Abstract
An increasing number of studies have shown that quantitative trait loci (QTLs) at the end of chromosome 1 identified in different chicken breeds and populations exert significant effects on growth traits in chickens. Nevertheless, the causal genes underlying the QTL effect remain poorly understood. Using an updated gene database, a novel lncRNA (named LncFAM) was found at the end of chromosome 1 and located in a growth and digestion QTL. This study showed that the expression level of LncFAM in pancreas tissues with a high weight was significantly higher than that in pancreas tissues with a low weight, which indicates that the expression level of LncFAM was positively correlated with various growth phenotype indexes, such as growth speed and body weight. A polymorphism screening identified four polymorphisms with strong linkage disequilibrium in LncFAM: a 5-bp indel in the second exon, an A/G base mutation, and 7-bp and 97-bp indels in the second intron. A study of a 97-bp insertion in the second intron using an F2 chicken resource population produced by Anka and Gushi chickens showed that the mutant individuals with genotype II had the highest values for body weight (BW) at 0 days and 2, 4, 6, 8, 10 and 12 weeks, shank girth (SG) at 4, 8 and 12 weeks, chest width (CW) at 4, 8 and 12 weeks, body slant length (BSL) at 8 and 12 weeks, and pelvic width (PW) at 4, 8 and 12 weeks, followed by ID and DD genotypes. The amplification and typing of 2,716 chickens from ten different breeds, namely, the F2 chicken resource population, dual-type chickens, including Xichuan black-bone chickens, Lushi green-shell layers, Dongxiang green-shell layers, Changshun green-shell layers, and Gushi chickens, and commercial broilers, including Ross 308, AA, Cobb and Hubbard broilers, revealed that II was the dominant genotype. Interestingly, only genotype II existed among the tested populations of commercial broilers. Moreover, the expression level in the pancreas tissue of Ross 308 chickens was significantly higher than that in the pancreas tissue of Gushi chickens (P < 0.001), which might be related to the conversion rates among different chickens. The prediction and verification of the target gene of LncFAM showed that LncFAM might regulate the expression of its target gene FAM48A through cis-expression. Our results provide useful information on the mutation of LncFAM, which can be used as a potential molecular breeding marker.
Keywords: chicken, indel, chromosome 1, QTL, growth traits, LncFAM
Introduction
Growth traits are important economic traits in the poultry industry, and many researchers have performed QTL mapping of chicken growth traits and identified QTLs located on chicken chromosome 2 (GGA2) (Honkatukia et al., 2005), GGA3 (Wang et al., 2012), GGA4 (Nassar et al., 2012) and GGAZ (Wang et al., 2012). Among these chromosomes, GGA1, which is the largest chromosome in chickens, contains many QTLs affecting body weight (Zhang et al., 2013), growth (Gao et al., 2010a), food intake (Yuan et al., 2015), chest muscle weight (Ikeobi et al., 2004) and fat deposition (Ikeobi et al., 2003). Different worldwide research groups have identified a QTL at the end of GGA1 that exerts substantial effects on growth traits in different chicken breeds and populations (Hee-Bok et al., 2006; Liu et al., 2008; Uemoto et al., 2009; Besnier et al., 2011; Podisi, et al., 2013; Sheng et al., 2013). However, due to the limited recombination events investigated by most research groups, the epistasis effect is seriously disturbed, the genetic marker density is insufficient, and the progress of fine mapping in this region is slow. These issues represent some obstacles to the study of candidate genes for growth traits. With the update of the NCBI genome database on March 28, 2018, many new genes have been discovered. Among these genes, LOC107052041 (named LncFAM according to its target gene), which is a novel lncRNA located at 173.5 Mb on GGA1, was annotated. LncFAM is located in QTLs that were associated with growth in an AIL pedigree from two outbred chicken lines and a Chinese indigenous × commercial broiler chicken cross population (Besnier et al., 2011; Sheng et al., 2013) and with daily feed intake and efficiency in a White Leghorn and Dongxiang reciprocal cross population (Yuan et al., 2015). Therefore, LncFAM is likely the major functional gene involved in chicken growth and development. In this study, four polymorphisms with strong linkage disequilibrium in LncFAM (a 5-bp indel in the second exon, an A/G base mutation, and 7-bp and 97-bp indels in the second intron) were found. To facilitate detection, we used 97 bp as the detection target. The aims of this study were to explore whether this gene is a major gene affecting chicken growth and to develop valuable molecular genetic markers for broiler breeding. The associations between the polymorphisms caused by a 97-bp indel and various growth traits of chickens, the differences in the genotype distributions among different breeds and the gene expression profile in different tissues were analyzed.
Materials and Methods
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All the animal experiments were performed according to the Regulations of the Chinese National Research Council (1994) and approved by the Henan Agricultural University Institutional Animal Care and Use Committee (Permit Number: 11–0085).
Laboratory animals and data collection
The F2 resource population was described by Han et al. (2011). In detail, the phenotypic and genotypic associations investigated in this experiment were selected from the Gushi and Anka chicken families (4 Anka ♂ × 24 Gushi ♀, orthogonal, 2 Gushi ♂ × 12 Anka ♀, reverse cross, 70 F1 individuals). A rooster was then selected from the F1 offspring of each pedigree. To best select the F1 hens with the widest segregation of F2 traits, hens distributed in various families were selected, and individuals with a rich appearance and heterozygosity were identified. According to a male:female ratio of 1:9, the F2 generation was produced by mating with hens belonging to another family hens (no relationship between the male and female chickens). The F2 generation consisted of seven families, and Anka chickens were the male parents of four orthogonal lines. Gushi chickens represented the male parents of the three reverse cross, which comprised 42 grandparents, 70 F1 parents, and 800 F2 chickens. The F2 resource population of hybrid progenies was raised in the same environment, and the animals were provided free access to food and water. At the age of 84 days, each bird was euthanized by cervical dislocation followed by decapitation. The following growth traits were measured during this period: body weight (BW) at 0 days and 2, 4, 6, 8, 10 and 12 weeks; shank length (SL) at 0, 4, 8 and 12 weeks; and shank girth (SG), chest width (CW), body slant length (BSL) and pelvis width (PW) at 4, 8 and 12 weeks. Two samples of blood were collected from the jugular vein during slaughter. One sample was placed in an anticoagulant tube for DNA extraction and then stored at -20℃. The other sample was placed in a centrifuge tube for separation of the serum and stored at -40℃. After blood collection, 800 individuals were slaughtered to determine the carcass weights, such as the semi-evisceration weight (SEW), evisceration weight (EW), sebum weight (SW), sebum weight percentage (SWP), breast muscle weight (BMW), leg muscle weight (LMW), and carcass weight (cW) (Han et al., 2011).
The typing of the novel lncRNAs was studied among different varieties, such as dual-type chickens, including Xichuan black-bone chickens (XC, n = 266, 6 weeks), Lushi green-shell layers (LS, n = 143, 6 weeks), Dongxiang green-shell layers (DX, n = 129, 16 weeks), Changshun green-shell layers (CS, n = 92, 6 weeks), and Gushi chickens (GS, n = 143, 16 weeks), which were obtained from Henan, Henan, Jiangxi, Henan, and Guizhou, respectively, and commercial broilers, namely, Ross 308 (n = 172), AA (n = 300), Cobb (n = 212) and Hubbard (n = 459) broilers. Blood samples were collected from these varieties in the laboratory for DNA extraction and typing. The genomic information for Kauai feral chickens, red jungle fowls, Tibetan chickens and Fighting chickens was obtained from published data in the NCBI database (Gering et al., 2015; Wang et al., 2015).
Genomic DNA extraction and PCR
Genomic DNA was extracted from blood using a DNA extraction kit (TaKaRa MiniBEST Whole Blood Genomic DNA Extraction Kit), and its quality was assessed. The primers used to amplify the target fragments were designed by Premier Primer 6.0 software (Premier Biosoft International, Palo Alto, CA, USA) and synthesized by Sangon Biotech Company (Shanghai, China). The primer pairs used in this study were listed in Table 1.
Table 1.
Details of primer pairs for the chicken LncFAM gene
| Gene | Primer, 5'-3' | Size, bp | Annealing temperature, °C | Accession number |
|---|---|---|---|---|
| LncFAM 97bp-insertion detection | F: TCTCCACCCATTTTTATGCTGC R: TGTTGCCAGAAATATGCTTGCT | 206/303 | 60 | NC_006088 |
| LncFAM polymorphic sites detection | F: CCCTACTCCCCTTTCCCTTTT R: GGAAACCCTGAGTCTGAAGATAGT | 1689 | 60 | NC_006088 |
| RT- LncFAM | F: TGTGCAAGGGATTGGCATCA R: CATTTCCAGAAAAGGGCCGTG | 157 | 60 | XR_003073445 |
| RT-FAM48A | F: TGAAAGTGCTCGTCAGAGACC R: AGACATGACAACGTCTCCTGC | 180 | 60 | NM_001006275 |
| RT-POSTN | F: CCACCACATGGTAAACAAGCG R: AACAGCAGTTAGGACACGGT | 202 | 60 | NM_001030541 |
| GAPDH | F: GAACATCATCCCAGCGTCCA R: CGGCAGGTCAGGTCAACAAC | 132 | 60 | NM_204305 |
DNA from each individual was subjected to indel identification and genotype analysis using LncFAM primers. Sequencing was performed by Sangon Biotech Company (Shanghai, China), and the differences in the distribution of different genotypes among populations were analyzed using SPSS 22.0 software (version 22.0; Statistical Product and Service Solutions, IBM Corporation, Armonk, NY, USA). The allele frequency and genotype frequency of each mutation point were calculated using GENEPOP software (http://genepop.curtin.edu.au/), and the polymorphism information content (PIC), effective allele numbers (Ne) and expected heterozygosity (He) were calculated simultaneously.
Data analysis for correlation analysis
Prior to statistical analysis, the non-normally distributed variables were logarithmically transformed to approximate a normal distribution. The body weights and body sizes were analyzed by repeated measures and multivariate tests of a GLM. Two traits, namely, the chest width (CW) and body slanting length (BSL), satisfied Mauchly’s test of sphericity and were thus analyzed by ANOVA. An association analysis was performed based on a GLM, and the additive and dominance effects of a gene were calculated through regression analysis (SAS Institute Inc, 2000; SPSS, Chicago, IL, USA). A Bonferroni test was performed for multiple comparisons. Model I was used for growth traits, meat quality traits and serum variables, whereas based on the effect of the carcass weight on carcass traits, the carcass traits were analyzed using Model II with carcass weight as a covariate.
The analytical models were as follows:
Our model was designed based on the least-squares method. In these models, Yijklm was the observed value, μ was the overall population mean, Gi was the fixed effect of the genotype (i=3), including the additive and dominance effects of the gene (additive effect values of -1, 0 and 1 represent the II, ID and DD genotypes, respectively, and dominant effect values of 1, -1 and 1 represent the II, ID and DD genotypes, respectively), Sj was the fixed effect of sex (j=2), Hk was the fixed effect of hatching (k=1, 2), fl was the fixed effect of family (l=1, 7), eijklm represented random error, b was the regression coefficient for the carcass weight, Wijklm was the individual slaughter weight, and was the average slaughter weight. In this study, all the data are expressed as the means ± SEM. P values less than 0.05 were considered to indicate statistical significance, and the Bonferroni test was used for multiple comparisons (Li et al., 2018b; Ren et al., 2019).
RNA extraction, cDNA synthesis and fluorescence quantitative PCR
To study the expression of LncFAM in different varieties, we extracted total RNA from the heart, liver, spleen, lung, kidney, abdominal fat, stomach muscle, glandular stomach, duodenum, jejunum, ileum, caecum, pectoral and leg muscle from four GS laying hens at 20 and 30 weeks. In addition, pancreas tissues of Ross 308 at the age of 6 weeks, and the pancreas tissues of Gushi chickens at different stages (including 1day, 6, 14, 20 and 30weeks) were also taken. Then they were treated with the TRIzol reagent (Takara, Otsu, Japan) following the manufacturer’s recommended protocol. The RNA concentration and integrity were estimated spectrophotometrically using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and verified electrophoretically using an agarose gel. Only samples with an optical density (OD) absorption ratio (OD 260 nm/OD 280 nm) between 1.9 and 2.0 and no signs of degradation were used for further analysis. cDNA synthesis was performed using a PrimeScript RT reagent kit with gDNA Eraser (Takara).
All the samples were analyzed by real-time fluorescence-based quantitative PCR (qPCR) using quantitative primers, and qPCR was performed using the SYBR Green method and a Roche Light Cycler 96 instrument. The real-time PCR efficiency for each primer set was determined using a standard curve of pooled cDNA (Gonzalez et al., 2008). The efficiency of the primers used in this study ranged from 99% to 105.2%. The thermal cycling parameters included the preincubation stage (95°C/5 min), 35 cycling stages (95°C/30 s, 60°C/30 s and 72°C/30 s), melting stage (95°C/10 s, 65°C/60 s and 97°C/1 s) and cooling stage (37°C/30 s).
In order to reduce the result deviation caused by the potential tissue differential expression of internal reference genes, we first analyzed the expression trend of LncFAM with GAPDH as the housekeeping gene, and then repeated the test with β-actin as internal reference gene to verify the results obtained by GAPDH (Only the normalization with GAPDH was shown). The relative expression levels and the significance of the differences in expression among different tissues and developmental stages were analyzed using the 2-ΔΔCt method and one-way analysis of variance (ANOVA) followed by Duncan’s test, respectively (Liang et al., 2019). The quantitative real-time PCR analysis yielded a cycle threshold (Ct) value for each sample, and the CT values generated for each sample were used for the calculation of ΔCT (CT target–CT reference) and ΔΔCT [ΔCT test sample–ΔCT (the specific organization or individual)]. Data obtained from the real-time RT-PCR was presented as means ± SEM.
The significance of the data was analyzed using SPSS software. In the analysis, 0.01 ≤ P ≤ 0.05 indicated a significant difference between the experimental and control groups, P ≤ 0.01 indicated an extremely significant difference between the experimental and control groups, and P > 0.05 indicated no difference between the experimental and control groups. The figures were drawn using GraphPad Prism 6 (GraphPad Software Inc., 2007, San Diego, CA, USA).
Results
Detection of the expression level of LncFAM
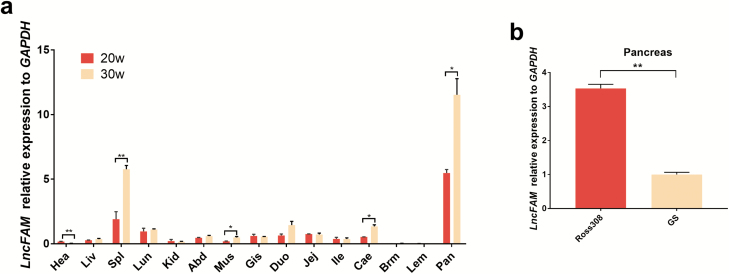
Because LncFAM is within the growth QTLs and is a novel discovered lncRNA, we speculated that LncFAM might also be related to the growth characteristics of chickens. Tissue samples from GS chickens at 20 and 30 weeks (w) were analyzed by qRT-PCR, and the results are shown in Figure 1a.
Figure 1.
Relative expression patterns of LncFAM. a Relative expression patterns of LncFAM in Gushi chicken tissues at 20 weeks and 30 weeks of age. The horizontal axis and vertical axis represent various tissues and their relative mRNA level values. Hea, heart; Liv, liver; Spl, spleen; Lun, lung; Kid, kidney; Abd, abdominal fat; Mus, stomach muscle; Gis, glandular stomach; Duo, duodenum; Jej, jejunum; Ile, ileum; Cae, caecum; Brm, breast muscle; Lem, leg muscle; and Pan, pancreas. *, P < 0.05, **, P < 0.01. b Expression profiles of LncFAM in the pancreas in a low-weight local chicken breeds, GS, and a high-weight commercial broiler, Ross 308. GS = Gushi chickens, 6 weeks, n = 6; Ross 308, 6 weeks, n = 6, *P < 0.05, **P < 0.01.
The expression of LncFAM in various tissues of GS chickens showed that LncFAM was mainly expressed in pancreas tissues (Figure 1a). To further confirm the relationship between the expression level of LncFAM and the growth phenotype, the expression of this lncRNA in pancreas tissue from a low-weight local chicken breed, GS, and a high-weight commercial broiler, Ross 308, was detected (Figure 1b). The expression of LncFAM in the pancreas of Ross 308 broilers at 6 weeks was significantly higher than that in dual-purpose chickens (GS), which indicated that the expression level of LncFAM might be related to growth phenotype indexes, such as growth speed and body weight. To explore potential DNA markers for growth improvement, we subsequently focused on the identification of polymorphisms in LncFAM.
Detection of genetic polymorphisms in LncFAM
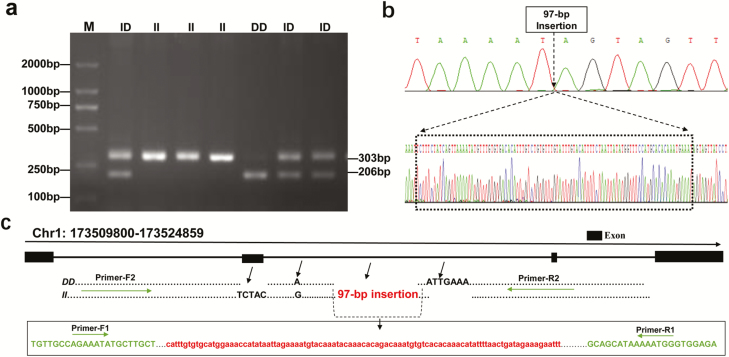
The expression analysis identified LncFAM as an important candidate gene related to the growth phenotype. To screen effective genetic variation loci and analyze the effects of genetic variation, we first identified polymorphic loci in LncFAM and found four sequence variants, namely, a 5-bp indel (TCTAC) in the second exon, an A/G base mutation, and a 7- bp indel (ATTGAAA) and a 97-bp indel in the second intron. Further analysis revealed complete linkage disequilibrium among these loci (Figure 2c and S2).
Figure 2.
Structural information for LncFAM. a Agarose gel electrophoresis pattern for the 97 bp insertion in LncFAM. b Sequencing for the 97 bp insertion in LncFAM. c Distribution of four polymorphic loci involving the 97 bp insertion in LncFAM in individuals with genotypes II and DD. Among the presented components, the green portion represents the 97 bp insertion. The structure of four polymorphic sites in LncFAM. Primer 1 and Primer 2 represent the primers used to detect the LncFAM 97 bp insertion and the LncFAM polymorphic site, respectively.
Online predictions revealed that the lncRNA has no protein coding function (data not shown). To more easily analyze the genetic effects of these mutation sites, we selected the 97-bp insertion for further study. Primers were designed to amplify the target fragment of LncFAM. An illustration of the indel region was presented in Figure 2a-b. The electrophoresis results revealed three genotypes, which were denoted II, ID and DD (Figure 2a), and the insertion locations are shown in Figure 2bS1 and S2.
Genotypic and allelic frequencies of LncFAM among F2 resource populations and different varieties
To detect the variations among dual-type chickens (including XC, LS, DX, CS, and GS) and commercial broilers (including Ross 308, AA, Cobb and Hubbard), we performed a PCR analysis on ten resource groups, including the F2 generation. In addition, a population genetic analysis was performed by electrophoresis-based typing. As shown in Table 2, the allele frequency of I in all chicken breeds was higher than that of D, and the DD genotype frequency in all breeds was lower than those of ID and II. Interestingly, only genotype II was found in commercial broilers.
Table 2.
Genotypic and allelic frequencies and related genetic parameters for the chicken LncFAM gene1
| Genotypic and allelic frequencies | P-value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Breeds/n | II | ID | DD | I | D | He | Ne | PIC | (HWE) | |
| F2 generation resource population | F2/800 | 0.63 | 0.33 | 0.04 | 0.80 | 0.20 | 0.320 | 1.480 | 0.272 | 0.256 |
| Dual-purpose chickens | XC/266 | 0.85 | 0.14 | 0.01 | 0.92 | 0.08 | 0.147 | 1.166 | 0.132 | 0.717 |
| LS/143 | 0.85 | 0.15 | 0.00 | 0.92 | 0.08 | 0.147 | 1.166 | 0.132 | 0.319 | |
| DX/129 | 0.61 | 0.36 | 0.03 | 0.79 | 0.21 | 0.332 | 1.495 | 0.276 | 0.380 | |
| CS/92 | 0.67 | 0.31 | 0.02 | 0.83 | 0.17 | 0.282 | 1.403 | 0.246 | 0.570 | |
| GS/143 | 0.82 | 0.16 | 0.02 | 0.90 | 0.10 | 0.180 | 1.223 | 0.166 | 0.160 | |
| Commercial broilers | Hubbard /459 | 1.00 | -/- | -/- | 1.00 | -/- | -/- | -/- | -/- | -/- |
| Ross308 /172 | 1.00 | -/- | -/- | 1.00 | -/- | -/- | -/- | -/- | -/- | |
| Cobb/212 | 1.00 | -/- | -/- | 1.00 | -/- | -/- | -/- | -/- | -/- | |
| AA/300 | 1.00 | -/- | -/- | 1.00 | -/- | -/- | -/- | -/- | -/- |
1F2, F2 resource population, XC, Xichuan black-bone chicken, LS, Lushi green eggshell chicken, DX, Dongxiang green eggshell chicken, CS, Changshun green-shell chicken, and GS, Gushi chicken. He, expected heterozygosity; Ne, effective allele numbers; PIC, polymorphism information content, PIC >0.50 represents high polymorphism, while 0.25 < PIC <0.50 represents moderate polymorphism, and PIC < 0.25 represents low polymorphism; P-value (HWE): P-value of Hardy–weinberg equilibrium.
PIC and He are not only indicators for measuring the degree of allelic polymorphism and gene mutation but also indicators of genetic variation within the population. In this study, no polymorphism was detected in the different breeds of commercial broilers, and only genotype II was found, which might be due to the high pressure of artificial selection. Based on the analysis of gene polymorphisms in the F2 generation resource population and dual-purpose chickens, this indel was found to be in HWE in the F2 resource and dual-purpose chickens (P > 0.05). In addition, with the exception of the F2 generation resource population and Dongxiang chickens, which were moderately polymorphic, other breeds exhibited low polymorphism, which indicated that the F2 generation and dual-purpose chicken population exhibited greater genetic variation and selection potential than commercial broilers.
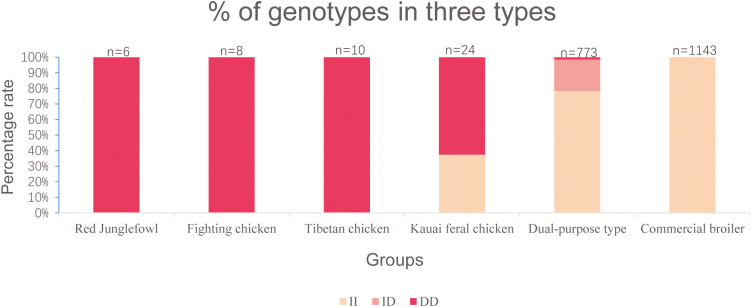
To detect the different distribution patterns of LncFAM in domesticated chickens, the percentages of the three genotypes in red jungle fowls (reference genome), Fighting chickens (reference genome), Tibetan chickens (reference genome), Kauai chickens (reference genome), dual-purpose chickens and commercial broiler hens were detected. The results showed that the different varieties of LncFAM polymorphisms were significantly different among breeds. Red jungle fowls, the wild ancestors of domestic chickens, Tibetan chickens and Fighting chickens, which survived at high altitudes under severe climatic conditions with little traffic between regions and no blood exchange among populations, only exhibited genotype DD in their genomes. Genotype II was found in chickens with a low degree of domestication that lived on wild islands, such as Kauai chickens. Subsequently, the proportion of genotype II increased successively in dual-purpose chickens derived from local chickens. Finally, only genotype II was present in highly selected commercial broiler chickens (Figure 3).
Figure 3.
Percentages of different genotypes in different populations of dual-purpose chickens (including Xichuan, Dongxiang, Changshun, Lushi, and Gushi) and commercial broilers (Ross 308, AA broilers, Cobb and Hubbard). Note: The genomic information for Red jungle fowl, Fighting chickens, Tibetan chickens and Kauai chickens was obtained from published data in the NCBI database (Gering et al., 2015; Wang et al., 2015). The genotyping data were analyzed by Blast software.
Associations between the indel in LncFAM and growth and carcass traits
The correlation results obtained for the F2 population are shown in Tables 2 and 3. The 97-bp insertion mutation exerted a significant effect on the BW at 0, 2, 6 and 8 weeks, SL at 0 weeks, SG at 4, 8 and 12 weeks, CW at 8 and 12 weeks, and PW at 4 and 12 weeks (P < 0.05). Among these traits, the BW at 6 weeks, SG at 4, 8, and 12 weeks, CW at 8 weeks, and PW at 4 weeks were extremely positively correlated with the 97-bp indel (P < 0.01). In addition, with the exception of the SW, SWR, and SL at 8 and 12 weeks and the BSL at 4 weeks, genotype II exhibited the highest values for BW, SL, SG, CW, BSL, PW, SEW, EW, BMW, LMW, and cW, whereas the DD genotype was associated with the lowest values for BW, SL, SG, CW, BSL, PW, SEW, EW, BMW, LMW, and cW.
Table 3.
Least squares means (SE) for carcass traits at 84 d for 97 bp indel mutation of the LncFAM gene in F2 chickens from a Gushi-broiler cross
| Genotypes, Mean ±SE | |||||
|---|---|---|---|---|---|
| Growth Traits1 | Age, week | II | ID | DD | P-Value |
| BW, g | 0 | 30.87±0.41a | 30.44±0.43a | 29.36±0.63b | 0.004 |
| 2 | 123.46±2.21a | 121.40±2.35a | 113.67±4.21b | 0.026 | |
| 4 | 325.09±6.38 | 317.63±6.65 | 311.64±10.2 | 0.054 | |
| 6 | 570.59±11.81a | 550.79±12.32ab | 544.15±19.03b | 0.007 | |
| 8 | 826.48±16.61a | 803.15±17.46ab | 785.84±28.2b | 0.036 | |
| 10 | 1119.70±22.17 | 1106.79±23.13 | 1067.84±35.77 | 0.181 | |
| 12 | 1362.87±24.64 | 1342.68±25.91 | 1305.09±42.11 | 0.170 | |
| SL, cm | 0 | 2.58±0.01a | 2.58±0.01ab | 2.54±0.02b | 0.039 |
| 4 | 5.53±0.06 | 5.45±0.06 | 5.37±0.14 | 0.249 | |
| 8 | 7.91±0.05 | 7.93±0.06 | 7.88±0.13 | 0.835 | |
| 12 | 9.41±0.06 | 9.42±0.07 | 9.31±0.12 | 0.631 | |
| SG, cm | 4 | 2.72±0.02a | 2.68±0.03ab | 2.63±0.04b | 0.009 |
| 8 | 3.44±0.03a | 3.40±0.03ab | 3.35±0.05b | 0.010 | |
| 12 | 3.87±0.02a | 3.82±0.02ab | 3.73±0.05b | 0.004 | |
| CW, cm | 4 | 4.12±0.03 | 4.08±0.04 | 3.95±0.09 | 0.147 |
| 8 | 5.74±0.03ab | 5.61±0.04b | 5.54±0.11a | 0.007 | |
| 12 | 6.38±0.06a | 6.28±0.06ab | 6.10±0.13b | 0.023 | |
| BSL, cm | 4 | 11.40±0.09 | 11.42±0.10 | 11.22±0.17 | 0.404 |
| 8 | 16.28±0.08 | 16.17±0.09 | 16.20±0.22 | 0.418 | |
| 12 | 19.84±0.07 | 19.72±0.08 | 19.49±0.20 | 0.108 | |
| PW, cm | 4 | 5.20±0.03a | 5.09±0.03b | 5.06±0.08ab | 0.005 |
| 8 | 6.90±0.05 | 6.85±0.05 | 6.70±0.13 | 0.219 | |
| 12 | 8.74±0.04a | 8.61±0.05b | 8.42±0.15b | 0.020 |
1BW, Body weight, SL, Shank length, SG, Shank girth, CW, Chest width, BSL, Body slant length, PW, Pelvic width.
a,bMeans with different superscripts show significant differences (P < 0.05) and the same letters indicating no difference (P > 0.05).
The development trends in F2 chickens with the three LncFAM genotypes at different ages were analyzed (Figure S3). The growth rates of genotype II individuals were faster in terms of the BW, SG, CW, BSL, and PW, followed by the ID genotype, whereas the DD genotype grew slowly, which indicated that individuals with genotype II exhibit better growth performance than those individuals with genotype ID or DD (P < 0.05).
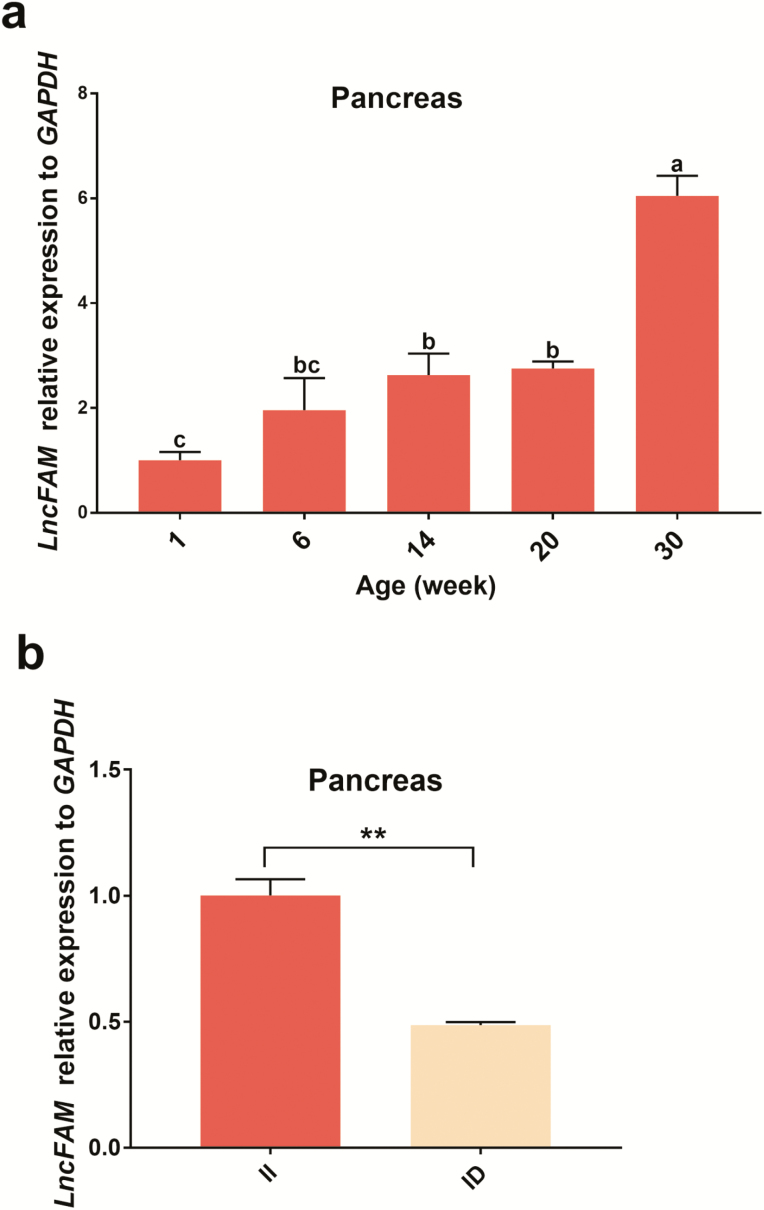
Expression patterns of the LncFAM gene in the pancreas
An examination of the pancreas at various times revealed that the total expression level significantly increased during the first 30 weeks of growth and development (all pancreatic tissues from GS chickens were labeled as genotype II, P < 0.05) (Figure 4a). To further confirm the relationships between the expression level of LncFAM and different genotypes, the different genotype expression levels in the pancreas of GS chickens at 6 weeks were detected, and the expression level obtained for genotype II (1.33 ± 0.05) was significantly higher than that found for genotype ID (0.65 ± 0.01, P < 0.01, Figure 4b).
Figure 4.
Relative expression patterns of LncFAM in the pancreas. a The expression of LncFAM in the pancreas at different ages. All individuals used in the experiment were genotype II Gushi chickens, and different letters indicate significant differences (P < 0.05), whereas the same letters indicate the absence of significant differences (P > 0.05). b Expression patterns of different genotypes of LncFAM in the pancreas of Gushi chickens at 6 weeks of age. II, n = 4; ID, n = 4. Error bars represent the SEM. *P < 0.05, **P < 0.01.
LncFAM cis-regulation of the FAM48A gene
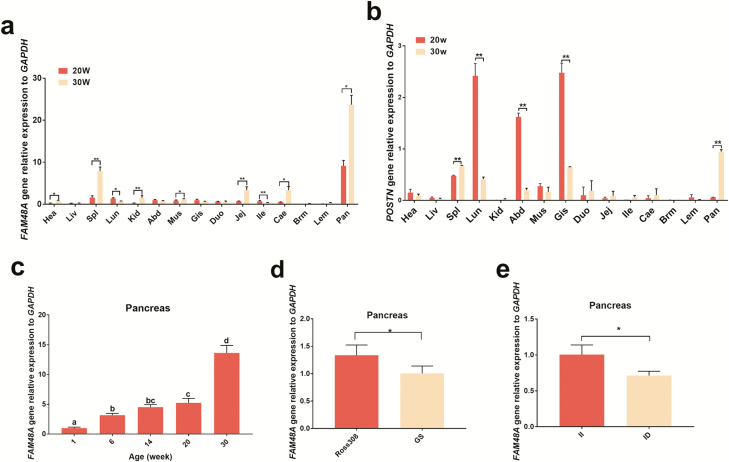
The expression levels of the FAM48A and POSTN genes adjacent to the LncFAM gene were analyzed, and the results revealed that LncFAM and FAM48A genes showed a coexpression trend and that both genes exhibited their highest expression level in the pancreas (Figure 5a). In contrast, the POSTN gene did not show the same expression trend as the LncFAM gene (Figure 5b and S4). An analysis of the FAM48A collinearity showed that FAM48A expression was relatively consistent among different species, which suggested that FAM48A is an important gene (Figure S5). Transcriptome sequencing showed that the highest expression of FAM48A was found in the spleen (Figure S6), which is consistent with the tissue expression profiling results. The expression of FAM48A in the pancreas was detected, and the expression levels were consistent with the expression trend obtained for LncFAM (Figure 5c-e). The correlations between FAM48A and LncFAM expression in individual tissues were tested by Pearson’s correlation, and the two genes were significantly correlated in the tested tissues (P < 0.05, Table S1).
Figure 5.
Expression patterns of the FAM48A gene and POSTN gene in the pancreas. a Relative expression patterns of the FAM48A gene in Gushi chicken tissues at 20 weeks and 30 weeks of age. b Relative expression patterns of the POSTN gene in Gushi chicken tissues at 20 weeks and 30 weeks of age. Hea, heart; Liv, liver; Spl, spleen; Lun, lung; Kid, kidney; Abd, abdominal fat; Mus, stomach muscle; Gis, glandular stomach; Duo, duodenum; Jej, jejunum; Ile, ileum; Cae, caecum; Brm, breast muscle; Lem, leg muscle; and Pan, pancreas. *P < 0.05, **P < 0.01. c The expression of the FAM48A gene in the pancreas at different ages. All individuals used in the experiment were genotype II Gushi chickens. Different letters indicate significant differences (P < 0.05), whereas the same letters indicate the absence of significant differences (P > 0.05). d Expression profiles of the FAM48A gene in the pancreas of different chicken breeds. GS, Gushi chicken, 6 weeks, n = 6; Ross 308, 6 weeks, n = 6. *P < 0.05, **P < 0.01. e Expression patterns of different genotypes of the FAM48A gene in the pancreas of Gushi chickens at 6 weeks of age. II, n = 6; ID, n = 6.
Discussion
MAS breeding is a type of technology that utilizes molecular markers at the DNA level to genetically improve the biological population. In other words, molecular biology technology has been used for molecular-level breeding (Fang et al., 2010; Fang et al., 2010; Chen et al., 2019; Kang et al., 2019; Liu et al., 2019).
The term indel refers to the insertion or deletion of nucleotide fragments of different sizes at the same site in genomes between close species or different individuals of the same species (Weber et al., 2002). Indel markers exhibit high accuracy, good stability and simple classification systems and have been applied in animal and plant population genetic analyses, molecular-assisted breeding, medical diagnosis and other research fields, which is conducive to the further development and utilization of good genes (Yang et al., 2017; Li et al., 2018a; Li et al., 2018b). These studies have greatly enriched the available markers for the MAS of livestock and poultry and have provided important clues regarding the molecular mechanisms through which genetic elements affect traits.
ENCODE’s research shows that approximately 80% of human genome sequences can be transcribed, whereas less than 2% of the human genome is used for protein translation, and most of the remaining transcripts are non-coding RNAs (ncRNAs) (Alessandro and Irene, 2014). The important role of ncRNAs in gene regulation is becoming a hotspot in biomedical research. lncRNAs can control gene activities through multiple mechanisms, such as direct or indirect regulation of gene expression via cis-/trans-action, functioning as protein baits in the nucleus and affecting the stability and translational process of mRNA (Markus et al., 2013). Recent studies have highlighted the significance of lncRNAs in development and diseases and their potential in future clinical applications (Chiara et al., 2015).
Because LncFAM is located within the QTL associated with growth characteristics and is a novel discovered lncRNA, we speculated that LncFAM might also be related to the growth characteristics of chickens. Tissue expression profile analysis is effective for investigating gene function and provides valuable reference information for obtaining a more in-depth understanding of gene expression, the target tissues of genes and possible functional sites (An and Liu, 2009; He et al., 2018). The pancreas exhibits the highest specific expression of LncFAM (Figure 1a). Many studies on lncRNAs have reported that the tissue specificity of lncRNAs reveals their specific biological functions in regulating growth and development (Guttm an et al., 2009; Cabili et al., 2011; Mercer et al., 2011). We speculated that this gene may play a significant role in the growth and development of the pancreas and spleen. The pancreas is the largest digestive gland in animals and is significantly related to digestion and absorption functions in animals; in fact, the main function of the pancreas is to synthesize and secrete pancreatic juice containing digestive enzymes (Kokue and Hayama, 1972; Almirall et al., 1995; Jones 1995). In addition, LncFAM is located in QTLs associated with daily food intake (Gao et al., 2010b; Yuan et al., 2015). Xu reported a close correlation between the activity of digestive enzymes and changes in feed intake. Increased feed intake is helpful for stimulating the weight of related digestive organs, and the secretion of pancreatic juice also increases with increases in feed intake during the compensation period Xu et al., 1994). A previous study showed that ages of 20 and 30 weeks correspond to the early stage and peak period of laying eggs, respectively Li et al., 2015). Therefore, we speculated that the expression of LncFAM in the chicken pancreas increases gradually during development because the feed intake of chickens gradually increases during growth and development, resulting in increased secretion of pancreatic juice. Thus, the expression of LncFAM increased gradually until reaching a peak at the time corresponding to that related to egg production (Figures 1a and 4a).
The detection of polymorphic loci related to this 97-bp indel in chickens of different breeds showed that genotype II was the main genotype, and interestingly, only genotype II was found in commercial broilers (Table 2). This phenomenon might be due to the fact that the feed conversion rate of commercial broilers is higher than that of dual-purpose chickens (Savory, 1975). Another explanation is that during the directed breeding of commercial broilers, genotype DD might not be conducive to the growth of broiler chickens, which would result in the gradual selection of genotype II during this process, and thus, genotype II ultimately becomes fixed in the population. The expression of genotype II in the pancreas was significantly higher than that of genotype ID, which suggested that the digestion and absorption capacity of genotype II was higher than that of genotype ID, and this finding might also explain why genotype II was fixed in different breeds of chickens. The expression of LncFAM in the pancreas of broilers was higher than that found in local broilers, which might be related to the higher digestive capacity of broilers (Figure 4b).
Both natural and artificial selection can cause changes in the gene frequency of biological populations under human and natural intervention, and the genes in a certain population are transmitted from generation to generation. The phenomenon in which the proportion of individuals with a certain genotype changes is called selection, and genetic variation, such as mutation and selection, leads to increased linkage imbalance (Todorov, 2002). The complete linkage disequilibrium between these loci also indirectly proves that the locus or interval that contains these sites is important and naturally or artificially selected (Figure 2c).
A genotypic percentage analysis of the 97-bp insert in red jungle fowls, Fighting chickens, Tibetan chickens, Kauai chickens, dual-purpose chickens and commercial broilers revealed that a high percentage of DD individuals was the original genetic state of chickens. However, over time, genotype II began to appear, and the proportion of genotype II increased slowly. We speculate that human intervention in the evolution of chickens resulted in the artificial selection of genotype II individuals to promote growth and development, and increases in the intensity of artificial breeding also increased the proportion of genotype II individuals (Figure 2).
The chicken body weight is not only an important economic trait but also a reference for molecular MAS (Georges and Michel, 2007; Tsudzuki, et al., 2007; Goddard and Hayes, 2009). The development of the body is often reflected in changes in morphometric traits, the ecological environment and the breeding mode. For example, the width of the chest is a good indicator of the meat quality of most poultry species (Scheuermann et al., 2003), and a large degree of correlation is generally observed between the body size and body weight, which indicates that the body size can be used as a reference indicator of body weight (Ankra-Badu et al., 2010). In addition, morphometric traits such as BW, SL, BL and SG can serve as useful indicators for predicting the fertility of roosters during broiler breeding (Ankra-Badu et al., 2010; Gao et al., 2010b). These results indicated that the morphometric traits of chickens are as important as the body weight of chickens and should not be ignored. In this study, the different genotypes generated with the 97-bp insertion were significantly correlated with most body size traits in the F2 generation (Tables 3 and 4). Individuals with genotype II exhibited the largest growth and carcass traits, which suggests that genotype II is the dominant genotype.
Table 4.
Least squares means (SE) for carcass traits at 84 d for 97 bp indel mutation of the LncFAM gene in F2 chickens from a Gushi-broiler cross
| Genotypes, Mean ±SE | ||||
|---|---|---|---|---|
| Carcass Traits1 | II | ID | DD | P-Value |
| SEW, g | 1108.31±22.06 | 1092.25±23.08 | 1071.23±37.08 | 0.298 |
| EW, g | 81.41±0.20 | 81.27±0.22 | 81.05±0.42 | 0.534 |
| SW, g | 8.05±1.11a | 7.76±1.21a | 13.30±2.36b | 0.046 |
| SWP, % | 0.87±0.13a | 0.86±0.14a | 1.50±0.25b | 0.020 |
| BMW, g | 71.13±2.22 | 69.25±2.31 | 67.28±3.49 | 0.160 |
| LMW, g | 100.38±2.54 | 98.19±2.64 | 94.51±4.00 | 0.095 |
| cW, g | 1226.54±23.70 | 1202.28±24.72 | 1168.32±38.51 | 0.069 |
1SEW, Semi-evisceration weight, EW, Evisceration weight, SW, Sebum weight, SWE, Sebum weight percentage, BMW, Breast muscle weight, LMW, Leg muscle weight, cW, Carcass weight.
a,bMeans with different superscripts show significant differences (P < 0.05) and the same letters indicating no difference (P > 0.05).
Many studies have reported that lncRNAs can exert cis- or trans-regulatory effects on adjacent genes (Guil et al., 2012; Cai et al., 2017), and we speculated that LncFAM would exert this regulatory role on surrounding genes. FAM48A is subunit Spt20 of the SAGA complex core structure, which is responsible for 10% of gene transcription in vivo (Lee et al., 2000; Koutelou et al., 2010), and transcriptome sequencing has indicated that this gene is highly expressed in the spleen, followed by the intestine, testicles, lungs and kidneys, and exhibits relatively low expression in muscle tissue (Jason et al., 2012; Barbosa-Morais et al., 2013) (Figure S6). This result was consistent with the expression trend of FAM48A in various tissues of chickens at 20 and 30 weeks (Figure 5a). In addition, the expression of the FAM48A gene was consistent with the expression trend found for LncFAM in the pancreas during different periods and those found for different genotypes (Figure 5c-e, Table S1). These findings suggested that this phenomenon might be caused by the cis-regulation of adjacent genes by LncFAM. Yuan et al. (2015) found that FAM48A is related to QTLs associated with daily food intake. This finding might provide an explanation for the high expression of the FAM48A gene in the pancreas. Because LncFAM is also located in this QTL, we speculated that LncFAM might regulate the FAM48A gene, and as a result, the function of the FAM48A might be similar to that of LncFAM. Therefore, we hypothesize that the FAM48A gene is cis-regulated by LncFAM, but whether FAM48A is the target gene of LncFAM needs to be further studied.
Conclusions
We determined that a 97-bp insertion in LncFAM was related to most growth traits in chickens and that the mutant individuals exhibited the dominant genotype. We speculated that mutant individuals exhibit stronger growth performance or better digestive function. By studying the adjacent genes of LncFAM, we determined that LncFAM might exert a cis-regulatory effect on its adjacent gene, FAM48A. In conclusion, our results provide a reference for candidate genes in growth QTLs on GGA1 and a potential molecular marker for chicken breeding. The coexpression trend obtained with FAM48A suggests that this study might also provide a reference for future studies of LncFAM target genes.
Supplementary Data
Supplementary data are available online at the Journal of Animal Science website.
Table S1. The correlations between FAM48A and LncFAM expression in the corresponding tissue were tested by Pearson’s correlation.
Figure S1. Sequencing for the indel mutation in the LncFAM gene. (A) Wild type, DD. (B) II. The lowercase letters in the figure represent 97 bp indel sequence, the red letters represent primer pairs.
Figure S2. Polymorphic locus screening for individuals with genotype II and genotype DD. (A): Agarose gel electrophoresis pattern for individuals with genotype II and genotype D. (B): Sequence alignment of polymorphic loci between genotype II and genotype DD.
Figure S3. Developmental changes in the growth traits of different genotypes of LncFAM in the F2 generation at different weeks. (A), (B), (C), (D), (E), (F) represent the association between the 97 bp insertion mutation and body weight, body slant length, shank length, chest width, pelvic width, and shank girth, respectively.
Figure S4. Expression patterns of the POSTN gene in the pancreas. (A) The expression of the POSTN gene in the pancreas at different ages. All individuals used in the experiment were genotype II Gushi chickens, the different lowercase letters indicate P < 0.05, and the same letters indicate no difference (P > 0.05). (B) Expression patterns of different genotypes of the POSTN gene in the pancreas of Gushi chickens at 6 weeks of age. (II, n = 6; ID, n = 6). *P < 0.05, **P < 0.01. (C) Expression profiles of the POSTN gene in the pancreas of different chicken breeds. All the individuals used in the experiment were genotype II. GS = Gushi chicken, 6 weeks, n = 6; Ross 308, 6 weeks, n = 6, *P < 0.05, **P < 0.01.
Figure S5. Collinearity analysis of the FAM48A gene. All the data on extant species displayed in this browser are from Ensembl, JGI, and Genoscope. http://www.genomicus.biologie.ens.fr/genomicus-71.01/cgi-bin/search.pl
Figure S6. Expression level in FPKM of the FAM48A and POSTN genes (A) FAM48A, (B) POSTN. The data come from the Expression Atlas update—an integrated database of gene and protein expression in humans, animals and plants. http://www.ensembl.org.
Acknowledgments
This study was supported by National Natural Science Foundation of China-Henan joint grant (U1804107), the Key Science and Technology Research Project of Henan Province (202102110085), National Natural Science Foundation of China (31872987), National Natural Science Foundation of China -Henan joint grant (U1704233) and the Earmarked Fund for Modern Agro-Industry Technology Research System (No. CARS-40-K04).
Conflicts of interest
The authors declare that they have no conflicts of interest.
Literature Cited
- Alessandro F., and Irene B. 2014. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 15:7–21. doi: 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Almirall M., Francesch M., Perez-Vendrell A. M., Brufau J., and Esteve-Garcia E.. . 1995. The differences in intestinal viscosity produced by barley and beta-glucanase alter digesta enzyme activities and ileal nutrient digestibilities more in broiler chicks than in cocks. J. Nutr. 125:947–955. doi: 10.1093/jn/125.4.947 [DOI] [PubMed] [Google Scholar]
- An Q. C., and Liu G. Y.. . 2009. Molecular cloning, sequence identification, and tissue expression profile analysis of three novel porcine genes: SDHB, SNRPA and CRYBB1. Mol. Biol. Rep. 36:683–690. doi: 10.1007/s11033-008-9229-0 [DOI] [PubMed] [Google Scholar]
- Ankra-Badu G. A., Le Bihan-Duval E., Mignon-Grasteau S., Pitel F., Beaumont C., Duclos M. J., Simon J., Carré W., Porter T. E., Vignal A., . et al. 2010. Mapping QTL for growth and shank traits in chickens divergently selected for high or low body weight. Anim. Genet. 41:400–405. doi: 10.1111/j.1365-2052.2009.02017.x [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais N. L., Manuel I., Qun P., Hui Y., Xiong G. Serge L. J. Lee S. Valentina K. Claudia W. Stephen, and Recep C.. . 2013. The evolutionary landscape of alternative splicing in vertebrate species. Science. 338:1587–1593. doi: 10.1126/science.1230612 [DOI] [PubMed] [Google Scholar]
- Besnier F., Wahlberg P., Rönnegård L., Ek W., Andersson L., Siegel P. B., and Carlborg O.. . 2011. Fine mapping and replication of QTL in outbred chicken advanced intercross lines. Genet. Sel. Evol. 43:3. doi: 10.1186/1297-9686-43-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., and Rinn J. L.. . 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25:1915–1927. doi: 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B., Li Z., Ma M., Wang Z., Han P., Abdalla B. A., Nie Q., and Zhang X.. . 2017. LncRNA-Six1 encodes a micropeptide to activate Six1 in Cis and is involved in cell proliferation and muscle growth. Front. Physiol. 8:230. doi: 10.3389/fphys.2017.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wang J., Liu N., Cui W., Dong W., Xing B., and Pan C.. . 2019. Pig SOX9: expression profiles of Sertoli cell (SCs) and a functional 18 bp indel affecting testis weight. Theriogenology 138:94–101. doi: 10.1016/j.theriogenology.2019.07.008 [DOI] [PubMed] [Google Scholar]
- Chiara T., Pier Paolo D. F., and Francesco N.. . 2015. The role of non-coding RNAs in the regulation of stem cells and progenitors in the normal mammary gland and in breast tumors. Front Genet. 6:72. doi: 10.3389/fgene.2015.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M., Nie Q., Luo C., Zhang D., and Zhang X.. . 2010. Associations of GHSR gene polymorphisms with chicken growth and carcass traits. Mol. Biol. Rep. 37:423–428. doi: 10.1007/s11033-009-9556-9 [DOI] [PubMed] [Google Scholar]
- Gao Y., Du Z. Q., Feng C. G., Deng X. M., Li N., Da Y., and Hu X. X.. . 2010a. Identification of quantitative trait loci for shank length and growth at different development stages in chicken. Anim. Genet. 41:101–104. doi: 10.1111/j.1365-2052.2009.01962.x [DOI] [PubMed] [Google Scholar]
- Gao Y., Du Z. Q., Wei W. H., Yu X. J., Deng X. M., Feng C. G., Fei J., Feng J. D., Li N., and Hu X. X.. . 2010b. Mapping quantitative trait loci regulating chicken body composition traits. Anim. Genet. 40:952–954. doi: 10.1111/j.1365-2052.2009.01911.x [DOI] [PubMed] [Google Scholar]
- Georges, and Michel. 2007. Mapping, fine mapping, and molecular dissection of quantitative trait loci in domestic animals. Annu Rev Genomics Hum Genet. 8:131. doi: 10.1146/annurev.genom.8.080706.092408 [DOI] [PubMed] [Google Scholar]
- Gering E., Johnsson M., Willis P., Getty T., and Wright D.. . 2015. Mixed ancestry and admixture in Kauai’s feral chickens: invasion of domestic genes into ancient Red Junglefowl reservoirs. Mol. Ecol. 24:2112–2124. doi: 10.1111/mec.13096 [DOI] [PubMed] [Google Scholar]
- Goddard M. E., and Hayes B. J.. . 2009. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 10:381–391. doi: 10.1038/nrg2575 [DOI] [PubMed] [Google Scholar]
- Gonzalez J. M., Dijkhuis R. D., Johnson D. D., Carter J. N., and Johnson S. E.. . 2008. Differential response of cull cow muscles to the hypertrophic actions of ractopamine-hydrogen chloride. J. Anim. Sci. 86:3568–3574. doi: 10.2527/jas.2008-1049 [DOI] [PubMed] [Google Scholar]
- Guil S., and Esteller M.. . 2012. Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 19:1068–1075. doi: 10.1038/nsmb.2428 [DOI] [PubMed] [Google Scholar]
- Guttman M., Amit I., Garber M., French C., Lin M. F., Feldser D., Huarte M., Zuk O., Carey B. W., Cassady J. P., . et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227. doi: 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R. L., Li Z. J., Li M. J., Li J. Q., Lan X. Y., Sun G. R., Kang X. T., and Chen H.. . 2011. Novel 9-bp indel in visfatin gene and its associations with chicken growth. Br. Poult. Sci. 52:52–57. doi: 10.1080/00071668.2010.537310 [DOI] [PubMed] [Google Scholar]
- He X., Zhang C., Shi C., and Lu Q.. . 2018. Meta-analysis of mRNA expression profiles to identify differentially expressed genes in lung adenocarcinoma tissue from smokers and non-smokers. Oncol Rep. 39:929–938. doi: 10.3892/or.2018.6197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hee-Bok P., Lina J., Per W., Siegel P. B., and Leif A.. . 2006. QTL analysis of body composition and metabolic traits in an intercross between chicken lines divergently selected for growth. Physiol Genomics. 25:216–223. doi: 10.1152/physiolgenomics.00113.2005 [DOI] [PubMed] [Google Scholar]
- Honkatukia M., Tuiskula-Haavisto M., de Koning D. J., Virta A., Mäki-Tanila A., and Vilkki J.. . 2005. A region on chicken chromosome 2 affects both egg white thinning and egg weight. Genet. Sel. Evol. 37:563–577. doi: 10.1186/1297-9686-37-6-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeobi C. O., Woolliams J. A., Morrice D. R., Law A., Windsor D., Burt D. W., and Hocking P. M.. 2003. Quantitative trait loci affecting fatness in the chicken. Anim. Genet. 33:428–435. doi: 10.1046/j.1365-2052.2002.00911.x [DOI] [PubMed] [Google Scholar]
- Ikeobi C. O. N., Woolliams J. A., Morrice D. R., Law A., Windsor D., Burt D. W., and Hocking P. M.. . 2004. Quantitative trait loci for meat yield and muscle distribution in a broiler layer cross. Livest Prod Sci. 87:143–151. doi: 10.1016/j.livprodsci.2003.09.020 [DOI] [Google Scholar]
- Jason M., Caitlin R., Ping C., and Burge C. B.. . 2012. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 338:1593–1599. doi: 10.1126/science.1228186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. P. 1995. Manipulation of organ growth by early-life food restriction: its influence on the development of ascites in broiler chickens. Br. Poult. Sci. 36:135–142. doi: 10.1080/00071669508417759 [DOI] [PubMed] [Google Scholar]
- Kang Z., Zhang S., He L., Zhu H., Wang Z., Yan H., Huang Y., Dang R., Lei C., Chen H., . et al. 2019. A 14-bp functional deletion within the CMTM2 gene is significantly associated with litter size in goat. Theriogenology. 139:49–57. doi: 10.1016/j.theriogenology.2019.07.026 [DOI] [PubMed] [Google Scholar]
- Kokue E., and Hayama T.. . 1972. Effects of starvation and feeding on the exocrine pancreas of the chicken. Poult. Sci. 51:1366–1370. doi: 10.3382/ps.0511366 [DOI] [PubMed] [Google Scholar]
- Koutelou E., Hirsch C. L., and Dent S. Y.. . 2010. Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 22:374–382. doi: 10.1016/j.ceb.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Causton H. C., Holstege F. C., Shen W. C., Hannett N., Jennings E. G., Winston F.. . 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 405:701–704. doi: 10.1038/35015104 [DOI] [PubMed] [Google Scholar]
- Li J., Erdenee S., Zhang S., Wei Z., Zhang M., Jin Y., Wu H., Chen H., Sun X., and Xu H.. . 2018a. Genetic effects of PRNP gene insertion/deletion (indel) on phenotypic traits in sheep. Prion. 12:1. doi: 10.1080/19336896.2017.1405886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Liu D., Tang S., Li D., Han R., Tian Y., Li H., Li G., Li W., Liu X., . et al. 2018b. A multiallelic indel in the promoter region of the Cyclin-dependent kinase inhibitor 3 gene is significantly associated with body weight and carcass traits in chickens. Poult Sci. 98:556–565. doi: 10.3382/ps/pey404 [DOI] [PubMed] [Google Scholar]
- Li H., Wang T., Xu C., Wang D., Ren J., Li Y., Tian Y., Wang Y., Jiao Y., Kang X., . et al. 2015. Transcriptome profile of liver at different physiological stages reveals potential mode for lipid metabolism in laying hens. BMC Genomics 16:763. doi: 10.1186/s12864-015-1943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K., Wang X., Tian X., Geng R., Li W., Jing Z., Han R., Tian Y., Liu X., Kang X., . et al. 2019. Molecular characterization and an 80-bp indel polymorphism within the prolactin receptor (PRLR) gene and its associations with chicken growth and carcass traits. 3 Biotech 9:296. doi: 10.1007/s13205-019-1827-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Han R., Wang X., Li W., Tang S., Li W., Wang Y., Jiang R., Yan F., Wang C., . et al. 2019. A novel 86-bp indel of the motilin receptor gene is significantly associated with growth and carcass traits in Gushi-Anka F2 reciprocal cross chickens. Br. Poult. Sci. 60:649–658. doi: 10.1080/00071668.2019.1655710 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhang H., Li H., Li N., Zhang Y., Zhang Q., Wang S., Wang Q., and Wang H.. . 2008. Fine-mapping quantitative trait loci for body weight and abdominal fat traits: effects of marker density and sample size. Poult. Sci. 87:1314–1319. doi: 10.3382/ps.2007-00512 [DOI] [PubMed] [Google Scholar]
- Markus K., Zurab S., Ci C., Dan E., Webster Z. Ashley Q. Kun C. S. Lee R. J. Flockhart A. F. Groff, et al. 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 493:231–U245. doi: 10.1038/nature11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Gerhardt D. J., Dinger M. E., Crawford J., Trapnell C., Jeddeloh J. A., Mattick J. S., and Rinn J. L.. . 2011. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 30:99–104. doi: 10.1038/nbt.2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar M. K., Goraga Z. S., and Brockmann G. A.. . 2012. Quantitative trait loci segregating in crosses between New Hampshire and White Leghorn chicken lines: II. Muscle weight and carcass composition. Anim. Genet. 43:739–745. doi: 10.1111/j.1365-2052.2012.02344.x [DOI] [PubMed] [Google Scholar]
- Podisi B. K., Knott S. A., Burt D. W., and Hocking P. M.. . 2013. Comparative analysis of quantitative trait loci for body weight, growth rate and growth curve parameters from 3 to 72 weeks of age in female chickens of a broiler-layer cross. BMC Genet. 14:22. doi: 10.1186/1471-2156-14-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T., Li W., Liu D., Liang K., Wang X., Li H., Jiang R., Tian Y., Kang X., and Li Z.. . 2019. Two insertion/deletion variants in the promoter region of the QPCTL gene are significantly associated with body weight and carcass traits in chickens. Anim. Genet. 50:279–282. doi: 10.1111/age.12741 [DOI] [PubMed] [Google Scholar]
- Savory J. C. 1975. A growth study of broiler and layer chicks reared in single‐strain and mixed‐strain groups. Brit Poult. Sci. 16:315–318. doi: 10.1080/00071667508416192 [DOI] [Google Scholar]
- Scheuermann G. N., Bilgili S. F., Hess J. B., and Mulvaney D. R.. . 2003. Breast muscle development in commercial broiler chickens. Poult. Sci. 82:1648–1658. doi: 10.1093/ps/82.10.1648 [DOI] [PubMed] [Google Scholar]
- Sheng Z., Pettersson M. E., Hu X., Luo C., Qu H., Shu D., Shen X., Carlborg O., and Li N.. . 2013. Genetic dissection of growth traits in a Chinese indigenous × commercial broiler chicken cross. BMC Genomics 14:151. doi: 10.1186/1471-2164-14-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov A. A. 2002. Genetic dissection of complex traits. Am. J Hum Genet. 71:209–210. doi: 10.1086/341033 [DOI] [Google Scholar]
- Tsudzuki M., Onitsuka S., Akiyama R., Iwamizu M., Goto N., Nishibori M., Takahashi H., and Ishikawa A.. . 2007. Identification of quantitative trait loci affecting shank length, body weight and carcass weight from the Japanese cockfighting chicken breed, Oh-Shamo (Japanese Large Game). Cytogenet. Genome Res. 117:288–295. doi: 10.1159/000103190 [DOI] [PubMed] [Google Scholar]
- Uemoto Y., Sato S., Odawara S., Nokata H., Oyamada Y., Taguchi Y., Yanai S., Sasaki O., Takahashi H., and Nirasawa K.. . 2009. Genetic mapping of quantitative trait loci affecting growth and carcass traits in F2 intercross chickens. Poult Sci. 88:477–482. doi: 10.3382/ps.2008-00296 [DOI] [PubMed] [Google Scholar]
- Wang S. Z., Hu X. X., Wang Z. P., Li X. C., Wang Q. G., Wang Y. X., Tang Z. Q., and Li H.. . 2012. Quantitative trait loci associated with body weight and abdominal fat traits on chicken chromosomes 3, 5 and 7. Genet Mol Res. 11:956. doi: 10.4238/2012.April.19.1 [DOI] [PubMed] [Google Scholar]
- Wang M. S., Li Y., Peng M. S., Zhong L., Wang Z. J., Li Q. Y., Tu X. L., Dong Y., Zhu C. L., Wang L., . et al. 2015. Genomic analyses reveal potential independent adaptation to high altitude in tibetan chickens. Mol. Biol. Evol. 32:1880–1889. doi: 10.1093/molbev/msv071 [DOI] [PubMed] [Google Scholar]
- Weber J. L., David D., Heil J., Fan Y., Zhao C., and Marth G.. . 2002. Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet. 71:854–862. doi: 10.1086/342727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R. J., Mellor D. J., Birtles M. J., Reynolds G. W., and Simpson H. V.. . 1994. Impact of intrauterine growth retardation on the gastrointestinal tract and the pancreas in newborn pigs. J. Pediatr. Gastroenterol. Nutr. 18:231–240. doi: 10.1097/00005176-199402000-00018 [DOI] [PubMed] [Google Scholar]
- Yang Q., Yan H., Li J., Xu H., Wang K., Zhu H., Chen H., Qu L., and Lan X.. . 2017. A novel 14-bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size. Anim. Genet. 48:499–500. doi: 10.1111/age.12551 [DOI] [PubMed] [Google Scholar]
- Yuan J., Wang K., Yi G., Ma M., Dou T., Sun C., Qu L. J., Shen M., Qu L., and Yang N.. . 2015. Genome-wide association studies for feed intake and efficiency in two laying periods of chickens. Genet. Sel. Evol. 47:82. doi: 10.1186/s12711-015-0161-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Nie Q., and Zhang X.. . 2013. Overview of genomic insights into chicken growth traits based on genome-wide association study and microRNA regulation. Curr Genomics. 14:137–146. doi: 10.2174/1389202911314020006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.