Abstract
Flaviviruses, including dengue virus and Zika virus, contain a single-stranded positive sense RNA genome that encodes viral proteins essential for replication and also serves as the template for new genome synthesis. As these processes move in opposite directions along the genome, translation must be inhibited at a defined point following infection to clear the template of ribosomes to allow efficient replication. Here, we demonstrate in vitro and in cell-based assays that the viral RNA polymerase, NS5, inhibits translation of the viral genome. By reconstituting translation in vitro using highly purified components, we show that this translation block occurs at the initiation stage and that translation inhibition depends on NS5-RNA interaction, primarily through association with the 5′ replication promoter region. This work supports a model whereby expression of a viral protein signals successful translation of the infecting genome, prompting a switch to a ribosome depleted replication-competent form.
INTRODUCTION
Members of the Flaviviridae, including dengue virus (DENV), Zika virus (ZIKV), West Nile virus (WNV) and yellow fever virus are some of the most important emerging and re-emerging pathogens (1–3). Habitat range expansion of their mosquito vectors has resulted in increasing human populations at risk of infection. Although typically self-limiting, flavivirus infection can cause fatal complications such as haemorrhagic fever, while ZIKV infection in pregnant women has been linked with neonatal microcephaly (4).
Flaviviruses possess single-stranded positive sense RNA genomes, ∼11 kb in length, that have a 5′ cap structure similar to cellular mRNAs but lack a 3′ polyA tail (5). A long open reading frame, flanked by highly structured 5′ and 3′ untranslated regions (UTR), encodes structural proteins required for virus assembly and non-structural proteins involved in replication of the viral genome (Figure 1A). Following release into the cytoplasm the genome is translated as a long polyprotein that is cleaved by viral and cellular proteases to produce mature viral proteins. The viral genome is then copied in a 3′-5′ direction by the virally encoded RNA-dependent RNA polymerase (RdRp). This negative-sense strand, in turn, serves as a template for new positive-strand synthesis for further translation or packaging into progeny virions. As translation and replication inherently move in opposite directions, viral genome usage must be tightly controlled. However, the mechanism of so-called ‘lifestyle-switching’ between translation and replication of the flaviviral genome is not well understood.
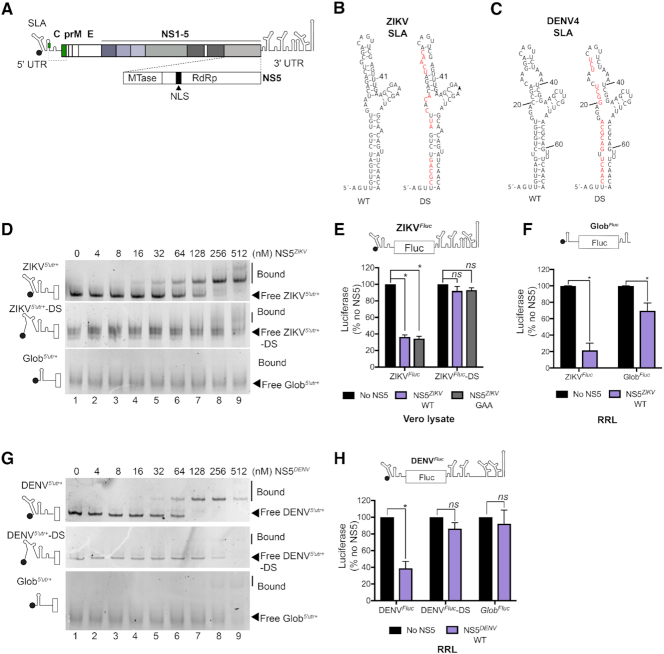
Figure 1.
NS5 inhibits translation. (A) Schematic of the ZIKV genome. The start codon of the long open reading frame is a green box. C, capsid sequence, partially shown as a solid line to show conserved RNA structure, and in green, full capsid sequence represented by the dotted line. prM, precursor membrane; E, envelope; NS, non-structural; SLA, stem-loop A; nuclear localization sequence (NLS). (B) Nucleotide sequence of ZIKV PE243 SLA. SLA was mutated to destabilize (DS) SLA by replacing the wildtype sequence with the red nucleotides shown in the right panel. (C) Nucleotide sequence of DENV4 SLA. Mutations introduced to destabilize (DS) DENV4 SLA are indicated in red in the right panel. (D) EMSA of the first 359 nt of wildtype (ZIKV5′utr+; upper panel) or SLA-destabilized (ZIKV5′utr+-DS; middle panel) ZIKV RNA or Glob5′utr+ RNA (all shown schematically on left) with increasing concentrations of NS5ZIKV. Free and bound RNA are indicated. (E) Luciferase production after incubation of ZIKVFluc (schematic on top) RNA in Vero lysate in the absence or presence of 256 nM NS5ZIKV. (F) Luciferase production after incubation of ZIKVFluc RNA or GlobFluc RNA (schematic on top) in the absence or presence of 256 nM NS5ZIKV in rabbit reticulocyte lysate (RRL). (G) EMSA of the first 400 nt of wildtype (DENV5′utr+; upper panel) or SLA-destabilized (DENV5′utr+-DS; middle panel) DENV4 RNA or Glob5′utr+ RNA (all shown schematically on left) with increasing concentrations of NS5DENV. Free and bound RNA are indicated. (H) Luciferase production after incubation of DENVFluc RNA (schematic on top) or GlobFluc RNA in the absence or presence of 256 nM NS5DENV in RRL. (E, F and H) Values, normalized to the no polymerase control, are mean +/− SEM from three independent experiments. Statistical significance was determined by a Student's unpaired t-test with significant values indicated with an asterisk. *P<0.05. **P<0.001.
The flavivirus non-structural protein 5 (NS5) is the largest mature viral protein expressed during infection (Figure 1A). It harbours an N-terminal methyltransferase (MTase) domain responsible for generating the cap1 structure at the 5′ end of newly synthesized viral genomes which promotes translation (6) and shields viral RNA from immune detection (7,8). The MTase domain is separated by a short linker from a C-terminal RdRp domain that adopts a characteristic right-handed fingers-palm-thumb fold (9–13). NS5 binds to a conserved RNA structural feature, stem loop A (SLA), at the 5′ extremity of the flaviviral genome (14–16) which serves as a promoter for viral replication (14). Flavivirus genome replication critically depends on hybridization between complementary RNA sequences in the 5′ and 3′ ends (6,17–20). Hybridization of these sequences, termed the upstream of AUG region (UAR), the downstream of AUG region (DAR) and cyclization sequence (CS), is thought to facilitate transfer of NS5 bound to SLA to the 3′ end of the genome to initiate replication (14,21). Recently, we demonstrated that the large-scale structural rearrangements around the translation start site that accompany 5′-3′ hybridization disrupt flavivirus genome translation (6), promoting a replication-competent state.
Aside from its main roles as the viral RdRp and MTase, the flavivirus NS5 protein has been ascribed other functions that promote viral replication. The best characterized of these is the role of NS5 in disrupting host immune signalling pathways (reviewed in 22). DENV and ZIKV NS5 both target signal-transducer and activator of transcription (STAT)2 for proteasomal degradation (23–25), disrupting interferon signalling and stalling the cellular antiviral response. NS5 from different flaviviruses has also been reported to enter the nucleus due to the presence of a well conserved nuclear localization sequence (NLS) (26–29). Although the role and importance of nuclear localization remains to be clarified (30), DENV NS5 was reported to interact with components of the U5 snRNP complex and interfere with splicing during infection (31).
Here, we investigated the interplay between the NS5/SLA interaction and flaviviral genome translation. Using in vitro and cell-based translation experiments, we demonstrate that NS5 binding to SLA directly blocks translation of the viral genome. This inhibition occurs at the translation initiation stage as determined by detailed in vitro reconstitution analysis. Our findings support a model whereby, following initial rounds of translation, recruitment of newly synthesized NS5 to SLA inhibits viral protein synthesis, priming the viral RNA for replication.
MATERIAL AND METHODS
Plasmids and reagents
A pCC1BAC vector containing the open reading frame of the ZIKV BeH819015 isolate flanked by the 5′ and 3′ UTRs of the ZIKV PE243 isolate and bearing an inline duplicate copy of the capsid protein fused to a Nluc or mCherry gene and 2A peptide sequence (32) were kindly provided by Andres Merits. The existing SP6 promotor in the Nluc containing plasmid was replaced with a T7 promoter as previously described (6). A plasmid containing the first 359 nt of ZIKV PE243 isolate (ZIKV5′utr+) was previously described (6). A plasmid expressing Fluc flanked by the 5′ and 3′ regions of ZIKV PE243 isolate (ZIKVFluc) was previously described (33). ZIKV RNA expressing plasmids with mutations in SLA (ZIKV5′utr+-DS, ZIKVFluc-DS) were generated by site directed mutagenesis. Gene fragments containing the first 400 nt of DENV4 (DENV5′utr+) or Fluc flanked by the first 148 nt and complete 3′ UTR of DENV4 (DENVFluc) bearing a 5′ T7 promoter and EcoRI and XhoI sites were synthesized by Integrated DNA Technologies (IDT) and cloned into pUC57. GeneBank number FJ196850 was used for the DENV4 sequence. DENV RNA expressing plasmids containing mutations in SLA (DENV5′utr-DS, DENVFluc-DS) were also synthesized by IDT and cloned as described for the wild type sequences. A plasmid containing Fluc flanked by the 5′ and 3′ UTRs of human globin was previously described (8). A plasmid containing the 5′ and 3′ regions of ZIKV PE243 joined by a short linker sequence (ZIKVmini) and a mutant version in which the 3′ UAR sequence was replaced with the 5′ UAR sequence (ZIKVmini- Δ3′ UAR) were previously described (6). The tRNAiMet transcription vector has been described (34). Recombinant human eIF expression plasmids have been described previously: eIF1 and eIF1A (35), eIF4A and eIF4B (36) and methionyl-tRNA synthetase (37). Wildtype and GAA mutant ZIKV NS5 sequences were amplified by PCR from the full-length ZIKV or full-length ZIKV-NS5-GAA plasmids previously described (6). The full length or amino acids 270–900 comprising the polymerase domain of ZIKV NS5 coding region was subcloned into pET28b (Novagen) between NheI and HindIII to generate a bacterial expression construct for NS5ZIKV with an N-terminal His6-tag or into pcDNA 3.1+ (ThermoFisher) between NheI and NotI with an N-terminal FLAG tag for mammalian cell expression. A gene fragment encoding NS5 from DENV4 (GeneBank number FJ196850), codon optimized for expression in E. coli was synthesized by IDT and cloned into pET28b between NheI and HindIII to generate a bacterial expression construct for NS5DENV with an N-terminal His6-tag. Hippuristanol was generously shared by Jerry Pelletier.
In vitro transcription
Plasmids were linearized with HindIII (ZIKV5′utr+, ZIKVFluc and ZIKVmini), EcoRV (DENV5′utr+ and DENVFluc), AgeI (ZIKVNluc, ZIKVmCherry), FspI (GlobFluc) or BstN1 (tRNAiMet). The globin 5′ UTR and first 97 nt of Fluc with a T7 promoter sequence were amplified from GlobFluc by PCR to generate a double stranded template to transcribe Glob5′utr+ RNA. ZIKVmCherry RNA was transcribed using the SP6 RiboMax transcription kit (Promega). All other RNAs were transcribed with recombinant T7 polymerase (50 ng/μl) in buffer containing 40 mM HEPES pH 7.5, 32 mM MgOAc, 40 mM DTT, 2 mM Spermidine, 10 mM each NTP and 0.2 U/μl RNaseOUT (Invitrogen) for 2 h at 37°C. ZIKVNluc and ZIKVmCherry RNA were purified using TRI Reagent (Sigma) before ethanol precipitation. All other transcription reactions were treated with DNaseI and RNA was extracted with acidic phenol/chloroform and ethanol precipitated. Residual nucleotides were removed with Illustra MicroSpin G-50 columns (GE Healthcare). RNA was capped using the ScriptCap system (CellScript).
Bacterial protein expression and purification
Recombinant His-tagged NS5ZIKV and NS5DENV were expressed in Rosetta 2 (DE3) pLysS Escherichia coli (Novagen). Cells were grown to an OD600 of 0.6 in 2 × TY media at 37°C. Expression was induced by adding 0.5 mM isopropyl β-d-1-thiogalactopyranoside. The induced culture was incubated at 20°C for 16 h. Cells were harvested and lysed in a buffer containing 20 mM Tris pH 7.5, 400 mM KCl, 5% glycerol, 1 mM DTT and 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mg/ml lysozyme (from hen egg) and 20 mM imidazol. His-tagged proteins were isolated by affinity chromatography on Ni-NTA Agarose beads (Qiagen) and additionally purified by FPLC on a Superdex 200 Increase 10/300 GL size exclusion column (GE Healthcare) in 20 mM Tris pH 7.5, 300 mM KCl, 5% glycerol and 1 mM DTT.
Vero lysate preparation
Vero cell lysate were prepared as described (38) with some minor modifications. Briefly, Vero cells (ATCC) were grown in T150 flasks until 80% confluency, harvested and washed twice with PBS and a short wash with a hypotonic lysis buffer (10 mM HEPES, pH 7.5, 10 mM KOAc, 0.5 mM MgOAc, 5 mM DTT and EDTA free protease inhibitor cocktail (ROCHE)). The cells were subsequently lysed in 2 ml lysis buffer by mechanically passaging in a 27G needle on ice at least 20 times. The lysate was cleared by centrifugation at 10 000 × g for 5 min. 0.75 mM CaCl2 and 15 U/ml of micrococcal nuclease was added for 10 min at 25°C before 3 mM EGTA was added to inhibit nuclease activity. The final lysate was stored at −80°C.
In vitro translation and luciferase measurement
In vitro translation was performed using the Flexi RRL System (Promega). For Vero cell lysate translation reactions, 5 μl of Vero lysate prepared as in (38) was supplemented with 1.6 mM HEPES, pH7.6, 2 mM creatine phosphate, 0.01 μg μl−1 creatine kinase, 0.01 mM spermidine, 100 mM KCl, 2 mM MgOAc, 4 mM DTT, 25 μM amino acids (Promega). After 90 min at 30°C, reactions were terminated by addition of 50 volumes of passive lysis buffer (Promega) before luciferase signal was measured by GloMax (Promega). Luciferase values were normalized to the no NS5 control for each experiment.
48S complex assembly
In vitro reconstitution of 48S complex assembly was performed as previously described (6). 0.2 pmol RNA was incubated with the indicated eIFs (2 pmol 40S subunit, 4 pmol Met-tRNAiMet, 4 pmol eIF2, 3 pmol eIF3, 10 pmol eIF4A, 5 pmol eIF4B, 2.5 pmol eIF4F, 10 pmol eIF1, 10 pmol eIF1A) at 37°C for 10 min in a reaction volume of 20 μl Buffer A (20 mM Tris pH 7.5, 100 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 0.25 mM spermidine, 1.6 U/μl RNaseOUT (Invitrogen), 0.4 mM guanosine triphosphate (GTP) and 2 mM adenosine triphosphate (ATP)). Purified NS5ZIKV was included at the concentrations described in the figure legends. Assembled complexes were analysed by primer extension inhibition using 2.5 U avian myeloblastosis virus reverse transcriptase (Promega) in the presence of 32P-labelled primer, 8 mM MgCl2 and 0.5 mM dNTPs. cDNA products were phenol/chloroform extracted and ethanol precipitated before being resolved on denaturing 6% polyacrylamide sequencing gels and detected by autoradiography using an FLA7000 Typhoon scanner (GE).
Electrophoresis mobility shift assay
In vitro transcribed capped RNA (8.6 pmol) was heated at 75°C and snap cooled on ice in the presence of RNA refolding buffer (50 mM Tris, pH 7.5, 100 mM KCl and 5 mM MgCl2). RNA (860 fmol) was subsequently incubated with the indicated proteins (at a final concentration range of 0 nM to 512 nM) at 30°C for 15 min in a reaction volume of 10 μl containing Buffer A supplemented with 5 μg BSA (Sigma) and 2 μg yeast tRNA (Ambion). Reaction mixtures were preincubated at 30°C for 5 min prior to RNA addition. Following addition of 10× native RNA loading dye (0.05% bromophenol blue, 0.05% xylene cyanol FF, 50% glycerol), reactions were analysed by native PAGE on 0.5× TBE, 5% polyacrylamide gels containing 5% glycerol on ice at 4°C. Gels were stained with 1 μg/ml ethidium bromide in 0.5× TBE for 30 min prior to visualization using a UV transilluminator. Image analysis was performed using ImageJ.
Immunofluorescence microscopy
In a six-well plate, 1 × 106 Vero cells were transfected using lipofectamine 2000 (Invitrogen) in 0.2 ml Opti-MEM (Gibco) with 200 fmol FLAG-NS5ZIKV plasmid and fixed at the indicated time points with 4% PFA for 15 min and washed with PBS. Fixed cells were permeabilized by treating with 0.2% (v/v) Triton X-100/PBS for 5 min at room temperature before washing three times with PBS and then blocked by incubation with 5% (w/v) milk in PBS/0.1%Tween-20. Primary antibody against FLAG (M2 Mouse, Sigma, F1804), and secondary antibody was coupled to Alexa-Fluor 488 (Invitrogen). Cell nuclei were stained using DAPI (Sigma, 1:10 000 in water). Immunofluorescence microscopy was performed on an EVOS FL Auto 2 (Invitrogen).
Viral genome nanoluciferase translation measurement
Capped, in vitro transcribed ZIKVNluc RNA (1.5 pmol) was electroporated into 3 × 106 Vero cells suspended in 100 μl of Opti-MEM (Gibco) using a NEPA21 electroporator (Nepagene). Cells were seeded sub-confluently in 24-well plates and samples were harvested for luciferase assays and RT-qPCR analysis at the indicated time points by washing with phosphate-buffered saline and lysis in passive lysis buffer (Promega). Nanoluciferase activity was measured using the Nano-Glo luciferase assay system (Promega) by GloMax (Promega).
Virus production and titration
The capped ZIKVmCherry RNA was transfected in Vero cells using lipofectamine 2000 (Invitrogen) in 0.2 ml Opti-MEM (Gibco). 24 h post-transfection the cells were washed and supplemented with fresh media and monitored daily until significant cytopathic effect was observed, at which point supernatants were harvested, centrifuged at 1000 × g for 10 min to remove cellular debris and aliquots stored at −80°C. Virus was then titred by plaque assay. 100 μl of tenfold serial dilutions of virus samples were added to a six-well plate containing a confluent monolayer of Vero cells and incubated for 1 h with shaking after every 15 min. The cells were overlaid with 1 ml of DMEM supplemented with 2% FBS and 1% final concentration of low melting agarose (Sigma). After 4–7 days of incubation, the cells were fixed with 2% formaldehyde and stained with toluidine blue. Plaques were counted and viral titres in plaque forming units per ml calculated.
mCherry virus detection- fluorescence microscopy and flow cytometry
In a six-well plate, 1 × 106 Vero cells were transfected using lipofectamine 2000 (Invitrogen) in 0.2 ml Opti-MEM (Gibco) with 200 fmol of N-terminal FLAG NS5ZIKV or empty vector prior to infection with ZIKVmCherry virus (MOI:10). The cells were incubated with virus for 45 min and then washed twice with PBS and once with DMEM to remove the unadsorbed virus. 2 ml of complete medium was added and cells were incubated for 72 h. For the 0 h post infection (HPI) sample, cells were washed three times with PBS to remove the unadsorbed virus and harvested directly by treating with Accutase (BioLegend), washed again and resuspended in PBS. Cells were fixed with 2% paraformaldehyde (PFA) for 15 min. For other timepoints, cells were washed three times with PBS, harvested and fixed as above. For all time points after washing with PBS but before cell harvest, fluorescence microscopy was performed to monitor mCherry expression on an EVOS FL Auto 2 (Invitrogen). For quantification of mCherry signal, fixed cells were resuspended in PBS and analysed by flow cytometry using a BD Biosciences LSR Fortessa analyser and data was analysed using FACSDIVA software (BD Biosciences). A minimum of 10 000 cells were collected and analysed for each sample in three independent experiments.
Puromycylation assay
In a six-well plate, 1 × 106 Vero cells were transfected with 500 fmol NS5 expression plasmids or empty pCDNA3.1, using lipofectamine 2000 (Invitrogen) in 0.2 ml Opti-MEM (Gibco). After 1 h, cells were overlaid with 0.8 ml antibiotic-free DMEM then, after 7 h, 1 ml of media was added containing 10 μg/ml puromycin (5 μg/ml final concentration). Cells were harvested after 2 h in passive lysis buffer (Promega) for analysis by immunoblotting. Membranes were blocked in 5% milk PBST before probing with anti-puromycin (kindly shared by Prof. Ian Goodfellow, University of Cambridge—1:20 in Odyssey blocking buffer, LiCor), then anti-tubulin (1:1000 in 5% BSA PBST, Abcam ab-6046), visualized using IRDye 800CW goat anti-mouse and IRDye 680RD goat anti-rat secondary antibodies (LiCor), respectively. Protein signals were detected on an Odyssey CLx Imaging System (Li-Cor). Puromycylation signal was quantified using ImageJ.
RESULTS
ZIKV and DENV NS5 inhibit translation
To investigate the effect of NS5 on viral translation, we bacterially expressed and purified His-tagged full-length ZIKV NS5 (NS5ZIKV) (Supplementary Figure S1) and confirmed specific interaction with ZIKV SLA using an electrophoretic mobility shift assay (EMSA). NS5ZIKV bound an RNA fragment containing the first 359 nt of the ZIKV genomic RNA, encompassing the 5′ UTR and first 252 nt of the capsid sequence (ZIKV5′utr+), with nanomolar affinity (Figure 1D, upper panel). This affinity is consistent with that previously reported for DENV NS5 and DENV SLA (14). In contrast, NS5ZIKV failed to shift an RNA (Figure 1D, middle panel) mutated so as to destabilize SLA (ZIKV5′utr+-DS), as shown in Figure 1B. We next examined the impact of the NS5ZIKV/SLA interaction on translation in vitro. An in vitro transcribed and capped mRNA, comprising the first 158 nt of the ZIKV genomic RNA (contains SLA, SLB, cHP and 5′CS) followed by firefly luciferase (Fluc) in frame with the authentic viral translation start site and the complete ZIKV 3′ UTR (ZIKVFluc, Figure 1E top schematic), was incubated with NS5ZIKV in a translation-competent Vero cell lysate. Translation was monitored by measuring luminescence from the luciferase reporter and normalized to the no polymerase control reaction. Both wildtype NS5ZIKV, and a mutant bearing a G664DD → G664AA substitution (NS5ZIKV-GAA) that disrupts the polymerase active site, reduced translation of ZIKVFluc RNA (by 64% and 66%, respectively; Figure 1E). In contrast, addition of either protein failed to inhibit translation of the ZIKVFluc-DS RNA in which SLA is mutated (as in Figure 1B), suggesting that the interaction between NS5 and SLA, but not the polymerase activity, is important for the translation inhibition observed. NS5ZIKV inhibited ZIKVFluc RNA translation in rabbit reticulocyte lysate (RRL) to the same extent as the Vero lysate (Figure 1F) and so RRL was used for subsequent experiments.
We next examined if the translation inhibitory effect of NS5 is conserved in different flaviviruses. We expressed and purified His-tagged full-length DENV NS5 (NS5DENV) (Supplementary Figure S1) and confirmed its ability to bind to DENV RNA in an EMSA experiment (Figure 1G). Like NS5ZIKV, NS5DENV bound DENV5′utr+ RNA with low nanomolar affinity (Figure 1G, upper panel) but did not bind a mutated version of DENV5′utr+ (Figure 1G, middle panel) in which destabilizing mutations were introduced in SLA (DENV5′utr+-DS) as shown in Figure 1C. NS5DENV inhibited translation of a reporter RNA bearing the DENV SLA, SLB and cHP with an in frame Fluc reporter and the full DENV 3′ UTR (DENVFluc, Figure 1H, upper schematic) but not a SLA destabilized mutant RNA (DENVFluc-DS).
In EMSA experiments, NS5ZIKV only weakly associated with a short RNA fragment containing the 5′ UTR of human globin (Glob5′utr+) at the highest concentrations tested (Figure 1D, lower panel), while no interaction between NS5DENV and Glob5′utr+ was detected (Figure 1G, lower panel). Consistently, NS5ZIKV and NS5DENV had a smaller inhibitory effect on the translation of a reporter RNA bearing the 5′ and 3′ UTRs of human globin (GlobFluc, Figure 1F, upper schematic) than on the corresponding viral 5′ UTR containing Fluc reporter RNA (Figure 1F and H). The weak but significant inhibition of GlobFluc translation in the presence of NS5ZIKV (Figure 1H) may result from addition of high concentrations of an RNA-binding protein to the reaction mixture, although this effect was not observed in the presence of NS5DENV (Figure 1H). These results demonstrate that NS5 can specifically inhibit translation in a manner consistent with its ability to bind SLA RNA.
NS5 inhibits translation at the initiation stage
As NS5 binds to the highly conserved SLA at the 5′ extremity of the flaviviral genomic RNA, we hypothesized that it may interfere with translation factor recruitment and inhibit translation at the initiation stage. We recently reported an in vitro reconstitution method for examining flavivirus cap-dependent translation initiation (6). Using this system (39), individually purified small ribosomal subunits, eukaryotic translation initiation factors (eIFs) and initiator tRNA are combined to study the role of specific factors in regulating translation of a particular RNA. During translation initiation the 40S ribosomal subunit and associated eIFs are recruited to the 5′ cap of the mRNA (40). After ribosomal scanning of the 5′ UTR and translation start site selection, a stable 48S complex is assembled at the initiation site. Following start codon recognition, the 48S complex is poised for 60S ribosomal subunit joining and translation initiation. 48S complex assembly in the in vitro reconstitution system is monitored by detection of truncated cDNA products on a denaturing PAGE gel caused by the inhibition of reverse transcriptase (RT). The position of RT stops on the RNA are determined by comparison to a Sanger sequencing reaction. 48S complex assembly results in the appearance of truncated cDNA products +15–17 nt downstream of the start codon (shown schematically in Figure 2A). The secondary structure of the ZIKV 5′ region is shown in Figure 2B.
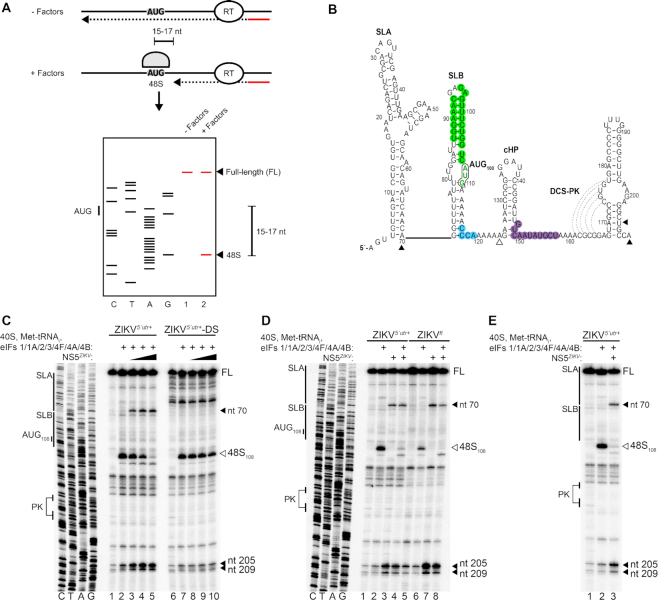
Figure 2.
NS5ZIKV inhibits translation at the initiation stage. (A) Schematic of the in vitro reconstitution and toeprinting assay described in the text (from 6). (B) Nucleotide sequence and secondary structure of the ZIKV 5′ region. The start codon AUG108, (green font and black outline), UAR (green), DAR (blue) and 5′ CS (purple) are highlighted. Dotted lines indicate base pairing in the downstream of CS pseudoknot (DCS-PK). (C–E) Toeprinting analysis of 48S complex assembly on (C) capped ZIKV5′utr+ or ZIKV5′utr+-DS RNA, (D) capped ZIKV5′utr+ or full-length (ZIKVfl) RNA and (E) capped ZIKV5′utr+, in the presence of the indicated factors. The start codon and/or structural elements are labelled on the left. Toeprints caused by 48S complex assembly (open arrowhead) or NS5ZIKV binding (closed arrowhead) are marked on the right and also indicated in (B). NS5ZIKV was preincubated with the RNA before addition to the reconstitution reaction (C and D) or added to the reconstitution reaction at the same time as the translation factors (E). (C) NS5ZIKV was included at 64 nM (lanes 3 and 8), 128 nM (lanes 4 and 9) or 256 nM (lanes 5 and 10). (D) NS5ZIKV was included at 256 nM in the indicated lanes. FL, full length.
Addition of small ribosomal subunits, the canonical eIFs (1,1A, 2, 3, 4F, 4A, 4B) and initiator tRNAi to ZIKV5′utr+ resulted in a strong RT stop +16–17 nt downstream of AUG108, the start codon of the long viral-RNA open reading frame, characteristic of 48S complex assembly at this site (Figure 2C, compare lanes 1 and 2). Preincubation of ZIKV5′utr+ RNA with NS5ZIKV before the addition of eIFs led to a dose dependent decrease in 48S complex assembly (Figure 2C, compare lane 2 to lanes 3–5). As a control addition of the eIF4A inhibitor hippuristanol caused a similar disappearance of the 48S dependent RT stop (Supplementary Figure S2). In contrast, preincubation of the SLA destabilized ZIKV5′utr+-DS RNA with the same concentrations of NS5ZIKV had a much weaker inhibitory effect on 48S complex assembly (Figure 2C, compare lane 7 to lanes 8–10), consistent with our in vitro translation data (Figure 1E). As apparent in Figure 2C, there is a difference in the RT arrest pattern in the region corresponding to SLA between the wild type and destabilized SLA RNAs providing evidence of a change in the RNA structure. This change is evident in the RNA only lanes as well as lanes containing translation factors and/or NS5. In contrast, no changes in the RT arrest pattern was observed in the rest of the RNA indicating that gross structural changes introduced by the SLA destabilizing mutations were restricted to the SLA region. NS5ZIKV preincubation also inhibited 48S complex formation on capped, full-length genomic ZIKV RNA (Figure 2D, compare lanes 6 and 8). To determine if NS5ZIKV could directly compete with the translation initiation machinery we performed the in vitro reconstitution experiment without prior incubation of the polymerase and the template RNA. As shown in Figure 2E, NS5ZIKV efficiently inhibits 48S complex assembly on ZIKV5′utr+ RNA without prior incubation.
Inclusion of NS5ZIKV to the reconstitution system caused increased RT arrest at two positions independently of the translation components (Figure 2D, compare lane 1 to lanes 3 and 4). The first, at nt 71, is adjacent to the 3′ boundary of SLA, and the second at nt 205/209 occurs at the 3′ boundary of the first stem following the cHP in the capsid coding region. This stem forms part of the downstream of 5′ CS pseudoknot (DCS-PK) (Figure 2C, compare lane 2 to lanes 3–5). Disruption of the DCS-PK was previously reported to affect genome circularization and replication in DENV (41). Our data are consistent with previous results from WNV (15), indicating that a second binding site is conserved between at least some flaviviruses. Interestingly, competition EMSAs with a truncated WNV 5′ region lacking SLA indicated that the second site of NS5 interaction was of lower affinity as it could not outcompete binding to a SLA-containing RNA (15). Consistent with this, NS5ZIKV did not induce RT arrest at nucleotides 205/209 in the ZIKV5′utr+ RNA in which the higher affinity SLA binding site was mutated (Figure 2C, compare lanes 5 and 10).
Although the mode of interaction between NS5 and SLA remains to be fully elucidated, previous studies have demonstrated that differing regions of NS5 contribute to interaction with SLA depending on which virus is being examined. For example, the polymerase domain of NS5DENV was reported as sufficient to bind SLA RNA in EMSA studies (14,21) whereas the polymerase domain of NS5 from WNV was not sufficient to interact with SLA (15). We generated a truncated His-tagged version of ZIKV NS5, NS5ZIKV-Pol containing amino acids 270–900, that includes the polymerase domain but lacks the methyl transferase domain. Consistent with the findings from WNV (15), the polymerase domain of NS5ZIKV did not associate with SLA containing RNA as examined by EMSA (Supplementary Figure S3A). NS5ZIKV-Pol did not inhibit 48S complex assembly when included in the in vitro reconstitution system and also failed to induce the appearance of the NS5ZIKV dependent RT arrest observed with the full-length protein (Supplementary Figure S3C). These results are consistent with a requirement for NS5 RNA binding to inhibit translation.
NS5 inhibits translation of the viral genome in cells
To test the translation inhibitory activity of NS5 in cells we generated an N-terminally FLAG-tagged NS5ZIKV (FLAG-NS5ZIKV) expression construct and examined its effect on translation of a viral reporter RNA. To promote cytoplasmic retention of our overexpressed NS5 so that it could encounter the transfected reporter RNA, we introduced point mutations in the NLS (42) of NS5ZIKV (Lys390 and Arg393 to Ala) to generate FLAG-NS5ZIKV-NLS. In this background, we also mutated the active site G664DD → G664AA to minimize aberrant RNA replication activity in the FLAG-NS5ZIKV-NLS/GAA construct. Expression of FLAG-NS5ZIKV-NLS and FLAG-NS5ZIKV-NLS/GAA in Vero cells was determined by immunofluorescence following staining with an anti-FLAG antibody over the time course shown in Figure 3A. At 10 h post transfection, accumulation of NS5 in the cytoplasm was clearly visible and so we chose this time point for further experiments.
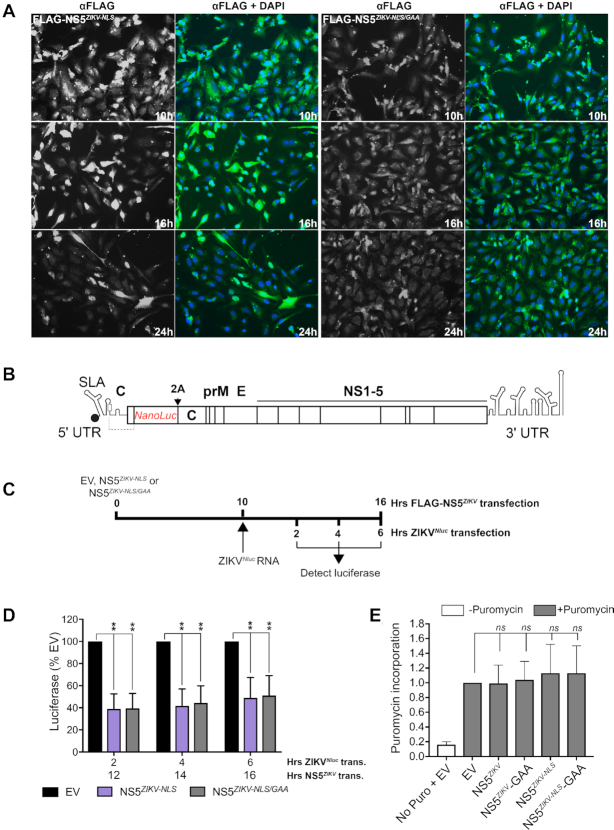
Figure 3.
NS5ZIKV inhibits viral genomic RNA translation in cells. (A) Expression of FLAG-tagged NS5ZIKV was detected by immunofluorescence using an anti-FLAG antibody over a 24 h period following transfection of Vero cells. For each FLAG-NS5ZIKV expression construct tested anti-FLAG is shown in greyscale on the left while nuclei are stained blue with DAPI and anti-FLAG is green in the associated panel to the right. (B) Schematic of the reporter ZIKV bearing nanoluciferase (Nluc) (32). The duplicate capsid sequence is separated from the marker with a 2A StopGo sequence (indicated with 2A). (C) Experimental strategy used for D. (D) Vero cells were electroporated with capped, in vitro transcribed ZIKVNluc marker RNA 10 h after transfection with the indicated FLAG-NS5ZIKV or empty vector (EV). Luciferase signal was quantified at the indicated time points. Values are normalized to the EV control at each time point and are mean ± SEM from three independent experiments. Time post NS5ZIKV transfection (trans.) and time post electroporation (elec.) of ZIKVNluc RNA is included on the X-axis for clarity. Statistical significance was determined by a Student's unpaired t-test with significant values indicated with an asterisk. *P< 0.05, **P< 0.01. (E) Vero cells were transfected with empty vector (EV) or the indicated wild type or mutant FLAG-NS5ZIKV expressing plasmids and after 10 h puromycin was added as indicated and the cells incubated for a further 2 h. Protein lysates were analysed by western blotting using an anti-puromycin antibody. The signal was quantified for each lane and normalized to that for tubulin which was used as a loading control. A representative blot is shown in Supplementary Figure S4. Data are mean ± SEM from three independent experiments. Wildtype and mutant NS5 data were compared to EV by a Student's unpaired t-test and no significant change was observed.
We investigated the impact of NS5 overexpression on translation in Vero cells of an in vitro transcribed and capped full-length ZIKV genomic RNA that contains a nanoluciferase reporter (ZIKVNluc), shown schematically in Figure 3B. We transfected Vero cells with FLAG-NS5ZIKV-NLS or NS5ZIKV-NLS/GAA and 10 hours later introduced the ZIKVNluc RNA by electroporation (timeline of experiment shown in Figure 3C). Translation of the viral genomic RNA, as measured by nanoluciferase production, was decreased by 60% in the FLAG-NS5ZIKV expressing cells when compared to the empty vector control (Figure 3D). The luciferase signal remained lower for 6 hours post transfection (Figure 3D) after which point the control luciferase signal began to decrease (data not shown) in line with our previous results (6). The presence of NS5 did not affect global protein synthesis in the cell as measured by puromycin incorporation (Figure 3E and Supplementary Figure S4). Together, our results identify a previously undescribed role for flavivirus NS5 as a regulator of virus translation.
Overexpression of NS5ZIKV inhibits viral replication
As our data demonstrated that NS5 inhibits viral translation we next examined the effect of NS5 expression on viral replication. We generated high titre stocks of ZIKV containing an mCherry reporter (ZIKVmCherry, shown schematically in Figure 4A). The production of this tagged ZIKV has been previously reported (32). Using a similar strategy to our nanoluciferase experiments we first overexpressed FLAG-NS5ZIKV-NLS or FLAG-NS5ZIKV-NLS/GAA for 8 hours in Vero cells before infecting with a high multiplicity of infection (MOI:10) of ZIKVmCherry (Figure 4B). Viral replication was then monitored by detection of the mCherry signal on a fluorescence microscope. Transfection of either FLAG-NS5ZIKV constructs resulted in a marked delay in replication of the mCherry tagged ZIKV as compared to transfection with empty vector, as evidenced by the lower fluorescence signal detected at each time point analysed (Figure 4C). The NS5-induced delay in ZIKV replication observed in our fluorescence microscopy experiments was also confirmed by flow cytometry analysis. The number of mCherry positive cells increased at each time point examined following ZIKVmCherry infection of non-transfected or empty vector control plasmid transfected cells (Supplementary Figure S5 and Table S1, 72 hour time point shown in Figure 4D). In contrast, transfection of FLAG-NS5ZIKV-NLS or FLAG-NS5ZIKV-NLS/GAA prior to infection resulted in a decrease in the number of mCherry positive cells at each time point examined (72 h time point shown in Figure 4D, see also Supplementary Figure S5 and Table S1). Our finding that the presence of NS5 before infection impairs ZIKV replication is consistent with its ability to inhibit translation of the viral genome and has important implications for establishing trans-replicase plasmid-based systems for examining flavivirus replication as recently developed for other positive sense RNA viruses (43).
Figure 4.
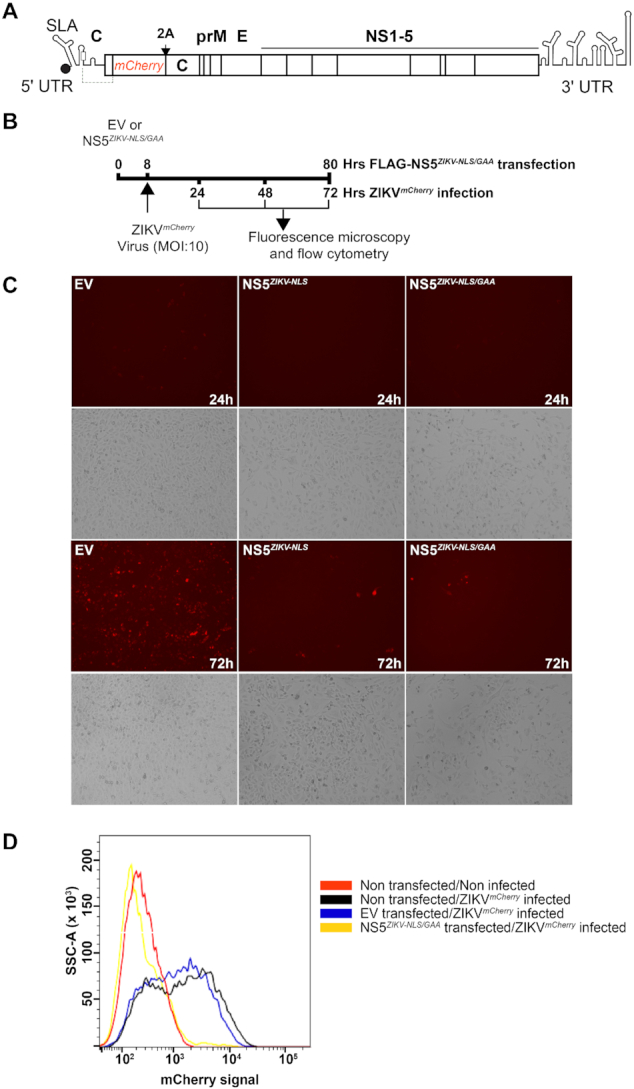
NS5ZIKV pre-expression disrupts viral replication. (A) Schematic of the reporter ZIKV bearing nanoluciferase (Nluc) (32). The duplicate capsid sequence is separated from the marker with a 2A StopGo sequence. (B) Experimental strategy used for C–E. (C and D) Vero cells were infected with mCherry reporter virus (MOI:10) 8 h after transfection with FLAG-NS5ZIKV-NLS/GAA or empty vector (EV). (C) mCherry signal was detected directly using a fluorescence microscope at the indicated time points. The bright field view is shown below each fluorescence image. (D) Cells were fixed and analysed by flow cytometry 72 hours after infection. See also Supplementary Figure S5.
DISCUSSION
Viruses with limited genome lengths frequently encode proteins with multiple functions. RNA viruses, in particular, also evolve complex genome structures that regulate various stages of the replication cycle, thus maximizing the usage of coding and non-coding regions of viral RNA. Here we have shown that, along with serving as the well described replication promoter (14), the interaction of the flavivirus NS5 protein with the structured 5′ UTR serves a secondary role as a mechanism to control translation of the viral genome.
Genome circularization through hybridization of complementary RNA sequences in the 5′ and 3′ regions is essential for flavivirus replication (44). It was previously reported that NS5 binds more efficiently to SLA in the linear form of a DENV4 minigenome RNA in vitro (45), although the difference detected in EMSA experiments was small. We recently demonstrated that the circularized, replication-competent form of the viral RNA is translated less efficiently than the linear form due to structural rearrangements that occur around the translation start site (6), providing a mechanism to maintain a ribosome clear template for replication. Although the difference in affinity was small, we also detected enhanced binding of ZIKV NS5 to the linear form of ZIKV RNA (Supplementary Figure S6), supporting the hypothesis that NS5 is recruited to RNA in the linear conformation (45). This is consistent with NS5 binding to and inhibiting protein synthesis on the translation-competent form of the genomic RNA. As multiple viral proteins are involved in establishing a replication complex (46) and have been reported to promote genome circularization (47,48), locating the NS5 RdRp at the C-terminus of the large open reading frame, and sequestration within the viral polyprotein, may delay initiation of replication until other viral factors have been synthesized. Future work will investigate the contribution of NS5 translation inhibition to the temporal switch to signal that sufficient viral proteins have been synthesized to initiate replication.
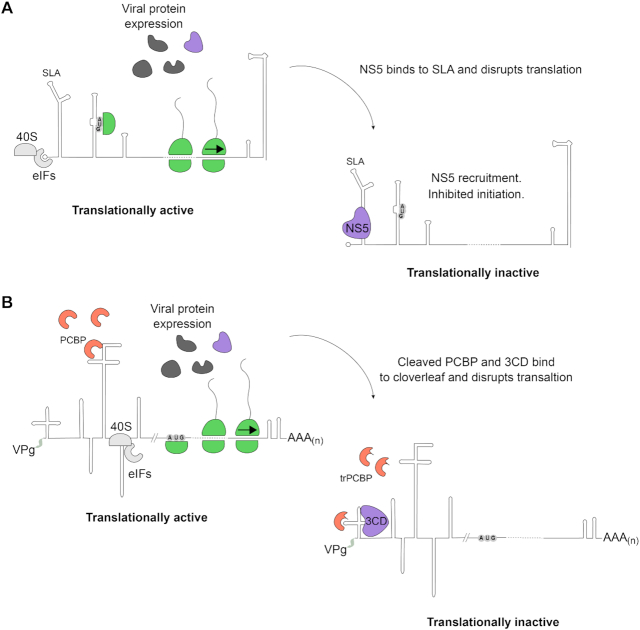
Our findings support a model for flavivirus replication (Figure 5) whereby the viral genome is efficiently translated after release into the cytoplasm in the linear form but, after sufficient NS5 is expressed, NS5 binds to the cap-proximal SLA structure and inhibits further translation of the viral genome. Subsequent circularization of the genome, promoted by viral and cellular factors (47–51), mediates transfer of NS5 to the viral genome 3′ end to initiate replication. It is possible that genome circularization depends upon ribosome clearance from the RNA template as, by analogy with bacteria, the ribosome has been described as a major helicase activity within the cell (52,53). As such, NS5 recruitment to the 5′ end, together with genome circularization (6), may provide distinct mechanisms for decreasing ribosome occupancy on the viral RNA by inhibiting translation initiation thus facilitating efficient genome replication.
Figure 5.
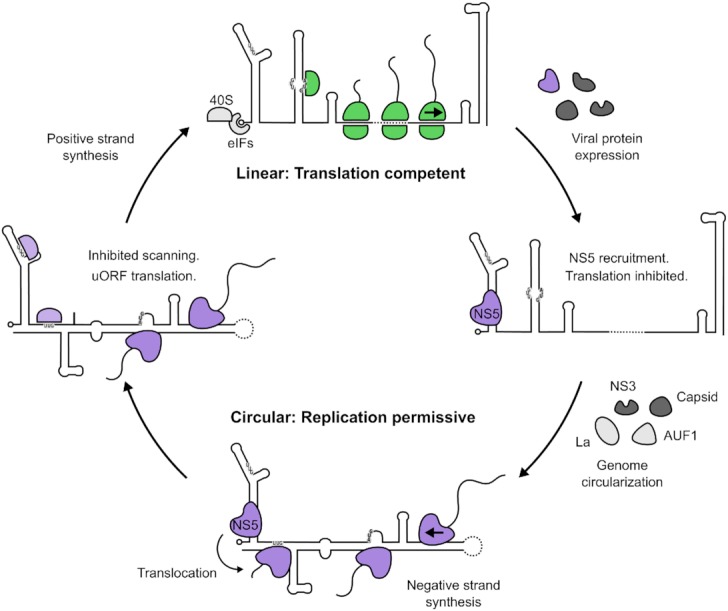
Model of flavivirus genome translation regulation. Linear genomic RNA recruits eIFs and 40S ribosomal subunits in a cap-dependent manner (6). Secondary structure in the 5′ UTR is effectively resolved by eIFs and the scanning ribosome to allow efficient translation from the authentic initiation codon, leading to the accumulation of viral proteins. NS5 is subsequently recruited to SLA and antagonizes translation initiation, priming the RNA for replication. Following genome circularization promoted by viral and host factors (47,48,50,51,58), NS5 translocates to the 3′ end of the genome to begin negative strand synthesis. The circularized conformation of the viral genome acts as a barrier to prevent further ribosome loading by impeding scanning ribosomes (6), indicated by the red stop sign, thus keeping the replicating template free of ribosomes.
This proposed role for NS5 in temporally regulating translation of 5′ capped flaviviral RNA is reminiscent of the similar role postulated for the picornaviral 3CD protease/polymerase precursor protein during poliovirus (PV) replication (54,55) (Figure 6). Like flaviviruses, picornaviruses possess a single-stranded positive sense RNA genome, translation of which must be inhibited to promote efficient replication (54). Binding of 3CD to the cloverleaf structure at the 5′ of the PV genomic RNA was reported to regulate PV translation (54) which is driven by a cap-independent, internal ribosomal entry site (56,57). Although the mechanism of PV 3CD-IRES translation control has not been fully elucidated, together with our results, these findings point to a conserved system of controlling the timing of the translation–replication switch in different positive-sense single-stranded RNA virus families exploiting the RNA-binding capabilities of the viral polymerase.
Figure 6.
Model for conserved mechanisms of temporal translation control in positive strand RNA viruses. (A) Flavivirus RNA is translated in a cap-dependent manner following genome release into the cytoplasm. As the viral polymerase NS5 is expressed it binds to the stem loop A (SLA) replication promoter at the 5′ end of the genome preventing translation initiation. (B) Poliovirus (PV) translation, driven by an internal ribosomal entry site (IRES), occurs after release of the genome to the cytoplasm. The cellular polyC-binding protein (PCBP) is required for PV IRES activity (57). As the viral protease/polymerase precursor 3CD is expressed, it can subsequently bind to the 5′ cloverleaf structure and disrupt IRES dependent translation. PCBP is also cleaved by the viral protease so it can no longer promote IRES translation. In both cases, viral translation arrest clears the template for efficient replication.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Andres Merits for sharing the ZIKV reverse genetics system and to Tatyana Pestova and Christopher Hellen for sharing expression constructs.
Author contributions: T.J.S. and T.R.S. conceived the study. T.F., T.J.S., H.V.M., D.M. and T.R.S. designed experiments. T.F., T.J.S., H.V.M., A.J., S.S. and T.R.S. performed experiments. All authors interpreted data. T.R.S. supervised the research and wrote the paper which all authors commented on.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Royal Society/Wellcome Trust Sir Henry Dale Fellowship [202471/Z/16/Z to T.R.S.]; BBSRC Project Grant [BB/S007350/1 to T.R.S.]; Wellcome Trust PhD Studentship [105389/Z/14/Z to T.J.S.]; University of Cambridge, Department of Pathology PhD Studentship (to H.V.M.); Wellcome Trust PhD Studentship [203893/Z/15/A to S.S.]; A.J. was supported by an Amgen Scholarship. Funding for open access charge: Wellcome Trust grant (to T.R.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Pierson T.C., Diamond M.S.. The emergence of Zika virus and its new clinical syndromes. Nature. 2018; 560:573–581. [DOI] [PubMed] [Google Scholar]
- 2. Gutiérrez-Bugallo G., Piedra L.A., Rodriguez M., Bisset J.A., Lourenço-de-Oliveira R., Weaver S.C., Vasilakis N., Vega-Rúa A.. Vector-borne transmission and evolution of Zika virus. Nat. Ecol. Evol. 2019; 3:561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Silva J.V.J., Lopes T.R.R., Oliveira-Filho E.F., Oliveira R.A.S., Durães-Carvalho R., Gil L.H.V.G.. Current status, challenges and perspectives in the development of vaccines against yellow fever, dengue, Zika and chikungunya viruses. Acta Trop. 2018; 182:257–263. [DOI] [PubMed] [Google Scholar]
- 4. Mlakar J., Korva M., Tul N., Popović M., Poljšak-Prijatelj M., Mraz J., Kolenc M., Resman Rus K., Vesnaver Vipotnik T., Fabjan Vodušek V. et al.. Zika virus associated with microcephaly. N. Engl. J. Med. 2016; 374:951–958. [DOI] [PubMed] [Google Scholar]
- 5. Barrows N.J., Campos R.K., Liao K.C., Prasanth K.R., Soto-Acosta R., Yeh S.C., Schott-Lerner G., Pompon J., Sessions O.M., Bradrick S.S. et al.. Biochemistry and molecular biology of flaviviruses. Chem. Rev. 2018; 118:4448–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanford T.J., Mears H.V., Fajardo T., Locker N., Sweeney T.R.. Circularization of flavivirus genomic RNA inhibits denovo translation initiation. Nucleic Acids Res. 2019; 47:9789–9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H. et al.. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010; 468:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fleith R.C., Mears H.V., Leong X.Y., Sanford T.J., Emmott E., Graham S.C., Mansur D.S., Sweeney T.R.. IFIT3 and IFIT2/3 promote IFIT1-mediated translation inhibition by enhancing binding to non-self RNA. Nucleic Acids Res. 2018; 46:5269–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duan W., Song H., Wang H., Chai Y., Su C., Qi J., Shi Y., Gao G.F.. The crystal structure of Zika virus NS5 reveals conserved drug targets. EMBO J. 2017; 36:919–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Godoy A.S., Lima G.M.A., Oliveira K.I.Z., Torres N.U., Maluf F.V., Guido R.V.C., Oliva G.. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat. Commun. 2017; 8:14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Upadhyay A.K., Cyr M., Longenecker K., Tripathi R., Sun C., Kempf D.J.. Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallogr. Sect. Struct. Biol. Commun. 2017; 73:116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao B., Yi G., Du F., Chuang Y.C., Vaughan R.C., Sankaran B., Kao C.C., Li P.. Structure and function of the Zika virus full-length NS5 protein. Nat. Commun. 2017; 8:14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dubankova A., Boura E.. Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses. Antiviral Res. 2019; 169:104536. [DOI] [PubMed] [Google Scholar]
- 14. Filomatori C.V., Lodeiro M.F., Alvarez D.E., Samsa M.M., Pietrasanta L., Gamarnik A.V.. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006; 20:2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dong H., Zhang B., Shi P.Y.. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology. 2008; 381:123–135. [DOI] [PubMed] [Google Scholar]
- 16. Bujalowski P.J., Bujalowski W., Choi K.H.. Interactions between the dengue virus polymerase NS5 and stem-loop A. J. Virol. 2017; 91:e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alvarez D.E., De Lella Ezcurra A.L., Fucito S., Gamarnik A.V.. Role of RNA structures present at the 3′ UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005; 339:200–212. [DOI] [PubMed] [Google Scholar]
- 18. Friebe P., Harris E.. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. J. Virol. 2010; 84:6103–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khromykh A.A., Meka H., Guyatt K.J., Westaway E.G.. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001; 75:6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang B., Dong H., Stein D.A., Iversen P.L., Shi P.Y.. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology. 2008; 373:1–13. [DOI] [PubMed] [Google Scholar]
- 21. Filomatori C.V., Iglesias N.G., Villordo S.M., Alvarez D.E., Gamarnik A.V.. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J. Biol. Chem. 2011; 286:6929–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Best S.M. The many faces of the flavivirus NS5 protein in antagonism of Type I interferon signaling. J. Virol. 2017; 91:e01970-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grant A., Ponia S.S., Tripathi S., Balasubramaniam V., Miorin L., Sourisseau M., Schwarz M.C., Sánchez-Seco M.P., Evans M.J., Best S.M. et al.. Zika virus targets human STAT2 to inhibit Type I interferon signaling. Cell Host Microbe. 2016; 19:882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ashour J., Laurent-Rolle M., Shi P.-Y., Garcia-Sastre A.. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 2009; 83:5408–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morrison J., Laurent-Rolle M., Maestre A.M., Rajsbaum R., Pisanelli G., Simon V., Mulder L.C.F., Fernandez-Sesma A., García-Sastre A.. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize Type I interferon signaling. PLoS Pathog. 2013; 9:e1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hertzog J., Dias Junior A.G., Rigby R.E., Donald C.L., Mayer A., Sezgin E., Song C., Jin B., Hublitz P., Eggeling C. et al.. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks Type I IFN induction and signaling. Eur. J. Immunol. 2018; 48:1120–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Forwood J.K., Brooks A., Briggs L.J., Xiao C.Y., Jans D.A., Vasudevan S.G.. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem. Biophys. Res. Commun. 1999; 257:731–737. [DOI] [PubMed] [Google Scholar]
- 28. Buckley A., Gaidamovich S., Turchinskaya A., Gould E.A.. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J. Gen. Virol. 1992; 73:1125–1130. [DOI] [PubMed] [Google Scholar]
- 29. Hannemann H., Sung P.Y., Chiu H.C., Yousuf A., Bird J., Lim S.P., Davidson A.D.. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J. Biol. Chem. 2013; 288:22621–22635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jans D.A., Martin A.J.. Nucleocytoplasmic trafficking of dengue non-structural protein 5 as a target for antivirals. Adv. Exp. Med. Bio. 2018; 1062:199–213. [DOI] [PubMed] [Google Scholar]
- 31. De Maio F.A., Risso G., Iglesias N.G., Shah P., Pozzi B., Gebhard L.G., Mammi P., Mancini E., Yanovsky M.J., Andino R. et al.. The dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog. 2016; 12:e1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mutso M., Saul S., Rausalu K., Susova O., Žusinaite E., Mahalingam S., Merits A.. Reverse genetic system, genetically stable reporter viruses and packaged subgenomic replicon based on a Brazilian Zika virus isolate. J. Gen. Virol. 2017; 98:2712–2724. [DOI] [PubMed] [Google Scholar]
- 33. Chavali P.L., Stojic L., Meredith L.W., Joseph N., Nahorski M.S., Sanford T.J., Sweeney T.R., Krishna B.A., Hosmillo M., Firth A.E. et al.. Neurodevelopmental protein Musashi-1 interacts with the Zika genome and promotes viral replication. Science. 2017; 357:83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pestova T, Hellen C.U.. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation invitro. RNA. 2001; 7:1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pestova T.V., Borukhov S.I., Hellen C.U.T.. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998; 394:854–859. [DOI] [PubMed] [Google Scholar]
- 36. Pestova T.V, Hellen C.U.T., Shatsky I.N.. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996; 16:6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lomakin I.B., Shirokikh N.E., Yusupov M.M., Hellen C.U.T., Pestova T.V.. The fidelity of translation initiation: Reciprocal activities of elF1, IF3 and YciH. EMBO J. 2006; 25:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rakotondrafara A.M., Hentze M.W.. An efficient factor-depleted mammalian invitro translation system. Nat. Protoc. 2011; 6:563–571. [DOI] [PubMed] [Google Scholar]
- 39. Pisarev A.V., Unbehaun A., Hellen C.U.T., Pestova T.V.. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007; 430:147–177. [DOI] [PubMed] [Google Scholar]
- 40. Jackson R.J., Hellen C.U.T., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Z.-Y., Li X.-F., Jiang T., Deng Y.-Q., Zhao H., Wang H.-J., Ye Q., Zhu S.-Y., Qiu Y., Zhou X. et al.. Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. J. Virol. 2013; 87:6804–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ng I.H.W., Chan K.W., Tan M.J.A., Gwee C.P., Smith K.M., Jeffress S.J., Saw W.G., Swarbrick C.M.D., Watanabe S., Jans D.A. et al.. Zika virus NS5 forms supramolecular nuclear bodies that sequester importin-α and modulate the host Immune and pro-inflammatory response in neuronal cells. ACS Infect. Dis. 2019; 5:932–948. [DOI] [PubMed] [Google Scholar]
- 43. Bartholomeeusen K., Utt A., Coppens S., Rausalu K., Vereecken K., Ariën K.K., Merits A.. A chikungunya virus trans-replicase system reveals the importance of delayed nonstructural polyprotein processing for efficient replication complex formation in mosquito cells. J. Virol. 2018; 92:e00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Garcia-Blanco M.A., Vasudevan S.G., Bradrick S.S., Nicchitta C.. Flavivirus RNA transactions from viral entry to genome replication. Antiviral Res. 2016; 134:244–249. [DOI] [PubMed] [Google Scholar]
- 45. Liu Z.Y., Li X.F., Jiang T., Deng Y.Q., Ye Q., Zhao H., Yu J.Y., Qin C.F.. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. Elife. 2016; 5:e17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klema V.J., Padmanabhan R., Choi K.H.. Flaviviral replication complex: coordination between RNA synthesis and 5′-RNA capping. Viruses. 2015; 7:4640–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ivanyi-Nagy R., Darlix J.L.. Core protein-mediated 5′-3′ annealing of the West Nile virus genomic RNA invitro. Virus Res. 2012; 167:226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gebhard L.G., Kaufman S.B., Gamarnik A.V.. Novel ATP-independent RNA annealing activity of the dengue virus NS3 helicase. PLoS One. 2012; 7:e36244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vashist S., Bhullar D., Vrati S.. La protein can simultaneously bind to both 3′- and 5′-noncoding regions of Japanese encephalitis virus genome. DNA Cell Biol. 2011; 30:339–346. [DOI] [PubMed] [Google Scholar]
- 50. Friedrich S., Schmidt T., Geissler R., Lilie H., Chabierski S., Ulbert S., Liebert U.G., Golbik R.P., Behrens S.-E.. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J. Virol. 2014; 88:11586–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Friedrich S., Engelmann S., Schmidt T., Szczepankiewicz G., Bergs S., Liebert U.G., Kümmerer B.M., Golbik R.P., Behrens S.-E.. The host factor AUF1 p45 supports flavivirus propagation by triggering the RNA switch required for viral genome cyclization. J. Virol. 2018; 92:e01647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takyar S., Hickerson R.P., Noller H.F.. mRNA helicase activity of the ribosome. Cell. 2005; 120:49–58. [DOI] [PubMed] [Google Scholar]
- 53. Qu X., Wen J.D., Lancaster L., Noller H.F., Bustamante C., Tinoco I.. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011; 475:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gamarnik A.V., Andino R.. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998; 12:2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gamarnik A.V., Andino R.. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 2000; 74:2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pelletier J., Sonenberg N.. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988; 334:320–325. [DOI] [PubMed] [Google Scholar]
- 57. Sweeney T.R., Abaeva I.S., Pestova T.V., Hellen C.U.T.. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014; 33:76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vashist S., Anantpadma M., Sharma H., Vrati S.. La protein binds the predicted loop structures in the 3′ non-coding region of Japanese encephalitis virus genome: role in virus replication. J. Gen. Virol. 2009; 90:1343–1352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.