Abstract
The study determined incidence, enterotoxigenecity and antimicrobial susceptibility profiles of Bacillus cereus isolated from ready-to-eat (RTE) milk products (n = 80), RTE meat products (n = 40), beverages (n = 40) and water samples (n = 60, from food preparing and serving outlets/restaurants) collected from eight different tourist places of Himachal Pradesh. 11.4% (25/220) samples were contaminated with Bacillus and isolates were identified as B. cereus (76.0%, n = 19), B. alvei (12.0%, n = 3), B. polymyxa (8.0%, n = 2) and B. firmus (4.0%, n = 1) by conventional and molecular methods. B. cereus incidence was highest in cheese based foods (25.0%) followed by vegetable soups (16.7%), khoa based foods (14.0%), milk based beverages (10.5%), paneer based foods (8.6%), cream based foods (8.3%) and water (8.3%) samples. Multiplex polymerase chain reaction detected enterotoxigenic genes only in B. cereus isolates. nhe complex (encoding non-haemolytic enterotoxins, ABC) genes were detected only in B. cereus isolates. 57.6% (11/19), 36.8% (7/19) and 5.3% (1/19) harboured all three (nheA, nheB, nheC), two (nheB, nheC) and one (nheC) nhe gene, respectively. Among hbl complex genes (encoding haemolytic enterotoxins CAD), only hblC (36.8%, 7/19) was detected. Incidence B. cereus cytK (encoding cytotoxin enterotoxin) was 52.6% (10/19). Each B. cereus isolate harboured two or more enterotoxigenic genes. Seven isolates had at least one gene from haemolytic and non-haemolytic complexes along with cytK. High levels (> 50%) of antimicrobial resistance were recorded for penicillin, amoxicillin, ampicillin cefixime and ceftazidine in tested B. cereus isolates. Two isolates were identified as multidrug resistant isolates with resistance to ≥ 3 antibiotic classes.
Electronic supplementary material
The online version of this article (10.1007/s13197-020-04267-y) contains supplementary material, which is available to authorized users.
Keywords: Enterotoxins, Antimicrobial susceptibility, nhe complex, hlb complex, cytK
Introduction
Bacillus are Gram-positive, facultative anaerobic and spore-forming rods. Bacillus genus includes both food borne pathogens and food spoilage-associated bacteria, such as B. cereus, B. subtilis, B. licheniformis,B. pumilus, B. weihenstephanensis and B. sporothermodurans (Kotiranta et al. 2000; Gopal et al. 2015). B. cereus is most commonly detected food pathogen from this genus (Logan 2011; Tewari and Abdullah 2015; Gopal et al. 2015). Among other members, B. licheniformis can cause an enteric disease and food poisoning in humans. Similarly, strains of B. subtilis may occasionally cause food poisoning outbreaks involving foods such as milk powder (Fernández-No et al. 2011; Gopal et al. 2015). Bacillus spp. are widely distributed in the environment with soil as the natural habitat (Kramer and Gilbert 1989; Tewari and Abdullah 2015).
Bacillus spores are devoid of metabolic activity and are refractory to extreme environmental conditions such as heat, freezing, drying and radiation (Bottone 2010). These spores can be transmitted through processed, pasteurized and heat-treated food products (Kotiranta et al. 2000). Bacillus are frequently isolated from ready-to-eat (RTE) foods (Aruwa and Akinyosoye 2015), packaged spices and herbs (Aksu et al. 2000), meat and meat products (Bashir et al. 2017), milk and milk products (Yusuf et al. 2018) and fried rice (Perera and Ranasinghe 2012). In India, Bacillus species had been isolated from foods of plant and animal origin including fermented and RTE foods (Roy et al. 2007; Tewari and Abdullah 2015; Chettri and Tamang 2015; Yusuf et al. 2018). In a recent study, all the analyzed fermented soybean food products marketed in Northeast India were found to be contaminated with B. cereus (Keisam et al. 2019).
B. cereus is ubiquitous in nature and spores and vegetative forms of the bacterium can contaminate foods of plant and animal origin (Tewari and Abdullah 2015; Kramer and Gilbert 1989). B. cereus causes two types of food poisoning syndromes, diarrheal and emetic. Bacteria/spores ingested with contaminated food, release diarrheal enterotoxin in the intestinal tract of the host (Berthold-Pluta et al. 2015; Rajkovic 2014). The diarrheal syndrome has been associated with a wide variety of foods including meats, milk, vegetables and fish (Aruwa and Akinyosoye 2015). Emetic syndrome is characterized by vomiting which occurs due to ingestion of food containing pre-formed emetic toxin (Rajkovic 2014). The emetic syndrome has been generally associated with rice products, starchy foods such as potato, pasta, noodles, spaghetti, pastry and cheese products (Aruwa and Akinyosoye 2015). Rarely, emetic and diarrheal syndromes can occur simultaneously (Kramer and Gilbert 1989; Tewari and Abdullah 2015). B. cereus is an established food borne pathogen but the illness caused is under-diagnosed and is generally not reported (Tewari et al. 2015; Kotiranta et al. 2000). This under-reporting can be attributed to the lack of proper diagnostic tools, similarity of symptoms to other common food borne infections such as stapphyloccocal gastroenteritis and Clostridium perfringens food poisoning and self limiting (24–48 h) nature of the disease (Tewari and Abdullah 2015).
Himachal Pradesh is a hilly state and thousands of people every year visit different tourist destinations of the region such as Shimla, Kullu–Manali, Dharamsala Mcleodganj, Chamba–Dalhousie etc. This all around the year tourist activity results in relatively higher consumption of RTE or street foods at these popular destinations compared to other places. Systematic studies are required to determine incidence of various food borne pathogens in Himachal Pradesh as not much data is available on their occurrence (Lakhanpal et al. 2019; Ahmadi and Panda 2015). The present study was designed to determine incidence of B. cereus in RTE food products, beverages and water samples collected from different tourist places of Himachal Pradesh. We also ascertained the gene profiles of enterotoxins (haemolytic BL, non-haemolytic NHE and cytotoxin-K) and emetic toxin (cereulide) frequently associated with B. cereus food poisoning outbreaks (Yang et al. 2005; Owusu-Kwarteng et al. 2017). Antibiograms of B. cereus isolates recovered in this study were also determined.
Materials and methods
Sample collection
A total of 220 samples, comprising RTE milk products (n = 80), RTE meat products (n = 40), water (n = 60) and beverages (n = 40) were collected from eight different tourist places of Himachal Pradesh to determine the incidence of Bacillus spp. (Table 1). Water samples analysed in this study were from RTE food preparing and serving outlets/restaurants from where food samples were collected. RTE foods included, chicken momo (n = 25), paneer based foods (n = 23), khoa based foods (n = 21), cheese based foods (n = 16), mutton momos (n = 15), cream based foods (n = 12) and curd (n = 8). Beverages tested for Bacillus spp. included, chicken soups (n = 15), milk beverages [n = 19; lassi (n = 10) and milk shakes (n = 9)] and vegetable soups (n = 6). Samples were aseptically collected and transported to laboratory in ice box for immediate processing or kept at 4 °C till processing was done. All the samples tested in this study were collected between August 2017 and April 2018. Except for Shimla and Chamba/Dalhousie, samples were collected two or more times from different sampling locations with a maximum of four times from Dharamsala–Mcleodganj and Palampur.
Table 1.
Incidence of Bacillus spp. in ready to eat foods, beverages and water samples collected from different places of Himachal Pradesh
| Places | Type of samples (no. of samples collected) | Number of samples collected (%) | Samples contaminated with Bacillus spp. (%) |
|---|---|---|---|
|
Baijnath/Bir–Billing (24) |
Khoa based (5), paneer based (3), cheese based (3), curd (2), lassi (1), chicken soup (1), water (9) | 24 (10.9) | 2 (8.3) (p = 1.0) |
|
Chamba/Dalhousie (20) |
Chicken momo (4), mutton momo (4), cheese based (1), cream based (2), khoa based (2), lassi (1), milk shake (2), water (4) | 20 (9.1) | 1 (5.0) (p = 0.7) |
|
Dharamsala–Mcleodganj (53) |
Paneer based (5), cheese based (8), cream based (4), chicken momo (4), mutton momo (3), curd (2), lassi (5), chicken soup (3), vegetable paneer soup (3), milk shake (1), water (15) | 53 (24.1) | 5 (9.4) (p = 0.8) |
|
Kangra (24) |
Mutton momo (4), paneer based (2), cream based (2), khoa based (2), curd (2), chicken momo (1), vegetable soup (3), chicken soup (2), milk shake (1), water (5) | 24 (10.9) | 3 (12.5) (p = 0.7) |
|
Kullu–Manali (37) |
Paneer based (4), chicken momo (4), cheese based (2), khoa based (2), cream based (2), chicken soup (6), milk shake (5), lassi (1), water (11) | 37 (16.8) | 8 (21.6) (p= 0.04) |
|
Mandi (25) |
Paneer based (5), chicken momo (5), curd (2), khoa (2), cream based (1)) chicken soup (3), lassi (2), water (5) | 25 (11.4) | 1 (4.0) (p = 0.3) |
|
Palampur (23) |
Khoa based (5), paneer based (4), cream based (1), chicken momo (3), mutton momo (3), water (7) | 23 (10.5) | 5 (21.7) (p = 0.15) |
|
Shimla (14) |
Chicken momo (4), khoa based (3), cheese based (2), mutton momo (1), water (4) | 14 (6.7) | 0 (0.0) (p = 0.38) |
| Total | 220 | 25 (11.4) | |
Values in bold indicate significant associations between incidence of Bacillus spp. in ready to eat foods and place (p ≤ 0.05)
Isolation and characterization of Bacillus species
Isolation of Bacillus spp. was done as described previously (Public Health England 2018; Tewari et al. 2015). A portion (25 g or 25 ml) from the centre of each sample was collected aseptically and was homogenised with 225 ml of alkaline peptone water (CDH Lab., Delhi, India). 10 ml of this homogenate was enriched in 90 ml of sterile alkaline peptone water at 37 °C for 24 h. 100 µl of enriched samples were streaked on polymyxin pyruvate egg yolk mannitol bromothymol blue (PEMBA, HiMedia Lab, Mumbai, India) agar plates in duplicate and incubated at 37 °C for 24 h (Tewari et al. 2015). The fimbriate peacock blue-coloured colonies surrounded by a blue zone of egg yolk hydrolysis showing Bacillus like growth characteristics were purified for identification by biochemical tests and carbohydrate utilization reactions (Vos et al. 2009; Public Health England 2018). All the chemicals and reagents used in biochemical characterization and carbohydrate fermentation tests of Bacillus isolates were purchased from HiMedia laboratories, Mumbai, India. B. cereus strains, ATCC 13061 (HiMedia Lab, Mumbai, India) and VTCC BAA 737 (National Centre for Veterinary Type Cultures Collections Hisar, Haryana, India) were used as positive controls.
DNA extraction and amplification reactions
DNA was extracted from 1 ml of overnight (12–16 h at 37 °C) cultures of Bacillus isolates grown in brain heart infusion broth (HiMedia Lab, Mumbai, India). This overnight growth was centrifuged at 12,000 rpm for 5 min and supernatant was discarded. DNA was extracted from the pellet using HiPurA bacterial genomic DNA purification kit (HiMedia Lab, Mumbai, India) as per manufacturer’s instructions. The extracted DNA was stored at − 20 °C till further use.
Polymerase chain reaction (PCR) amplifications (GeneAmp PCR System 9700, LABINDIA, India) were done to reconfirm biochemically characterized Bacillus isolates and to ascertain toxigenic profiles. All primers used for DNA amplification reactions were purchased from Integrated DNA Technologies, Iowa, USA. Bacillus isolates characterized biochemically were reconfirmed by PCR targeting 16SrRNA (Gomaa and Momtaz 2006). PCR mixture (25 µl) contained, template DNA (5 µl), GoTaq Green Master Mix (12.5 µl, Thermo Fisher Scientific, Mumbai, India), nuclease free water (6.5 µl, Thermo Fisher Scientific, Mumbai, India), 0.5 µl of forward primer (10 pmol/µl), 0.5 µl of reverse primer (10 pmol/µl). Amplifications were done by using primers (PA-F: 5′-AGAGTTTGATCCTGGCTCAG-3′ and PH-R: 3′-AAGGAGGTGATCCAGCCGCA-5′) and amplification parameters as described by Gomaa and Momtaz (2006). DNA amplifications were carried out using PCR conditions; initial denaturation at 95 °C for 5 min, followed by 30 cycles of amplification at 95 °C for 1 min (denaturation), 55 °C for 1 min (annealing), 72 °C for 2 min (extension) and final extension at 72 °C for 7 min (Gomaa and Momtaz 2006).
A multiplex PCR was performed to detect the genes encoding haemolytic BL (hblA, hblC and hblD), non-haemolytic NHE (nheA, nheB and nheC) and cytotoxic-K (cytK) enterotoxins in Bacillus spp. isolates recovered in this study (Ngamwongsatit et al. 2008). Primer pairs used for the detection of enterotoxigenic and emetic toxin genes are given in Supplementary Table 1. Amplification conditions for enterotoxin genes were; initial denaturation at 95 °C for 5 min, followed by 30 cycles of 94 °C for 45 s, 54 °C for 1 min, 72 °C for 2 min and a final extension at 72 °C for 5 min (Ngamwongsatit et al. 2008). Detection of Ces, encoding emetic toxin cereulide was done using amplification parameters; initial denaturation at 95 °C for 10 min followed by 35 cycles of 94 °C for 1 min (denaturation), 54 °C for 1 min (annealing), 72 °C for 1 min (extension), and final extension at 72 °C for 5 min (Kim et al. 2011). For all toxin genes, PCR reaction mixture contained (25 µl) DNA template (2 μl), GoTaq Green Master Mix (12.5 µl), nuclease free water (9.5 µl) and 0.5 µl (primer (10 pmol/µl) each of forward and reverse primers for hblA, hblC, hblD, nheA, nheB, nheC, cytK, and ces amplification (Supplementary Table 1).
PCR amplicons for 16S rRNA and toxin genes were analyzed by electrophoresis on 1% and 2% agarose gels using 1 kb and 100 bp DNA ladders (Thermo Fisher Scientific, Mumbai, India), respectively. Agrose gels were stained with ethidium bromide (10 µg/100 ml) and were visualized under UV trans-illuminator (Alphalmager, Alpha Innotech Co., SanLeandro, USA).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using standard disc diffusion method on Mueller–Hinton agar (Bauer et al. 1966) for seven different classes of antimicrobials comprising 15 antibiotics including aminoglycosides [gentamicin (120 mcg) and amikacin (30 mcg)], cephaems [cefixime (5 mcg), cefotaxime (30 mcg), ceftazidime (30 mcg) and ceftriaxone (30 mcg)], lincosamides [clindamycin (2 mcg)], macrolides [erythromycin (15 mcg) and azithromycin (15 mcg)], penicillins [penicillin G (10U), ampicillin (10 mcg) and amoxicillin (30 mcg)], phenicols [chloramphenicol (30 mcg)] and quinolones [norfloxacin (10 mcg) and gatifloxacin (5 mcg)].
Antibiotic discs were placed on Mueller–Hinton agar plates inoculated with B. cereus isolates at 37 °C and zones of inhibition were measured after 24 h. Antibiotic concentrations of the discs used and antimicrobial susceptibility interpretation criteria were as recommended by the Clinical and Laboratory Standards Institute (CLSI 2013). Isolates resistant to 3 or more unique antimicrobial classes were classified as multi drug resistant (MDR) strains (Waters et al. 2011).
Statistical analysis
Fisher’s exact test was used to determine the association between incidence of B. cereus and its enterotoxins with and types of RTE foods tested in this study. Significance was set at a two tailed p value < 0.05.
Results and discussion
B. cereus is one of 22 World Health Organization prioritized food borne pathogens for assessing the load of food borne illnesses (Kirk et al. 2015). This investigation determined the burden of B. cereus and its enterotoxins in RTE foods from popular tourist destination of Himachal Pradesh. In general, consumption of RTE foods remains higher at tourist destinations compared to other places. These foods are generally prepared and stored in an uncontrolled environment without following good hygienic and manufacturing practices. High levels of food safety standards should be followed while preparing and serving RTE/street foods at such tourist places since visiting populations after getting infections can carry pathogens to susceptible populations.
Of 220 samples tested, Bacillus isolates were recovered from 25 samples with an overall incidence of 11.4%. PCR amplification of 16S rRNA (~ 1500 bp, Gomaa and Momtaz 2006; Sadashiv and Kaliwal 2014) confirmed all (n = 25) biochemically (Supplementary Table 2) characterized isolates as Bacillus spp. (Fig. 1). Organji et al. (2015) and Hadithi et al. (2016) had reported higher Bacillus incidence rates of 17.3% and 24.76%, respectively in RTE foods comprising cooked rice, pasteurized milk, yogurt, infant milk powder and white cheese. Incidence of Bacillus was highest in samples collected from Palampur (21.7%, 5/23,) followed by those from Kullu–Manali (8/37, 21.6%, p = 0.04), Kangra (12.5%, 3/24), Dharamsala–Mcleodganj (9.4%, 5/53), Baijnath/Bir–Billing (8.3%, 2/24), Chamba–Dalhousie (5%, 1/20), Mandi (4%, 1/25). None of the samples (n = 14) tested from Shimla was found positive for Bacillus spp. (Table 1). Absence of Bacillus spp. in samples collected from Shimla reflects following of good personnel and preparatory hygiene practices while preparing and serving the foods.
Fig. 1.
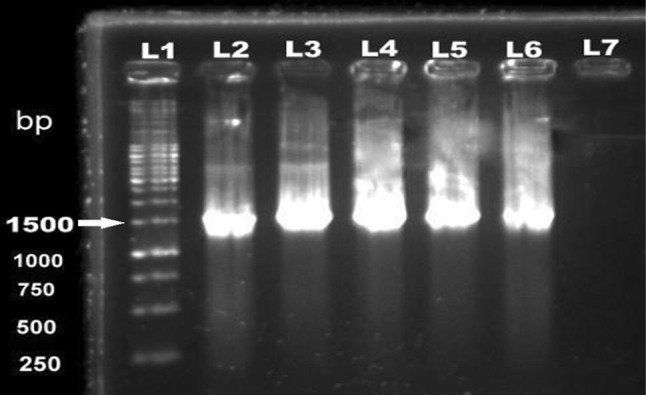
Molecular characterization of Bacillus isolates by targeting 16SrRNA (1500 bp). Lane 1—1 kb ladder, Lanes 2, 3, 4, 5—Bacillus isolates from this study, Lane 6—B. cereus (ATCC 13061) positive control, Lane 7: nuclease free water as negative control
The recovered Bacillus isolates were identified as B. cereus (76.0%, 19/25), B. alvei (12.0%, 3/25), B. polymyxa (8.0%, 2/25) and B. firmus (4.0%, 1/25). Bacillus contamination was significantly higher (31.3%, 5/16, p = 0.02) in cheese based RTE foods compared to other RTE foods, beverages and water samples (Table 2). B. cereus is major food borne pathogen from genus Bacillus, whereas B. alvei, B. polymyxa and B. firmus are more associated with food spoilage (Hanlin 1998). All identified B. cereus isolates were Gram negative rods, motile and were sporulated. These isolates were catalase, Voges Proskauer and citrate positive and showed β-haemolysis on blood agar. None of the B. cereus isolates utilized mannitol and arabinose where as all were glucose and trehalose positive (Vos et al. 2009; Public Health England 2018). We recorded an overall B. cereus incidence of 8.6% (19/220) whereas incidence in RTE foods, beverages and water was 9.2% (11/120), 7.5% (3/40) and 8.3% (5/60), respectively. Tewari et al. (2015) and Fayaz et al. (2017) reported 30.9% and 36.7% incidence of B. cereus in meat and meat products, respectively. Yusuf et al. (2018) reported B. cereus in 28.4% samples of milk and milk products.
Table 2.
Incidence of different Bacillus species in ready to eat food, beverages and water samples tested
| Types of samples collected (no. of samples) | Type of products | No. of samples tested (%) | No. of Bacillus positive samples (%) | Bacillus species identified | Place of sample collection | |||
|---|---|---|---|---|---|---|---|---|
| B. cereus na (%) | B. alvei n (%) | B. polymyxa n (%) | B. firmus n (%) | |||||
| Ready to eat foods (n = 120) | Chicken momo | 25 (11.4) | 2 (8.0) | 1 (4.0) | 0 (0.0) | 1 (4.0) | 0 (0.0) | B. cereus (Palampur, n = 1), B. polymyxa (Kullu Manali, n = 1) |
| Paneer based | 23 (10.5) | 4 (17.4) | 2 (8.7) | 1 (4.3) | 1 (4.3) | 0 (0.0) | B. cereus (Palampur, n = 2), B. alvei (Baijnath/Bir–Biling, n = 1), B. polymyxa (Palampur, n = 1) | |
| Khoa based | 21 (9.5) | 3 (14.3) | 3 (14.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | B. cereus (Kangra, n = 2; Mandi, n = 1) | |
| Cheese based | 16 (7.3) | 5 (31.3) | 4 (25.0) | 1 (6.3) | 0 (0.0) | 0 (0.0) | B. cereus (Dharamsala–Mcleodganj, n = 2; Manali, n = 2), B. alvei (Dharamsala–Mcleodganj, n = 1) | |
| Mutton momo | 15 (6.8) | 1 (6.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (6.7) | B. firmus (Chamba/Dalhousie, n = 1) | |
| Cream based | 12 (5.5) | 2 (16.7) | 1 (8.3) | 1 (8.3) | 0 (0.0) | 0 (0.0) | B. cereus (Manali, n = 1), B. alvei (Palampur, n = 1) | |
| Curd | 8 (3.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | Nil | |
| Beverages (n = 40) | Chicken soup | 15 (6.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | Nil |
| Milk beveragesb | 19 | 2 | 2 | 0 (0.0) | 0 (0.0) | 0 (0.0) | B. cereus (Dharamsala–Mcleodganj, n = 1; Kangra, n = 1) | |
| Vegetable soup | 6 (2.7) | 1 (16.7) | 1 (16.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | B. cereus (Dharamsala–Mcleodganj n = 1) | |
| Water (n = 60) | Water | 60 (27.3) | 5 (8.3) | 5 (8.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | B. cereus (Bir–Billing, n = 1; Kullu, Manali, n = 4) |
| Total | 220 | 25 (11.4) | 19 (8.6) | 3 (1.4) | 2 (0.9) | 1 (0.5) | ||
Values in bold indicate significant association (p ≤ 0.05)
aNumber of isolates
bMilk beverages included lassi (n = 10) and milk shake (n = 9) samples
B. cereus recovery was highest from cheese based foods (25.0%, 4/16) followed by vegetable soups (16.7%, 1/6), khoa based foods (14.0%, 3/21), milk based beverages (10.5%, 2/19), paneer based foods (8.6%, 2/23), cream based foods (8.3%, 1/12) and water (8.3%, 5/60) samples (Table 2). B. cereus contamination in cheese based RTE foods (25%, 4/16) was significantly higher compared to other RTE foods (6.7%, 7/104, p = 0.03) and overall (7.4%, 15/204, p = 0.04) all the samples analysed in this study. Abbas et al. (2014) reported 16.7% incidence of B. cereus in soft cheese, lower than this study. Salem et al. (2015) reported 20% samples each of white cheese and Kareesh cheese contaminated with B. cereus in Egypt. Iurlina et al. (2006) reported 50% incidence of B. cereus in Port Salut Argentino cheeses. Singh et al. (2015) found 52.9% paneer samples contaminated with B. cereus, much higher than recorded in this study for paneer based RTE foods (8.6%). Yusuf et al. (2018) found 16% paneer samples contaminated with B. cereus. High incidence of B. cereus in cheeses and other dairy products can be due to high hydrophobicity of the spores, the low spore surface charge and the spore morphology which results in their strong adhesion to different surfaces and survival during processing of the milk and milk products (Andersson et al. 1995; Iurlina et al. 2006).
None of the tested chicken soup (n = 15) samples were contaminated with Bacillus spp. This might be due to the combined effect of boiling and spices along with hygienic preparation of soups. Steaming under pressure, roasting, frying and grilling destroys spores and vegetative forms of B. cereus (Schneider et al. 2004). All the tested curd samples (n = 8) were also negative for Bacillus spp. This can be attributed to low pH and the presence of natural micro flora (Tirloni et al. 2017).
All 25 Bacillus isolates were screened by multiplex PCR for the presence of enterotoxin genes; hblD (1018 bp), nheB (935 bp), hblA (884 bp), nheA (759 bp), hblC (695 bp), nheC (618 bp) and cytK (565 bp). We detected enterotoxin encoding genes only in B. cereus isolates. None of the B. alvei, B. polymyxa and B. firmus isolates carried any of the enterotoxin producing genes. ces (emetic toxin) was not present in any of the Bacillus isolates (Fig. 2).
Fig. 2.
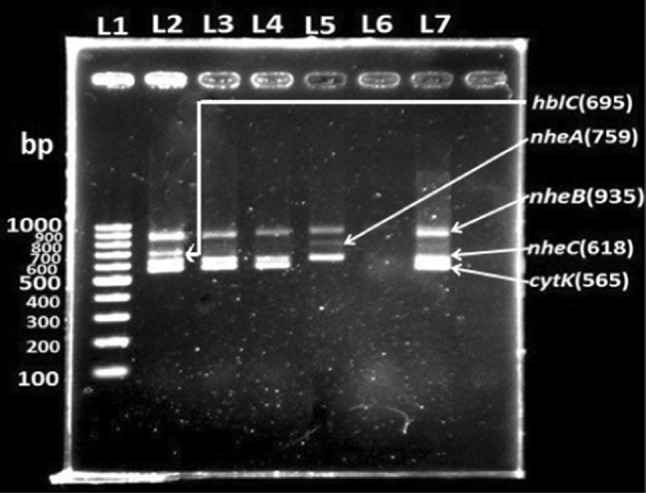
Multiplex PCR products of enterotoxigenic genes amplified from genomoic DNA of Bacillus cereus isolates. Lane 1—100 bp ladder; Lane 2—amplified nheB, hblC, nheC, cytK products; Lane 3—amplified nheB, nheC, cytK products; Lane 4—amplified nheB, nheC, cytK products; Lane 5—amplified nheB, nheA, nheC products; Lane 6—nuclease free water as negative control; Lane 7—B. cereus (ATCC 13061) positive control
Each B. cereus isolate was found to harbour at least two enterotoxin encoding genes (Table 3). One or more nhe (encoding non-haemolytic enterotoxins) complex genes were present in all (19/19, 100%) the tested isolates. Owusu-Kwarteng et al. (2017) reported 92% B. cereus isolates from dairy farms and traditional dairy food products positive for one or more nhe complex encoding genes. Similarly, Tewari et al. (2015) reported 89.7% of B. cereus isolates with at least one of the genes of nhe complex. Among nhe complex genes, we observed highest incidence of nheC gene (100%, 19/19) followed by nheB (94.7%, 18/19) and nheA (57.9%, 11/19). Abbas et al. (2014) reported higher presence of nheA (90.3%) compared to nheB (58.1%) and nheC (54.8%) in B. cereus isolated from milk and milk products. 57.9% B. cereus isolates in this study contained all three nhe genes, 36.8% contained two genes (nheB, nheC) and 5.3% had only one nhe gene i.e. nheC. Using qPCR/MiSeq sequencing data, high incidence of nheA (135/145), nheB (113/145) and nheC (145/145) was detected in 147 fermented soybean products contaminated with B. cereus (Keisam et al. 2019). In our study, incidence of B. cereus cytK was (52.6%, n = 10/19). Earlies studies had reported incidence ranging from 41.4% to 88.0% (Owusu-Kwarteng et al. 2017; Tewari et al. 2015; Rather et al. 2011; Ngamwongsatit et al. 2008).
Table 3.
Enterotoxigenic genes detected in B. cereus (n = 19) isolates
| Type of products (na/Nb) | Isolates positive for hbl gene (%) | Isolates positive for nhe gene (%) |
cytK n (%) |
||||
|---|---|---|---|---|---|---|---|
|
hblA n (%) |
hblC n (%) |
hblD n (%) |
nheA n (%) |
nheB n (%) |
nheC n (%) |
||
| Chicken momo (1/25)c | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (4.0) | 1(4.0) | 1(4.0) | 0 (0.0) |
| Paneer based (2/23) | 0 (0.0) | 2 (8.7) | 0 (0.0) | 1 (4.3) | 2 (8.7) | 2(8.7) | 1(4.3) |
| Khoa based (3/21) | 0 (0.0) | 1 (4.8) | 0 (0.0) | 3 (14.3) | 3 (14.3) | 3(14.3) | 2 (9.5) |
| Cheese based (4/16) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (12.5) | 4 (25.0) | 4 (25.0) | 2 (12.5) |
| Cream based (1/12) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (8.3) | 1 (8.3) | 0 (0.0) |
| Milk beveragesd (2/19) | 0 (0.0) | 2 (10.5) | 0 (0.0) | 1 (5.3) | 2 (10.5) | 2 (10.5) | 2 (10.5) |
| Vegetable soup (1/6) | 0 (0.0) | 1 (16.7) | 0 (0.0) | 0 (0.0) | 1 (16.7) | 1(16.7) | 0 (0.0) |
| Water (5/60) | 0 (0.0) | 1 (1.7) | 0 (0.0) | 3 (5.0) | 4 (6.7) | 5 (8.3) | 3 (5.0) |
| Total | 0 (0.0) | 7 | 0 (0.0) | 11 | 18 | 19 | 10 |
None of the B. alvei (n = 3), B. polymyxa (n = 2) and B. firmus (n = 1) were carrying any of the enterotoxin encoding gene
Values in bold indicate significant association (p ≤ 0.05)
aNumber of samples positive for one or more one type of B. cereus enterotoxins
bTotal number of samples tested for type of food product
cNo B. cereus isolate was detected in mutton momo (n = 15), curd (n = 8) and chicken soup (n = 15) samples (refer Table 2)
dMilk beverages included lassi (n = 10) and milk shake (n = 9) samples
Among hbl (encoding haemolytic enterotoxins) complex genes, only hblC was detected in 36.8% (7/19) B. cereus isolates. Fernandes et al. (2014) detected hblC gene in 40% B. cereus isolates, higher than recorded in this study. Owusu-Kwarteng et al. (2017) detected one or more hblACD complex genes in 75% of B. cereus isolates from dairy farms and traditional food products. hblC alone was found only in 10% isolates. In soyabean fermented products from Northeast India, occurence of hblA and hblD genes was as high as high as 98.6% (Keisam et al. 2019). Incidence of hlbC was not investigated in this study. We observed lower incidence of hbl complex compared to nhe complex genes. This observation is in agreement to previous studies (Al-Khatib et al. 2007; Tewari et al. 2015; Owusu-Kwarteng et al. 2017).
B. cereus enterotoxigenic genes in an operon (hblCDA and nheABC) can occur independently of each other. Tewari et al. (2015) had reported independent occurrence of hbl, nhe and cytK enterotoxin in B. cereus. In the present study, seven isolates were devoid of two genes in hbl complex (Table 3). Similarly, eight isolates showed the absence of one or two genes in nhe complex. Ngamwongsatit et al. (2008) had reported simultaneous presence of all the three genes in hbl and nhe operons. It is suggested that all the three genetic components of hbl and nhe complex in B. cereus are necessary for maximal enterotoxin activity (Beecher et al. 1995; Lindbäck et al. 2004). Yang et al. (2005) reported presence of at least one of the enterotoxin genes (hblACD complex, nheABC complex, cytK and entFM) in B. cereus isolates recovered from samples collected food poisoning outbreaks.
Statistical analysis revealed significant (p = 0.038) presence of nheA (25.0%, 4/16,) and nheB (25%, 4/16) in B. cereus isolates recovered from cheese based RTE foods compared to those from other foods, beverages and water samples (Table 3). Abbas et al. (2014) reported highest nheA incidence (80%) in soft cheese products followed by nheB (60%), nheC (40%) and hblC (20%).
On the basis of enterotoxins present, B. cereus isolates were divided into 8 groups (Supplementary Table 3). 36.8% (7/19) isolates had at least one gene encoding haemolytic (hbl) and non-haemolytic (nhe) complex along with cytotoxic-K gene. These groups were G1 (3/19, 15.8%), G2 (1/19, 5.3%), G4 (1/19, 5.3%) and G5 (2/19, 10.5%). Group-1 (G1) isolates had all the five enrerotoxigenic genes i.e. hblC/nheA/nheB/nheC/cytK detected in this study. Tewari et al. (2015) reported two isolates (6.9%, 2/29) carrying these genes (hbl/nheA/nheB/nheC/cytK) along with entFM but combination devoid of any of the hbl genes. Group 3 (G3) isolates (4/19, 21.1%) carried all the genes encoding non-haemolytic complex and for cytotoxic enterotoxin (nheA/nheB/nheC/cytK). Group 7 (G7, 4/19, 21.1%) and Group-8 (G8, 3/19, 15.8%) isolates carried only nhe complex genes, nheA/nheB/nheC and nheB/nheC, respectively. Earlier studies have reported 10.4% to 68.4% presence of all three nhe complex genes in B. cereus isolates (Cadirci et al. 2013; Tewari et al. 2015; Yibar et al. 2017).
Antimicrobial susceptibility profiles of B. cereus isolates were determined for seven different antibiotic classes comprising 15 antibiotics recommended by CLSI for B. cereus antimicrobial susceptibility testing (Table 4). All (n = 19) the tested isolates were susceptible to lincosamides (clindamycin), quinolones (gatifloxacin and norfloxacin) and phenicols (choloramphenicol). Susceptibility levels varied between 95 and 100% for aminoglycosides [gentamicin (18/19, 95%) and amikacin (19/19, 100%)] and macrolides [erythromycin (18/19, 95%) and azithromycin (19/19, 100%)]. Among cephems, resistance levels ranged from 5% (1/19, cefotaxime) to 100% (19/19, cefixime). In penicillin class of tested antibiotics, resistance levels were 100% (19/19), 74% (14/19) and 58% (11/19) for penicillin G, amoxicillin with clavulanic acid and ampicillin, respectively. Sood et al. (2017) and Owusu-Kwarteng et al. (2017) had also reported resistance to ampicillin, carbenicillin, cefepime, cephalothin, and penicillin G. 100% resistance against penicillin G as observed in this study was also recorded by other (Bashir et al. 2017; Owusu-Kwarteng et al. 2017; Yusuf et al. 2018). In the present study, 100% susceptibility was recorded for gentamicin and chloramphenicol, in accordance to Agwa et al. (2012) and Owusu-Kwarteng et al. (2017). Other workers had also shown high susceptibility of B. cereus to chloramphenicol (Hadithi et al. 2016; Owusu-Kwarteng et al. 2017).
Table 4.
Antimicrobial susceptible profiles of 19 isolates of B. cereus
| Antimicrobial class | Antibiotic | Concentration (mcg) | Susceptible isolates na (%) | Resistant isolates n (%) |
|---|---|---|---|---|
| Aminoglycosides | Amikacin (AK) | 30 | 18 (95) | 1 (5) |
| Gentamicin (HLG) | 120 | 19 (100) | – | |
| Cephems | Cefixime (CFM) | 5 | – | 19 (100) |
| Cefotaxime (CTX) | 30 | 18 (95) | 1 (5) | |
| Ceftazidime (CAZ) | 30 | 7 (37) | 12 (63) | |
| Ceftriaxone (CTR) | 30 | 16 (84) | 3 (16) | |
| Lincosamides | Clindamycin (CD) | 2 | 19 (100) | – |
| Macrolides | Erythromycin (ERY) | 15 | 18 (95) | 1 (5) |
| Azithromycin (AZM) | 15 | 19 (100) | – | |
| Quinolones | Gatifloxacin (GAT) | 5 | 19 (100) | 0 (0) |
| Norfloxacin (NX) | 10 | 19 (100) | 0 (0) | |
| Penicillins | Penicillin G (PEN) | 10 | 0 (0) | 19 (100) |
| Amoxicillin/clavulanic acid (AMC) | 30 | 5 (26) | 14 (74) | |
| Ampicillin (AMP) | 10 | 8 (42) | 11 (58) | |
| Phenicols | Chloramphenicol (C) | 30 | 19 (100) | 0 (0) |
Antibiotic concentrations and interpretation criteria were as per CLSI (2013)
aNumber of isolates
We observed ten different resistance patterns for 19 B. cereus isolates recovered in this study (Supplementary Table 4). All the isolates were resistant to two to six antibiotics tested in this study (Supplementary Table 3). Two isolates (10.5%, 2/19) were identified as MDR showing resistance to ≥ 3 antibiotic classes. One MDR isolate was resistant to penicillins, cephems, and macrolides whereas other one was resistant to penicillin, cephems and aminoglycosides.
Conclusion
This study is the first scientific investigation to determine the incidence of food and water borne B. cereus in Himachal Pradesh. RTE food and water samples were collected from different tourist places of Himachal Pradesh and were analyzed to detect the presence of Bacillus cereus. Incidence of food borne B. cereus observed in this study was lower compared to studies conducted earlier. Low incidence of B. cereus can be attributed to the screening of cooked or processed food samples and use of conventional culture and biochemical methods for the identification of bacteria. Molecular techniques such as qPCR and MiSeq sequence analysis have been shown to be more effective in ascertaining prevalence of food borne pathogens and their toxigenic profiles (Keisam et al. 2019). We recorded higher levels of B. cereus contamination in cheese based RTE foods compared to other RTE foods, beverages and water samples. Each B. cereus isolate recovered in this study harboured at least two enterotoxigenic genes and two isolates were identified as MDR strains. These findings indicate B. cereus is a potential public health burden associated with RTE foods in Himachal Pradesh.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are thankful to CSK Himachal Pradesh Agricultural University for financial and infrastructure support for this research. The necessary help and technical support provided by Head, Department of Veterinary Microbiology, Dr. GC Negi College of Veterinary and Animal Sciences, CSK Himachal Pradesh Agricultural is duly acknowledged.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abbas BA, Khudor MH, Saeed BMS. Detection of hbl, nhe and bceT toxin genes in Bacillus cereus isolates by multiplex PCR. Int J Curr Microbiol Appl Sci. 2014;3:1009–1016. [Google Scholar]
- Agwa OK, Uzoigwe CI, Wokoma EC. Incidence and antibiotic sensitivity of Bacillus cereus isolated from ready to eat foods sold in some markets in Portharcourt, Rivers state, Nigeria. Asian J Microbiol Biotechnol Environ Sci. 2012;14:13–18. [Google Scholar]
- Ahmadi SA, Panda AK. Prevalence of Escherichia coli and Salmonella spp. in ready-to-eat milk and milk products in Himachal Pradesh. J Vet Public Health. 2015;13:25–29. [PubMed] [Google Scholar]
- Aksu H, Bostan K, Ergün Ö. Presence of Bacillus cereus in packaged some spices and herbs sold in Istanbul. Pak J Biol Sci. 2000;3:710–712. [Google Scholar]
- Al-Khatib MS, Khyami-Horani H, Badran E, Shehabi AA. Incidence and characterization of diarrheal enterotoxins of fecal Bacillus cereus isolates associated with diarrhea. Diag Microbiol Infect Dis. 2007;59:383–387. doi: 10.1016/j.diagmicrobio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Andersson A, Ronner U, Granum PE. What problems does the food industry have with the spore-forming pathogens Bacillus cereus and Clostridium perfringens? Int J Food Microbiol. 1995;28:145–155. doi: 10.1016/0168-1605(95)00053-4. [DOI] [PubMed] [Google Scholar]
- Aruwa CE, Akinyosoye FA. Microbiological assessment of ready-to-eat foods (RTEs) for the presence Bacillus species. J Adv Biol Biotechnol. 2015;3:145–152. [Google Scholar]
- Bashir M, Malik MA, Moien J, Badroo GA, Mohd Bhat A, Singh M. Prevalence and characterization of Bacillus cereus in meat and meat products in and around Jammu region of Jammu and Kashmir, India. Int J Curr Microbiol Appl Sci. 2017;6:1094–1106. [Google Scholar]
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- Beecher DJ, Schoeni JL, Wong AC. Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect Immun. 1995;63:4423–4428. doi: 10.1128/iai.63.11.4423-4428.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold-Pluta A, Puta A, Debevere J. The effect of selected factors on the survival of Bacillus cereus in the human gastro-intestinal tract. Microb Pathog. 2015;82:7–14. doi: 10.1016/j.micpath.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Bottone EJ. B. cereus, a volatile human pathogen. Clin Microbiol Rev. 2010;23:382–398. doi: 10.1128/CMR.00073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadirci O, Gucukoglu A, Goknur T, Kevenk O, Mustafa A. Determination of enterotoxigenic gene profiles of Bacillus cereus isolates isolated from dairy desserts by multiplex PCR. Kafkas Univ Vet Fak Derg. 2013;19:869–874. [Google Scholar]
- Chettri R, Tamang JP. Bacillus species isolated from Tungrymbai and Bekang, naturally fermented soybean foods of India. Int J Food Microbiol. 2015;197:72–76. doi: 10.1016/j.ijfoodmicro.2014.12.021. [DOI] [PubMed] [Google Scholar]
- CLSI . Performance standards for antimicrobial susceptibility testing. Wayne: Clinical and Laboratory Standards Institute; 2013. [Google Scholar]
- Fayaz S, Badroo GA, Ahmad A, Rasool U, Mustafa R, Mudasir M. Molecular characterization of enterotoxigenic Bacillus cereus species isolated from meat using conventional PCR and multiplex PCR. Int J Curr Microbiol Appl Sci. 2017;6:324–328. [Google Scholar]
- Fernandes MDS, Fujimoto G, Schneid I, Kabuki DV, Kuaye AY. Enterotoxigenic profile, antimicrobial susceptibility and biofilm formation of Bacillus cereus isolated from ricotta processing. Int Dairy J. 2014;38:16–23. [Google Scholar]
- Fernández-No IC, Guarddon M, Böhme K, Cepeda A, Calo-Mata P, Barros-Velázquez J. Detection and quantification of spoilage and pathogenic Bacillus cereus, Bacillus subtilis and Bacillus licheniformis by real-time PCR. Food Microbiol. 2011;28:605–610. doi: 10.1016/j.fm.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Gomaa OM, Momtaz OA. 16S rRNA characterization of a Bacillus isolate and its tolerance profile after subsequent subculturing. Arab J Biotechnol. 2006;10:107–116. [Google Scholar]
- Gopal N, Hill C, Ross PR, Beresford TP, Fenelon MA, Cotter PD. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front Microbiol. 2015;6:1418. doi: 10.3389/fmicb.2015.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadithi AL, Hadeel T, Entesar AK. Incidence of Bacillus cereus in food and environmental sources in Basrah/Iraq. Antibiotic resistance profiles of recovered isolates. Int J Res Acad Rev. 2016;4:117–127. [Google Scholar]
- Hanlin JH. Spoilage of acidic products by Bacillus species. Dairy Food Environ Sanit. 1998;18:655–659. [Google Scholar]
- Iurlina MO, Saiz AI, Fuselli SR, Fritz R. Prevalence of Bacillus spp. in different food products collected in Argentina. LWT. 2006;39:105–110. [Google Scholar]
- Keisam S, Tuikhar N, Ahmed G, Jeyaram K. Toxigenic and pathogenic potential of enteric bacterial pathogens prevalent in the traditional fermented foods marketed in the Northeast region of India. Int J Food Microbiol. 2019;296:21–30. doi: 10.1016/j.ijfoodmicro.2019.02.012. [DOI] [PubMed] [Google Scholar]
- Kim JM, Forghani F, Jung-Beom K, Yong-Bae P, Myoung-Su P, Wang J, Park JH, Deog-Hwan O. Improved multiplex PCR assay for simultaneous detection of Bacillus cereus emetic and enterotoxic isolates. Food Sci Biotechnol. 2011;21:1439–1444. [Google Scholar]
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 2015;12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotiranta A, Lounatmaa K, Haapasalo M. Epidemiology and pathogenesis of B. cereus infections. Microbes Infect. 2000;2:189–198. doi: 10.1016/s1286-4579(00)00269-0. [DOI] [PubMed] [Google Scholar]
- Kramer JM, Gilbert RJ. B. cereus and other Bacillus species. In: Doyle MP, editor. Foodborne bacterial pathogens. New York: Marcel Dekker; 1989. pp. 21–77. [Google Scholar]
- Lakhanpal P, Panda AK, Rajesh Chahota, Shivani Choudhary, Thakur SD. Incidence and antimicrobial susceptibility of Staphylococcus aureus isolated from ready-to-eat animal foods from tourist destinations of north western Himalayas, Himachal Pradesh, India. J Food Sci Technol. 2019;56:1078–1083. doi: 10.1007/s13197-018-03556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbäck T, Fagerlund A, Rødland MS, Granum PE. Characterization of the Bacillus cereus nhe enterotoxin. Microbiology. 2004;150:3959–3967. doi: 10.1099/mic.0.27359-0. [DOI] [PubMed] [Google Scholar]
- Logan NA. Bacillus and relatives in food borne illness. J Appl Microbiol. 2011;112:417–429. doi: 10.1111/j.1365-2672.2011.05204.x. [DOI] [PubMed] [Google Scholar]
- Ngamwongsatit P, Buasri W, Pianariyanon P, Pulsrikarn C, Ohba M, Assavanig A, Panbangred W. Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK and entFm) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int J Food Microbiol. 2008;121:352–356. doi: 10.1016/j.ijfoodmicro.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Organji SR, Abulreesh HH, Elbanna K, Ebrahim G, Osman H, Khider M. Occurrence and characterization of toxigenic Bacillus cereus in food and infant feces. Asian Pac J Trop Biomed. 2015;5:515–520. [Google Scholar]
- Owusu-Kwarteng J, Wuni A, Akabanda F, Tano-Debrah K, Jespersen L. Prevalence, virulence factor genes and antibiotic resistance of Bacillus cereus sensu lato isolated from dairy farms and traditional dairy products. BMC Microbiol. 2017;17:65. doi: 10.1186/s12866-017-0975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera ML, Ranasinghe GR. Prevalence of Bacillus cereus and associated risk factors in Chinese-style fried rice available in the city of Colombo, Sri Lanka. Foodborne Pathog Dis. 2012;9:125–131. doi: 10.1089/fpd.2011.0969. [DOI] [PubMed] [Google Scholar]
- Public Health England (2018) UK standards for microbiology investigations. Identification of Bacillus species. Standards Unit, Microbiology Services, Public Health England. Bacteriology-identification 3.1, pp 1–27. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/697260/ID_9i3.1.pdf. Accessed on 12 May 2018
- Rajkovic A. Microbial toxins and low level of food borne exposure. Trends Food Sci Technol. 2014;38:149–157. [Google Scholar]
- Rather MA, Aulak RS, Gill JPS, Rao TS, Hassan MN. Direct detection of Bacillus cereus and its enterotoxigenic genes in meat and meat products by polymerase chain reaction. J Adv Vet Res. 2011;1:99–104. [Google Scholar]
- Roy A, Moktan B, Sarkar PK. Characteristics of Bacillus cereus isolates from legume-based Indian fermented foods. Food Control. 2007;18:1555–1564. [Google Scholar]
- Sadashiv SO, Kaliwal BB. Isolation, characterization and antibiotic resistance of Bacillus spp. from bovine mastitis in the region of north Karnataka, India. Int J Curr Microbiol Appl Sci. 2014;3:360–373. [Google Scholar]
- Salem NA, Jakee JE, Nasef SA, Badr H. Prevalence of Bacillus cereus in milk and milk products. Anim Health Res J. 2015;3:168–172. [Google Scholar]
- Schneider KR, Parish M E, Goodrich RM, Cookingham T (2004) Preventing food borne illness: B. cereus and Bacillus anthracis. Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. FSHN04-05
- Singh VK, Shukla S, Chaturvedi A. Study the incidence of Bacillus cereus isolates from dairy foods. Pharm Innov J. 2015;3:41–43. [Google Scholar]
- Sood B, Sahota PP, Hunjan M. Multidrug resistant Bacillus cereus in fresh vegetables: a serious burden to public health. Int J Curr Microbiol Appl Sci. 2017;6:649–661. [Google Scholar]
- Tewari A, Abdullah S. Bacillus cereus food poisoning: international and Indian perspective. J Food Sci Technol. 2015;52:2500–2511. doi: 10.1007/s13197-014-1344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari A, Singh SP, Singh R. Incidence and enterotoxigenic profile of Bacillus cereus in meat and meat products of Uttarakhand, India. J Food Sci Technol. 2015;52:1796–1801. doi: 10.1007/s13197-013-1162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirloni E, Ghelardi E, Celandroni F, Bernardi C, Stella S. Effect of dairy product environment on the growth of Bacillus cereus. J Dairy Sci. 2017;100:7026–7034. doi: 10.3168/jds.2017-12978. [DOI] [PubMed] [Google Scholar]
- Vos PD, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, William BW. Bergey’s manual of systematic bacteriology. 2. Dordrecht: Springer; 2009. [Google Scholar]
- Waters AE, Contente-Cuomo T, Buchhagen J, Liu CM, Watson L, Pearce K, Foster JT, Bowers J, Driebe EM, Engelthaler DM, Keim PS, Price LB. Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin Infect Dis. 2011;52:1227–1230. doi: 10.1093/cid/cir181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang IC, Shih DY, Huang TP, Huang YP, Wang JY, Pan TM. Establishment of a novel multiplex PCR assay and detection of toxigenic strains of the species in the Bacillus cereus group. J Food Prot. 2005;68:2123–2130. doi: 10.4315/0362-028x-68.10.2123. [DOI] [PubMed] [Google Scholar]
- Yıbar A, Çetinkaya F, Soyutemiz E, Yaman G. Prevalence, enterotoxin production and antibiotic resistance of Bacillus cereus isolated from milk and cheese. Kafkas Univ Vet Fak Derg. 2017;23(4):635–642. [Google Scholar]
- Yusuf U, Kotwal SK, Gupta S, Ahmed T. Identification and antibiogram pattern of Bacillus cereus from the milk and milk products in and around Jammu region. Vet World. 2018;11:186–191. doi: 10.14202/vetworld.2018.186-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


