Abstract
The phosphorylation pattern of Pol2 CTD Y1S2P3T4S5P6S7 repeats comprises an informational code coordinating transcription and RNA processing. cis–trans isomerization of CTD prolines expands the scope of the code in ways that are not well understood. Here we address this issue via analysis of fission yeast peptidyl-prolyl isomerase Pin1. A pin1Δ allele that does not affect growth per se is lethal in the absence of cleavage-polyadenylation factor (CPF) subunits Ppn1 and Swd22 and elicits growth defects absent CPF subunits Ctf1 and Dis2 and termination factor Rhn1. Whereas CTD S2A, T4A, and S7A mutants thrive in combination with pin1Δ, a Y1F mutant does not, nor do CTD mutants in which half the Pro3 or Pro6 residues are replaced by alanine. Phosphate-acquisition genes pho1, pho84 and tgp1 are repressed by upstream lncRNAs and are sensitive to changes in lncRNA 3′ processing/termination. pin1Δ hyper-represses PHO gene expression and erases the de-repressive effect of CTD-S7A. Transcriptional profiling delineated sets of 56 and 22 protein-coding genes that are down-regulated and up-regulated in pin1Δ cells, respectively, 77% and 100% of which are downregulated/upregulated when the cis-proline-dependent Ssu72 CTD phosphatase is inactivated. Our results implicate Pin1 as a positive effector of 3′ processing/termination that acts via Ssu72.
INTRODUCTION
The carboxyl-terminal domain (CTD) of the Rpb1 subunit of RNA polymerase II (Pol2) consists of tandem heptapeptide repeats of consensus sequence Y1S2P3T4S5P6S7. The CTD provides a scaffold for recruitment of proteins that regulate transcription, modify chromatin structure, and catalyze or regulate RNA capping, splicing, and polyadenylation (1–4). The intrinsically plastic CTD structure is tuned by dynamic phosphorylation and dephosphorylation of the heptad serine, threonine, and tyrosine residues and by isomerization of the prolines between trans and cis configurations. These variations in the primary structure comprise a CTD code that conveys informational cues about the transcription machinery that are read by CTD-binding proteins (1–3). Insights into CTD coding principles have been gained by: (i) elucidating how individual proteins recognize the CTD, and (ii) genetically manipulating the CTD primary structure and assessing effects on cell physiology.
The fission yeast Schizosaccharomyces pombe CTD has 29 repeats. The junction CTD segment to the body of Rpb1 consists of 4 repeats that deviate in size and/or sequence from the consensus heptad; this segment is referred to as the CTD ‘rump’. Distal to the rump is an array of 25 heptad repeats that conform perfectly to the YSPTSPS consensus, with the single exception of an alanine in lieu of Pro3 in the fifth heptad downstream of the rump. By introducing alanine and conservative substitutions in lieu of Tyr1, Ser2, Pro3, Thr4, Ser5, Pro6, and Ser7 of every heptad of the CTD array (5–7), we have shown that phenylalanine is functional in lieu of Tyr1 and that Ser5 is the only strictly essential phosphorylation site. The ability of S. pombe to grow when the Tyr1, Ser2, Thr4, or Ser7 residues are uniformly replaced by a non-phosphorylatable side chain resonates with transcriptome analysis showing that only a small fraction of fission yeast mRNAs are dysregulated by CTD phospho-site mutations (8). Recent studies show that the effects of mutating these four CTD phospho-sites on cell growth are genetically buffered by RNA 3′ processing/termination factors that are functionally redundant to the phospho-mark or the side-chain hydroxyl (7). The singular essentiality of Ser5-PO4 in fission yeast is linked to recruitment of the mRNA capping apparatus to the Pol2 elongation complex, insofar as the requirement for Ser5-PO4 can be bypassed by covalently fusing the capping enzyme to Pol2 (5,9).
Our knowledge of the requirements for Pro3 and Pro6 in fission yeast is relatively limited and summarized as follows. First, whereas replacing every Pro3 or Pro6 by alanine is lethal, replacing every other Pro3 or Pro6 with alanine is benign, signifying that reduced proline content is tolerated and that Pro3 and Pro6 need not be present in consecutive heptads (5,9). Second, the essentiality of Pro6, but not that of Pro3, is linked to capping enzyme recruitment. To wit, the lethality of rpb1-P6A (but not that of rpb1-P3A) can be rescued by fusion of the capping enzyme to the mutated CTD (9). Third, Pro6 is essential for the deposition of the Ser5-PO4 and Ser7-PO4 marks, but is not needed for acquisition of the Ser2-PO4 mark (9). These results highlight functional distinctions between the Pro3 and Pro6 letters of the fission yeast CTD code, but do not provide clues to what cellular transactions (other than capping) require or are affected by a CTD proline.
Here we approach this problem obliquely via a genetic analysis in fission yeast of Pin1 (10), a peptidyl-prolyl isomerase that catalyzes interconversion of cis and trans proline conformations. Pin1 isomerase (known as Ess1 in budding yeast) consists of: (i) an N-terminal WW domain that binds Ser/Thr-PO4•Pro dipeptide-containing ligands; and (ii) a C-terminal catalytic domain that effects isomerization of Ser/Thr-PO4•Pro containing substrates (11–13). Ess1 is essential for Saccharomyces cerevisiae viability and extensive genetic analyses (via conditional and hypomorphic ess1 alleles) and biochemical studies of yeast Ess1 established its many connections to Pol2 transcription and the ability of Ess1 to bind and isomerize the phosphorylated CTD, preferentially at the Ser5-PO4•Pro6 site (reviewed in 14). Several studies implicate Ess1 in RNA 3′ processing and transcription termination (14–16). A role for Ess1 in promoting CTD dephosphorylation emerged with the discovery that the CTD Ser5 phosphatase Ssu72 specifically recognizes its substrate when Pro6 is in the cis conformation (17,18). The budding yeast termination factor Nrd1 also selectively binds to a Ser5-phosphorylated CTD when Pro6 is in the cis conformation (19).
The Pin1 genetic landscape in fission yeast is attractive insofar as Pin1 is inessential for growth (10). The fission yeast CTD phosphatase Ssu72, which is a putative target of Pin1 regulation, is also inessential for growth (9). We presume that the effects of eliminating the Pin1 CTD prolyl isomerase are genetically buffered by cellular factors that collaborate with the CTD to promote specific events in fission yeast RNA biogenesis. S. pombe Ssu72 is a core component of the 13-subunit Cleavage and Polyadenylation Factor (CPF) complex responsible for 3′ processing of Pol2 transcripts (20). Ssu72 and four other CPF subunits (Ctf1, Dis2, Swd22, and Ppn1) are constituents of a recently identified genetic interactome connecting Pol2 CTD phospho-sites to RNA 3′ processing/termination and the transcriptional control of fission yeast phosphate homeostasis (7). We focus here on potential involvement of Pin1 and the CTD prolines in this genetic interaction network. We find that a pin1Δ null allele is synthetically lethal in the absence of CPF subunits Ppn1 and Swd22 and elicits conditional growth defects in the absence of CPF subunits Ctf1 and Dis2 and termination factor Rhn1. By contrast, pin1Δ displays no apparent mutational synergy with a phosphatase-dead ssu72-C13S allele. Whereas CTD S2A, T4A, and S7A mutants thrive in combination with pin1Δ, a Y1F mutant does not, nor do chimeric CTD mutants in which half of the Pro3 or Pro6 residues are replaced by alanine.
In light of these new genetic connections, we query the role of Pin1 in fission yeast phosphate homeostasis, a transcriptional program that is known to be governed by the Pol2 CTD code and the 3′ processing/termination machinery. The S. pombe phosphate regulon comprises three genes that specify, respectively, a cell surface acid phosphatase Pho1, an inorganic phosphate transporter Pho84, and a glycerophosphate transporter Tgp1 (21). Expression of pho1, pho84, and tgp1 is actively repressed during growth in phosphate-rich medium by the transcription in cis of a long noncoding (lnc) RNA from the respective 5′ flanking genes prt, prt2, and nc-tgp1 (6,22–27). A Pol2 CTD-S7A allele that prevents installation of the Ser7-PO4 mark de-represses pho1 and pho84 in phosphate-replete cells. By contrast, prevention of the Thr4-PO4 mark hyper-represses pho1 and pho84 under phosphate-rich conditions (6). A model for the repressive arm of fission yeast phosphate homeostasis is that transcription of the upstream lncRNA interferes with expression of the downstream mRNA genes by displacing the activating transcription factor Pho7 from its binding site(s) in the mRNA promoters that overlap the lncRNA transcription units (22,24,28–30). This model is supported by recent findings that: (i) mutations of CPF subunits and Rhn1—proteins that normally promote 3′ processing/termination—result in hyper-repression of pho1 under phosphate-replete conditions; and (ii) the de-repression of pho1 elicited by the CTD-S7A allele is erased by CPF and Rhn1 mutations (7).
Here, we find that pin1Δ hyper-represses the PHO regulon in phosphate-replete cells and erases the de-repressive effect of CTD-S7A. RNA-seq identified a set of 56 protein-coding RNAs that were down-regulated in pin1Δ cells, 43 of which (77%) were downregulated when the cis-proline-dependent Ssu72 CTD phosphatase was inactivated. All 22 protein-coding RNAs that were upregulated in pin1Δ cells were concordantly upregulated when Ssu72 was inactivated. Our findings point to Pin1 prolyl isomerase as a positive agent of 3′ processing and transcription termination that likely functions via its effects on Ssu72.
MATERIALS AND METHODS
Deletion of pin1
PCR amplification and standard cloning methods were employed to construct a plasmid in which the pin1 coding sequence from –8 to +517 (relative to the translational start codon +1) is replaced by a kanMX antibiotic resistance cassette. The disruption cassette was excised from the plasmid and transfected into diploid S. pombe cells. G418-resistant transformants were selected and analyzed by Southern blotting to confirm correct integration at one of the pin1 loci. Heterozygous diploids were sporulated and G418-resistant pin1Δ haploids were isolated.
Allelic exchange at the pin1 locus
Strains harboring marked wild-type and mutated pin1 alleles were constructed as follows. An integration cassette for wild-type pin1 consisted of five elements in series from 5′ to 3′: (i) a 615-bp segment of genomic DNA 5′ of the pin1+ start codon; (ii) an open reading frame (ORF) encoding wild-type Pin1; (iii) a 268-bp segment including polyA/termination signals from the nmt1+ gene; (iv) a kanMX gene conferring resistance to G418 and (v) a 593-bp segment of genomic DNA 3′ of the pin1+ stop codon. Two-stage PCR overlap extension with mutagenic primers was used to introduce missense mutations into the pin1 ORF and mutated DNA restriction fragments were inserted in the integration cassette in lieu of the wild-type pin1 ORF. All inserts were sequenced to exclude the presence of unwanted mutations. The integration cassettes were transfected into diploid S. pombe cells. G418-resistant transformants were selected and correct integrations at the target locus were confirmed by Southern blotting. A segment of the pin1::kanMX allele was amplified by PCR and sequenced to verify that the desired mutations were present. The heterozygous diploids were then sporulated and G418-resistant haploids were isolated.
Mutational effects on fission yeast growth
Cultures of S. pombe strains were grown in YES liquid medium until A600 reached 0.6–0.8. The cultures were adjusted to A600 of 0.1 and 3 μl aliquots of serial 5-fold dilutions were spotted on YES agar. The plates were photographed after incubation for 2 days at 34°C, 2.5 days at 30°C and 37°C, 4 days at 25°C, and 6 days at 20°C.
Tests of mutational synergies
Standard genetic methods were employed to generate haploid strains harboring mutations/deletions in two (or three) differently marked genes. In brief, pairs of haploids with missense or null mutations were mixed on malt agar to allow mating and sporulation and then the mixture was subjected to random spore analysis. Spores (∼1500) were plated on YES agar and on media selective for marked mutant alleles; the plates were incubated at 30°C for up to 5 days to allow slow growing progeny to germinate and form colonies. At least 500 viable progeny were screened by replica-plating for the presence of the second marker gene, or by sequentially replica-plating from YES to selective media. A finding that no haploids with both marker genes were recovered after 6–8 days of incubation at 30°C was taken to indicate synthetic lethality. By sequentially replica-plating and gauging the numbers of colonies at each step, we ensured that wild-type (unmarked) and the differentially marked single mutant alleles were recovered at the expected frequencies (unless specifically stated otherwise). Growth phenotypes of viable double-mutants were assessed in parallel with the individual mutants and wild-type cells at different temperatures (20–37°C) as described above.
Acid phosphatase activity
Cells were grown at 30°C in YES liquid medium. Aliquots of exponentially growing cultures were harvested, washed with water, and resuspended in water. To quantify acid phosphatase activity, reaction mixtures (200 μl) containing 100 mM sodium acetate (pH 4.2), 10 mM p-nitrophenyl phosphate, and cells (ranging from 0.01 to 0.1 A600 units) were incubated for 5 min at 30°C. The reactions were quenched by addition of 1 ml of 1 M sodium carbonate, the cells were removed by centrifugation, and the absorbance of the supernatant at 410 nm was measured. Acid phosphatase activity is expressed as the ratio of A410 (p-nitrophenol production) to A600 (cells). The data shown in graphs are averages (±SEM) of at least three assays using cells from three independent cultures.
Western blotting
S. pombe strains were grown in YES medium at 30°C until A600 reached 0.6 to 0.8. Aliquots (10 A600 units) of cells were collected by centrifugation and lysed in 20% (w/v) trichloroacetic acid. Total acid-insoluble protein was recovered by centrifugation. The pellets were washed with ethanol and resuspended in 1 M Tris–HCl (pH 8.0). Aliquots of the samples were adjusted to 2% SDS and 0.1 mM dithiothreitol (DTT) to contain the same amount of total protein based on the A280 of the extracts. The protein samples were analyzed by electrophoresis through 6% (for Rpb1 and Spt5 immunoblots) and 12% (for Pin1 immunoblots) polyacrylamide gels containing 0.1% SDS. The gel contents were transferred to a PVDF membrane (0.2 μm pore; Bio-Rad). The membranes were probed with either rabbit polyclonal anti-Pin1 antibody (raised against purified recombinant S. pombe Pin1 at Pocono Rabbit Farm, Canadensis, PA, USA), rabbit polyclonal anti-RNA Pol2 (phospho-CTD-Ser2) antibody (Bethyl laboratories; catalog number A300-654A; lot number A300-654A-3), rabbit polyclonal anti-RNA Pol2 (phospho-CTD-Thr4) antibody (Active Motif; catalog number 61307; lot number 13912001), rabbit polyclonal anti-RNA Pol2 (phospho-CTD-Ser5) antibody (Bethyl laboratories; catalog number A304-408A; lot number A304-408A-2), rat monoclonal anti-RNA Pol2 (phospho-CTD-Ser7) antibody (Millipore; catalog number 04-1570; lot number NG1840150), or anti-Spt5 antibody (31). Immune complexes were visualized by using horseradish peroxidase-linked anti-rabbit IgG (GE Healthcare; catalog number NA934) or anti-rat IgG (Santa Cruz Biotechnology; catalog number sc-2006), and an ECL prime Western Blotting detection system (GE Healthcare; catalog number RPN2232).
RNA analysis
Total RNA was extracted via the hot phenol method (32) from 15–20 A600 units of yeast cells that had been grown exponentially to A600 of 0.6–0.8 at 30°C. For analysis of specific transcripts by primer extension, aliquots (10 μg) of total RNA were used as templates for M-MuLV reverse transcriptase-catalyzed extensions of 5′ 32P-labeled oligodeoxynucleotide primers complementary to the pho1, pho84, tgp1 or act1 mRNAs. The primer extension reactions were performed as described previously (33) and the products were analyzed by electrophoresis of the reaction mixtures through a 22-cm 8% polyacrylamide gel containing 7 M urea in 80 mM Tris-borate, 1.2 mM EDTA. The 32P-labeled primer extension products were visualized by autoradiography of the dried gel. The primer sequences were as follows: pho1 5′-GTTGGCACAAACGACGGCC; pho84 5′-AATGAAGTCCGAATGCGGTTGC; tgp1 5′-GATTCATCCCAGCCACCAGAC; act1 5′-GATTTCTTCTTCCATGGTCTTGTC.
Transcriptome profiling by RNA-seq
RNA was isolated from S. pombe wild-type and pin1Δ cells grown in liquid YES medium at 30°C to an A600 of 0.5 to 0.6. Cells were harvested by centrifugation and total RNA was extracted via the hot phenol method. The integrity of total RNA was gauged with an Agilent Technologies 2100 Bioanalyzer. The Illumina TruSeq stranded mRNA sample preparation kit was used to purify poly(A)+ RNA from 500 ng of total RNA and to carry out the subsequent steps of poly(A)+ RNA fragmentation, strand-specific cDNA synthesis, indexing, and amplification. Indexed libraries were normalized and pooled for paired-end sequencing performed by using an Illumina HiSeq 4000 system. FASTQ files bearing paired-end reads of length 51 bases were mapped to the S. pombe genome (ASM294v2.28) using HISAT2-2.1.0 with default parameters (34). The resulting SAM files were converted to BAM files using Samtools (35). Count files for individual replicates were generated with HTSeq-0.10.0 (36) using exon annotations from Pombase (GFF annotations, genome-version ASM294v2; source ‘ensembl’). RPKM analysis and pairwise correlations (Supplementary Figures S4 and S5) were performed as described previously (8). Differential gene expression and fold change analysis was performed in DESeq2 (37). Cut-off for further evaluation was set for genes that had an adjusted P-value (Benjamini–Hochberg corrected) of ≤0.05, had an average normalized count across all samples of ≥100, and were up or down by at least 2-fold in comparison to wild-type.
RESULTS
Deletion of pin1 does not affect CTD serine/threonine phosphorylation globally
Fission yeast pin1Δ cells grew well on YES agar at 30°C to 37°C; pin1Δ colony size was slightly smaller than pin1+ at lower temperatures (Figures 1 and 2). Whole-cell extracts from pin1+ (WT) and pin1Δ strains growing logarithmically at 30°C were resolved by SDS-PAGE and subjected to Western blotting with polyclonal CTD Ser2, Thr4, Ser5 phospho-specific antibodies, a monoclonal Ser7 phospho-specific antibody, and a polyclonal antibody against transcription factor Spt5 (as a loading control) (Supplementary Figure S1). Ablation of Pin1 did not globally impact the levels of the four phospho-CTD marks.
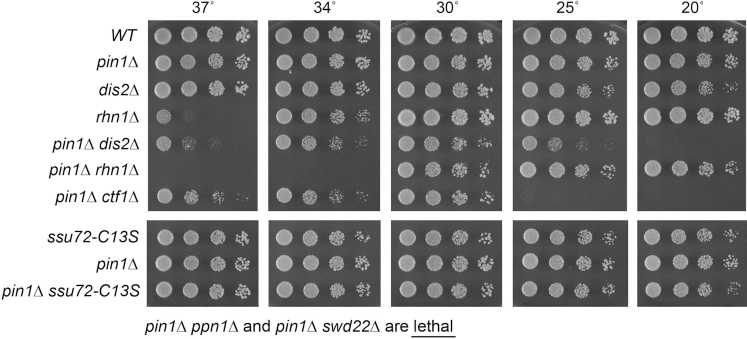
Figure 1.
Synthetic genetic interactions of Pin1 with CPF subunits and Rhn1. S. pombe strains with genotypes as specified on the left were grown in liquid culture at 30°C. Serial 5-fold dilutions were spotted to YES agar and incubated at the indicated temperatures. As noted below the panels, pin1Δ was lethal in the absence of CPF subunits Ppn1 and Swd22.
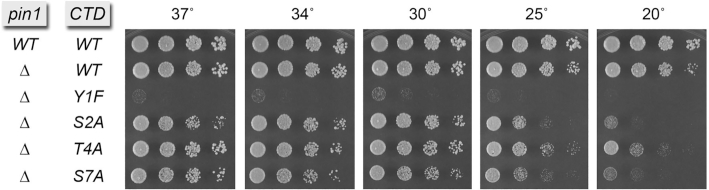
Figure 2.
Genetic interactions of pin1Δ with CTD phospho-site mutants. S. pombe strains bearing the indicated pin1 alleles in combination with rpb1-CTD alleles as specified were grown in liquid culture at 30°C. Serial 5-fold dilutions were spotted to YES agar and incubated at the indicated temperatures.
Synthetic genetic interactions of Pin1 with CPF subunits and Rhn1
To test for potentially instructive mutational synergies with proteins involved in RNA 3′ processing/termination, the pin1Δ strain was mated to a series of fission yeast knockout strains lacking CPF subunits Ctf1, Ppn1, Swd22 or Dis2, and to a strain with a catalytically dead (C13S) version of Ssu72. Ctf1 and Ssu72 are constituents of the 10-subunit CPF core complex; Dis2, Ppn1 and Swd22 comprise a heteromeric subassembly (the DPS module) that associates with the core but is not necessary for core assembly (20). Ssu72 and Dis2 are protein phosphatases of the cysteinyl-phosphatase and binuclear metallophosphoesterase families, respectively. We also crossed pin1Δ to a strain lacking the CTD-binding transcription termination factor Rhn1 (7). The resulting heterozygous diploids were sporulated and, for each allelic pair, a random collection of 500–1000 viable haploid progeny were screened by serial replica-plating for the presence of mutant allele-linked drug-resistance markers. A failure to recover any viable haploids with both markers, while recovering the three other haploid progeny (unmarked and the two singly marked haploids) with the expected frequency, was taken as evidence of synthetic lethality.
We thereby found that pin1Δ was lethal in the absence of DPS module components Ppn1 and Swd22 (Figure 1). The double mutant haploids that passed selection were spotted on YES agar at 20°C to 37°C in parallel with the component single mutants. pin1Δ was synthetically sick in the absence of the third DPS subunit Dis2, i.e. whereas dis2Δ grew at all temperatures, the pin1Δ dis2Δ strain was slow growing at 30°C (as gauged by colony size), formed pinpoint colonies at 37°C, and failed to grow at 20°C (Figure 1). Mutants of CPF core subunits Ctf1 and Ssu72 diverged in their effects on pin1Δ. Whereas ctf1Δ cells grow at all temperatures (7), the pin1Δ ctf1Δ double-mutant was severely cold-sensitive (failing to grow at 20°C and 25°C) and also slow growing at 37°C (Figure 1). By contrast, the pin1Δ ssu72-C13S strain displayed no synthetic growth defect vis-à-vis the pin1Δ and ssu72-C13S single mutants (Figure 1). The latter result is consistent with the idea that Pin1 promotes the cis conformation of CTD Pro6 that is required for recognition by Ssu72, i.e. if Pin1 and Ssu72 act in the same biochemical pathway, their simultaneous inactivation would not have additive effects. With respect to Rhn1, absence of which results in a conditional growth defect at 37°C (7), combination with pin1Δ exacerbates the temperature sensitivity so that pin1Δ rhn1Δ cells fail to grow at 34°C (Figure 1). Collectively, these results point to Pin1 participating in RNA 3′ processing in a manner genetically redundant to CPF subunits Ppn1, Swd22, Dis2 and Ctf1 and termination factor Rhn1.
Genetic interactions of pin1Δ with CTD phospho-site mutants
We crossed the pin1Δ strain with a series of S. pombe rpb1 mutant strains in which the native CTD length was maintained as 29 heptads (4 rump and 25 consensus repeats) and Tyr1, Ser2, Thr4 or Ser7 in every heptad was replaced by Phe, Ala, Ala or Ala, respectively. We reported previously that: (i) all of the full-length rpb1-CTD mutant strains grew well on YES agar medium at 30°C; (ii) Y1F, S2A, T4A and S7A cells grew slowly at 20°C and (iii) the S2A mutant grew slowly at 37°C (7). Here, we find that pin1Δ was viable in combination with rpb1-CTD alleles S2A, T4A and S7A and that the double mutants displayed conditional growth defects (cs or ts) similar to the rpb1-CTD single mutants (Figure 2). By contrast, the pin1Δ Y1F double mutant that was recovered after 4-day incubation on selective agar medium and then grown out in YES broth was unable to form macroscopic colonies on YES agar at 20–37°C, leading to the conclusion that this allelic combination was severely deleterious.
Opposite effects of pin1Δ and CTD-S7A mutations on phosphate regulon expression
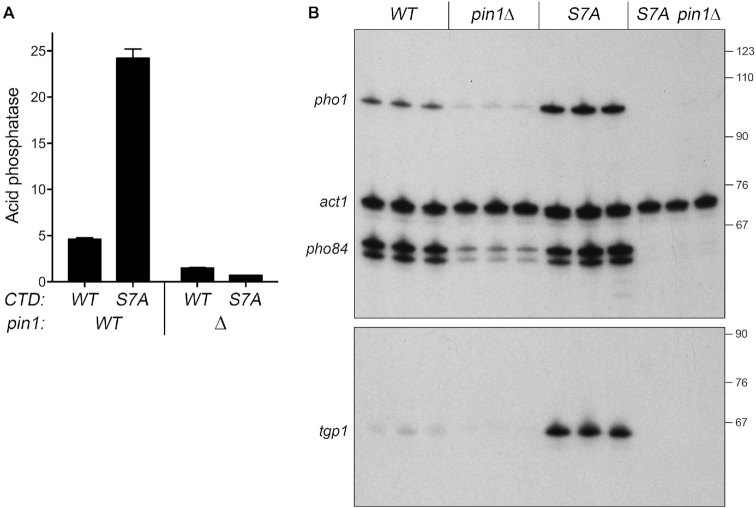
We queried the effect of pin1Δ on pho1 expression during exponential growth at 30°C in liquid culture under phosphate-replete conditions (Figure 3A). Acid phosphatase activity, a gauge of Pho1 enzyme level, was quantified by incubating suspensions of serial dilutions of the phosphate-replete pin1+ and pin1Δ cells for 5 min with p-nitrophenylphosphate and assaying colorimetrically the formation of p-nitrophenol. The basal Pho1 activity of wild-type cells was hyper-repressed by 3-fold in pin1Δ cells (Figure 3A). By performing primer extension analysis of pho1, pho84, and tgp1 mRNA levels in phosphate-replete wild-type and pin1Δ cells, we found that all three transcripts of the PHO regulon were hyper-repressed in the pin1Δ strain (Figure 3B).
Figure 3.
Opposite effects of pin1Δ and CTD-S7A mutations on phosphate regulon expression. (A) S. pombe strains bearing the indicated pin1 and rpb1-CTD alleles were grown to A600 of 0.5 to 0.8 in liquid culture in YES medium at 30°C. Cells were then harvested, washed with water, and assayed for Pho1 acid phosphatase activity by conversion of p-nitrophenylphosphate to p-nitrophenol. Activity is expressed as the ratio of A410 (p-nitrophenol production) to A600 (input cells). Each datum in the bar graph is the average of assays using cells from at least three independent cultures ± SEM. (B) Total RNA from fission yeast cells with the indicated genotypes was analyzed by reverse transcription primer extension using a mixture of radiolabeled primers complementary to the pho84, act1, and pho1 mRNAs (top panel) or the tgp1 mRNA (bottom panel). The reaction products were resolved by denaturing PAGE and visualized by autoradiography. The positions and sizes (nt) of DNA markers are indicated on the right.
Hyper-repression of pho1 by pin1Δ echoes the effect of CPF subunit and rhn1Δ mutations and the rpb1-CTD-T4A alleles that negatively impact prt lncRNA termination and thereby increase prt interference with the pho1 promoter. This contrasts with the de-repression of pho1 elicited by rpb1-CTD-S7A (Figure 3A) (6,8), an effect attributed to precocious termination of prt transcription (7). rpb1-CTD-S7A cells bearing inactivating mutations of CPF subunits or Rhn1 maintain a repressed pho1 status (7), implying that precocious prt termination elicited by S7A requires CPF and Rhn1 (7). A pertinent question is whether pin1Δ exerts a similar effect on the pho1 de-repression seen in CTD-S7A cells. The instructive findings were that: (i) de-repression of Pho1 acid phosphatase activity by S7A was erased in the pin1Δ S7A strain (Figure 3A) and (ii) the increased pho1, pho84, and tgp1 mRNA levels elicited by S7A were eliminated in the pin1Δ S7A double mutant (Figure 3B).
Structure–function analysis of fission yeast Pin1
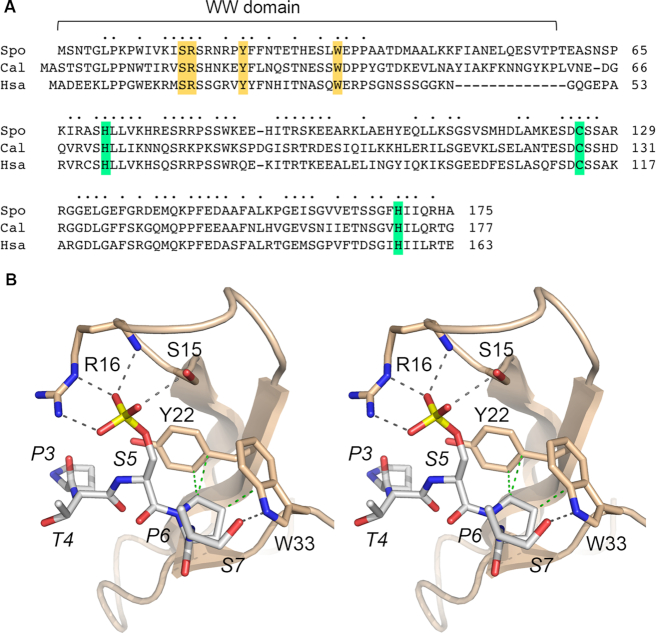
We exploited synthetic lethal phenotypes and effects on phosphate homeostasis to probe the contributions of amino acids at the CTD interface in the WW domain and in the active site of the prolyl isomerase domain. Mutagenesis was guided by primary structure conservation between S. pombe Pin1 and the orthologous proteins from Candida albicans and Homo sapiens (Figure 4A) and by the crystal structure of human Pin1 with a CTD Ser5-phosphopeptide engaged in the WW domain (12). Figure 4B shows a stereo view of the Pin1 WW•CTD-Ser5-PO4 complex, with conserved CTD-interacting amino acids numbered according to their position in S. pombe Pin1. The equivalents of Ser15 and Arg16 donate hydrogen bonds to the three Ser5-PO4 oxygens. The counterparts of Tyr22 and Trp33 make van der Waals interactions with CTD Pro6; Trp33 also donates a hydrogen bond from its indole nitrogen to CTD Ser7 Oγ (Figure 4B). Double alanine mutations S15A-R16A and Y22A-W33A in the WW domain were introduced at the chromosomal pin1 locus. For the catalytic domain, we made single alanine mutations at conserved amino acids His71, Cys125 and His169 (highlighted in green in Figure 4A) that are located in the active site (11). Previous mutational studies of human Pin1 showed that changing the conserved Cys113 to Ala drastically reduced peptidyl prolyl isomerase activity in vitro and eliminated Pin1 activity in vivo, as gauged by complementation of a yeast ess1-ts mutant (11,38). By contrast, changing the conserved upstream His59 or downstream His157 in human Pin1 to alanine maintained prolyl isomerase activity and ess1Δ complementation (38,39). The S. pombe H71A, C125A and H169A alleles were introduced at the chromosomal pin1 locus. Immunoblotting of whole cell extracts with rabbit antibody raised against recombinant S. pombe Pin1 detected a ∼19 kDa polypeptide in pin1 WT, S15A-R16A, Y22A-W33A, H71A, C125A, and H169A cells that was absent in pin1Δ cells (Supplementary Figure S2).
Figure 4.
Structure-function analysis of fission yeast Pin1. (A) Primary structure alignment of Pin1 protein from S. pombe (Spo), Candida albicans (Cal) and Homo sapiens (Hsa). Positions of amino acid side chain identity/similarity in all three proteins are denoted by dots above the alignment. Gaps in the alignment are indicated by dashes. Conserved amino acids in the N-terminal WW domain and the C-terminal prolyl isomerase domain of S. pombe Pin1 that were subjected to alanine mutagenesis in the present study are highlighted in gold and green shading, respectively. (B) Stereo view of the structure of human Pin1 with a CTD Ser5-phosphopeptide ligand engaged in the WW domain (from pdb 1F8A). The CTD peptide is rendered as a stick model with gray carbons. Conserved Pin1 amino acids that engage the CTD are shown as stick models with beige carbons, and are numbered according to their position in S. pombe Pin1. Atomic contacts between Pin1 and the CTD are indicated by dashed lines (hydrogen bonds are colored black and van der Waals interactions are colored green).
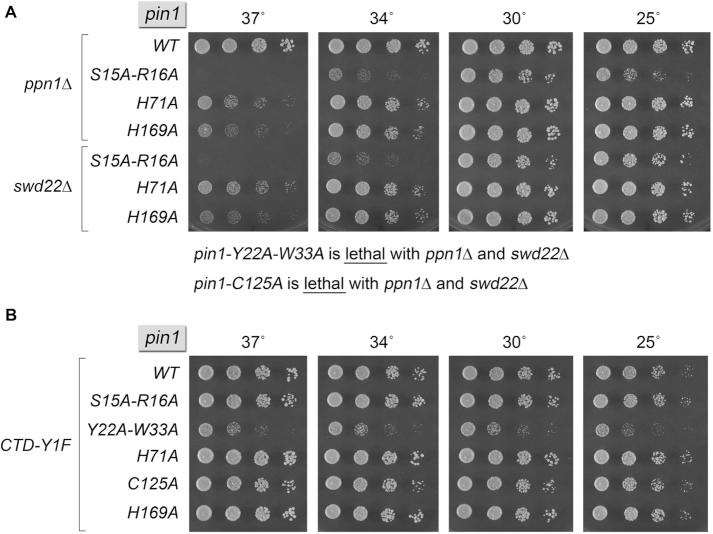
The pin1-Ala strains were crossed to ppn1Δ and swd22Δ to see if the pin1-Ala alleles were functional in genetic backgrounds in which Pin1 is essential for viability. The key findings were that pin1 alleles Y22A-W33A and C125A were synthetically lethal with ppn1Δ and swd22Δ (Figure 5A). We infer that WW domain binding to the Ser5-phosphorylated CTD (and/or other Ser/Thr-PO4•Pro containing ligands) and catalysis of proline isomerization by the C-terminal module are both essential for Pin1 activity in vivo in the absence of CPF subunits Ppn1 and Swd22. The S15A-R16A allele supported growth of ppn1Δ and swd22Δ cells at 30°C but elicited a severe temperature-sensitive growth phenotype at 34 and 37°C (Figure 5A), indicative of the importance of WW domain contacts to the phosphorylated amino acid preceding proline. It is conceivable that the main-chain hydrogen bond from Pin1 residue 16 to the phosphate of the phosphopeptides (Figure 4B) allows for residual activity of the apparently hypomorphic S15A-R16A mutant. The pin1 H71A and H169A alleles also displayed ts growth phenotypes in the ppn1Δ and swd22Δ backgrounds, albeit not as severe as S15A-R16A (Figure 5A).
Figure 5.
Synthetic genetic interactions of Pin1-Ala mutants. S. pombe strains bearing the indicated pin1 alleles in combination with ppn1Δ or swd22Δ (A) and rpb1-CTD-Y1F (B) were spot-tested for growth on YES agar at the indicated temperatures. As noted below panel A, pin1-Y22A-W33A and pin1-C125A were lethal in the ppn1Δ and swd22Δ backgrounds.
We also tested the pin1-Ala alleles for mutation synergy with rpb1-CTD-Y1F. Here, we found that pin1-Y22A-W33A elicited a severe growth defect (Figure 5B), akin to that of pin1Δ (Figure 2). By contrast, pin1-C125A displayed no growth defect in combination with CTD-Y1F (Figure 5B). These results imply that WW domain binding to the Y1F mutant phospho-CTD is critical for Pin1 function, but catalysis of proline isomerization is not. The S15A-R16A, H71A and H169A alleles that behaved as hypomorphs in the ppn1Δ and swd22Δ backgrounds did not adversely affect growth in combination with CTD-Y1F (Figure 5B).
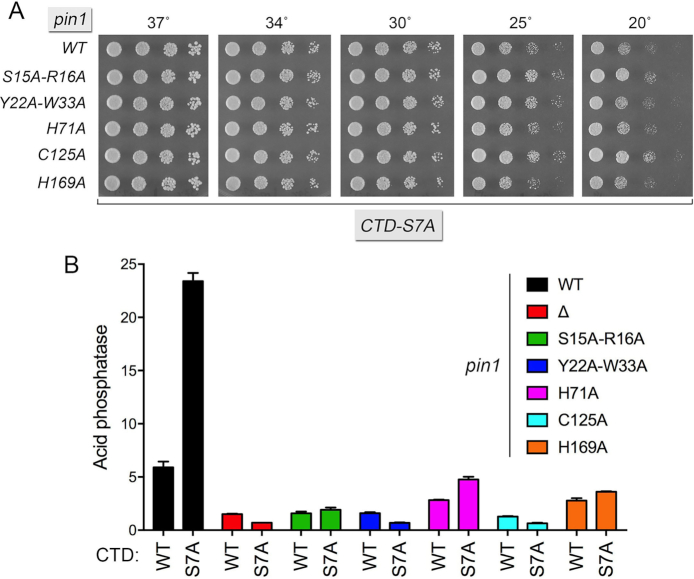
Finally, we tested the pin1-Ala alleles for their effect on pho1 expression under phosphate-replete conditions, per se and in combination with the pho1 de-repressive CTD-S7A mutation. The pin1-Ala CTD-S7A double mutants grew as well as the CTD-S7A single mutant on YES agar at 20–37°C (Figure 6A). Each of the pin1-Ala alleles elicited hyper-repression of Pho1 activity in CTD-WT cells and effaced or attenuated the de-repression of Pho1 activity in CTD-S7A cells (Figure 6B). The pin1 Y22A-W33A and C125A mutations mimicked pin1Δ with respect to their severe effect on pho1 expression (Figure 6B). The S15A-R16A, H169A and H71A alleles exerted incrementally less severe repression in the CTD-S7A background.
Figure 6.
Effect of Pin1-Ala mutants on Pho1 expression. (A) S. pombe rpb1-CTD-S7A strains bearing the indicated pin1 alleles were spot-tested for growth on YES agar at the indicated temperatures. (B) S. pombe strains bearing the indicated pin1 alleles in the context of the wild-type or S7A Pol2 CTD backgrounds were grown in liquid culture at 30°C and assayed for acid phosphatase activity.
A nexus between Pin1 and inositol pyrophosphate (IPP) status
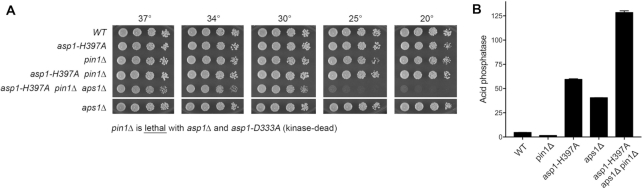
Recent studies implicate the inositol pyrophosphate signaling molecule IP8 as a new player in the interactome of the CTD code with 3′ processing and transcription termination (40). The evidence rests on biochemical phenotypes and mutational synergies elicited by genetic manipulations of Asp1, a bifunctional enzyme composed of an N-terminal IPP kinase domain and a C-terminal IPP pyrophosphatase domain (40,41). The in vivo effect of an asp1Δ null allele or a kinase-dead asp1-D333A allele is to eliminate intracellular IP8 and to increase the level of IP7; the in vivo effect of a pyrophosphatase-dead asp1-H397A allele is to increase the level of IP8 without affecting the level of IP7 (41). Thus, the function of the Asp1 kinase is to generate IP8 via phosphorylation of its substrate IP7 and the function of the Asp1 pyrophosphatase is to convert its substrate IP8 back to IP7. A failure to synthesize IP8 (in asp1Δ cells) results in hyper-repression of the PHO regulon, thereby phenocopying mutations of CPF subunits, Rhn1 and CTD mutant T4A (40). The synthetic lethality of asp1Δ (no IP8) with mutations of CPF subunits Ppn1, Swd22, and Ssu72 argues that IP8 plays an important role in promoting essential 3′ processing/transcription termination events in fission yeast, albeit in a manner that is genetically redundant to CPF. Moreover, asp1Δ erases the de-repression of pho1 in CTD-S7A cells (40). These similarities between the effects of eliminating IP8 and ablating Pin1 prompted us to test for mutational synergies between pin1Δ and alleles asp1Δ, kinase-dead asp1-D333A, and pyrophosphatase-dead asp1-H397A. Viable pin1Δ asp1-H397A double-mutants were recovered after mating the differentially marked pin1Δ and asp1-H397A strains and screening large numbers of random spores, albeit at lower frequency than expected based on random segregation, owing to the fact that the pin1+ and asp1+ genes are both located on chromosome III and separated by only 93 kbp. pin1Δ asp1-H397A cells grew as well as wild-type cells on YES agar at 20–37°C (Figure 7A). By contrast, no viable pin1Δ asp1Δ or pin1Δ asp1-D333A double-mutants were recovered after mating and screening large numbers of random spores, signifying that absence of Pin1 is synthetically lethal with absence of IP8 (Figure 7A). These results indicate that Pin1 and IP8 promote 3′ processing/termination in a genetically redundant manner, such that loss of both is lethal.
Figure 7.
Nexus between Pin1 and inositol pyrophosphate (IPP) status. (A) Pin1 is essential in the absence of IP8. S. pombe strains with genotypes as specified were spot-tested for growth on YES agar at the indicated temperatures. (B) Lethality of asp1-H397A aps1Δ is rescued by pin1Δ. Cells with the indicated genotypes were grown in liquid culture in YES medium at 30°C and assayed for Pho1 acid phosphatase activity.
We proceeded to test the pin1-Ala alleles for function in the asp1Δ and asp1-D333A backgrounds, where pin1 is essential (Supplementary Figure S3). As noted previously (40), cells lacking Asp1 kinase activity form smaller colonies than WT cells on YES agar medium. The pin1-Y22A-W33A mutant was synthetically lethal with asp1Δ and asp1-D333A, whereas the C125A allele was synthetically very sick (Supplementary Figure S3A, B). These results signify that WW domain binding and prolyl isomerization by Pin1 are crucial when IP8 is not present. The H71A, H169A and S15A-R16A alleles did not cause significant synthetic defects in asp1Δ and asp1-D333A cells.
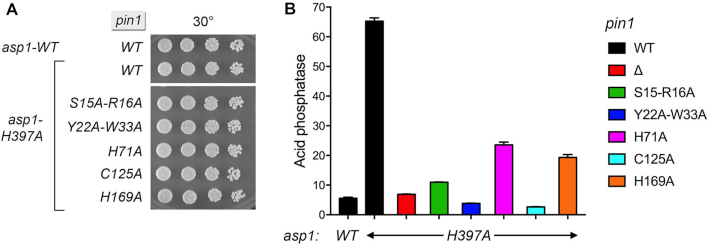
Previous studies showed that pho1 is de-repressed in asp1-H397A cells by virtue of precocious termination of prt lncRNA transcription (40). This de-repression is erased by mutations of CPF subunits, Rhn1, and the Thr4 letter of the CTD code (40). Here we find that pho1 de-repression in asp1-H397A cells is also severely attenuated in the pin1Δ background (Figure 8B). That the repressive effect of Pin1 absence on pho1 expression wins out over the de-repressive effect of increased IP8 (or increased IP8:IP7 ratio) in an asp1-H397A mutant indicates that Pin1 is required for asp1-H397A action in eliciting precocious termination of prt lncRNA transcription.
Figure 8.
Effect of pin1-Ala alleles on pho1 de-repression by asp1-H397A. (A) S. pombe strains bearing the indicated asp1 and pin1 alleles were spot-tested for growth on YES agar at 30°C. (B) Cells with the indicated asp1 and pin1 genotypes were grown in liquid culture in YES medium at 30°C and assayed for Pho1 acid phosphatase activity.
We then tested the effect of the pin1-Ala mutants on pho1 expression in the de-repressed asp1-H397A background. Each of the pin1-Ala asp1-H397A strains grew as well as wild-type cells at 30°C (Figure 8A). The pin1 Y22A-W33A and C125A alleles effaced the de-repression of pho1 elicited by asp1-H397A (Figure 8B). pin1-S15A-R16A also blunted pho1 de-repression by asp1-H397A, whereas pin1-H169A and H71A were less effective in this regard.
Lethality of asp1-H397A aps1Δ is rescued by pin1Δ
S. pombe Aps1 is an IPP pyrophosphatase belonging to the Nudix hydrolase family (42). An aps1Δ null mutation has no effect on fission yeast growth (Figure 7A) but does elicit a de-repression of pho1 expression in phosphate-replete medium (Figure 7B) (40). Our recent observation that the IPP pyrophosphatase-dead alleles asp1-H397A and aps1Δ are synthetically lethal implies that accumulation of too much IP8 is in some way toxic to fission yeast (40). Genetic evidence that this toxicity arises from unconstrained precocious transcription termination emerged from the findings that the synthetic lethality of asp1-H397A aps1Δ was rescued by CPF mutations ppn1Δ, swd22Δ, ctf1Δ and ssu72-C13S (40). Here, we queried whether ablating Pin1 would also suppress the toxicity of too much IP8. We crossed asp1-H397A pin1Δ with aps1Δ pin1+, then sporulated the resulting diploids and screened random spores for each of the differentially marked loci of interest. In this way, we recovered viable asp1-H397A aps1Δ pin1Δ haploids that grew on YES agar at 30–37°C, but not at 20 and 25°C (Figure 7A). Assays of Pho1 expression (Figure 7B) showed that: (i) Pho1 levels were 2-fold higher in the asp1-H397A aps1Δ pin1Δ strain than in the asp1-H397A single-mutant and (ii) adding the aps1Δ allele to the asp1-H397A pin1Δ double-mutant overrode the antagonistic effects of pin1Δ on pho1 de-repression by asp1-H397A (compare Figures 7B and 8B). Thus, pin1Δ phenocopied ssu72-C13S (40) with respect to both suppression of asp1-H397A aps1Δ synthetically lethality and de-repression of pho1 in the double IPP pyrophosphatase-dead asp1-H397A aps1Δ background.
Genetic interactions of CTD proline mutants
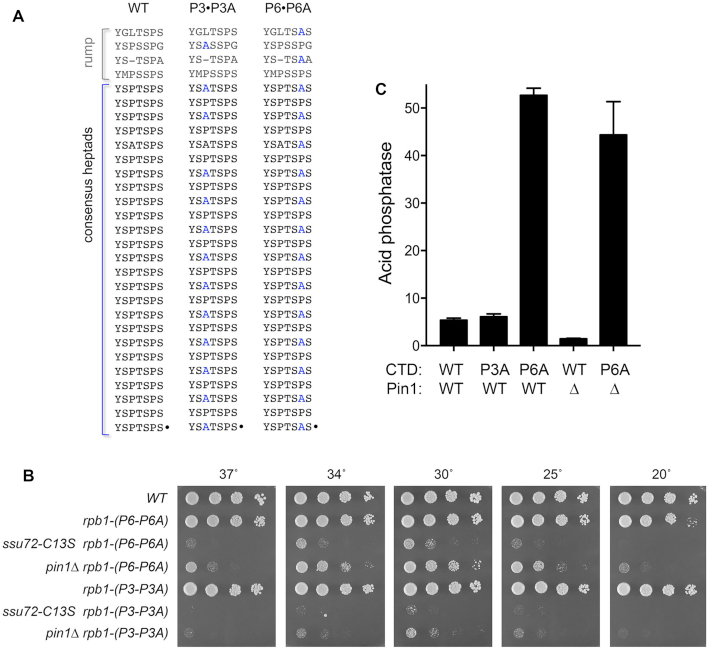
To begin to probe which of the CTD prolines might be affected by Pin1, we constructed two new rpb1-CTD mutants—P3•P3A and P6•P6A—in which Pro3 or Pro6 was changed to alanine in every other heptad of the full-length CTD repeat array (Figure 9A). The rpb1-CTD-(P3•P3A) strain grew as well as wild-type on YES agar at all temperatures tested (Figure 9B). rpb1-CTD-(P6•P6A) cells grew like wild-type at 25°C to 37°C, but formed smaller colonies at 20°C (Figure 9B). Mating differentially marked pin1Δ and rpb1-CTD-(P3•P3A) strains yielded a pin1Δ rpb1-CTD-(P3•P3A) double-mutant that was barely viable on YES agar at 30°C (forming pinpoint colonies) and failed to grow at higher and lower temperatures (Figure 9B). A pin1Δ rpb1-CTD-(P6•P6A) double-mutant grew more slowly than the parental single-mutants at 30°C (as gauged by colony size) and displayed strong ts and cs growth defects at 37°C and 20°C, respectively (Figure 9B). Thus, Pin1 becomes virtually essential when the number of Pro3-containing heptads in the Pol2 CTD is reduced by half. Reducing CTD Pro6 content by half impacted cell growth in the absence of Pin1, albeit not as severely as an equivalent reduction in Pro3 content. A noteworthy finding was that the CTD phosphatase-dead ssu72-C13S mutant was even more deleterious than pin1Δ in combination with the CTD-(P3•P3A) and CTD-(P6•P6A) alleles (Figure 9B), raising the prospect that the elimination of Pin1-mediated cis-trans isomerization of CTD proline residues is a forme fruste of the phenotype elicited by inactivation of the Ssu72 CTD phosphatase that depends on a cis proline conformation.
Figure 9.
Genetic interactions with CTD proline mutants. (A) Chimeric P3•P3A and P6•P6A CTD mutants. The amino acid sequences of the CTD heptad arrays of WT and proline mutant full-length rpb1-CTD alleles are shown, with mutated residues indicated in blue font. (B) S. pombe strains bearing the indicated rpb1-CTD, pin1, and ssu72 alleles were spot-tested for growth on YES agar at the indicated temperatures. (C) Cells with the indicated rpb1-CTD and pin1 genotypes were grown in liquid culture in YES medium at 30°C and assayed for Pho1 acid phosphatase activity.
The CTD-(P3•P3A) and CTD-(P6•P6A) mutations had divergent effects on pho1 expression, whereby reducing Pro6 content de-repressed pho1 whereas reducing Pro3 content did not (Figure 9C). The de-repressive P6A effect is likely attributable to a proportional reduction in Ser5 phosphorylation by proline-directed CTD kinases. We had shown previously that: (i) complete replacement of Pro6 by alanine (lethality of which was rescued by fusion of the CTD to mRNA capping enzyme) resulted in elimination of the Ser5-PO4 mark and de-repression of pho1 (9) and (ii) serial reduction in the Ser5 content of the CTD by Ser-to-Ala changes elicited a progressive increase in pho1 expression in phosphate-replete cells, i.e. Ser5-PO4 status is a tunable determinant of pho1 expression (6). An instructive finding here was that the de-repression of pho1 by CTD-(P6•P6A) was not reversed in the pin1Δ background (Figure 9C). The observations that CTD-(P6•P6A) wins out over the hyper-repressive action of pin1Δ, whereas pin1Δ hyper-repression wins out over the pho1 de-repressive effects of CTD-S7A (Figure 3A) suggest that Pro6 (and, by inference, Ser5 phosphorylation status) exerts a stronger influence on transcription interference with pho1 expression than does Ser7.
RNA-seq analysis defines a Pin1-responsive gene set
The concordant reduction in expression of the PHO regulon genes upon genetic ablation of Pin1 raised the prospect that other genes might be similarly dysregulated in the pin1Δ genetic background. To explore this idea, we performed RNA-seq on poly(A)+ RNA isolated from wild-type and pin1Δ cells. cDNAs obtained from three biological replicates (using RNA from cells grown to mid-log phase in YES medium at 30°C) were sequenced for each strain. In the datasets, 97–98% of the reads were mapped to unique genomic loci (Supplementary Figure S4). Read densities (RPKM) for individual genes were highly reproducible between biological replicates (Pearson coefficients of 0.97 to 0.98; Supplementary Figure S5). As an internal control, we affirmed that there were no reads for the deleted pin1 coding sequence in the pin1Δ strain. A cutoff of ±2-fold change in normalized transcript read level and an adjusted P-value of ≤0.05 were the criteria applied to derive an initial list of differentially expressed annotated loci in the mutant versus wild-type. We then focused on differentially expressed coding genes with average normalized read counts across all samples of ≥100 (DESeq2 baseMean parameter) in order to eliminate the many, mostly non-coding, transcripts that were expressed at very low levels in vegetative cells.
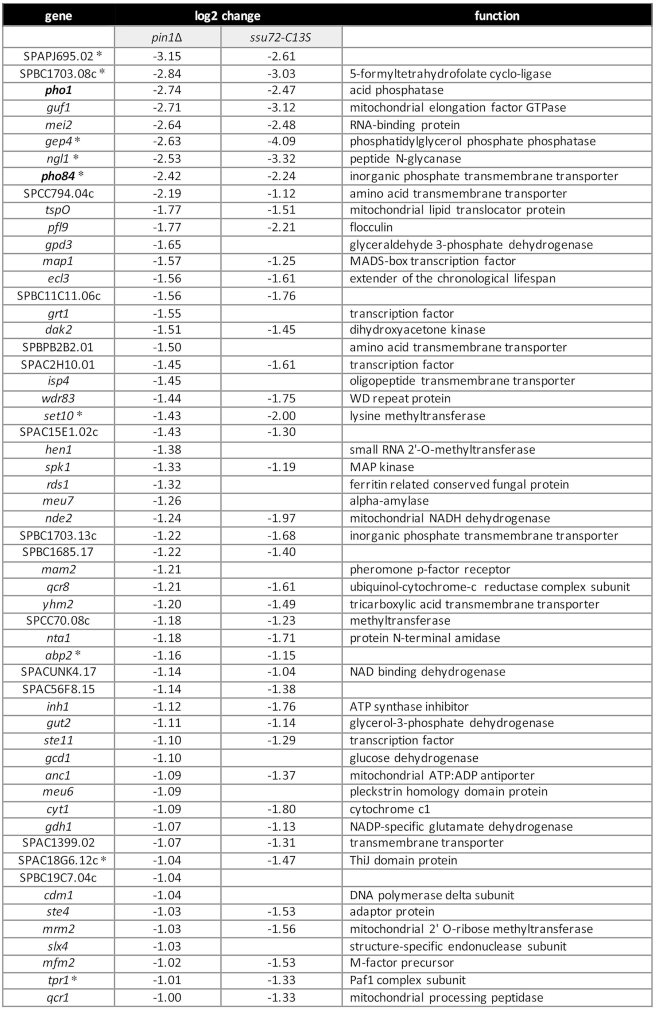
Figure 10 shows the list of 56 annotated protein-coding genes that were down-regulated by these criteria in pin1Δ cells. This gene set includes two of the three phosphate homeostasis genes: pho1 (down 7-fold) and pho84 (down 4-fold). The log2 (pin1Δ/WT) value for the tgp1 transcript (which is expressed at lower steady-state levels in phosphate-replete cells than the pho1 or pho84 RNAs) was –0.94 (1.9-fold decrement), which was just below our 2-fold criterion. Thus, the RNA-seq data affirms the conclusions from assays of Pho1 enzyme activity and of mRNA levels by primer extension that the PHO regulon is hyper-repressed in the absence of Pin1. RNA-seq revealed no decrement in pin1Δ cells of the mRNA encoding Pho7, the transcription factor that drives pho1, pho84, and tgp1 mRNA synthesis.
Figure 10.
Transcription profiling identifies a Pin1-dependent gene set. List of 56 annotated protein-coding genes that were down-regulated at least two-fold in pin1Δ cells, 43 of which (77%) were also down-regulated at least 2-fold in ssu72-C13S cells. The log2 fold changes versus wild-type are shown. Nine pin1Δ down-regulated genes that were also downregulated in CTD-Y1F cells are denoted by asterisks.
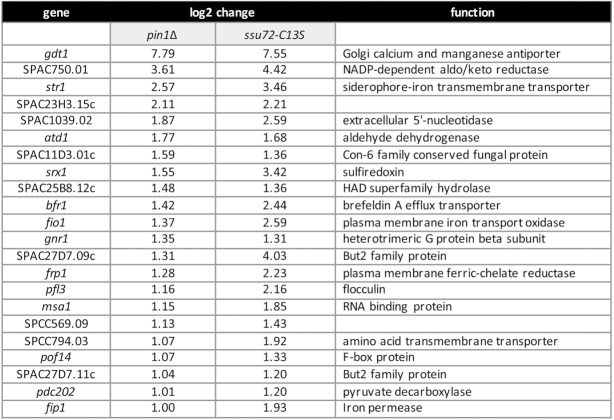
Figure 11 features the 22 annotated protein-coding genes that were up-regulated in pin1Δ cells. The set of up-regulated transcripts includes those encoding several proteins involved in fission yeast iron homeostasis: the siderophore Str1; sulfiredoxin Srx1; iron oxidase-permease Fio1; ferric reductase Frp1, and iron permease Fip1. Expression of these genes is normally repressed during growth in rich medium by the iron-sensing DNA-binding GATA-family transcriptional repressor Fep1 (43). RNA-seq showed no effect of pin1Δ on the level of fep1 mRNA.
Figure 11.
Transcription profiling identifies genes upregulated in the absence of Pin1. List of 22 annotated protein-coding genes that were up-regulated at least two-fold in pin1Δ cells, 22 of which (100%) were also up-regulated at least two-fold in ssu72-C13S cells. The log2 fold changes versus wild-type are shown.
Overlapping transcriptional signatures of pin1Δ and CTD-Y1F
The up-regulation of five iron homeostasis genes in pin1Δ cells echoes the up-regulation of the iron regulon seen in a CTD-Y1F mutant, where only 17 protein-coding transcripts were elevated by ≥2-fold (8). [The str3 iron siderophore gene that was upregulated in CTD-Y1F was also up-regulated in pin1Δ (log2 of 3.24), but its expression level was below the criteria for inclusion in Figure 11.] This correlation of the transcriptional signatures of pin1Δ and CTD-Y1F is noteworthy in light of the severe growth defect of the pin1Δ rpb1-CTD-Y1F double-mutant reported in the present study (Figure 2). Our results suggest that Pin1 (via its interaction with CTD pSer-Pro dipeptides) and the Tyr1 letter of the CTD code have overlapping roles in transducing repression of the iron uptake regulon. Moreover, two other genes are upregulated in both the pin1Δ and CTD-Y1F mutants: SPAC23H3.15c and atd1 (8; Figure 11), for a total of seven coordinately up-regulated mRNAs (P < 4.48e–9).
The correlation of pin1Δ and CTD-Y1F RNA-seq data extends to the sets of down-regulated protein-coding genes, whereby nine of the mRNAs down-regulated in pin1Δ cells were also down-regulated by ≥2-fold in the CTD-Y1F mutant (8) (P < 1.87e–13). These are denoted by asterisks in Figure 10 and include five of the top nine pin1Δ down-regulated mRNAs.
Highly concordant transcriptional signatures of pin1Δ and ssu72-C13S
If the lack of cis-trans isomerization of CTD proline residues by Pin1 exerts its effects on gene expression by affecting the activity of the cis-proline-dependent Ssu72 CTD phosphatase, then we would expect to see similar or overlapping patterns of gene dysregulation in pin1Δ and ssu72-C13S fission yeast mutants. We recently accrued RNA-seq data for the ssu72-C13S strain versus wild-type (deposited in NCBI GEO data base; accession GSE127550; ref. 40) and compared those data to the pin1Δ dataset reported presently. The concordance between the two transcription profiles was stunning. To wit: (i) of the 56 protein-coding genes that were down-regulated at least two-fold in pin1Δ cells, 43 (77%) were also down-regulated at least 2-fold in ssu72-C13S cells (P < 3.67e–64) (Figure 10 and Supplementary Figure S6A); and (ii) all 22 protein-coding genes that were up-regulated at least two-fold in pin1Δ cells were also up-regulated at least two-fold in ssu72-C13S cells (100% overlap) (P < 6.33e–38) (Figure 11 and Supplementary Figure S6B).
Overlapping transcriptional signatures of pin1Δ and asp1-D333A
Our experiments here indicate that Pin1 and the IP8-synthesizing Asp1 kinase act in parallel pathways of 3′ processing/termination, in which case the patterns of gene dysregulation in pin1Δ cells and cells lacking IP8 might overlap. We recently obtained RNA-seq data for the asp1-D333A strain versus wild-type (deposited in NCBI GEO data base; accession GSE131237; 40) and compared those data to the present pin1Δ data. The salient findings were that: (i) 21 of the 56 protein-coding genes that were down-regulated in pin1Δ cells were also down-regulated at least 2-fold in asp1-D333A cells (P < 1.01e–22) and (ii) 15 of the 22 protein-coding genes that were up-regulated in pin1Δ cells were also up-regulated at least 2-fold in asp1-D333A cells (P < 1.13e–21) (Supplementary Figure S7).
DISCUSSION
The genetic and transcriptional profiling experiments conducted here implicate fission yeast Pin1 peptidyl prolyl isomerase as an agent of 3′ processing and transcription termination that exerts its effects on gene expression through the cis-proline-directed CTD phosphatase Ssu72. The concordant synthetic genetic interactions of Pin1 and Ssu72 loss-of-function mutants are summarized in Figure 12A and discussed below.
Figure 12.
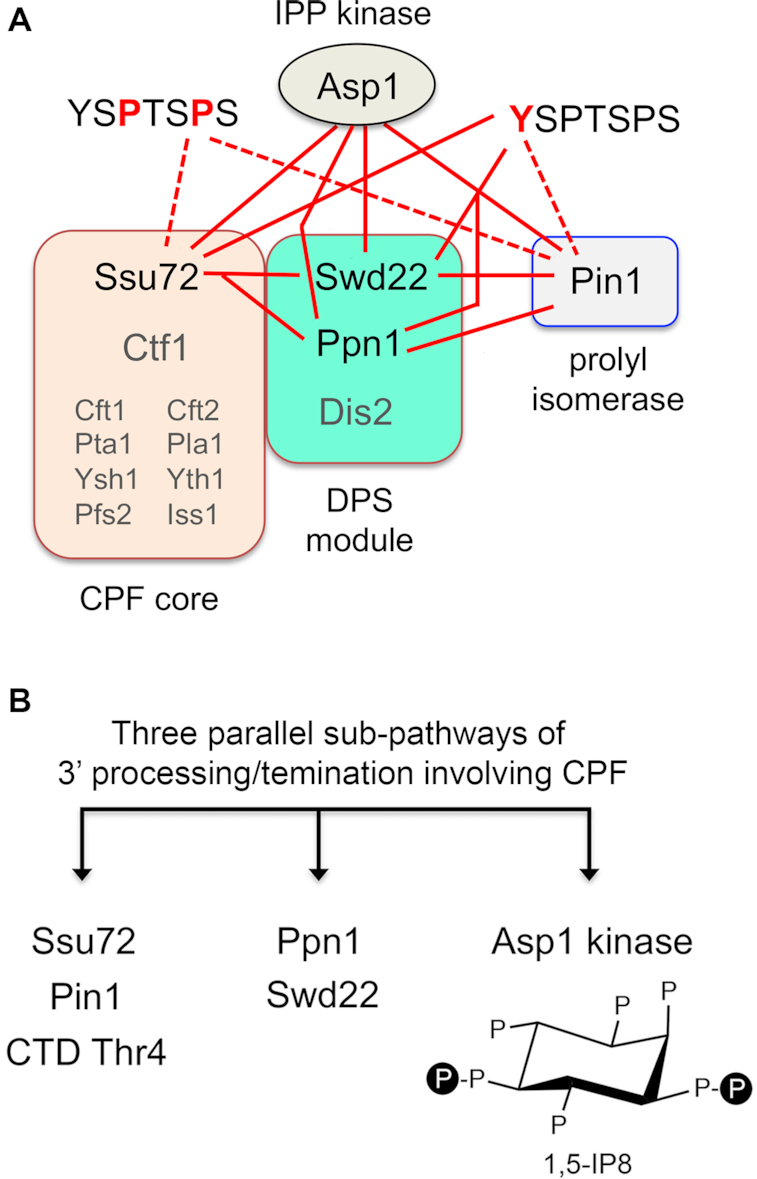
Synthetic genetic interactions of Pin1 with CPF, Asp1, and Pol2 CTD accord with those of Ssu72 and suggest three parallel sub-pathways of 3′ processing/termination via CPF. (A) The fission yeast CPF complex promotes 3′ processing and termination directed by poly(A) signals. CPF consist of a 10-subunit core (comprising 8 essential subunits in small font and two inessential subunits—Ssu72 and Ctf1—in large font) and a 3-subunit DPS module as shown. Pairwise synthetic lethalities of null or active site mutations in the CPF subunits Ssu72, Ppn1 and Swd22 with each other, with a null mutant of the Pin1 peptidyl prolyl isomerase, and with a deletion or IPP kinase-dead mutant of Asp1 (resulting in absence of IP8) are indicated by solid red lines. Also shown are synthetic lethalities (solid red lines) or severe synthetic sickness (dashed red lines) with mutations of Pol2 CTD YSPTSPS heptads at positions Tyr1 (depicted at right) or Pro3 and Pro6 (at left). (B) Three parallel sub-pathways. Inactivating mutations of the components of one sub-pathway are synthetically lethal with mutations in components of the other sub-pathway, but not with mutations of factors in the same sub-pathway. S. pombe viability depends on any two of the three sub-pathways being intact. With respect to PHO gene regulation, mutations in any one of the three sub-pathways result in pho1 hyper-repression in phosphate-replete cells.
Because neither Pin1 nor Ssu72 is essential for vegetative growth of S. pombe (unlike the case of S. cerevisiae in which both are essential), we were able to test genetically the hypothesis that the effects of ablating Pin1 are buffered by other cellular factors that are functionally redundant to cis-trans proline isomerization of the Pol2 CTD. Earlier studies had identified a network of pair-wise synthetic lethalities of ssu72-C13S with null mutations of CPF subunits Ppn1, Swd22 and Dis2 and synthetic sickness with CPF subunit Ctf1 and termination factor Rhn1 (7). Our present findings that pin1Δ was synthetically lethal with ppn1Δ and swd22Δ (indicated by solid red lines in Figure 12A) and synthetically sick with ctf1Δ, dis2Δ and rhn1Δ, but displayed no synthetic defect with ssu72-C13S, provides evidence of the participation of Pin1 and Ssu72 in a common sub-pathway of 3′ processing/termination. This conclusion is fortified by the concordant effects of pin1Δ and ssu72-C13S on the termination-sensitive PHO regulon, whereby inactivation of either enzyme hyper-repressed PHO gene expression in phosphate-replete cells and negated the PHO de-repressive effects of the CTD-S7A and asp1-H397A mutations.
The inference from these results is that: (i) Pin1-mediated isomerization of CTD prolines is important for Ssu72 phosphatase activity; (ii) absence of Pin1/Ssu72 per se is benign with respect to vegetative growth, but is lethal when other components of the 3′ processing/termination machinery are missing. Indeed, the available genetic interaction data point to at least two parallel sub-pathways of 3′ processing/termination involving CPF. One sub-pathway embraces CPF core subunit Ssu72, Pin1, and the Thr4 letter of the CTD code. The other involves CPF DPS module subunits Ppn1 and Swd22 (Figure 12B). Inactivating mutations of the components of one pathway are synthetically lethal with mutations in components of the other sub-pathway, but not with mutations of factors in the same sub-pathway (7). We envision a third parallel sub-pathway that relies on IP8 synthesis by the Asp1 kinase (Figure 12B). Because asp1Δ and the kinase-defective asp1-D333A mutant are synthetically lethal with inactivating mutations in both of the first two sub-pathways—i.e., ppn1Δ, swd22Δ and ssu72-C13S (38) and pin1Δ (present study) (Figure 12A), and CTD-T4A (not shown)—we surmise that fission yeast viability depends on any two of the three sub-pathways being intact. With respect to PHO gene expression, mutations in any one of the three sub-pathways result in pho1 hyper-repression in phosphate-replete cells.
Several observations here underscore the CTD as a relevant target of the Pin1/Ssu72 effect. First, we find that pin1Δ has a severe synthetic growth defect in combination with a Y1F mutation in every heptad repeat of the Pol2 CTD (denoted by the dashed red line in Figure 12A); this echoes the synthetic lethality of ssu72-C13S in the CTD-Y1F background (7) (solid red line in Figure 12A). It was suggested previously (based on mutational analysis of Rhn1 guided by the structure of the budding yeast ortholog Rtt103 bound to a CTD-Thr-PO4 peptide) that CTD Tyr1 contributes positively to transcription termination via hydrogen bonding of the Tyr1 hydroxyl to a conserved asparagine side chain in Rhn1, i.e., mutating Tyr1 to Phe is expected to weaken Rhn1 affinity for the CTD (7). Second, we see that pin1Δ and ssu72-C13S display strong synthetic growth defects when the Pro3 or Pro6 content of the full-length Pol2 CTD is reduced by half in the rpb1-(P3•P3A) and -(P6•P6A) strains (Figure 12A, dashed lines). The P3A and P6A heptads comprising 50% of the CTD necessarily adopt a trans-peptide conformation at the Ser2-Ala3 and Ser5-Ala6 positions and cannot be isomerized by Pin1. A plausible scenario is that this situation now demands that Pin1 be available to isomerize prolines in the remaining half of the heptads that have the wild-type consensus sequence and/or the single proline residues in the Pro-Ala mutant heptads. Third, our transcription profiling experiments uncovered significant overlap in the mRNAs that are dysregulated in pin1Δ and CTD-Y1F cells.
Our identification of pin1Δ synthetic lethalities enabled a structure-guided mutational analysis of fission yeast Pin1 function, by testing pin1-Ala alleles for complementation in the Pin1-requiring ppn1Δ, swd22Δ, asp1Δ, asp1-D333A and rpb1-CTD-Y1F genetic backgrounds. We observed that a Y22A-W33A mutation of the WW domain interface with the phospho-CTD eliminated Pin1 function in every genetic context tested. An inactivating mutation C125A in the prolyl isomerase active site (11,40) eliminated Pin1 activity in cells lacking Ppn1, Swd22 and Asp1 kinase, but did not affect complementation of rpb1-CTD-Y1F. Other Pin1 mutations behaved as hypomorphs or were benign with respect to complementation of synthetic lethality. Pin1’s contribution to pho1 expression, particularly in the de-repressed state in CTD-S7A and asp1-H397A cells, appeared to be a more sensitive gauge of mutational effects than complementation of synthetic lethality. We infer that WW domain binding to Ser-PO4•Pro peptides (e.g. in the CTD) and isomerase activity are both important for most of Pin1’s biological activities in fission yeast.
Transcriptional profiling via RNA-seq highlighted the concordant effects of inactivating Pin1 and Ssu72 on fission yeast gene expression during vegetative growth. We identified sets of Pin1-reponsive mRNAs down-regulated (n = 56) or up-regulated (n = 22) in pin1Δ cells, of which 77% and 100% were concordantly dysregulated in ssu72-C13S cells. There was also significant agreement between the pin1Δ transcriptional signature and that of asp1-D333A cells that fail to synthesize IP8 (36% overlap for down-regulated mRNAs and 68% for up-regulated mRNAs) as well as that of rpb1-CTD-Y1F cells. These results reinforce our inferences from genetic interactions that Pin1/Ssu72, IP8 status, and the CTD code collectively perform functions essential for 3′ processing/termination, while individually impacting a limited set of overlapping gene expression programs, e.g. phosphate homeostasis and iron homeostasis. The sensitivity of PHO gene expression to manipulations affecting 3′ processing/termination of upstream lncRNA synthesis makes sense in the context of the transcriptional interference paradigm. It will be of interest to pinpoint the characteristics of the other Pin1/Ssu72-responsive genes that underlie their down-regulation or up-regulation.
DATA AVAILABILITY
The RNA-seq data in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE144092 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144092).
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01-GM52470, R35-GM126945, R01-GM134021]. Funding for open access charge: NIH [R35-GM126945].
Conflict of interest statement. None declared.
REFERENCES
- 1. Eick D., Geyer M.. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013; 113:8456–8490. [DOI] [PubMed] [Google Scholar]
- 2. Corden J.L. RNA polymerase II C-terminal domain: tethering transcription to transcript and template. Chem. Rev. 2013; 113:8423–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jeronimo C., Bataille A.R., Robert F.. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem. Rev. 2013; 113:8491–522. [DOI] [PubMed] [Google Scholar]
- 4. Yurko N.M., Manley J.L.. The RNA polymerase II CTD “orphan” residues: emerging insights into the functions of Tyr-1, Thr-4, and Ser-7. Transcription. 2018; 9:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwer B., Shuman S.. Deciphering the RNA polymerase II CTD code in fission yeast. Mol. Cell. 2011; 43:311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwer B., Sanchez A.M., Shuman S.. RNA polymerase II CTD phospho-sites Ser5 and Ser7 govern phosphate homeostasis in fission yeast. RNA. 2015; 21:1770–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanchez A.M, Shuman S., Schwer B.. RNA polymerase II CTD interactome with 3′ processing and termination factors in fission yeast and its impact on phosphate homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E10652–E10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwer B., Bitton D.A., Sanchez A.M., Bähler J., Shuman S.. Individual letters of the RNA polymerase II CTD code govern distinct gene expression programs in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:4185–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwer B., Sanchez A.M., Shuman S.. Punctuation and syntax of the RNA polymerase II CTD code in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:18024–18029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang H.K., Forsburg S.L., John U.P., O’Connell M.J., Hunter T.. Isolation and characterization of the Pin1/Ess1p homologue in Schizosaccharomycespombe. J. Cell. Sci. 2001; 114:3779–3788. [DOI] [PubMed] [Google Scholar]
- 11. Ranganathan R., Lu K.P., Hunter T., Noel J.P.. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997; 89:875–886. [DOI] [PubMed] [Google Scholar]
- 12. Verdecia M.A., Bowman M.E., Lu K.P., Hunter T., Noel J.P.. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 2000; 7:639–643. [DOI] [PubMed] [Google Scholar]
- 13. Li Z., Li H., Devasahayam G., Gemmill T., Chaturvedi V., Hanes S.D., Van Roey P.. The structure of the Candidaalbicans Ess1 prolyl isomerase reveals a well-ordered linker that restricts domain mobility. Biochemistry. 2005; 44:6180–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanes S.D. The Ess1 prolyl isomerase: traffic cop of the RNA polymerase II transcription cycle. Biochim. Biophys. Acta. 2014; 1839:316–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krishnamurthy S., Ghazy M.A., Moore C., Hampsey M.. Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries. Mol. Cell. Biol. 2009; 29:2925–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singh N., Ma Z., Gemmill T., Wu X., Defiglio H., Rossettini A., Rabeler C., Beane O., Morse R.H., Palumbo M.J. et al.. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol. Cell. 2009; 36:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiang K., Nagaike T., Xiang S., Kilic T., Beh M.M., Manley J.L., Tong L.. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010; 467:729–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Werner-Allen J.W., Lee C.J., Liu P., Nicely N.I., Wang S., Greenleaf A.L., Zhou P.. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 2011; 286:5717–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kubicek K., Cerna H., Holub P., Pasulka J., Hrossova D., Loehr F., Hofr C., Vanacova S., Stefl R.. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Dev. 2012; 26:1891–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vanoosthuyse V., Legros P., van der Sar S.J., Yvert G., Toda K., Le Bihan T., Watanabe Y., Hardwick K., Bernard P.. CPF-associated phosphatase activity opposes condensin-mediated chromosome condensation. PLoS Genet. 2014; 10:e1004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carter-O’Connell I., Peel M.T., Wykoff D.D., O’Shea E.K.. Genome-wide characterization of the phosphate starvation response in Schizosaccharomycespombe. BMC Genomics. 2012; 13:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chatterjee D., Sanchez A.M., Goldgur Y., Shuman S., Schwer B.. Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under phosphate-replete conditions in fission yeast. RNA. 2016; 22:1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanchez A.M., Shuman S., Schwer B.. Poly(A) site choice and Pol2 CTD Serine-5 status govern lncRNA control of phosphate-responsive tgp1 gene expression in fission yeast. RNA. 2018; 24:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garg A., Sanchez A.M., Shuman S., Schwer B.. A long noncoding (lnc) RNA governs expression of the phosphate transporter Pho84 in fission yeast and has cascading effects on the flanking prt lncRNA and pho1 genes. J. Biol. Chem. 2018; 293:4456–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shah S., Wittmann S., Kilchert C., Vasiljeva L.. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014; 28:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee N.N., Chalamcharia V.R., Reyes-Turce F., Mehta S., Zofall M., Balachandran V., Dhakshnamoorthy J., Taneja N., Yamanaka S., Zhou M. et al.. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013; 155:1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ard R., Tong P., Allshire R.C.. Long non-coding RNA-mediate transcriptional interference of a permease gene confers drug tolerance in fission yeast. Nat. Commun. 2014; 5:5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwer B., Sanchez A.M., Garg A., Chatterjee D., Shuman S.. Defining the DNA binding site recognized by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis. mBio. 2017; 8:e01218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garg A., Goldgur Y., Schwer B., Shuman S.. Distinctive structural basis for DNA recognition by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis. Nucleic Acids Res. 2018; 46:11262–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garg A., Goldgur Y., Sanchez A.M., Schwer B., Shuman S.. Structure of fission yeast transcription factor Pho7 bound to pho1 promoter DNA and effect of Pho7 mutations on DNA binding and phosphate homeostasis. Mol. Cell. Biol. 2019; 39:e00132-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwer B., Schneider S., Pei Y., Aronova A., Shuman S.. Characterization of the Schizosaccharomycespombe Spt5-Spt4 complex. RNA. 2009; 15:1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herrick D., Parker R., Jacobson A.. Identification and comparison of stable and unstable RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990; 10:2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwer B., Mao X., Shuman S.. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 1998; 26:2050–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pyl P.T., Anders S., Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou X.Z., Kops O., Werer A., Lu P.J., Shen M., Stoller G., Küllertz G., Stark M., Fischer G., Lu K.P.. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and Tau proteins. Mol. Cell. 2000; 6:873–883. [DOI] [PubMed] [Google Scholar]
- 39. Bailey M.L., Shilton B.H., Brandl C.J., Litchfield D.W.. The dual histidine motif in the active site of Pin1 has a structural rather than catalytic role. Biochemistry. 2008; 47:11481–11489. [DOI] [PubMed] [Google Scholar]
- 40. Sanchez A.M., Garg A., Shuman S., Schwer B.. Inositol pyrophosphates impact phosphate homeostasis via modulation of RNA 3′ processing and transcription termination. Nucleic Acids Res. 2019; 47:8452–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pascual-Ortiz M., Saiardi A., Walla E., Jakopec V., Künzel N.A, Span I., Vangala A., Fleig U.. Asp1 bifunctional activity modulates spindle function via controlling cellular inositol pyrophosphate levels in Schizosaccharomycespombe. Mol. Cell Biol. 2018; 38:e00047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Safrany S.T., Ingram S.W., Cartwright J.L., Falck J.R., McLennan A.G., Barnes L.D., Shears S.B.. The diadenosine hexaphosphate hydrolase from Schizosaccharomycespombe and Saccharomyces cerevisiae are homologues of the human disphoshoinositol polyphosphate phosphohydrolase: overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 1999; 274:21735–21740. [DOI] [PubMed] [Google Scholar]
- 43. Labbé S., Pelletier B., Mercier A.. Iron homeostasis in the fission yeast Schizosaccharomyces pombe. Biometals. 2007; 20:523–537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE144092 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144092).