Abstract
Mammalian cells have evolved multiple pathways to repair DNA double strand breaks (DSBs) and ensure genome stability. In addition to non-homologous end-joining (NHEJ) and homologous recombination (HR), cells evolved an error-prone repair pathway termed microhomology-mediated end joining (MMEJ). The mutagenic outcome of MMEJ derives from the activity of DNA polymerase theta (Polθ) – a multidomain enzyme that is minimally expressed in normal tissue but overexpressed in tumors. It has become evident that Polθ expression is critical for the proliferation of HR deficient cancer cells. As a result, this mutagenic repair emerged as an attractive target for cancer therapy, and inhibitors are currently in pre-clinical development. Here we review the multifunctionality of this enigmatic polymerase, focusing on its role during DSB repair in mammalian cells and its impact on cancer genomes.
Keywords: DNA damage, DNA Polymerase theta, Polθ dysfunctional telomeres, DSB, MMEJ, alt-EJ
DNA double-strand break repair in mammalian cells
DNA double-strand breaks (DSBs) are highly deleterious lesions that arise due to exogenous agents, including ionizing radiation and chemotherapeutic drugs. In addition, DSBs accumulate as a result of DNA replication, meiosis, and the assembly and diversification of antigen receptor genes by Class Switch and V(D)J recombination1. The ends of linear chromosomes may also be recognized as DSBs when telomeres are rendered dysfunctional following telomerase deficiency or upon the removal of the protective Shelterin complex2. Different sources of DSBs lead to diverse chemistry at DNA ends that can be resolved using various pathways to ensure genome stability.
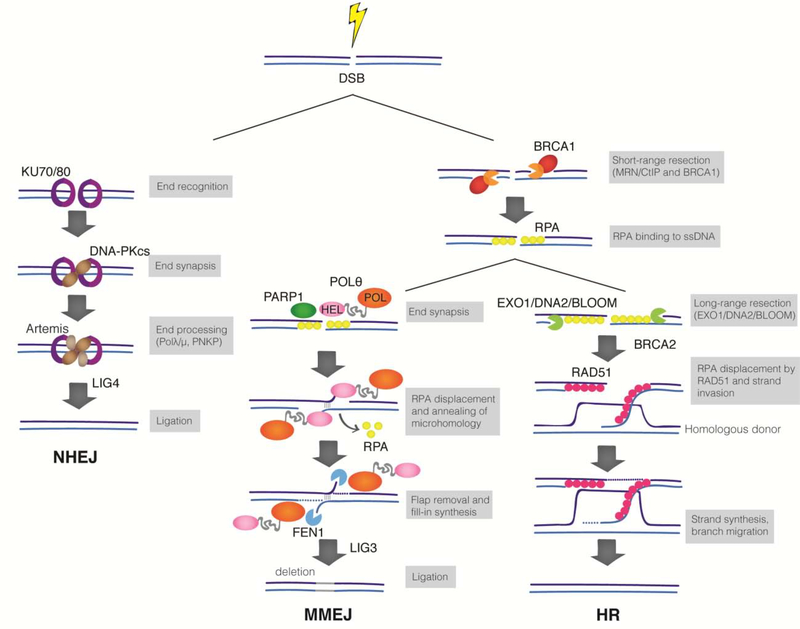
The most studied DSB repair mechanisms include non-homologous end-joining (NHEJ) that uses little or no homology to seal DNA ends, and homologous recombination (HR) that employs the sister chromatid as template to promote error-free repair (Figure 1). NHEJ is the main DSB repair pathway in mammalian cells. This canonical end-joining pathway is initiated when the Ku70/80 (Ku) heterodimer recognizes broken DNA ends and recruits DNA-dependent protein kinase catalytic subunit (DNA-PKcs). The DNA-PK complex then phosphorylates numerous factors to promote end-joining by Ligase 4 (Lig4)3. NHEJ employs additional end-processing enzymes that are essential for the joining of chemically incompatible ends, including Artemis, polynucleotide kinase 3’ phosphatase (PNKPT1), and two family X-Polymerase – Polλ and Polμ3. Although NHEJ can act throughout the cell cycle, its activity is predominant in G1. On the other hand, the activity of HR is restricted to S and G2 phases of the cell cycle4. HR requires DNA resection, where nucleolytic degradation of a DSB generates a 3’ single-stranded (ss) DNA overhang. This process is initiated by the MRE11-RAD50-NBS1 complex (MRN) in conjunction with CtIP. Extended resection is then catalyzed redundantly by EXO1 and DNA2-BLM5. Following resection, the 3’ ssDNA tail is bound by RPA which is subsequently exchanged for RAD51 through the action of BRCA2 and PALB26. RAD51 nucleofilaments mediate strand invasion and homology search on the sister chromatid. Ultimately, missing nucleotides are filled-in by copying the undamaged chromatid and repair is completed with minimal alterations to the original sequence6.
Figure 1. Major DSB repair pathways in mammalian cells.
DSBs in G1 are primarily repaired by NHEJ. The Ku heterodimer recognizes broken DNA and recruits DNA-PKcs to orchestrate end-joining by Lig4. During NHEJ, minimal processing of DNA ends leads to repair with minimal alteration to the original sequence. DSBs in S/G2 phases of the cell cycle are subjected to end-resection by MRN/CtIP, leading to short ssDNA that is rapidly coated by RPA. Resected DSBs are substrates for MMEJ and HR, and the choice between these pathways is poorly understood. Polθ-helicase displaces RPA to promotes the synapsis of the opposing ends. If annealing occurs using internal microhomology, flaps are generated and are processed by FEN1. Polθ fills-in the gapped DNA and hands over the substrate to Lig3 to seal the end. When resected DNA is subject to long end-resection by EXO1/DNA2 and BLM, RPA1 is exchanged for Rad51 to promote strand invasion and copying from the sister chromatid.
Microhomology-mediated end joining, an intrinsically mutagenic repair pathway
In addition to the well-characterized NHEJ and HR, mammalian cells employ a mechanistically distinct, yet less-understood pathway termed Microhomology-Mediated End Joining (MMEJ). MMEJ is part of a broader and error-prone mechanism of end joining, known as alternative end joining (alt-EJ) (Figure 1). Repair by MMEJ is driven by the annealing of micro-homologous sequences flanking the DNA ends, and its outcome is mutagenic due to deletions and insertions that scar break sites. MMEJ activity has been detected in all kingdoms of life and studied in bacteria7,8, yeast9, flies10, worms11, plants12, fish13 and mammals14,15. Its activity was first described in NHEJ defective S. cerevisiae mutants9 and found to be dependent on the presence microhomology9. Early evidence for MMEJ in mammalian cells emerged from the analysis of Class Switch Recombination (CSR) in NHEJ-deficient B cells14. MMEJ was initially viewed as a back-up pathway for canonical repair. Consistent with substrates being rerouted to MMEJ when HR and NHEJ are compromised, cells lacking BRCA1/2, Ku, and Lig4 rely on MMEJ for survival16–18. However, it has become evident that in certain contexts, MMEJ operates even when NHEJ and HR are proficient. For example, MMEJ was found to be essential to repair DSBs in the developing zebrafish embryos13. Furthermore, MMEJ activity was detected during V(D)J recombination in NHEJ-proficient B cells that carry mutations in the RAG recombination genes15. Accordingly, MMEJ is no longer viewed as a back-up repair mechanism. Nevertheless, it remains unclear as to when MMEJ prevails over other repair pathways and what prevents it from accessing substrates that are typically repaired by HR and NHEJ.
Evidence for MMEJ activity at deprotected telomeres emerged from the analysis of telomerase-deficient mice, where chromosome end-to-end fusions persisted in the absence of Lig4 and DNA-PKcs19. Subsequently, sequence analysis of telomere fusions in cells derived from chronic lymphocytic leukemia (CLL) and ataxia-telangiectasia-like disorder (ATLD) patients highlighted an MMEJ signature comprising frequent microhomology, deletions, and insertions20– 22. Furthermore, depletion of MMEJ factors compromised the ability of cancer cells to escape telomere crisis20, indicating that this mutagenic repair can foster genome instability in the early stage of tumorigenesis to promote cancer progression. Mechanistic insight into how mammalian telomeres suppress MMEJ was obtained through genetic experiments in mouse cells 23,24 which revealed that mutagenic repair is fully unleashed upon the depletion of all subunits of the Shelterin complex and in the absence of Ku. Interestingly, the function of MMEJ at telomeres is not confined to the very tip of chromosomes. DSBs internal to telomeric repeats are also repaired by MMEJ as opposed to canonical NHEJ, potentially implicating MMEJ during the repair of specialized loci such as telomere repeats25.
Mechanistic basis of microhomology-mediated end joining
Our current understanding of the mechanistic basis of MMEJ derives primarily from genetic studies in model organisms and based on different sources of DSBs. The first step in MMEJ is shared with HR and involves MRE11/CtIP dependent resection. This exposes flanking microhomology and allows base pairing of ssDNA ends26. Evidences point to a role for PARP1 in facilitating annealing of resected ends27,28. Following synapsis, DNA polymerase theta (Polθ) fills-in the flanking ssDNA regions. This then stabilizes paired intermediates and prevents long-range resection that would otherwise promote HR. MMEJ is completed when Ligase 3 seals DNA ends29. Annealing can also be driven by internal microhomology which then leads to 3’ ssDNA flaps that must be removed prior to fill-in synthesis and ligation. Recent work implicated the flap endonuclease, FEN1 during this processing step. Specifically, FEN1 was identified in a genome-wide CRISPR-Cas9 screen for genes that are synthetic lethal with mutant BRCA genes and found to promote MMEJ using a reporter system30.
Only a handful of MMEJ factors have been characterized so far, and efforts are currently underway to unveil the full genetic make-up of this pathway. A better understanding of factors upstream of Polθ, including ones that shape the chromatin landscape of DSBs, will provide insight into when and how cells opt for mutagenic MMEJ over HR and NHEJ. A recent study identified HMCES (5-Hydroxymethylcytosine binding, embryonic stem cell-specific protein) as a novel MMEJ factor31 that promotes efficient CSR in mature B cells. In vitro assays detected HMCES binding at resected DNA substrates, suggesting that it could act prior to the annealing step. However, the precise function of HMCES during MMEJ and whether it is active beyond CSR remains to be determined.
MMEJ mutagenicity is attributed to the promiscuous activity of Polθ
A distinguishing feature of MMEJ in higher eukaryotes is the presence of nucleotide insertions at break sites. Insertions, which are highly mutagenic, have been attributed to the activity of Polθ that is encoded by the POLQ gene. Polθ was identified in D. melanogaster through the analysis of mus308 mutants that displayed hypersensitivity to interstrand cross-links inducing agents32. Thereafter, Polθ activity was linked to MMEJ during the repair of DSBs induced by P-element transposition in flies33. Since then, the role of Polθ in MMEJ has been characterized in several multicellular organisms including worms, fish, mammals, and plants, whereas its homologs are lacking in yeast and other fungi34. In mammals, inhibition of Polθ sensitizes cells to DSB inducing agents, including bleomycin, etoposide, camptothecin and irradiation35,36. Its function in MMEJ-mediated repair has been established based on the analysis of endonuclease-cleavage reporter constructs, CRISPR-Cas9 induced breaks in human and mouse cells16,17,36–39, and chromosomal translocation in mouse cells7 (Figure 2). In all cases, Polθ activity scarred repair junctions by means of nucleotide insertions at break sites. Notably, Polθ-driven MMEJ activity was also found to be critical during the random integration of foreign DNA into host genomes in plants and mammals12,37,40. Work from our laboratory established the role of Polθ during the processing of dysfunctional telomeres17. Sequence analysis of telomere fusions in cells lacking NHEJ revealed breakpoints with non-telomeric nucleotide insertions that were diminished upon Polθ inhibition17. Taken together, these studies underscore templated insertions by Polθ as an evolutionary conserved feature of MMEJ. This raises the question of why would cells retain a mutagenic repair mechanism when more accurate pathways such as HR and NHEJ are in place. Emerging evidence from C. elegans suggests that repair by MMEJ could promote genome diversification. Specifically, insertions and small deletions indicative of Polθ activity in lab strains are also evident in the genomes of wild isolates41. It would be interesting to investigate whether Polθ is a source of genome diversity in higher eukaryotes including human germ cells.
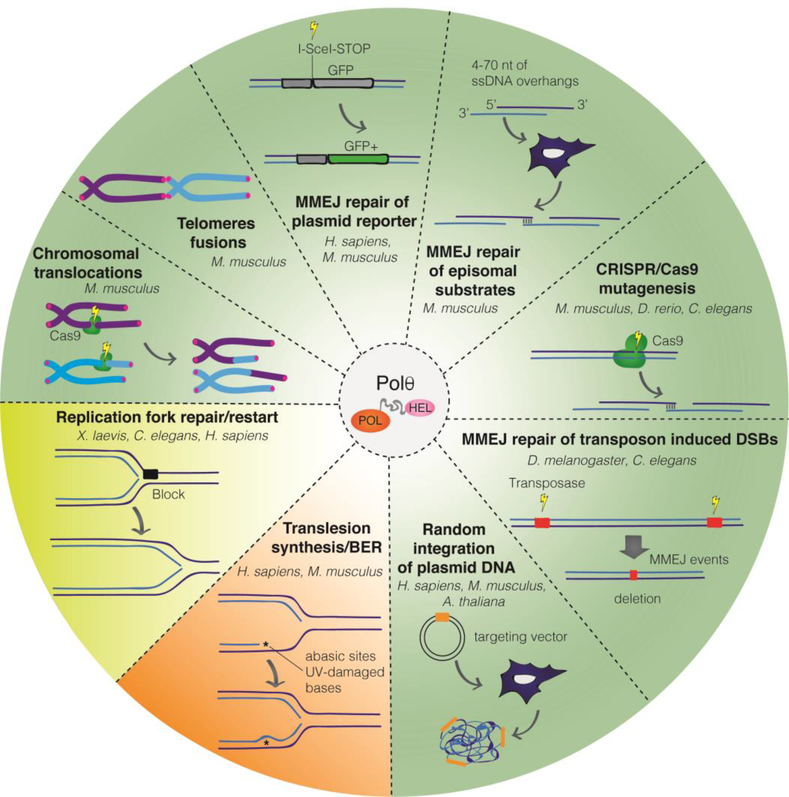
Figure 2. Overview of all reported Polθ activities.
Representation of the various activities that are carried out by Polθ. The major and well-established Polθ function is during DSB repair by MMEJ and has been studied using different substrates and in various model systems (green). Polθ has also been linked to the repair of breaks associated with replication forks (yellow). Lastly, Polθ employs its translesion synthesis activity to bypass UV-damaged bases and abasic sites (orange).
DNA polymerase Polθ as a multifunctional enzyme
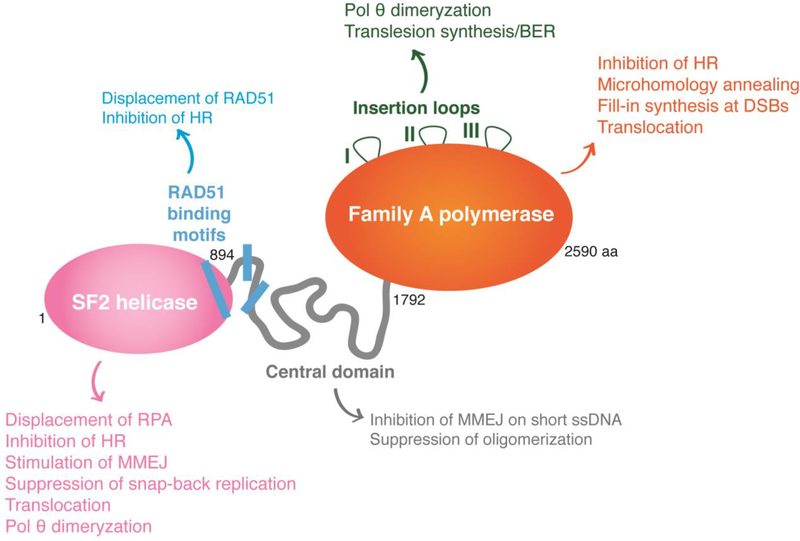
Polθ is a unique enzyme, as it is the only DNA polymerase that contains a helicase-like domain at its N-terminus (Figure 3), and the modalities are separated by a large unstructured central domain. Structural analysis of the different enzymatic domains confirmed that they form dimers and possibly multimers in solution. It has been proposed that dimerization helps Polθ tether DNA ends together and stabilize synapsed intermediates. Whether Polθ dimerization occurs in vivo is yet to be established42–44.
Figure 3. Polθ is a unique multidomain enzyme.
Schematic representation of the different domains of human Polθ, depicting the structure and function of its helicase domain (pink), the unstructured central domain (grey) and the family-A polymerase domain (orange). The helicase domain drives MMEJ activity during chromosomal translocations, promotes Polθ dimerization, and suppresses HR and snap-back replication. The central domain contains three predicted RAD51 binding motifs that are not conserved in mouse. These motifs are implicated in suppression of HR through the inhibition of RAD51 nucleofilament formation. The primary and unequivocal function of the polymerase domain is to perform fill-in synthesis during MMEJ. The polymerase domain has also been reported to participate in tethering DNA ends. The polymerase domain comprised three insertion loops that are essential for Polθ to bypass bulky lesions and abasic sites, and were proposed to promote dimerization.
Polθ-polymerase domain belongs to the A-family DNA polymerases but lacks proofreading activity45. Polθ-polymerase activity has been investigated biochemically using a range of DSB substrates and tested genetically in several model systems. In vitro characterizations of human Polθ-polymerase identified template-dependent as well as template-independent DNA synthesis46,47. Templated nucleotides are copied from regions flanking the break sites in trans and in cis. The latter employs a unique “snap-back” reaction that self-copies the ssDNA end47. On the other hand, non-templated insertions have been mainly characterized in vitro using full-length protein as well as the polymerase domain and are attributed to a terminal transferase activity that is stimulated by Mn++48. However, template-independent activity is abolished in the presence of Mg++ and when ssDNA substrates that cannot self-anneal were used49. Of note, Polθ-dependent nucleotide insertions not matching flanking DNA sequences were detected at chromosomal translocations as well as telomere fusions in mouse cells47. These nucleotides could either be inserted by a template-independent activity or through iterative cycles of templated copying, slippage, and re-priming. The identification of a separation of function mutant would be critical to ascertain the relevance of a terminal transferase Polθ activity in vivo.
Polθ-helicase belongs to the SF2 family of helicases that includes Hel308 and RecQ50. Inhibition of Polθ-helicase manifested in reduced microhomology at repair junctions in flies10 and impaired chromosomal translocation in mouse cells51. In vitro assays uncovered a strong ATPase activity that is stimulated by ssDNA43,50. Recent work from our laboratory revealed that Polθ-helicase exhibits an ATP-dependent DNA annealing activity that facilitate base-pairing of ssDNA, even when pre-bound by RPA. In effect, we showed that Polθ employs its ATPase activity to counteract RPA binding and promote the annealing of resected DNA substrates. This would ultimately favor repair by MMEJ while preventing HR51. Consistent with a role for Polθ-helicase in suppressing HR, Ceccaldi and colleagues found that inhibition of Polθ-helicase increases RAD51 loading and HR-mediated repair of I-SceI-based reporters16. Furthermore, inhibition of Polθ-helicase in mouse cells enhanced the efficiency of HR-mediated CRISPR-Cas9 gene targeting51.
The function of Polθ in other repair pathways
In addition to its critical role during MMEJ, Polθ has overlapping functions with other repair pathways, including translesion synthesis, base-excision repair (BER), and replication repair (Figure 2). Polθ catalyzes DNA synthesis across abasic sites and possesses a mild 5’-dRP lyase activity45,50,52, both relevant for BER. Genetic data suggest that Polθ could serve as a back-up for Polβ during BER. Specifically, DT40 cells lacking both Polθ and Polβ are sensitive to MMS due to reduced BER activity53. Furthermore, a CRISPR-Cas9 genome-wide screen in Polq deficient mouse cells identified multiple genes involved in the BER pathway as synthetic lethal interactors with Polθ54. Biochemical studies have determined that Polθ is unable to bypass pyrimidine dimers, a type of UV photoproduct. However, the polymerase domain is capable of extending mismatched DNA termini and could therefore indirectly contribute to repairing UV-induced lesions. In support of this hypothesis, Polq−/− mice have an increased risk of developing UV-induced skin cancers, and the incidence raises dramatically when combined with Polh deficiency55.
MMEJ activity is maximal in S phase and it is therefore not surprising that Polθ has been implicated in the repair of replication forks. The first evidence for Polθ function in replication repair came upon examining G4 DNA stability in C. elegans. Worms lacking FANCJ, which ensures replication across G4 sites, rely on Polθ to repair collapsed forks and prevent the accumulation of large deletions11. Furthermore, Polθ depletion in human cells reduces replication speed and compromises replication fork restart in the presence of hydroxyurea16. Direct evidence for Polθ role at collapsed forks is based on a recent study using Xenopus egg extracts. Specifically, supplementing frog extracts with mitotic kinases to trigger replisome disassembly led to fork breakage and Polθ dependent rearrangement56. The observation that Polθ mediated repair is not inhibited by mitotic kinases raises the interesting possibility that MMEJ is the predominant DSB repair pathway in mitosis when NHEJ and HR are known to be greatly repressed4.
Polθ inhibition during cancer therapy
Expression of Polθ is tightly regulated; the polymerase is largely repressed in somatic cells and upregulated in several human cancers, including lung, gastric, and colorectal57. In addition, Polθ levels are especially high in HR deficient breast58 and ovarian cancers16 and associated with poor clinical outcomes. The underlying mechanism that regulates Polθ expression remains unknown. Nevertheless, its expression pattern and the reliance of BRCA mutated cells on MMEJ for survival16,17 renders this unique polymerase an appealing target for cancer treatment59. In fact, efforts are currently underway to develop Polθ inhibitors for the treatment of HR defective tumors.
Exploiting synthetic lethality in DNA repair to eliminate cancer cells is best exemplified by the PARP1-BRCA genetic interaction. The profound impact of PARP1 inhibition on the survival of a sizeable fraction HR defective tumors led to the development of five PARP inhibitors that are currently in clinical trials. Despite the clinical benefits achieved in several BRCA-mutated cancers, resistance to therapy is a common theme among all PARP inhibitors. Resistance mechanisms are not fully understood, but evidence suggests that at least a subset of these tumors involve mutations in NHEJ factors that would respond to Polθ inhibition60,61.
While Polθ synthetic lethality manifests in the context of HR and NHEJ defective tumors, rearrangements consistent with MMEJ activity have been detected in wide range of cancers62. Furthermore, MMEJ footprint is found at telomere fusions in CLL21, translocations that drive lymphomas63, and during chromothripsis64. Mutagenic repair by Polθ could drive genome plasticity that fuels cancer progression and therefore, its inhibition could limit the mutational rate that renders tumors resistant to cytotoxic therapies. In conclusion, targeting Polθ has substantial clinical potential and this can only be realized upon a complete understanding of the mechanistic basis of MMEJ.
Acknowledgements
We thank Michael Smith and Marco Tigano for providing feedbacks on this manuscript. Work on Polθ in the Sfeir lab is supported by a grant from NIH/NCI (R01CA229161) and a fellowship from the Pershing Square Sohn Cancer Research Alliance.
Biography
Schimmel et al., 2017. This study reveals that in mouse embryonic stem cells Polθ acts in parallel and redundantly with NHEJ to repair DSBs and that virtually all DSBs are repaired through these two pathways. The author induced a blunt DSB with CRISPR-Cas9 in a reporter gene and analyzed the repair products revealing the mutation spectra of Polθ and NHEJ factors. The study supports the notion that Polθ repair relies on microhomology. Surprisingly, in the case of long 3’ or 5’ ssDNA overhangs, NHEJ dominates over MMEJ.
Deng et al., 2019. This is the first study that provides evidence of Polθ-mediated MMEJ events during mitosis. The authors showed that in X. laevis extracts the replisome disassembly during mitosis causes replication fork breakage that form rearrangements driven Polθ and characterized by microhomology.
Balagere et al., 2019. This study shows that HMCES, a protein reported to protect stalled replication forks, plays an additional role in the MMEJ pathway during class switch recombination in murine B cells. The author demonstrates the ability of HMCES to interact with DNA overhangs of different lengths.
Black et al., 2019. The authors used a biochemical approach to characterize MMEJ at the molecular level using full-length human Polθ. This study shows that Polθ-helicase domain is essential for MMEJ. In addition, its central domain was proposed to regulate of Polθ multimerization and DNA substrates requirements during MMEJ.
Feng et al., 2019. This study expands the synthetic lethality interactions of Polθ. Through a CRISPR-Cas9 screen in mouse cells, the authors identified 140 genes that are required for the survival of Polθ deficient cells. These genes belong to several pathways, including DNA repair, DNA damage signaling, and chromatin structure. Breast cancers with alterations in many of these genes had increased Polθ and had an MMEJ genomic footprint.
Footnotes
Declaration of interests
Agnel Sfeir is a co-founder, consultant, and shareholder in REPARE Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jackson SP & Bartek J The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Lange T Shelterin-Mediated Telomere Protection. Annu. Rev. Genet. 52, 223–247 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Pannunzio NR, Watanabe G & Lieber MR Nonhomologous DNA End Joining for Repair of DNA Double-Strand Breaks. J. Biol. Chem. jbc.TM117.000374 (2017). doi: 10.1074/jbc.TM117.000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orthwein A et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Symington LS & Gautier J Double-Strand Break End Resection and Repair Pathway Choice. Annu. Rev. Genet. 45, 247–271 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Wright WD, Shah SS & Heyer W-D Homologous recombination and the repair of DNA Double-Strand Breaks. J. Biol. Chem. jbc.TM118.000372 (2018). doi: 10.1074/jbc.TM118.000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aniukwu J, Glickman MS & Shuman S The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 22, 512–527 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chayot R, Montagne B, Mazel D & Ricchetti M An end-joining repair mechanism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107, 2141–2146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulton SJ & Jackson SP Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15, 5093–5103 (1996). [PMC free article] [PubMed] [Google Scholar]
- 10.Chan SH, Yu AM & McVey M Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6, 1–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koole W et al. A polymerase theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 5, 1–10 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Van Kregten M et al. T-DNA integration in plants results from polymerase-θ-mediated DNA repair. Nat. Plants 2, (2016). [DOI] [PubMed] [Google Scholar]
- 13.Thyme SB & Schier AF Polq-Mediated End Joining Is Essential for Surviving DNA Double-Strand Breaks during Early Zebrafish Development. Cell Rep. 15, 707–714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan CT et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Corneo B et al. Rag mutations reveal robust alternative end joining. Nature 449, 483–486 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Ceccaldi R et al. Homologous-recombination-deficient tumours are dependent on Polθ - mediated repair. Nature 518, 258–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mateos-Gomez PA et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt DW et al. Essential Roles for Polymerase θ-Mediated End Joining in the Repair of Chromosome Breaks. Mol. Cell 63, 662–673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maser RS et al. DNA-Dependent Protein Kinase Catalytic Subunit Is Not Required for Dysfunctional Telomere Fusion and Checkpoint Response in the Telomerase-Deficient Mouse. Mol. Cell. Biol. 27, 2253–2265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones RE et al. Escape from telomere-driven crisis is DNA ligase III dependent. Cell Rep. 8, 1063–1076 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Lin TT et al. Telomere dysfunction and fusion during the progression of a human malignancy. Blood 44, 1899–1908 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Tankimanova M et al. Mre11 modulates the fidelity of fusion between short telomeres in human cells. Nucleic Acids Res. 40, 2518–2526 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rai R et al. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 29, 2598–2610 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sfeir A & De Lange T Removal of shelterin reveals the telomere end-protection problem. Science (80-. ). 336, 593–597 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doksani Y & de Lange T Telomere-Internal Double-Strand Breaks Are Repaired by Homologous Recombination and PARP1/Lig3-Dependent End-Joining. Cell Rep. 17, 1646–1656 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truong LN et al. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 110, 7720–7725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Audebert M, Salles B & Calsou P Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 279, 55117–55126 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Wang M et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang L et al. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 36, 3297–3310 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mengwasser KE et al. Genetic Screens Reveal FEN1 and APEX2 as BRCA2 Synthetic Lethal Targets. Mol. Cell 73, 885–899.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balagere S et al. Article HMCES Functions in the Alternative End-Joining Pathway of the DNA DSB Repair during Class Switch Recombination in B Cells Article HMCES Functions in the Alternative End-Joining Pathway of the DNA DSB Repair during Class Switch Recombination in B C. Mol. Cell 1–11 (2020). doi: 10.1016/j.molcel.2019.10.031 [DOI] [Google Scholar]
- 32.Harris PV et al. Molecular Cloning of Drosophila mus308, a Gene Involved in DNA Cross-Link Repair with Homology to Prokaryotic DNA Polymerase I Genes †. MOLECULAR AND CELLULAR BIOLOGY 16, (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan SH, Yu AM & McVey M Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6, 1–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sfeir A & Symington LS Microhomology-Mediated End Joining : A Back-up Survival Mechanism or Dedicated Pathway ? Trends Biochem. Sci. 40, 701–714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goff JP et al. Lack of DNA Polymerase Q Radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiaton. Radiat. Res. 172, 165–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yousefzadeh MJ et al. Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ. PLoS Genet. 10, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito S, Maeda R & Adachi N Dual loss of human POLQ and LIG4 abolishes random integration. Nat. Commun. 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schimmel J et al. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 36, e201796948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z et al. DNA polymerase (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem. 294, 3909–3919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zelensky AN, Schimmel J, Kool H, Kanaar R & Tijsterman M Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun. 8, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Schendel R, Roerink SF, Portegijs V, Van Den Heuvel S & Tijsterman M Polymerase θ is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat. Commun 6, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zahn KE et al. Human DNA polymerase ?? grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 22, 304–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman JA, Cooper CDO, Aitkenhead H & Gileadi O Structure of the Helicase Domain of DNA Polymerase Theta Reveals a Possible Role in the Microhomology-Mediated End-Joining Pathway. Structure 23, 2319–2330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Black SJ et al. Molecular basis of microhomology-mediated end-joining by purified full-length Polθ. Nat. Commun. 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seki M et al. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 23, 4484–4494 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hogg M, Sauer-Eriksson AE & Johansson E Promiscuous DNA synthesis by human DNA polymerase θ. Nucleic Acids Res. 40, 2611–2622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter J, Kent T, Mateos-Gomez PA, Sfeir A & Pomerantz RT Polymerase is a robust terminal transferase that oscillates between three different mechanisms during end-joining. Elife 5, 1–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walter J, Kent T, Mateos-Gomez PA, Sfeir A & Pomerantz RT Polymerase is a robust terminal transferase that oscillates between three different mechanisms during end-joining. (2016). doi: 10.7554/eLife.13740.001 [DOI] [PMC free article] [PubMed]
- 49.He P & Yang W Template and primer requirements for DNA Pol θ-mediated end joining. Proc. Natl. Acad. Sci. U. S. A. 115, 7747–7752 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seki M, Marini F & Wood RD POLQ (Pol ??), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 31, 6117–6126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mateos-Gomez PA et al. The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol. (2017). doi: 10.1038/nsmb.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon JH, Choudhury JR, Park J, Prakash S & Prakash L A role for DNA polymerase θ in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. J. Biol. Chem. 289, 13177–13185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshimura M et al. Vertebrate POLQ and POLβ Cooperate in Base Excision Repair of Oxidative DNA Damage. Mol. Cell 24, 115–125 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng W et al. Genetic determinants of cellular addiction to DNA polymerase theta. Nat. Commun. 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon JH et al. Error-Prone Replication through UV Lesions by DNA Polymerase θ Protects against Skin Cancers. Cell 176, 1295–1309.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng L et al. Mitotic CDK promotes replisome disassembly, fork breakage, and complex DNA rearrangements. Mol. Cell 73, 428433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawamura K et al. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer 109, 9–16 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Lemee F et al. DNA polymerase up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. 107, 13390–13395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higgins BGS & Boulton SJ Beyond PARP—POL Q as an anticancer target. Science (80-. ). 359, 1217–1219 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Gupta R et al. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell 173, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noordermeer SM et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stephens PJ et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu C et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109, 811–821 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Kloosterman WP et al. Constitutional Chromothripsis Rearrangements Involve Clustered Double-Stranded DNA Breaks and Nonhomologous Repair Mechanisms. Cell Rep. 1, 648–655 (2012). [DOI] [PubMed] [Google Scholar]