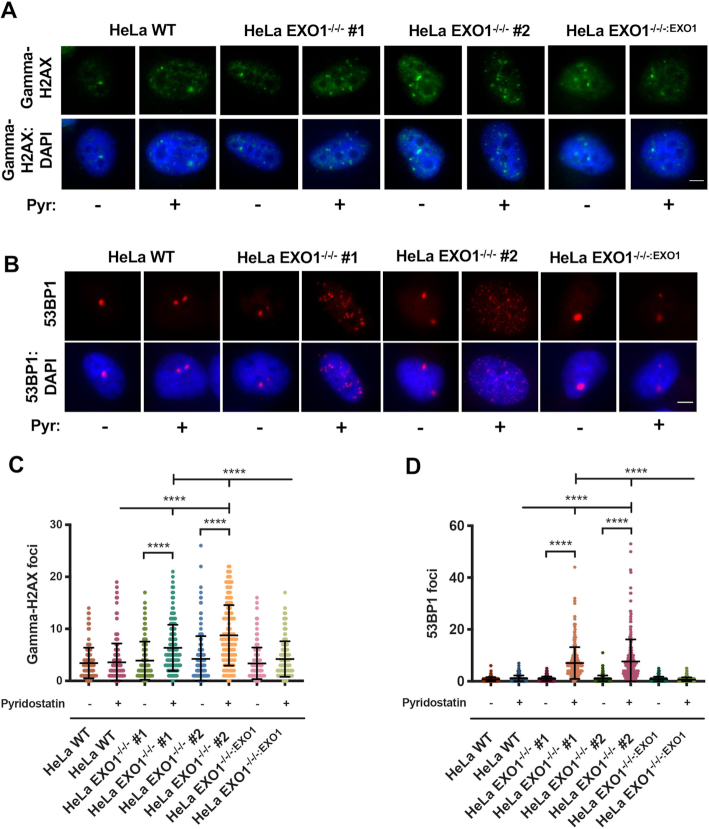
To determine the fate of prolonged fork stalling at G4s in the absence of EXO1, we first assayed for the level of DNA DSBs. Immunofluorescent staining of gamma-H2AX, a marker for DSBs (40), revealed no difference in the levels of DSBs in all backgrounds under untreated conditions (Figure 4A and C), the increase in TIFs described earlier (Figure 1B and C; Supplementary Figure S9A) notwithstanding. However, in the presence of pyridostatin, DSBs were significantly elevated in the EXO1-null, but not in the WT or complemented cell lines (Figure 4A and C). DNA DSBs can occur throughout the genome and are typically repaired by either error-free HDR or error-prone non-homologous end joining (NHEJ). Importantly, extensive resection at sites of DSBs is recognized as a commitment step to the HDR pathway (41). Thus, if resection is lacking or diminished as is the case in the EXO1-null cells (Figure 3), then error-prone NHEJ may be preferentially utilized. To probe for this possibility, we assessed the levels of 53BP1, a marker for the NHEJ pathway and a factor that is known to impede resection (42). In unchallenged conditions, there was no difference in 53BP1 foci between EXO1-proficient and -deficient backgrounds (Figure 4B and D; Supplementary Figure S9B). However, in the presence of pyridostatin there was a striking increase in the levels of 53BP1 foci in EXO1-null cells, whereas no change was observed in EXO1-proficient cells (Figure 4B and D). Further, this increase in 53BP1 was most pronounced in S-phase cells, implying 53BP1 deposition occurs in response to replicative stress or fork collapse in the absence of EXO1 (Supplementary Figure S7A and B). To assess the magnitude of the contribution of NHEJ in G4-induced break repair, we inhibited DNA-PKcs in EXO1-proficient and -deficient cells and assessed the levels of gamma-H2AX. DNA DSB accumulation was elevated 2-fold in the absence of functional NHEJ and EXO1 compared to EXO1 loss alone (Supplementary Figure S8A and B). This increase in damage was not mirrored by DNA-PKcs inhibition in the presence of functional EXO1 and NU7441 treatment alone had no effect on viability across EXO1-proficient and -deficient cell lines (Supplementary Figure S8C). Altogether, these data imply that EXO1-mediated fork resection is critical for the rescue of replication forks impeded by G4s. This fork rescue is presumably carried out by the HDR machinery. In the absence of EXO1, we propose that reduced resection at G4s instead leads to NHEJ-mediated repair of the collapsed forks.
Figure 4.
G4 stabilization results in increased DSBs repaired by NHEJ when EXO1 is absent. (A) Representative images of gamma-H2AX staining in HeLa WT, EXO1−/−/− #1 and #2, and EXO1−/−/−:EXO1 with and without 7.5 μM pyridostatin treatment for 24 h. Nuclei are DAPI stained; scale bar = 10 μm. (B) Representative images of 53BP1 staining in HeLa WT, EXO1−/−/− #1 and #2, and EXO1−/−/−:EXO1 with and without 7.5 μM pyridostatin treatment for 24 h. Nuclei are DAPI stained; scale bar = 10 μm. (C) Quantification of (A). Gamma-H2AX foci were scored for EXO1 WT −/+ pyridostatin, EXO1−/−/− #1 and #2 −/+ pyridostatin, and EXO1−/−/−:EXO1 −/+ pyridostatin; n values are 181, 303, 208, 217, 158, 154, 169 and 167; respectively. One-way ANOVA analysis followed by Tukey's post-hoc test was used for statistical analysis. (D) Quantification of (B). 53BP1 foci were scored for EXO1 WT −/+ pyridostatin, EXO1−/−/− #1 and #2 −/+ pyridostatin, and EXO1−/−/−:EXO1 −/+ pyridostatin; n values are 290, 220, 262, 225, 250, 221, 272 and 229; respectively. One-way ANOVA analysis followed by Tukey's post-hoc test was utilized to evaluate statistical significance.
G4 stabilization exacerbates telomere aberrations in the absence of EXO1
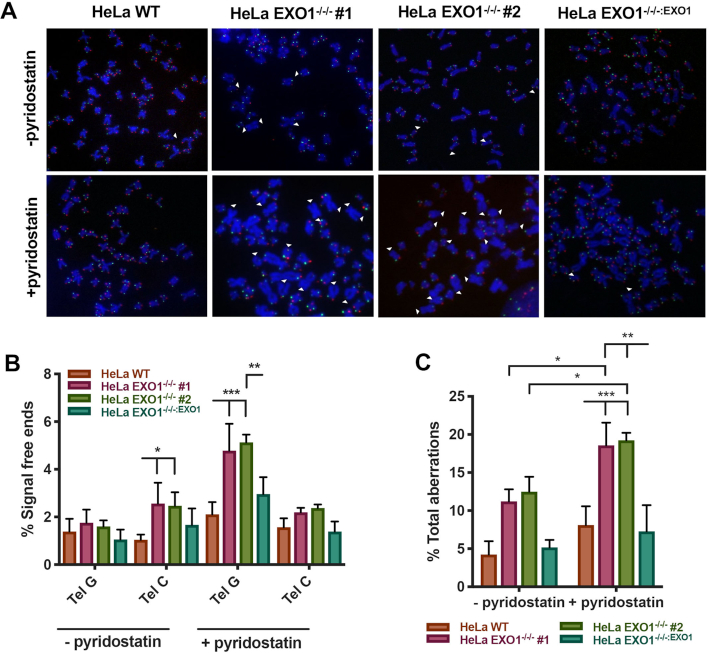
Impaired telomere replication events can, in many cases, lead to severe telomere dysfunction. Indeed, telomere fusion events, telomere breaks, and telomere loss have all been associated with faulty telomere replication (7,43). Thus, cells lacking EXO1 and challenged by a G4 stabilizer should accrue even more genomic instability at their chromosome ends than they normally possess (Figure 1). To rigorously assess telomere integrity, we performed chromosome orientation FISH (CO-FISH) analyses (44). In this protocol, the G-rich and the C-rich telomere strand of a metaphase chromosome are labeled with different colored probes such that strand-specific differences can be assessed. In unchallenged conditions, telomere dysfunction was elevated in the nuclease-deficient background, specifically on the C-rich template strand (‘Tel C’; Figure 5A and B). After treatment with pyridostatin, however, signal-free ends increased significantly in the EXO1-deficient cells and preferentially on the G-rich template strand (‘Tel G’; Figure 5A and B). These data imply that EXO1’s already established role at a telomere (G-overhang formation) may perhaps be distinct from its roles in G4 resolution, as evidenced by the stranded switch in telomere defects in the absence and presence of the G4 stabilizer. Additionally, pyridostatin treatment induced damage at telomeres as evidenced by TIFs in the absence of EXO1 (Supplementary Figure S10A and B). These data imply telomeres are indeed being lost to inappropriate repair events. Importantly, however, in all cases, an EXO1 deficiency synergized with G4 stabilization to increase total telomere abnormalities (inclusive of signal-free ends, telomere fusions, dysfunction and chromosome breaks) and thus unequivocally demonstrated that the absence of EXO1 leads to G4 stabilizer-induced genomic instability (Figure 5C).
Figure 5.
G4 stabilization exacerbates telomere aberrations in the absence of EXO1. (A) Representative images of CO-FISH analysis with and without 7.5 μM pyridostatin treatment for 12 h. Lagging telomeres are displayed green and leading telomeres displayed red. (B) Quantification of signal-free ends in HeLa WT, EXO1−/−/− #1 and #2, and EXO1−/−/−:EXO1 cells with and without pyridostatin treatment. Statistical significance was determined by one-way ANOVA analysis followed by Tukey's post-hoc test; n = 4500. (C) Quantification of all aberrations in HeLa WT, EXO1−/−/− #1 and #2, and EXO1−/−/−:EXO1 with and without pyridostatin. Aberrations include signal-free ends, sister chromatid fusions, and chromosome:chromosome fusions. One-way ANOVA analysis followed by Tukey's post-hoc test was used for statistical analysis; n = 4500.
EXO1 expression as a marker for cancer prognosis
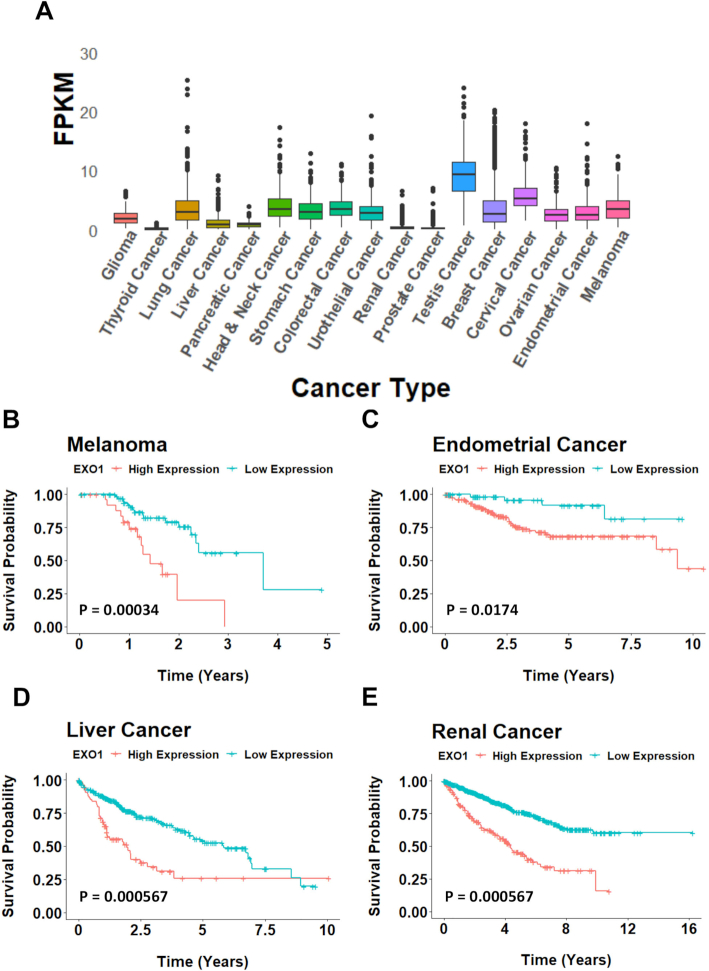
G4 stabilizers have recently been exploited as a potential cancer therapeutic in several DNA damage repair-deficient cancers (34). Here, we have established that an EXO1 deficiency leads to a hypersensitivity to G4 small molecule stabilizers. Together, these data suggest that the expression of EXO1 in a given cancer may serve as a marker to determine whether or not G4 stabilization would be an efficacious treatment option. In other words, if a patient's cancer expresses high levels of EXO1, they likely possess a robust backup pathway to resolve G4s when helicases fail. Alternatively, if a cancer expresses low levels of EXO1, it lacks a backup pathway for G4 resolution and will likely be sensitive to G4 stabilization. This treatment approach is, of course, dependent on the deregulation of EXO1 expression in a cancer cell. Thus, to assess the EXO1 expression profile across a variety of tissue-specific cancers we utilized existing TCGA datasets. EXO1 expression was variable in regard to an affected organ or tissue type being, for example, extremely abundant in testis cancer and virtually undetectable in renal or prostate cancers (Figure 6A). However, this diverse expression of EXO1 across cancer subsets also suggested that its expression may be linked to certain cancers' progression. To determine whether EXO1 expression correlated with patient prognosis, we analyzed 17 different cancer types and grouped patients by either high expression or low expression of EXO1. Among these 17 cancers, four types showed significant association between EXO1 expression and outcome (false discovery rate <0.05; after Benjamini–Hochberg multiple testing correction): melanoma, liver cancer, endometrial cancer and renal cancer (Figure 6B–E). In all four cases, high EXO1 expression was associated with poor survival outcomes. Intriguingly, renal cancer, where the average EXO1 expression across tumors is among the lowest cancer types (Figure 6A), also displayed the same marked association between high EXO1 expression levels and survival outcome (Figure 6E). Thus, G4 stabilization therapy coupled with EXO1 inhibition or endogenously low levels of EXO1 expression presents a putative cancer treatment option for cancers with diverse current standards of care. In toto, these data also suggest that low EXO1 expression in these cancers confers a survival advantage.
Figure 6.
EXO1 expression as a marker for cancer prognosis. (A) FPKM expression values across 16 cancer subtypes. (B–E) Kaplan–Meier survival curve of cancers sorted by high (orange) and low (blue) expression of EXO1. Melanoma: low expression, n = 74; high expression, n = 28; P = 2e−04. Endometrial cancer: low expression, n = 136; high expression, n = 405; P = 0.0041. Liver cancer: low expression, n = 289; high expression, n = 76; P < 0.0001. Renal cancer: low expression, n = 694; high expression, n = 183; P < 0.0001.
DISCUSSION
In this study, we document a bona fide role for EXO1 in safeguarding DNA replication forks through G4-rich regions. Specifically, we have demonstrated that telomeres, which are particularly G4-rich regions, are hypersensitive to G4-stabilizing compounds in the absence of EXO1. Without EXO1, elevated levels of replication fork stalling within the telomere occur in G4-stabilized conditions and this ultimately results in increased telomeric fusions and loss. Mechanistically, we also demonstrate that the replication forks stalled at G4 structures have decreased levels of resection proximal to these sites without EXO1. As a consequence, these forks are not properly restarted and they have the propensity to collapse, where the resulting DNA DSBs are repaired via error-prone NHEJ rather than the more error-free (but EXO1-dependent) HDR pathway. Thus, an EXO1 deficiency coupled with G4 stabilization leads to increased DSBs at G4-enriched sites and this, in turn, leads to elevated levels of genomic instability due to error-prone NHEJ-mediated repair (Figure 7).
Replicating through G4 structures becomes especially difficult under G4-stabilized conditions as these structures become harder to dismantle. It is generally accepted that helicases typically unwind G4s and that their failure to do so can lead to replication fork stalling (45). Intuitively, under G4-stabilizing conditions, helicases will likely have a higher propensity to fail to unwind at G4s, giving rise to the need for a parallel or backup mechanism to resolve the structure and restart replication. Intriguing, this hypothesis suggests that there may be natural conditions (i.e. in the absence of G4-stabilizing compounds), where G4s may be stabilized and/or resistant to helicase activity. What these conditions may be and/or whether there is a specialized subset of G4s that require a parallel/backup pathway are issues that warrant further investigations. Interestingly, however, in the absence of EXO1, there is not an increase in the helicase RTEL1 associated with the G4s (Supplementary Figure S6), suggesting perhaps that RTEL1 may be limiting for G4 resolution. Importantly, EXO1-mediated resection around these impediments is a critical step for the repair and rescue process when G4s fail to be unwound. EXO1 resection occurs in a 5′ → 3′ direction. Thus, EXO1-mediated resection at a stalled fork would presumably target nascent lagging-strand DNA as part of the fork recovery process and it would, perforce, leave the template G-rich strand single-stranded (Figure 7, left panel). Indeed, in assessing tracts of ssDNA in the genome, we observed that a large number contained a G4 structure, indicating that resection had exposed the G-rich template strand (Figure 3B). Further, we showed that these G4s lie on the ends of ssDNA tracts, consistent with resection being unidirectional on one side of the structure rather than on both sides, as is the case at DNA DSBs. Finally, our data corroborate a recent study (36) showing that resection can take place around these structures when breaks occur proximal to them, indicating that this is part of a general repair process and not necessarily telomere-specific.
Indeed, our data imply that EXO1 resection plays a dual role in safeguarding these replication forks. First, EXO1 resection may facilitate restart, circumventing fork collapse entirely, which in turn promotes genome stability (Figure 7, left panel). This function is best exemplified by EXO1-proficient cells, which do not display increased DNA damage and prolonged fork stalling in response to G4 stabilization. Second, if the replication fork does, e.g. after extensive stalling, collapse, then EXO1-mediated resection can act as a commitment step in HDR-mediated rescue (Figure 7, left panel). This interpretation is supported by the increase in 53BP1 foci (indicative of NHEJ-mediated repair) in EXO1-deficient cells after pyridostatin treatment, which was not mirrored in EXO1-proficient cells (Figure 4B and D). Thus, the absence of EXO1 results in a proverbial ‘double whammy’ for cells: it precludes the proper repair of stalled replication forks and it then funnels these same broken forks into an error-prone process (Figure 7, right panel). Finally, it should be noted that previous work has indicated that EXO1 plays a role in fork reversal (25,46). Our data do not exclude this possibility, which indeed may occur to some extent as a necessary prerequisite to fork restart (Figure 7, left panel). Moreover, other reports have identified the ability of another 5′ → 3′ nuclease, DNA2, to clip out G4 structures from dsDNA (29). Our data do not directly support a similar role for EXO1, but it remains to be seen whether or not the endonuclease ability of EXO1 is capable of such a function. Future in vitro studies and separation-of-function mutations will be necessary to confirm or refute this possibility although we note that DNA2 loss-of-function human cells are not viable (unpublished data) and thus the complete loss-of-function EXO1 and DNA2 mutants are not functionally equivalent.
Interestingly, EXO1-null cells displayed small, but significant, telomeric C-rich strand-specific defects as detected by CO-FISH in unchallenged conditions (Figure 5B). We speculate that this may be due to EXO1’s role in 3′ overhang generation. Thus, the leading C-rich template strand is usually replicated to the end of the chromosome and then the C-strand is resected by the combined action of the nucleases Apollo and EXO1 (24). Because of this, loss of EXO1 activity in the mouse is associated with shorter G-overhangs (24). We have observed an identical phenotype in our human HeLa (Supplementary Figure S2) and HCT116 (data not shown) EXO1-null cell lines. Thus, a parsimonious interpretation of these results is that, in unperturbed conditions, there is a higher incidence of aberrant events (signal-free ends and aberrant chromosomes) caused by the shorter G-overhangs on leading chromosome ends. While certainly non-trivial, we would like to emphasize that the G-rich strand defects under G4-stabilized conditions that we have described here for EXO1-null human cells dominate the cellular phenotype. This activity constitutes a new role for EXO1 at the telomere.
Recently, G4s have emerged as a promising therapeutic target for cancer treatment (47). It is well established that DNA repair pathway dysregulation is a major driver of cancer development and thus a possible therapeutic target as well (48). Proper resolution of G4s requires specialized replication and repair proteins that may be either mutated and/or variably expressed in specific cancers. Thus, stabilization of these structures for certain cancer profiles is being exploited as a favorable treatment strategy (34). EXO1 is commonly mutated in cancers, but it remains unclear whether or not these mutations yield a non-functional protein, result in gain-of-function or have no effect. Additionally, EXO1 is frequently overexpressed in a variety of cancer types (49). This upregulation of EXO1 may represent a cancer cell's method of overcoming difficult G4s when other pathways have become incapable of resolving these impediments. It follows that a cancer cell that expresses EXO1 at high levels may be particularly resistant to G4 stabilization as a form of therapy as it should be able to initiate a robust backup response to G4 helicase failure. Interestingly, high expression of EXO1 in many cancer backgrounds also correlates with poor prognosis in comparison to low EXO1 expression (Figure 6B–E). These data suggest two complementary avenues of therapeutic intervention. Thus, in cancer patients expressing low levels of EXO1 (who already have a relatively better prognosis; Figure 6B), survival might be even more enhanced by including treatments with G4-stabilizing compounds. In a reciprocal fashion, cancer patients expressing high levels of EXO1 (who currently have a relatively poor prognosis) might benefit from small molecule inhibitors of EXO1 (potentially in combination with G4-stabilizing compounds). Although, to our knowledge, there are currently no approved EXO1 inhibitors available, our data would predict that such compounds could be therapeutically useful.
The data we have presented here demonstrate that EXO1 contributes to telomeric G4 resolution, which is normally a hurdle for the replication and repair machinery. When a replication fork stalls at a G4, we postulate that EXO1 is recruited to that G4 (Supplementary Figure S6) and facilitates resection and ultimately repair or restart of the fork to ensure proper completion of replication and survival (Figure 7, left panel). In contrast, the absence of EXO1 equates with a decreased ability to resolve, repair and replicate through telomeric G4 structures leading to fork collapse and NHEJ-mediated repair (Figure 7, right panel). The aberrant NHEJ repair events, in turn, result in telomere loss, genomic instability and death as the loss of telomeric DNA can ultimately lead to apoptosis/senescence (50,51). Finally, EXO1 expression profiles in cancer may make it a favorable biomarker for the potential efficacy of small molecule G4 stabilizers as a therapeutic treatment.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the imaging center at the University of Minnesota for providing use of and assistance with the Typhoon FLA, Zeiss TIRF and Nikon TiE instrumentation. This publication was made possible by an NIA-funded pre-doctoral fellowship to S.S. (T32 AG029796).
Author contributions: S.S. and E.A.H. conceived and designed the overall project. S.S. and K.K. generated the cell lines used, and performed CO/T-FISH and immunofluorescence assays. S.S. performed all DNA combing experiments, western blots and toxicity analyses. S.K. performed image analysis and quantification. K.L. generated the cancer expression and survival data under the guidance of A.-K.B. and C.L.M. E.A.H. and S.S. prepared and edited the manuscript, which was critiqued by A.-K.B. and C.L.M.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences [GM088351, in part, to E.A.H.)]; National Cancer Institute [CA190492, in part, to E.A.H.]; NIH [GM074917 to A.-K.B.]; NSF [MCB 1818293 to A.-K.B.]. Funding for open access charge: National Cancer Institute [CA190492].
Conflict of interest statement. E.A.H sits on the Scientific Advisory Boards of Horizon Discovery, Ltd and Intellia Therapeutics.
REFERENCES
- 1. de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harb. Symp. Quant. Biol. 2010; 75:167–177. [DOI] [PubMed] [Google Scholar]
- 2. O'Sullivan R.J., Karlseder J.. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010; 11:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shay J.W., Wright W.E.. Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 2019; 20:299–309. [DOI] [PubMed] [Google Scholar]
- 4. Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T.. Mammalian telomeres end in a large duplex loop. Cell. 1999; 97:503–514. [DOI] [PubMed] [Google Scholar]
- 5. Van Ly D., Low R.R.J., Frolich S., Bartolec T.K., Kafer G.R., Pickett H.A., Gaus K., Cesare A.J.. Telomere loop dynamics in chromosome end protection. Mol. Cell. 2018; 71:510–525. [DOI] [PubMed] [Google Scholar]
- 6. Maestroni L., Matmati S., Coulon S.. Solving the telomere replication problem. Genes. 2017; 8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mason-Osann E., Gali H., Flynn R.L.. Resolving roadblocks to telomere replication. Methods Mol. Biol. 2019; 1999:31–57. [DOI] [PubMed] [Google Scholar]
- 8. Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T.. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009; 138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ozer O., Hickson I.D.. Pathways for maintenance of telomeres and common fragile sites during DNA replication stress. Open Biol. 2018; 8:180018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Valton A.L., Prioleau M.N.. G-quadruplexes in DNA replication: a problem or a necessity?. Trends Genet. 2016; 32:697–706. [DOI] [PubMed] [Google Scholar]
- 12. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 14. Valton A.L., Hassan-Zadeh V., Lema I., Boggetto N., Alberti P., Saintome C., Riou J.F., Prioleau M.N.. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014; 33:732–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De S., Michor F.. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat. Struct. Mol. Biol. 2011; 18:950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chavez A., Tsou A.M., Johnson F.B.. Telomeres do the (un)twist: helicase actions at chromosome termini. Biochim. Biophys. Acta. 2009; 1792:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pasero P., Vindigni A.. Nucleases acting at stalled forks: how to reboot the replication program with a few shortcuts. Annu. Rev. Genet. 2017; 51:477–499. [DOI] [PubMed] [Google Scholar]
- 19. Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J.. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004; 306:1951–1953. [DOI] [PubMed] [Google Scholar]
- 20. Sarek G., Kotsantis P., Ruis P., Van Ly D., Margalef P., Borel V., Zheng X.F., Flynn H.R., Snijders A.P., Chowdhury D. et al.. CDK phosphorylation of TRF2 controls t-loop dynamics during the cell cycle. Nature. 2019; 575:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vannier J.B., Pavicic-Kaltenbrunner V., Petalcorin M.I., Ding H., Boulton S.J.. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012; 149:795–806. [DOI] [PubMed] [Google Scholar]
- 22. Jacob N.K., Kirk K.E., Price C.M.. Generation of telomeric G strand overhangs involves both G and C strand cleavage. Mol. Cell. 2003; 11:1021–1032. [DOI] [PubMed] [Google Scholar]
- 23. Keijzers G., Liu D., Rasmussen L.J.. Exonuclease 1 and its versatile roles in DNA repair. Crit. Rev. Biochem. Mol. Biol. 2016; 51:440–451. [DOI] [PubMed] [Google Scholar]
- 24. Wu P., Takai H., de Lange T.. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012; 150:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cotta-Ramusino C., Fachinetti D., Lucca C., Doksani Y., Lopes M., Sogo J., Foiani M.. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell. 2005; 17:153–159. [DOI] [PubMed] [Google Scholar]
- 26. Karanja K.K., Cox S.W., Duxin J.P., Stewart S.A., Campbell J.L.. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle. 2012; 11:3983–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J.L., Kowalczykowski S.C.. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010; 467:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nanbu T., Nguyễn L.C., Habib A.G.K., Hirata N., Ukimori S., Tanaka D., Masuda K., Takahashi K., Yukawa M., Tsuchiya E. et al.. Fission yeast Exo1 and Rqh1-Dna2 redundantly contribute to resection of uncapped telomeres. PLoS One. 2015; 10:e0140456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin W., Sampathi S., Dai H., Liu C., Zhou M., Hu J., Huang Q., Campbell J., Shin-Ya K., Zheng L. et al.. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 2013; 32:1425–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Masuda-Sasa T., Polaczek P., Peng X.P., Chen L., Campbell J.L.. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J. Biol. Chem. 2008; 283:24359–24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salic A., Mitchison T.J.. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sternberg S.H., Doudna J.A.. Expanding the biologist's toolkit with CRISPR–Cas9. Mol. Cell. 2015; 58:568–574. [DOI] [PubMed] [Google Scholar]
- 33. Myung K., Ghosh G., Fattah F.J., Li G., Kim H., Dutia A., Pak E., Smith S., Hendrickson E.A.. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol. Cell. Biol. 2004; 24:5050–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O’Neil N.J., Santos N.D., Silvester J., Wei V., Garcia J. et al.. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017; 8:14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Margalef P., Kotsantis P., Borel V., Bellelli R., Panier S., Boulton S.J.. Stabilization of reversed replication forks by telomerase drives telomere catastrophe. Cell. 2018; 172:439–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jimeno S., Camarillo R., Mejias-Navarro F., Fernandez-Avila M.J., Soria-Bretones I., Prados-Carvajal R., Huertas P.. The helicase PIF1 facilitates resection over sequences prone to forming G4 structures. Cell Rep. 2018; 25:3543. [DOI] [PubMed] [Google Scholar]
- 37. Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M. Jr, Sen D., Lansdorp P.M.. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014; 42:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huertas P., Cruz-Garcia A.. Single molecule analysis of resection tracks. Methods Mol. Biol. 2018; 1672:147–154. [DOI] [PubMed] [Google Scholar]
- 39. Raderschall E., Golub E.I., Haaf T.. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kinner A., Wu W., Staudt C., Iliakis G.. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008; 36:5678–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heyer W.D., Ehmsen K.T., Liu J.. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010; 44:113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zimmermann M., de Lange T.. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014; 24:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martínez P., Blasco M.A.. Replicating through telomeres: a means to an end. Trends Biochem. Sci. 2015; 40:504–515. [DOI] [PubMed] [Google Scholar]
- 44. Bailey S.M., Cornforth M.N., Kurimasa A., Chen D.J., Goodwin E.H.. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001; 293:2462–2465. [DOI] [PubMed] [Google Scholar]
- 45. Mendoza O., Bourdoncle A., Boule J.B., Brosh R.M. Jr, Mergny J.L.. G-quadruplexes and helicases. Nucleic Acids Res. 2016; 44:1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lemacon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N. et al.. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017; 8:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miller K.M., Rodriguez R.. G-quadruplexes: selective DNA targeting for cancer therapeutics?. Expert Rev. Clin. Pharmacol. 2011; 4:139–142. [DOI] [PubMed] [Google Scholar]
- 48. Curtin N.J. DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer. 2012; 12:801–817. [DOI] [PubMed] [Google Scholar]
- 49. Zhang M., Zhao D., Yan C., Zhang L., Liang C.. Associations between nine polymorphisms in EXO1 and cancer susceptibility: a systematic review and meta-analysis of 39 case-control studies. Sci. Rep. 2016; 6:29270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ramirez R., Carracedo J., Jimenez R., Canela A., Herrera E., Aljama P., Blasco M.A.. Massive telomere loss is an early event of DNA damage-induced apoptosis. J. Biol. Chem. 2003; 278:836–842. [DOI] [PubMed] [Google Scholar]
- 51. Bernadotte A., Mikhelson V.M., Spivak I.M.. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging. 2016; 8:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.