Abstract
Background:
Lead (Pb) is a ubiquitous environmental contaminant with an array of detrimental health effects in children and adults, including neurological and immune dysfunction. Emerging evidence suggests that Pb exposure may alter the composition of the gut microbiota, however few studies have examined this association in human populations. The purpose of this study was to examine the association between urinary Pb concentration and the composition of the adult gut microbiota in a population-based sample of adults.
Methods:
Data used in this study were collected as part of the Survey of the Health of Wisconsin (SHOW) and its ancillary microbiome study. The SHOW is a household-based health examination survey of Wisconsin residents, collecting a variety of survey data on health determinants and outcomes, as well as objective measurements of body habitus, and biological specimens including urine. The ancillary microbiome study added additional questions and biological specimen collection, including stool, from participants age 18 +. Pb concentration was analyzed in urine samples, and gut microbiota composition was assessed using DNA sequencing of the 16S rRNA V4 region, extracted from stool samples. Data processing and statistical analyses were performed in mothur, Python, R, and SAS.
Results:
Of 696 participants, urinary Pb concentration was highest in those age 70 +, females, those with a high school diploma or lower, current and former smokers, and those without indoor pets. In adjusted models, increasing urinary Pb levels were associated with increases in microbial a-diversity (p = 0.071) and richness (p = 0.005). Differences in microbial β-diversity were significantly associated (p = 0.003) with differences in urinary Pb level. Presence of Proteobacteria, including members of the Burkholderiales, was significantly associated with increased urinary Pb.
Conclusion:
These results suggest that Pb exposure is associated with differences in the composition of the adult gut microbiota in a population-based human sample. Further investigation of this association is warranted.
Keywords: Heavy metals, Lead, Microbiome, Microbiota, 16S rRNA, Epidemiology
1. Introduction
Xenobiotics, including lead (Pb) remain persistent public health problems worldwide. The bacteria present throughout the body, known as the microbiota, and their collective genomes, the microbiome, may play a role in mediating the relationship between human Pb exposure and its downstream health outcomes. The gut is a particularly rich site for bacterial colonization, and the gut microbiota play key functions in human metabolism and health (Cénit et al., 2014; Sekirov et al., 2010). The gut microbiota typically maintains a symbiotic relationship with its host, and when that system is in a state of imbalance, or dysbiosis, host health can be affected through changes in metabolic function (Le Chatelier et al., 2013) and the production of signaling molecules (Schirmer et al., 2016), as well as infection (Buffie et al., 2015). Gut dysbiosis has been associated with a range of acute and chronic diseases including infection (Lozupone et al., 2013), digestive conditions (Morgan et al., 2012), and mental health (Jiang et al., 2015). One facet of gut microbial balance is diversity, a combined measure of the number of species present and the abundance of individuals within each species (Hughes et al., 2001). Changes in microbial diversity are interpreted as changes in microbial composition and can be used as a signal of dysbiosis. Many factors affect the composition of the gut microbiota, and investigations of the complex interactions between environmental pollutants and the microbiome are only recently underway.
Pb has been a pervasive environmental contaminant for many years, with no known necessary biological function for humans and most bacteria (Jarosławiecka and Piotrowska-Seget, 2014). Pb has been causally linked to myriad detrimental health effects in both children and adults, including neurological disorders, kidney malfunction, anemia, and reduced immune function, and no “safe” threshold of Pb exposure exists (Abadin et al., 2007). Health effects are more prominent in children than adults, but immune effects are seen in adults even at low levels of exposure (Abadin et al., 2007; Dietert and Piepenbrink, 2006; Tong et al., 2000). Immune effects of Pb include a shift from Type 2 helper (Th2) cells to Th1 cells, and an increase in both interleukin 4 and IgE secretion, all of which reduce the immune system’s ability to fight infection, while increasing autoimmune tendencies (Dietert and Piepenbrink, 2006). In addition to the direct toxic effect of Pb on bacteria, these immune effects can alter the gut microbiome by reducing the body’s ability to maintain its balance, thus allowing greater potential for opportunistic species to become overly abundant.
Animal studies have found that both chronic and acute Pb exposure alter microbial diversity, composition of bacterial taxa, and metabolic function within the gut (Breton et al., 2013b; Gao et al., 2017; Wu et al., 2016; Xia et al., 2018a, 2018b; Zhai et al., 2017). Not only do gut microbial composition and function shift with Pb exposure in mice (Gao et al., 2017), but the gut microbiota also alter absorption of Pb into the blood stream by acting as a barrier to absorption, and by altering host gene expression of proteins involved in metal metabolism (Breton et al., 2013a). However, very little has been done to determine if these relationships translate to human populations. Two small studies in children have examined the relationship between Pb and the gut microbiome as exploratory analyses outside of their main study aims, with mixed results (Bisanz et al., 2014; Zhai et al., 2019). No study to date has examined the relationship between Pb exposure and the microbiota of adults with more typical levels of Pb exposure.
The objective of this study was to improve our understanding of the association between Pb exposure and the composition of the gut microbiota in an adult human population. Our goals were to examine associations between urinary Pb levels and gut microbial diversity in a sample of 696 adults recruited from the Survey of the Health of Wisconsin (SHOW) and its ancillary microbiome study. We hypothesize that increasing urine Pb levels would be associated with a significant decrease in a-diversity, and significant shifts in β-diversity within the gut microbiome, driven by differences in specific bacterial taxa.
2. Material and methods
2.1. Data source
Data and biological specimens used in this analysis came from the SHOW and its ancillary microbiome study, both described in previous publications (Eggers et al., 2018; Nieto et al., 2010). The SHOW is a yearly statewide health examination survey that collects a wide range of health exposure and outcome data addressing all major determinants of health including health care access, social determinants, lifestyle and behavioral factors. Modeled after the National Health and Nutrition Examination Survey (NHANES), the SHOW began in 2008, and uses a three tier clustered randomization scheme to select participants from around the state of Wisconsin. The SHOW collects survey data, as well as objective measures of body habitus and biological specimens including urine, plasma, and serum. In 2016 the microbiome ancillary study was added to the SHOW protocol, recruiting participants age 18 and older (Eggers et al., 2018). Participants completed the standard SHOW components plus additional survey components, in addition to submitting swabs of the skin, nose, and mouth, and samples of saliva and stool. The analysis for the current study uses survey data, urine samples, and stool samples collected by the SHOW and the microbiome study in 2016 and 2017. The SHOW protocol and the protocol for this study have both been approved by the University of Wisconsin Institutional Review Board, and all participants completed written consent to participation.
2.2. Variables
The main predictor variable was creatinine-adjusted urinary Pb concentration (original measurement in μg/L, after creatinine adjustment units are pg/L) as a measure of Pb exposure (Sakai, 2000). Urinary Pb measurement was used in this study because whole blood samples, the typical matrix for Pb exposure measurement, were not available for the majority of study participants. Urinary Pb concentration was measured using inductively coupled plasma mass spectrometry. One value was below the limit of detection (LOD), and was replaced with the LOD/√2. Pb exposure was adjusted for urinary creatinine by standardizing the units and dividing Pb by creatinine to account for variation in urinary output and kidney function. For descriptive tables, creatinine-adjusted urinary Pb was then categorized by quartiles. For regression analyses, creatinine-adjusted Pb was log transformed for normality. Whole blood Pb was also measured in a subset of 110 individuals from whom samples were collected in order to examine correlations between urinary and blood Pb levels.
The main outcome variables include gut microbial α-diversity, measured using the inverse-Simpson index (Simpson, 1949), richness, measured using the abundance-based coverage estimator (ACE) (Gotelli and Colwell, 2001), and β-diversity measured using the Bray-Curtis dissimilarity index (Bray and Curtis, 1957), as well as taxonomic units from phylum to genus levels. Richness is an estimate of the number of different species, or in this case operational taxonomic units (OTUs), present within the gut of an individual, and α-diversity is a combination of richness and evenness of abundance among the OTUs present. Both measures can be interpreted as higher estimates representing a richer or more diverse gut microbiota. The β-diversity measurement is a distance matrix comparing the similarity (or dissimilarity in the case of Bray-Curtis) of the OTU composition of each sample to each other. Samples that are more distant from each other are more different in composition from samples that are less distant.
Variables that were hypothesized a priori as potential confounders were examined using a directed acyclic graph (DAG) and eligible for inclusion in statistical models. Demographic variables include age, gender, income, poverty status, and race-ethnicity. Income was operationalized using self-reported total household income, which was then calculated as a percentage of the Federal Poverty Level (FPL) based on the Health and Human Services guidelines for the number of people in the household. Race and ethnicity were self-reported and then collapsed into four categories of non-Hispanic White, non-Hispanic Black, Hispanic, non-Hispanic other.
Other potential confounding factors include behavioral variables such as smoking, antibiotic use in the last year, and dietary components. Diet was analyzed using the Diet History Questionnaire (DHQ-II), which asks about usual diet consumption over the last year (Diet History Questionnaire DHQ-II and, 2019). Dietary components were included as consumption per 1000 Kcals consumed. Antibiotic use in the last year was self-reported as either yes or no. Although it was unlikely that antibiotic use was directly associated with Pb exposure, it is an important variable due to its strong effects on microbial composition. Since the gut microbiome can return to a normal state in less than a year following antibiotics, we did not exclude these participants and, instead examined history of antibiotic use an important covariate.
Additional physiological and environmental factors were also considered as covariates, including indoor pet ownership, body mass index (BMI), urbanicity, and length of residence in current home. BMI was calculated based on measured height and weight, using the equation: weight (kg)/height(cm)^2. BMI was categorized as follows for some components of the analysis: underweight (< 18.5), normal weight (18.5–24.9), overweight (25–29.5), and obese (≥30). Urbanicity was defined using the Rural-Urban Commuting Area (RUCA) Codes.
2.3. Microbiota analysis
Genomic DNA was extracted from stool samples as described previously (Eggers et al., 2018). Briefly, chemical, heat, and mechanical disruption were used to lyse the bacterial cells. DNA was purified by phenol-chloroform-isoamyl alcohol extraction, and further purified using NucleoSpin Gel and PCR clean-up kit (Mcherey-Nagel, Germany). Purified DNA was quantified using PicoGreen in a microplate reader. The 16S rRNA V4 region of the extracted DNA was barcoded and amplified using custom PCR primers following the protocol by Kozich et al. (Kozich et al., 2013).The PCR reaction consisted of 5 μL (25 ng) sample DNA, 0.5 μL (10 μM) of each primer, 12.5 μL of 2× KAPA Hotstart Ready Mix (Kapa Biosystems, Wilmington, MA, United States), and water to 25 μL total volume. Amplification conditions were 95 °C for min, 25 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 5 min. After PCR, samples were run through 1.0% low melt agarose gel (National Diagnostics, Atlanta, GA) electrophoresis to further remove unwanted DNA. Bands of the correct length were then extracted using Zymo Gel DNA Recovery Kit (Zymo Research, Irvine, CA, United States). Once the gel was removed, samples were quantified by Qubit® Fluorometer (Invitrogen, San Diego, CA, United States) and pooled to 4 nM. A DNA sequencing control of 10% PhiX was added to the aliquot and sequenced on an Illumina MiSeq sequencer using a MiSeq v2 (2×250bp) Reagent Kit (Illumina, Inc., San Diego, CA) per manufacturer’s instructions. The samples for this analysis were sequenced in three runs of equal size. To avoid sequencing batch effects, samples collected in 2016 were stratified by urinary Pb concentration, age, and gender and randomized in a 1:1 ratio to each of two plates. The samples collected in 2017 were run on a third plate with no randomization.
Raw sequencing data was processed using mothur v. 1.39 (Kozich et al., 2013) using the Standard Operating Procedure for MiSeq data (MiSeq SOP, n.d.). Briefly, contigs (overlapping sequences) were aligned using the SILVA v.132 16S rRNA gene reference database (Pruesse et al., 2007), and low quality reads were removed. Sequences of the wrong length were removed, and chimeras were detected and removed using UCHIME (Edgar et al., 2011), Sequences were assigned to operational taxonomic units (OTUs) at the species level (97% similarity) using the GreenGenes database v. gg_13_8_99 (DeSantis et al., 2006). Coverage was assessed by Good’s index, as calculated in mothur. OTU counts were normalized to 10,000 sequences per sample. Normalized OTU counts were used for α-diversity and richness calculations, which were performed in R. Additional data processing was done using Python v.3.7.2, SAS v. 9.4, and R v 3.5.2.
2.4. Statistical analysis
Statistical analysis was performed in SAS and R. Frequency tables were calculated for all potential confounders by creatinine-adjusted Pb quartiles. Univariate analyses including P-values based on χ2 are shown for categorical variables, and p for trend is shown for continuous variables to test for predictors of Pb exposure. Univariate analysis of all potential confounding variables with ACE and inverse-Simpson as the outcome was also conducted to determine level of association for inclusion in regression models (results not shown). Linear regression of log creatinine-adjusted urinary Pb was performed for inverse-Simpson and ACE to determine if increased Pb exposure was associated with α-diversity and richness. For each analysis, univariate regression estimates are shown as well as a fully adjusted model, and models stratified by relevant effect modifiers. Linear regression models were adjusted for clustering by household. The fully adjusted models were adjusted for age, gender, BMI, antibiotic use, race/ethnicity, education, smoking, fiber consumption (g/1000 Kcal), urbanicity, and indoor pet. Variables used to stratify models examining effect modification were age, smoking, BMI, urbanicity, and fiber consumption. P-values ≤0.05 were considered statistically significant.
The β-diversity distances (Bray-Curtis) were calculated in R using the vegan package (Oksanen et al., 2016). Permutational Analysis of Variance (PERMANOVA) was then used to estimate associations between log creatinine-adjusted urinary Pb and β-diversity. Adjusted PERMANOVA models included the same covariates as the adjusted linear regression models.
To examine specific taxa that contributed to differences in β-diversity, the Quasi-Conditional Association Test using General Estimating Equations (QCAT-GEE) in the miLineage R package was used, which includes a Benjamini-Hochberg correction for multiple comparisons (FDR) (Tang et al., 2017). The QCAT-GEE has three parts: the zero-part, which assesses differences in presence/absence of each taxa, the positive-part, which assesses differences in abundance of each taxa, and the two-part, which combines the zero and positive-parts. This test is adjusted for the same covariates as the adjusted linear models of a-diversity and richness, and outputs the names of taxa that are significantly different by Pb level after adjustment for covariates and multiple comparisons. To determine direction of association, logistic regression was used for taxa significant in the zero-part, and linear regression was used for taxa significant in the positive-part.
As a sensitivity analysis, and to explore different biological matrices of exposures, correlation statistics were run to compare the creatinine-adjusted urinary Pb measurement to more traditional whole blood Pb measurements in a subset of 110 participants.
3. Results
3.1. Population characteristics and Pb exposure
The study population was age 18 and over, with 48.7% between the ages of 50 and 69, 57.3% female, and 83.0% Non-Hispanic White. The population was relatively evenly distributed between low, middle, and high income categories, with 26.9% of participants receiving a high school diploma or less. 56.5% of participants have never been smokers, 34.9% have used antibiotics in the past year, 54.7% own an indoor pet, and 76.1% were overweight or obese. Participants resided in urban (66.2%), suburban (10.9%), and rural areas (22.9%), with 54.7% of participants living in their homes for > 10 years. The geometric mean urinary Pb for all samples was 0.30 μg/L (SE 1.04). Creatinine-adjusted urinary Pb concentration was found to increase with age, was higher in women, among those with a high school diploma or less, current or former smokers, those who do not own an indoor pet, and those who have been living in their current residence for > 10 years (Table 1). Increasing dietary fiber intake was also significantly associated with increasing urinary Pb concentration (Table 1). In our sensitivity analysis, creatinine-adjusted urinary Pb and blood Pb were strongly correlated (Pearson = 0.621, p < 0.0001).
Table 1.
Distribution of demographics and potential covariates by creatinine-adjusted urinary Pb quartile.
| Total | Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | |
|---|---|---|---|---|---|
|
| |||||
| Exposure | N | Range (N) | Range (N) | Range (N) | Range (N) |
| Creatinine-adjusted urinary Pb (pg/L) | 696 | 0.03–0.24 (174) | 0.24–0.39 (174) | 0.39–0.60 (174) | 0.60–15.31 (174) |
| Categorical covariates | N | n (%) | n (%) | n (%) | n (%) |
| Age§ | 696 | ||||
| 18–29 | 58 | 36 (62.1) | 19 (32.6) | 2 (3.4) | 1 (1.7) |
| 30–49 | 172 | 84 (48.8) | 49 (28.4) | 19 (11.0) | 20 (11.6) |
| 50–69 | 339 | 49 (14.5) | 78 (23) | 115 (33.9) | 97 (28.6) |
| ≥ 70 | 127 | 5 (3.9) | 28 (22) | 38 (29.9) | 56 (44.1) |
| Gender† | 696 | ||||
| Female | 399 | 87 (21.8) | 90 (22.6) | 106 (26.6) | 116 (29.1) |
| Male | 297 | 87 (29.3) | 84 (28.3) | 68 (22.9) | 58 (19.5) |
| Race/ethnicity | 695 | ||||
| Non-Hispanic White | 577 | 138 (23.9) | 145 (25.1) | 153 (26.5) | 141 (24.4) |
| Non-Hispanic Black | 70 | 21 (30.0) | 20 (28.6) | 14 (20.0) | 15 (21.4) |
| Hispanic | 24 | 9 (37.5) | 6 (25.0) | 3 (12.5) | 6 (25.0) |
| Non-Hispanic other | 24 | 5 (20.8) | 3 (12.5) | 4 (16.7) | 12 (50.0) |
| Family income | 696 | ||||
| Low income | 204 | 49 (24.0) | 52 (25.5) | 47 (23.0) | 56 (27.5) |
| Middle income | 220 | 62 (28.2) | 53 (24.1) | 51 (23.2) | 54 (24.5) |
| High income | 272 | 63 (23.2) | 69 (25.4) | 76 (27.9) | 64 (23.5) |
| Education* | 695 | ||||
| ≤ High School | 187 | 34 (18.2) | 49 (26.2) | 47 (25.1) | 57 (30.5) |
| Some college | 247 | 56 (22.7) | 65 (26.3) | 64 (25.9) | 62 (25.1) |
| ≥ Bachelor’s degree | 261 | 84 (32.2) | 60 (23.0) | 63 (24.1) | 54 (20.7) |
| Smoking§ | 685 | ||||
| Current | 93 | 14 (15.1) | 30 (32.3) | 21 (22.6) | 28 (30.1) |
| Former | 205 | 33 (16.1) | 51 (24.9) | 59 (28.8) | 62 (30.2) |
| Never | 387 | 123 (31.8) | 93 (24.0) | 91 (23.5) | 80 (20.7) |
| Antibiotic use | 653 | ||||
| Yes | 228 | 58 (25.4) | 58 (25.4) | 51 (22.4) | 61 (26.8) |
| No | 425 | 104 (24.5) | 109 (25.6) | 117 (27.5) | 95 (22.4) |
| Indoor pet† | 693 | ||||
| Yes | 379 | 115 (30.3) | 97 (25.6) | 84 (22.2) | 83 (21.9) |
| No | 314 | 58 (18.5) | 77 (24.5) | 90 (28.7) | 89 (28.3) |
| BMI | 690 | ||||
| Underweight/normal | 165 | 40 (24.2) | 38 (23.0) | 42 (25.5) | 45 (27.3) |
| Overweight/obese | 525 | 133 (25.3) | 136 (25.9) | 131 (25.0) | 125 (23.8) |
| Urbanicity | 695 | ||||
| Urban | 460 | 119 (25.9) | 124 (27.0) | 109 (23.7) | 108 (23.5) |
| Suburban | 76 | 23 (30.3) | 15 (19.7) | 20 (26.3) | 18 (23.7) |
| Rural | 159 | 32 (20.1) | 34 (21.4) | 45 (28.3) | 48 (30.2) |
| Length of residence§ | 688 | ||||
| < 1 year | 62 | 23 (37.1) | 19 (30.6) | 12 (19.5) | 8 (12.9) |
| 1–3 years | 105 | 35 (33.3) | 31 (29.5) | 20 (19.0) | 19 (18.1) |
| 3–10years | 145 | 50 (34.5) | 42 (29.0) | 27 (18.6) | 26 (17.9) |
| > 10years | 376 | 64 (17.0) | 82 (21.8) | 112 (29.8) | 118 (31.4) |
| Continuous covariates | N | Mean ± SE | Mean ± SE | Mean ± SE | Mean ± SE |
| Dietary iron (mg/1000 Kcal) | 615 | 7.6 ± 0.24 | 7.3 ± 0.17 | 7.9 ± 0.2 | 7.6 ± 0.18 |
| Dietary calcium (mg/1000 Kcal) | 615 | 719.4 ± 42.56 | 732.8 ± 32.94 | 771.5 ± 39.57 | 799.2 ± 43.84 |
| Dietary fiber* (g/1000 Kcal) | 615 | 9.5 ± 0.3 | 10.2 ± 0.36 | 12.3 ± 0.41 | 11.9 ± 0.41 |
| Dietary vitamin C (mg/1000 Kcal) | 615 | 52 ± 2.81 | 59 ± 2.94 | 66.3 ± 3.34 | 64.2 ± 3.17 |
Data come from the microbiome study sample of the Survey of the Health of Wisconsin 2016–2017. Urinary Pb geometric mean statistics are not adjusted for creatinine concentration or household clustering. Categorical distribution statistics calculated using frequency tables adjusted for household clustering, including p-values from the χ2 statistic. Continuous covariate statistics calculated including adjustment for household clustering, including p for trend.
P≤0.05
P≤0.01.
P≤0.0001.
3.2. Gut microbial composition
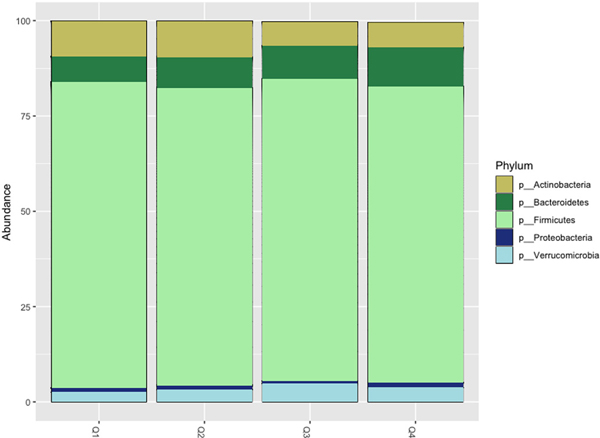
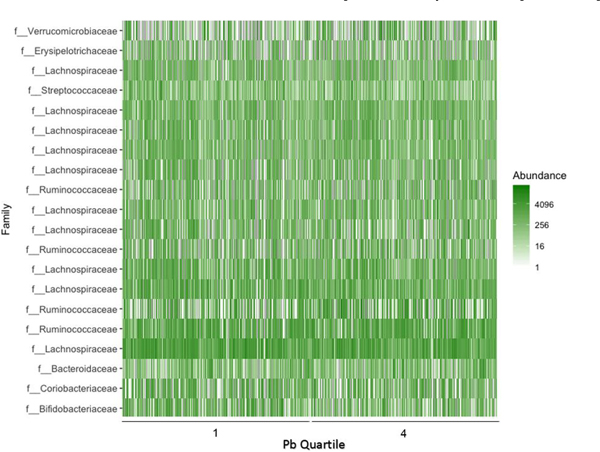
The number of high quality, processed sequences per sample ranged from 10,599–589,443 with an average of 32,765. Sequencing produced Goods coverage of 99.2% or higher for all samples, indicating that < 1% of sequencing reads in each sample are from OTUs that only appear once. Firmicutes were the most prevalent bacterial phylum across all four creatinine-adjusted urinary Pb quartiles, followed by the Bacteriodetes and Actinobacteria (Fig. 1). A heat map comparing individuals from the first and fourth quartiles of Pb exposure demonstrates the wide variability in microbial composition even within Pb exposure groups (Fig. 2).
Fig. 1.
Gut Bacterial Phyla by Urinary Pb Quartile.
Relative abundance of the five most abundant bacterial phyla found in the study samples by creatinine-adjusted urinary Pb quartiles.
Fig. 2.
Most Abundant OTUs by Urinary Pb Quartile.
Abundance of the top 20 most abundant OTUs in participants from the first and fourth quartiles of creatinine-adjusted urinary Pb level.
3.3. Pb and α-diversity, richness
Inverse-Simpson values ranged from 1.0 to 44.9, with a mean of 14.3 In models of α-diversity (inverse-Simpson) the main effect of log creatinine-adjusted urinary Pb was a slight increase in diversity in both the unadjusted (p = 0.023) and adjusted (p = 0.071) models (Table 2). Other factors significantly associated with greater α-diversity were more education, suburban residence, and not owning an indoor pet. Stratified analysis (Table 3) showed that the positive association between Pb and α-diversity was strongest for those age 50 and above (p = 0.04), former smokers (p = 0.008), and suburban (p = 0.002) and rural (p = 0.02) residents, compared to urban residents.
Table 2.
Effect of Urinary Lead concentration on α-diversity (Inverse-Simpson) and richness (ACE).
| Outcome: | Inverse-Simpson | ACE | ||
|---|---|---|---|---|
|
|
|
|||
| Unadjusted | Adjusted | Unadjusted | Adjusted | |
|
| ||||
| Variable | β (95% CI) | β (95% CI) | β (95% CI) | β (95% CI) |
|
| ||||
| Urine Pba | 0.82 (0.12, 1.53)* | 0.93 (−0.08, 1.93) | 5.11 (14.67, 34.76)§ | 17.43 (5.23, 29.63)† |
| Age | 0.03 (−0.01, 0.07) | 0.39 (−0.2, 0.97) | ||
| Gender (Female) | −1.00 (−2.1, 0.11) | 4.37 (−10.88, 19.61) | ||
| BMI | −0.05 (−0.13, 0.04) | −1.59 (−2.71, −0.46)† | ||
| Antibiotics (Yes) | −0.89 (−2.04, 0.26) | −22.55 (−39.8, −5.30)* | ||
| Race/ethnicity | ||||
| Non-Hispanic White | Reference | Reference | ||
| Non-Hispanic Black | 0.80 (−0.91, 2.52) | 34.31 (3.51, 65.11)* | ||
| Hispanic | 1.72 (−1.14, 4.58) | 24.6 (−20.53, 69.72) | ||
| Non-Hispanic other | −0.58 (−3.27, 2.11) | −19.23 (−58.15, 19.70) | ||
| Education | ||||
| ≤ High School | Reference | Reference | ||
| Some college | 2.21 (0.95, 3.46)‡ | 14.58 (−5.56, 34.72) | ||
| ≥ Bachelor’s Degree | 2.75 (1.39, 4.11)§ | 21.57 (0.02, 43.13)* | ||
| Smoking | ||||
| Never | Reference | Reference | ||
| Current | −1.37 (−2.98, 0.25) | −11.87 (−38.93, 15.19) | ||
| Former | −0.39 (−1.67, 0.9) | 2.93 (−17.03, 22.90) | ||
| Fiber (g/1000 Kcal) | 0.07 (−0.06, 0.2) | 2.67 (0.64, 4.7)† | ||
| Urbanicity | ||||
| Urban | Reference | Reference | ||
| Suburban | 1.79 (0.07, 3.5)* | 17.38 (−3.80, 38.56) | ||
| Rural | 0.52 (−1.06, 2.1) | 10.52 (−12.43, 33.46) | ||
| Indoor pet (Yes) | −1.70 (−2.84, −0.55)† | −10.41 (−27.37, 6.56) | ||
Data come from the microbiome study sample of the Survey of the Health of Wisconsin 2016–2017.
Abbreviations: Pb = lead; ACE = abundance-based coverage estimator; BMI = body mass index.
Results are shown from linear regression models, unadjusted and adjusted for the covariates shown above.
Creatinine-adjusted and log transformed.
P≤0.05.
P≤0.01.
P≤0.001.
P≤0.0001
Table 3.
Association of urinary lead with α-diversity (Inverse-Simpson), richness (ACE), stratified by effect modifiers.
| Outcome: | Inverse-Simpson | ACE |
|---|---|---|
|
| ||
| Stratification variable | Urine Pba β (9S% CI) | Urine Pba β (9S% CI) |
| Age | ||
| < 50 | 0.31 (−1.18, 1.80) | −2.5 (−26.1, 21.1) |
| ≥50 | 1.33 (0.03, 2.63)* | 25.0 (10.5, 39.6)‡ |
| Smoking | ||
| Current | 1.42 (−0.44, 3.29) | −16.6 (− 58.0, 24.8) |
| Former | 3.19 (0.83, 5.54)† | 47.8 (17.6, 78.1)† |
| Never | 0.34 (−0.85, 1.52) | 16.2 (2.5, 29.9)* |
| BMI | ||
| Underweight/normal | 0.48 (−1.27, 2.23) | 19.1 ( –0.7, 39.0) |
| Overweight/obese | 1.07 (−0.03, 2.18) | 18.4 (3.4, 33.3)* |
| Urbanicity | ||
| Urban | −0.29 (−1.40, 0.81) | 8.5 (−7.4, 24.5) |
| Suburban | 2.66 (0.95, 4.38)† | 26.8 (8.8, 44.8)† |
| Rural | 1.10 (0.44, 4.76)* | 31.3 (2.6, 59.9)* |
| Fiber consumption | ||
| Tertile 1 | 0.58 (−1.19, 2.35) | 5.5 (−20.1, 31.1) |
| Tertile 2 | 0.73 (−0.67, 2.13) | 26.4 (10.0, 42.7)† |
| Tertile 3 | 0.86 (−0.99, 2.71) | 7.2 (−17.1, 31.4) |
| Antibiotic use | ||
| Yes | 0.92 (−0.68, 2.51) | 12.7 (−6.7, 32.0) |
| No | 0.75 (−0.42, 1.92) | 19.6 (4.5, 34.7)* |
Data come from the microbiome study sample of the Survey of the Health of Wisconsin 2016–2017.
Abbreviations: Pb=lead; ACE=abundance-based coverage estimator; BMI=body mass index.
Linear regression estimates and confidence intervals from separate stratified analysis models for each variable. All models were adjusted by age, gender, BMI, race/ethnicity, education, smoking, fiber, urbanicity, and pets, EXCEPT for the stratification variable.
P-values shown are testing whether the adjusted regression estimate for urinary Pb in each subgroup analysis is significantly different than 0.
Creatinine-adjusted and log transformed.
P ≤ 0.05.
P ≤ 0.01.
P≤ 0.001.
ACE richness estimates ranged from 26.1 to 654.8, with a mean of 265.6. Models of richness (ACE) showed a positive effect of increased urinary Pb levels, although effect size varied between models, and was significant in both the unadjusted (p < 0.0001) and adjusted (p = 0.005) models (Table 2). Other significant predictors of increased richness were Non-Hispanic Black race compared to Non-Hispanic White race, completing a bachelor’s degree or higher education, and increasing dietary fiber consumption. Significant predictors of reduced richness were increasing BMI and antibiotic use. Stratified analysis (Table 3) demonstrated stronger positive association between urinary Pb levels and richness for those age 50 and above (p = 0.0008), former (p = 0.008) and never smokers (p = 0.02), suburban (p = 0.004) and rural (p = 0.03) residents, those in the second tertile of fiber consumption (p = 0.002), and those who have not used antibiotics in the past year (p = 0.01).
3.4. Pb and β-diversity
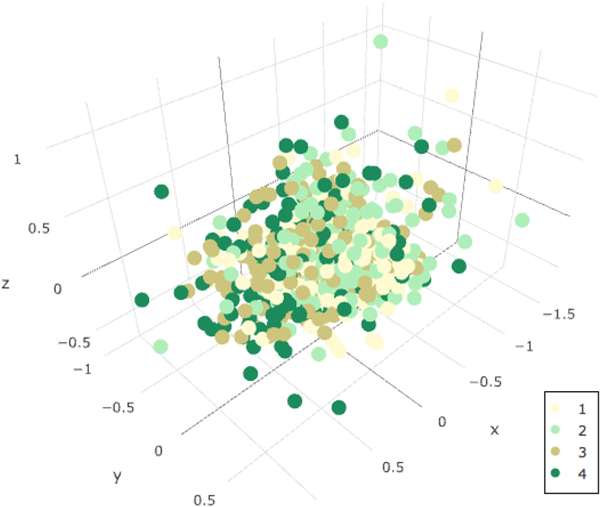
The β-diversity distance (Bray-Curtis) was calculated and displayed by quartiles of creatinine-adjusted urine Pb level in a non-metric multidimensional scaling (NMDS) plot in Fig. 3. Although there is not clear visual separation of groups within the study population in Fig. 3, the PERMANOVA analysis indicated small but significant differences in β-diversity by level of log creatinine-adjusted urine Pb level (Table 4) in both the unadjusted and adjusted models. Other factors significantly associated with changes in β-diversity in the sample were age, gender, BMI, antibiotic use, identifying as Black, having a bachelor’s degree or higher, being a current smoker, fiber intake, and living in a rural area.
Fig. 3.
β-Diversity distance by Urinary Pb Quartile.
Bray-Curtis dissimilarity distances, colored by quartile of creatinine-adjusted urinary Pb level. Distance between dots represents the difference in OTU composition between samples.
Table 4.
Association of urinary lead concentration with β-diversity (Bray-Curtis), unadjusted and adjusted for covariates.
| Outcome: | Bray-Curtis | |
|---|---|---|
|
| ||
| Unadjusted | Adjusted | |
|
| ||
| Variable | R2% (P) | R2% (P) |
| Urine Pba | 0.60 (0.001) | 0.35 (0.003) |
| Age | 1.03 (0.001) | |
| Gender (Female) | 0.49 (0.001) | |
| BMI | 0.35 (0.006) | |
| Antibiotics | ||
| Yes | 0.40 (0.003) | |
| Don’t know | 0.31 (0.020) | |
| No | Reference | |
| Race/ethnicity | ||
| Non-Hispanic White | Reference | |
| Non-Hispanic Black | 0.46 (0.001) | |
| Hispanic | 0.21 (0.177) | |
| Non-Hispanic other | 0.16 (0.495) | |
| Education | ||
| ≤ High School | Reference | |
| Some college | 0.19 (0.260) | |
| ≥ Bachelor’s Degree | 0.36 (0.005) | |
| Smoking | ||
| Never | Reference | |
| Current | 0.33 (0.010) | |
| Former | 0.23 (0.103) | |
| Fiber (g/1000 Kcal) | 0.74 (0.001) | |
| Urbanicity | ||
| Urban | Reference | |
| Suburban | 0.19 (0.305) | |
| Rural | 0.39 (0.001) | |
| Indoor pet (Yes) | 0.19 (0.279) | |
Data come from the microbiome study sample of the Survey of the Health of Wisconsin 2016–2017.
Abbreviations: Pb=lead; BMI=body mass index.
Results shown from PERMANOVA models, unadjusted and adjusted for covariates shown above.
Creatinine-adjusted and log transformed.
3.5. Pb and bacterial taxa
Several bacterial taxa were found to be significantly different according to levels of log creatinine-adjusted urine Pb level before correction for multiple comparisons (Table 5). After FDR correction, only taxa within the phylum Proteobacteria and the order Burkholderiales remained significant at the < 0.05 level. The odds of Proteobacteria colonization with increasing log creatinine-adjusted urinary Pb were 2.36 (95% CI = 1.25–4.44) unadjusted, and 2.49 (95% CI = 1.05–5.87) when adjusted for confounders. The odds of Burkholderiales colonization with increasing Pb were 1.47 (95% CI = 1.13–1.90) unadjusted, and 1.98 (95% CI = 1.39–2.82) when adjusted for age, gender, body mass index, antibiotic use, race/ethnicity, education, smoking, fiber consumption, urbanicity, and indoor pet ownership.
Table 5.
Specific bacterial taxa associated with level of urinary Pb concentration, adjusted for covariates.
| Uncorrected P-value | Direction | |||
|---|---|---|---|---|
|
|
||||
| Two-part | Zero-part | Positive-part | ||
|
| ||||
| Phylum | ||||
| Proteobacteria | 0.0236 | 0.0006* | 0.9620 | ↑ |
| Class | ||||
| Alphaproteobacteria | 0.6436 | 0.0198 | 0.6436 | ↑ |
| Betaproteobacteria | 0.5050 | 0.0099 | 0.5050 | ↑ |
| Deltaproteobacteria | 0.0119 | 0.0119 | 1.0000 | ↑ |
| Gammaproteobacteria | 0.0317 | 0.7030 | 0.0307 | ↑ |
| Order | ||||
| Burkholderiales | 0.0044 | 0.0002* | 0.7822 | ↑ |
| Clostridiales | 0.0008 | 0.1582 | 0.0008 | ↓ |
| Desulfovibrionales | 0.0921 | 0.0277 | 0.8208 | ↑ |
| Rhizobiales | 0.1683 | 0.0099 | 0.1683 | ↑ |
| Family | ||||
| Alcaligenaceae | 0.4356 | 0.0099 | 0.4356 | ↑ |
| Barnesiellaceae | 0.2574 | 0.0495 | 0.2574 | ↑ |
| Brucellaceae | 0.0248 | 0.0248 | 1.0000 | ↑ |
| Clostridiaceae | 0.0218 | 0.0406 | 0.1416 | ↓ |
| Desulfovibrionaceae | 0.0644 | 0.0178 | 0.4772 | ↑ |
| Enterococcaceae | 0.0129 | 0.9307 | 0.0129 | ↑ |
| Lactobacillaceae | 0.1881 | 0.0396 | 0.5446 | ↓ |
| Oxalobacteraceae | 0.3267 | 0.0495 | 0.3267 | ↑ |
| Rikenellaceae | 0.0584 | 0.0208 | 1.0000 | ↑ |
| Genus | ||||
| Clostridium | 0.0455 | 0.7406 | 0.0455 | ↓ |
| Coprococcus | 0.0238 | 0.0099 | 0.3178 | ↓ |
| Desulfovibrio | 0.8119 | 0.0396 | 0.8119 | ↑ |
| Eubacterium | 0.0149 | 0.0525 | 0.0663 | ↑ |
| Pediococcus | 0.0124 | 0.0124 | 1.0000 | ↓ |
| Ruminococcus | 0.0208 | 0.0614 | 0.0347 | ↑ |
Data come from the microbiome study sample of the Survey of the Health of Wisconsin 2016–2017.
Analysis was performed using Quasi Conditional Association Test using Generalized Estimating Equations (QCAT-GEE), adjusted for age, gender, body mass index, antibiotic use, race/ethnicity, education, smoking, fiber consumption, urbanicity, and indoor pets.
Indicates p < 0.05 after FDR correction.
4. Discussion
In this analysis of the association between urinary Pb exposure and gut microbial composition, significant differences in α-diversity and richness were found with increasing levels of urinary Pb concentrations. In contrast with preliminary hypotheses, Pb exposure, as measured in urine, was associated with an increase in the number of species present within the gut, driving an increase in overall diversity. Associations between urinary Pb exposure and measures of increased richness and α-diversity were strongest for those ages 50 and above, former smokers, and those living in suburban and rural areas. Urinary Pb concentration was also associated with significant differences in β-diversity, indicating that the composition of the gut microbiota was different with increasing Pb exposure. Pb level was associated with significantly increased colonization by Proteobacteria, specifically Burkholderiales.
This study has important implications for translational microbiome research because it is one of the few human studies to date to consider the relationship between heavy metal exposures, such as Pb, on the gut microbiota. While this study’s results are not a direct translation of previously published experimental animal studies, some findings were consistent. For example, Gao et al. (2017) found significant differences in the abundance of several bacterial taxa upon Pb exposure in mice, including reductions in the Clostridiales and Coprococcus. These taxa were similarly significantly reduced in this study, prior to correction for multiple comparisons. Among mice orally exposed to Pb, Gao et al. (2017) also found significant changes in the trajectories of α and β-diversity, and metabolic function with Pb exposure that became stronger over time with continued exposure, although they saw a decrease in α-diversity with increased Pb. Collectively these findings underscore the importance of examination of chronic Pb exposure in composition of the microbiota over time. Future research is needed to determine how this impacts overall metabolic function and chronic health.
Previous epidemiologic studies have examined Pb exposure and changes in the gut microbiome only as secondary analyses in studies among children, with mixed results. Zhai et al. (2019), found no evidence of association, and Bisanz et al. (2014) found increased prevalence of Succinivibrionaceae and Gammaproteobacteria in children with highly elevated blood Pb levels. Similarly, Gammaproteobacteria were significantly associated with Pb in this study of adults prior to FDR correction. Gammaproteobacteria are members of the phylum Proteobacteria, and changes in the Gammaproteobacteria composition may contribute to changes in Proteo-bacteria found in this study. While the findings of this study are somewhat in line with those of Bisanz et al. (2014), the differences in results are not surprising given the vastly different population composition, sample size, exposure level, exposure measurement type, and primary study purpose and design. Several investigations of different animal models also found the abundance of varying taxa to be associated with increased Pb exposure (Breton et al., 2013b; Gao et al., 2017; Xia et al., 2018a, 2018b; Zhai et al., 2017). When considered within the context of previous work, our study suggests that Pb exposure is associated with gut microbial composition, even in an adult human population with relatively low levels of exposure.
The findings that α-diversity and richness were associated with higher urinary Pb concentrations among a sample of adults were not consistent with our hypothesis that both measures of microbial diversity would decrease with Pb exposure. Possible explanations include that the source of Pb exposure may also be introducing different bacteria, or increasing the survival ability of bacteria that would not otherwise thrive in that environment. Or, Pb exposure could be reducing the abundance of highly abundant bacteria and alter the evenness of abundance across all taxa. Alternatively, the immune suppressing effects of Pb may outweigh the direct effects of Pb on the gut bacteria, allowing for increased colonization.Ultimately, α-diversity and richness results are composite measures of microbial gut composition and significant shifts, regardless of direction, point to the need for additional investigations that explore the changes in bacterial composition and function associated with Pb exposure in more detail.
Although Proteobacteria make up a small proportion of the overall composition of the gut microbiota (Fig. 1), their increased levels may also contribute to this finding. Proteobacteria are a phylum of Gramnegative bacteria that include the classes α-, β-, δ-, and γ-proteo-bacteria, order Burkholderiales, and the families Alcaligenaceae and Oxalobacteraceae, all of which were significant either before or after FDR correction in our QCAT-GEE analysis. The order Burkholderiales contains a wide variety of bacterial species that perform a plethora of metabolic functions. Some members of the Alcaligenaceae family are known to be respiratory pathogens, including Bordatella pertussis, the causative agent of whooping cough (Austin, 2014). A species of clinical relevance within the Oxalobacteraceae family of the Burkholderiales, is Oxalobacter formigens, a bacteria commonly found in the gastrointestinal tract that uses oxalate as its primary source of carbon and energy (Baldani et al., 2014). Oxalate is a dietary component that typically binds with Ca2+, and in excess can lead to calcium oxalate kidney stones (Holmes and Assimos, 2004). In a study using data from the American Gut Project, the abundance of O. formigens was found to be associated with increased a-diversity and resilience of the gut microbiota to disturbances (Liu et al., 2017). This may further explain our finding that increasing α-diversity was associated with increasing urine Pb concentration. Oxalate can also bind with Pb2+ to form lead oxalate. When oxalate binds to Ca or Pb, it reduces the amount free and available for absorption into the blood stream. Thus, it is possible that the presence of O. formigens in the gut microbiota could increase urine Pb levels, rather than Pb exposure resulting in an increase in O. formigens presence or abundance. However, there is not strong evidence that this is the case in our study. Further investigation of the association between O. formigens and human biomarkers of Pb is needed.
An important considerations in interpreting results is that exposures in this sample are relatively low, with a lower geometric mean urinary Pb concentration than the NHANES representative sample (0.45 μg/L) (Buser et al., 2016), however, there is overlap in exposure levels between the two study populations. Additionally, approximately 76% of this study population was overweight or obese based on objectively measured height and weight. This is consistent with previous estimates of a representative sample of the Wisconsin population (Eggers et al., 2016). This skew towards a higher BMI may be contributing to the difference in Firmicute to Bacteriodetes ratio identified in this study population compared to previous studies of healthy adults (The Human Microbiome Project Consortium, 2012). We also acknowledge that differences in the DNA extraction method used for these analyses, including heat, chemical, and mechanical lysis, may additionally contribute to this finding. However, the high level of variability in community structure between individuals is consistent with previous findings (The Human Microbiome Project Consortium, 2012).
Eating a healthy diet has been previously suggested as an intervention to mitigate the toxicity of Pb exposure (Kordas et al., 2018; Zhai et al., 2015). Mechanisms include competition by essential metals, reduction of oxidative stress, and improved immune function, counteracting the negative effects of heavy metals (Zhai et al., 2015). In our analysis, stratifying the analysis by tertiles of dietary fiber consumption modified the effects of Pb on gut microbial composition, with those in the middle tertile showing the largest increase in α-diversity and richness with increase Pb level. This may be a previously unconsidered pathway through which healthy diets ameliorate the toxic effects of Pb. Given the ongoing need for strategies to reduce Pb exposure and its toxic effects (Cassidy-Bushrow et al., 2017; Song et al., 2017), this area is promising for further investigation including both observational and clinical studies.
Although this analysis contributes novel insights to what is known about Pb and the gut microbiota, it has some limitations. Fecal samples are a useful matrix for examining the contents of the gut microbiota, however, some aspects of sample collection could have been improved upon in this study. For example, the SHOW ancillary microbiome study collects only one stool sample per participant, which gives a cross-sectional snapshot of the microbiota but may not accurately represent the normal composition. Moreover, the cross-sectional nature of this study allows for investigation of association, not causality. An expansion of the microbiome study began in 2018 and collects a second fecal sample (along with several environmental samples) from the participants in this study. Future analysis will be able to examine gut microbiota composition longitudinally as well as urinary Pb level over time, and provide additional insights into the relationship between the two.
We also note that extracting DNA from fecal samples that have not been frozen and thawed is ideal for getting the most accurate sequencing results, as some bacterial DNA is damaged with each freeze-thaw cycle (Cardona et al., 2012). Fecal samples from the SHOW ancillary microbiome study go through at least one freeze-thaw cycle before sequencing. Although this is a known limitation, the PCR used to amplify DNA prior to sequencing would still be able to identify non-viable organism as long as the DNA encoding the 16S rRNA is undamaged. The gut microbiota analysis is further limited by the use of 16S rRNA amplicon sequencing as opposed to whole genome sequencing. Sequencing entire genomes would allow for more accurate identification of bacteria, and in-depth analysis of potential metabolic function within the gut microbiome. This type of analysis is useful given the interpersonal variation in taxonomic composition of the gut microbiota (Jovel et al., 2016). Using metagenomic data would allow for the examination of how the functional capacity of the microbiome is altered upon exposure to Pb.
The use of urine samples for the measurement of Pb exposure, while useful in measuring exposure, may also induce some misclassification, as it is not the gold standard for Pb exposure measurement. Because this study was observational, determination of Pb exposure was limited to post-exposure measurement. This poses challenges for addressing this particular research question as the gut microbiota play a role in the metabolism of Pb within the gut, as postulated in the case of O. formigens, which may affect the level of Pb that is absorbed into the bloodstream and later exits the body through the urine.
5. Conclusions
This study found that levels of adult urinary Pb concentration are associated with significant differences in gut microbial composition, even at Pb levels below the national average, including changes in a and β-diversity, as well as differences in colonization by Proteobacteria, specifically Burkholderiales. This study also sets a basis for comparison of future studies of Pb exposure and the human microbiota. Further examination of this association and its downstream health effects is warranted.
Supplementary Material
Acknowledgements
The authors would like to thank the study participants, and staff at the Survey of the Health of Wisconsin, the University of Wisconsin Survey Center, Dr. Safdar’s Infectious Disease Research Lab, the Suen lab, and Noel Stanton at the Wisconsin State Laboratory of Hygiene.
Funding
Funding for this work was provided by the Wisconsin Partnership Program and the Department of Medicine Pilot Award Program at the University of Wisconsin School of Medicine and Public Health, USA. S.E. was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, USA (T32 HD049311). AEK was supported by a National Library of Medicine training grant to the Computation and Informatics in Biology and Medicine Training Program, USA (NLM 5T15LM007359). Authors also acknowledge support from the National Institutes of Health core grant to the Center for Demography and Ecology at the University of Wisconsin-Madison, USA (P2C HD047873). No funding agency had any role in the design of the study, or in the collection, analysis, and interpretation of data, or in writing the manuscript.
Abbreviations:
- Pb
Lead
- Th2
Type 2 helper
- SHOW
Survey of the Health of Wisconsin
- OTUs
Operational Taxonomic Units
- BMI
Body Mass Index
- NMDS
Non-Metric Multidimensional Scaling
- PERMANOVA
Permutational Analysis of Variance
- QCAT-GEE
Quasi-Conditional Association Test using General Estimating Equations
- FDR
Benjamini-Hochberg correction for multiple comparisons
- NHANES
National Health and Nutrition Examination Survey
- LOD
Limit of Detection
- DAGs
Direct Acyclic Graphs
- FPL
Federal Poverty Level
- DHQ-II
Diet History Questionnaire
- RUCA
Rural-Urban Commuting Area
Footnotes
Appendix A. Supplementary Data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2019.105122.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abadin H, Ashizawa A, Stevens Y-W, Llados F, Diamond G, Sage G, Citra M, Quinones A, Bosch SJ, Swarts SG, 2007. Toxicological Profile for Lead. [PubMed] [Google Scholar]
- Austin B, 2014. The family alcaligenaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (Eds.), The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. Springer, Berlin, pp. 729–757. [Google Scholar]
- Baldani J, Rouws L, Cruz L, Olivares F, Schmid M, Hartmann A, 2014. The family oxalobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (Eds.), The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. Springer, Berlin, pp. 919–974. [Google Scholar]
- Bisanz JE, Enos MK, Mwanga JR, Changalucha J, Burton JP, Gloor GB, Reid G, 2014. Randomized open-label pilot study of the influence of probiotics and the gut microbiome on toxic metal levels in Tanzanian pregnant women and school children. mBio 5, e01580–14. 10.1128/mBio.01580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray JR, Curtis JT, 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. [Google Scholar]
- Breton J, Daniel C, Dewulf J, Pothion S, Froux N, Sauty M, Thomas P, Pot B, Foligné B, 2013a. Gut microbiota limits heavy metals burden caused by chronic oral exposure. Toxicol. Lett. 222, 132–138. 10.10167/j.toxlet.2013.07.021. [DOI] [PubMed] [Google Scholar]
- Breton J, Massart S, Vandamme P, De Brandt E, Pot B, Foligné B, 2013b. Ecotoxicology inside the gut: impact of heavy metals on the mouse microbiome. BMC Pharmacol. Toxicol. 14, 62. 10.1186/2050-6511-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG, 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser MC, Ingber SZ, Raines N, Fowler DA, Scinicariello F, 2016. Urinary and blood cadmium and lead and kidney function: NHANES 2007–2012. Int. J. Hyg. Environ. Health 219, 261–267. 10.1016/j.ijheh.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona S, Eck A, Cassellas M, Gallart M, Alastrue C, Dore J, Azpiroz F, Roca J, Guarner F, Manichanh C, 2012. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 12, 158. 10.1186/1471-2180-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Bushrow AE, Sitarik AR, Havstad S, Park SK, Bielak LF, Austin C, Johnson CC, Arora M, 2017. Burden of higher lead exposure in African-Americans starts in utero and persists into childhood. Environ. Int 108, 221–227. 10.1016/j.envint.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cénit MC, Matzaraki V, Tigchelaar EF, Zhernakova A, 2014. Rapidly expanding knowledge on the role of the gut microbiome in health and disease. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis 1842, 1981–1992. 10.1016/j.bbadis.2014.05.023. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL, 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietert RR, Piepenbrink MS, 2006. Lead and immune function. Crit. Rev. Toxicol. 36, 359–385. 10.1080/10408440500534297. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R, 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S, Remington PL, Ryan K, Nieto FJ, Peppard P, Malecki K, 2016. Obesity prevalence and health consequences: findings from the survey of the health of Wisconsin. WMJ 115, 238–243. [PMC free article] [PubMed] [Google Scholar]
- Eggers S, Malecki KM, Peppard P, Mares J, Shirley D, Shukla SK, Poulsen K, Gangnon R, Duster M, Kates A, Suen G, Sethi AK, Safdar N, 2018. Wisconsin microbiome study, a cross-sectional investigation of dietary fibre, microbiome composition and antibiotic-resistant organisms: rationale and methods. BMJ Open 8, e019450. 10.1136/bmjopen-2017-019450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Chi L, Mahbub R, Bian X, Tu P, Ru H, Lu K, 2017. Multi-omics reveals that lead exposure disturbs gut microbiome development, key metabolites, and metabolic pathways. Chem. Res. Toxicol. 30, 996–1005. 10.1021/acs.chemrestox.6b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotelli NJ, Colwell RK, 2001. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4, 379–391. [Google Scholar]
- Holmes RP, Assimos DG, 2004. The impact of dietary oxalate on kidney stone formation. Urol. Res. 32, 311–316. 10.1007/s00240-004-0437-3. [DOI] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM, 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67, 4399–4406. 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosławiecka A, Piotrowska-Seget Z, 2014. Lead resistance in micro-organisms. Microbiology 160, 12–25. 10.1099/mic.0.070284-0. [DOI] [PubMed] [Google Scholar]
- Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, Wang W, Tang W, Tan Z, Shi J, Li L, Ruan B, 2015. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 48, 186–194. 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, Wong GK-S, 2016. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol 7. 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordas K, Burganowski R, Roy A, Peregalli F, Baccino V, Barcia E, Mangieri S, Ocampo V, Mañay N, Martínez G, Vahter M, Queirolo EI, 2018. Nutritional status and diet as predictors of children’s lead concentrations in blood and urine. Environ. Int. 111, 43–51. 10.1016/j.envint.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD, 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diet History Questionnaire (DHQ-II) and Canadian Diet History Questionnaire (C-DHQ II) [WWW Document], 2019. https://epi.grants.cancer.gov/dhq2/about/ (accessed 8. 12.19).
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, MetaHIT Consortium, Guedon E, Delorme C, Layec S, Khaci G, van de Guchte M, Vandemeulebrouck G, Jamet A, Dervyn R, Sanchez N, Maguin E, Haimet F, Winogradski Y, Cultrone A, Leclerc M, Juste C, Blottiere H, Pelletier E, LePaslier D, Artiguenave F, Bruls T, Weissenbach J, Turner K, Parkhill J, Antolin M, Manichanh C, Casellas F, Boruel N, Varela E, Torrejon A, Guarner F, Denariaz G, Derrien M, van Hylckama Vlieg JET, Veiga P, Oozeer R, Knol J, Rescigno M, Brechot C, M’Rini C, Mérieux A, Yamada T, Bork P, Wang J, Ehrlich SD, Pedersen O, 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Liu M, Koh H, Kurtz ZD, Battaglia T, PeBenito A, Li H, Nazzal L, Blaser MJ, 2017. Oxalobacter formigenes-associated host features and microbial community structures examined using the American gut project. Microbiome 5. 10.1186/s40168-017-0316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, Palmer BE, 2013. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 14, 329–339. 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MiSeq SOP Mothur [WWW document]. https://www.mothur.org/wiki/MiSeq_SOP, Accessed date: 14 November 2016.
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C, 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79. 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto FJ, Peppard PE, Engelman CD, McElroy JA, Galvao LW, Friedman EM, Bersch AJ, Malecki KC, 2010. The Survey of the Health of Wisconsin (SHOW), a novel infrastructure for population health research: rationale and methods. BMC Public Health 10, 785. 10.1186/1471-2458-10-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H, 2016. Vegan: Community Ecology Package. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO, 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, 2000. Biomarkers of Lead exposure. Ind. Health 38, 127–142. 10.2486/indhealth.38.127. [DOI] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, ter Horst R, Jansen T, Jacobs L, Bonder MJ, Kurilshikov A, Fu J, Joosten LAB, Zhernakova A, Huttenhower C, Wijmenga C, Netea MG, Xavier RJ, 2016. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167, 1125–1136.e8. 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LCM, Finlay BB, 2010. Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Simpson EH, 1949. Measurement of diversity. Nature 163 (4148), 688–688. 10.1038/163688a0. [DOI] [Google Scholar]
- Song B, Zeng G, Gong J, Liang J, Xu P, Liu Z, Zhang Y, Zhang C, Cheng M, Liu Y, Ye S, Yi H, Ren X, 2017. Evaluation methods for assessing effectiveness of in situ remediation of soil and sediment contaminated with organic pollutants and heavy metals. Environ. Int. 105, 43–55. 10.1016/j.envint.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Tang Z-Z, Chen G, Alekseyenko AV, Li H, 2017. A general framework for association analysis of microbial communities on a taxonomic tree. Bioinforma. Oxf. Engl 33, 1278–1285. 10.1093/bioinformatics/btw804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium, 2012. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Schirnding YE von, Prapamontol T, 2000. Environmental lead exposure: a public health problem of global dimensions. Bull. World Health Organ. 78, 1068–1077. 10.1590/S0042-96862000000900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wen XW, Faulk C, Boehnke K, Zhang H, Dolinoy DC, Xi C, 2016. Perinatal lead exposure alters gut microbiota composition and results in sex-specific body-weight increases in adult mice. Toxicol. Sci. 151, 324–333. 10.1093/toxsci/kfw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Jin C, Pan Z, Sun L, Fu Z, Jin Y, 2018a. Chronic exposure to low concentrations of lead induces metabolic disorder and dysbiosis of the gut microbiota in mice. Sci. Total Environ. 631–632, 439–448. 10.1016/j.scitotenv.2018.03.053. [DOI] [PubMed] [Google Scholar]
- Xia J, Lu L, Jin C, Wang S, Zhou J, Ni Y, Fu Z, Jin Y, 2018b. Effects of short term lead exposure on gut microbiota and hepatic metabolism in adult zebrafish. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol 209, 1–8. 10.1016/j.cbpc.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Narbad A, Chen W, 2015. Dietary strategies for the treatment of cadmium and lead toxicity. Nutrients 7, 552–571. 10.3390/nu7010552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Li T, Yu L, Xiao Y, Feng S, Wu J, Zhao J, Zhang H, Chen W, 2017. Effects of subchronic oral toxic metal exposure on the intestinal microbiota of mice. Sci. Bull 62, 831–840. 10.1016/j.scib.2017.01.031. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Cen S, Jiang J, Zhao J, Zhang H, Chen W, 2019. Disturbance of trace element and gut microbiota profiles as indicators of autism spectrum disorder: a pilot study of Chinese children. Environ. Res. 171, 501–509. 10.1016/j.envres.2019.01.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.