Abstract
Box C/D RNA protein complexes (RNPs) catalyze site-specific 2′-O-methylation of RNA with specificity determined by guide RNAs. In eukaryotic C/D RNP, the paralogous Nop58 and Nop56 proteins specifically associate with terminal C/D and internal C'/D' motifs of guide RNAs, respectively. We have reconstituted active C/D RNPs with recombinant proteins of the thermophilic yeast Chaetomium thermophilum. Nop58 and Nop56 could not distinguish between the two C/D motifs in the reconstituted enzyme, suggesting that the assembly specificity is imposed by trans-acting factors in vivo. The two C/D motifs are functionally independent and halfmer C/D RNAs can also guide site-specific methylation. Extensive pairing between C/D RNA and substrate is inhibitory to modification for both yeast and archaeal C/D RNPs. N6-methylated adenine at box D/D' interferes with the function of the coupled guide. Our data show that all C/D RNPs share the same functional organization and mechanism of action and provide insight into the assembly specificity of eukaryotic C/D RNPs.
INTRODUCTION
Box C/D RNAs constitute a large group of non-coding RNAs in eukaryotes and archaea. They assemble with multiple proteins into RNA–protein complexes (RNPs) that catalyze site-specific 2′-O-methylation of ribosomal RNAs, snRNAs and tRNAs in an RNA-guided manner and also assist assembly of eukaryotic ribosomes (1–5). The majority of eukaryotic C/D RNAs are localized in the nucleolus, hence known as small nucleolar RNA (snoRNA). Other functions have also been documented for C/D snoRNAs, such as regulation of mRNA splicing and processing and maintaining chromatin structure (6–8).
Box C/D RNAs are characterized by a bipartite, pseudo-symmetric structure that contains box C (RUGAUGA) and D (CUGA) at the terminus, the related box C' and D' at the internal region and two spacers. The spacer can pair with complementary substrates, selecting the substrate's fifth nucleotide upstream of box D'/D for modification (9,10). Box C and D combine into a kink-turn structure comprised of a non-canonical stem with tandem sheared GA pairs, a 3-nt bulge and a canonical Watson-Crick paired stem (11–13). The internal C' and D' motif can form a kink-loop structure without a canonical stem (14).
Archaeal C/D RNPs have been reconstituted in vitro and extensively analyzed biochemically and structurally (15–22). Archaeal C/D RNAs associate with three proteins: Nop5, fibrillarin (Fib) and L7Ae. Nop5, which consists of an N-terminal domain (NTD), a coiled-coil domain and a C-terminal domain (CTD), forms the scaffold for complex assembly. The coiled-coil domain mediates dimerization of Nop5. The NTD associates with the methyltransferase fibrillarin to form a flexible catalytic module. The CTD of Nop5 and L7Ae function as the RNA binding module and sandwich the k-turn motif formed by box C and D. The structure of an entire substrate-bound C/D RNP illustrates the organization of a monomeric RNP (mono-RNP), where box C/D and C'/D' of a C/D RNA are anchored on opposite sides of the Nop5 dimer (16). The guide-substrate duplex is capped by two CTDs of Nop5, which limits the length of paired substrate to a maximum of 10 nucleotides (nt) during modification (20).
In vitro reconstituted archaeal C/D RNPs can also adopt a dimeric state (di-RNP). The C/D RNA is proposed to be exchanged between two protein complexes in the di-RNP, rather than associating with the same protein complex as in the mono-RNP (23). However, this model has not gained direct structural support (24,25) and is incompatible with recognition of substrate length (20). The recently determined cryo-EM structures of 90S pre-ribosome clearly show that eukaryotic U3 C/D snoRNP is a mono-RNP (26–30).
Eukaryotic C/D RNPs are more complex and asymmetric in structure than archaeal complexes. Archaeal C/D RNAs have a compact size of ∼60 nt, conserved box C' and D' and spacers restricted to 12 nt long (31,32). By contrast, eukaryotic C/D RNAs have a longer and more diverse size of 60–300 nt, often imperfect box C' and D' motifs and longer spacers that can potentially form 10 to 20 base pairs (bp) with substrates (9,33,34). Eukaryotic C/D RNAs are associated with four core proteins: Nop56, Nop58, fibrillarin (Nop1 in Saccharomyces cerevisiae) and Snu13 (15.5K or NHP2L1 in humans). Nop58 and Nop56 are paralogs of archaeal Nop5 and specifically bind the terminal C/D and internal C'/D' motifs of guide RNAs, respectively (35,36).
A functional eukaryotic C/D RNP has not been reconstituted using recombinant proteins, which greatly limits the study of its structure and mechanism of action. Biogenesis of eukaryotic C/D RNP is a complicated, multiple-step process that requires a number of trans-acting factors (37). It is unknown whether assembly factors are required for in vitro reconstitution of eukaryotic C/D RNP.
In this study, we have reconstituted eukaryotic C/D RNPs from the thermophilic yeast Chaetomium thermophilum (Ct). The reconstituted Ct RNP is active in site-specific methylation but lacks the assembly specificity present in endogenous complexes. We analyzed the structure-function relationship of the eukaryotic Ct and archaeal Sulfolobus solfataricus (Ss) C/D RNPs in parallel and found that they share the same functional organization and mechanism of action. We have consistently found that the bipartite structure of the C/D RNA is not required for function and that the halfmer C/D RNAs can guide site-specific modification.
MATERIALS AND METHODS
Gene cloning
Gene cloning was conducted with the In-Fusion (Kakara), QuikChange (Stratagene) and Transfer-PCR (38) approaches. The full-length genes of Nop58 (CTHT_0006520, 582 residues), Nop56 (CTHT_0018510, 523 residues), Fib (CTHT_0067980, 313 residues) and Snu13 (CTHT_0066810, 127 residues) were PCR-amplified from the Ct genomic DNA, cloned into the Multiple Cloning Site (MCS) 1 of plasmid pETDuet-1 (Novagen) and fused to an N-terminal His6-Smt3 protein. The introns (two in Nop58, one in Nop56, eight in Fib, three in Snu13) of each gene were removed with QuikChange. For plasmids encoding Nop56 and Nop58, the His6-tag was removed and the intron-free His6-Smt3-Fib gene was inserted at MCS 2. Smt3-Nop56 was also cloned into plasmid pET28a. All four proteins were fused to an N-terminal Smt3 protein. Snu13 and Fib additionally contain a His6-tag at the N terminus. To create helix 9′ deletion mutants, residues 303–317 of Ss Nop5, residues 327–341 of Ct Nop58 and residues 344–358 of Ct Nop56 were removed with QuikChange.
Protein expression and purification
For co-expression of Nop58, Nop56 and Fib, the pETDuet-1 plasmid encoding Smt3-Nop58 and His6-Smt3-Fib and the pET28a plasmid encoding Smt3-Nop56 were co-transformed into Escherichia coli BL21(DE3) strain and selected with antibiotics kanamycin and ampicillin. The transformed cells were grown in LB medium at 37°C to OD600 = 0.8–1.0. The culture was cooled to 18°C and then induced for protein expression with 0.2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 16 h. Harvested cells were resuspended in buffer T300 (300 mM NaCl and 50 mM Tris–Cl, pH 8.0) supplemented with 1 mM PMSF and 30 mM imidazole and lysed in a high pressure JN-3000 PLUS cell disruptor (JNBIO). After the lysate was clarified by centrifugation at 39 000 g for 1 h at 4°C, the supernatant was filtered through a 0.45 μm membrane (Merck Millipore) and loaded onto a HisTrap column (GE Healthcare). The column was washed with 50 mM imidazole in T300 and eluted with 200 mM imidazole in T300. The three eluted proteins were digested with Ulp1 protease for 2 h at 4°C to remove the Smt3 tags. The sample was loaded onto a heparin column (GE Healthcare). The three proteins were eluted at 1 M NaCl in a 0.3 to 1 M NaCl linear gradient in 50 mM Tris–HCl (pH 8.0), concentrated and stored at −80°C.
The Nop58/Fib or Nop56/Fib complex was co-expressed from respective pETDuet-1 plasmids. The His6-Smt3-Snu13 fusion protein was individually expressed. These proteins were expressed, purified and removed of Smt3 in a similar way as described above. Snu13 was eluted at about 0.5 M NaCl from the heparin column. The Ss Nop5/Fib complex and L7Ae were purified as previously described (18).
In vitro transcription of RNA
The Ct snR61 gene was amplified from the Ct genomic DNA and cloned downstream of the T7 promoter in plasmid pBCSK. The sR1c gene was derived from sR1, which was chemically synthesized, and cloned in plasmid pUC19. Mutations were introduced with QuikChange and confirmed by DNA sequencing. The templates for in vitro transcription were PCR-amplified from the plasmids. For C/D-like snoRNAs, DNA templates including a T7 promoter were chemically synthesized. RNAs were in vitro transcribed according to standard protocol and purified by 8 M urea denaturing polyacrylamide gel electrophoresis (PAGE).
Chemical synthesis of RNA
Unmodified and pre-methylated substrate RNAs were purchased from Takara. The C/D RNAs for m6A experiments were chemically synthesized on a 394 DNA/RNA synthesizer (Applied Biosystems) Ribonucleotide phosphoramidites with 2′O-tert-butyldimethyl-silyl (t-BDMS) protection were purchased from Link Technologies and t-BDMS-protected N6-methyl-A-CE phosphoramidite was purchased from Glen Research. RNA was deprotected in a 25% ethanol/ammonia solution at room temperature for 3 h and evaporated to dryness. To remove t-BDMS groups, all oligoribonucleotides were re-dissolved in 100 μl of anhydrous DMSO and 125 μl of triethylamine trihydrofluoride (Aldrich) and agitated at 65°C in dark for 2.5 h. After cooling on ice for 10 min, the RNA was precipitated with 1 ml of butanol, washed twice with 70% ethanol and suspended in double-distilled water. RNA was further purified by 7% urea PAGE.
Ligation of full length U46
Full length U46 RNA was chemically synthesized in two pieces: a 5′ fragment of nt 1–54 and a 3′ fragment of nt 55–98, which were then joined by splint DNA-assisted ligation. The 3′ fragment contains either an unmodified or N6-methylated adenine in box D. The 3′ fragment was 5′ phosphorylated by T4 polynucleotide kinase (NEB) and purified with phenol extraction and ethanol precipitation. The two fragments of 800 pmol each were annealed with 1000 pmol of splint DNA (5′- TTGCATTGGCAAGTGGCACACAGGTGACCAAGACGGCCAC-3′) in 30 μl of water by heating at 95°C for 5 min and cooling down to room temperature. The ligation reactions contained 50 units of T4 RNA ligase 2 (NEB), T4 RNA ligase buffer, 1 mM ATP and 50 units of RNase inhibitor (Invitrogen) in a 50 μl volume and were incubated for 2 h at 25°C and for 12 h at 37°C. The ligation products were purified by 8% urea PAGE.
Activity assay
Protein concentrations were quantified by measuring absorbance at 280 nm and using theoretical absorbance coefficients of proteins, assuming ideal molecular ratios of 1:1:2 for the co-purified Nop56/Nop58/Fib complex, 1:1 for the Nop56/Fib complex and 1:1 for the Nop58/Fib complex. Due to impurities and unequal protein stoichiometry, protein amounts were only roughly estimated.
A 10-μl reaction containing 20 mM HEPES-Na, pH 8.0, 150 mM NaCl and 1.5 mM MgCl2 was prepared on ice in PCR tubes. The Ss RNP was assembled from 1 μM guide RNA, 3 μM L7Ae, 2μM Nop5-Fib complex and 30 μM substrate RNA. The Ct RNP was assembled from 2 μM guide RNA, 6 μM Snu13, 2 μM of the Nop58/Nop56/Fib ternary complex or 4 μM of the Nop56/Fib or Nop58/Fib binary complexes and 30 μM substrate RNA. After incubation for 10 min at 20°C, the modification was initiated by addition of 30 μM unlabeled S-adenosylmethionine (SAM) and 0.25 μCi [methyl-3H] SAM (83 Ci mmol−1; PerkinElmer). After incubation for 20 min at 70°C for Ss RNP and at 50°C for Ct RNP, each reaction was mixed with an equal volume of RNA loading buffer that contains 95% (v/v) deionized formamide, 0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol FF and 5 mM EDTA, pH 8.0. RNAs were heated at 95°C for 2 min and separated on a 10% (w/v) polyacrylamide gel containing 8 M urea and TBE. The gel was fixed in 10% (v/v) acetic acid, 25% (v/v) isopropanol and 65% (v/v) water for 30 min and then soaked in Amplify solution (GE Healthcare) with agitation for 15–30 min. The gel was dried using a Model 583 Gel Dryer (Bio-Rad) and exposed to X-ray films for 3–4 days at −80°C.
EMSA
RNA was assembled with proteins in 10 μl of buffer containing 25 mM HEPES-K, pH 7.6, 300 mM NaCl, 0.01% NP-40, 10% glycerol and 2 mM MgCl2. The sample was mixed with 1 μl of loading buffer containing 50% glycerol, 0.25% (w/v) bromophenol blue and 0.25% (w/v) xylene cyanol FF, and separated on 5% native polyacrylamide gels in Tris-glycine, pH 8.3 buffer. RNA was stained with SYBR GOLD (Invitrogen) and visualized with a GelDoc XR+ imaging system.
RESULTS
Reconstitution of a functional Ct C/D RNP
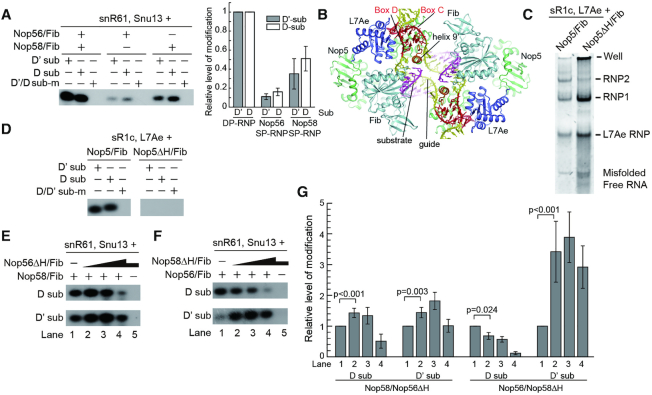
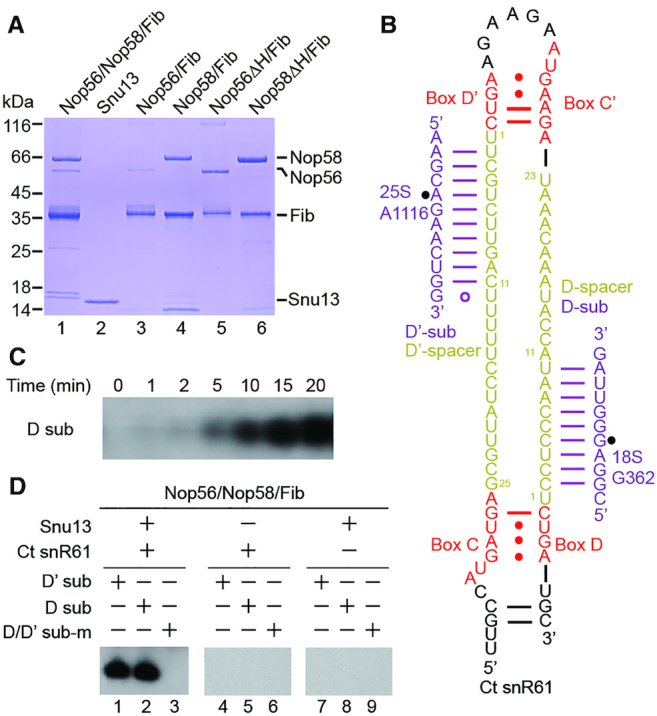
Our initial attempts to reconstitute eukaryotic C/D RNP with recombinant Saccharomyces cerevisiae (Sc) proteins and in vitro transcribed C/D RNA yielded no functional enzyme. Efforts to assemble Sc C/D RNP in bacteria were not successful either (39). Therefore we turned to proteins derived from the thermophilic yeast C. thermophilum that frequently display good stability and solubility in vitro (40). To enhance protein solubility, each of the four C/D RNP proteins was fused to a Smt3 protein (Sumo protein in S. cerevisiae) at the N-terminus. Nop56, Nop58 or both proteins were co-expressed with Fib in E. coli. The ternary complex of Nop56/Nop58/Fib and the binary complexes of Nop56/Fib and Nop58/Fib could be co-purified via a single six-His tag attached to Fib (Figure 1A). However, these proteins were not stoichiometric in the co-purified complexes, with free Fib protein often in excess and Nop56 in low abundance. The Snu13 protein was expressed and purified separately. The Ct genome encodes an snR61 snoRNA that is predicted to target A1116 of 25S rRNA and G362 of 18S rRNA with its D' and D spacer, respectively (Figure 1B).
Figure 1.
Reconstitution of Ct C/D RNP. (A) An SDS-PAGE gel showing co-purified Ct C/D RNP proteins. Positions of molecular markers are labeled. Nop56ΔH and Nop58ΔH refer to helix 9′ deletion mutants. (B) Secondary structure of Ct snR61 snoRNA. Boxes C, D, C' and D' are colored in red and spacers in yellow. Two predicted substrates (sub) are shown in purple and the modification sites are marked by circles. (C) Time course of modification of the D substrate. (D) 2′-O-methylation activity of Ct C/D RNP. The enzyme was assembled from snR61 RNA (2 μM), co-purified Nop56, Nop58 and Fib proteins (∼2 μM) and Snu13 (6 μM), and incubated with the substrates (30 μM) complementary to the D or D' spacer or mixed pre-methylated substrates (sub-m) in the presence of 30 μM cold SAM and trace amount of 3H-labeled SAM for 20 min at 50°C. Modified RNAs were resolved in denaturing PAGE and visualized by 3H autoradiography.
The Ct C/D RNP was reconstituted from Snu13, co-purified Nop56, Nop58 and Fib proteins and an in vitro transcribed Ct snR61 snoRNA. To measure modification activity, the assembled C/D RNP was incubated with 15-fold excess of cognate substrates and 3H-labeled S-adenosylmethionine (SAM) at 50°C, an optimal growth temperature for C. thermophilum. The modification reaction was resolved in denaturing gels and the 3H-methylated substrate RNA was detected by autoradiography. Based on the time course of the D substrate modification, a reaction time of 20 min was set for all subsequent experiments (Figure 1C). We found that both substrates complementary to the D and D' spacers were efficiently modified (Figure 1D). The modification was site-specific since the substrates pre-methylated at the target sites could not be modified anymore. Furthermore, the modification was dependent on Snu13 and the guide RNA (Figure 1D). We conclude that the reconstituted Ct C/D RNP is a functional RNA-guided modification enzyme.
Both Nop56 and Nop58 are required for optimal activity
In eukaryotic C/D RNPs, Nop58 and Nop56 form a heterodimer and specifically bind the terminal C/D and internal C'/D' RNA motifs, respectively. To examine if both Nop56 and Nop58 are needed for activity, the individual Nop56/Fib and Nop58/Fib binary complexes were purified and assembled with Snu13 and snR61 RNA (Figure 2A). Interestingly, the RNPs containing a single paralog (SP) of Nop56 or Nop58 were also active in site-specifically modifying both D and D' substrates (Figure 2A). Nevertheless, the SP-RNPs appear to be less active than the RNPs assembled with dual paralogs (DP), suggesting that both Nop56 and Nop58 are required for optimal activity of the enzyme.
Figure 2.
Both Nop56 and Nop58 are required for optimal activity of C/D RNP. (A) Activities of Ct RNPs containing single or dual paralogs of Nop56 and Nop58. RNPs were assembled from snR61 RNA (2 μM), Snu13 (6 μM) and either one of Nop56/Fib and Nop58/Fib binary complexes (4 μM) or both complexes (2 μM each). Other reaction conditions were the same as Figure 1C. The modification catalyzed by SP-RNPs was normalized against those catalyzed by DP-RNP. The mean and standard deviation determined from three experiments are drawn on the left panel. (B) Structure of substrate-bound Ss C/D RNP. Nop5 (green), Fib (light cyan) and L7Ae (blue) are represented as ribbons, the guide RNA (yellow and red) and substrate RNA (purple) are shown as sticks. Helix 9′ deleted in Nop5ΔH mutant is colored in orange. (C) Gel shift assay of Ss C/D RNP. The RNP was assembled from sR1c RNA, L7Ae and Nop5/Fib or Nop5ΔH/Fib and resolved on a native gel. RNA was stained by SYBR GOLD. RNP1 and RNP2 refer to monomeric and dimeric species of C/D RNP. (D) Deletion of helix 9′ of Nop5 inactivates Ss C/D RNP. The Ss RNP was assembled from sR1c RNA (1 μM), L7Ae (3 μM) and Nop5/Fib or Nop5ΔH/Fib (2 μM). Substrates (30 μM) were modified for 20 min at 70°C. (E) Titration of Nop56ΔH mutant increases modification activity of Nop58. Nop56Δ/Fib concentration is at 0, 0.5, 2, 8 and 2 μM in lanes 1–5. Nop58/Fib concentration is at 2 μM in lanes 1–4 and zero in lane 5. (F) Titration of Nop58ΔH mutant increases modification activity of Nop56. The reactions are the same as (E) other than swapping the wild-type and mutant proteins. (G) Quantification of titration experiments in (E, F). The level of modified substrate was normalized to that in lane 1. The mean and standard deviation were determined from four experiments. P-values between lanes 1 and 2 were calculated by the randomized block ANOVA method on the raw data.
To further study the synergistic interaction between Nop56 and Nop58, we sought a mutation of the proteins that abolishes the modification activity, but does not interfere with the assembly, of C/D RNP. The α-helix 9′ in the CTD of archaeal Nop5 contacts the guide-substrate duplex (16), is highly conserved (Supplementary Figure S1) and appears to be important for substrate placement (Figure 2B). As helix 9′ is disordered in the RNA-free Nop5 structure (18,19), its deletion may minimally impact the folding of Nop5. Indeed, deletion of helix 9′ and its linker to the CTD in Nop5 (Nop5ΔH) did not prevent the assembly of archaeal Ss C/D RNP (Figure 2C), but completely abolished the modification activity (Figure 2D). Deletion of the equivalent region in Nop56 and Nop58 (Nop56ΔH and Nop58ΔH) also eliminated the activity of respective Ct SP-RNPs (Figure 2E, F, lane 5). These results indicate that helix 9′ is essential for the modification function of Nop5 family proteins.
Remarkably, titration of the inactive Nop56ΔH mutant into the Nop58 SP-RNP increased modification of both the D and D' substrates (Figure 2E, lane 2, 2G). This suggests that Nop56ΔH forms a heterodimer with Nop58 and the resulting DP-RNP is more active than the Nop58 SP-RNP. Reverse titration of the inactive Nop58ΔH mutant into the Nop56 SP-RNP similarly enhanced modification of the D' substrate, although decreased modification of the D substrate (Figure 2F, lane 2, 2G). As the Nop56 and Nop58 mutants would sequester the guide RNA into inactive SP-RNPs, they also compete with the assembly of active RNPs and inhibit the modification. The inhibitory effect was mostly clear when the mutant proteins were added in excess (Figure 2E, F, lanes 4). During the titration of Nop58ΔH into Nop56 SP-RNP, modification of the D substrate was surprisingly reduced from the beginning (Figure 2F, lane 2). The protein level of Nop58ΔH was actually much higher than that of Nop56 (Figure 1A, lanes 3 and 6), which would lead to an early onset of inhibition. Moreover, the D substrate appeared to be more sensitive to the inhibition than the D' substrate (Figure 2E, lane 4). These data indicate that both Nop56 and Nop58 are required for optimal activity of C/D RNP.
Ct C/D RNPs recognize a restricted length of substrate
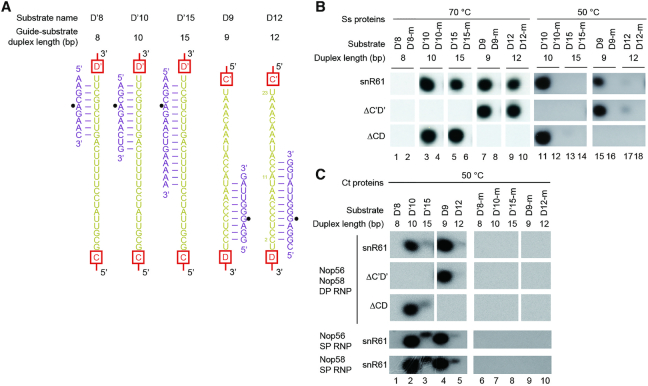
Using two-piece guide RNAs we have previously shown that archaeal Ss C/D RNPs recognize a maximum of 10 nt of substrate during modification (20). Eukaryotic and archaeal C/D RNAs differ significantly in spacer length. The spacer is constrained to ∼12 nt in archaeal C/D RNA (31,32), whereas in eukaryotes it is much longer where it can potentially form 10–20 bp with substrates (9). To examine whether eukaryotic Ct RNPs also recognize substrate length, several substrates were designed to form 8 to15 bp with the D' or D spacers of snR61 (Figure 3A).
Figure 3.
Yeast C/D RNPs recognize a restricted length of substrate. (A) Sequences of analyzed substrates (purple) and their base pairing interactions with the D' or D spacer of Ct snR61 (yellow). Box motifs are shown schematically. Modification sites are indicated by circles. The D'8, D'10 and D'15 substrates form 8, 10 and 15 bp with the D' spacer, respectively. The D9 and D12 substrates form 9 and 12 bp with the D spacer, respectively. Position 1 of the spacer is not counted for duplex formation as it is not involved in substrate binding. (B, C) Substrates described in A and their pre-methylated versions (-m) were modified by Ss (B) or Ct (C) RNPs assembled with Ct snR61 and its ΔC’D' and ΔCD mutants. The Ct RNPs containing single- (SP) or dual-paralogs (DP) of Nop56 and Nop58 were assembled as in Figure 2A. Reactions were conducted at 70°C and 50°C for Ss RNPs and at 50°C for Ct RNPs.
These substrates were first modified by archaeal Ss RNPs assembled on snR61 (Figure 3B, row 1). The substrates that form 9–10 bp with the guide RNA were efficiently modified at both 70°C and 50°C (Figure 3B, lanes 3, 7, 11 and 15). The substrates that form 12 and 15 bp with the guide RNA were efficiently modified at 70°C (Figure 3B, lanes 5 and 9), but poorly at 50°C (Figure 3B, lanes 13 and 17). This indicates that excessive base pairing (>10 bp) between guides and substrates inhibits modification at low temperature, where unwinding of duplex terminus is slower. Our data confirm that the archaeal Ss RNP assembled with a natural one-piece box C/D RNA recognizes the length of substrate.
Similarly to Ss RNPs, Ct RNPs efficiently modified the substrates that form 9–10 bp with the guide (Figure 3C, row 1, lanes 2 and 4), but not the substrates that form longer duplexes with the guide at 50°C (lanes 3 and 5). We conclude that the Ct C/D RNP recognizes a restricted length of substrate in vitro. The activities of Ct SP-RNPs assembled with single paralog of Nop56 or Nop58 were also inhibited by extensive guide-substrate pairing (Figure 3C, row 4 and 5). As the size of the guide-substrate duplex is measured by the capping interactions of two CTDs of Nop5 family proteins, Nop56 or Nop58 in the SP-RNPs likely adopts homodimeric arrangement. The substrate that forms 8-bp with the D' spacer could not be modified by all RNPs likely due to low stability of short substrate-guide duplexes. Finally, the varied extent of pairing did not change the specificity of modification in all cases (Figure 3B, C).
Box C/D and C'/D' are functionally independent in both archaeal Ss and eukaryotic Ct RNPs
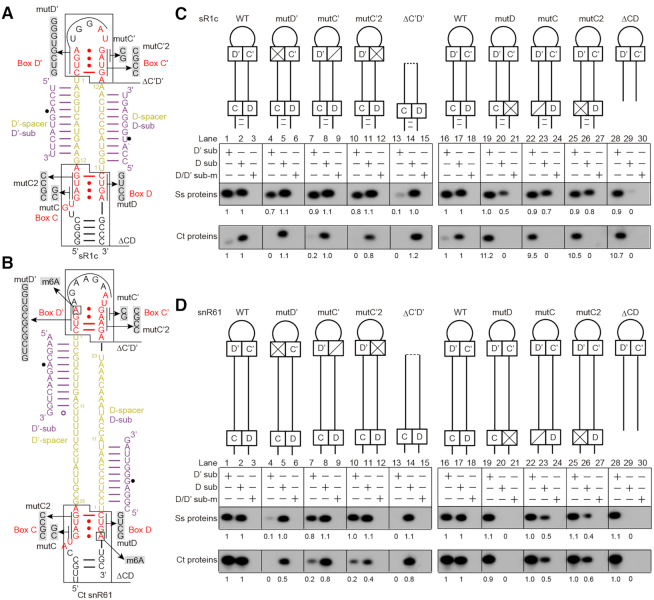
Box C/D RNAs are characterized by a bipartite structure with two sets of C/D motif and spacer. To study the functional interdependence between the C/D and C'/D' motifs, the Ct snR61 RNA and an archaeal sR1c RNA were subjected to mutagenesis (Figure 4A, B). sR1c is a typical archaeal C/D RNA that contains a k-turn formed by the terminal C and D box, a k-loop formed by the internal C' and D' box and two functional 12-nt-long spacers. sR1c is derived from a sR1 RNA that has only one functional guide upstream of box D (15,41). The individual C, D, C' and D' motifs of snR61 and sR1c were mutated to disrupt the tandem sheared GA base pairs or the entire C/D or C'/D' motifs were deleted (Figure 4A, B). Since a C/D RNA contains two copies of C/D motifs, disruption of one motif would in principle affect assembly of one L7Ae, but not the entire complex. Electrophoretic mobility shift assays (EMSA) show that the binding of the second copy of L7Ae to sR1c was reduced by deletion of the C/D or C'/D' motifs, but quite tolerant to individual box mutations (Supplementary Figure S2A). Importantly, all mutant RNAs still assemble into multiple RNPs, including the major monomeric and dimeric species. The mutant RNPs displayed some variations in the degree of oligomerization, which however do not correlate with and hence are unlikely to account for, the changes in the activity of D and D' guides. The reconstituted Ct RNPs failed to enter native gels and apparently formed soluble aggregates (Supplementary Figure S2B). Since both the monomeric and dimeric Ss RNPs are active (16) and the aggregated Ct RNP displayed robust activity, different oligomerization states of C/D RNP appeared to affect its activity very little.
Figure 4.
Box C/D and C'/D' are functionally independent. (A, B) Secondary structure of archaeal sR1c (A) and eukaryotic Ct snR61 (B) RNAs. The analyzed mutations (mut) and deletions (Δ) are illustrated. The locations of m6A modification in box D and D' of snR61 are marked. (C, D) Modification activity of C/D RNPs assembled with sR1c (C) and snR61 (D) mutant RNAs. Ct RNPs were assembled with either C/D RNA (2 μM), Snu13 (6 μM), Nop56/Fib (2 μM) and Nop58/Fib (2 μM). Ss RNPs were assembled with either C/D RNA (1 μM), co-purified Nop5/Fib complex (2 μM) and L7Ae (3 μM). The D and D' substrates and mixed pre-methylated substrates (sub-m) were reacted for 20 min by Ct RNPs at 50°C and by Ss RNPs at 70°C. The relative level of modification shown under each band was normalized to that guided by wild-type sR1c or snR61.
Assembly of wild-type (WT) sR1c and snR61 RNAs with Ss and Ct proteins all yielded active enzymes that site-specifically modified the cognate D and D' substrates (Figure 4C, D, lanes 1–3 and 16–18). For each protein/RNA combination, deletion of box C'/D' (ΔC’D') eliminated modification of the D' substrates, but did not affect modification of the D substrates (Figure 4C, D, lanes 13–14). Similarly, deletion of box C/D (ΔCD) specifically abolished modification of the D substrates, but did not affect modification of the D' substrates (Figure 4C, D, lanes 28–29). These results clearly demonstrate that box C/D and C'/D' are independently required for the function of the D and D' guides, respectively, in both Ss and Ct C/D RNPs.
Mutations of individual motif generally, specifically inhibited the activity of the coupled guides. However, mutations in C'/D' motif of snR61 within Ct RNP also moderately reduced the modification of the uncoupled D substrate (Figure 4D, lanes 5 and 11). Mutations of individual motifs displayed more variable consequences than complete deletion of C/D motifs, depending on the specific RNA/protein combination. For sR1c RNA, the motif mutants strongly inhibited the coupled activity of Ct RNP (Figure 4C, lower panel, lanes 4, 7, 10, 20, 23 and 26), but were quite well tolerated in the Ss RNP (Figure 4C, upper panel, lanes 4, 7, 10, 20, 23, 26). For snR61 RNA, mutations of the D and D' motifs blocked the modification of coupled substrates in both Ss and Ct RNPs (Figure 4D, lanes 4 and 20), but mutations of the C and C' motifs only moderately affected modification (Figure 4D, lanes 7, 10, 23 and 26). Two kinds of box C/C' mutants were analyzed and both showed nearly identical phenotypes. Finally, none of the mutations or deletions of C/D RNAs altered the site-specificity of modification.
We notice that the D' substrate of sR1c was inefficiently modified by the Ct RNP (Figure 4C, lanes 1 and 16). Surprisingly, the modification was enhanced by approximately 10-fold upon mutations of the C or D motif (Figure 4C, lower panel, lanes 19, 22, 25 and 28). The C'/D' motif of sR1c appeared to be improperly assembled in the Ct RNP and became better organized after the C/D motif was mutated. In addition, Ct RNP was more sensitive to mutations of sR1c than Ss RNP (Figure 4C, lanes 4, 7, 10, 20, 23 and 26). By contrast, the Ct and Ss RNPs respond similarly to mutations of snR61 RNA (Figure 4D). These findings suggest that sR1c is not perfectly loaded into the Ct RNP.
The Ss and Ct RNPs assembled with snR61 halfmers (ΔC’D' and ΔCD mutants) poorly modified the substrates that form 12–15 bp with guides at 50°C (Figure 3B, lanes 13 and 17; Figure 3C, lanes 3 and 5). These data indicate that halfmer C/D RNAs also recognize the length of substrate, just like bipartite C/D RNAs.
Human C/D box-like halfmer snoRNAs can function as methylation guides
The above data show that halfmer C/D RNAs that contain a single set of box C/D or C'/D' and one closed- or open-ended spacer is functional in guiding site-specific modification in vitro. One important question is whether halfmer C/D RNAs exist and function in cells. Recent studies by sRNA-seq and CLIP-seq of C/D RNP proteins have identified a group of C/D box-like snoRNAs in human cells (42,43). They contain box C and D linked by a short spacer and apparently lack the internal box C' and D'. To test if they function as methylation guides, four human C/D-box like snoRNAs were assembled with Ct proteins for modification assay (Figure 5A-B). Many C/D-box like snoRNAs have predicted targets on rRNAs and snRNAs (42,43), but the predicted guide-substrate duplexes often contain mismatches. Both predicted targets and perfectly paired substrates were assayed for modification (Figure 5A). The snoID_0741 and snoID_0426 RNAs were inactive in guiding modification, probably because their spacers (11 and 15 nt) are too short to form an effective duplex with substrates. The snoID_0666 RNA, with an 18-nt spacer, was functional in guiding site-specific modification of substrates. Another RNA snoID_0427, with a 21-nt spacer, was inactive, probably due to lack of a terminal stem. Our data show that at least some C/D-like halfmer snoRNAs are functional as modification guide.
Figure 5.
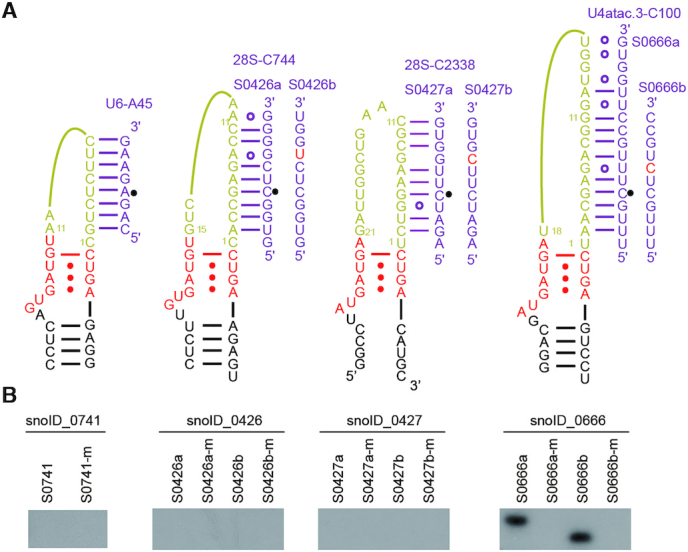
Modification activity of Ct C/D RNPs assembled with human C/D-like snoRNAs. (A) Secondary structural models of human C/D-like snoRNAs. The predicted substrates on rRNAs and snRNAs (a series) and perfectly paired substrates (b series) are shown. (B) Activity of C/D RNPs assembled with human C/D-like snoRNA (2 μM), Snu13 (6 μM), Nop56/Fib(2 μM) and Nop58/Fib (2 μM). Unmethylated and pre-methylated (-m) substrates were modified for 20 min at 50°C.
N6-methyl adenine in box D interferes with guide function
A subset of human box C/D snoRNAs were recently shown to contain an m6A at the last position of box D or D' (44). This modification occurs when a cytosine is located 3′ of box D/D', the resulting GAC sequence may become a target for m6A modification enzymes. Since the last adenine in box D forms a trans sugar-Hoogsteen GA pair with a guanine in box C, its N6-methylation has been shown to interfere with the k-turn formation and binding of 15.5K (equivalent to Snu13 in yeast) protein (44). To examine whether the activity of C/D snoRNP is affected by m6A, we measured the modification activity of Ct RNP assembled with Ct snR61 or a human snoRNA U46 (SNORD46) that were modified by inclusion of an m6A in box D or D' (Figures 4B and 6A). U46 has been experimentally shown to contain an m6A in box D (45,46). Moreover, the cytosine 3′ of box D, which is part of the m6A methylation target sequence, is highly conserved (∼90%) in U46 homologs (44). U46 is predicted to guide 2′-O-methylation of A3739 of 28S rRNA with the D spacer (Figure 6A) and has no recognizable box C' and D'. These C/D snoRNAs were chemically synthesized to include incorporation of an m6A at box D or D'. Because of its long length (98 nt), U46 was synthesized in two pieces that were joined by splint-mediated ligation. A shortened version of U46 (called U46-S) that lacks a 39-nt internal region was also synthesized as a single oligonucleotide.
Figure 6.
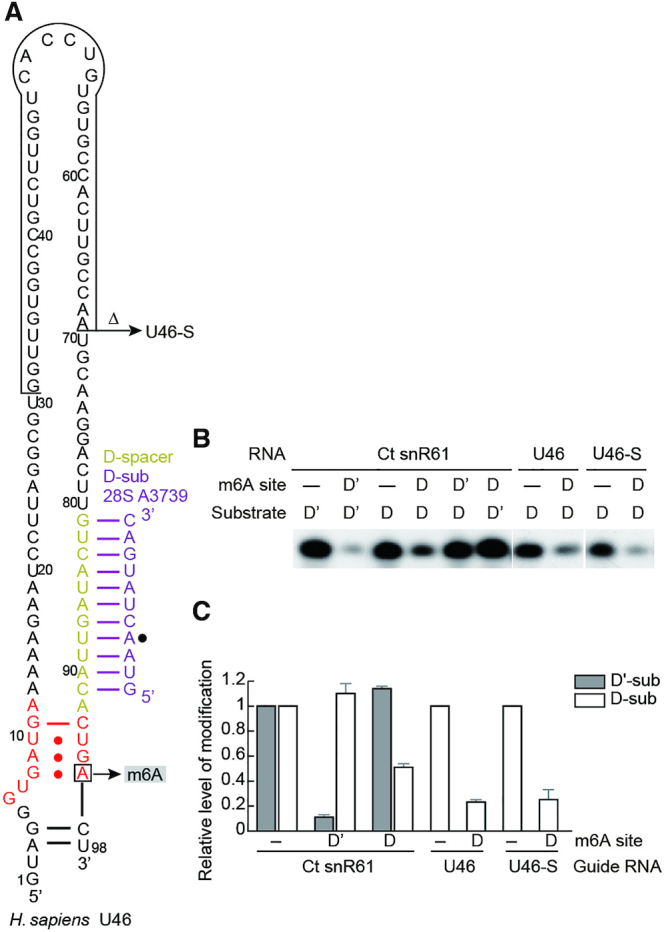
m6A in box D/D' inhibits activity of Ct C/D RNP. (A) Secondary structure of human U46 snoRNA. Nucleotides 31–69 are deleted in U46-S. The m6A site is indicated. (B) Modification activity of Ct RNPs assembled with snR61, U46 and U46-S that contain an m6A or unmodified adenine in box D or D'. (C) Quantification of modification. The level of 2′-O-methylation guided by m6A-containing C/D RNA is normalized to that guided by unmodified C/D RNAs. The mean and standard deviation determined from three experiments are shown.
In all tested snoRNAs, N6-methylation at box D or D' significantly reduced the modification of substrate compared to that guided by the unmodified snoRNAs (Figure 6B, C). In case of snR61 where both spacers are functional, m6A only affected the modification at the coupled site, as expected for the functional independence between the C/D and C'/D' motifs. We conclude that m6A in box D/D' interferes with the activity of the coupled guide.
DISCUSSION
Eukaryotic C/D snoRNPs are more complex than archaeal counterparts in protein composition, function and biogenesis pathway. Here, we have, for the first time, reconstituted an active eukaryotic C/D RNP with recombinant proteins, which allows us to dissect its functional organization and compare with archaeal C/D RNP.
In vitro reconstituted yeast C/D RNP lacks assembly specificity
Eukaryotic C/D RNPs are characterized by specific association between Nop58 and box C/D, and between Nop56 and box C'/D'. One possible mechanism for the specific assembly could be that Nop58 and Nop56 intrinsically distinguish the terminal and internal box motifs. However, we find that the SP-RNPs containing only Nop56 or Nop58 can modify both D' and D substrates. Both C/D and C'/D' motifs should be assembled in these SP-RNPs, otherwise the substrate coupled to the unassembled motifs would not be modified. In addition, the DP-RNPs with one of Nop56 or Nop58 inactivated by deletion of helix 9′ also modify both substrates (Figure 2D-E), suggesting that the guide RNAs adopt mixed orientations in the DP-RNPs (Figure 7A). These observations suggest that the reconstituted Ct C/D RNPs lack the specific organization found in natural C/D RNPs.
Figure 7.
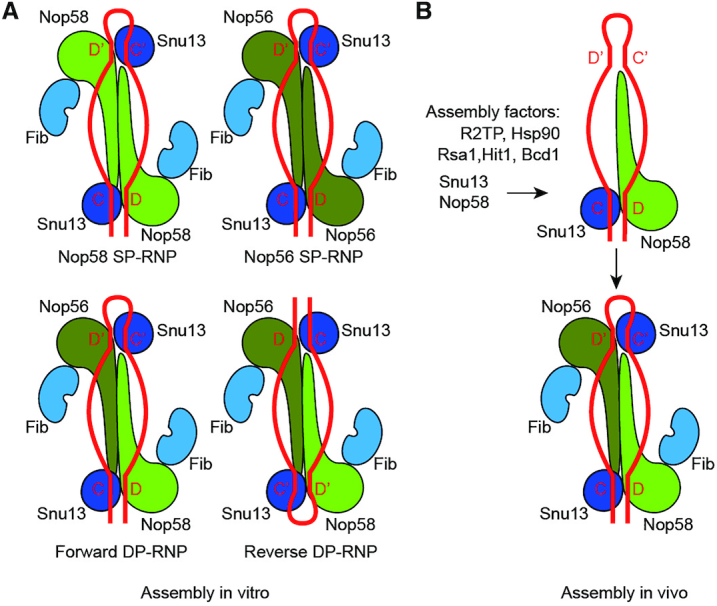
Assembly model of eukaryotic C/D RNP. (A) In vitro assembly. Active enzymes can be assembled from single and dual paralog (SP, DP) of Nop56 and Nop58. The SP-RNPs are less active than the DP-RNPs. Due to the lack of binding specificity between the box C/D motifs and Nop56/Nop58 proteins, the C/D RNA adopts both forward and reverse orientations in reconstituted DP-RNPs. (B) In vivo assembly. With assistance of assembly factors, Nop58 and Snu13 are first assembled on box C/D, establishing the asymmetric and specific organization of C/D RNP.
Nop56 or Nop58 most likely forms a homodimer in SP-RNPs as the activity of SP-RNPs is inhibited by extensive pairing between substrates and guides. This feature structurally requires the presence of two CTDs of Nop5 family proteins in SP-RNPs that clamp at both ends of the substrate-guide duplex. We also show that the DP-RNP is more active than the SP-RNPs. Compared to homodimers of Nop56 or Nop58, the heterodimer of Nop56 and Nop58 may have a higher association affinity and load substrate into a more optimal position for modification.
Our data suggest that Nop58 and Nop56 cannot structurally distinguish the terminal C/D and internal C'/D' motifs by themselves. We speculate that the asymmetric organization of endogenous C/D RNP is determined by its assembly machinery in cells (Figure 7B). Biogenesis of eukaryotic C/D snoRNP is a complicated and conserved process that requires a number of trans-acting factors, including the R2TP complex, which is composed of Rvb1, Rvb2, Tah1 and Pih1, Hsp90, Rsa1, Hit1 and Bcd1(37). Assembly of C/D snoRNP initiates at the C/D motif. Unassembled Nop58, bound to the R2TP complex, is loaded onto the nascent snoRNA transcript (47–49). Assembly of Snu13 to box C/D likely occurs at the time as Nop58 and requires the RT2P complex, Hsp90, Rsa1 and Hit1 (50–52). The assembly machinery of Nop58 and Snu13 may recognize some specific features of box C/D, such as the terminal stem that close box C and D (53,54) and proximity to RNA polymerase. Lack of assembly factors and specific assembly order would lead to mixed orientations of snoRNA in in vitro reconstituted C/D RNPs.
Box C/D and C'/D' are functionally independent
One key feature of C/D RNA is its bipartite structure with two sets of box C/D and spacer. The functional interdependence between box C/D and C'/D' have been investigated using reconstituted archaeal complexes (21,22,41,55,56) and yeast genetics (33,57). Both dependence and independence between two box C/D motifs were observed in the previous studies. A popular view in the field is that box C/D and C'/D' motifs are functionally coupled and both required for efficient methylation. This view is appealing given that C/D RNA and the dimeric complex of Nop5 family proteins are both pseudo-symmetric.
Our data consistently show that box C/D and C'/D' are functionally independent from each other on both archaeal and eukaryotic RNPs. The halfmer C/D RNAs (ΔCD, ΔC’D') can also guide site-specific modification, no matter whether box C/D or C'/D' is present and the guide is in closed- or open-end configuration. A previous study in yeast using an engineered snoRNA found that box C/D can function independently of box C'/D', but the function of box C'/D' depends on box C/D (57). The result on box C'/D' is inconsistent with our data and could be complicated by the assembly problem of halfmer C/D RNAs that contain only the internal box C'/D' (10,58,59). With the simplified in vitro reconstitution system, we show that the C'/D' and C/D motifs are equivalent at the structural level and function independently of each other. In addition, a halfmer C/D RNA was previously shown to guide non-specific modification (55). This RNA forms a highly stable 15-bp duplex (containing 12 GC pairs) with the substrate. The long duplex likely prevents loading of the substrate into the size-limiting protein channel, hence causing non-specific modification.
The structure of substrate-bound C/D RNP had suggested that anchoring of two box C/D motifs at opposite ends of the Nop5 dimer is important for correct placement of the substrate-guide duplex in the enzyme (16). The functional independence between box C/D and C'/D' indicates that the covalent linkage between the 5′ end of the spacer and box C/C' is actually not required for substrate loading, and is, in fact, is loosely connected, allowing for extra spacer sequences to be accommodated by looping out (20). Halfmer C/D RNAs also recognize the length of paired substrate (Figure 3B-C), suggesting that the substrate is bound and loaded in the same manner for enzymes assembled with halfmer and bipartite C/D RNAs.
The recently identified box C/D-like snoRNAs containing only box C and D and a short spacer represent examples of halfmer C/D RNAs (42,43). The C/D-box like snoRNAs commonly show weak expression levels and poor conservation and their function is still unknown. Our data suggest that they are capable of guiding site-specific methylation provided that they have a spacer of sufficient length (∼18 nt) and possibly a stable terminal stem. Because box C/D is essential for snoRNA accumulation, whereas box C'/D' is dispensable (10,58,59), halfmer RNAs containing only box C'/D' are unlikely to be assembled in cells.
Conservation and variations between eukaryotic and archaeal C/D RNPs
Although eukaryotic and archaeal C/D RNPs differ in protein composition and RNA features, they share essentially the same functional organization. In both types of C/D RNPs, box C/D and C'/D' are functionally independent, and the guide RNA pairs with a restricted length substrate during modification. These similar properties are based on the same mono-RNP structure adopted by eukaryotic and archaeal C/D RNPs. The substrate-guide duplex is capped by the two CTDs of dimeric Nop5 in the mono-RNP structure, which would restrict the length of substrates, paired to the guide during modification, to a maximum of 10 nt. All analyzed active C/D RNPs, whether assembled from archaeal Ss or eukaryotic Ct proteins, single or dual Nop56 and Nop58 proteins, or bipartite or halfmer C/D RNAs, are invariantly inhibited by extensive substrate pairing (Figure 3). All of these RNPs should adopt the same basic organization of mono-RNP and load substrates in the same manner.
Halfmer C/D RNAs are monovalent in protein binding and cannot induce dimerization by bridging two complexes. In fact, C/D RNA mutants with one of either C/D or C'/D' motifs deleted assemble into both monomeric and dimeric RNP species, just as bipartite C/D RNAs (Supplementary Figure S2A). This strongly suggests that the dimeric form of archaeal C/D RNP is a result of protein association, rather than RNA bridging. The assembly pattern and guiding activity of halfmer C/D RNAs further refute the RNA-swapped model of di-RNP (23).
Archaeal and eukaryotic C/D RNAs display different profiles of spacer length. The spacer is restricted to 12 nt in archaeal C/D RNAs but has a longer and more variable length in eukaryotic C/D RNAs. We show that the archaeal Ss and eukaryotic Ct proteins assemble with each of two typical archaeal and eukaryotic C/D RNAs into active enzymes, indicating that the spacer length of C/D RNA is not distinguished by Ss and Ct RNPs. In particular, the Ss RNP is fully active with snR61 that has 21-nt and 23-nt spacers, which is in contradiction with the previous observation showing that the activity of an archaeal RNP is inhibited when the spacer exceeds 12 nt (31). Although restricted spacer length is characteristic of archaeal C/D RNAs, it is not required for function. Eukaryotic C/D snoRNAs tend to have long spacers, potentially giving an advantage in making extra pairing interactions with substrates (34).
We show that extensive base pairing with substrate is inhibitory to snR61-guided modification. Eukaryotic C/D snoRNAs are predicted to form 10–21 base pairs with substrate (9). In light of our findings, the potential long duplex may not actually form in cells. Analyses of the predicted duplex between rRNA and snoRNA show that the core pairing region encompasses the 3rd to 11th position upstream box D/D' (60). The pairing outside the core region is not, or even negatively, selected during the course of evolution. Alternatively, a long substrate-guide duplex could initially form to facilitate the recruitment of substrate, but needs to be disrupted for substrate loading and catalysis. The extent of pairing is probably a fine balance between the requirements for substrate recruitment and loading into the active channel.
Identification of the m6A modification in box D/D' adds another layer of regulation for human snoRNA assembly and function (44). We show that the m6A modification, like a mutation in box D/D' (Figure 4), significantly reduces activities of reconstituted C/D RNPs (Figure 6). The m6A would interfere with the formation of the GA pair in k-turns and affect the binding of Snu13 (44). Given the different roles of box C/D and C'/D' during in vivo assembly of C/D snoRNP, the m6A modification on box D would directly disrupt the assembly of C/D snoRNP and the modification on box D' would affect the function of the D' guide.
Despites these similarities, the Ct and Ss RNPs do show some degree of specificity for C/D RNA. The eukaryotic snR61 functions well in both Ct and Ss RNPs, but the archaeal sR1c RNA is not fully active at the D' side in Ct RNPs. In addition, Ct RNPs are highly sensitive to box mutations of sR1c that causes only mild effect on Ss RNPs (Figure 4C). This suggests the Ct RNP does not accommodate the sR1c RNA well. It needs further investigation what features in the C/D RNA are recognized differently by Ct and Ss proteins.
CONCLUSION
We have shown that the reconstituted Ct C/D RNP is active, yet lacks the assembly specificity in natural C/D RNPs. All information for assembling eukaryotic C/D RNP is not entirely encoded in its components. Despite some differences, archaeal and eukaryotic C/D snoRNPs share the same functional organization. The two C/D box motifs are functionally independent, rather than coupled as traditionally viewed. All C/D RNPs recognize a restricted length of substrate during modification.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Hongjie Zhang for help in radioactivity experiments. Author contributions: K.Y. and D.L. conceived the study, Z.Y. and W.J. conducted experiments, L.H. synthesized RNA and all authors contribute to manuscript writing and editing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [91940302, 91540201, 31430024, 31325007]; National Key R&D Program of China [2017YFA0504600]; Strategic Priority Research Program of Chinese Academy of Sciences [XDB08010203]; 100 Talents Program of CAS. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Watkins N.J., Bohnsack M.T.. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2011; 3:397–414. [DOI] [PubMed] [Google Scholar]
- 2. Liang B., Li H.. Structures of ribonucleoprotein particle modification enzymes. Q. Rev. Biophys. 2011; 44:95–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001; 20:3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henras A.K., Plisson-Chastang C., Humbert O., Romeo Y., Henry Y.. Synthesis, function, and heterogeneity of snoRNA-guided posttranscriptional nucleoside modifications in eukaryotic ribosomal RNAs. Enzymes. 2017; 41:169–213. [DOI] [PubMed] [Google Scholar]
- 5. Vitali P., Kiss T.. Cooperative 2′-O-methylation of the wobble cytidine of human elongator tRNA(Met)(CAT) by a nucleolar and a Cajal body-specific box C/D RNP. Genes Dev. 2019; 33:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang C., Shi J., Guo Y., Huang W., Huang S., Ming S., Wu X., Zhang R., Ding J., Zhao W. et al.. A snoRNA modulates mRNA 3′ end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res. 2017; 45:8647–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falaleeva M., Pages A., Matuszek Z., Hidmi S., Agranat-Tamir L., Korotkov K., Nevo Y., Eyras E., Sperling R., Stamm S.. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1625–E1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schubert T., Pusch M.C., Diermeier S., Benes V., Kremmer E., Imhof A., Langst G.. Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Mol. Cell. 2012; 48:434–444. [DOI] [PubMed] [Google Scholar]
- 9. Kiss-Laszlo Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T.. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996; 85:1077–1088. [DOI] [PubMed] [Google Scholar]
- 10. Cavaille J., Nicoloso M., Bachellerie J.P.. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996; 383:732–735. [DOI] [PubMed] [Google Scholar]
- 11. Winkler W.C., Grundy F.J., Murphy B.A., Henkin T.M.. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001; 7:1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klein D.J., Schmeing T.M., Moore P.B., Steitz T.A.. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001; 20:4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang L., Lilley D.M.J.. The kink-turn in the structural biology of RNA. Q. Rev. Biophys. 2018; 51:e5. [DOI] [PubMed] [Google Scholar]
- 14. Nolivos S., Carpousis A.J., Clouet-d’Orval B.. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005; 33:6507–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Omer A.D., Ziesche S., Ebhardt H., Dennis P.P.. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin J., Lai S., Jia R., Xu A., Zhang L., Lu J., Ye K.. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature. 2011; 469:559–563. [DOI] [PubMed] [Google Scholar]
- 17. Xue S., Wang R., Yang F., Terns R.M., Terns M.P., Zhang X., Maxwell E.S., Li H.. Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle. Mol. Cell. 2010; 39:939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ye K., Jia R., Lin J., Ju M., Peng J., Xu A., Zhang L.. Structural organization of box C/D RNA-guided RNA methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:13808–13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aittaleb M., Rashid R., Chen Q., Palmer J.R., Daniels C.J., Li H.. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 2003; 10:256–263. [DOI] [PubMed] [Google Scholar]
- 20. Yang Z., Lin J., Ye K.. Box C/D guide RNAs recognize a maximum of 10 nt of substrates. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:10878–10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tran E.J., Zhang X., Maxwell E.S.. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C'/D' RNPs. EMBO J. 2003; 22:3930–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bortolin M.L., Bachellerie J.P., Clouet-d’Orval B.. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp). Nucleic Acids Res. 2003; 31:6524–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bleichert F., Gagnon K.T., Brown B.A. 2nd, Maxwell E.S., Leschziner A.E., Unger V.M., Baserga S.J.. A dimeric structure for archaeal box C/D small ribonucleoproteins. Science. 2009; 325:1384–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yip W.S., Shigematsu H., Taylor D.W., Baserga S.J.. Box C/D sRNA stem ends act as stabilizing anchors for box C/D di-sRNPs. Nucleic Acids Res. 2016; 44:8976–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lapinaite A., Simon B., Skjaerven L., Rakwalska-Bange M., Gabel F., Carlomagno T.. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature. 2013; 502:519–523. [DOI] [PubMed] [Google Scholar]
- 26. Sun Q., Zhu X., Qi J., An W., Lan P., Tan D., Chen R., Wang B., Zheng S., Zhang C. et al.. Molecular architecture of the 90S small subunit pre-ribosome. Elife. 2017; 6:e22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng J., Kellner N., Berninghausen O., Hurt E., Beckmann R.. 3.2-A-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017; 24:954–964. [DOI] [PubMed] [Google Scholar]
- 28. Chaker-Margot M., Barandun J., Hunziker M., Klinge S.. Architecture of the yeast small subunit processome. Science. 2017; 355:eaal1880. [DOI] [PubMed] [Google Scholar]
- 29. Barandun J., Chaker-Margot M., Hunziker M., Molloy K.R., Chait B.T., Klinge S.. The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 2017; 24:944–953. [DOI] [PubMed] [Google Scholar]
- 30. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Kos-Braun I., Kos M., Thoms M., Berninghausen O., Beckmann R. et al.. Architecture of the 90S Pre-ribosome: A structural view on the birth of the Eukaryotic ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 31. Tran E., Zhang X., Lackey L., Maxwell E.S.. Conserved spacing between the box C/D and C'/D' RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA. 2005; 11:285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Omer A.D., Lowe T.M., Russell A.G., Ebhardt H., Eddy S.R., Dennis P.P.. Homologs of small nucleolar RNAs in Archaea. Science. 2000; 288:517–522. [DOI] [PubMed] [Google Scholar]
- 33. Kiss-Laszlo Z., Henry Y., Kiss T.. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998; 17:797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Nues R.W., Granneman S., Kudla G., Sloan K.E., Chicken M., Tollervey D., Watkins N.J.. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J. 2011; 30:2420–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cahill N.M., Friend K., Speckmann W., Li Z.H., Terns R.M., Terns M.P., Steitz J.A.. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 2002; 21:3816–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Granneman S., Kudla G., Petfalski E., Tollervey D.. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Massenet S., Bertrand E., Verheggen C.. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017; 14:680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Erijman A., Dantes A., Bernheim R., Shifman J.M., Peleg Y.. Transfer-PCR (TPCR): a highway for DNA cloning and protein engineering. J. Struct. Biol. 2011; 175:171–177. [DOI] [PubMed] [Google Scholar]
- 39. Peng Y., Yu G., Tian S., Li H.. Co-expression and co-purification of archaeal and eukaryal box C/D RNPs. PLoS One. 2014; 9:e103096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amlacher S., Sarges P., Flemming D., van Noort V., Kunze R., Devos D.P., Arumugam M., Bork P., Hurt E.. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011; 146:277–289. [DOI] [PubMed] [Google Scholar]
- 41. Omer A.D., Zago M., Chang A., Dennis P.P.. Probing the structure and function of an archaeal C/D-box methylation guide sRNA. RNA. 2006; 12:1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jorjani H., Kehr S., Jedlinski D.J., Gumienny R., Hertel J., Stadler P.F., Zavolan M., Gruber A.R.. An updated human snoRNAome. Nucleic Acids Res. 2016; 44:5068–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kishore S., Gruber A.R., Jedlinski D.J., Syed A.P., Jorjani H., Zavolan M.. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol. 2013; 14:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang L., Ashraf S., Wang J., Lilley D.M.. Control of box C/D snoRNP assembly by N(6)-methylation of adenine. EMBO Rep. 2017; 18:1631–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen K., Lu Z., Wang X., Fu Y., Luo G.Z., Liu N., Han D., Dominissini D., Dai Q., Pan T. et al.. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed. Engl. 2015; 54:1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bizarro J., Charron C., Boulon S., Westman B., Pradet-Balade B., Vandermoere F., Chagot M.E., Hallais M., Ahmad Y., Leonhardt H. et al.. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J. Cell Biol. 2014; 207:463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kakihara Y., Makhnevych T., Zhao L., Tang W., Houry W.A.. Nutritional status modulates box C/D snoRNP biogenesis by regulated subcellular relocalization of the R2TP complex. Genome Biol. 2014; 15:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prieto M.B., Georg R.C., Gonzales-Zubiate F.A., Luz J.S., Oliveira C.C.. Nop17 is a key R2TP factor for the assembly and maturation of box C/D snoRNP complex. BMC Mol. Biol. 2015; 16:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rothe B., Saliou J.M., Quinternet M., Back R., Tiotiu D., Jacquemin C., Loegler C., Schlotter F., Pena V., Eckert K. et al.. Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction. Nucleic Acids Res. 2014; 42:10731–10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rothe B., Back R., Quinternet M., Bizarro J., Robert M.C., Blaud M., Romier C., Manival X., Charpentier B., Bertrand E. et al.. Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly. Nucleic Acids Res. 2014; 42:2015–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jady B.E., Rothe B., Pescia C., Robert M.C., Kiss T. et al.. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 2008; 180:579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Darzacq X., Kiss T.. Processing of intron-encoded box C/D small nucleolar RNAs lacking a 5′,3′-terminal stem structure. Mol. Cell. Biol. 2000; 20:4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deschamps-Francoeur G., Garneau D., Dupuis-Sandoval F., Roy A., Frappier M., Catala M., Couture S., Barbe-Marcoux M., Abou-Elela S., Scott M.S.. Identification of discrete classes of small nucleolar RNA featuring different ends and RNA binding protein dependency. Nucleic Acids Res. 2014; 42:10073–10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hardin J.W., Batey R.T.. The bipartite architecture of the sRNA in an archaeal box C/D complex is a primary determinant of specificity. Nucleic Acids Res. 2006; 34:5039–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rashid R., Aittaleb M., Chen Q., Spiegel K., Demeler B., Li H.. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 2003; 333:295–306. [DOI] [PubMed] [Google Scholar]
- 57. Qu G., van Nues R.W., Watkins N.J., Maxwell E.S.. The spatial-functional coupling of box C/D and C'/D' RNPs is an evolutionarily conserved feature of the eukaryotic box C/D snoRNP nucleotide modification complex. Mol. Cell. Biol. 2011; 31:365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Watkins N.J., Leverette R.D., Xia L., Andrews M.T., Maxwell E.S.. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996; 2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 59. Cavaille J., Bachellerie J.P.. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996; 78:443–456. [DOI] [PubMed] [Google Scholar]
- 60. Chen C.L., Perasso R., Qu L.H., Amar L.. Exploration of pairing constraints identifies a 9 base-pair core within box C/D snoRNA-rRNA duplexes. J. Mol. Biol. 2007; 369:771–783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.