Abstract
Pangolins, or scaly anteaters, have recently been flagshiped as one of the most illegally traded mammals, and as a corollary, as potential intermediate hosts at the origin of the COVID-19 pandemic. In order to improve the traceability of their trade, we developed 20 polymorphic microsatellite loci for the white-bellied pangolin (Phataginus tricuspis), the species most frequently found on African bushmeat markets. We genotyped 24 white-bellied pangolins from the Douala market, Cameroon, originating from the Ebo forest c. 75 km north-east of Douala. The number of alleles per locus ranged from 4 to 12 (mean = 6.95), and mean observed and expected heterozygosities were 0.592 (0.208–0.875) and 0.671 (0.469–0.836), respectively. Genetic diversity was higher than that cross-estimated from microsatellite loci developed for other species of pangolins. Two loci deviated from Hardy–Weinberg equilibrium and two loci showed linkage disequilibrium. Genetic variance (PCoA) was increased with the addition of 13 pangolins of unknown origin, possibly suggesting that the Douala market is fed from differentiated source populations of white-bellied pangolins. Each of the 37 individuals had a unique multilocus genotype. The unbiased probability of identity (uPI) and the probability of identity among siblings (PIsibs) were both very low (uPI = 8.443 e−21; PIsibs = 1.011 e−07). Only five microsatellite loci were needed to reach the conservative value of PIsibs < 0.01, overall indicating a powerful discriminating power of our combined loci. These 20 newly developed microsatellite loci might prove useful in tracing the local-to-global trade of the white-bellied pangolin, and will hopefully contribute to the DNA-assisted implementation of future conservation strategies at reasonable costs.
Electronic supplementary material
The online version of this article (10.1007/s11033-020-05511-6) contains supplementary material, which is available to authorized users.
Keywords: Pangolins, Phataginus tricuspis, Bushmeat, Microsatellite loci, Illegal wildlife trade, Cameroon
Introduction
The illegal wildlife trade is a flourishing, parallel economy that threatens the worldwide biodiversity [1]. Pangolins, or scaly anteaters, have recently been flagshiped as one of the most illegally traded mammals, being literally eaten to extinction [2]. Based on reports from the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), a minimum number of c. 895,000 African and Asian pangolins were trafficked during the last decade (2010–2019). They were mainly destined to Asian markets including China and Vietnam [3], where they feed the growing demand from the Traditional Chinese Medicine market [4]. As a corollary, the pangolin trade has very recently been pinpointed as posing a serious threat to human health, pangolins being potentially involved in the COVID-19 pandemic [5, 6]. Pangolins are also part of the traditional wild meat intake and pharmacopeia in most of their range countries [7, 8], and as such suffer from the ‘bushmeat crisis’ that threaten the survival of many vertebrates in the tropics [9].
The traceability of the pangolin trade is rendered difficult by the elusive, international criminal networks that fuel the market, but also by the various forms under which pangolins are traded. Indeed, a large spectrum of pangolin-related ‘items’ are involved in the trade, from scales alone to smoked skins, boiled and ‘pealed’ carcasses, chopped meat, scale powder and embryo soups [10–12]. The societal demand for mitigating the pangolin trade [13] implies to develop tools capable of accurately identifying the trafficked species of pangolins, tracing their geographic origins (sources) and estimating the number of pangolins when body parts and scales are seized.
The genetic ‘tool box’ may constitute an efficient support to reach these objectives. To date, a certain number of mitochondrial, nuclear and Y-borne genes have been sequenced in pangolins [14–17], providing useful resources for the accurate ‘tagging’ of the species involved in the trade [18]. On the other hand, our ability to trace the geographic origins of seized pangolins has remained limited because of a lack of exhaustive DNA registers and the reduced geographic resolution of traditional gene sequencing [19–23]. Although microsatellite loci were developed in an Asian (Manis javanica) and an African species (Smutsia temminckii) [24, 25], their application to the issue of the traceability of the trade remains limited or pending [26, 27]. Recently, single nucleotide polymorphic markers (SNPs) have successfully been used to assign seizures of M. javanica to potential geographic sources in South-East Asia [28]. So far, no genotyping approach has been applied to assess the number of individuals involved in large (scale) seizures, despite the potential advantage this would have for seizure processing [29].
Pangolins are highly sensitive to habitat degradation and have slow reproduction rates [30]. They are one of the mammalian lineages most prone to extinction (https://www.edgeofexistence.org/). In the last years, there have been pressing initiatives for increasing awareness on the unsustainability of the pangolin trade and for promoting law enforcement at the international level. All eight species were moved to the Appendix I of CITES in 2016 [31] and have recently been upgraded to “Vulnerable”, “Endangered” or “Critically Endangered” on the IUCN Red List of Threatened Species [32].
Recent seizures indicate that, as Asian pangolins are becoming rarer, traffickers are now sourcing from Africa possibly via the same criminal networks used for the trade of ivory [33, 34]. The white-bellied pangolin (Phataginus tricuspis) is the most abundant species of pangolin in Africa, with a widespread sub-Saharan distribution covering the African lowland rainforests [30]. It is one of the most frequent mammalian species observed on the stalls of the range countries’ bushmeat markets [35–38]. The species is also likely undergoing heavy international trade pressure as it has been the constituent of large seizures both in Africa and Asia since 2010 [39].
Recently, an unexpected level of cryptic diversity was revealed across the species range through the existence of six geographically traceable genetic lineages [17]. Although those results might constitute valuable support to delimitate the sources of traded pangolins at the regional scale, deeper resolution is needed to improve our ability to trace the trade at the local scale. We therefore developed and characterized 20 microsatellite loci from the genome of P. tricuspis, with the objective of providing a tool applicable at reasonable costs to range countries and international agencies that would be keen to implement the genetic tracing of the pangolin trade.
Materials and methods
A total of 37 samples of white-bellied pangolins were collected from the Douala market network. A first sample set included 24 samples from the Ebo forest (population ‘Ebo’), c. 75 km north-east of Douala, as ascertained by circumscribing the supply area that receives bushmeat from Ebo together with interviews with bushmeat sellers (data not shown). A second sample set included 13 samples likely originating from diverse geographic sources, possibly including Ebo (‘altEbo’), according to the same interviews. Samples consisted of fresh and smoked tissues (tongue or muscle) collected from dead animals sold on the market. Sampling was done, after explaining the aim of our study, with the initial agreement of the seller and following the guidelines of the Comité d'Ethique Institutionnel de la recherche pour la Santé humaine de l'Université de Douala (CEI-UDo). All the samples were preserved in 90% ethanol.
Genomic DNA was extracted with the NucleoSpin® Tissue Kit (MACHEREY–NAGEL, Hoerdt, France), following the manufacturer’s recommendations. The elution step was repeated twice in 50 μL BE to maximize DNA yield and concentration. DNA concentration was quantified using the NanoDrop 1000 Spectrophotometer (ThermoFisher Scientific). All the DNA extracts were stored at -20 °C.
Microsatellite markers were developed at Ecogenics, Balgach, Switzerland (https://www.ecogenics.ch/home.html). An Illumina TruSeq nano library was built from a single DNA extract, which was enriched for simple sequence repeat content using magnetic streptavidin beads and biotin‐labeled CT and GT repeat oligonucleotides. The library was sequenced on an Illumina MiSeq sequencing platform using a nano 2 (500 cycles) sequencing chip. The resulting paired-end reads which passed Illumina’s chastity filter were subject to de-multiplexing and trimming of Illumina adaptor residuals. Subsequently, the quality of the surviving reads was checked with the software FastQC 0.117 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc). In a next step, the paired-end reads were merged with the software USEARCH 10.0.240 [40] to reform in silico the sequenced molecule. The resulting reads were screened with the software Tandem Repeats Finder 4.09 [41]. After this process, 8,488 merged reads contained a microsatellite insert with a tetra- or a trinucleotide of at least six repeat units or a dinucleotide of at least 10 repeat units. Primer design was performed with Primer 3 4.1.0 [42]. Suitable primer design was possible in 6,025 microsatellite candidates. Individual tests for amplification and polymorphism of the 48 best primer-designed loci were performed on 14 individuals (from the western central African and Gabon lineages; see [17]), after which 20 polymorphic markers were retained (Table 1). Overall, eight loci were di-nucleotide, three tri-nucleotide and nine tetra-nucleotide repeats.
Table 1.
Characterization of 20 polymorphic microsatellite loci in the white-bellied pangolin (Phataginus tricuspis) from Cameroon
| Locus | Primer sequences (5′→3′) | Repeat Motif | Na | Size range—bp | Ho | He | AR | PHW | Genbank Acc. No | Dye | Multiplex |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PT_34432 |
F: AGATATGGCACCCCAATGTTTC R: AAGTGTATGCACCCCTTTGC |
(AC)21 | 9 | 107–141 | 0.667 | 0.608 | 8.498 | 0.950 | MT109130 | VIC | II |
| PT_276641 |
F: TGAGTCCTCCAGATCAGCAC R: GAACTCAGGGAAATGGTGGC |
(TGAG)9 | 5 | 182–198 | 0.542 | 0.549 | 4.75 | 0.397 | MT109129 | VIC | IV |
| PT_308752 |
F: CTGCAACGGGATTTCCCTAC R: TGTTTCCCCAGGGCTTAGAG |
(TTCC)16 | 8 | 144–208 | 0.625 | 0.784 | 7.749 | 0.391 | MT109128 | NED | II |
| PT_338821 |
F: AGTCAACGATGCAATAGGAAAC R: TGGCATCTTTCTAGGCGTGG |
(TG)22 | 9 | 232–254 | 0.571 | 0.836 | 9.000 | 0.000a | MT109127 | PET | III |
| PT_353755 |
F: GGTCACATGCTGAAGTGCTC R: TGCAATGAGGTAGGGACACG |
(TTA)9 | 8 | 206–242 | 0.75 | 0.76 | 7.847 | 0.021 | MT109126 | 6-FAM | IV |
| PT_378852 |
F: CCGCTGTCTTTCCAGAATGC R: GTCCCCACGGAGCTTACTAC |
(CCT)7 | 3 | 163–175 | 0.5 | 0.546 | 2.999 | 0.389 | MT109125 | 6-FAM | IV |
| PT_464918 |
F: TGGCAATACTGGTGTTTAGGAAC R: CTGTCATGGGCTAGTCACAC |
(AAT)10 | 8 | 110–131 | 0.667 | 0.755 | 7.847 | 0.428 | MT109124 | 6-FAM | II |
| PT_619913 |
F: AGCGGAAGGGTCACTAATGG R: TCTGTGCTGCTTCATAAAAGAGG |
(TGAA)8 | 7 | 202–226 | 0.5 | 0.637 | 6.737 | 0.728 | MT109123 | NED | III |
| PT_739516 |
F: ACATGTGGCATAGTCTGTTGG R: AGAGGCCTATGGGACAGTAAG |
(TTTG)8 | 5 | 226–246 | 0.417 | 0.545 | 4.750 | 0.691 | MT109122 | VIC | III |
| PT_796077 |
F: GGGACAATGCCAAAGGGTAG R: CAAACATTTGGGGCGAGTTG |
(TTTA)13 | 9 | 157–201 | 0.542 | 0.829 | 8.871 | 0.105 | MT109121 | VIC | I |
| PT_839522 |
F: CAGAGCTTGTACGCCATCAG R: CAGTGACAGTGTAATCCTGCG |
(AC)20 | 6 | 203–217 | 0.435 | 0.783 | 5.988 | 0.000a | MT109120 | PET | I |
| PT_1162028 |
F: CCTTTGTTATCCACCGACCC R: AGAACAGTGCCTTATGGAAGC |
(CATC)9 | 7 | 142–166 | 0.708 | 0.619 | 6.624 | 0.842 | MT109119 | 6-FAM | I |
| PT_1225378 |
F: GGGACTTGATAATAGGGGGTC R: AGCTGGGAATCTCAAACTGG |
(AATT)8 | 5 | 227–247 | 0.542 | 0.469 | 4.873 | 0.978 | MT109118 | 6-FAM | III |
| PT_1453906 |
F: GACCCCCATCTTTATGGAAGC R: AGTATGCCACTTGATGGATGG |
(ATCT)9 | 4 | 177–193 | 0.208 | 0.489 | 3.874 | 0.003 | MT109117 | 6-FAM | II |
| PT_1594892 |
F: AAAACGGAAGTGAAATTAAAGCC R: TGTTGTTCAAGGGCCAACTG |
(TG)12 | 6 | 202–218 | 0.458 | 0.624 | 5.999 | 0.012 | MT109116 | VIC | II |
| PT_1669238 |
F: ACCCACAGGATGTGCTACTC R: AGATTTCCCATCTACCCCGC |
(GT)14 | 8 | 178–204 | 0.667 | 0.715 | 7.623 | 0.780 | MT109115 | PET | II |
| PT_1753627 |
F: GTCCATCTACGCAACCCAAG R: GTTTAACCAGCTCCCACAATTTC |
(TG)16 | 7 | 234–252 | 0.75 | 0.782 | 6.848 | 0.797 | MT109114 | 6-FAM | I |
| PT_1849728 |
F: TCAAGGAACTTAGCTTCAGTTTG R: AGACTTTCGGTGTCATCCCC |
(TG)11 | 5 | 112–120 | 0.583 | 0.526 | 4.998 | 0.560 | MT109113 | 6-FAM | IV |
| PT_1973508 |
F: GGCTAATGTACAGTCTGATGTTCTC R: GGGCCCACGTGATTCTATGG |
(CA)23 | 12 | 221–249 | 0.833 | 0.827 | 11.36 | 0.962 | MT109111 | NED | I |
| PT_2019332 |
F: CAGTGTAACCAAAAAGAGTCTGC R: ACAGTTTTGGCACCTTCAATTTATC |
(ATCT)10 | 8 | 190–222 | 0.875 | 0.731 | 7.625 | 0.967 | MT109112 | PET | IV |
The number of individuals was 24 for all loci
Na number of alleles, Ho observed heterozygosity, He expected heterozygosity, AR allelic richness, and PHW P-values for tests of Hardy–Weinberg equilibrium deviation after Bonferroni correction
aLoci with significant departure from Hardy–Weinberg equilibrium (adjusted P-value = 0.0025)
We used MULTIPLEX MANAGER 1.2 [43] to optimize a design into four PCR multiplexes (4–6 loci) using four different ABI fluorescent dyes (Table 1). PCR amplifications for each multiplex were carried out in 20 μL reaction mixture containing approximately 20 ng of genomic DNA, 1 × Multiplex PCR Master Mix (QIAGEN Multiplex PCR Plus Kit; Qiagen, Courtaboeuf, France) and 0.2 μM of each primer pair. PCR thermoprofiles included an initial denaturation step (95 °C for 5 min), followed by 32 cycles of denaturation (95 °C for 30 s)–annealing (60 °C for 90 s)–elongation (72 °C for 30 s), and a final elongation step (60 °C for 30 min).
The PCR products were run on an ABI 3730 DNA Analyser (Thermo Fisher Scientific) at GeT-PlaGe (Génotoul, Institut National de Recherche Agronomique, Castanet-Tolosan, France; https://get.genotoul.fr/). Allele scoring and final extraction of genotypes were performed in Geneious 9.0.5 [44] with the Microsatellites plugin (https://www.geneious.com/features/microsatellite-genotyping/).
As a geographically coherent population, Ebo (N = 24) was used for the validation of our 20 microsatellite loci. We ran the detection of potential scoring errors and null alleles in MICROCHECKER 2.2.3 [45]. Deviations from the Hardy–Weinberg equilibrium were tested for each locus with GenAlEx 6.503 [46]. We used a permutation test under GENETIX 4.05.2 [47] to estimate linkage disequilibrium (LD) between each pair of loci (1000 permutations). The Bonferroni correction was applied to each of those statistical procedures.
The number of alleles per loci (Na) and the observed (Ho) and expected heterozygosities (He) were estimated in GenAlEx. Allelic richness (AR) was calculated in FSTAT 2.9.3.2 [48].
We used the whole sample set (Ebo + altEbo; N = 37) to conduct a Principal Coordinates Analysis (PCoA) in GenAlEx with pairwise population matrix unbiased genetic distances [49] in order to explore genetic variance among individuals sold in the Douala markets.
Values of unbiased probability of identity and probability of identity among siblings (uPI and PIsibs) were calculated in Gimlet 1.3.3 [50]. We used the Multilocus tagging option in GenAlex to detect identical genotypes in our dataset (sub-option ‘Matches’).
Results and discussion
The 20 loci were successfully amplified (with 100% PCR success rate), with only 0.84% missing data in our final multilocus dataset (Table 1). Null alleles were detected in five loci (PT_338821, PT_796077, PT_839522, PT_1453906, PT_1594892). Two of those loci significantly deviated from the Hardy–Weinberg equilibrium (Table 1). Two loci were subjected to linkage disequilibrium (PT_353755, PT_1225378). Deviation from Hardy–Weinberg equilibrium and linkage disequilibrium among loci can be due to genetic differentiation [51, 52], as was also suspected from loci developed for other pangolin species [24, 25]. Our results may suggest that we genotyped several populations from the Ebo forest, the latter covering > 1500 km2, although we cannot totally discard the possibility that the Ebo sample set was not as accurately circumscribed as expected from our interviews.
The mean number of alleles per locus was 6.95 (from 4 to 12 alleles) and mean AR was 6.74. Mean He was 0.671 (0.469–0.836) and mean Ho was 0.592 (0.208–0.875). The number of alleles and levels of genetic diversity were higher than in previous cross-amplification studies (mean number of alleles = 2.61–4; [24, 25]). Those results were expected given the reduced sample set (N = 2–10) used in the previous studies and the bias related to cross-amplification among phylogenetically divergent species of pangolins [18].
The 20 newly developed microsatellite loci represent the first markers isolated from the genome of P. tricuspis and may prove useful in tracing the local-to-global trade of the species. Each of the 37 individuals had a unique multilocus genotype. The unbiased probability of identity (uPI) and the probability of identity among siblings (PIsibs), that is, the probability that two individuals drawn at random from a population, including or not including siblings, will have the same genotype, were both very low (uPI = 8.443 e−21; PIsibs = 1.011 e−07; Fig. S1). Only five microsatellite loci were needed to reach the conservative value of PIsibs < 0.01 [53], overall indicating a powerful discriminating power of our combined loci. This could have important conservation implications, as notably regarding the individual-based management of in situ and ex situ populations [54, 55] and the accurate assessment of the number of individuals contained in large seizures of scales [29]. Although China has temporarily banned the wildlife trade, the durable effect and efficiency of law enforcement on this illegal market remains questionable [56] and accurate tools will be needed to trace the consumption of pangolins.
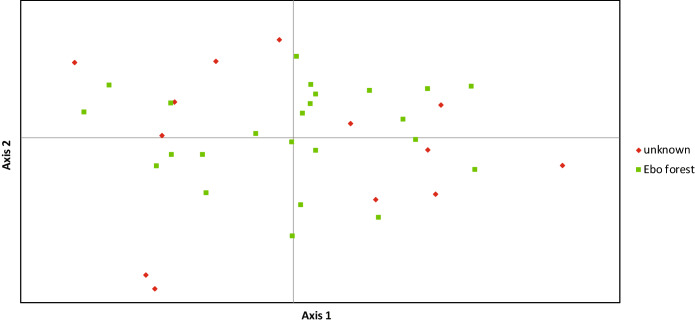
Genetic variance was increased through the addition of pangolins with unknown origin (altEbo), involving the potential presence of ‘outliers’ along the axes 1 and 2 of the PCoA (14.88% cumulated variation; Fig. 1), in agreement with the interviewees’ answers regarding the origin of pangolins. Although our results are preliminary, this may suggest that the Douala market network is fed from differentiated source populations of white-bellied pangolins, but this working hypothesis will have to be tested further with an accurate geographic representation of pangolin populations. The set of microsatellite loci that we have developed significantly adds to the molecular toolbox available for the species, and will hopefully contribute to the fine-scale implementation of future conservation strategies at reasonable costs. Because the previously published, cross-amplified loci were not validated on a sufficient number of geographically traceable samples, we recommend the use of our 20 newly developed microsatellite loci for population genetic studies on the white-bellied pangolin and related conservation and forensic applications.
Fig. 1.
Principal Coordinates Analysis (PCoA) based on microsatellite data in white-bellied pangolins from the Douala market originating from Ebo forest and unknown sources. Percentages of variance explained: axis 1 = 7.79%; axis 2 = 7.09%
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file1 Fig. S1. Unbiased probability of identity (uPI) and probability of identity among siblings (PIsibs) for increasing, optimized locus combinations. (PDF 16 kb)
Acknowledgements
We are grateful to the staff of the “Plateau technique – Biologie moléculaire et microbiologie” at EDB for assistance during lab work. SA and PG received support from the Agence Nationale de la Recherche (ANR-17-CE02-0001; PANGO-GO). ADD received a grant”séjours scientifiques de haut niveau” (No. 939537G) from the Service de Coopération et d’Action Culturelle (SCAC) de l’Ambassade de France au Cameroun. Research permit in Cameroon was issued by the Ministère de la Recherche Scientifique et de l’Innovation (000094/MINRESI/B00/C00/C10/nye). We thank two reviewers for their fruitful comments on an earlier version of the manuscript.
Author contributions
EL, ADM, MT and PG designed the study. SA and ADD did the lab work. SA, EL and PG ran the data analysis. ADD and ADM collected the genetic samples. All co-authors participated to the writing of the manuscript.
Funding
This study was funded by the Agence Nationale de la Recherche (ANR-17-CE02-0001; PANGO-GO).
Data availability
The microsatellite sequences were submitted to Genbank.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
No animal testing was performed during this study. This study received the approval of the "Comité d’Ethique Institutionnel de la recherche pour la santé humaine de l’Université de Douala (CEI-UDo)".
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mt S-R, Challender DWS, Hinsley A, Verissimo D, Milner-Gulland EJ. Illegal wildlife trade: scale, processes, and governance. Annu Rev Environ Resour. 2019;44:201–228. doi: 10.1146/annurev-environ-101718-033253. [DOI] [Google Scholar]
- 2.Liu Y, Weng Q. Fauna in decline: plight of the pangolin. Science. 2014;345:884–884. doi: 10.1126/science.345.6199.884-a. [DOI] [PubMed] [Google Scholar]
- 3.Challender DWS, Heinrich S, Shepherd CR, Katsis LKD. Chapter 16—International trade and trafficking in pangolins, 1900–2019. In: Challender DWS, Nash HC, Waterman C, editors. Pangolins. New York: Academic Press; 2020. pp. 259–276. [Google Scholar]
- 4.Cheng W, Xing S, Bonebrake TC. Recent pangolin seizures in China reveal priority areas for intervention: network analysis of China pangolin market. Conserv Lett. 2017;10:757–764. doi: 10.1111/conl.12339. [DOI] [Google Scholar]
- 5.Xiao K, Zhai J, Feng Y et al (2020) Isolation and characterization of 2019-nCoV-like Coronavirus from Malayan Pangolins. bioRxiv 10.1101/2020.02.17.951335
- 6.Lam TTY, Shum MHH, Zhu HC et al (2020) Identification of 2019-nCoV relation coronaviruses in Malayan pangolins in southern China. bioRxiv 10.1101/2020.02.13.945485
- 7.Soewu DA, Sodeinde OA. Utilization of pangolins in Africa: fuelling factors, diversity of uses and sustainability. Int J Biodivers Conserv. 2015;7:1–10. doi: 10.5897/IJBC2014.0760. [DOI] [Google Scholar]
- 8.Xing S, Bonebrake TC, Cheng W, et al. Chapter 14—Meat and medicine: historic and contemporary use in Asia. In: Challender DWS, Nash HC, Waterman C, et al., editors. Pangolins. New York: Academic Press; 2020. pp. 227–239. [Google Scholar]
- 9.Nasi R, Brown D, Wilkie D et al (2008) Conservation and use of wildlife-based resources: the bushmeat crisis. Secrétariat de la Convention sur la diversité biologique, Montréal et Centre pour la recherche forestière (CIFOR), Bogor, Indonesia. Technical Series 33, p 50
- 10.Mohapatra RK, Panda S, Nair MV, Acharjyo LN, Challender DWS. A note on the illegal trade and use of pangolin body parts in India. Traffic Bull. 2015;27:33–40. [Google Scholar]
- 11.Xu L, Guan J, Lau W, Xiao Y. An overview of pangolin trade in China. Traffic Brief Pap. 2016;2016:1–10. [Google Scholar]
- 12.Zhang MX, Gouveia A, Qin T, Quan R, Nijman V. Illegal pangolin trade in northernmost Myanmar and its links to India and China. Glob Ecol Conserv. 2017;10:23–31. doi: 10.1016/j.gecco.2017.01.006. [DOI] [Google Scholar]
- 13.Harrop SR. Chapter 17—Conserving pangolins through international and national regulation and effective law enforcement. In: Challender DWS, Nash HC, Waterman C, editors. Pangolins. New York: Academic Press; 2020. pp. 283–292. [Google Scholar]
- 14.Yu HT, Ma GC, Lee DJ, et al. Molecular delineation of the Y-borne Sry gene in the Formosan pangolin (Manis pentadactyla pentadactyla) and its phylogenetic implications for Pholidota in extant mammals. Theriogenology. 2011;75:55–64. doi: 10.1016/j.theriogenology.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Gaubert P, Antunes A. What’s behind these scales? Comments to “The complete mitochondrial genome of Temminck’s ground pangolin (Smutsia temminckii; Smuts, 1832) and phylogenetic position of the Pholidota (Weber, 1904)”. Gene. 2015;563:106–108. doi: 10.1016/j.gene.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Hassanin A, Hugot JP, Van Vuuren BJ. Comparison of mitochondrial genome sequences of pangolins (Mammalia, Pholidota) CR Biol. 2015;338:260–265. doi: 10.1016/j.crvi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Gaubert P, Njiokou F, Ngua G, et al. Phylogeography of the heavily poached African common pangolin (Pholidota, Manis tricuspis) reveals six cryptic lineages as traceable signatures of Pleistocene diversification. Mol Ecol. 2016;25:5975–5993. doi: 10.1111/mec.13886. [DOI] [PubMed] [Google Scholar]
- 18.Gaubert P, Antunes A, Meng H, et al. The complete phylogeny of pangolins: scaling up resources for the molecular tracing of the most trafficked mammals on Earth. J Hered. 2018;109:347–359. doi: 10.1093/jhered/esx097. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh HM, Lee JCI, Wu JH, et al. Establishing the pangolin mitochondrial D-loop sequences from the confiscated scales. Forensic Sci Int. 2011;5:303–307. doi: 10.1016/j.fsigen.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Miller MP, Yang F, et al. Molecular tracing of confiscated pangolin scales for conservation and illegal trade monitoring in Southeast Asia. Glob Ecol Conserv. 2015;4:414–422. doi: 10.1016/j.gecco.2015.08.002. [DOI] [Google Scholar]
- 21.Luczon AU, Ong PS, Quilang JP, Fontanilla IKC. Determining species identity from confiscated pangolin remains using DNA barcoding. Mitochondrial DNA Part B. 2016;1:763–766. doi: 10.1080/23802359.2016.1238752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar VP, Rajpoot A, Shkula M, et al. Illegal trade of Indian Pangolin (Manis crassicaudata): genetic study from scales based on mitochondrial genes. Egyptian J Forensic Sci. 2016;6:524–533. doi: 10.1016/j.ejfs.2016.06.008. [DOI] [Google Scholar]
- 23.Mwale M, Dalton DL, Jansen R, et al. Forensic application of DNA barcoding for identification of illegally traded African pangolin scales. Genome. 2017;60:272–284. doi: 10.1139/gen-2016-0144. [DOI] [PubMed] [Google Scholar]
- 24.Luo SJ, Cai QX, David VA, et al. Isolation and characterization of microsatellite markers in pangolins (Mammalia, Pholidota, Manis spp.) Mol Ecol Notes. 2007;7:269–272. doi: 10.1111/j.1471-8286.2006.01577.x. [DOI] [Google Scholar]
- 25.Du Toit Z, Dalton DL, Du Plessis M, et al. Isolation and characterization of 30 STRs in Temminck’s ground pangolin (Smutsia temminckii) and potential for cross amplification in other African species. J Genet. 2020;99(1):1–6. doi: 10.1007/s12041-019-1160-8. [DOI] [PubMed] [Google Scholar]
- 26.Luo SJ, Antunes A, Yang FF et al (2009) A pilot study of genetic surveillance of illegal pangolin trade in Asia. In: Pantel S, Chin SY (eds) Proceedings of the workshop on trade and conservation of Pangolins Native to South and Southeast Asia, 30 June–2 July 2008, Singapore Zoo, Singapore. TRAFFIC Southeast Asia, Petaling Jaya, Selangor, Malaysia, p 194
- 27.Saifei G, Dongmei Y, Ying W, Jianjun P. Application of microsatellite marking to individual identification for pangolins ( Manis spp.) Biotechnol Bull. 2010;12:32. [Google Scholar]
- 28.Nash HC, Low GW, Choo SW, et al. Conservation genomics reveals possible illegal trade routes and admixture across pangolin lineages in Southeast Asia. Conserv Genet. 2018;19:1083–1095. doi: 10.1007/s10592-018-1080-9. [DOI] [Google Scholar]
- 29.Ullmann T, Verissimo D, Challender D. Evaluating the application of scale frequency to estimate the size of pangolin scale seizures. Glob Ecol Conserv. 2019;20:e00776. doi: 10.1016/j.gecco.2019.e00776. [DOI] [Google Scholar]
- 30.Gaubert P. Family manidae (pangolins) In: Wilson DE, Mittermeier RA, editors. Handbook of the mammals of the world Hoofed mammals. Barcelona: Lynx Edicions; 2011. pp. 82–103. [Google Scholar]
- 31.Challender DWS, O’Criodain C. Chapter 19—addressing trade threats to pangolins in the convention on international trade in endangered species of Wild Fauna and Flora (CITES) In: Challender DWS, Nash HC, Waterman C, editors. Pangolins. New York: Academic Press; 2020. pp. 305–320. [Google Scholar]
- 32.Hoffmann R, Challender DWS. Chapter 33—conservation strategies and priority actions for pangolins. In: Challender DWS, Nash HC, Waterman C, editors. Pangolins. New York: Academic Press; 2020. pp. 531–535. [Google Scholar]
- 33.Challender DWS, Hywood L. African pangolins under increased pressure from poaching and intercontinental trade. Traffic Bull. 2012;24:53–55. [Google Scholar]
- 34.Lale-Demoz A, Lewi G (2013) Transnational organized crime in East Asia and the Pacific: a threat assessment United Nations Office on Drugs and Crime, Bangkok, Thailand
- 35.Sodeinde OA, Adedipe SR. Pangolins in south-west Nigeria—current status and prognosis. Oryx. 1994;28:43–50. doi: 10.1017/S0030605300028283. [DOI] [Google Scholar]
- 36.Fa JE, Seymour S, Dupain J, et al. Getting to grips with the magnitude of exploitation: bushmeat in the Cross-Sanaga rivers region, Nigeria and Cameroon. Biol Conserv. 2006;129:497–510. doi: 10.1016/j.biocon.2005.11.031. [DOI] [Google Scholar]
- 37.Boakye MK, Kotzé A, Dalton DL, Jansen R. Unravelling the pangolin bushmeat commodity chain and the extent of trade in Ghana. Hum Ecol. 2016;44:257–264. doi: 10.1007/s10745-016-9813-1. [DOI] [Google Scholar]
- 38.Ingram DJ, Coad L, Abernethy KA, et al. Assessing Africa-wide pangolin exploitation by scaling local data. Conserv Lett. 2017;11:e12389. doi: 10.1111/conl.12389. [DOI] [Google Scholar]
- 39.Jansen R, Sodeinde O, Soewu D, et al. Chapter 9—White-bellied pangolin Phataginus tricuspis (Rafinesque, 1820) In: Challender DWS, Nash HC, Waterman C, et al., editors. Pangolins. New York: Academic Press; 2020. pp. 139–156. [Google Scholar]
- 40.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 41.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Untergasser A, Cutcutache I, Koressaar T, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holleley C, Geerts PG. Multiplex Manager 1,0: a cross-platform computer program that plans and optimize multiplex PCR. Biotechniques. 2009;46(7):511. doi: 10.2144/000113156. [DOI] [PubMed] [Google Scholar]
- 44.Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- 46.Peakall R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996–2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier (France)
- 48.Goudet J. FSTAT, version 2.9.3. A program to estimate and test gene diversities and fixation indices. Lausanne: Lausanne University; 2001. [Google Scholar]
- 49.Nei M. Genetic distance between populations. Am Nat. 1972;106:283–291. doi: 10.1086/282771. [DOI] [Google Scholar]
- 50.Valière N. GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes. 2002;2:377–379. [Google Scholar]
- 51.Rosa AL, Lima-Rezende CA, Rodrigues FP, Caparroz R. Development and characterization of 19 polymorphic microsatellite loci from the Red-cowled Cardinal (Paroaria dominicana, Passeriformes, Aves) using next-generation sequencing. Mol Ecol Rep. 2019;46:5531–5536. doi: 10.1007/s11033-019-04919-z. [DOI] [PubMed] [Google Scholar]
- 52.Ostrowski MF, David J, Santoni S, et al. Evidence for a large-scale population structure among accessions of Arabidopsis thaliana: possible causes and consequences for the distribution of linkage disequilibrium. Mol Ecol. 2006;15:1507–1517. doi: 10.1111/j.1365-294X.2006.02865.x. [DOI] [PubMed] [Google Scholar]
- 53.Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol. 2001;10:249–256. doi: 10.1046/j.1365-294X.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- 54.Hua L, Gong S, Wang F, et al. Captive breeding of pangolins: current status, problems and future prospects. ZooKeys. 2015;507:99–114. doi: 10.3897/zookeys.507.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pietersen DW, Challender DWS. Chapter 34—research needs for pangolins. In: Challender DWS, Nash HC, Waterman C, editors. Pangolins. New York: Academic Press; 2020. pp. 537–543. [Google Scholar]
- 56.Zhou AM, Buesching CD, Macdonal DW, Newman C. China: clamp down on violations of wildlife trade ban. Nature. 2020;578:217. doi: 10.1038/d41586-020-00378-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Fig. S1. Unbiased probability of identity (uPI) and probability of identity among siblings (PIsibs) for increasing, optimized locus combinations. (PDF 16 kb)
Data Availability Statement
The microsatellite sequences were submitted to Genbank.