Abstract
One primary objective of synthetic biology is to improve the sustainability of chemical manufacturing. Naturally occurring biological systems can utilize a variety of carbon sources, including waste streams that pose challenges to traditional chemical processing, such as lignin biomass, providing opportunity for remediation and valorization of these materials. Success, however, depends on identifying micro-organisms that are both metabolically versatile and engineerable. Identifying organisms with this combination of traits has been a historic hindrance. Here, we leverage the facile genetics of the metabolically versatile bacterium Acinetobacter baylyi ADP1 to create easy and rapid molecular cloning workflows, including a Cas9-based single-step marker-less and scar-less genomic integration method. In addition, we create a promoter library, ribosomal binding site (RBS) variants and test an unprecedented number of rationally integrated bacterial chromosomal protein expression sites and variants. At last, we demonstrate the utility of these tools by examining ADP1’s catabolic repression regulation, creating a strain with improved potential for lignin bioprocessing. Taken together, this work highlights ADP1 as an ideal host for a variety of sustainability and synthetic biology applications.
INTRODUCTION
A critical element of sustainability is creating a closed and efficient carbon cycle by designing processes that utilize renewable resources and minimize or reclaim waste streams (1,2). The emergence of synthetic biology has provided revolutionary new opportunities to perform sustainable and green chemistry (3). In addition to operating at ambient reaction conditions and without the need for harsh reagents, such as heavy metals, a key advantage for natural systems is their ability to utilize and adapt to a wide variety of feedstocks as carbon sources. This metabolic flexibility is exemplified by the conversion of waste C1 gases to ethanol with remarkable tolerance to real-time changes in feed gas content and quality using acetogens (4,5). Such demonstrations highlight the potential for synthetic biology to expand and improve waste remediation processes.
To integrate additional waste streams, though, metabolically versatile micro-organisms must be identified that are engineerable and capable of quickly undergoing many Design-Build-Test-Learn (DBTL) cycles. Recent advances in synthetic biology have identified numerous hosts with advantageous traits such as solvent tolerance (6), novel metabolic capabilities (7) and fast growth rates (8), but progress with these hosts is often slowed by their genetic intractability. Unlike model organisms such as Escherichia coli and Saccharomyces cerevisiae, emerging hosts have comparatively few genetic tools and require lengthy DBTL cycles. Ideally, novel hosts would be identified that expand current functional capabilities and are easily manipulated.
For these reasons, the bacterium Acinetobacter baylyi ADP1 represents an exciting emerging synthetic biology host (9,10). ADP1 has a small genome, grows quickly (doubling time of as little as 35 min in rich medium) (11) and is naturally competent (i.e. capable of taking up DNA during standard growth conditions). The frequency of natural transformation and allelic replacement in this bacterium is exceptionally high and unrivaled in any other known microbe, a feature that has long been touted as an ideal basis for genetic engineering (12,13). Moreover, by way of the β-ketoadipate pathway, ADP1 is able to convert renewable, lignin derived aromatic compounds to simple carbon building blocks (14,15). Such catabolic versatility is significant as lignin is a major and notably underutilized component of non-food biomass (16). The heterogeneity and complexity of lignin have precluded simple approaches for its upgrading (17), and metabolic engineering has been proposed as a solution (18). Though other bacteria including Pseudomonas and Rhodococcus species have been identified with similar lignin consumption attributes to ADP1 (14), and advances have been made to their genetic toolsets (19–21), these hosts can be challenging to engineer. ADP1 provides an opportunity to advance lignin bioprocessing through accelerated engineering cycles.
Although the genetic tractability of ADP1 has been appreciated for decades, the previously insufficient development of genetic tools and reliable procedures for high-throughput workflow has proven to be an obstacle to the widespread adoption of ADP1 as a platform organism. To address this critical need, we established simple and rapid cloning workflows that show notable reductions in experimental time compared to even established hosts such as E. coli. We created a promoter library, RBS variants and characterized an unprecedented number of targeted locations and promoter variants for chromosomal protein expression. In addition, we established a one-step marker-less chromosomal integration technique leveraging Cas9. These tools, which were reproducibly useful in different laboratories, were instrumental in studying the regulated consumption of aromatic monomers in ADP1. As demonstrated by this application, strains with improved lignin bioprocessing attributes can be created with tools that allow fine-tuned gene expression. Our work highlights ADP1’s amenability for biofoundry workflows (22) and should establish this strain as a preferred option for numerous synthetic biology applications, including lignin-based metabolic engineering.
MATERIALS AND METHODS
Cell culturing
In this study wild-type A. baylyi (ADP1) (23,24) and E. coli DH5α were used for cloning and cell culture work. ADP1 was used for all expression tests and genetic part validation. Liquid cultures were grown at 30°C, 250 rpm, unless otherwise noted. Plate-based cultivation was carried out at 30°C, 150 rpm. Cultivations were run in Fisher's LB Broth Miller (LB) or Difco M9 Minimal Medium (M9), unless otherwise stated. M9 was supplemented with various carbon sources as indicated. Kanamycin (kan) at 25 μg/ml or Spectinomycin at 25 μg/ml were used for antibiotic selection. Agar plates were made with 15 g of Teknova Agar per liter of medium. Glycerol stocks were stored at −80°C and were created by adding 750 μl of overnight cell culture medium to 250 μl of 60% (v/v) glycerol for a final glycerol concentration of 15% (v/v).
Cloning
Polymerase chain reactions (PCRs) were carried out with PrimeSTAR Max 2× master mix from Takara Bio, following manufacturer instructions for annealing temperatures and extension times using the three-step protocol (denature, anneal, extend). An Excel file with a list of all primers used in this study can be found with the supplementary information [Supplementary File S2 (Primers)]. In addition, cloning files representing the final parts used for this study can found with the Supplementary Data [Supplementary File S1 (Tools Strains) and Supplementary File S3 (Tools Plasmids)]. General cloning, primer design and in silico Gibson assembly design were carried out with Benchling. Gibson assemblies were run using NEB 2× Gibson master mix, following manufacturer protocols. All mutagenic primers were designed on the NEBaseChanger web interface (https://nebasechanger.neb.com/). All blunt end ligations following primer-based mutagenesis were carried out using gel purified DNA and the ‘KLD’ mix from the NEB Q5 Site-Directed Mutagenesis Kit, following manufacturer protocols.
PCR templates for colony PCRs were obtained by picking individual colonies and resuspending them into 20 μl of nuclease-free water, while patching them to a separate agar plate for secondary validation and to obtain clonally pure strains. For colony PCR, master mixes were prepared and ∼10 μl colony PCRs were run with 9.8 μl of the master mix and 0.5 μl of the water in which the colony had been resuspended as the template. All final genomic integrations were sequenced via their colony PCR product.
Fluorescent measurement experiments
Fluorescent measurements were taken with a Synergy H1 Microplate Reader from BioTek. mCherry readings were taken at both the top and bottom with excitation at 585 nM and emission at 615 nM. Gain was set at 100. The read height was set at 7 mm. Reported fluorescent values were exclusively taken from the top measurements. Plate reader cell density measurements were taken by reading the OD600. Plate reader experiments were carried out in biological triplicate, except for the pBAV1k promoter library, pBAV1k RBS (BCD) variants, chromosomal integration mapping, chromosomally integrated promoter library and chromosomally integrated RBS (BCD) variants, which were carried out as biological triplicates run on three separate days (nine replicates total).
To prepare plate reader fluorescent experiments, overnight cultures were carried out in sterile 0.4 ml 96-well flat bottom plates (Fisher Cat. No. 267576) at 30°C and 150 rpm, scraping glycerol stocks for inoculation into 200 μl of LB with appropriate antibiotic. Sub-culturing (1:100 dilution) was carried out the following morning by successive dilutions of first 40 μl of LB overnight culture added to 160 μl of the final medium (LB or M9, with corresponding antibiotic and carbon source, which was 8 mM protocatechuate (PCA) for the M9 medium) to make an initial 40:200 (1:5) dilution plate. Then, 10 μl of this 1:5 dilution was added to 190 μl of the final medium (1:100 final dilution). The final cultures were run in sterile 0.4 ml optical bottom black 96-well plates. After 3 h, these cultures were induced with 1 mM IPTG (5 μl of 40 mM IPTG stock). Fluorescent measurements were taken the next morning after gentle resuspension by pipetting up and down, being careful to not introduce any bubbles to the culture. All cultures were covered with sterile AeraSeal films. Films were replaced with a fresh sterile film each time they were removed (for induction and for fluorescent measurements).
Time course experiments were carried out similarly, but with reads taken every 15 min after induction. Time course experiments were carried out in both LB medium and an adaptation of the Averhoff laboratory Acinetobacter Minimal Succinate Medium from the Barrick laboratory at University of Texas at Austin. The recipe for this medium is available online (https://barricklab.org/twiki/bin/view/Lab/ProtocolsRecipesABMS). Briefly, in a 1 L container, 880 ml of deionized water, 20 ml of 1 M sodium succinate, 50 ml of a 20× mineral solution and 50 ml of a 20× phosphate buffer is prepared. The 20× mineral solution consists of 10 g NH4Cl, 5.8 g MgSO4 heptahydrate, 1 g KNO3, 0.67 g CaCl2 dihydrate, 20 mg of (NH4)6Mo7O24 tetrahydrate and 10 ml of SL9 solution added to 500 ml of deionized water. The SL9 consists of 800 ml deionized water, 12.8 g nitrilotriacetic acid, 2 g FeSO4 heptahydrate dissolved in 2 M HCl, 104 mg anhydrous CoCl2, 122 mg MnCl2 tetrahydrate, 70 mg ZnCl2, 36 mg Na2MoO4 dihydrate, 13 mg NiCl2 and 1 ml of solution B. Where solution B represents 60 mg H3BO3 and 20 mg CuCl2 dihydrate dissolved into 10 ml of deionized water. This SL9 solution is pH adjusted to 6.5 with 1 M NaOH before being brought to volume (1 l). The phosphate solution includes 68 g KH2PO4 and 132.5 g Na2HPO4 hydrate per 500 ml of deionized water. The pH of the phosphate solution should be around 7.0 and can be adjusted with 1 M NaOH.
Fluorescence per OD600 [FL/OD (AU)] was calculated by first subtracting the values of fluorescence and OD600 for the blank medium from all wells. Next, fluorescence was divided by OD600. Following, the wild-type ADP1 average FL/OD value for that medium context was subtracted from all wells. Averages and standard deviations or standard error were calculated, as indicated.
Natural transformation
Natural transformation was carried out by first inoculating wild-type ADP1 from a glycerol stock into a 3 ml overnight culture, grown at 30°C and 250 rpm in LB medium in a culture tube. The following day, 70 μl of this culture was added to 1 ml of fresh LB medium in a culture tube and incubated for 3 h along with the transforming DNA (plasmid, PCR product, Gibson product, ligation product). Transforming DNA did not need to be purified by gel extraction or PCR clean up kit before direct addition to the ADP1 culture, and as little as 25 ng of transforming DNA added to the 1 ml of culture was sufficient to obtain transformants. After the 3 h, 150 μl of the medium was plated on the appropriate selective plate, spread with glass beads, and allowed to dry briefly before moving to the incubator.
Promoter library creation
The promoter library was created using the above natural transformation workflow. A single set of mutagenic primers was purchased with ‘NNN’ diversity 5 bp upstream of the bacterial consensus −35 box (TTGACA), in the 17 bp in between the −35 and −10 box (TATAAT), and for the 5 bp after the −10 box [Supplementary File S2 (Primers), primers BWB361, BWB362]. A single PCR was run with the pBWB162 vector as the template. The product was gel extracted and added into the KLD NEB mix (DpnI, kinase, ligase) and allowed to react for 5 min at room temperature. This product was then directly added into the ADP1 culture for transformation. After 3 h, 150 μl of culture was plated on two separate plates. Plates were incubated at 30°C. The next day, 96 colonies were picked and inoculated into a 96-well plate for fluorescence screening.
qPCR
qPCR was run on a Bio-Rad CFX96 Real-time System C1000 Thermal Cycler using iQ SYBR Green Supermix, Bio-Rad Hard-Shell® 96-Well PCR plates (high profile, semi skirted, green/clear), and Bio-Rad Microseal ‘B’ adhesive film covers. qPCRs were run using manufacturers protocols for iQ SYBR Green Supermix, specified for the CFX96 System. Based on previous qPCR work in ADP1 with the pBAV1k plasmid (25), the single copy gene spoT/relA was used as a genomic reference with previously identified primers (here BWB903 and BWB904). For plasmid qPCR of pBWB162, the primers BWB905, BWB906 were used. BWB905 and BWB906 were designed following manufacturing guidelines in for annealing temperature and qPCR product size.
To obtain template DNA, cells were cultured overnight in LB, and then diluted 1:100 into either LB or M9 with 8 mM PCA medium and grown overnight, mimicking the expression testing workflow. The next morning, the cultures were placed on ice and spun down at 4000 rpm for 10 min at 4°C. After decanting and aspirating the culture, a pipette tip was touched to the cell pellet and resuspended into 100 μl of nuclease free water. Following, 1 μl of this water was added to 9 μl of a master mix of iQ SYBR Green Supermix and the relevant primers to make a 10 μl PCR reaction. Relative plasmid copy numbers were derived by dividing the Ct value of the plasmid by the Ct value of the genomic PCR for the same culture.
Genomic integration
For the genomic integration mapping, an insulated integration cassette was first constructed by the addition of two upstream terminators and one additional downstream terminator. These insulators were added by successive PCR mutagenesis via overhangs to create pBWB206. Once the genomic integration cassette was created, knock in cassettes were constructed by overlap PCR. Overlap PCRs were run in two stages. First, three PCRs were run to obtain two ∼500 bp oligos with homology matching the upstream and downstream regions of the ADP1 gene to be knocked out, respectively, and the insulated genomic integration cassette. Each part had 20–30 bp of overhang homology to the part to which it would be assembled by overlap PCR. Approximately 1 μl of each gel extracted part was added to a 45 μl PCR reaction and run for 14 cycles to create the full-length knock in cassette. Following, outer primers were added to amplify the full-length knock in cassette for an additional 19 cycles. Knock in cassettes were either directly added to the ADP1 medium or gel extracted first. Approximately 5 μl of either PCR product, gel extracted or not, was added to the medium. The same workflow was used for the promoter library and RBS/BCD variant integrations, but these cassettes did not bear the insulator regions.
Flow cytometry
A BD LSRFortessa was used for flow cytometry analysis. mCherry fluorescence was detected with a 552-nm laser using the Texas Red setting (610/620 nm). Cells were first gated by forward and side scatter and then gated based on fluorescence such that the wild-type strain with no fluorescent marker was only collected at 0.2–0.3%. A total of 100 000 events were captured for each run.
Cells were prepared by growth overnight (30°C, 250 rpm) from a glycerol stock in LB medium with appropriate antibiotic. The next morning, the cultures were diluted 1:100 and again grown at 30°C, 250 rpm. Cultures were induced with 1 mM IPTG after 3 h. After overnight growth of induced cells, cultures were place on ice, ODs measurements were taken, the cells were spun down (4000 rpm, 10 min) and resuspended such that in 500 μl of cold 1× phosphate-buffered saline there were ∼1 000 000 cells with an assumed OD of 1 equal to 8 × 108 cell/ml. Strains sorted included wild-type ADP1 (BWB062), pBAV1k-lacI-Trc-mCherry (pBWB162) for plasmid-based expression and ΔvanAB::kan-lacI-Trc-mCherry (BWB335) for chromosomal expression.
Cas9-based marker-less integration method
The Cas9-based marker-less, scar-less integration method utilized a modified version of the RSF1010-based broad-host-range CRISPRi-ME plasmid developed by the Barrick laboratory (26), for which dCas9 was reverted back to an active Cas9. In addition, various substitutions for the guide RNA (gRNA) were made. Marker-less integrations were carried out in the ISx strain developed by the Barrick lab where six of ADP1’s insertion sequences have been removed (11). For this method a similar natural transformation workflow was utilized as described above, but with two primary changes. First, ≥500 ng of DNA for both the linear genomic transformation cassette and the Cas9 and gRNA bearing plasmid (RSF1010) were added to the culture for transformation. Second, after adding the transforming DNA, the culture was incubated for 6 instead of 3 h. Because of the significantly lower efficiency for co-transforming two elements (∼10 cfu/ml-μg), 900 μl of the LB medium was plated as three separate 300 μl aliquots on LB agar plates with spectinomycin. These plates were allowed to grow for 2 days at 30°C to observe colonies. Smaller colonies generally correlated with successful integrations.
Colonies were screened by colony PCR and patched on LB agar with spectinomycin and then screened by colony PCR a second time to confirm clonal purity for the integration. After clonal purity was confirmed by colony PCR, hits were streaked on LB agar plates with no antibiotic. The following day, colonies were patched to both LB agar without antibiotic and with spectinomycin to determine which colonies had cleared the plasmid. Those that grew only on LB agar without antibiotic were examined by colony PCR to confirm presence of desired integration. The PCR product of this colony PCR was sent for sequencing, and these hits were inoculated into LB medium and grown at 30°C, 250 rpm overnight. The next morning strains were archived as glycerol stocks.
gRNAs were designed using the ATUM design tool (https://www.atum.bio/eCommerce/cas9/input) with the settings for E. coli K12 MG1655, NGG PAM and wild-type Cas9, searching on a supplied target sequence for the genes to be knocked out of the ADP1 genome (here vanAB, pcaHG and hcaB).
Media and growth conditions for studies of aromatic compound consumption
Because catabolic studies were designed for comparison with past results, the media and growth conditions matched previous investigations. A defined minimal medium was used for culturing A. baylyi strains (27). This medium was supplemented with 2 mM benzoate and 2 mM p-hydroxybenzoate (POB) for carbon source consumption experiments and 20 mM pyruvate for natural transformation culturing. Escherichia coli XLI Blue cells (Stratagene) were used as plasmid hosts and grown in LB. All cultures were incubated at 37°C with aeration (shaking at 250 rpm).
Strain and plasmid construction for studies of aromatic compound consumption
DNA for plasmid construction was PCR-amplified with a high-fidelity polymerase, PrimeSTAR Max (Takara Biosciences) or Phusion (New England Biosciences), using the primers listed in Supplementary File S2 (Primers). Plasmids were assembled using an overlapping sequence method conducted in vivo in E. coli XL1-Blue competent cells (Stratagene) (28). In some cases, standard restriction digestion was followed by ligation (Quick Ligation Kit, New England Biosciences). Plasmids are described further in Supplementary Table S2. Chromosomal changes in A. baylyi were constructed by allelic replacement (29,30) and are listed in Supplementary Table S3. After transforming recipients with donor DNA, desired transformants were selected for drug resistance or growth in the presence of sucrose as previously described (30). Alternatively, cells were screened to detect transformants that lost the ability to metabolize POB by plating on solid minimal medium with 2 mM succinate and 2 mM POB. Cells that can grow on both carbon sources form larger colonies than those that can grow on succinate but not POB. The small colonies were then patched to test for growth on 5 mM POB as the sole carbon source. Isolates that were unable to grow on POB as the sole carbon source were further evaluated. All plasmids and strains were confirmed by PCR and/or regional DNA sequencing.
To create strains with altered PcaK expression, the pcaK coding sequence was first deleted from its normal operonic position (to generate ACN2451). ACN2451 was then used as the transformation recipient for linearized plasmids carrying pcaK controlled by different constitutive promoters. Each pcaK allele was introduced between convergently oriented chromosomal genes (quiA and ACIAD1717) with independent transcription ensured by a transcriptional terminator engineered downstream of quiA, as depicted in Supplementary Figure S44. A drug resistance cassette (ΩK52468) downstream of the plasmid-borne pcaK was used to select transformants that had undergone allelic replacement.
Creating random changes in the CatM/BenM binding site
To prevent CatM and BenM from binding to a site upstream of pcaU, changes were introduced by PCR with a mutagenic primer (SRB368) designed to eliminate two conserved features of this site important for interactions with CatM and BenM: (31) (i) a conserved sequence (T-N11-A) and (ii) a small region of dyad symmetry surrounding this sequence. The mutagenic primer was synthesized with random nucleotides (NNNN) in place of the chromosomal sequences in ADP1 at positions 1 708 958–1 708 961 and 1 708 969–1 708 972 (NCBI entry NC_005966). PCR products were cloned to generate plasmids (pBAC1794, pBAC1795, pBAC1797, pBAC1798, pBAC1799 and pBAC1800) in which there is an ΩK cassette downstream of pcaU. These plasmids were linearized and used to transform ADP1 to create strains in which the plasmid-borne alleles replaced the corresponding wild-type regions (ACN2529, ACN2496, ACN2498, ACN2530, ACN2526 and ACN2499, respectively). These strains were initially selected by the drug resistance conferred by ΩK. DNA sequencing confirmed the specific mutations that were introduced in these plasmids and strains. In all cases, the two key features of the wild-type BenM/CatM binding site (noted earlier) were eliminated (see Supplementary Tables S2 and 3). These strains retained the ability to use POB as the sole carbon source.
Alteration of sequences to prevent homologous recombination between two different synthetic promoters used on the genome of a single strain
DNA is identical in the regions surrounding the critical promoter sequences of different transcriptional constructs in the tool kit. To use two promoters (F6C5 and T5) to control the transcription of two adjacent cat-gene chromosomal regions, the DNA surrounding the promoter sequence of one promoter was altered to avoid the possibility of recombination between identical sequences. The lac promoter from pBTL-2 (32) was changed by site directed mutagenesis with primers SRB441 and SRB442 to generate the F6C5 promoter on plasmid pBAC1682.
Carbon source consumption experiments
For each strain tested for aromatic compound catabolism, cells were first grown in 5 ml minimal medium in test tubes overnight and sub-cultured 100 μl into 5 ml fresh medium to be grown overnight again. Then 1 ml of the second culture was used to inoculate 50 ml fresh medium in a 250 ml Erlenmeyer flask from which hourly samples were taken. For aromatic monomer co-consumption experiments, the inoculating cultures were grown on 2 mM POB or 2 mM benzoate and 2 mM POB, as indicated. After experimental samples were taken, cells were removed by filtration, and the supernatant was analyzed to monitor metabolites by high performance liquid chromatography (HPLC) as previously described (33,34). Retention times for POB and benzoate were 6 and 13.8 min, respectively. Data were plotted with the 0 time-point defined as being 1 h before depletion of any carbon source was first detected. Experiments were performed in biological triplicate. The optical density was measured at a wavelength of 600 nm on a spectrophotometer (Beckman DU 640 or Eppendorf Biophotometer Plus 6132).
RESULTS
Evaluating and optimizing ADP1 transformations
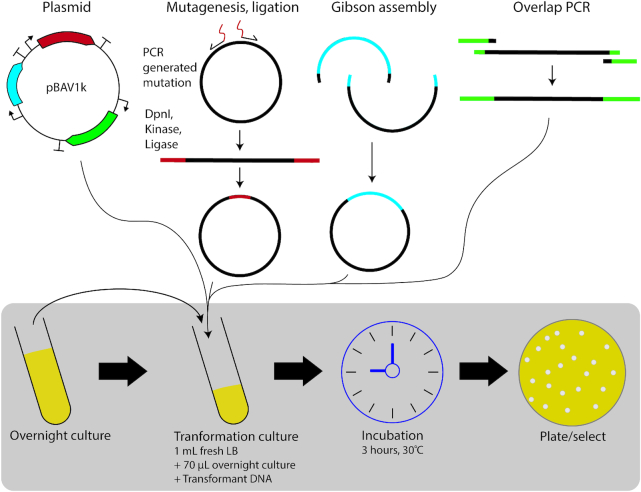
To begin, we wanted to validate a simple standardized cloning workflow. Previous works established varied transformation protocols (11–12,35–37), a complete single-gene deletion library (36) and an E. coli compatible broad-host-range vector (pBAV1k) (25), but lacked consensus for molecular cloning steps. Owing to ADP1’s natural competency and native homologous recombination machinery, transformation workflows follow a general outline of: (i) incubation of ADP1 with transforming DNA in either liquid or on solid medium and (ii) selection. Using a modified version of the available pBAV1k vector (pBWB162, pBAV1k-kanR-lacI-Trc-mCherry), we tested the simplest possible conditions (LB, liquid medium, short incubation times) and identified 30°C, 3-h incubation with as little as 25 ng of transforming DNA as efficient (Supplementary Figure S1). Overnight incubations provided no improvement in transformation efficiency compared to 3 h (Supplementary Figure S2). In addition, experiments run at 37°C negatively impacted transformation rates compared to 30°C (Supplementary Figure S3). Using this workflow, we successfully transformed plasmid DNA, ligation products, Gibson products and PCR products by natural competency (summarized in Figure 1). Owing to ADP1’s growth rate (11), these protocols not only require much less experimental effort (∼10 min of experimental time) compared to E. coli transformation protocols but provide colonies on the same timeframe as E. coli (overnight).
Figure 1.
ADP1 cloning workflow. Transformation of ADP1 follows a simple process of sub-culturing an overnight culture grown in LB from a glycerol stock, adding transforming DNA, incubating for a short period of time (3 h) at 30°C and then selecting. With this exact workflow for all cases listed, we were able to transform plasmid DNA (pBWB162 transformations), ligation products (promoter library creation), Gibson products (initial construction to insert mCherry into pBAV1k) and linear PCR products (chromosomal integration mapping).
Interestingly, ethanol precipitated pBWB162 miniprepped from ADP1 gave a 4.8-fold improvement in transformation efficiency compared to a miniprep from E. coli (Supplementary Figure S4), with ∼95 000 cfu/ml-μg for this 5 kb plasmid using our 3-h incubation workflow. In addition, we found that ADP1, which has no native plasmids, provides strong plasmid yields from a standard miniprep kit (>3–4 μg for a 5 ml overnight culture). Worth noting, to obtain clarified lysates for a standard miniprep kit, we needed to extend the centrifugation step after neutralization to two >10 min spins at 17 000 g (max speed) to obtain suitable DNA purity. In all, ADP1 transformation requires no specialized preparation or purchasing of competent cells, involves a single 10-min hands-on step compared to five steps of equal or greater complexity for E. coli heat shock, gives high transformation efficiencies, does not require extended time for colony growth and could be easily parallelized with simple liquid handling equipment.
Creating a promoter library and ribosome-binding site (RBS) variants
Precise control of gene expression is critical for engineering micro-organisms, so we next moved to construct expression tools (38). We first sought to validate previous work (39) with a set of common synthetic biology promoters. In addition, we wanted to compare these promoters to the bacterial components of a newly developed set of broad-spectrum promoters (40) along with potentially strongly expressing native ADP1 promoters chosen based on ADP1 transcriptomic data (41). Each promoter was put into the context of pBWB162, which was first tested for potential readthrough and inducibility and found to have minimal leakiness and to be stably induced with 1 mM IPTG (Supplementary Figures S5 and 6).
Testing this group of promoters (Supplementary Figure S7), we found that bacterial consensus-based promoters (Trc, Tac, T5) all gave strong expression. This result is perhaps expected as ADP1 carries an RpoD/σ70 homolog, and previous in vitro work showed that E. coli σ70 RNA polymerase gave transcription of an ADP1 gene with its native promoter and transcription factor (42). T7 did not give expression, in contrast to a previous study (39). The synthetic broad-spectrum promoters gave expression, but the relative expression strength of the four promoters did not match order of strength found in the original work (40) (Supplementary Figure S7). Though the native ADP1 promoter for gltI, potentially the strongest native promoter based on transcriptomic data (41), gave strong expression, it was still not as strong as Trc (Supplementary Figure S7). To confirm the expression results were not simply due to the specific conditions for promoter screening, such as endpoint measurement or the medium conditions, we ran time course experiments in both LB and an ADP1 minimal medium with succinate as the sole carbon source. These experiments confirmed expression from only the putative gltI promoter among the native ADP1 promoters tested (Supplementary Figures S8–11).
Because of the expression results, in combination with the promoter architecture of Trc being well understood while the architecture of gltI is not, we chose to construct a promoter library based on Trc. Leveraging ADP1’s natural competency, promoter library construction followed a simple workflow (Supplementary Figure S12). Mutagenic primers were used to introduce variability flanking and in between the bacterial consensus −35 and −10 boxes, and clones with a range of expression strength were isolated (Supplementary Figures S13 and 14). Sequences for the promoter library can be found in Supplementary Table S1, and full plasmid sequence files are available with the Supplementary Materials.
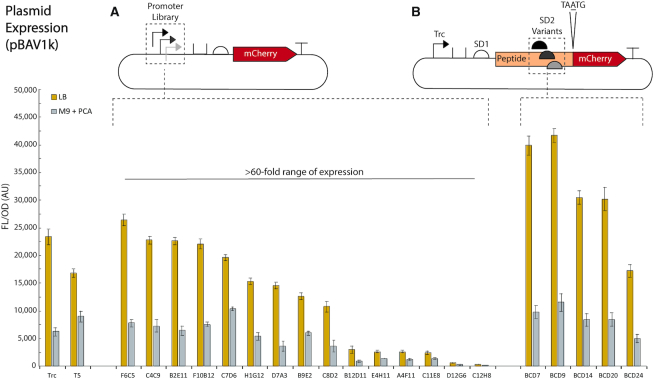
We sought to identify genetic parts with robustness to different conditions, and, as promoter expression characteristics can change depending on the growth medium, we rescreened the promoters in both LB and unmodified M9 with 8 mM PCA as the sole carbon source (Figure 2, Supplementary Figure S15 for OD dot overlay). Unless otherwise stated, all M9 cultures used PCA as the only carbon source. We chose PCA because it is a common intermediate in the breakdown of lignin via the β-ketoadipate pathway. Our 15-member promoter library shows 73-fold expression range in LB and 60-fold expression range in M9 (Figure 2A).
Figure 2.
Plasmid expression of promoter library and RBS variants. Built by introducing ‘N’ variability surrounding the −35 and −10 boxes of Trc in the context of pBWB162, our promoter library (A) shows 73-fold expression range for LB and 60-fold expression range for M9 with PCA. (B) Utilizing the BCD design to create RBS variants, with the promoter held constant (Trc), we observe expression variance related to ribosomal binding strength. All experiments were performed as biological triplicate on three separate days (nine replicates), except LB expression of D12G6, which only had eight replicates. Error bars represent SEM. SD = Shine-Delgarno.
Next, to test the effect of RBS modulation on expression, we picked a range of RBS variants from the previously developed bicistronic design (BCD) library (43). Briefly, this design involves two RBSs, where the first RBS initiates the translation of a leader peptide for which the final nucleotide of the stop codon overlaps with the start codon of the gene of interest (TAATG). This design is proposed to alleviate RBS performance variability due to mRNA secondary structure by utilizing the helicase activity of the ribosome. These RBS/BCD variants were successfully applied in ADP1 and showed a 2.4-fold range of expression in LB and 2.3-fold range of expression in M9 (Figure 2B).
Certain promoter and BCD variants performed differently in LB compared to M9-based expression, highlighting the need to screen in each medium before applying genetic parts to metabolic engineering contexts. Examples include Trc and T5 switching order of strength (P-value for difference in LB 0.00064, P-value for difference in M9 0.040), and C7D6 and B9E2 showing relatively greater expression strength in M9. To determine if plasmid copy number influences the expression differences, we carried out real-time qPCR for pBWB162 for both conditions (Supplementary Figures S16–18). The results showed pBWB162 has 3.16-fold higher relative copy number in LB compared to M9 (Supplementary Figure S18). Interestingly, averaging over all promoter and RBS parts in Figure 2, the FL/OD for LB is 3.17-fold greater than M9. This suggests plasmid copy number likely has an influence, although it may not fully account for the difference in expression. In addition, as discussed in the following sections, for chromosomal expression where the template copy number should be normalized to approximately one, LB and M9 conditions show much closer expression values. In summary, our promoter library and RBS variants together provide access to >100-fold range of expression strength in ADP1 in both media conditions.
Mapping genomic integrations
For industrial metabolic engineering applications, genomic integration of a metabolic pathway is typically desirable for stable long-term expression without a selective pressure (44). Because ADP1 readily incorporates DNA into the genome, genomic integration was straightforward. Accordingly, we tested a variety of chromosomal locations in ADP1 for their capacity for heterologous protein expression. To demonstrate the ease of ADP1 genomic integration, we chose 30 non-essential gene sites evenly spaced throughout the genome based upon a previous complete single-gene knockout study (36). In this approach our knock in cassette directly replaced the non-essential gene (GenBank files with the reporter cassette in a given locus are available with the Supplementary Materials). This scale of genomic integration, though not to the extent of random integration techniques such as transposon insertion (45), is greater than any other rational and targeted bacterial chromosomal expression screen, including previously constructed E. coli and Bacillus subtilis libraries (46,47).
To generate homologous recombination cassettes, we utilized an overlap PCR strategy to incorporate 500 bp of homology on either side of an insertion cassette (Supplementary Figure S19). Multiple attempts to use either 50 or 100 bps of flanking homology, introduced by PCR, for knock ins using this 3.7 kb insertion cassette gave no transformants in our hands. In addition, the λ-red system (48) appeared too toxic in ADP1, as multiple different plasmid constructs for gam-β-exo expression could not be stably maintained. To prevent transcriptional readthrough from biasing analyzed expression, our integration cassette bore three additional terminators to act as insulators (pBWB206). These insulators consisted of two upstream terminators and one downstream terminator following the vector's pre-existing terminator. Insulators were chosen based on a previously designed orthogonal terminator library (49). The inclusion of these insulators dampened expression from this vector, lowering plasmid-based expression 20-fold compared to the parent plasmid pBWB162 (Supplementary Figure S20). After overlap PCR assembly, linear PCR product was either directly added to the medium or gel extracted and then added to the medium following the established transformation workflow.
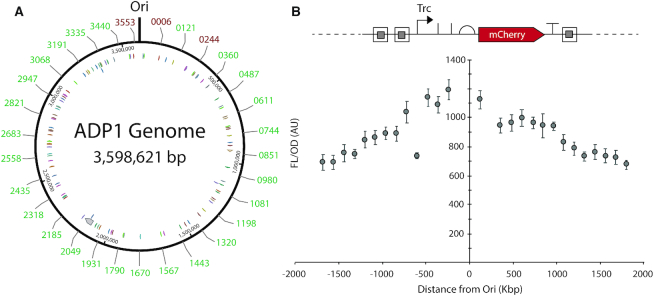
Of the 30 sites tested, 27 had correct transformants from a single attempt, which were identified by screening eight colonies by colony PCR for each. We tested these sites for expression, plotting expression with respect to distance from the origin of replication (Figure 3 for M9, Supplementary Figure S21 for LB). When a regression was plotted for expression with respect to distance from the origin of replication it gave an R2 0.787, and all data points fell within one standard deviation except the integration at ACIAD3068, which represents the stringent response protein RelA (Supplementary Figure S22). For consistency, the orientation of all expression cassettes was away from the origin of replication, and interestingly the (−) strand generally gave higher expression (Figure 3B). This was also true for two locations where we integrated the expression cassette in both orientations, although the expression difference was only statistically significant in LB and not M9 medium (Supplementary Figure S23). Overall, this mapping provides numerous and diverse integration sites for stable and effective protein expression in ADP1.
Figure 3.
ADP1 chromosomal expression mapping. (A) Representation of the ADP1 genome, highlighting sites of integration by their ACIAD number. We integrated our insulated mCherry expressing cassette (pBWB206) (B) at 30 locations evenly spaced throughout ADP1’s genome, exactly replacing the gene in that location, unless its reading frame overlapped with an upstream or downstream gene. In those cases, the upstream or downstream gene was left intact. Expression orientation was always away from the origin of replication (Ori). Of the 30 integration sites, 27 (A, green [lighter] numbers) produced a typical growth phenotype. The three red (darker) numbers represent integrations that were not successful (ACIAD006, ACIAD0244, and ACIAD3553). The 27 successful sites were assayed for mCherry expression. (B) Expression shows an obvious correlation to distance from the Ori. One notable exception is ACIAD3068, which corresponds to relA, a stringent response related gene. While this integration did not impact growth, lack of stringent response regulation may impact protein expression. All cultures were carried out in M9 with 8 mM PCA and in biological triplicate on three separate days (nine replicates). Error bars represent SEM. Circular ADP1 genome representation originally generated using the NCBI ADP1 genome viewed by Benchling.
Integration of promoter library and RBS variants into the genome
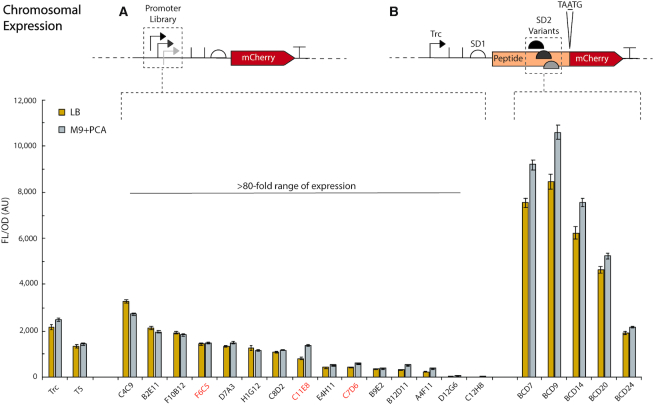
To further highlight the ease of genomic manipulation in ADP1, we integrated the entire promoter library and set of BCD variants at a single locus (ACIAD0980, genes vanAB). These knock ins were constructed with a similar overlap PCR strategy as the genomic integration mapping cassettes, but on a cassette without additional insulators. Typically, eight colonies were picked for screening by colony PCR and at least four of the eight had successful integration. Upon screening, the promoter library was found to give >80-fold range of expression for both LB and M9 (Figure 4A, Supplementary Figure S24 for OD dot overlay). M9-based expression was closer to LB expression when tested on the genome compared to the plasmid, which may be due to a more normalized copy number of the gene. In fact, the BCD variants actually showed greater expression per OD600 in M9 than LB (Figure 4B, Supplementary Figure S24 for OD dot overlay). There was a more significant decrease in expression by moving to the chromosome for the promoter library than for the BCD variants (Supplementary Figures S25–28). This is potentially due to lower copy numbers of mRNA being generated from chromosomal expression and the ability of the strong BCDs to maintain high translation per mRNA in both circumstances, although this hypothesis was not examined directly in our work. Chromosomal integration also afforded examination of the heterogeneity of expression between plasmid and genomically integrated cassettes by way of flow-cytometry, showing that expression from the chromosome gives a lower mean expression, narrower distribution of expression and fewer non-expressing cells compared to plasmid expression (Supplementary Figures S29–37). Worth noting, three promoters in the library changed their position in terms of expression strength (F6C5, C11E8, C7D6, all highlighted with orange [lighter color] on Figure 4A) upon chromosomal integration. To our knowledge, this is the most thorough comparison between plasmid and chromosomal integration expression among a consistent set of promoters in the literature.
Figure 4.
Chromosomal expression of promoter library and RBS variants. By integrating the entire library at ACIAD0980 (vanAB), we were able to examine the chromosomal expression behavior of our promoter library and set of BCD variants. (A) The promoter library performs similarly to plasmid expression, albeit with lower overall expression. However, the library still gives a >80-fold expression range for both LB and M9. In addition, M9 expression more closely matches LB expression on the chromosome compared to plasmid-based expression. (B) The BCD variants show less of a decrease in expression when moved to the chromosome, particularly M9 BCD expression. A few promoter library variants behaved differently with respect to relative expression within the library, highlighted in orange (lighter color, promoters F6C5, C11E8, and C7D6). All experiments were run in biological triplicate on three separate days (nine replicates total). Error bars represent SEM.
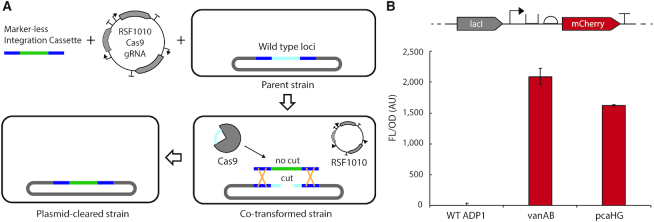
One-step, Cas9-based marker-less and scar-less genomic integration
At last, to enable facile, antibiotic-free successive genomic integrations, we tested a method to obtain marker-less genomic modification. Though both sacB (50) and tdk (11–12,36) counter-selection approaches have been successfully applied in ADP1, we hypothesized that a Cas9-based approach may be more efficient. As ADP1 has no known non-homologous end joining machinery, Cas9 genomic cutting should be lethal. Therefore, co-transforming a marker-less genomic integration cassette that replaces the original locus, along with a plasmid bearing active Cas9 and a gRNA targeting the wild-type locus, could provide a one-step method for marker-less genomic integration (Supplementary Figure S38).
By modifying a previously developed dCas9 bearing vector that replicates in ADP1, generously provided by the Barrick laboratory (26), we created a vector with constitutively expressed active Cas9 (pBWB415). After exchanging the gRNA to target-specific ADP1 loci, we tested the specificity of Cas9 to cut the target of interest. We found that only strains with the target locus removed allowed transformation of the plasmid (Supplementary Figure S39). No colony growth was observed from wild-type ADP1 transformed with a pBWB419, pBWB424 and pBWB431, with gRNAs that targeted vanAB, pcaHG and hcaB respectively, indicating an exceptionally low false positive rate.
Next we tested the co-transformation of a marker-less integration cassette along with a plasmid bearing constitutively expressed Cas9 and a gRNA targeting the locus to be exchanged (Figure 5A). After modifying our transformation protocol to improve efficiency (≥500 ng DNA for each part, 6-h incubation), we observed transformants. Though the transformation efficiency is low (∼10 cfu/ml-μg), nearly all colonies screened were positive hits. For the integration of a marker-less lacI-Trc-mCherry cassette at pcaHG (Supplementary Figure S40), seven of the eight colonies screened by colony PCR were clonally pure after patching. Following the integration, colonies are streaked onto non-selective LB agar to clear the Cas9-bearing plasmid. The constitutively expressed Cas9 represents a burden sufficient to clear the plasmid by the next day (Supplementary Figure S40).
Figure 5.
Cas9-based marker-less, scar-less genomic integration. (A) Shows workflow, where first a marker-less integration cassette with flanking regions of 500 bp of homology to the chromosome and the RSF1010-Cas9-gRNA plasmid are co-transformed into a parent strain. If co-transformation is successful, the native homologous recombination machinery of ADP1 will exchange alleles, and Cas9 will be unable to cut the target locus, allowing survival for the RSF1010 plasmid. Following, streaking on non-selective medium clears the plasmid. (B) Shows fluorescent expression from marker-less integration of lacI-Trc-mCherry at both the vanAB and pcaHG loci. Error bars are standard deviation for biological triplicate.
Subsequent patching indicated clearance of the Cas9 plasmid, and sequencing of a colony PCR product verified the correct integration. After this process, the integration is retained and protein expression is observed from the marker-less cassette (Figure 5B). It is worth noting that the full clearance and validation process occurs after three separate patching or streaking procedures and over the course of several days. In our hands, the cassette is maintained throughout this process and still functionally expresses. In summary, this Cas9-based approach provides a simple and rapid one-step integration, with a subsequent single day plasmid clearance, that should enable accelerated ADP1 genomic engineering.
Tool application for lignin valorization
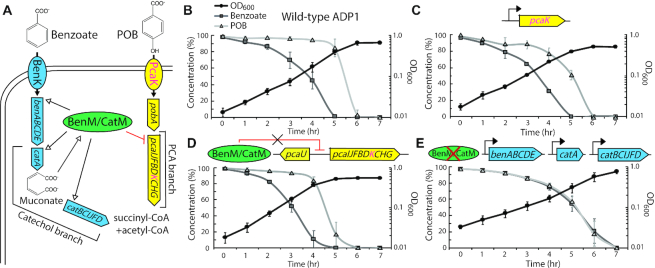
To demonstrate the applicability of these tools and their reliability and reproducibility in different labs (at Northwestern University and at the University of Georgia), we studied aspects of ADP1’s catabolite repression. The ultimate goal is to enable simultaneous consumption of lignin-derived aromatic compounds in mixtures generated by the pretreatment of biomass (14,18). This type of consolidated bioprocessing depends on engineering bacteria for rapid production of valuable end-products and may require overriding catabolite repression, which imposes preferential carbon source utilization (51,52). In ADP1, all aromatic compounds are consumed via one of two branches of the β-ketoadipate pathway (Supplementary Figure S41). ADP1 preferentially consumes benzoate, degraded via the catechol branch of this pathway before POB degraded via the PCA branch (33). Studies of benzoate and POB co-metabolism can, therefore, serve as a proxy for more complex mixtures, including lignin hydrolysates.
The mechanisms for hierarchical consumption of individual aromatic compounds in a mixture remain unclear, in part, because hypotheses generated from past studies have been difficult to test. The genetic tools described in this report provide new avenues for investigating the significance of two key regulatory observations. The first such observation is that the production of a metabolite, muconate, is essential for benzoate-mediated inhibition of POB degradation (52). The second is that two paralogous transcriptional regulators, BenM and CatM, play an important role in controlling the preferential consumption of benzoate in the presence of POB (33). BenM and CatM serve as activators and as repressors, and both respond to muconate (53). As activators, BenM and CatM are required for muconate-dependent transcription of the ben and cat genes for benzoate consumption. In contrast, they interact with muconate to bind DNA in a position that can prevent the transcription of a large pca operon (33). One hypothesis to explain the basis of preferential carbon source consumption is that repression of a gene in this operon (pcaK, which encodes a POB transporter), prevents POB consumption by lowering its uptake and thereby preventing its further catabolism (Figure 6A and Supplementary Figure S42). To investigate this hypothesis, we used the new gene expression tools to determine how the following affect co-consumption: (i) constitutive expression of PcaK, (ii) ablation of BenM/CatM binding sites that could be used to repress pcaK transcription and (iii) constitutive expression of genes for benzoate degradation in the absence of both regulators (Supplementary Figure S43).
Figure 6.
Co-consumption of benzoate and POB. (A) Schematic depicting regulation and metabolism of POB and benzoate. These two growth substrates, which enter the PCA and catechol branches of the β-ketoadipate pathway, respectively, are funneled to central carbon metabolism. POB uptake and degradation are encoded by the pob and pca genes, whereas benzoate uptake and degradation are encoded by the ben and cat genes. Benzoate consumption requires two transcriptional regulators (BenM and CatM) that, in response to muconate, activate ben- and cat-gene transcription. These regulators also bind upstream of the pcaIJFBDKCHG operon in a muconate-dependent fashion at a site that should repress transcription (33). Consumption patterns are shown for: (B) the wild-type strain, ADP1, (C) a strain (ACN2469) where the transporter gene pcaK is transcribed constitutively from a synthetic promoter (D5H6), (D) a strain (ACN2596) where the binding site for BenM/CatM upstream of the pca operon has been scrambled to prevent binding of these regulators and (E) a strain (ACN2569) where benM has been inactivated, catM has been deleted and where the genes for benzoate degradation, which are normally activated by BenM and CatM, are under constitutive expression. In ACN2569 the ben genes are controlled by a promoter mutation (PbenA5147) and the cat genes are transcribed from synthetic promoters (T5 for catA and F6C5 for catBCIJFD). Data shown are average of at least three biological replicates. Error bars represent standard deviation.
To facilitate uptake of POB in the presence of benzoate, the PcaK transporter was constitutively expressed under the control of several synthetic promoters such that its transcription would not be repressed by BenM or CatM. Despite using different strength promoters for PcaK expression, these attempts to increase POB transport did not accelerate its degradation (Supplementary Figure S44). Sequential consumption of benzoate before POB was observed as with the wild-type (Figure 6C compared to 6B). Second, to alleviate BenM and CatM-based repression of pcaK, the repressive binding sites upstream of the pca operon were scrambled. Again, this change was not sufficient to allow co-consumption (Figure 6D and Supplementary Figure S45). However, deletion of BenM and CatM, coupled with constitutive expression of benzoate degrading enzymes, led to co-consumption of POB and benzoate (Figure 6E). Collectively, these results imply that repression of the pca operon by BenM and CatM is insufficient to account for muconate-dependent inhibition of POB degradation. Since the removal of both regulators alleviated this type of catabolite repression, despite the production of muconate during benzoate catabolism, it is likely that BenM and CatM control carbon source preferences by acting at multiple loci. These studies, and additional results and discussions in a Supplementary Note, demonstrate the utility and ease of implementation for these tools, including the chromosomal use of multiple synthetic promoters in the same strain. This methodology not only contributes to improved understanding of multiple carbon source consumption but also delivers a strain that simultaneously consumes benzoate and POB that can serve as a starting point for further improvements to optimize the consumption of lignin-derived mixtures.
DISCUSSION
The cumulative tools developed in this study, along with other recent works that have established an additional plasmid vector (26), CRISPRi (26) and ADP1 chromosomal evolution tools (34), effectively equip ADP1 as a model organism for lignin-based metabolic engineering and other synthetic biology applications. ADP1 has significant advantages over other micro-organisms for DBTL cycles. The simplicity associated with natural transformation and facile allelic replacement lends itself to automation, liquid handling and incorporation into biofoundry workflows (22), especially as advances in DNA synthesis have alleviated the bottleneck of gene synthesis. Modifying the genome is straightforward, on par with the simplicity of plasmid transformation, highlighted by the >50 targeted chromosomal integrations carried out in this study. As the chromosomal tools developed in this study, specifically locations for targeted integration for successful heterologous expression and the testing of a promoter library and RBS variants for their chromosomal expression, exceed more established hosts such as E. coli and B. subtilis, we expect this work to promote ADP1’s use as a host for stable and robust engineering.
Our validation in two different laboratories indicates the tool set is reproducible and scientifically rigorous. Further highlighting the importance of these tools is the ease with which regulatory hypotheses were tested, revealing new information about preferential carbon source utilization and generating strains for use in lignin valorization. Milestone plasmids from this study have been deposited for easy access on Addgene, and cloning files are available with the Supplementary Materials. In addition, we have attempted to document approaches that were unsuccessful as a service to others that might work with ADP1.
Because of its ease of engineering, the metabolic versatility of ADP1 may expand to metabolic engineering applications beyond lignin-derived aromatics to more traditional glucose-based processes. As ADP1 exclusively utilizes the cofactor-generating Entner-Doudoroff pathway while maintaining high growth rates, as opposed to E. coli's preferred use for maximal growth of the Embden–Meyerhof–Parnas glycolytic pathway (54), there may be benefit to using ADP1 to produce products with heavy cofactor needs. Indeed, Santala and coworkers have already provided demonstration of glucose-based metabolic engineering in ADP1 (55). Ultimately, we envision this host will be adopted beyond the lignin community and will play a role in the future of sustainability and synthetic biology.
DATA AVAILABILITY
Cloning files for all genetic constructs are provided with the supplementary materials as Genebank files. Milestone plasmids strains have been deposited to Addgene. [Addgene numbers: pBWB162 (#140634), pBWB206 (#140635), pBWB294 (#140636), pBWB419 (#140637), pBWB424 (#140638)].
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Gregg T. Beckham, Dr Isabel Pardo and Dr Christopher W. Johnson for their helpful conversations about this manuscript. We would like to thank Prof. Joshua Leonard for the use of the CFX96 for the RT-qPCR experiments. We acknowledge the generous gift of the CRISPRi-ME plasmid and ADP1 ISx strain provided by Prof. Jeffrey Barrick We gratefully acknowledge the contributions of Melissa Tumen-Velasquez to the construction and design of some ACN strains and pBAC plasmids. Northwestern University NUSeq Core Facility; Northwestern Flow Core.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH Biotechnology Training Grant [T32-GM008449-23 to B.W.B., E.A.]; National Science Foundation (NSF), Collaborative Grant [MCB 1614953 to K.E.J.T., MCB 1615365 to E.L.N.]. Funding for open access charge: NSF Grant [MCB 1614953].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sheldon R.A. Fundamentals of green chemistry: efficiency in reaction design. Chem. Soc. Rev. 2012; 41:1437–1451. [DOI] [PubMed] [Google Scholar]
- 2. Keijer T., Bakker V., Slootweg J.C.. Circular chemistry to enable a circular economy. Nat. Chem. 2019; 11:190–195. [DOI] [PubMed] [Google Scholar]
- 3. Keasling J.D. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 2008; 3:64–76. [DOI] [PubMed] [Google Scholar]
- 4. Liew F.M., Martin M.E., Tappel R.C., Heijstra B.D., Mihalcea C., Köpke M.. Gas Fermentation-A flexible platform for commercial scale production of low-carbon-fuels and chemicals from waste and renewable feedstocks. Front. Microbiol. 2016; 7:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bomgardner M.M. LanzaTech raises funds and expands its footprint. C&EN. 2018; 96: [Google Scholar]
- 6. Nikel P.I., Chavarría M., Danchin A., de Lorenzo V.. From dirt to industrial applications: Pseudomonas putida as a Synthetic Biology chassis for hosting harsh biochemical reactions. Curr. Opin. Chem. Biol. 2016; 34:20–29. [DOI] [PubMed] [Google Scholar]
- 7. Köpke M., Held C., Hujer S., Liesegang H., Wiezer A., Wollherr A., Ehrenreich A., Liebl W., Gottschalk G., Dürre P.. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:13087–13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weinstock M.T., Hesek E.D., Wilson C.M., Gibson D.G.. Vibrio natriegens as a fast-growing host for molecular biology. Nat. Methods. 2016; 13:849–851. [DOI] [PubMed] [Google Scholar]
- 9. Young D.M., Parke D., Ornston L.N.. Opportunities for genetic investigation afforded By Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu. Rev. Microbiol. 2005; 59:519–551. [DOI] [PubMed] [Google Scholar]
- 10. de Berardinis V., Durot M., Weissenbach J., Salanoubat M.. Acinetobacter baylyi ADP1 as a model for metabolic system biology. Curr. Opin. Microbiol. 2009; 12:568–576. [DOI] [PubMed] [Google Scholar]
- 11. Suárez G.A., Renda B.A., Dasgupta A., Barrick J.E.. Reduced mutation rate and increased transformability of Transposon-Free Acinetobacterbaylyi ADP1-ISx. Appl. Environ. Microbiol. 2017; 83:e01025-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Metzgar D., Bacher J.M., Pezo V., Reader J., Döring V., Schimmel P., Marlière P., de Crécy-Lagard V.. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 2004; 32:5780–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elliott K.T., Neidle E.L.. Acinetobacter baylyi ADP1: transforming the choice of model organism. IUBMB Life. 2011; 63:1075–1080. [DOI] [PubMed] [Google Scholar]
- 14. Salvachúa D., Karp E.M., Nimlos C.T., Vardon D.R., Beckham G.T.. Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem. 2015; 17:4951–4967. [Google Scholar]
- 15. Harwood C.S., Parales R.E.. The β-Ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996; 50:553–590. [DOI] [PubMed] [Google Scholar]
- 16. Schutyser W., Renders T., Van den Bosch S., Koelewijn S.-F., Beckham G.T., Sels B.F.. Chemicals from lignin: An interplay of lignocellulose fractionation, depolymerisation, and upgrading. Chem. Soc. Rev. 2018; 47:852–908. [DOI] [PubMed] [Google Scholar]
- 17. Beckham G.T., Johnson C.W., Karp E.M., Salvachúa D., Vardon D.R.. Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 2016; 42:40–53. [DOI] [PubMed] [Google Scholar]
- 18. Linger J.G., Vardon D.R., Guarnieri M.T., Karp E.M., Hunsinger G.B., Franden M.A., Johnson C.W., Chupka G., Strathmann T.J., Pienkos P.T. et al.. Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:12013–12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elmore J.R., Furches A., Wolff G.N., Gorday K., Guss A.M.. Development of a high efficiency integration system and promoter library for rapid modification of Pseudomonas putida KT2440. Metab. Eng. Commun. 2017; 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cook T.B., Rand J.M., Nurani W., Courtney D.K., Liu S.A., Pfleger B.F.. Genetic tools for reliable gene expression and recombineering in Pseudomonas putida. J. Ind. Microbiol. Biotechnol. 2018; 45:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DeLorenzo D.M., Rottinghaus A.G., Henson W.R., Moon T.S.. Molecular toolkit for gene expression control and genome modification in Rhodococcus opacus PD630. ACS Synth. Biol. 2018; 7:727–738. [DOI] [PubMed] [Google Scholar]
- 22. Hillson N., Caddick M., Cai Y., Carrasco J.A., Chang M.W., Curach N.C., Bell D.J., Le Feuvre R., Friedman D.C., Fu X. et al.. Building a global alliance of biofoundries. Nat. Commun. 2019; 10:2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Juni E., Janik A.. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J. Bacteriol. 1969; 98:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vaneechoutte M., Young D.M., Ornston L.N., Baere T. De, Nemec A., Van Der Reijden T., Carr E., Tjernberg I., Dijkshoorn L.. Naturally transformable Acinetobacter sp. Strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl. Environ. Microbiol. 2006; 72:932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bryksin A.V., Matsumura I.. Rational design of a plasmid origin that replicates efficiently in both Gram-Positive and Gram-Negative bacteria. PLoS One. 2010; 5:e13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geng P., Leonard S.P., Mishler D.M., Barrick J.E.. Synthetic genome defenses against selfish DNA elements stabilize engineered bacteria against evolutionary failure. ACS Synth. Biol. 2019; 8:521–531. [DOI] [PubMed] [Google Scholar]
- 27. Singh A., Bedore S.R., Sharma N.K., Lee S.A., Eiteman M.A., Neidle E.L.. Removal of aromatic inhibitors produced from lignocellulosic hydrolysates by Acinetobacter baylyi ADP1 with formation of ethanol by Kluyveromyces marxianus. Biotechnol. Biofuels. 2019; 12:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kostylev M., Otwell A.E., Richardson R.E., Suzuki Y.. Cloning should be simple: Escherichia coli DH5α-mediated assembly of multiple DNA fragments with short end homologies. PLoS One. 2015; 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neidle E.L., Hartnett C., Ornston L.N.. Characterization of Acinetobacter calcoaceticus catM, a repressor gene homologous in sequence to transcriptional activator genes. J. Bacteriol. 1989; 171:5410–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seaton S.C., Elliott K.T., Cuff L.E., Laniohan N.S., Patel P.R., Neidle E.L.. Genome-wide selection for increased copy number in Acinetobacter baylyi ADP1: Locus and context-dependent variation in gene amplification. Mol. Microbiol. 2012; 83:520–535. [DOI] [PubMed] [Google Scholar]
- 31. Alanazi A.M., Neidle E.L., Momany C.. The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators. Acta Crystallogr. D Biol. Crystallogr. 2013; 69:1995–2007. [DOI] [PubMed] [Google Scholar]
- 32. Lynch M.D., Gill R.T.. Broad host range vectors for stable genomic library construction. Biotechnol. Bioeng. 2006; 94:151–158. [DOI] [PubMed] [Google Scholar]
- 33. Brzostowicz P.C., Reams A.B., Clark T.J., Neidle E.L.. Transcriptional Cross-Regulation of the catechol and protocatechuate branches of the β-ketoadipate pathway contributes to carbon source-dependent expression of the Acinetobacter sp. Strain ADP1 pobA gene. Appl. Environ. Microbiol. 2003; 69:1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tumen-Velasquez M., Johnson C.W., Ahmed A., Dominick G., Fulk E.M., Khanna P., Lee S.A., Schmidt A.L., Linger J.G., Eiteman M.A. et al.. Accelerating pathway evolution by increasing the gene dosage of chromosomal segments. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:7105–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santala S., Efimova E., Kivinen V., Larjo A., Aho T., Karp M., Santala V.. Improved triacylglycerol production in Acinetobacter baylyi ADP1 by metabolic engineering. Microb. Cell Fact. 2011; 10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Berardinis V., Vallenet D., Castelli V., Besnard M., Pinet A., Cruaud C., Samair S., Lechaplais C., Gyapay G., Richez C. et al.. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 2008; 4:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luo J., Lehtinen T., Efimova E., Santala V., Santala S.. Synthetic metabolic pathway for the production of 1-alkenes from lignin-derived molecules. Microb. Cell Fact. 2019; 18:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Biggs B.W., De Paepe B., Santos C.N.S., de Mey M., Kumaran Ajikumar P.. Multivariate modular metabolic engineering for pathway and strain optimization. Curr. Opin. Biotechnol. 2014; 29:156–162. [DOI] [PubMed] [Google Scholar]
- 39. Murin C.D., Segal K., Bryksin A., Matsumura I.. Expression vectors for Acinetobacter baylyi ADP1. Appl. Environ. Microbiol. 2012; 78:280–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang S., Liu Q., Zhang Y., Du G., Chen J., Kang Z.. Construction and characterization of broad-spectrum promoters for synthetic biology. ACS Synth. Biol. 2018; 7:287–291. [DOI] [PubMed] [Google Scholar]
- 41. Stuani L., Lechaplais C., Salminen A.V., Ségurens B., Durot M., Castelli V., Pinet A., Labadie K., Cruveiller S., Weissenbach J. et al.. Novel metabolic features in Acinetobacter baylyi ADP1 revealed by a multiomics approach. Metabolomics. 2014; 10:1223–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bundy B.M., Collier L.S., Hoover T.R., Neidle E.L.. Synergistic transcriptional activation by one regulatory protein in response to two metabolites. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7693–7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.-A., Tran A.B., Paull M., Keasling J.D., Arkin A.P. et al.. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013; 10:354–360. [DOI] [PubMed] [Google Scholar]
- 44. Tyo K.E.J., Ajikumar P.K., Stephanopoulos G.. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat. Biotechnol. 2009; 27:760–765. [DOI] [PubMed] [Google Scholar]
- 45. Scholz S.A., Diao R., Wolfe M.B., Fivenson E.M., Lin X.N., Freddolino P.L.. High-resolution mapping of the Escherichia coli chromosome reveals positions of high and low transcription. Cell Syst. 2019; 8:212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bryant J.A., Sellars L.E., Busby S.J.W., Lee D.J.. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014; 42:11383–11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sauer C., Syvertsson S., Bohorquez L.C., Cruz R., Harwood C.R., van Rij T., Hamoen L.W.. Effect of genome position on heterologous gene expression in Bacillus subtilis: An unbiased analysis. ACS Synth. Biol. 2016; 5:942–947. [DOI] [PubMed] [Google Scholar]
- 48. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen Y.-J., Liu P., Nielsen A.A.K., Brophy J.A.N., Clancy K., Peterson T., Voigt C.A.. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods. 2013; 10:659–664. [DOI] [PubMed] [Google Scholar]
- 50. Jones R.M., Williams P.A.. Mutational analysis of the critical bases involved in activation of the AreR-regulated σ54-dependent promoter in Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 2003; 69:5627–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bleichrodt F.S., Fischer R., Gerischer U.C.. The β-ketoadipate pathway of Acinetobacter baylyi undergoes carbon catabolite repression, cross-regulation and vertical regulation, and is affected by Crc. Microbiology. 2010; 156:1313–1322. [DOI] [PubMed] [Google Scholar]
- 52. Gaines G.L.I., Smith L., Neidle E.L.. Novel nuclear magnetic resonance spectroscopy methods demonastrate preferential carbon source utilization by Acinetobacter calcoaceticus. J. Bacteriol. 1996; 178:6833–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Craven S.H., Ezezika O.C., Momany C., Neidle E.L.. Gerisher U. LysR homologs in Acinetobacter: insights into a diverse and prevalent family of transcriptional regulators. Acinetobacter Molecular Biology. 2008; Norfolk: Caister Academic Press; 163–202. [Google Scholar]
- 54. Ng C.Y., Farasat I., Maranas C.D., Salis H.M.. Rational design of a synthetic Entner-Doudoroff pathway for improved and controllable NADPH regeneration. Metab. Eng. 2015; 29:86–96. [DOI] [PubMed] [Google Scholar]
- 55. Lehtinen T., Efimova E., Santala S., Santala V.. Improved fatty aldehyde and wax ester production by overexpression of fatty acyl-CoA reductases. Microb. Cell Fact. 2018; 17:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cloning files for all genetic constructs are provided with the supplementary materials as Genebank files. Milestone plasmids strains have been deposited to Addgene. [Addgene numbers: pBWB162 (#140634), pBWB206 (#140635), pBWB294 (#140636), pBWB419 (#140637), pBWB424 (#140638)].