Abstract
Food safety issues across the global food supply chain have become paramount in promoting public health safety and commercial success of global food industries. As food regulations and consumer expectations continue to advance around the world, notwithstanding the latest technology, detection tools, regulations and consumer education on food safety and quality, there is still an upsurge of foodborne disease outbreaks across the globe. The development of the Electronic nose as a noninvasive technique suitable for detecting volatile compounds have been applied for food safety and quality analysis. Application of E-nose for pathogen detection has been successful and superior to conventional methods. E-nose offers a method that is noninvasive, fast and requires little or no sample preparation, thus making it ideal for use as an online monitoring tool. This manuscript presents an in-depth review of the application of electronic nose (E-nose) for food safety, with emphasis on classification and detection of foodborne pathogens. We summarise recent data and publications on foodborne pathogen detection (2006–2018) and by E-nose together with their methodologies and pattern recognition tools employed. E-nose instrumentation, sensing technologies and pattern recognition models are also summarised and future trends and challenges, as well as research perspectives, are discussed.
Keywords: Sensors, Pattern recognition, Foodborne pathogens, Volatile organic compounds (VOCs), Electronic nose
Introduction
Food pathogens characterise a special form of microbial pathogens, which are acquired and spread through food. Foodborne Pathogens (Campylobacter, Clostridium botulinum, Escherichia coli O157: H7, Listeria monocytogenes, Norovirus, Salmonella, Staphylococcus aureus, Shigella, Toxoplasma gondii, Vibrio vulnificus) are a significant source of foodborne illnesses, hospitalization and deaths in the world (Havelaar et al. 2015).
The global incidence of foodborne related diseases is on the rise with a reported 600 million illnesses and 420,000 deaths every year, leading to the loss of 33 million healthy life years measured in Disability-Adjusted Life Years-DALY’s (Franz et al. 2018).
Foodborne illnesses usually occur through the contamination of surfaces, oral-faecal route and improper food storage (Nygren et al. 2013). The detection of foodborne pathogens is a critical component in the elimination of pathogens in the food supply chain. Current detection methods include conventional cell culture standards, immunological assays, DNA based methods, Biosensor based methods, as well as emerging spectroscopic methods and spectral imaging techniques.
Recent developments in sensor technologies have led to innovative analytical approaches such as the electronic nose (E-nose) that been developed and applied in the food industry in reaction to emerging food safety issues. E-nose provides a rapid, non-invasive online monitoring tool for food safety and can be used for qualitative and semi-quantitative detection (Chen et al. 2013).
An E-nose is a device capable of identifying simple or complex odours by combining a chemical sensor array system with partial specificity and a suitable pattern recognition system (Gardner and Bartlett 1994). E-nose can analyse volatile organic compounds (VOCs) produced by microorganisms is employed as a possible alternative method in the identification and classification of different chemicals and bacteria.
E-nose has gained widespread application in the food industry and has been applied for the detection of food spoilage bacteria (Pattarapon et al. 2018; Wang et al. 2012) and total volatile basic nitrogen (Li et al. 2016), trimethylamine (Ampuero et al. 2002), fungal infections (Lippolis et al. 2018; Liu et al. 2018; Pallottino et al. 2012). E-nose presents numerous advantages over conventional and other non-invasive methods such as vibrational spectroscopic methods and hyperspectral imaging.
In this article, the following specific objectives are discussed:
The use of electrochemical sensors for monitoring microbial growth.
E-nose Instrumentation, sampling and pattern recognition methods.
The application of E-nose will be discussed as a non-destructive analytical tool for food safety analysis (foodborne pathogen).
Methodology
Electrochemical techniques for monitoring microbial growth
Electrochemical (EC) techniques such as electrochemical impedance spectroscopy, voltammetry, potentiometry, and coulometry have made substantial contributions to the food analysis. EC techniques directly convert chemical processes that occur in a solution at the electrode/electrolyte interface into quantifiable electronic signals such as altered conductive properties (conductometric), current (amperometric), and potential or charge accumulation (potentiometric) (Niu et al. 2014).
The application of electrochemical sensors and detectors for food analysis is expanding rapidly due to their inherent sensitivity, selectivity, and speed of detection. Electrochemistry provides a noninvasive method for monitoring microbial activity as well as for monitoring electron flow within microbial communities (Martin et al. 2018). The theoretical basis of electrochemical gas sensor operation involves interactions between gaseous molecules and sensor-coating materials. Electrochemical gas sensors will be described during the course of this article.
Electronic nose instrumentation
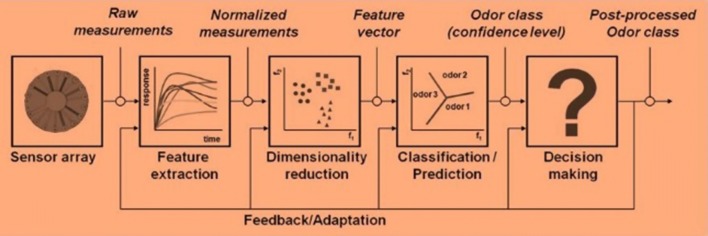
An electronic nose (Fig. 1) is an artificial olfaction system that comprises of units for gas/odour sampling, sensing, signal preprocessing, pattern recognition, and odour expression (Jia et al. 2018). E-nose allows for capturing volatile chemical compounds into an array of sensors through a sampling system. Signal response is generated and subsequently transmitted to a computer system for processing and pattern recognition.
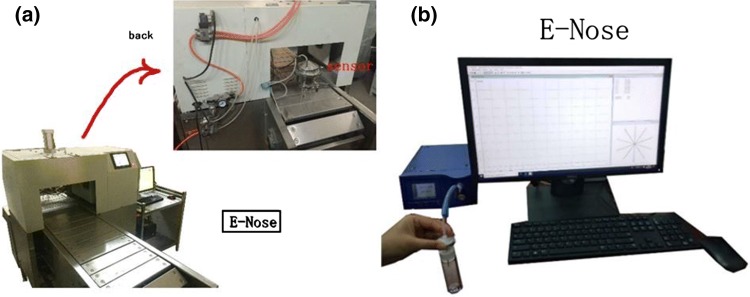
Fig. 1.
a A fabricated E-nose machine for online detection at Jiangsu University, b commercial E-nose machine, Airsense PEN3 (Airsense Analytics GmbH, Schwerin, Germany)
Electronic nose sensor types
Electronic noses employ an array of chemical sensors with varying specificities which reacts and respond to volatile organic compounds present in the gases collected from samples (Jiang and Chen 2014; Zohora et al. 2013). The selection of sensors to employ is quite large and have been classified into broader classes they include quartz crystal microbalance sensors, surface acoustic wave sensors, electrochemical sensors, optical sensors, and calorimetric sensors. A detailed description of these sensors together with their merits and demerits is comprehensively discussed by Wilson and Baietto (2009).
Some chemical-based sensors (catalytic, semiconducting metal oxide, solid electrolyte, polymer and field-effect transistor-based sensors) have been used for E-nose. Metal oxide semiconductors (MOS) have been commonly used as sensing elements in the electronic nose by researchers due to their availability, high sensitivity and their ability to respond to oxidising and reducing compounds. The MOS sensor is based on the adsorption of gas molecules to incite change in conductivity. The measured change in conductivity corresponds to the amount of volatile organic compounds adsorbed. One disadvantage of MOS is its susceptibility to poisoning by sulphur compounds present in the odorant mixture.
Another conductivity sensor is the polymer sensor made up of polypyrroles, thiophenes, indoles polyaniline, furan material polymers (Ghasemi-Varnamkhasti et al. 2018). Chemicals form either ionic or covalent bonds when exposed to the polymers. Changes in conductivity occur due to the transfer of electrons along the polymer chain. Polymer sensors operate at ambient temperatures and do not require heating. They are suitable for use as portable instruments having a simple electronic interface. Polymer sensors are however susceptible to humidity which can mask the responses of VOCs.
Electronic nose sampling systems
To effectively design a pattern recognition and analysis system for electronic nose data, the processes involved in analyzing the data generated must be studied as shown in Fig. 2.
Fig. 2.
Stages of classic signal processing of electronic nose data by Gutierrez-Osuna and Nagle (1999)
Selecting a suitable sampling of the volatile fractions and conveying it to the sensor array is a major challenge when designing the analytical methodology for microbial volatile profiling with electronic nose. The sampling technique for bacteria used usually depend on the sample state of matter (liquid, solid, semi-solid), food matrix and the level of concentration or bacterial load. Sampling systems that allow for agitation and the use of a longer sampling period to generate more volatiles are mostly preferred.
Headspace methods, analytical distillation methods or direct extraction methods are usually employed for sampling odour active analytes where adequate isolation is required. The most widely used method is the static headspace (SHS) sampling technique. It comprises of placing the microbial sample in a hermetically sealed vial after equilibrium between the matrix and the gaseous phase is established, the headspace is sampled (Peris and Escuder-Gilabert 2009). Therefore sampling techniques are designed to be stable and be able to withstand environmental effects (Rayappan et al. 2017).
A detailed description of E-nose sampling methods is described by Majchrzak et al. (2018).
Feature extraction and dimensionality reduction
Preprocessing is the significant first step in E-nose data analysis and is usually performed to remove irrelevant information from the signal data. Preprocessing of multivariate signals is usually performed to prepare the for obtained multivariate pattern analysis. These methods are used for baseline manipulation, compression, noise reduction, detection and removal of outliers and normalisation (Sanaeifar et al. 2017).
The influence of preprocessing methods on class recognition of chemical compounds is described in several studies with Gardner et al. (1998) and Gutierrez-Osuna and Nagle (1999) using 36 and 48 different pre-processing algorithms respectively. Sensor data preprocessing methods include Scaling (dimensional auto-scaling, mean centring, relative scaling, vector auto-scaling, logarithmic scaling and power scaling), and Baseline correction (fractional difference) (Jha et al. 2019). In other to remove background noise from the raw sensor responses, Fractional techniques deduct the baseline value of the sensor response value and then divide by the baseline value, to yields a per unit response. The application of fractional changes in conductance delivers the most suitable pattern-recognition performance for MOS (Hierlemann and Gutierrez-Osuna 2008).
In Vector auto-scaling, each example vector is normalized with the mean and variance computed for each example across the different dimensions whereas, in dimensional auto-scaling, the mean and variance are computed for each dimension along with all the examples in the database. The coordinates have zero mean and unit variance (Gutierrez-Osuna and Nagle 1999). In Mean centring constant terms are removed in the data so as to make the data compatible with the model. It is applied to center on the inconsistent part of the data, and to leave only the relevant variation between the samples for analysis (van den Berg et al. 2006).
Logarithmic scaling involves extracting the logarithm of previous measures. It has been applied to prevent the influence of large variations in chemical vapor concentration on sensor responses (Watson et al. 1993). Power scaling, on the other hand, applies power law for nonlinearity class separation in feature space. The raw sensor signal is scaled by suitable inverse power law to linearize the sensor output (Sunil Kumar Jha and Yadava 2011).
Autoscaling is considered the most effective preprocessing method applied to the data before feature extraction. Autoscaling involves mean centring of individual datasets and dividing it by the standard deviation for rescaling with unit variance. The main advantage is to preclude high sensor responses from dominating the analysis. The data produced by an E-nose is made up of a set of semi-independent variables from the sensor array and a set of dependent variables (Scott et al. 2006).
The general improvement of an E-nose system usually involves the optimisation of feature extraction and selection method, pattern recognition method as well as sensitive material selection and sensor array optimization for homemade E-nose devices. The latter refers to hardware selection and optimization. The primary goal of feature extraction is to extract robust information from sensor responses with less redundancy. This would ensure the overall effectiveness of the pattern recognition algorithm applied subsequently (Carmel et al. 2003).
Feature extraction methods can be grouped into three according to the source of features extraction. Firstly from curve fitting which fits the response curves based on a particular model and extracts a set of fitting parameters as the features, examples include polynomial model, exponential model, fractional function model and the S function model (Yan et al. 2015).
Secondly, from original response curves of sensors by the extraction piecemeal signal features examples include secondary derivatives, maximum values, differences, primary derivatives integrals, the adsorption slope, and the maximum adsorption slope and lastly, from applying transforms such as fast Fourier transform (FFT) and discrete wavelet transform (DWT) (Huang et al. 2006).
In addition to the above conventional feature extraction techniques, new methods have been applied in recent years. Energy vector (EV) is a vector of energy, which contains the energy of each sensor and all the mutual energies and is useful when studying the relationship between signals of sensors of the same array.
Parallel factor analysis (PARAFAC) as a multi-way data decomposition method, PARAFAC simultaneously determines the pure contributions to the dataset and optimizing each factor as a time, in trilinear systems (Zhang et al. 2014). Dynamic moments (DM) and Phase space (PS) are usually applied in dynamical systems whereas the power density spectrum (PSD) describes the distribution of power into frequency components composing that signal. The statistical average of a signal is examined in terms of its frequency content and windowed time slicing (WTS) is another recent method based on window functions. It multiplies the time response of each sensor by time windows to obtain the area values and these values are further used as features (Kaur et al. 2012; Yan et al. 2015).
Dimension reduction is achieved through principal component analysis (PCA) or independent component analysis (ICA) for uncorrelated and independent factors respectively. PCA is the most commonly used method for dimensionality reduction and feature extraction, also known as the Karhunen–Loève transform, PCA employs orthogonal transformations to eliminate colinearity in variables and the sensor array response matrix is transformed along the virtual axes of minimum correlation (Jolliffe 2014).
ICA is a linear method capable of identifying hidden factors of random variables. ICA attempts to fragment a multivariate signal into independent non-Gaussian signals (Hyvärinen et al. 2001). In ICA the sensor array response matrix is transformed along the virtual axes with minimum correlation and statistical dependency (Jha et al. 2019). Other methods employed for dimension reduction and feature extraction include wavelet transform, independent component analysis (ICA), principal kernel component analysis (KPCA) and linear discriminant analysis (LDA). In addition to the visual discrimination of chemical compounds during these processes, the resulting data is used as input for qualitative classification and quantitative estimation of chemical concentration.
Pattern recognition methods for electronic nose
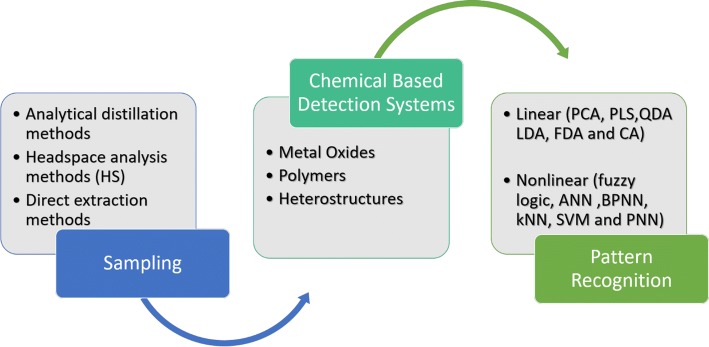
Pattern recognition methods are then applied to analyse and classify the processed data and can be classified as linear or nonlinear (Fig. 3). These methods can also be classified as supervised (k-nearest neighbor, Linear discriminant analysis, naïve Bayes, Backpropagation artificial neural networks, adaptive resonance theory map and support vector machine) and unsupervised (k-means clustering, self-organizing map, fuzzy clustering and hierarchical cluster analysis). A supervised learning algorithm learns from labelled training data while unsupervised learning deals with the unlabelled data (Sizochenko et al. 2019).
Fig. 3.
Sampling, detection and analysis of volatiles by E-nose
Other classification categories include reinforcement learning (Reinforcement learning neural network) approaches neighborhood approaches (RMSE Neighbourhood and Similarity measure), neural networks (Feedforward neural networks, Spiking Neural Networks and Learning Vector Quantization), and decision/bagged trees algorithms.
Artificial neural network (ANN) and support vector machine (SVM) are predominantly employed for E-nose data classification due to their robustness and high accuracy. E-nose data is also known for demonstrating strong nonlinearity. Artificial Neural Networks (ANN) is another supervised learning model employed for classification and regression analysis. ANN is ambiguously inspired by the biological neural networks in the human brain and usually consist of three layers an input, output and hidden layers (Siswantoro et al. 2017).
SVM is a supervised learning model employed for classification and regression analysis. The algorithm was created by Hava Siegelmann and Vladimir Vapnik (Vapnik 2000). SVM is a powerful tool used for function estimation, nonlinear classification, and density estimation and has formed the bases for the development of kernel-based methods. A detailed theoretical discussion of SVM and its application to E-nose datasets is described (Acevedo et al. 2007; Distante et al. 2003; El Barbri et al. 2008; Laref et al. 2018; Pardo and Sberveglieri 2005). Back Propagation (BP) learning is a method usually employed for training most of the applied ANNs with multilayer perceptron (MLP) trained by the error back-propagation algorithm the most commonly used ANN in food analysis and classification (Dębska and Guzowska-Świder 2011). A detailed overview of ANN and its application to E-nose datasets is described by (Balasubramanian et al. (2008); Luo et al. 2004). E-nose data requires training for odor discrimination or differentiation, and this is usually performed by correlating E-nose responses with chromatography (GC, HPLC), sensory analysis or calibrating with known samples (Ghasemi-Varnamkhasti et al. 2018).
Discussions
Volatiles associated with the microbial growth
Volatile organic compounds (VOCs) are a diverse group of carbon-based chemicals that are volatile at ambient temperature and can be detected through smell. VOCs have low-molecular-weights with high vapour pressures that are easily volatilized (Tait et al. 2014).
During food spoilage, pathogenic and spoilage microorganisms act upon food substrates and emit specific VOCs. This odor active molecules are generated during the process of breaking down food. Electronic nose with the requisite training program is able to discriminate amongst several volatile profiles (Giungato et al. 2018). Microorganisms have their unique characteristic volatile compounds they emit during growth (Avalos et al. 2018). Some of these volatiles provide unique odor fingerprints for a particular microorganism and can be employed for pathogen identification and discrimination without the use of conventional food analytical techniques.
In another study, Berna et al. (2013) stated that various characteristic odours are associated with pathogenic bacteria; E. coli is associated with an amino acid (indole) distinctive odour, Salmonella typhimurium is associated with methyl ketones, primary and secondary alcohols (Balasubramanian et al. 2016). There are no clear reasons why microorganism produces VOCs but researchers have hypothesised and attributed their production to signalling or defence mechanisms (Selim et al. 2017) as well as for growth monitoring (Kai et al. 2009).
Data of VOCs in (Table 1) reveal basic information about microbial activities at the molecular level (Robin Michael Statham and John 2012). Physiological conditions such as moisture content, oxygen, pH, and temperature affect the composition and amount of volatiles produced by a particular microorganism. Another factor that affects the composition and amount of volatiles produced are the carbon-energy sources present for the microbes to act upon (Romoli et al. 2014). VOCs produced mainly by bacteria are produced through primary metabolism (metabolites necessary for development, growth and reproduction such as DNA, amino acids, fatty acids synthesis) and secondary metabolism (organic metabolites not involved directly in normal growth and reproduction and are intermediates of the primary metabolism). Fatty acids, acetic acid, keto acids, and amino acids act as precursors during metabolic oxidation of glucose resulting in the production of some Microbial Volatile organic compounds (MVOCs) (Selim et al. 2017).
Table 1.
A summary of volatile organic compounds produced by foodborne pathogens
| Foodborne pathogen | VOCs | References |
|---|---|---|
| Escherichia coli | Indole, 1-decene (E. coli O157:H7 in TSYA), Dimethyl disulfide, ethanol, 2-nonanone, 2-heptanone, indole, pentyl cyclopropane (E. coli in tryptone-yeast NaCl super-broth), 2,5-dimethyl tetrahydrofuran, dimethyl disulfide, 2-heptanone, 2 undecanone, indole, unknown, 2-tridecane, 2,5 dimethyl pyrazine, benzaldehyde, dimethyl trisulfide, 2-nonanone, nonanal, decanal (Escherichia coli O157:H7 and a nonpathogenic strain of E. coli) | Siripatrawan (2008a) and Senecal et al. (2002) |
| Listeria monocytogenes |
Acetaldehyde, Ethanol, Acetone, 2-Methyl-propanal, 2,3-Butanedione, 2-Butanone Acetic acid, 1-Butanol,3-Methyl-butanal,2-Methyl-butanal,3-Methyl-3-buten-1-ol 3-Hydroxy-2-butanone, Dimethyl disulfide, Pyrazine, Pyrrole, Hexanal, Butyl ester acetic acid,3-Methyl-2-butenal, Methyl-pyrazine, Methoxy-phenyl-oxime 2,5-Dimethyl-pyrazine,4,6-Dimethyl-pyrimidine, D-Limonene, 6-Methyl-5-hepten-2-one, Octanal 2-Ethyl-1-hexanol, Benzaldehyde, 2-Ethyl-6-methyl-pyrazine,2-Ethyl-5-methyl-pyrazine Pentyl-cyclopropane, Nonanal, Benzeneacetaldehyde, Acetophenone,1-Nonanol Phenylethyl alcohol, Decanal, Tetradecane,1-Ethylidene-1H-Indene,1,5-Dimethyl-naphthalene, Butylated Hydroxytoluene (Tryptone soy broth) |
Yu et al. (2014) |
| Salmonella |
Primary alcohols (1-octanol, 1-decanol), secondary alcohols (2-undecanol, 2-tridecanol), methyl ketones (2-nonanone,2-undecanone), 3-methyl-1-butanol (S. typhimurium in tryptic soy yeast agar), Hydrogen sulfide, ethanol, carbon disulfide, dimethyl cyclopropane, 1-propanol (S. typhimuriumin tryptone-yeast NaCl super-broth), Dimethyl sulfide, carbon disulfide, heptane, acetic acid, ethyl acetate, methyl alcohol, ethyl benzene, 1-pentanol, 3-octanone, 3-octanol, 1-hepten-3-ol (S. typhimuriumin alfalfa sprouts—glass vial) |
Senecal et al. (2002), Siripatrawan and Harte (2007) and Siripatrawan (2008a) |
| Staphylococcus aureus | Isovaleric acid, 2-methyl butyric acid, isobutyric acid, 1-hydroxy 2-propanone, 1-hydroxy 2-butanone, butyric acid, 4-methylhexanoic acid (S. aureus in blood agar) | Preti et al. (2009) |
| E. coli, S. sonnei, S. typhimurium, Bacillus cereus, L. monocytogenes, S. aureus |
1 Octanol, 1-decanol, dodecanol, 2 undecanone, 2-tridecenone, indole (E. coli), 1 octanol, 1-decanol, dodecanol, 2-nonanone,1-undecene, 2-undecanone, 2-tridecanone (S. sonnei), 1 octanol,1-decanol, dodecanol (S. typhimurium), 2-undecanone, dimethyl disulfide (B. cereus), 2-undecanone, 2-tridecenone, vdimethyl trisulfide (L. monocytogenes), 2-tridecenone, dimethyl disulfide (S. aureus |
Elgaali et al. (2002) |
| Shigella sonnei | Methanethiol, dimethyl sulfide (TSA) | Warren et al. (2007) |
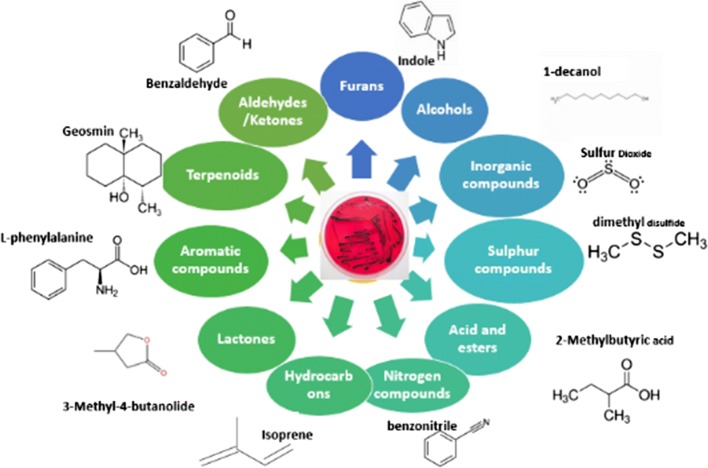
Bacterial volatiles (Fig. 4) is usually dominated by alcohols, furans, alkenes, aldehydes and ketones, terpenoids, sulphur compounds, acids and esters (Piechulla and Degenhardt 2014). About 346 known volatile compounds have been reported as bacterial VOCs. Reported volatile compounds classification is shown in Fig. 4 with an example of volatile compounds in each category. This volatiles may differ in species and that allow for species differentiation, although researchers for same species reported different Volatile compounds due to different substrates and detection times. Volatile compounds produced by foodborne pathogens during microbial growth in food samples are employed to characterise these pathogens.
Fig. 4.
Categories of chemical classes of VOCs for bacterial identification
Application of electronic nose technology for foodborne pathogen detection
As described previously, several authors have reported the detection and measurements of VOCs associated with bacterial foodborne pathogens, this had led to the attempts to create a profile of microbial VOCs (MVOCs) for a particular pathogen. The application of E-nose for pathogen detection is described in detail in this section and summarised in Table 2.
Table 2.
Recent studies on food pathogen detection by electronic nose from 2013 to 2018
| Pathogens | Matrix | Sensor types | Chemometric analysis | References |
|---|---|---|---|---|
| E. coli O157:H7, Salmonella typhimurium 857, Staphylococcus aureus 29213, Pseudomonas aeruginosa 27853 | Beef | 32-polymer sensor nose chip | Abdallah et al. (2013) | |
| Salmonella typhimurium | Beef | 8 MOS sensors | LDA, QDA | Balasubramanian et al. (2012) |
| Salmonella typhimurium | Fresh alfalfa sprouts | 12 MOS sensors | PCA | Siripatrawan and Harte (2015) |
| E. coli, Salmonella typhimurium | Super broth | 12 MOS sensors | PCA, BPNN | Siripatrawan (2008a) |
| E. coli, Listeria innocua. |
Lysogeny broth, Brain–Heart Infusion media |
12 MOS sensors | ULDA | Green et al. (2011) |
| E. coli DH5α, Listeria innocua, Enterococcus faecalis, E. coli Biotype I | Brain–heart infusion | 12 MOS sensors | PCA, ULDA | Green et al. (2014) |
| E. hormaechei and E. coli | Mixed vegetable soups | 4 MOS sensors | LDA | Gobbi et al. (2015) |
| Escherichia coli | Processed tomatoes | 6 SMO sensors | PCA, | Concina et al. (2009) |
| Staphylococcus. Salmonella, Shigella | Apple | 6 SMO sensors | PCA, HCA | Ezhilan et al. (2018) |
| Escherichia coli, Listeria monocytogenes, Salmonella typhimurium | Brain Heart Infusion |
4 MOX thin film gas sensors and 2 MOX nanowires gas sensors |
PCA | Sberveglieri et al. (2015) |
| Listeria monocytogenes, Staphylococcus lentus, Bacillus cereus | – | 18 MOS sensors | PCA, HCA, DA, ANN | Yongxin and Zhao (2012) |
| Escherichia coli | Goat meat | 32 polymer sensors | PCA | Ding et al. (2010) |
| Escherichia coli O157:H7, Salmonella spp. | Lettuce | PCA | Berna et al. (2013) | |
| L. monocytogenes standard strains | Brain heart infusion broth | 18 MOS sensors | PCA, HCA, ANN | Xue et al. (2012) |
| Enterococcus faecalis, Escherichia coli and Staphylococcus aureus | Street foods | 9 MOS sensors | SVM | Balbin et al. (2017) |
| Salmonella enterica | Poultry manure | 12 MOS sensors | ANN | Kizil et al. (2015) |
| Salmonella typhimurium | Beef | 7 MOS sensors | ICA, PCA | Balasubramanian et al. (2008) |
| Escherichia coli (ATCC 25922) | Alfalfa Sprouts | 12 MOS sensors | ANN | Siripatrawan et al. (2006) |
| Escherichia coli (ATCC 25922) | Packaged fresh vegetable | 12 MOS sensors | SOM | Siripatrawan (2008b) |
Strain and species discrimination
Green et al. (2011) successfully distinguished between E. coli and Listeria innocua in phosphate-buffered saline by employing electronic nose based on metal oxide sensor (MOS) and uncorrelated linear discriminant (ULDA) analysis with a classification accuracy of 92.4%. This method was established on odor signature processing of single bacteria colonies removed directly from the surface of the agar medium. Bacteria identification based on the E-nose response of single colonies allows for rapid results and reduces the need for culturing serological and biochemical tests.
Green et al. (2014) investigated the reliability of using E-nose for bacterial identification at the genus level using individual colonies. Four non-pathogenic bacteria species (E. coli DH5α, Listeria innocua, Enterococcus faecalis, E. coli Biotype I) were used for this study, achieving a classification accuracy of > 80%, with a higher classification of 96.7% when E-nose sampling was repeated for the same colony and using all existing odor responses for sample characterization. Rapid identification of L. monocytogenes, Staphylococcus lentus, and Bacillus cereus was carried out by integrating E-nose data with chemometrics (Yongxin and Zhao 2012). Results from the study showed discrimination of four different strains of Vibrio parahaemolyticus with PCA explaining nearly 99% of the variance and discrimination of four different species of Pseudomonas by cluster analysis (CA) and PCA.
Xue et al. (2012) proposed E-nose together with chemometrics for strain and species-level differentiation. L. monocytogenes cultured on brain heart infusion broth. In this study PCA integrated with ANN for feature, extraction was successful in the identification of volatile metabolites of nine strains of L. monocytogenes and four species of Listeria spp.
These results showed E-nose has clear potential as an accurate early diagnostic screening tool for bacterial foodborne pathogen detection since discrimination between individual bacterial colonies at both species and strain level was possible. This is crucial since virulence and pathogenicity are often associated with only a subset of bacterial strains and it is importance for a technique to have the ability to distinguish between pathogenic and nonpathogenic strains during a foodborne outbreak.
Food matrix detection
Meat products Balasubramanian et al. (2008) achieved successful prediction of Salmonella typhimurium in contaminated beef using E-nose data and independent component analysis (ICA). A stepwise linear regression prediction (SLRP) model was built with the independent component (IC) and principal components (PC) with a prediction accuracy of 69.64% and 82.99%, and a root mean squared error (RMSE) of 1.358 and 0.803 for PCA and ICA respectively. The results showed that ICA performed better than PCA on the E-nose dataset, ICA which is higher-order statistical techniques can explore higher-order information of the original inputs than PCA (Cao et al. 2003).
Balasubramanian et al. (2012) compared two different gas sensor-based artificial olfactory systems i.e. conducting polymer-based and metal oxide-based sensors to successfully screen Salmonella typhimurium in beef. LDA and QDA classification models achieved varying levels of success for polymer E-nose (69%), metal oxide E-nose (≥ 70%) and a fusion of the sensors (> 80%) for classifying “No Salmonella” (microbial counts < 0.7 log10 cfu/g) and “Salmonella inoculated” (microbial counts ≥ 0.7 log10 cfu/g) in meat samples stored at 10 °C. The use of only relevant sensors (through Fisher Criteria Ranking of sensors) and sensor fusion approaches proved important in achieving higher classification accuracies.
Ding et al. (2010) employed a Cyranose-320 E-nose based on 32 polymer sensors for the rapid detection of E. coli in goat meat samples with preliminary results showing 18–77% detection accuracies for cultured bacteria. There was no differentiation between PCA data generated for contaminated and uncontaminated meat samples due to overlapping or very close marking, also the sensor was responsive to lower concentrations of the bacteria.
Street foods are a major source of foodborne illnesses, Balbin et al. (2017) applied SVM on E-nose signals for the identification and classification of E. coli and Staphylococcus aureus in street foods. Results from the study revealed successful detection of the pathogens in the street foods (Kwek–Kwek, pork barbeque and isaw) before and after cooking showing the use of E-nose as an online tool for process monitoring during food preparations.
An E-nose with a 32-sensor nose chip was applied by Abdallah et al. (2013) to detect E. coli O157: H7, Salmonella typhimurium 857, and S. aureus 29213 in fresh and frozen beef. Results from the study showed a strong correlation (p < 0.005) in gas concentration before and after the samples were contaminated with the pathogens.
Fruits and vegetables Concina et al. (2009) applied E-nose for the detection of microbial contaminants in processed tomatoes. E. coli with both KNN pattern recognition method showing good classification scores of 83% after 48 h from inoculation. The study revealed the influence of microorganism metabolic kinetics, on the headspace composition during microbial growth.
Siripatrawan et al. (2006) collected volatile metabolites produced by E. coli using an E-nose with 12 metal oxide electronic sensor. The data generated was employed to predict the E. coli numbers in packaged alfalfa sprouts using the ANN model with a regression coefficient (R2) = 0.903.
Siripatrawan (2008a) developed a rapid method for differentiating E. coli and Salmonella typhimurium by combining E-nose data with PCA and BPNN models. PCA was employed for data exploration and dimensionality reduction and to successfully visualized class separation amongst sample subgroups. BPNN achieved successful prediction with a regression coefficient R2 = 0.96 between true and predicted data.
A Self-organizing map () algorithm was applied for the classification of E. coli in packaged fresh vegetable by Siripatrawan (2008b). The SOM algorithm combined with the data from E-nose successfully classified E. coli above higher than 105 cfu/g in the vegetable samples. In a more recent study, Siripatrawan and Harte (2015) applied the Kohonen network for data visualization of Salmonella typhimurium present in packaged fresh alfalfa sprouts. The Kohonen network could visually distinguish different levels of S. typhimurium contamination on the self-organising map (SOM). The Kohonen network was valuable and better at visualizing multi-dimensional nonlinear data and showed a much more perfect separation of different sample groups than a conventional linear principal component analysis (PCA) approach.
Gobbi et al. (2015) achieved a rapid diagnosis of E. coli in vegetable soups. E-nose with four metal oxide sensors together with LDA analysis achieved a classification performance of 98% for E. coli contamination at a detection threshold of 8 and 3 cells/100 ml. The discrimination of bacterial contamination in this study was independent of the initial and final microbial concentrations. The study showed the possibility of diagnosing bacterial contamination during growth however it must be noted that the release of VOCs from bacteria changes during their growth is unknown.
Ezhilan et al. (2018), a trilayer approach, based on a homemade E-nose was used to study the presence of Staphylococcus, Salmonella and Shigella bacteria in delicious royal apple from the order of zero, 102, 103–104 cfu/mL. Voltage responses for E-nose sensors together with PCA and wards HCA was applied to analyze the samples. The developed E-nose combining data classification schemes, bacterial culture study, and GC–MS analysis successfully assessed freshness or contamination levels of the apple samples.
Others Salmonella enterica is a pathogen usually associated with poultry and S. enterica is primarily transferred via manure contamination during processing. Kizil et al. (2015) applied E-nose to detect the presence of S. enterica poultry manure with the ANN model achieving a classification accuracy of 94% for both training and validation sets. E-nose application in food quality control was investigated by Sberveglieri et al. (2015) for the detection of microorganism in water and different food matrices by employing 6 MOX gas sensors and PCA. E. coli, Salmonella typhimurium and L. monocytogenes at a concentration of 9 × 108 bacteria/ml.
Future trends and perspectives
Optimization of pattern recognition algorithms using metaheuristic algorithms to improve E-nose detection
The development of intelligent algorithms for pattern recognition, feature extraction, and parameters optimization is crucial for the rapid application of E-nose for routine food analysis (Luo et al. 2018). Metaheuristic optimization algorithms have the ability to resolve complex large-scale nonlinear optimization problems, and cannot be handled other analytic approaches are beginning to gain recognition in improving E-nose pattern recognition algorithms to enhance the performance of E-noses by sensor selection (Guan et al. 2014; Jiang et al. 2017; Luo et al. 2018). Heuristic methods are applied to further enhance the performance of E-noses by sensor selection and to optimize the gas sensor array as well as dimensionality reduction of the feature matrix.
Sensor development approaches
The development of reliable drift free sensors as well as investigating new material for attaining improved selectivity is crucial in achieving commercial use of E-nose in the food industry. Performance degradation of E-nose data as a result of sensor drift (variation in the sensor response in identical measurement conditions) and noise have been widely reported (Tian et al. 2018; Wijaya et al. 2017).
A Noise filtering framework based discrete wavelet transform (DWT) for handling noisy signals generated by an E-nose sensor array was developed by Wijaya et al. (2019) with significant to existing methods. The development of low-cost disposable sensors would mitigate against the decrease in the sensitivity and specificity of sensors over time. Electronic nose as a non-invasive technique provides a better alternative for the detection of complex gas mixtures, issues such as odour identification at concentrations levels higher than those of the biological counterpart as well as providing answers with regards to the concentration of a particular compound in mixtures abound. This setback in the use of E-nose is associated with lack of adequate biomolecules to allow the system to fully mimic the biological sense of smell.
The development in the fields of genetic engineering, biotechnology and nanotechnology has led to the improved development of biomimetic electronic nose, bio-enose, b-enose, bioelectronic noses in recent years (Wasilewski et al. 2017). This new development of bioelectronics noses provides for a more precise mimicking of human smell imprints by applying highly selective and sensitive sensors.
Low-, mid-, and high-level data fusion
Applications such as multi-sensor data fusion are said to increase the probability of classification. Several researchers have applied E-nose data fusion with other non-invasive methods such as hyperspectral imaging (Liu et al. 2019), computer vision (Xu et al. 2019), electronic tongue (Banerjee et al. 2019). A fusion of electronic nose, electronic tongue, hyperspectral imaging and computer vision data at the low, intermediate, and high-level fusion models have shown to be effective and in some instances better results than single sensor models.
Potentialities of E-nose for bacterial pathogen detection
Although the application of the above mentioned future trends and perspectives for E-nose application in the food industry have been predominantly applied for food quality analysis especially for classification purposes, future prospects for applying this technology for bacterial pathogen detection remain feasible. Microorganism detection by E-nose has some drawbacks which include low sensitivity and specificity in comparison with some microbiological and molecular methods. The detection of volatile compounds is usually interfered by a complex mixture of water vapour and carbon dioxide in the background mixture (Sanaeifar et al. 2017).
Other setbacks include a high limit of detection (LOD) as Siripatrawan (2008b) reported detection limit of E. coli above 105 cfu/g with Gobbi et al. (2015) reported sensitivity as low as 3 cfu per 100 ml for E. coli. This setback, however, could be solved by the application enzyme substrates to liberate exogenous VOCs of foodborne bacterial pathogens to increase the diagnostic specificity of VOCs. This is achieved by modifying bacteria growth media with substrates that liberate unique VOCs through enzymatic metabolism in response to the presence of enzyme activity exhibited by a target pathogen. This methodology has been successfully studied using conventional detection methods such as gas chromatography-ion mobility spectrometry (GC–IMS) and gas chromatography-mass spectrometry (GC–MS) and can be improved with electronic nose application which is noninvasivee. Example substrates include 2-nitrophenyl-β-d-glucuronide (E. coli), 2-nitrophenyl-β-d-glucopyranoside (Listeria spp.) and 2-nitrophenyl-β-d-galactoside-6-phosphate (Staphylococcus aureus).
Conclusion
E-nose provides an ideal methodology for in-line process control with straight and rapid discrimination of numerous compounds requiring less or no sample preparation as well as reagent consumption. In as much as laboratory-based assessments have shown good classification rates a number of challenges with regards to humidity influence, selectivity, sensor drift, signal to noise ratio and redundancy of sensors must be resolved before the technology is moved into real-time industry application.
The influence of environmental conditions such as temperature and humidity is another drawback. Metal-oxide sensors exhibit swift response and recovery times but require high power levels to the sensors at elevated temperatures. Polymer sensors are cheap to use and operate at room temperature, however, they are sensitive to temperature and humidity.
Improved modelling and correlation between the existence of chemical markers and sensor responses together with a carefully selected sampling system and sensor arrays would greatly enhance the accuracy of results obtained from E-nose data analysis.
Acknowledgements
This work was sponsored by the National Natural Science Foundation of China (No. 31671932).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abdallah SA, Al-Shatti LA, Alhajraf AF, Al-Hammad N, Al-Awadi B. The detection of foodborne bacteria on beef: the application of the electronic nose. SpringerPlus. 2013;2:687. doi: 10.1186/2193-1801-2-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo FJ, Maldonado S, Domínguez E, Narváez A, López F. Probabilistic support vector machines for multi-class alcohol identification. Sens Actuators B Chem. 2007;122:227–235. doi: 10.1016/j.snb.2006.05.033. [DOI] [Google Scholar]
- Ampuero S, Zesiger T, Gustafsson V, Lundén A, Bosset J. Determination of trimethylamine in milk using an MS based electronic nose. Eur Food Res Technol. 2002;214:163–167. doi: 10.1007/s00217-001-0463-0. [DOI] [Google Scholar]
- Avalos M, van Wezel GP, Raaijmakers JM, Garbeva P. Healthy scents: microbial volatiles as new frontier in antibiotic research? Curr Opin Microbiol. 2018;45:84–91. doi: 10.1016/j.mib.2018.02.011. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Panigrahi S, Logue CM, Doetkott C, Marchello M, Sherwood JS. Independent component analysis-processed electronic nose data for predicting Salmonella typhimurium populations in contaminated beef. Food Control. 2008;19:236–246. doi: 10.1016/j.foodcont.2007.03.007. [DOI] [Google Scholar]
- Balasubramanian S, Amamcharla J, Panigrahi S, Logue CM, Marchello M, Sherwood JS. Investigation of different gas sensor-based artificial olfactory systems for screening Salmonella typhimurium contamination in beef. Food Bioprocess Technol. 2012;5:1206–1219. doi: 10.1007/s11947-010-0444-z. [DOI] [Google Scholar]
- Balasubramanian S, Amamcharla J, Shin J-E. Chapter 7-Possible application of electronic nose systems for meat safety: an overview. In: Rodríguez Méndez ML, editor. Electronic noses and tongues in food science. San Diego: Academic Press; 2016. pp. 59–71. [Google Scholar]
- Balbin JR, Sese JT, Babaan CVR, Poblete DMM, Panganiban RP, Poblete JG (2017) Detection and classification of bacteria in common street foods using electronic nose and support vector machine. In: 2017 7th IEEE international conference on control system, computing and engineering (ICCSCE), 24–26 Nov 2017, pp 247–252. 10.1109/iccsce.2017.8284413
- Banerjee MB, Roy RB, Tudu B, Bandyopadhyay R, Bhattacharyya N. Black tea classification employing feature fusion of E-nose and E-tongue responses. J Food Eng. 2019;244:55–63. doi: 10.1016/j.jfoodeng.2018.09.022. [DOI] [Google Scholar]
- Berna Z, Webb CC, Erickson MC (2013) Electronic nose and fast GC for detection of volatiles from Escherichia coli O157:H7 Escherichia coli and Salmonella in lettuce. In: International Society for Horticultural Science (ISHS), Leuven, Belgium, pp 1255–1261. 10.17660/ActaHortic.2013.1012.169
- Cao LJ, Chua KS, Chong WK, Lee HP, Gu QM. A comparison of PCA, KPCA and ICA for dimensionality reduction in support vector machine. Neurocomputing. 2003;55:321–336. doi: 10.1016/S0925-2312(03)00433-8. [DOI] [Google Scholar]
- Carmel L, Levy S, Lancet D, Harel D. A feature extraction method for chemical sensors in electronic noses. Sens Actuators B Chem. 2003;93:67–76. doi: 10.1016/S0925-4005(03)00247-8. [DOI] [Google Scholar]
- Chen S, Wang Y, Choi S. Applications and technology of electronic nose for clinical diagnosis. Open J Appl Biosensor. 2013;02(02):12. doi: 10.4236/ojab.2013.22005. [DOI] [Google Scholar]
- Concina I, Falasconi M, Gobbi E, Bianchi F, Musci M, Mattarozzi M, Pardo M, Mangia A, Careri M, Sberveglieri G. Early detection of microbial contamination in processed tomatoes by electronic nose. Food Control. 2009;20:873–880. doi: 10.1016/j.foodcont.2008.11.006. [DOI] [Google Scholar]
- Dębska B, Guzowska-Świder B. Application of artificial neural network in food classification. Anal Chim Acta. 2011;705:283–291. doi: 10.1016/j.aca.2011.06.033. [DOI] [PubMed] [Google Scholar]
- Ding N-y, Lan Y-b, Zheng X-z. Rapid detection of E. coli on goat meat by electronic nose. Adv Nat Sci. 2010 doi: 10.3968/931. [DOI] [Google Scholar]
- Distante C, Ancona N, Siciliano P. Support vector machines for olfactory signals recognition. Sens Actuators B Chem. 2003;88:30–39. doi: 10.1016/S0925-4005(02)00306-4. [DOI] [Google Scholar]
- El Barbri N, Llobet E, El Bari N, Correig X, Bouchikhi B. Application of a portable electronic nose system to assess the freshness of Moroccan sardines. Mater Sci Eng C. 2008;28:666–670. doi: 10.1016/j.msec.2007.10.056. [DOI] [Google Scholar]
- Elgaali H, Hamilton-Kemp TR, Newman MC, Collins RW, Yu K, Archbold DD. Comparison of long-chain alcohols and other volatile compounds emitted from food-borne and related Gram positive and Gram negative bacteria. J Basic Microbiol. 2002;42:373–380. doi: 10.1002/1521-4028(200212)42:6<373::AID-JOBM373>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Ezhilan M, Nesakumar N, Jayanth Babu K, Srinandan CS, Rayappan JBB. An electronic nose for royal delicious apple quality assessment—a tri-layer approach. Food Res Int. 2018;109:44–51. doi: 10.1016/j.foodres.2018.04.009. [DOI] [PubMed] [Google Scholar]
- Franz CMAP, den Besten HMW, Böhnlein C, Gareis M, Zwietering MH, Fusco V. Microbial food safety in the 21st century: emerging challenges and foodborne pathogenic bacteria. Trends Food Sci Technol. 2018;81:155–158. doi: 10.1016/j.tifs.2018.09.019. [DOI] [Google Scholar]
- Gardner JW, Bartlett PN. A brief history of electronic noses. Sens Actuators B Chem. 1994;18:210–211. doi: 10.1016/0925-4005(94)87085-3. [DOI] [Google Scholar]
- Gardner JW, Craven M, Dow C, Hines EL. The prediction of bacteria type and culture growth phase by an electronic nose with a multi-layer perceptron network. Meas Sci Technol. 1998;9:120–127. doi: 10.1088/0957-0233/9/1/016. [DOI] [Google Scholar]
- Ghasemi-Varnamkhasti M, Apetrei C, Lozano J, Anyogu A. Potential use of electronic noses, electronic tongues and biosensors as multisensor systems for spoilage examination in foods. Trends Food Sci Technol. 2018;80:71–92. doi: 10.1016/j.tifs.2018.07.018. [DOI] [Google Scholar]
- Giungato P, Di Gilio A, Palmisani J, Marzocca A, Mazzone A, Brattoli M, Giua R, de Gennaro G. Synergistic approaches for odor active compounds monitoring and identification: state of the art, integration, limits and potentialities of analytical and sensorial techniques. TrAC Trends Anal Chem. 2018;107:116–129. doi: 10.1016/j.trac.2018.07.019. [DOI] [Google Scholar]
- Gobbi E, Falasconi M, Zambotti G, Sberveglieri V, Pulvirenti A, Sberveglieri G. Rapid diagnosis of Enterobacteriaceae in vegetable soups by a metal oxide sensor based electronic nose. Sens Actuators B Chem. 2015;207:1104–1113. doi: 10.1016/j.snb.2014.10.051. [DOI] [Google Scholar]
- Green GC, Chan ADC, Dan H, Lin M. Using a metal oxide sensor (MOS)-based electronic nose for discrimination of bacteria based on individual colonies in suspension. Sens Actuators B Chem. 2011;152:21–28. doi: 10.1016/j.snb.2010.09.062. [DOI] [Google Scholar]
- Green GC, Chan ADC, Lin M. Robust identification of bacteria based on repeated odor measurements from individual bacteria colonies. Sens Actuators B Chem. 2014;190:16–24. doi: 10.1016/j.snb.2013.08.001. [DOI] [Google Scholar]
- Guan B, Zhao J, Lin H, Zou X. Characterization of volatile organic compounds of vinegars with novel electronic nose system combined with multivariate analysis. Food Anal Methods. 2014;7:1073–1082. doi: 10.1007/s12161-013-9715-4. [DOI] [Google Scholar]
- Gutierrez-Osuna R, Nagle HT. A method for evaluating data-preprocessing techniques for odour classification with an array of gas sensors. IEEE Trans Syst Man Cybern B (Cybern) 1999;29:626–632. doi: 10.1109/3477.790446. [DOI] [PubMed] [Google Scholar]
- Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, de Silva NR, Gargouri N, Speybroeck N, Cawthorne A, Mathers C, Stein C, Angulo FJ, Devleesschauwer B, on behalf of World Health Organization Foodborne Disease Burden Epidemiology Reference G World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLOS Med. 2015;12:e1001923. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierlemann A, Gutierrez-Osuna R. Higher-order chemical sensing. Chem Rev. 2008;108:563–613. doi: 10.1021/cr068116m. [DOI] [PubMed] [Google Scholar]
- Huang X-J, Choi Y-K, Yun K-S, Yoon E. Oscillating behaviour of hazardous gas on tin oxide gas sensor: Fourier and wavelet transform analysis. Sens Actuators B Chem. 2006;115:357–364. doi: 10.1016/j.snb.2005.09.022. [DOI] [Google Scholar]
- Hyvärinen A, Karhunen J, Oja E. Noisy ICA. In: Haykin S, editor. Independent component analysis. Hoboken: Wiley; 2001. [Google Scholar]
- Jha Sunil Kumar, Yadava RDS. Power scaling of chemiresistive sensor array data for odor classification. J Pattern Recogn Res JPRR. 2011 doi: 10.13176/11.247. [DOI] [Google Scholar]
- Jha SK, Yadava RDS, Hayashi K, Patel N. Recognition and sensing of organic compounds using analytical methods, chemical sensors, and pattern recognition approaches. Chemometr Intell Lab Syst. 2019;185:18–31. doi: 10.1016/j.chemolab.2018.12.008. [DOI] [Google Scholar]
- Jia W, Liang G, Wang Y, Wang J. Electronic noses as a powerful tool for assessing meat quality: a mini review. Food Anal Methods. 2018;11:2916–2924. doi: 10.1007/s12161-018-1283-1. [DOI] [Google Scholar]
- Jiang H, Chen Q. Development of electronic nose and near infrared spectroscopy analysis techniques to monitor the critical time in SSF process of feed protein. Sensors. 2014 doi: 10.3390/s141019441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Jia P, Luo R, Deng B, Duan S, Yan J. A novel electronic nose learning technique based on active learning: EQBC-RBFNN. Sens Actuators B Chem. 2017;249:533–541. doi: 10.1016/j.snb.2017.04.072. [DOI] [Google Scholar]
- Jolliffe I. Principal component analysis, Statistics reference online, Wiley StatsRef. Hoboken: Wiley; 2014. [Google Scholar]
- Kai M, Haustein M, Molina F, Petri A, Scholz B, Piechulla B. Bacterial volatiles and their action potential. Appl Microbiol Biotechnol. 2009;81:1001–1012. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- Kaur R, Kumar R, Gulati A, Ghanshyam C, Kapur P, Bhondekar AP. Enhancing electronic nose performance: a novel feature selection approach using dynamic social impact theory and moving window time slicing for classification of Kangra orthodox black tea (Camellia sinensis (L.) O. Kuntze) Sens Actuators B Chem. 2012;166–167:309–319. doi: 10.1016/j.snb.2012.02.067. [DOI] [Google Scholar]
- Kizil Ü, Genç L, Genç TT, Rahman S, Khaitsa ML. E-nose identification of Salmonella enterica in poultry manure. Br Poult Sci. 2015;56:149–156. doi: 10.1080/00071668.2015.1014467. [DOI] [PubMed] [Google Scholar]
- Laref R, Losson E, Sava A, Siadat M. Support vector machine regression for calibration transfer between electronic noses dedicated to air pollution monitoring. Sensors. 2018;18:3716. doi: 10.3390/s18113716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang H, Sun L, Zhao G, Huang X. Application of electronic nose for measuring total volatile basic nitrogen and total viable counts in packaged pork during refrigerated storage. J Food Sci. 2016;81:M906–M912. doi: 10.1111/1750-3841.13238. [DOI] [PubMed] [Google Scholar]
- Lippolis V, Cervellieri S, Damascelli A, Pascale M, Di Gioia A, Longobardi F, De Girolamo A. Rapid prediction of deoxynivalenol contamination in wheat bran by MOS-based electronic nose and characterization of the relevant pattern of volatile compounds. J Sci Food Agric. 2018;98:4955–4962. doi: 10.1002/jsfa.9028. [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhao N, Zhou D, Sun Y, Sun K, Pan L, Tu K. Discrimination and growth tracking of fungi contamination in peaches using electronic nose. Food Chem. 2018;262:226–234. doi: 10.1016/j.foodchem.2018.04.100. [DOI] [PubMed] [Google Scholar]
- Liu Q, Sun K, Zhao N, Yang J, Zhang Y, Ma C, Pan L, Tu K. Information fusion of hyperspectral imaging and electronic nose for evaluation of fungal contamination in strawberries during decay. Postharvest Biol Technol. 2019;153:152–160. doi: 10.1016/j.postharvbio.2019.03.017. [DOI] [Google Scholar]
- Luo D, Hosseini HG, Stewart JR. Application of ANN with extracted parameters from an electronic nose in cigarette brand identification. Sens Actuators B Chem. 2004;99:253–257. doi: 10.1016/j.snb.2003.11.022. [DOI] [Google Scholar]
- Luo H, Jia P, Qiao S, Duan S. Enhancing electronic nose performance based on a novel QPSO-RBM technique. Sens Actuators B Chem. 2018;259:241–249. doi: 10.1016/j.snb.2017.12.026. [DOI] [Google Scholar]
- Majchrzak T, Wojnowski W, Dymerski T, Gębicki J, Namieśnik J. Electronic noses in classification and quality control of edible oils: a review. Food Chem. 2018;246:192–201. doi: 10.1016/j.foodchem.2017.11.013. [DOI] [PubMed] [Google Scholar]
- Martin AL, Satjaritanun P, Shimpalee S, Devivo BA, Weidner J, Greenway S, Henson JM, Turick CE. In-situ electrochemical analysis of microbial activity. AMB Express. 2018;8:162. doi: 10.1186/s13568-018-0692-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Sun F, Xu Y, Cong Z, Wang E. Applications of electrochemical techniques in mineral analysis. Talanta. 2014;127:211–218. doi: 10.1016/j.talanta.2014.03.072. [DOI] [PubMed] [Google Scholar]
- Nygren BL, Schilling KA, Blanton EM, Silk BJ, Cole DJ, Mintz ED. Foodborne outbreaks of shigellosis in the USA, 1998–2008. Epidemiol Infect. 2013;141:233–241. doi: 10.1017/S0950268812000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallottino F, Costa C, Antonucci F, Strano MC, Calandra M, Solaini S, Menesatti P. Electronic nose application for determination of Penicillium digitatum in Valencia oranges. J Sci Food Agric. 2012;92:2008–2012. doi: 10.1002/jsfa.5586. [DOI] [PubMed] [Google Scholar]
- Pardo M, Sberveglieri G. Classification of electronic nose data with support vector machines. Sens Actuators B Chem. 2005;107:730–737. doi: 10.1016/j.snb.2004.12.005. [DOI] [Google Scholar]
- Pattarapon P, Zhang M, Bhandari B, Gao Z. Effect of vacuum storage on the freshness of grass carp (Ctenopharyngodon idella) fillet based on normal and electronic sensory measurement. J Food Process Preserv. 2018;42:e13418. doi: 10.1111/jfpp.13418. [DOI] [Google Scholar]
- Peris M, Escuder-Gilabert L. A 21st century technique for food control: electronic noses. Anal Chim Acta. 2009;638:1–15. doi: 10.1016/j.aca.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Piechulla B, Degenhardt J. The emerging importance of microbial volatile organic compounds. Plant Cell Environ. 2014;37:811–812. doi: 10.1111/pce.12254. [DOI] [PubMed] [Google Scholar]
- Preti G, Thaler E, Hanson CW, Troy M, Eades J, Gelperin A. Volatile compounds characteristic of sinus-related bacteria and infected sinus mucus: analysis by solid-phase microextraction and gas chromatography–mass spectrometry. J Chromatogr B. 2009;877:2011–2018. doi: 10.1016/j.jchromb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- Rayappan JBB, Kulandaisamy AJ, Ezhilan M, Srinivasan P, Mani GK. Developments in electronic noses for quality and safety control. Adv Food Diagn. 2017 doi: 10.1002/9781119105916.ch3. [DOI] [Google Scholar]
- Robin Michael Statham T, John G. Microbial volatile compounds in health and disease conditions. J Breath Res. 2012;6:024001. doi: 10.1088/1752-7155/6/2/024001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romoli R, Papaleo MC, De Pascale D, Tutino ML, Michaud L, LoGiudice A, Fani R, Bartolucci G. GC–MS volatolomic approach to study the antimicrobial activity of the antarctic bacterium Pseudoalteromonas sp. TB41. Metabolomics. 2014;10:42–51. doi: 10.1007/s11306-013-0549-2. [DOI] [Google Scholar]
- Sanaeifar A, ZakiDizaji H, Jafari A, Mdl Guardia. Early detection of contamination and defect in foodstuffs by electronic nose: a review. TrAC Trends Anal Chem. 2017;97:257–271. doi: 10.1016/j.trac.2017.09.014. [DOI] [Google Scholar]
- Sberveglieri V, Núñez Carmona E, Pulvirenti A. Detection of microorganism in water and different food matrix by electronic nose. In: Mason A, Mukhopadhyay SC, Jayasundera KP, editors. Sensing technology: current status and future trends III. Cham: Springer; 2015. pp. 243–258. [Google Scholar]
- Scott SM, James D, Ali Z. Data analysis for electronic nose systems. Microchim Acta. 2006;156:183–207. doi: 10.1007/s00604-006-0623-9. [DOI] [Google Scholar]
- Selim KA, El Ghwas DE, Selim RM, Abdelwahab Hassan MI. Microbial volatile in defense. In: Choudhary DK, Sharma AK, Agarwal P, Varma A, Tuteja N, editors. Volatiles and food security: role of volatiles in agro-ecosystems. Singapore: Springer; 2017. pp. 135–170. [Google Scholar]
- Senecal AG, Magnone J, Yeomans W, Powers EM (2002) Rapid detection of pathogenic bacteria by volatile organic compound (VOC) analysis. In: Environmental and industrial sensing, 2002. SPIE, p 11
- Siripatrawan U. Rapid differentiation between E. coli and Salmonella typhimurium using metal oxide sensors integrated with pattern recognition. Sens Actuators B Chem. 2008;133:414–419. doi: 10.1016/j.snb.2008.02.046. [DOI] [Google Scholar]
- Siripatrawan U. Self-organizing algorithm for classification of packaged fresh vegetable potentially contaminated with foodborne pathogens. Sens Actuators B Chem. 2008;128:435–441. doi: 10.1016/j.snb.2007.06.030. [DOI] [Google Scholar]
- Siripatrawan U, Harte BR. Solid phase microextraction/gas chromatography/mass spectrometry integrated with chemometrics for detection of Salmonella typhimurium contamination in a packaged fresh vegetable. Anal Chim Acta. 2007;581:63–70. doi: 10.1016/j.aca.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Siripatrawan U, Harte BR. Data visualization of Salmonella typhimurium contamination in packaged fresh alfalfa sprouts using a Kohonen network. Talanta. 2015;136:128–135. doi: 10.1016/j.talanta.2014.11.070. [DOI] [PubMed] [Google Scholar]
- Siripatrawan U, Linz JE, Harte BR. Detection of Escherichia coli in packaged alfalfa sprouts with an electronic nose and an artificial neural network. J Food Prot. 2006;69:1844–1850. doi: 10.4315/0362-028X-69.8.1844. [DOI] [PubMed] [Google Scholar]
- Siswantoro J, Hilman MY, Widiasri M. Computer vision system for egg volume prediction using backpropagation neural network. IOP Conf Ser Mater Sci Eng. 2017;273:012002. doi: 10.1088/1757-899X/245/1/012002. [DOI] [Google Scholar]
- Sizochenko N, Syzochenko M, Fjodorova N, Rasulev B, Leszczynski J. Evaluating genotoxicity of metal oxide nanoparticles: application of advanced supervised and unsupervised machine learning techniques. Ecotoxicol Environ Saf. 2019;185:109733. doi: 10.1016/j.ecoenv.2019.109733. [DOI] [PubMed] [Google Scholar]
- Tait E, Perry JD, Stanforth SP, Dean JR. Use of volatile compounds as a diagnostic tool for the detection of pathogenic bacteria. TrAC Trends Anal Chem. 2014;53:117–125. doi: 10.1016/j.trac.2013.08.011. [DOI] [Google Scholar]
- Tian X, Wang J, Shen R, Ma Z, Li M. Discrimination of pork/chicken adulteration in minced mutton by electronic taste system. Int J Food Sci Technol. 2018 doi: 10.1111/ijfs.13977. [DOI] [Google Scholar]
- van den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genom. 2006;7:142–142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik VN. Methods of pattern recognition. In: Vapnik VN, editor. The nature of statistical learning theory. New York: Springer; 2000. pp. 123–180. [Google Scholar]
- Wang D, Wang X, Liu T, Liu Y. Prediction of total viable counts on chilled pork using an electronic nose combined with support vector machine. Meat Sci. 2012;90:373–377. doi: 10.1016/j.meatsci.2011.07.025. [DOI] [PubMed] [Google Scholar]
- Warren BR, Rouseff RL, Schneider KR, Parish ME. Identification of volatile sulfur compounds produced by Shigella sonnei using gas chromatography–olfactometry. Food Control. 2007;18:179–182. doi: 10.1016/j.foodcont.2005.09.017. [DOI] [Google Scholar]
- Wasilewski T, Gębicki J, Kamysz W. Bioelectronic nose: current status and perspectives. Biosens Bioelectron. 2017;87:480–494. doi: 10.1016/j.bios.2016.08.080. [DOI] [PubMed] [Google Scholar]
- Watson J, Ihokura K, Coles GSV. The tin dioxide gas sensor. Meas Sci Technol. 1993;4:711–719. doi: 10.1088/0957-0233/4/7/001. [DOI] [Google Scholar]
- Wijaya D, Sarno R, Fathra Daiva A (2017) Electronic Nose for Classifying Beef and Pork using Naïve Bayes. 10.1109/ISSIMM.2017.8124272
- Wijaya DR, Sarno R, Zulaika E. Noise filtering framework for electronic nose signals: an application for beef quality monitoring. Comput Electron Agric. 2019;157:305–321. doi: 10.1016/j.compag.2019.01.001. [DOI] [Google Scholar]
- Wilson AD, Baietto M. Applications and advances in electronic-nose technologies. Sensors (Basel, Switzerland) 2009;9:5099–5148. doi: 10.3390/s90705099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Wang J, Gu S. Rapid identification of tea quality by E-nose and computer vision combining with a synergetic data fusion strategy. J Food Eng. 2019;241:10–17. doi: 10.1016/j.jfoodeng.2018.07.020. [DOI] [Google Scholar]
- Xue C, Lin Y, Zhao Y, Xitao Z (2012) The identification of Listeria monocytogenes based on the electronic nose. In: 2012 International conference on computer science and information processing (CSIP), 24–26 Aug 2012. pp 467–472. 10.1109/csip.2012.6308893
- Yan J, Guo X, Duan S, Jia P, Wang L, Peng C, Zhang S. Electronic nose feature extraction methods: a review. Sensors (Basel, Switzerland) 2015;15:27804–27831. doi: 10.3390/s151127804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongxin Y, Zhao Y. Electronic nose integrated with chemometrics for rapid identification of foodborne pathogen. IntechOpen. 2012 doi: 10.5772/32099. [DOI] [Google Scholar]
- Yu Y-x, Sun X-h, Liu Y, Pan Y-j, Zhao Y. Odor fingerprinting of Listeria monocytogenes recognized by SPME–GC–MS and E-nose. Can J Microbiol. 2014;61:367–372. doi: 10.1139/cjm-2014-0652. [DOI] [PubMed] [Google Scholar]
- Zhang WN, Qin GJ, Hu NQ. Parallel factor analysis for gas sensor array signals. Appl Mech Mater. 2014;494–495:955–959. doi: 10.4028/www.scientific.net/AMM.494-495.955. [DOI] [Google Scholar]
- Zohora SE, Khan AM, Hundewale N. Chemical sensors employed in electronic noses: a review. In: Meghanathan N, Nagamalai D, Chaki N, editors. Advances in computing and information technology. Berlin: Springer; 2013. pp. 177–184. [Google Scholar]