Abstract
Experimental information from microscopy, structural biology, and bioinformatics may be integrated to build structural models of entire cells with molecular detail. This integrative modeling is challenging in several ways: the intrinsic complexity of biology results in models with many closely packed and heterogeneous components; the wealth of available experimental data is scattered among multiple resources and must be gathered, reconciled, and curated; and computational infrastructure is only now gaining the capability of modeling and visualizing systems of this complexity. We present recent efforts to address these challenges, both with artistic approaches to depicting the cellular mesoscale, and development and application of methods to build quantitative models.
Seeing the Invisible
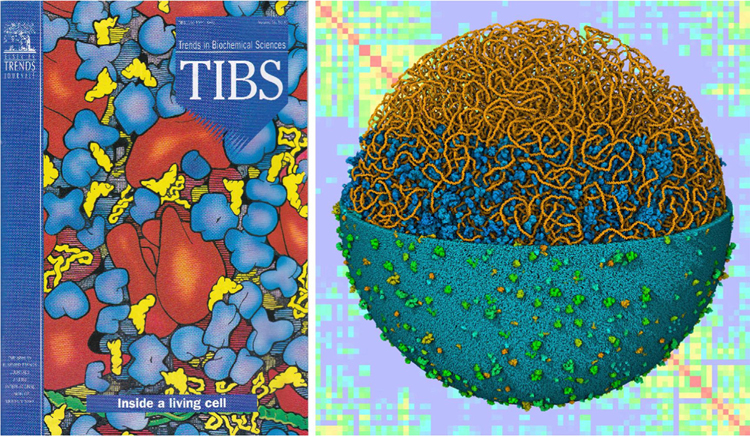
Almost 30 years ago, we presented a feature in Trends in Biochemical Sciences (TIBS) that created snapshots of the molecular structure of a bacterial cell by integrating the available information on ultrastructure, composition, and molecular structure [1] (Figure 1). At the time, there was barely enough information to support this challenge, and this information was difficult to find through resources such as the Citation Index. Since then, there has been an explosion of progress in experimental study of molecular and cellular biology, and a complementary revolution in the availability of scientific information. However, the scale range depicted in the 1991 TIBS article, the cellular mesoscale (see Glossary) bridging the atomic nanoscale and the cellular microscale, remains largely invisible to experiment and is currently the domain of integrative approaches.
Figure 1. Modeling and Visualizing the Cellular Mesoscale.
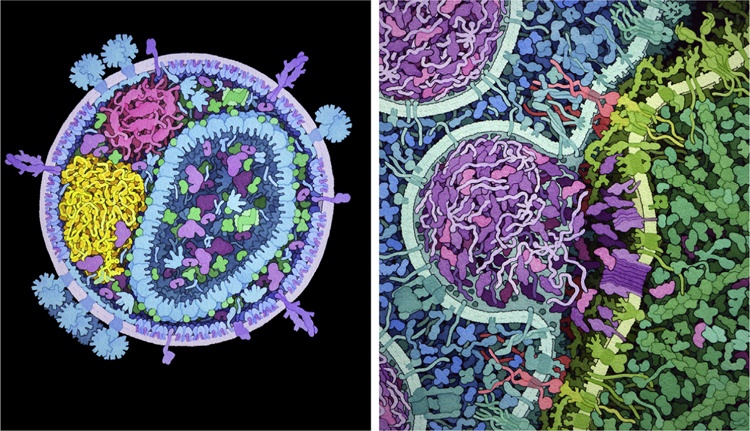
In 1991, traditional artistic methods (left) were used to create images of the molecular structure of a portion of bacterial cytoplasm, and today (right, foreground), computational methods can build integrative data-driven models of an entire bacterial cell and (right, background) simulate experimental data based on these models.
Today, the study of the cellular mesoscale is just as much art as science. Experimental techniques are being developed, from the top down and from the bottom up, to explore the atomic structure of cells. Some of these techniques are exploratory, harkening back to the early days of natural history. Methods in single particle cryoelectron microscopy (cryo-EM) have improved to the level where it is actively competing with X-ray crystallography as the way to determine atomic (or near atomic) structures of biomolecules and biomolecular assemblies [2]. Cryo-EM tomography is also allowing near-atomic exploration of large expanses (by fib-milled thin sections) of eukaryotic cells; ribosomes, microtubules, and other large assemblies are routinely localized and oriented, and methods are being explored to extend reach to the many other smaller biomolecules in cells [3].
Light microscopy is also in the midst of a revolution. Single-molecule methods allow for an unprecedented level of molecular quantification. GFP tagging and other fluorescence methods explore the properties of individual molecules within mesoscale environments, at the whole cell level, as evidenced by the ambitious efforts of the Allen Cell Institute [4] and Human Protein Atlas [5]. Much of current innovation in mesoscale science is driven by these new ways of seeing inside cells. We are also seeing a revolution in -omics studies that give us detailed and comprehensive recipes for genetic information, proteins, and their interactions in cells. These studies allow direct examination of the inner composition of cells throughout their lifecycle and across many environmental and disease states.
Taken together, these experimental approaches are narrowing in on the invisible mesoscale range, where all of these individual molecules come together and interact to create life. However, because no one experimental modality conveys all the needed information, these data must be synthesized into coherent representations of molecular structure and functions of whole living systems, which in turn need to be visualized, analyzed, and communicated for human understanding and insight. Integrative modeling approaches are currently the best way to explore the connections between these diverse data, and to synthesize hypothesis-driven views consistent with the available state-of-the-art in data. There is a growing toolbox of integrative methods to approach this challenge with different goals in mind.
The Challenge of the Mesoscale
The cellular mesoscale poses challenges that are testing the current limits of technology and understanding for mesoscale modeling and visualization. The intrinsic complexity of mesoscale biology (and indeed, biology in general) poses the first major challenge. Mesoscale environments are highly crowded, often with 20–30% of the space filled with macromolecular components [6]. These components are also highly heterogeneous in size and shape, often with functional modes of flexibility and highly specific modes of interaction.
The current archipelago of biological data poses an orthogonal challenge to this complexity. Many sources of data are available, often independent and with their own application program interfaces, formats, and data standards. Structural and sequence data is becoming increasingly interconnected through mature resources such as the Protein Data Bank (PDB)i and UniProtii, but it is still the ‘Wild West’ with much of proteomics and interactomics. Effective progress in mesoscale modeling will necessarily require effective methods for gathering, curating, and integrating these heterogeneous sources of information.
The limits of current computational infrastructures pose a third existential challenge. The amount of data involved in mesoscale systems strains the limits of current hardware and software, modalities of archiving, and dissemination, and they pose exciting new challenges for conceptualization and analysis. Current tools for modeling, simulation, and visualization have been designed for use on individual macromolecular structures or image/volume-based cellular data, and often fail when dealing with systems of this size and complexity.
What do we want? The field of mesoscale integrative modeling is currently exploratory, and researchers are inventing tools as new challenges appear on the structural horizon. This requires nimble and extensible design of software and methods to allow for rapid shifts to new lines of inquiry. The power of this approach has been demonstrated in the rise of modular languages such as Python, fostering the proliferation of specialized modules for well-defined functionalities. In addition, synergy with other disciplines, such as the very active computer gaming community, allows incorporation of highly-optimized methods into mesoscale modeling.
We also expect that progress will be driven by new ways of seeing, both in the experiments that probe the cellular mesoscale, and in the visualization software that we build to explore our integrative models. In the early days of macromolecular structure, creative innovations in visualization, such as Linus Pauling’s space-filling representation and Jane Richardson’s ribbon diagrams, revolutionized the way we think about biomolecules, crystallizing new modes of understanding [7]. We expect that mesoscale understanding will benefit from similar creative paradigm shifts.
Why do we care? In the following sections, we describe several overarching motivations for pursuing this work, and review some of the work that is being done to address these motivations.
Thinking Tools and Sanity Checks
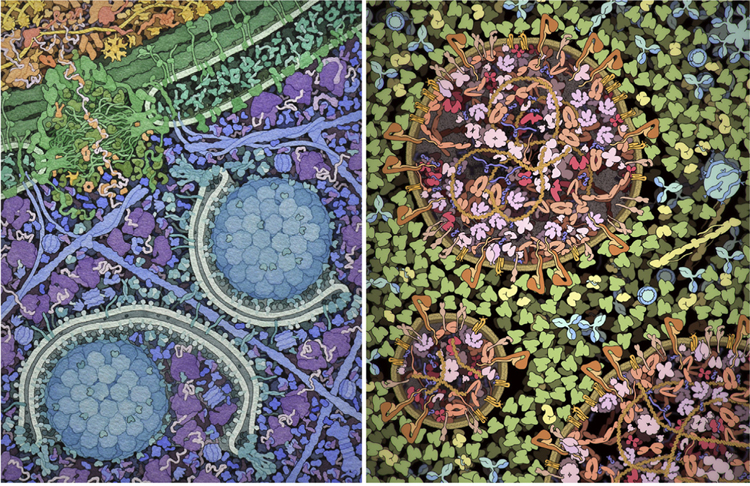
As we were gathering data for the 1991 TIBS review of bacterial inner structure, we did an informal survey of local researchers, asking a simple question: How far apart are the proteins in the cell? Not surprisingly, we got a huge variety of answers, ranging from close-packed molecules much like a protein crystal, to proteins being spaced many times their own diameters apart. One of the major goals of that TIBS article was to provide a mental image based on the existing body of knowledge, to give a foundation for further thought about the function of biomolecules in their cellular context. An artistic approach often provides a tractable path through a mesoscale synthesis, to capture the current state of the field and identify gaps in knowledge. For example, we have collaborated with several researchers on mesoscale subjects, including ongoing work on autophagy [8] and a summer internship exploring the structure of exosomes [9] (Figure 2). Artistic renderings are also widely used in education and dissemination [10]. For these nontechnical audiences, significant artistic license must often be used [11]; however, efforts such as the ‘Inner Life of the Cell’iii have proven their ability to inspire a whole generation of scientists.
Figure 2. Mesoscale Illustrations as Thinking Tools.
(Left) An illustration of the cytoplasm to vacuole targeting process of autophagy, created in collaboration with Daniel Klionsky with traditional media. (Right) Illustration of an exosome created by Julia Jiminez during a graduate internship, using CellPAINT software. In both cases, the collaboration benefited the scientist, by requiring a survey of the current state of knowledge, and our laboratory, by requiring development of methods to manage data-poor aspects of the systems.
Mesoscale modeling is also a natural extension of the integrative structural biology revolution. Currently, the term integrative structural biology refers to studies where structural models are built based on a variety of data, including near-atomic and lower resolution imaging (EM or light), crosslinking constraints, and proteomics information [12]. Methods, such as the integrative modeling platform [13], use this approach to determine structures of large targets such as the nuclear pore [14] and even larger systems [15]. For example, several laboratories are using chromosome conformation capture data and transcription frequencies to provide constraints for the modeling of entire bacterial chromosomes [16–18] and eukaryotic chromatin [19]. Integrative modeling and simulation are also exploring the structures of organelles such as synaptic vesicles [20] and photosynthetic chromatophore vesicles [21]. Additionally, interpretations of cryo-EM tomograms as structural models are actively being explored, for example, of synaptic boutons [22]. One of the central advantages of this type of work is to provide a sanity check on the underlying hypotheses connecting atomic structure with cellular structure, or as articulated by Covert [23]: confirming that the ‘aggregate behavior does not violate physical laws.’
Interacting with Invisible Realms
The ability to visualize and interact with structural models has been critical in creating our current understanding of biology. The advent of interactive computer graphics in the 1970s catalyzed the field of protein crystallography [24]. Tracking the exponential progress in structural biology in the following decades, a sophisticated toolbox of molecular graphics methods has been invented and developed, and today, dozens of turnkey programs are available for visualizing data from structural biology (Box 1). These hardware and software advances are now being expanded to produce computational and visualization tools that can meet the challenges of the biological mesoscale. Interactive mesoscale modeling focuses on the power of putting the human in the loop, promoting comprehension, guided physical manipulation, and conceptual synthesis.
Box 1. Tricks of the Trade.
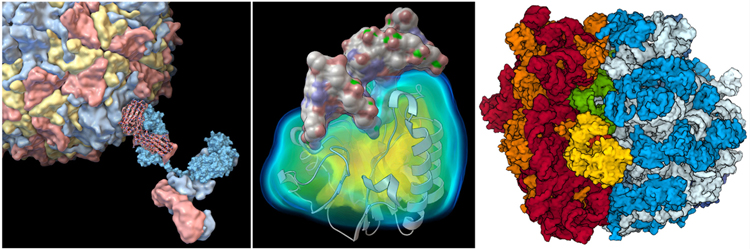
Molecular graphics programs are currently essential tools for biostructural research, education, and outreach, and numerous turnkey programs are available [24,68]. A few popular examples include powerful stand-alone methods such as Chimera [69], PyMolvi, VMD [70], and Python Molecular Viewer [71], which include comprehensive methods for visualization, analysis, and some modeling, as well as web-based methods such as Jmolvii, NGL [72], and Mol* [73], which focus on user-friendly visualization and push-button functionality (Figure I). Decades of development have refined the methods used in these programs to meet the challenges of biomolecular complexity and need for interactive performance. These methods include multiple representations to capture details of bonding, space occupancy, and interaction; selection, depth cuing, and clipping methods for highlighting regions of interest; and diverse methods for the dissemination of images, from static images to animations to 3D printed models. These programs are used in all aspects of biostructural research, from day-to-day working graphics for data analysis and dissemination, to scripted animations or interactives used in education, to highly-rendered realistic images used in editorial settings.
These traditional methods are currently being used and expanded to visualize mesoscale models, providing a solid foundation for development of new methods. In many respects, mesoscale visualization is much the same as molecular visualization, only larger and more complex. A recent development is the use of multiscale rendering approaches, where the level of detail of the representation is dynamically changed depending on how much of the viewer’s field of vision is occupied by a particular entity [25,74]. New approaches to user interaction, as described in the main text, are also being explored to streamline navigation through these complex models.
Figure I.
Illustrations of Biomolecular Assemblies Created by Each of the Authors with Freely Available Molecular Graphics Software. (Left) Poliovirus interacting with an antibody, using different representations to highlight hierarchical levels of structure and interaction. (Center) Volume rendering to display potentials used to model desolvation in docking studies in a DNA–endonuclease complex. (Right) Outlines and ambient occlusion used to create an illustrative illustration of a ribosome bound to tRNA and elongation factors. Ribosome illustration created online with Mol* at the RCSB Protein Data Bank, and the others were created with Python Molecular Viewer, using Protein Data Bank (PDB) IDs: 1IGT, 2PLV, 1DE8, and 4V5G.
Modern graphics processing units (GPUs) have revolutionized the creation of complex imagery, rendering mesoscale scenes containing billions of atoms at interactive rates [25]. With careful utilization of central processing units (CPUs), GPUs and data-oriented programming techniques, interactive scenes of over 1 trillion particles have been demonstrated [26]. More importantly, the utility of GPUs has expanded beyond image generation to speed up general computation, including molecular dynamics [27] and gaming physics engines such as FleX from Nvidia [28] to interactively handle 1 million particles. The GPU version of CellPACK, for example, can create structural models of whole bacteria in seconds [29], and a GPU implementation of procedural models of bacterial nucleoid structure of over 1 million base pairs, with supercoiled regions, can likewise be generated interactively [30]. While these hardware advances were mainly driven by the entertainment industries of film and gaming, scientists have been beneficiaries since real-time interactivity changes the nature of how we can model, navigate, and analyze complex structures and data.
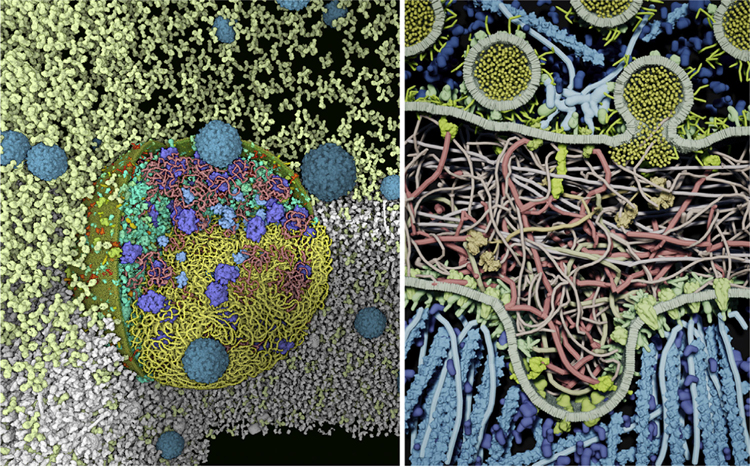
Importantly, scientific software development has also benefited from the commoditization of high-performance interaction, again driven by the gaming industry. Game engines, such as Unity3D, Torque, and Unreal Engine, have been developed to facilitate the production of gaming software for real-time interactions utilizing GPUs, fast rendering, physical simulations, and adaptable user interfaces. These development environments have been used in generating a variety of interactive mesoscale software applications, including CellPACKgpu, CellPAINT [31], and LifeBrush [32] (Figure 3).
Figure 3. Interaction in Mesoscale Visualization.
(Left) Atomic scale model of a mycoplasma and poliovirus in blood plasma created with CellPACKgpu. Interactive cutaways are used to highlight different aspects of the cell: DNA (yellow) is only shown below the horizontal clipping plane, and bacterial proteins (turquoise), ribosomes (blue), and mRNA (pink) are shown above, with one quadrant removed to show the interior. The surrounding plasma is shown in white, with only the antibodies (light yellow) shown above the clipping plane. (Right) Illustration of a neuromuscular junction, created and simulated interactively with LifeBrush (figure courtesy of Tim Davison and Christian Jacob).
New display and interaction technologies provide a third leg of the technology stool that supports mesoscale modeling, again benefitting from consumer level deployment. Virtual and augmented reality (VR and AR) systems are being driven by developments in devices ranging from smartphones to wearable displays, making 3D and stereoscopic viewing as well as novel interactive controllers both inexpensive and constantly improving in quality and capability. Since mesoscale structural models are complex systems with thousands of molecular components arrayed in a 3D space, amplifying a sense of presence and immersion can provide perceptual, cognitive, and operational enhancements for construction, navigation, manipulation, simulation, and analysis.
Driven by the wide commercial availability of devices and development environments, a number of biomolecule-level VR applications have appeared recently, such as the ChimeraX VR application [33] and collaborative drug design and development platforms by Nanomeiv. LifeBrush [32] is an illustrative simulation canvas for sketching, simulating, and visualizing the biological mesoscale that facilitates creation and visualization of real-time dynamic agent-based simulations of models of up to 10 000 biomolecular entities. Similarly, our VR version of CellPAINTv allows real-time experimentation by modifying placement and concentrations of the interacting biomolecules within a 3D mesoscale scene. In these applications, we have found that many operations within the perceived space feel more natural and easily accomplished than with a traditional display screen and mouse interface. The ability to sketch out structural spatial arrangements that would be difficult to produce in another way indicate that as the technology improves, these new environments will become an important part of mesoscale modeling and communication.
Quantifying Emergent Properties of the Mesoscale
The cellular mesoscale is a foreign environment that resists intuition. A host of properties, such as the large numbers, random motion, specific interactions, and ever-present bath of water, dominate the behavior and result in often-surprising emergent properties. As articulated in the review ‘The Middle Way’ [34], this is the domain of ‘collective organizing principles that formally grow out of the microscopic rules but are in a real sense independent of them.’ Study of the emergent properties of mesoscale biology relies on a dialog between simulation and experiment, and the central challenge is always to formulate a level of detail, both in experiment and in simulation, that probes the property of interest in an effective but achievable way.
For example, this combination of simulation and experiment has led to a growing understanding of the functional consequences of crowding in biological systems [35,36], described by Ellis as ‘obvious but underappreciated’ [37]. A wide range of simulation methods have been used [38], ranging from Minton’s pioneering hard-sphere models [39] to atomically detailed models of portions of bacterial cytoplasm [40,41] (Figure 4A). Study of biomolecular self-assembly has also benefited from this integrative simulation approach. Coarse-grain models have been used to explore all aspects of viral self-assembly, leading to the discovery of several general principles: highly directional interactions and reversible associations are necessary to evade traps leading to malformed capsids or starvation of assembly through overly stable intermediates [42].
Figure 4. Mesoscale Simulation.
(A) Close-up of a mesoscale molecular dynamics simulation of bacterial cytoplasm, revealing unexpected folding/unfolding dynamics promoted by interactions between proteins in the crowded environment. Image courtesy of Michael Feig. (B–D) Three applications of coarse-grained simulations to explore assembly of HIV-1: (B) simulation explores the balance of hexameric and pentameric protomers in formation of authentic and malformed capsids; (C) simulation of the assembly of gag in budding virions; (D) simulation of restriction of HIV-1 by TRIM5alpha. Image courtesy of Alex Pak, Alvin Yu, and Gregory Voth.
The presence (and possible functional importance) of quinary structure association, where many weak interactions lead to high-order assemblies, has also been a topic of much discussion and speculation [43–45]. While some features, such as the microtrabecular lattice [46], have fallen out of favor, many other examples, such as functional super-assemblies of aminoacyl-tRNA synthetases [47], are being discovered. Cryo-EM is being used to explore some of the best defined examples, including photosynthetic supercomplexes [48] and respirasomes [49]. The role of phase separation is an exciting new wrinkle on this topic, where many molecules form disordered assembles that partition function into a local microenvironment [50]. The entropic nature of this type of phase separation has been shown to lead to surprising properties, such as providing the impetus for segregation of bacterial chromosomes [51].
Simulation methods are also currently reaching the whole-cell level. Systems biology approaches are being used to quantify the detailed molecular composition and interaction over the life of the cell [52]. The ground-breaking WholeCellSim model of an entire mycoplasma cell [53] revealed ‘emergent behaviors that cross traditional network boundaries,’ including a novel emergent control on the mycoplasma cell cycle and pathological consequences of disrupting single genes [54]. This study also quantified the magnitude of the tools and underlying data management that are needed, and spurred experiments in data-poor domains of knowledge. In another study, the Lattice Microbes software uses a reaction–diffusion master equation to add a structural element to the process, following these reactions within a cellular environment [55]. This approach has been recently expanded in an ambitious study that performed Brownian dynamics within a crowded cellular environment to look at an inducible genetic switch in yeast, by using a structural model of cytoskeletal elements and nuclear pore from cryo-EM studies [56]. The study was able to quantify the dynamics of mRNA transport, revealing that the sparsity of nuclear pores has a greater impact than crowding by cytoskeletal obstacles.
New Paradigms in Drug Discovery
In many respects, the overarching goal of biological research is to explore and understand all steps from genotype to phenotype, and use this knowledge to identify key vulnerabilities to target with therapeutically relevant interventions. This is the grand challenge of mesoscale modeling: to fill in all of the gray areas in our understanding that bridge between molecular biology and cell biology, such that we can then apply this knowledge to health and welfare [57,58]. The field is in its infancy, but we are beginning to make our first steps towards this lofty goal.
The overall hypothesis is that characterization in the mesoscale context, structure and function will open the door to new avenues for drug development, targeting the structural and ultrastructural characteristics of cells. In fact, we are already doing this. For example, the interaction of a small molecule such as paclitaxel with specific sites in microtubules perturbs their dynamic assembly, ultimately blocking cell division. Associations between viruses and their cellular receptors are another point of intervention, often targeted with vaccines. Mesoscale modeling is beginning to take a rational approach to these types of assembly processes.
In some cases, the study begins by characterizing the mesoscale properties with the goal of identifying possible locations to target. For example, a structural model of influenza combined with Brownian dynamics was used to estimate the binding of sialic acid with hemagglutinin and neuraminidase proteins as a function of their arrangement on the viral surface [59], revealing how stalk height and secondary binding to neighboring molecules impact association rate constants. In this way, mesoscale models can provide insight on the relevant proximity of biological entities and properties that emerge as a result of packing within the crowded biological compartments, informing the desired characteristics of inhibitory molecules.
Conversely, we can take a bottom-up approach and design molecules to target a particular biomolecule, then explore how atomic scale perturbations propagate their effect and are amplified at larger scales. Given their tractable size, a number of drug-related mesoscale studies have been performed on viral systems with this goal in mind. For example, a coarse grain study of HIV-1 capsid assembly revealed that capsid inhibitors perturb the populations of small assembly intermediates, leading to the formation of aberrant forms of the capsid, suggesting that inhibitors that modify assembly pathways may reduce the requirement for a stoichiometric interaction of inhibitor with protein target [60] (Figure 4B). Further, modeling of integrase aggregation by allosteric integrase inhibitors (ALLINIs) [61], combined with mesoscale modeling of condensation of the HIV-1 ribonucleoprotein [62], are shedding light on the surprising emergent role of ALLINIs in viral maturation [63] (Figure 5A).
Figure 5. Examples of Intervention with Mesoscale Assemblies.
(Left) Treatment of HIV-1 with allosteric integrase inhibitors (ALLINIs) cause integrase (red) to aggregate and has the surprising mesoscale effect of corrupting the process of encapsidation of the viral RNA (yellow). (Right) Chimeric antigen receptors (red) recruit macrophages (left half of the image) to target cells (right half of the image), releasing perforins (magenta) and leading to destruction of the target. Illustrations from the HIVE Center and RCSB Protein Data Bank.
The current challenge is to use mesoscale modeling, and the understanding that comes from it, to spur rational development of therapeutic interventions. Several structure-based approaches may provide an effective place to start. Engineered chimeric molecules have been shown to be effective for making unnatural, but therapeutically useful, mesoscale connections. For example, chimeric antigen receptor therapy (CART, Figure 5B) uses an engineered molecule that combines two recognition elements to bring two cells together [64]. With PROTACs, a small molecule plays a similar role, linking a desired target to the machinery of ubiquitination [65]. A new class of highly selective covalent inhibitors may be used to perform a similar role in promoting desired linkages. To give this a mesoscale scope of action, new computational tools and protocols [66] are being developed that can be deployed to screen for suitable modifiable residues in whole proteomics screenings. In a particularly exciting recent development, these inhibitors have been used in an inverse discovery mode [67], screening a library of compounds against whole cells and fishing out the phenotype, and ultimately the protein target, for the mesoscale property we are trying to perturb. In an ideal iterative feedback loop, the inhibitor design effort would benefit dramatically by the explicit modeling of biological compartments, and in turn inform the mesoscale models with the new observed phenotype generated by the intervention.
Concluding Remarks
Integrative mesoscale research has the potential to revolutionize our understanding and application of cellular structure and function. For example, with drug phenotypes, this approach will allow us to answer the persistent question: How does a molecular intervention have organismic effect? Nearly all existing drugs block one metabolic or signaling step, providing an effective intervention, but one with little flexibility or robustness in the face of resistance mutation. The aspiration of the field of mesoscale modeling is to provide a holistic context for simulations, in which it is possible to study the long-range effects of drugs binding to their targets, including how effects propagate over cellular distances through perturbation of signaling and ultrastructural networks, and how effects propagate over time to affect assembly, morphology, and, ultimately, cellular function. The mesoscale approach, at least currently, is also highly exploratory and practical, with many new methods being developed as new hypotheses are conceived. Ultimately, mesoscale models are the ideal convergence point in which the large amount of data generated in the -omics approaches will be represented and contextualized with one another, providing a synergistic integrated insight that is not available from the pieces alone. We envision that as the field matures, we will have a comprehensive toolbox of methods for addressing features across the scale range from atoms to cells, and that researchers will have nimble solutions that allow simulation in tractable times when switching contexts from atomic to mesoscale, and back (see Outstanding Questions).
Outstanding Questions.
How do we create a comprehensive pipeline that integrates tools for data management, model generation, and analysis of mesoscale properties, in applications to cellular biology research?
What are the salient features and properties of the cellular mesoscale that impact the processes of life, and how can we target these properties in new pharmaceutical interventions?
How do we promote an effective dialog between experimentalists and computational biologists to further the goals of research into the cellular mesoscale?
What is the role of human interaction in integrative structural biology research?
Highlights.
Advances in integration of data and computational infrastructure together are enabling the modeling of large portions of cells, and in some cases, entire cells.
Mesoscale models of cellular environments promote research into cellular structure and function by providing thinking tools for hypothesis generation, by providing initial models for simulation and interpretation of experiments, and by providing a new window for the development of drugs.
New approaches to visualization and interaction are being developed to address the challenging needs of mesoscale research.
Acknowledgments
This work is supported by grant GM120604, GM069832, and GM103368 from the National Institutes of Health and the RCSB Protein Data Bank (National Science Foundation DBI-1832184, National Institutes of Health GM133198, and US Department of Energy DE-SC0019749). This is manuscript 29928 from the Scripps Research Institute.
Glossary
- Cellular mesoscale
scale level bridging the nanometer scale of atomic structure and the micrometer scale of cellular ultrastructure.
- Cryo-electron microscopy (cyro-EM)
electron microscopy of samples cooled to cryogenic temperatures and embedded in vitreous water.
- Game engines
software development environments for building and creating video games, including methods for rendering images, collision detection, memory management, and other tasks necessary for interactive performance.
- Integrative structural biology
currently refers to studies where structural models of biomolecules and biomolecular assemblies are built based on a variety of data from multiple disciplines, such as sub-atomic imaging (EM or light microscopy), crosslinking constraints, proteomics information, etc.
- Quinary structure
interaction (often transient) of molecules into higher-order assemblies, added to the traditional hierarchy of primary structure (sequence) to quaternary structure (oligomerization of subunits).
- Virtual and augmented reality (VR and AR)
virtual reality is the use of computer technology to simulate an environment, and in augmented reality, the real-world environment is enhanced by combination with computer-generated elements.
Resources
- i. www.wwpdb.org.
- ii. www.uniprot.org.
- iii. www.youtube.com/watch?v=FzcTgrxMzZk.
- iv. https://nanome.ai.
- v. https://sourceforge.net/projects/cell-paint.
- vi. www.pymol.org.
- vii. www.jmol.org.
References
- 1.Goodsell DS (1991) Inside a living cell. Trends Biochem. Sci 16, 203–206 [DOI] [PubMed] [Google Scholar]
- 2.Rout MP and Sali A (2019) Principles for integrative structural biology studies. Cell 177, 1384–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nievergelt AP et al. (2019) Towards a mechanistic understanding of cellular processes by cryoEM. Curr. Opin. Struct. Biol 58, 149–158 [DOI] [PubMed] [Google Scholar]
- 4.Horwitz R and Johnson GT (2017) Whole cell maps chart a course for 21st-century cell biology. Science 356, 806–807 [DOI] [PubMed] [Google Scholar]
- 5.Uhlen M et al. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 6.Fulton AB (1982) How crowded is the cytoplasm? Cell 30, 345–347 [DOI] [PubMed] [Google Scholar]
- 7.Goodsell DS and Jenkinson J (2018) Molecular illustration in research and education: past, present, and future. J. Mol. Biol 430, 3969–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodsell DS and Klionsky DJ (2010) Artophagy: the art of autophagy – the Cvt pathway. Autophagy 6, 3–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jimenez J et al. (2019) Integrative modeling and visualization of exosomes. J. Biocommunication 43, 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodsell DS et al. (2018) From atoms to cells: using mesoscale landscapes to construct visual narratives. J. Mol. Biol 430, 3954–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodsell DS and Johnson GT (2007) Filling in the gaps: artistic license in education and outreach. PLoS Biol 5, 2759–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alber F et al. (2008) Integrating diverse data for structure determination of macromolecular assemblies. Annu. Rev. Biochem 77, 443–477 [DOI] [PubMed] [Google Scholar]
- 13.Russel D et al. (2012) Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol 10, e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SJ et al. (2018) Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Im W et al. (2016) Challenges in structural approaches to cell modeling. J. Mol. Biol 428, 2943–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacker WC et al. (2017) Features of genomic organization in a nucleotide-resolution molecular model of the Escherichia coli chromosome. Nucleic Acids Res 45, 7541–7554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodsell DS et al. (2018) Lattice models of bacterial nucleoids. J. Phys. Chem. B 122, 5441–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildirim A and Feig M (2018) High-resolution 3D models of Caulobacter crescentus chromosome reveal genome structural variability and organization. Nucleic Acids Res 46, 3937–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosa A and Zimmer C (2014) Computational models of large-scale genome architecture. Int. Rev. Cell Mol. Biol 307, 275–349 [DOI] [PubMed] [Google Scholar]
- 20.Takamori S et al. (2006) Molecular anatomy of a trafficking organelle. Cell 127, 831–846 [DOI] [PubMed] [Google Scholar]
- 21.Singharoy A et al. (2019) Atoms to phenotypes: molecular design principles of cellular energy metabolism. Cell 179, 1098–1111.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilhelm BG et al. (2014) Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028 [DOI] [PubMed] [Google Scholar]
- 23.Macklin DN et al. (2014) The future of whole-cell modeling. Curr. Opin. Biotechnol 28, 111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson AJ (2018) Perspectives on structural molecular biology visualization: from past to present. J. Mol. Biol 430, 3997–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Muzic M et al. (2015) cellVIEW: a tool for illustrative and multi-scale rendering of large biomolecular datasets. Eurographics Workshop Vis. Comput. Biomed 2015, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schatz K et al. (2016) Interactive visual exploration of a trillion particles. In 2016 IEEE 6th Symposium on Large Data Analysis and Visualization (LDAV), pp. 56–64 [Google Scholar]
- 27.Kutzner C et al. (2015) Best bang for your buck: GPU nodes for GROMACS biomolecular simulations. J. Comput. Chem 36, 1990–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macklin M et al. (2014) Unified particle physics for real-time applications. ACM Trans. Graph 33, 153 [Google Scholar]
- 29.Klein T et al. (2018) Instant construction and visualization of crowded biological environments. IEEE Trans. Vis. Comput. Graph 24, 862–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein T et al. (2019) Parallel generation and visualization of bacterial genome structures. Comp. Graph. Forum 38, 57–68 [Google Scholar]
- 31.Gardner A et al. (2018) CellPAINT: interactive illustration of dynamic mesoscale cellular environments. IEEE Comput. Graph. Appl 38, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davison T et al. (2019) LifeBrush: painting, simulating, and visualizing dense biomolecular environments. Comput. Graph. UK 82, 232–242 [Google Scholar]
- 33.Goddard TD et al. (2018) Molecular visualization on the holodeck. J. Mol. Biol 430, 3982–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laughlin RB et al. (2000) The middle way. Proc. Natl. Acad. Sci. U. S. A 97, 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivas G and Minton AP (2016) Macromolecular crowding in vitro, in vivo, and in between. Trends Biochem. Sci 41, 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivas G and Minton AP (2018) Toward an understanding of biochemical equilibria within living cells. Biophys. Rev 10, 241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellis RJ (2001) Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci 26, 597–604 [DOI] [PubMed] [Google Scholar]
- 38.Elcock AH (2010) Models of macromolecular crowding effects and the need for quantitative comparisons with experiment. Curr. Opin. Struct. Biol 20, 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minton AP (1983) The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences. Mol. Cell. Biochem 55, 119–140 [DOI] [PubMed] [Google Scholar]
- 40.McGuffee SR and Elcock AH (2010) Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol 6, e1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feig M et al. (2015) Complete atomistic model of a bacterial cytoplasm for integrating physics, biochemistry, and systems biology. J. Mol. Graph. Model 58, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hagan MF (2014) Modeling viral capsid assembly. Adv. Chem. Phys 155, 1–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srere PA (2000) Macromolecular interactions: tracing the roots. Trends Biochem. Sci 25, 150–153 [DOI] [PubMed] [Google Scholar]
- 44.Gierasch LM and Gershenson A (2009) Post-reductionist protein science, or putting Humpty Dumpty back together again. Nat. Chem. Biol 5, 774–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chien P and Gierasch LM (2014) Challenges and dreams: physics of weak interactions essential to life. Mol. Biol. Cell 25, 3474–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clegg JS (2010) Revisiting the microtrabecular lattice. Cell Biol. Int 34, 1105–1107 [DOI] [PubMed] [Google Scholar]
- 47.Robinson JC et al. (2000) Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J. Mol. Biol 304, 983–994 [DOI] [PubMed] [Google Scholar]
- 48.Nevo R et al. (2012) Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J 70, 157–176 [DOI] [PubMed] [Google Scholar]
- 49.Letts JA and Sazanov LA (2017) Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol 24, 800–808 [DOI] [PubMed] [Google Scholar]
- 50.Hyman AA et al. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol 30, 39–58 [DOI] [PubMed] [Google Scholar]
- 51.Jun S and Wright A (2010) Entropy as the driver of chromosome segregation. Nat. Rev. Microbiol 8, 600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feig M and Sugita Y (2019) Whole-cell models and simulations in molecular detail. Annu. Rev. Cell Develop. Biol 35, 191–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karr JR et al. (2012) A whole-cell computational model predicts phenotype from genotype. Cell 150, 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrera J and Covert MW (2015) Why build whole-cell models. Trends Cell. Biol 25, 719–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts E et al. (2013) Lattice microbes: high-performance stochastic simulation method for the reaction-diffusion master equation. J. Comput. Chem 34, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Earnest TM et al. (2017) Challenges of integrating stochastic dynamics and cryo-electron tomograms in whole-cell simulations. J. Phys. Chem. B 121, 3871–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amaro RE and Mulholland AJ (2018) Multiscale methods in drug design bridge chemical and biological complexity in the search for cures. Nat. Rev. Chem 2, 0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singla J et al. (2018) Opportunities and challenges in building a spatiotemporal multi-scale model of the human pancreatic beta cell. Cell 173, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amaro RE et al. (2018) A computational assay that explores the hemagglutinin/neuraminidase functional balance reveals the neuraminidase secondary site as a novel anti-influenza target. ACS Cent. Sci 4, 1570–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pak AJ et al. (2019) Off-pathway assembly: a broad-spectrum mechanism of action for drugs that undermine controlled HIV-1 viral capsid formation. J. Am. Chem. Soc 141, 10214–10224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng N et al. (2016) Allosteric HIV-1 integrase inhibitors promote aberrant protein multimerization by directly mediating inter-subunit interactions: structural and thermodynamic modeling studies. Protein Sci 25, 1911–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodsell DS et al. (2019) Integrative modeling of the HIV-1 ribonucleoprotein complex. PLoS Comput. Biol 15, e1007150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kessl JJ et al. (2016) HIV-1 integrase binds the viral RNA genome and is essential during virion morphogenesis. Cell 166, 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang ZL and Chen YY (2017) CARs: synthetic immunoreceptors for cancer therapy and beyond. Trends Mol. Med 23, 430–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scheepstra M et al. (2019) Bivalent ligands for protein degradation in drug discovery. Comput. Struct. Biotechnol. J 17, 160–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Backus KM et al. (2016) Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mortenson DE et al. (2018) “Inverse drug discovery” strategy to identify proteins that are targeted by latent electrophiles as exemplified by aryl fluorosulfates. J. Am. Chem. Soc 140, 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Donoghue SI et al. (2010) Visualization of macromolecular structures. Nat. Methods 7, S42–S55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goddard TD et al. (2018) UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Humphrey W et al. (1996) VMD: Visual Molecular Dynamics. J. Mol. Graph 14, 27–28 [DOI] [PubMed] [Google Scholar]
- 71.Sanner MF (2005) A component-based software environment for visualizing large macromolecular assemblies. Structure 13, 447–462 [DOI] [PubMed] [Google Scholar]
- 72.Rose AS and Hildebrand PW (2015) NGL Viewer: a web application for molecular visualization. Nucleic Acids Res 43, W576–W579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sehnal D et al. (2018) Mol*: towards a common library and tools for web molecular graphics. In MolVA/EuroVis Proceedings June, 29–33 [Google Scholar]
- 74.Waldin N et al. (2019) Cuttlefish: color mapping for dynamic multi-scale visualizations. Comput. Graph. Forum 38, 150–164 [DOI] [PMC free article] [PubMed] [Google Scholar]