Abstract
Reversible lysine acetylation plays regulatory roles in diverse biological processes, including cell metabolism, gene transcription, cell apoptosis and ageing. Here, we show that lysine acetylation is involved in the regulation of chromosome segregation, a pivotal step during cell division in Streptomyces coelicolor. Specifically, deacetylation increases the DNA-binding affinity of the chromosome segregation protein ParB to the centromere-like sequence parS. Both biochemical and genetic experiments suggest that the deacetylation process is mainly modulated by a sirtuin-like deacetylase ScCobB1. The Lys-183 residue in the helix-turn-helix region of ParB is the major deacetylation site responsible for the regulation of ParB-parS binding. In-frame deletion of SccobB1 represses formation of ParB segregation complexes and leads to generation of abnormal spores. Taken together, these observations provide direct evidence that deacetylation participates in the regulation of chromosome segregation by targeting ParB in S. coelicolor.
INTRODUCTION
Chromosome segregation during cell division and differentiation is a fundamental cellular process in both prokaryotes and eukaryotes (1–3). Abnormal chromosome segregation induces genetic mutations and causes diverse disorders in humans, such as Down syndrome and Turner syndrome (4). Unlike the corresponding eukaryotic process, chromosome segregation in bacteria is a process during which chromosomes are separated rapidly as they are being replicated (5). Defects in the chromosome segregation system in bacteria result in abnormal cells that have no nucleoids or multiple nucleoids (6). In the model bacterium Caulobacter crescentus, depletion of the chromosome segregation protein blocks septum formation and affects cell viability (7). In Corynebacterium glutamicum and Bacillus subtilis, strains with deletions of genes encoding partitioning proteins are still viable but show increased anucleate cells and reduced growth rate (8).
Chromosome segregation in most bacteria (e.g. C. crescentus, B. subtilis and Mycobacterium smegmatis but not Escherichia coli) is mediated by ParA–ParB–parS or homologous systems (8–11). The ParA-ParB-parS system consists of an ATPase (ParA), a DNA-binding protein (ParB), and a cluster of centromeric DNA sequences (parS). In bacteria, parS is usually located adjacent to the chromosomal replication region (oriC) (10). During chromosome segregation, ParB binds to parS and the nearby sequence to generate a compact nucleoprotein complex that can further bind to the ParA subunits and stimulate ParA ATP hydrolysis. Consequently, the ParB–parS segregation complex follows a moving gradient of ATP-bound ParA in a Brownian ratchet mechanism that ultimately redistributes the oriC regions so that they flank cell division septa (12,13).
As a representative member of actinomycetes, Streptomyces coelicolor is known for its ability to differentiate and produce secondary metabolites (14). During chromosome segregation in S. coelicolor, multiple ParB molecules bind to more than 20 parS sites to form a massive nucleoprotein complex around oriC (15–17). Knocking out the parB coding sequence in S. coelicolor does not visibly affect cell growth but results in unevenly sized or anucleate spores (16). Hence, the formation of ParB complex foci in sporulating aerial hyphae is critical for proper chromosome segregation (16). While the transcriptional regulation of parAB is documented (18), no post-translational regulation has been reported for ParB.
Lysine acetylation (Figure 1A) is a reversible post-translational modification (PTM) that was shown to regulate diverse cellular processes widespread in organisms from E. coli to humans (19–22). In E. coli and Salmonella enterica, a wide variety of acetylation proteins enriched in central metabolism were reported (23–25). Acetylation alters the charge of lysine residues and changes protein structures, thereby affecting enzyme activity, DNA-binding affinity, or protein stability (26,27). In contrast to transcriptional regulation, PTMs such as acetylation usually fine-tune the activities of proteins rather than turn them on or off (28,29). In Streptomyces lividans, the activity of acetoacetyl-CoA synthetase was reported to be repressed by acetylation (30). Nevertheless, the understanding of acetylation regulation of prokaryotic cellular processes remains at an early stage.
Figure 1.
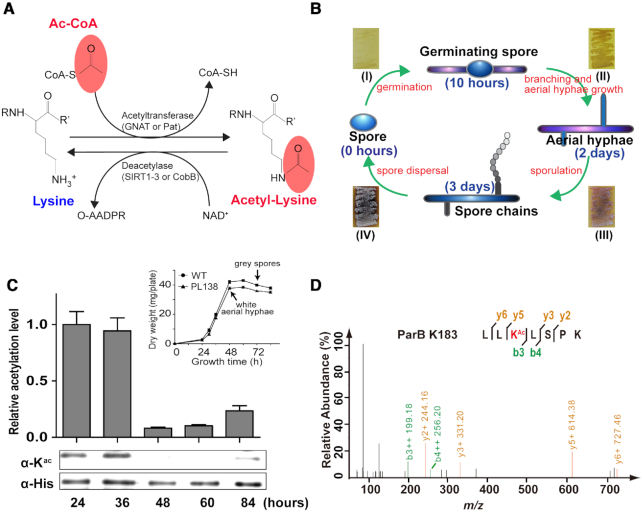
Identification of the chromosome partitioning protein ParB as an acetylation target in S. coelicolor. (A) Chemical structures of a lysine residue and an acetylated lysine. The corresponding substrates and the products of acetylation or deacetylation are shown. (B) Schematic representation of the S. coelicolor life cycle. S. coelicolor vegetative hyphae germinate from a single spore. They branch and develop into aerial filaments based on nutrient availability and other environmental signals. Aerial hyphae develop after ∼2 days of culture and then turn into spore chains. The life cycle restarts after the spores are mature and dispersed. Corresponding phenotypes (I–IV) of S. coelicolor on MS solid medium are shown as an example. (C) Analysis of the ParB acetylation level throughout the S. coelicolor cell cycle on MS solid culture. The His-tagged ParB proteins were isolated from the S. coelicolor PL138 strain at different time points (24, 36, 48, 60 and 84 h) and then quantified by western blotting. Their relative acetylation levels were normalized to the loading protein levels (see Materials and Methods). The results show that the ParB acetylation level was decreased after 48-h cultivation, which is at a growth phase that cultures are composed of both vegetative hyphae and white aerial hyphae (corresponding to growth phenotype III in Figure 1B). Similar growth curves were monitored between PL138 and wild-type S. coelicolor (upper right panel), ruling out possible impacts of the labelled ParB on cell growth. One representative result of triplicate measurements is shown. HPLC/MS-MS analysis identified Lys-183 (D) as one of the two ParB acetylation sites. The ParB protein was isolated from S. coelicolor after a 36-h cultivation and then digested and subjected to HPLC/MS-MS analyses.
In S. coelicolor, a previous in vitro study showed that SCO0452 exhibits deacetylase activity and deacetylates acetyl-CoA synthetase (31). By in-frame deletion of SCO0452, our preliminary results showed that ΔSCO0452 generated a few unevenly sized spores (Supplementary Figure S1). Further comparative acetylome analyses (unpublished data) found that among the cell development proteins, the chromosome segregation protein ParB is acetylation enriched in ΔSCO0452 cells. In this study, we confirmed that ParB is acetylated at Lys-183, which is regulated by the deacetylase SCO0452. Deacetylation of ParB enhances its binding affinity to the parS DNA sequences, while acetylation of ParB decreases the binding activity. By applying genetics and biochemical approaches, we observed that dysfunction in ParB acetylation results in the disturbance of segregation complex formation, which may lead to sporulation defects in S. coelicolor.
MATERIALS AND METHODS
Materials
All strains (Supplementary Table S1), plasmids (Supplementary Table S2), primers (Supplementary Table S3), and culture media used in this study are listed in the Supplemental Information. The pan anti-acetyllysine antibody used in this study was the same as that described in (23), and the anti-His-tag antibody was purchased from Abmart, Shanghai, China.
Determining the acetylation level of endogenous ParB in S. coelicolor
To determine the endogenous ParB acetylation level during various growth phases, the S. coelicolor PL138 strain [SCO3887::parB-his-aac(3)IV] was generated to express a His-tagged ParB by replacing the parB gene (SCO3887) with parB-his-aac(3)IV in the wild-type S. coelicolor chromosome (see Supplemental Information for details). Fresh spore suspension of PL138 was diluted to an OD600 of 1.5 and pre-germinated in TSB (tryptic soy broth) liquid medium at 50°C for 10 min. Five hundred microliters of spores were plated and grown on one MS (Mannitol Soya Flour) agar plate at 30°C. The MS plates (diameter: 150 mm) were covered with sterile plastic cellophane before spore plating. Cells were harvested from the MS plate using a bamboo scraper at the indicated time points (24, 30, 36, 48, 60, 72 and 84 h). One sample cell from each plate was collected for either the growth curve measurement or the endogenous ParB acetylation determination.
To determine the acetylation level of endogenous ParB, the samples were resuspended in cold Tris–HCl buffer (50 mM, pH 8.0) that contained NaCl (500 mM), imidazole (10 mM), EDTA (1 mM) and phenylmethanesulfonyl fluoride (PMSF, 1 mM) and were lysed using an EmulsiFlex-C5 cell disruptor (Avestin, Inc., Ottawa, Canada). The collected supernatants were loaded onto a 5-ml nickel resin column (GE Healthcare, USA) and purified with the ÄKTA™ FPLC System. After washing with buffer (50 mM Tris–HCl, 500 mM NaCl and 20 mM imidazole, pH 8.0), ParB proteins were obtained by elution from the system (elution buffer: 50 mM Tris–HCl, 500 mM NaCl and 500 mM imidazole, pH 8.0), resulting in >90% purity. To quantify the acetylation level of endogenous ParB, western blotting was performed using a pan anti-acetyl lysine antibody (23) for these ParB-His proteins. An anti-His (monoclonal antibody, Abmart, Supplementary Figure S14) western blot was employed as a loading control.
Determining ParB acetylation sites by mass spectrometry
The endogenous ParB acetylation sites were identified using protein purified from S. coelicolor PL138 after a 36-h cultivation in TSB medium. The purified ParB was digested by trypsin, and the corresponding peptides were analysed by nano-high-performance liquid chromatography (nano-HPLC)-tandem mass spectrometry (MS/MS) using an Agilent 1100 nanoflow system connected to a Q Exactive hybrid quadrupole-orbitrap mass spectrometer (Thermo Finnigan, CA, USA). The peptides were separated prior to mass spectral analysis using a C18 reversed-phase HPLC trap column (Zorbax 300SB-C18; 5.0 μm; 5.0 × 0.3 mm; Agilent). Solvent A was 0.1% (v/v) formic acid in water, while solvent B was 0.1% (v/v) formic acid in 84% (v/v) acetonitrile. The chromatographic gradient was as follows: 50 min for 4–50% (v/v) solvent B followed by 5 min for 50–100% (v/v) solvent B. The peptides eluted from the HPLC column/electrospray source were subjected to MS survey scans. Raw MS/MS data were used to search a user-defined amino acid sequence database with the Mascot 2.2 program. Cysteine carbamidomethylation was used as a fixed modification, while lysine acetylation and methionine oxidation were set as the variable modifications.
In vitro acetylation and deacetylation of ParB
The in vitro acetylation of ParB was performed at 30°C for 2 h in a 50-μl reaction mixture containing 50 mM Tris–HCl (pH 8.0), 0.55 μM ParB or ParB variants, 0.35 μM ScPat, and 0.2 mM acetyl-CoA. For the acetyl phosphate (AcP)-mediated acetylation reaction, a 50-μl mixture containing 50 mM Tris–HCl (pH 8.0) and 0.55 μM ParB or ParB variants with 1.0–3.0 mM AcP was incubated at 30°C for 2 h.
The in vitro deacetylation was performed at 30°C for 2 h in a 50-μl reaction mixture containing 50 mM Tris–HCl (pH 8.0), 6.0 mM MgCl2, 0.55 μM ParB or ParB variants, 1.2 μM ScCobB1 or 1.5 μM ScCobB2, and 1.0 mM NAD+.
The acetylated and deacetylated proteins were then analyzed in an electrophoretic mobility shift assay (EMSA) or a western blot assay.
ParB-parS electrophoretic mobility shift assay (EMSA)
A labelled parS probe containing two consecutive parS sequences was amplified from S. coelicolor genomic DNA using oligos with fluorescein amidite (FAM) attached at the 5′ end. The purified probe (50 ng) was then incubated with ParB or ParB variants at 30°C for 20 min in a 20-μl reaction mixture containing 50 mM Tris–HCl (pH 8.0), 25 mM KCl, 2.5 mM MgCl2, 1.0 mM DTT and 2.0 μg salmon sperm DNA. The mixtures were separated by 5% polyacrylamide gel electrophoresis and analysed with an ImageQuant LAS 4000 mini biomolecular imager (GE Healthcare, USA).
To quantify the ParB–parS interaction, the binding constant (KD) was determined using the same reaction system as above. ParB was treated with ScPat, ScCobB1 or ScCobB2 before the ParB–parS incubation, and ParB without any treatment was used as the control. Specifically, the parS probe (12.5 nM) was titrated with a series of different ParB proteins (0, 7, 14, 21, 28, 42, 56, 84, 112 nM). The fraction of probe bound was determined from the background-subtracted signal intensities in gels and calculated as bound/(bound + unbound). The data (fraction bound) in each reaction were plotted versus the concentration of ParB, and the results were fitted with a non-linear regression to the Hill equation in Prism 7 (GraphPad Software, CA, USA).
Analysing the acetylation or deacetylation of peptides by HPLC
Acetylated and non-acetylated peptides containing Lys-183 were synthesized by Guotai Inc. (Hefei, China). Two arginine residues were added to the C-terminus to ensure that the peptides were soluble in water (ParBK183Ac, TLRLLKAcLSPRR; ParBK183, TLRLLKLSPRR). The peptides were subjected to in vitro acetylation or deacetylation treatment as stated in the Supplemental Information and then analysed via HPLC using an Aeris peptide XB-C18 column (150 × 4.6 mm, 3.6 μm; Phenomenex, USA). The mobile phase consisted of solvent A (0.1% TFA in HPLC-grade H2O) and solvent B (0.1% TFA in HPLC-grade acetonitrile). Peptides were eluted in a linear gradient of 0–80% solvent B at 1 ml min−1 over 15 min. The column temperature was set at 25°C, and the peptides were monitored with UV light (215 nm).
Construction of ΔparB, ΔScpat, ΔSccobB1 and ΔSccobB2 S. coelicolor mutants
Monogenic S. coelicolor mutants (ΔparB, ΔScpat, ΔSccobB1 and ΔSccobB2) were generated using a PCR-based gene targeting method (32). Briefly, the target genes within cosmids were replaced in frame by an apramycin or kanamycin resistance cassette flanked by FLP recognition target (FRT) sites. Then, the resulting cosmids were propagated into S. coelicolor cells to induce a double exchange by homologous recombination. S. coelicolor clones in which the target gene was deleted were selected by flooding plates containing solid medium with apramycin (50 μg ml−1) or kanamycin (100 μg ml−1) for 16 h. The putatively positive clones were verified by PCR.
Confocal fluorescence microscopy
To monitor the formation of the ParB segregation complex, integrative plasmids were constructed and introduced into ΔparB, ΔScpat, ΔSccobB1 and ΔSccobB2 strains to generate fluorescent ParB-eGFP fusion proteins or variants (see Supplemental Information). Transgenic strains harboring fluorescent ParB or ParB variants were inoculated on MS agar with inserted coverslips. Samples were collected at the indicated times and fixed with a paraformaldehyde/glutaraldehyde mixture for 15 min. Then, samples were washed with PBS buffer and digested with 1 mg ml−1 lysozyme for 2 min. After another round of PBS wash, samples were blocked with 2% BSA for 10 min. For cell wall staining, samples were incubated with 10 μg ml−1 WGA-Texas red (Molecular Probes, USA) for 10 min; for DNA staining, samples were incubated with 1 μg ml−1 DAPI (Sigma-Aldrich, USA) for 10 min. After washing with PBS buffer, the samples were placed in antifade solution (Beyotime, Shanghai) and observed using a Leica TCS SP8 STED confocal microscope equipped with a 100× objective lens. The corresponding green filter sets (excitation and emission wavelengths of 488/20 and 530/20 nm, respectively) for eGFP observation and the red filter sets (excitation and emission wavelengths of 595/5 and 615/10 nm, respectively) for cell wall observation were used in this study. For the detection of chromosomes with DAPI staining, cells were observed using the corresponding DAPI filter sets (excitation and emission wavelengths of 380/30 and 460/50 nm, respectively).
Statistics
The results in the figures are displayed as the mean ± S.D. Comparisons between groups were made by unpaired two-tailed Student's t-test. Differences were considered statistically significant if the P-value was smaller than 0.05. Significance was indicated as *P ≤ 0.05; ***P ≤ 0.001; ns, P > 0.05, no significance.
RESULTS
Identification of ParB as an acetylation target in S. coelicolor
S. coelicolor is a model organism for studying bacterial cell development (Figure 1B) and secondary metabolite biosynthesis (14). In this study, we observed that the S. coelicolor chromosome segregation protein ParB is modified by acetylation and that its acetylation level is constantly changing throughout the cell cycle (Figure 1C, Supplementary Figure S2A). Remarkably, the ParB acetylation level decreased by ∼90% when S. coelicolor entered the sporulation phase after a 48-h cultivation (Figure 1C), hinting that ParB acetylation may be involved in S. coelicolor chromosome partitioning. To further investigate this possibility, ParB was purified from S. coelicolor and employed for HPLC–MS/MS analysis. After three rounds of independent analyses, two lysine sites in ParB [i.e. Lys-183 (Figure 1D) and Lys-187 (Supplementary Figure S2B)] were identified with reliable acetylation modification.
Deacetylation increases the binding affinity of ParB to parS
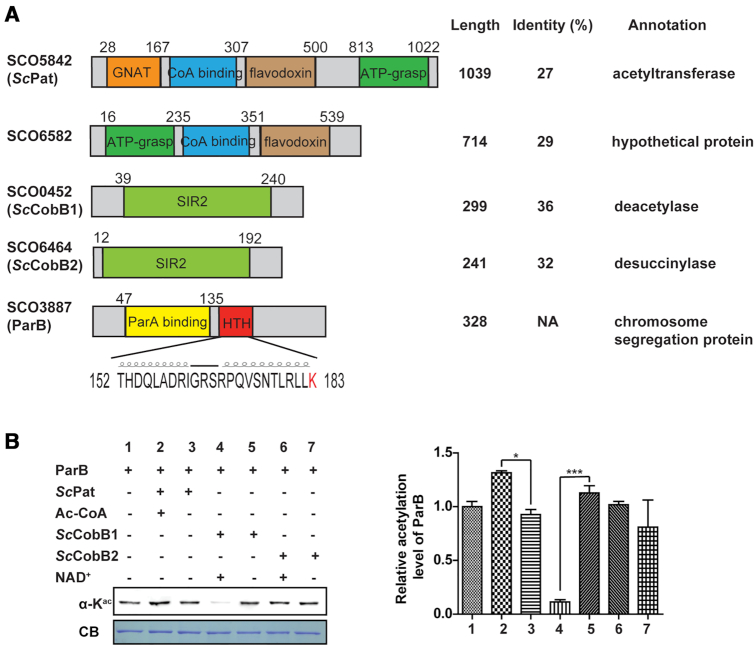
Although there are dozens of genes annotated as potential acetyltransferases, only GNAT (GNC5-type N-acetyltransferase)-family enzymes (33) are characterized as protein acetyltransferases in bacteria, e.g. Pat in S. enterica (34). Meanwhile, substantially fewer sirtuin proteins have been reported in prokaryotes than in mammalian cells (35). The sole copy of sirtuin named CobB was functionally characterized as both deacetylase and desuccinylase in E. coli (36). Collecting all the known lysine acetyltransferase and sirtuin deacetylase sequences as the query database allowed a putative acetyltransferase/deacetylase system to be predicted in the S. coelicolor proteome by the BLASTP program. As shown in Figure 2A, two putative protein acetyltransferases (SCO5842 and SCO6582) and two putative sirtuin deacetylases (SCO0452 and SCO6464) were identified because they share high similarities with E. coli Pat and CobB, respectively. Conserved motif analyses (Figure 2A) revealed a GNAT domain in SCO5842 and a sirtuin catalytic domain (SIR2) in both SCO0452 and SCO6464. However, no acetyltransferase domain was found in SCO6582. We therefore designated SCO5842, SCO0452 and SCO6464 as ScPat, ScCobB1 and ScCobB2, respectively, in this study (Figure 2A).
Figure 2.
ParB acetylation is mainly regulated by the sirtuin-like protein ScCobB1. (A) Acetyltransferase and deacetylase homologues were identified by bioinformatics analyses in S. coelicolor. Their similarities to the E. coli acetyltransferase Pat or deacetylase CobB are shown in the right panel. SCO5842 (ScPat) contains a GNAT domain (orange) and a very large region that is homologous to NDP-forming acyl-CoA synthetase, including a CoA-binding domain (blue), a flavodoxin domain (brown), and an ATP-grasp domain (green). SCO0452 (ScCobB1) and SCO6464 (ScCobB2) contain a conserved SIR2 (light green) domain. The predicted ParA-binding domain is at the N terminus (yellow), and an HTH domain is in the middle (red) of ParB. The amino acid sequence of the HTH motif with a highlighted K183 residue is provided. NA, not available. (B) The acetylation of ParB is mainly modulated by ScCobB1. Purified ParB protein was incubated with ScPat in the presence or absence of acetyl-CoA or treated with ScCobB1/ScCobB2 in the presence or absence of NAD+ for 2 h at 30°C. Then, the acetylation level was assessed by western blotting with a pan anti-acetyllysine antibody. The protein was stained with Coomassie blue (CB) and set as the loading control. ParB acetylation was decreased significantly after ScCobB1 treatment (sample 4). One representative result of triplicate measurements is shown. The relative acetylation levels of triplicates were further measured by ImageJ and normalized against the protein level (right panel). The value of the first sample was set as 100%. Mean values with standard deviations are presented.
To elucidate the possible acetylation system for ParB, recombinant ParB was treated in vitro with ScPat, ScCobB1 or ScCobB2. Western blot analysis showed that ScPat treatment slightly increased the ParB acetylation level, while treatment with ScCobB1 but not ScCobB2 decreased the ParB acetylation level (Figure 2B). These observations imply that ParB acetylation is regulated by ScCobB1 and ScPat in some ways. ScCobB2 does not have any significant effect on ParB deacetylation, which is consistent with our recent findings (37) that ScCobB2 is a specific desuccinylase in S. coelicolor. Here, we used ScCobB2 as a negative control in subsequent experiments.
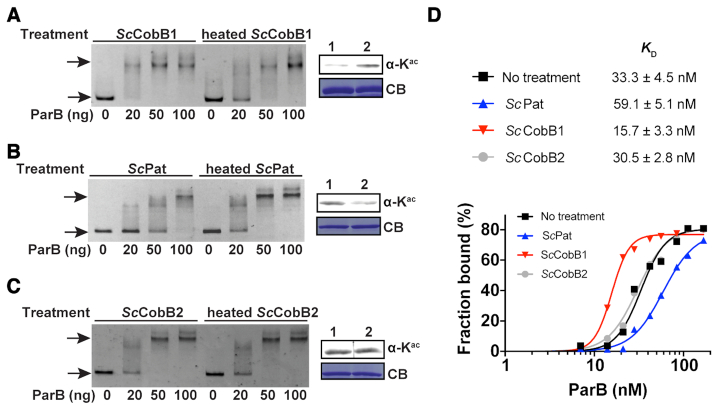
Because ParB is a central member of the ParA-ParB-parS system (8), the acetylation of ParB may affect its ability to bind parS sites. To test this hypothesis, an EMSA was conducted to detect the ParB binding affinity changes in different acetylation states. The ‘perfect’ parS, which contains two consecutive parS sequences (15), was chosen as the probe. As shown in Figure 3, the ParB binding affinity to parS increased when deacetylated by ScCobB1 (Figure 3A), while the opposite effect was observed after ScPat acetylation (Figure 3B). Consistent with our expectation, ScCobB2 treatment did not affect the DNA-binding ability of ParB (Figure 3C). The binding constants (KD) were further determined for ParB with different acetylation levels (Supplementary Figure S3). As shown in Figure 3D, the KD for the ParB control without any treatment was approximately 33.3 nM. However, the binding affinity of ParB treated with the deacetylase ScCobB1 was approximately 4-fold higher than that of ParB treated with the acetyltransferase ScPat (Figure 3D). No significant difference was observed between the ScCobB2-treated ParB and the control. Therefore, these results indicate that the DNA-binding affinity of ParB is positively regulated by ScCobB1 deacetylation in vitro.
Figure 3.
Deacetylation enhances the DNA-binding affinity of ParB in vitro. (A) An EMSA was performed by incubating a FAM-labelled parS sequence with varying amounts of ParB proteins that were pre-treated with active or heat-inactivated ScCobB1. Free DNA and DNA–protein complexes (indicated by arrows) were separated by 5% polyacrylamide gel electrophoresis. Compared to heated-ScCobB1-treated ParB, 20 ng (28 nM) deacetylated ParB can efficiently bind to parS fragments. The ParB acetylation level induced by active ScCobB1 (lane 1) or heated ScCobB1 (lane 2) was confirmed by western blot (right panel). (B) EMSA results of ParB that was pre-treated with active or heat-inactivated ScPat. (C) EMSA results of ParB treated with active or heat-inactivated ScCobB2. (D) Quantification of the DNA-binding affinity of ParB treated with ScCobB1, ScPat, or ScCobB2. The percentages of DNA bound [bound / (bound + free DNA)] in each reaction were plotted versus the concentrations of ParB (see Supplementary Figure S3 for details). The binding constants KD were determined by fitting a non-linear regression with the method of specific binding with a Hill slope.
We also assessed the possible effects of another acetylation donor, AcP (38), on ParB. Although ParB was acetylated after AcP treatment (up to 3 mM) for 2 h (Supplementary Figure S4A), the binding affinity of ParB to parS was unaffected (Supplementary Figure S4B). These observations imply that AcP-induced acetylation might not be specific to the lysine site(s) critical to ParB binding activity.
Identification of Lys-183 as a critical acetylation site in ParB
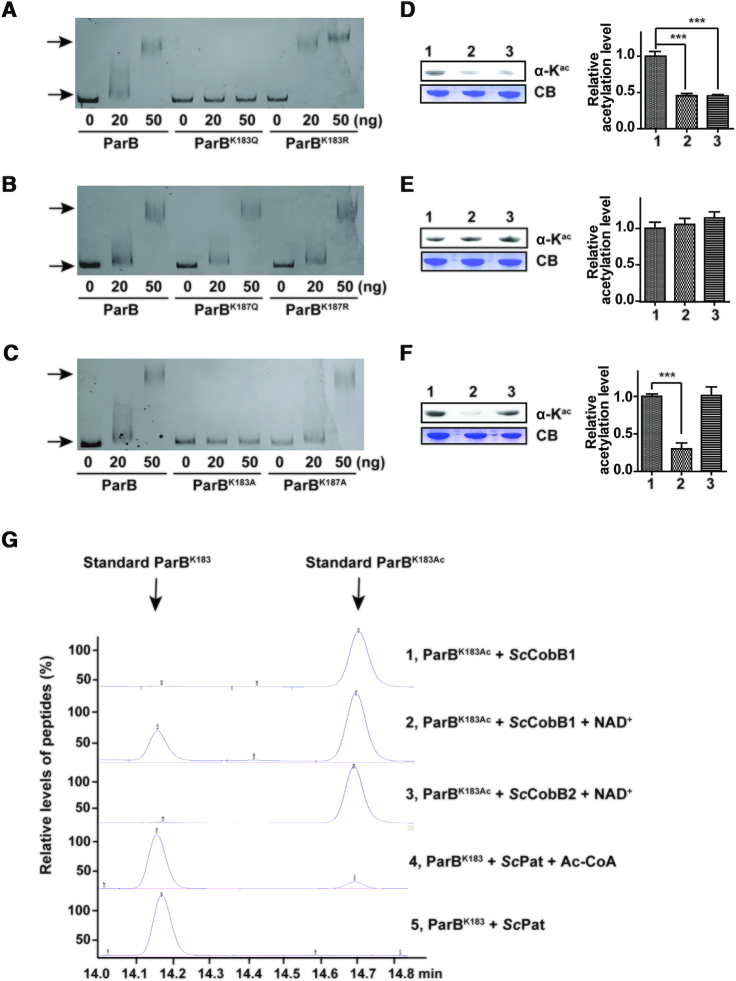
To clarify the regulatory role associated with the acetylation of Lys-183 and Lys-187 in S. coelicolor ParB, the two lysine sites were subjected to site-directed mutagenesis. Previous investigations (20,39) concluded that a Lys to Arg mutation (K to R) retains the positive charge, mimicking a deacetylated lysine, whereas a Lys to Gln mutation (K to Q) abolishes the positive charge, mimicking an acetylated lysine. The corresponding EMSAs were performed to examine the binding affinity of the ParB variants to parS DNA (Figure 4). Compared with wild-type ParB, the acetylated mimic ParBK183Q exhibited a very low DNA-binding affinity to parS, whereas the deacetylated mimic ParBK183R possessed a relatively higher affinity (Figure 4A). Both ParBK187Q and ParBK187R showed similar EMSA patterns to wild-type ParB (Figure 4B). These results indicate that Lys-183 rather than Lys-187 is responsible for the regulation of ParB DNA-binding activity. The mutagenesis of Lys to Ala (K to A) was further performed to generate ParBK183A and ParBK187A. ParBK183A exhibited no DNA-binding affinity, similar to ParBK183Q, while ParBK187A displayed similar affinity to wild-type ParB (Figure 4C). Protein motif analysis showed that Lys-183 but not Lys-187 is located at the tail end of the helix-turn-helix (HTH) DNA-binding motif of ParB (Figure 2A). As previously reported, the HTH motifs in diverse proteins are critical for these proteins to interact with DNA sequences (40,41). Taken together, these observations strongly suggest that Lys-183 is a critical site for ParB DNA-binding activity.
Figure 4.
The Lys-183 residue is the key acetylation site in ParB. (A) Mutation at Lys-183 alters the ParB DNA-binding affinity. An EMSA was performed using purified ParB, ParBK183Q, and ParBK183R as described in Figure 3. (B) The DNA-binding affinity of ParB was unaffected when Lys-187 was mutated. An EMSA was performed using purified ParB, ParBK187Q, and ParBK187R. (C) A lysine to alanine mutation (K to A) confirmed that Lys-183 but not Lys-187 alters the ParB DNA-binding affinity. An EMSA was completed using purified ParB, ParBK183A, and ParBK187A. The acetylation levels of ParB variants were determined by western blotting using a pan anti-acetyllysine antibody. The protein samples analysed in (D), (E) and (F) correspond to the ParB variants in (A), (B) and (C), respectively. One representative result of triplicate measurements is shown. The relative acetylation levels of ParB variants were quantified by ImageJ and normalized against their protein levels (right panels). (G) The regulation of ParB acetylation at Lys-183 was confirmed by HPLC. ParB peptides with an acetylated or non-acetylated Lys-183 residue were synthesized and incubated with ScPat, ScCobB1, or ScCobB2 at 30°C for 2 h. Reactions were terminated by the addition of 1% TFA, after which the samples were analysed by HPLC. Arrows indicate the retention time of the standard acetylated and non-acetylated peptides.
Compared to that of Lys-187 (Figure 4E), mutagenesis at Lys-183 resulted in ParB acetylation level changes (Figure 4D, F), indicating that Lys-183 is the pivotal acetylation site in ParB. However, both ParBK183Q and ParBK183R displayed low acetylation levels compared with those of wild-type ParB (Figure 4D). The relatively low acetylation level of ParBK183Q may have been because the structural change in ParBK183Q is not fully mimic the acetylation status of ParB, which was unrecognized by the specific antibody.
We also monitored the acetylation changes for these ParB variants in response to different treatments (i.e. ScPat or ScCobB1 treatment). In contrast to that of wild-type ParB, the acetylation of either ParBK183Q or ParBK183R was barely affected by ScPat or ScCobB1 treatment (Supplementary Figure S5A). Their DNA-binding affinities were also not changed by the treatments (Supplementary Figure S5B). These results imply that Lys-183 is the sole site in ParB regulated by the ScPat/ScCobB1 system.
With synthesized peptides harbouring the Lys-183 residue of ParB, we were able to recapitulate the acetylation/deacetylation reaction of ParB using an HPLC assay. As shown in Figure 4G, ScCobB1 reacted with the ParBK183Ac peptide and generated the deacetylated peptide ParBK183 in the presence of the cofactor NAD+. In contrast, ScPat acetylated the ParB peptide at the Lys-183 residue and produced a small fraction of ParBK183Ac. Meanwhile, ScCobB2 exhibited no deacetylation activity. The peptide was also treated with 3 mM AcP, but the results suggested that the ParBK183 peptide cannot be acetylated by AcP (Supplementary Figure S6).
ParB is mainly regulated by ScCobB1 deacetylation in vivo
To determine whether ParB is regulated by acetylation in vivo, we constructed monogenic mutants by in-frame knocking out Scpat (ΔScpat), SccobB1 (ΔSccobB1) or SccobB2 (ΔSccobB2) in S. coelicolor. Comparison of the whole protein extracts found that the ΔSccobB1 cells have an obviously increased acetylation level (Supplementary Figure S7). Conversely, ΔScpat has a slightly lower acetylation level than the wild-type control (Supplementary Figure S7). This observation indicates that the sirtuin-like deacetylase ScCobB1 may play a global regulatory role in S. coelicolor acetylation. Similar observations of global regulation by CobB have been reported in S. enterica (23) and M. smegmatis (42).
Based on S. coelicolor PL138, we were able to create another two ParB expression strains in the genetic background of ΔScpat or ΔSccobB1 (see Materials and Methods). For simplicity, we named these two strains PL139 and PL140, respectively. To observe the dynamics of ParB acetylation, ParB-His proteins were isolated at different time points (i.e. 24, 48 and 72 h) from PL138, PL139 and PL140 grown in TSB liquid medium and then subjected to western blot analysis (Figure 5, Supplementary Figure S8). The ParB acetylation level decreased when S. coelicolor PL138 entered into the stationary growth phase after a 48-h cultivation in liquid medium (Figure 5, samples 1, 4 and 7). These observations indicate that deacetylation may also occur in vegetative hyphae. By comparing the acetylation levels among PL138, PL139 and PL140, we observed a significant difference in PL140 (ΔSccobB1) since its ParB acetylation level was sustained and not decreased during the stationary growth phase (Figure 5, samples 3, 6 and 9). These results suggest that ScCobB1 plays a major regulatory role in ParB acetylation in vivo. In contrast, the regulation of ParB by ScPat was not significant most of the time, and it may be functional at only the early logarithmic growth phase (Figure 5, sample 2).
Figure 5.
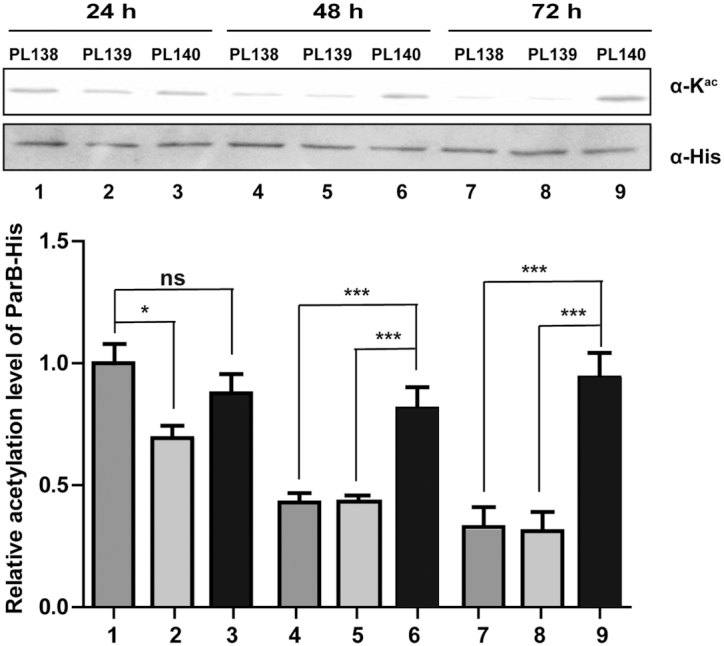
The acetylation level of ParB is regulated by ScCobB1 in vivo. ParB-His proteins were expressed by replacing the native parb with parb-his-aac(3)IV in the chromosomes of PL138 (WT), PL139 (ΔScpat), and PL140 (ΔSccobB1). Proteins were collected at different time points (i.e. 24 h, 48 h, 72 h) from these cells grown in TSB liquid medium. Their acetylation levels were assessed by western blotting using the pan anti-acetyllysine antibody. Compared to that in the WT strain, the in vivo acetylation level of ParB in ΔSccobB1 was sustained rather than being decreased after 48 h of cultivation. The relative acetylation levels of ParB-His proteins were quantified by ImageJ and normalized against their protein levels, which were determined by the anti-His antibody. The value of the first sample was set as 100%. One representative result of two replicate measurements is shown. Mean values with standard deviations are presented.
Deletion of SccobB1 represses the formation of ParB segregation complexes and leads to cell sporulation defects
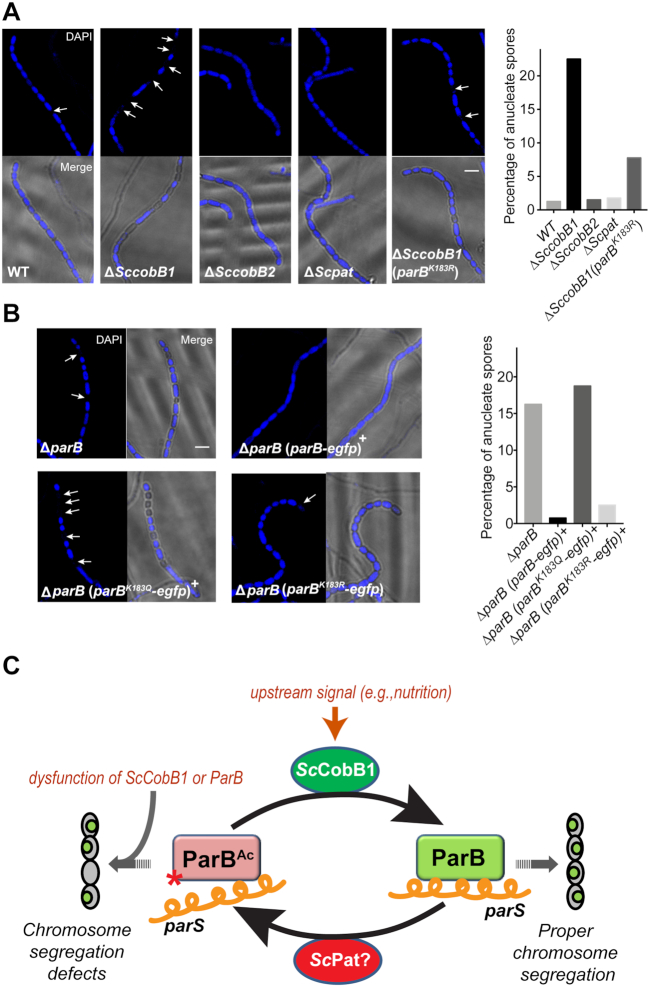
Previous studies suggested that mutations in the ParB DNA-binding motif lead to severe chromosome segregation defects (16). To determine whether the acetylation of ParB affects S. coelicolor chromosome segregation, fluorescence microscopy was employed to detect ParB segregation complex formation by labelling ParB with an enhanced GFP protein. The cells were inoculated onto MS solid agar for observation of chromosome segregation and cell division. Similar to previous reports (18), when wild-type S. coelicolor produced aerial hyphae after 48 h of cultivation, fluorescent ParB-eGFP foci were clearly observed in the cells (Figure 6A). However, no obvious focus was detected in ΔSccobB1 especially in its aerial hyphae (Figure 6B), indicating that the ParB segregation complex had not formed in ΔSccobB1 cells. ParB segregation foci can be visibly observed mainly in aerial hyphae and also in vegetative hyphae (16,43). To avoid missing the segregation foci in ΔSccobB1 cells, we monitored the fluorescence signal for a relatively long period to cover the growth stages from vegetative hyphae to sporulation (i.e. the growth stages from 24 to 72 h). Nevertheless, foci were not detected in ΔSccobB1 at any of these observation points (Supplementary Figure S9). On the other hand, ParB segregation foci were observed in both ΔSccobB2 (Figure 6C) and ΔScpat (Figure 6D), and the focus intensities were similar to those in wild-type S. coelicolor (Figure 6F). The expression level of ParB-eGFP was checked in ΔSccobB1, and we confirmed that it was not significantly different from that in other cells (Supplementary Figure S10). Taken together, these findings are consistent with the in vitro ParB-parS binding results, suggesting that ScCobB1 plays a dominant regulatory role in the formation of ParB segregation complexes in vivo.
Figure 6.
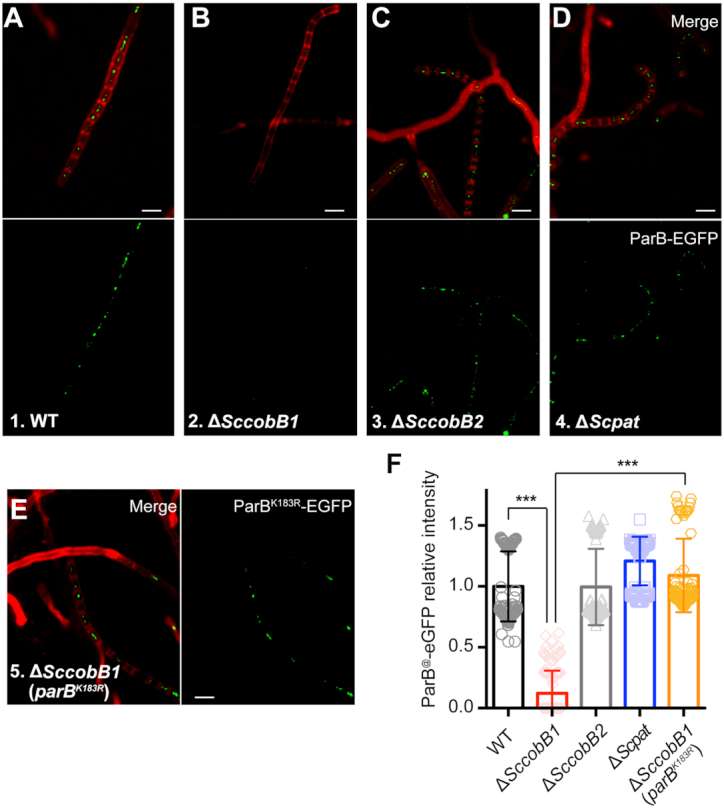
Deletion of SccobB1 represses the formation of ParB segregation complexes. The formation of ParB segregation complexes was analysed by fluorescence microscopy in wild-type (A), ΔSccobB1 (B), ΔSccobB2 (C), ΔScpat (D) and ΔSccobB1 (parBK183R) (E) cells after a 48-h cultivation on MS solid medium. Cell wall staining analyses (red) indicate that these cells were developed the aerial hyphae during this time, probably after aerial hyphae elongation but before spore maturation (53). While few foci were observed at the proximal regions of aerial hyphae and in the vegetative hyphae, ParB-eGFP foci were clearly observed at the distal regions of aerial hyphae. Very few ParB-eGFP foci were detected in ΔSccobB1 (B) compared with other strains. However, obvious foci for ParBK183R-eGFP were detected in ΔSccobB1, indicating the defects of ΔSccobB1 in segregation complex formation were restored. The ParB-eGFP (or ParBK183R-eGFP) focus intensities were further quantified by measuring the average fluorescence signal for at least 150 cells from each sample (F). The values of background-subtracted signal intensities from the 48-h cultivated aerial hyphae were calculated by ImageJ and normalized against the cell numbers. The signal intensity of the wild-type sample was set as 100%. Mean values with standard deviations are presented. All scale bars, 2 μm.
We also observed the formation of ParB segregation complexes using different ParB variants, i.e. the acetylated mimic ParBK183Q-eGFP, the deacetylated mimic ParBK183R-eGFP, and the wild-type ParB-eGFP, in the genetic background of ΔparB. The corresponding cell fluorescence was monitored after 48-h cultivation on MS solid agar (Supplementary Figure S11). Consistent with our in vitro data (Figure 4A), ParBK183R-eGFP formed foci in the aerial hyphae (Supplementary Figure S11) were slightly brighter than those of wild-type ParB-eGFP. In contrast, ParBK183Q-eGFP failed to form any visible foci, even after 72 h of cultivation (Supplementary Figure S11). In contrast to the absence of ParB-eGFP foci in ΔSccobB1, clear foci were detected for ParBK183R-eGFP in ΔSccobB1 (Figure 6E, F). This result suggests that the defects of ΔSccobB1 in segregation complex formation can be restored by the deacetylated mimic ParB, indicating that acetylation regulates chromosome segregation by targeting ParB.
Because chromosome segregation is necessary for proper cell division (16), we asked whether acetylation regulation affects S. coelicolor sporulation. Our scanning electron microscopy analyses indicated that the spores were uneven in size in ΔSccobB1 (Supplementary Figure S1). Similar results were observed in ΔparB in this (Supplementary Figure S1) and in previous reports (16). Therefore, chromosomal DNA of these mutant strains was stained with DAPI for further examination by fluorescence microscopy in this study (Figure 7). A total of 22.50% of spores were anucleate in ΔSccobB1 (Figure 7A), and 16.25% of spores were anucleate in ΔparB (Figure 7B). In contrast, the percentages of anucleate spores in wild-type S. coelicolor, ΔSccobB2, and ΔScpat were apparently lower (< 2%, Figure 7A). These observations confirm that ParB acetylation is involved in sporulation regulation. The proposal was further supported by the measurements of anucleate spores in the parB variant strains in which the parB gene was replaced by parBK183Q-egfp, parBK183R-egfp or parB-egfp (Figure 7B). Similar to that in ΔparB, the abundance of anucleate spores reached up to 18.75% in the acetylated mimic ParB (ParBK183Q-eGFP) strain. In contrast, the cells producing ParBK183R-eGFP, which mimicked the deacetylated ParB, have many fewer anucleate spores, nearly equal to that of the wild-type ParB-producing strains (ParB-eGFP) (Figure 7B). We also compared the sporulation efficiency of ΔSccobB1 (parBK183R-egfp) to the ΔSccobB1 (parB-egfp) cells. The data (Figure 7A) showed that ParBK183R partially restored the sporulation defects of SccobB1 mutant (64%), indicating that Lys-183 acetylation in ParB contributed to the cell sporulation regulation. Taken together, the fluorescence microscopy results suggest that dysfunction in ParB acetylation leads to segregation complex formation disturbances and may result in sporulation defects in S. coelicolor.
Figure 7.
Exorbitant acetylation of ParB leads to defective cell sporulation in S. coelicolor. Cells were harvested after a 96-h cultivation on MS solid medium to analyse their sporulation by fluorescence microscopy. DAPI staining of the chromosome revealed a higher percentage of anucleate spores (indicated with arrows) in ΔSccobB1 (A), ΔparB (B), and the acetylation mimicking ParB-producing (ParBK183Q) strain (B) than those in wild-type S. coelicolor. Moreover, the sporulation defects in ΔparB and ΔSccobB1 were fully (B) or partially (A) recovered by the deacetylation mimicking ParB (ParBK183R), respectively. The proportions of anucleate spores are shown in the right panels (400 spores calculated each). All scale bars, 2 μm. A model for the regulation of ParB acetylation in S. coelicolor is proposed in (C). The ParB binding affinity to the centromere-like parS sequence is repressed by acetylation, which is absolved by ScCobB1 deacetylation that triggered by the proper upstream signals (e.g. nutrition depletion). The exorbitant acetylation due to the dysfunction of ScCobB1 impedes the formation of chromosome segregation complexes and may lead to the generation of anucleate spores. Red star, acetylation at Lys-183 of ParB; orange helical line, parS; small green oval, S. coelicolor chromosome.
DISCUSSION
An increasing number of acetylation proteins have been characterized experimentally in recent years, e.g. acetyl-CoA synthetase (44), glutamine synthetase (45), isocitrate lyase (23) and DNA-binding protein Alba (46). However, most of these proteins are involved in cell metabolism or gene transcription. To the best of our knowledge, no study has reported that protein deacetylation facilitates chromosome segregation. Here, we present our analyses of a DNA-partitioning protein, ParB, whose activity is positively regulated by the sirtuin-like deacetylase ScCobB1 (Figure 7C). Deacetylation at Lys-183 of the HTH DNA-binding motif increases the ability of ParB to bind the centromere sequence, thereby activating ParB complex formation and facilitating cell sporulation (Figure 7C).
Transcriptional regulation of the parAB operon has been reported, and the parB gene is transiently upregulated by its second promoter, parABp2 (18). However, recent S. coelicolor transcriptome analyses revealed that total parB transcription is generally invariable throughout the cell cycle (47), implying that other regulatory mechanism(s) might be involved in chromosome segregation. The acetylation of ParB described here adds another layer to the elaborate regulation of chromosome segregation. Both western blot (Figures 1C) and ParB segregation complex formation (Figures 6) analyses show that deacetylation activates ParB once the sporulation phase is entered (an approximately 48-h cultivation on MS agar, Figures 1 and 6). The cell wall staining results (Figure 6) suggest that S. coelicolor developed aerial hyphae after the 48-h cultivation, and a ladder of uniformly spaced septa interspersed with chromosome segregation foci was observed. An aberrant highly acetylated ParB resulting from deletion of the ScCobB1 gene represses segregation complex formation (Figure 6) and leads to defective cell sporulation (Figure 7).
While the cellular signals of parB transcriptional regulation remain elusive, cells sense the nutrient status to dynamically regulate cellular acetylation (23). When glucose (or acetyl-CoA) is available, the cellular acetylation level is normally high, and cellular acetylation decreases after glucose is depleted (23,48). Interestingly, the initiation of cell sporulation is often triggered by nutritional downshift in Streptomyces (49,50). Hence, the diminishment in cell nutrients likely serves as an environmental signal (51) to increase ParB DNA-binding activity by deacetylation right before sporulation. Moreover, considering that ParB acetylation remains high during vegetative growth phase (Figure 1C), one plausible proposal is that inhibition of ParB may prevent interference by such large protein-DNA complexes on other DNA processes such as transcription. Nevertheless, more studies need to be executed to test these hypotheses. Our ParB work here provides evidence of an intimate connection between acetylation regulation and chromosome segregation.
Lys-183 localizes at the tail end of the second α-helix of the HTH motif (Figure 2A), which usually recognizes the major groove of the DNA double helix (52). The EMSA (Figure 3) results showed that acetylation of Lys-183 decreases the ParB interaction with parS, implying that Lys-183 contributes to the recognition of the parS sequence in vitro. Alignments of ParB sequences from different bacterial species (Supplementary Figure S12) suggest that Lys-183 is conserved among ParB orthologues in most actinobacteria and some representative proteobacteria (e.g. Mycobacterium, Amycolatopsis, Nocardia, and Agrobacterium), indicating a possible conserved regulatory role of acetylation in these organisms. However, this lysine residue is not conserved in the ParB orthologues in Caulobacter and Bacillus (Supplementary Figure S12), suggesting that other possible mechanisms might be employed by these microbes.
Both in vitro and in vivo results suggest that S. coelicolor ParB is dominantly regulated by the deacetylase ScCobB1 (Figures 2, 5 and 6). The in vitro data show that ScCobB1 increases the ParB DNA-binding affinity by ∼2-fold (Figure 3). Given that the ParB molecules need to bind to >20 parS sites in vivo (15–17), we speculate that a very small change in the local ParB concentration could affect the formation of massive segregation complexes. On the other hand, ScPat is the only GNAT family acetyltransferase in S. coelicolor (Figure 2A), our observations suggest that it may function only during the early vegetative growth (Figure 5). Considering that acetylation by ScPat is not very efficient both in vivo and in vitro, we still cannot exclude the possibility that other ParB acetyltransferases exist in the S. coelicolor proteome. Meanwhile, ScCobB2 has desuccinylase activity but no deacetylase activity (37), indicating that it functions as a divergent sirtuin, as in eukaryotic cells (35). Both our results (Supplementary Figure S13) and previous transcriptome data (47) showed that the transcriptional levels of SccobB1 and Scpat were barely changed on solid culture during the cell cycle. Therefore, the temporal availability of cellular co-factors (i.e. NAD+ for ScCobB1 and acetyl-CoA for ScPat) corresponding to nutrient alterations is likely to play a critical regulatory role in ParB acetylation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Zhong-jun Qin for generously providing the cosmids for all the S. coelicolor cloning and PCR targeting. We also appreciate Professor Shi-ming Zhao for the gift of Pan-anti-acetyllysine antibody, Professor Keith F. Chater for the providing of plasmids, Doctor Yi Zhong for the advice in manuscript preparation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2019YFA0904000]; National Natural Science Foundation of China [31400039, 31430004, 31421061]; Funding for open access charge: National Natural Science Foundation of China [31400039].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jallepalli P.V., Lengauer C.. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer. 2001; 1:109–117. [DOI] [PubMed] [Google Scholar]
- 2. Toro E., Shapiro L.. Bacterial chromosome organization and segregation. Cold Spring Harb. Perspect. Biol. 2010; 2:a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Badrinarayanan A., Le T.B., Laub M.T.. Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 2015; 31:171–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Santaguida S., Amon A.. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 2015; 16:473–485. [DOI] [PubMed] [Google Scholar]
- 5. Nielsen H.J., Li Y., Youngren B., Hansen F.G., Austin S.. Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 2006; 61:383–393. [DOI] [PubMed] [Google Scholar]
- 6. Webster M., Witkin K.L., Cohen-Fix O.. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 2009; 122:1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mohl D.A., Easter J. Jr, Gober J.W.. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 2001; 42:741–755. [DOI] [PubMed] [Google Scholar]
- 8. Mierzejewska J., Jagura-Burdzy G.. Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid. 2012; 67:1–14. [DOI] [PubMed] [Google Scholar]
- 9. Schumacher M.A. Bacterial plasmid partition machinery: a minimalist approach to survival. Curr. Opin. Struct. Biol. 2012; 22:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayes F., Barilla D.. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 2006; 4:133–143. [DOI] [PubMed] [Google Scholar]
- 11. Leonard T.A., Butler P.J., Lowe J.. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer - a conserved biological switch. EMBO J. 2005; 24:270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu L., Vecchiarelli A.G., Mizuuchi K., Neuman K.C., Liu J.. Directed and persistent movement arises from mechanochemistry of the ParA/ParB system. PNAS. 2015; 112:E7055–E7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang H., Schumacher M.A.. Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation. Genes Dev. 2017; 31:481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bentley S.D., Chater K.F., Cerdeno-Tarraga A.M., Challis G.L., Thomson N.R., James K.D., Harris D.E., Quail M.A., Kieser H., Harper D. et al.. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002; 417:141–147. [DOI] [PubMed] [Google Scholar]
- 15. Jakimowicz D., Chater K., Zakrzewska-Czerwinska J.. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequenceswithin the origin-proximal region of the linear chromosome. Mol. Microbiol. 2002; 45:1365–1377. [DOI] [PubMed] [Google Scholar]
- 16. Jakimowicz D., Gust B., Zakrzewska-Czerwinska J., Chater K.F.. Developmental-stage-specific assembly of ParB complexes in Streptomyces coelicolor hyphae. J. Bacteriol. 2005; 187:3572–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim H.J., Calcutt M.J., Schmidt F.J., Chater K.F.. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 2000; 182:1313–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jakimowicz D., Mouz S., Zakrzewska-Czerwinska J., Chater K.F.. Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomyces coelicolor. J. Bacteriol. 2006; 188:1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S.C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L. et al.. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006; 23:607–618. [DOI] [PubMed] [Google Scholar]
- 20. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H. et al.. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010; 327:1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chatterjee S., Senapati P., Kundu T.K.. Post-translational modifications of lysine in DNA-damage repair. Essays Biochem. 2012; 52:93–111. [DOI] [PubMed] [Google Scholar]
- 22. Fullgrabe J., Kavanagh E., Joseph B.. Histone onco-modifications. Oncogene. 2011; 30:3391–3403. [DOI] [PubMed] [Google Scholar]
- 23. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y. et al.. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010; 327:1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C.F., Grishin N.V., Zhao Y.. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics: MCP. 2009; 8:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weinert B.T., Scholz C., Wagner S.A., Iesmantavicius V., Su D., Daniel J.A., Choudhary C.. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013; 4:842–851. [DOI] [PubMed] [Google Scholar]
- 26. Hu L.I., Lima B.P., Wolfe A.J.. Bacterial protein acetylation: the dawning of a new age. Mol. Microbiol. 2010; 77:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Glozak M.A., Sengupta N., Zhang X., Seto E.. Acetylation and deacetylation of non-histone proteins. Gene. 2005; 363:15–23. [DOI] [PubMed] [Google Scholar]
- 28. Muller M.M. Post-Translational Modifications of Protein Backbones: Unique Functions, Mechanisms, and Challenges. Biochemistry. 2018; 57:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meek D.W., Anderson C.W.. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 2009; 1:a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tucker A.C., Escalante-Semerena J.C.. Acetoacetyl-CoA synthetase activity is controlled by a protein acetyltransferase with unique domain organization in Streptomyces lividans. Mol. Microbiol. 2013; 87:152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mikulik K., Felsberg J., Kudrnacova E., Bezouskova S., Setinova D., Stodulkova E., Zidkova J., Zidek V.. CobB1 deacetylase activity in Streptomyces coelicolor. Biochem. Cell. Biol. 2012; 90:179–187. [DOI] [PubMed] [Google Scholar]
- 32. Gust B., Challis G.L., Fowler K., Kieser T., Chater K.F.. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl Acad. Sci. U.S.A. 2003; 100:1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hentchel K.L., Escalante-Semerena J.C.. Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev. 2015; 79:321–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Starai V.J., Escalante-Semerena J.C.. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 2004; 340:1005–1012. [DOI] [PubMed] [Google Scholar]
- 35. Hirschey M.D. Old enzymes, new tricks: sirtuins are NAD(+)-dependent de-acylases. Cell Metab. 2011; 14:718–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Colak G., Xie Z., Zhu A.Y., Dai L., Lu Z., Zhang Y., Wan X., Chen Y., Cha Y.H., Lin H.. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell. Proteomics MCP. 2013; 12:3509–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang H., Li P., Ren S., Cheng Z., Zhao G., Zhao W.. ScCobB2-mediated Lysine desuccinylation regulates protein biosynthesis and carbon metabolism in Streptomyces coelicolor. Mol. Cell. Proteomics: MCP. 2019; 18:2003–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weinert B.T., Iesmantavicius V., Wagner S.A., Scholz C., Gummesson B., Beli P., Nystrom T., Choudhary C.. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell. 2013; 51:265–272. [DOI] [PubMed] [Google Scholar]
- 39. Megee P.C., Morgan B.A., Mittman B.A., Smith M.M.. Genetic-analysis of histone-H4 - essential role of lysines subject to reversible acetylation. Science. 1990; 247:841–845. [DOI] [PubMed] [Google Scholar]
- 40. Sanchez A., Rech J., Gasc C., Bouet J.Y.. Insight into centromere-binding properties of ParB proteins: a secondary binding motif is essential for bacterial genome maintenance. Nucleic Acids Res. 2013; 41:3094–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M.. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005; 29:231–262. [DOI] [PubMed] [Google Scholar]
- 42. Hayden J.D., Brown L.R., Gunawardena H.P., Perkowski E.F., Chen X., Braunstein M.. Reversible acetylation regulates acetate and propionate metabolism in Mycobacterium smegmatis. Microbiology. 2013; 159:1986–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kois-Ostrowska A., Strzalka A., Lipietta N., Tilley E., Zakrzewska-Czerwinska J., Herron P., Jakimowicz D.. Unique function of the bacterial chromosome segregation machinery in apically growing Streptomyces - Targeting the chromosome to new hyphal tubes and its anchorage at the tips. PLoS Genet. 2016; 12:e1006488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Starai V.J., Celic I., Cole R.N., Boeke J.D., Escalante-Semerena J.C.. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002; 298:2390–2392. [DOI] [PubMed] [Google Scholar]
- 45. You D., Yin B.C., Li Z.H., Zhou Y., Yu W.B., Zuo P., Ye B.C.. Sirtuin-dependent reversible lysine acetylation of glutamine synthetases reveals an autofeedback loop in nitrogen metabolism. PNAS. 2016; 113:6653–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bell S.D., Botting C.H., Wardleworth B.N., Jackson S.P., White M.F.. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002; 296:148–151. [DOI] [PubMed] [Google Scholar]
- 47. Yague P., Rodriguez-Garcia A., Lopez-Garcia M.T., Martin J.F., Rioseras B., Sanchez J., Manteca A.. Transcriptomic analysis of Streptomyces coelicolor differentiation in solid sporulating cultures: first compartmentalized and second multinucleated mycelia have different and distinctive transcriptomes. PLoS One. 2013; 8:e60665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Choudhary C., Weinert B.T., Nishida Y., Verdin E., Mann M.. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014; 15:536–550. [DOI] [PubMed] [Google Scholar]
- 49. Daza A., Martin J.F., Dominguez A., Gil J.A.. Sporulation of several species of Streptomyces in submerged cultures after nutritional downshift. J. Gen. Microbiol. 1989; 135:2483–2491. [DOI] [PubMed] [Google Scholar]
- 50. Chater K.F. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex. Curr. Opin. Microbiol. 2001; 4:667–673. [DOI] [PubMed] [Google Scholar]
- 51. Pietrocola F., Galluzzi L., Bravo-San Pedro J.M., Madeo F., Kroemer G.. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015; 21:805–821. [DOI] [PubMed] [Google Scholar]
- 52. Wharton R.P., Brown E.L., Ptashne M.. Substituting an alpha-helix switches the sequence-specific DNA interactions of a repressor. Cell. 1984; 38:361–369. [DOI] [PubMed] [Google Scholar]
- 53. Donczew M., Mackiewicz P., Wrobel A., Flardh K., Zakrzewska-Czerwinska J., Jakimowicz D.. ParA and ParB coordinate chromosome segregation with cell elongation and division during Streptomyces sporulation. Open Biol. 2016; 6:150263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.