Abstract
Secondary metabolites obtained from marine-derived fungi are rich sources of drug candidates. Three new sesquiterpenoids, chermesiterpenoids A–C (1–3), along with four known alkaloids (4–7), were isolated and identified from the marine red algal-derived fungus Penicillium chermesinum EN-480. The structures of these new sesquiterpenoids were elucidated using detailed analysis of the NMR data and their relative configurations were elucidated using nuclear overhauser effect spectroscopy (NOESY) spectra as well as gauge-independent atomic orbital (GIAO) NMR shift calculations and DP4+ probability analysis. Their absolute configurations were determined using electronic circular dichroism (ECD) calculations and modified Mosher’s method. Compounds 2 and 3 exhibited potent activities against human and aquatic pathogenic bacteria and plant pathogenic fungi.
Keywords: Penicillium chermesinum, algal-derived fungus, sesquiterpenoid, antimicrobial activity
1. Introduction
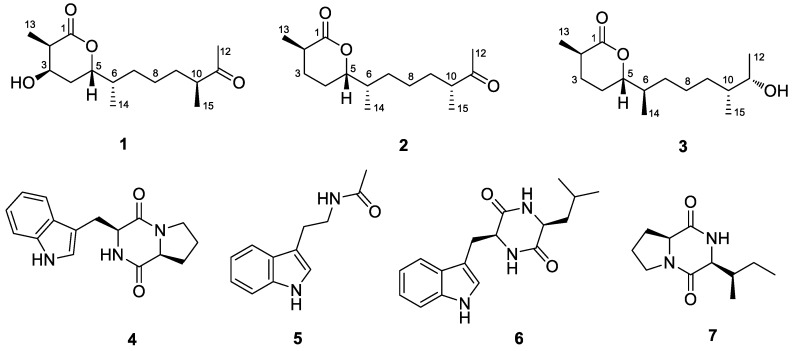
Marine-derived fungi are rich sources of diverse and bioactive secondary metabolites [1,2]. Among them, the species of the genus Penicillium play an important role and have increasingly attracted attention [3]. Our current research focuses on Penicillium chermesinum EN-480, a fungal strain isolated from the fresh tissue of the marine red alga Pterocladiella tenuis [4]. To date, meroterpenoids [4], cytotoxic metabolites [5], azaphilones [6], terphenyls [6], and plastatin [7] have been obtained from the species P. chermesinum. Some of them exhibited antibacterial [4] and enzyme inhibitory [6] activities. In our previous research, we obtained antimicrobial meroterpenoids from P. chermesinum EN-480 [4], and we recently tried to change the fermentation process to obtain more antibacterial secondary metabolites. As a result, we isolated three new sesquiterpenoids (1–3) as well as four known alkaloids, brevianmide F (4) [8], N-acetyltryptamine (5) [9], cyclo Trp-Ile (6) [10], and cyclo Ile-Pro (7) [11] (Figure 1). Compounds 1–3 were formed by esterification and cyclizing of catenulate farnesyl probably, and this is the first time this type of sesquiterpenoid from natural products has been described. The absolute configurations of this kind of secondary metabolites were difficult to determine because the alkyl chains were flexible and there were no conjugated chromophores in the compounds. Thus, we combined DP4+ probability analysis [12,13], electronic circular dichroism (ECD) calculations, and modified Mosher’s method [14,15] to solve this problem. Herein, details of the isolation, structure determination, and biological activities of these compounds are described.
Figure 1.
Structures of compounds 1–7.
2. Results and Discussion
2.1. Structure Elucidation of the New Compounds
Compound 1 was obtained as a yellowish solid. Its molecular formula was determined as C15H26O4 based on the (+)-HRESIMS (high resolution electrospray ionization mass spectroscopy) data (Figure S1, Supplementary Material), which has three degrees of unsaturation (index of hydrogen deficiency) [16]. The 13C NMR and DEPT (distortionless enhancement by polarization transfer) data (Table 1 and Figure S3) presented 15 carbon resonances, including four methyls, four methylenes, five methines (including two oxygenated), and two quaternary carbons (with one ketone carbonyl and one ester carbonyl). The 1H NMR data (Table 1 and Figure S2) revealed the presence of four methyl signals at δH 2.09 (H-12, 3H, s), 1.10 (H-13, 3H, d, J = 7.1 Hz), 0.84 (H-14, 3H, d, J = 6.9 Hz), and 0.98 (H-15, 3H, d, J = 6.8 Hz), as well as five methines (including two oxygenated at δH 4.44 (H-5, 1H, m) and 3.95 (H-3, 1H, m)) in compound 1. The 1H and 13C NMR data indicated that it is a sesquiterpenoid and the planar structure was further determined and fully supported using the COSY (homonuclear chemical shift correlation spectroscopy) and HMBC (heteronuclear multiple bond correlation) correlations (Figure 2, Figure S4 and S5). Compound 1 was possibly formed by the oxidation and esterification of farnesyl. This is the first time this type of sesquiterpenoid, which contains a pentanolactone moiety, has been described.
Table 1.
1H (125 MHz) and 13C NMR (500 MHz) data of compounds 1–3 (recorded in DMSO (dimethyl sulfoxide)-d6, δ in ppm).
| Pos. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| 1H (J in Hz) | 13C | 1H (J in Hz) | 13C | 1H (J in Hz) | 13C | |
| 1 | 173.5, C | 173.6, C | 173.6, C | |||
| 2 | 2.52, overlap | 41.0, CH | 2.40, dt (13.2, 6.7) | 35.5, CH | 2.40, m | 35.5, CH |
| 3 | 3.95, m | 65.8, CH |
α 1.90, m, β 1.56, overlap |
27.9, CH2 |
α 1.90, ddd (8.5, 6.2, 2.9) β 1.53, m |
27.7, CH2 |
| 4 |
α 1.73, m, β 1.82, m |
23.9, CH2 |
α 1.78, d (10.7) β 1.57, overlap |
27.6, CH2 |
α 1.78, m β 1.51, m |
24.5, CH2 |
| 5 | 4.44, m | 79.2, CH | 4.16, m | 84.4, CH | 4.17, m | 84.5, CH |
| 6 | 1.68, m, | 36.7, CH | 1.64, m | 37.0, CH | 1.64, m | 37.2, CH |
| 7 | 1.14, m | 31.3, CH2 | 1.08, m | 31.3, CH2 | 1.05, m | 31.8, CH2 |
| 8 | 1.26, m | 32.0, CH2 | 1.27, m | 24.6, CH2 | 1.40, m | 24.3, CH2 |
| 9 | α 1.14, m, β 1.55, m |
32.4, CH2 |
α 1.23, m, β 1.55, overlap |
32.4, CH2 |
α 1.03, m β 0.96, m |
32.4, CH2 |
| 10 | 2.52, overlap | 46.0, CH | 2.50, overlap | 46.0, CH | 1.31, m | 40.0, CH |
| 11 | 211.8, C | 211.8, C | 3.48, dt (10.7, 5.4) | 68.8, CH | ||
| 12 | 2.09, s | 27.9, CH3 | 2.09, s | 23.9, CH3 | 0.97, d (6.3) | 20.3, CH3 |
| 13 | 1.10, d (7.1) | 12.8, CH3 | 1.14, d (7.0) | 17.0, CH3 | 1.13, d (7.0) | 17.1, CH3 |
| 14 | 0.84, d (6.9) | 14.4, CH3 | 0.84, d (6.8) | 14.4, CH3 | 0.85, d (6.8) | 14.5, CH3 |
| 15 | 0.98, d (6.8) | 15.9, CH3 | 0.99, d (6.9) | 15.9, CH3 | 0.79, d (6.7) | 14.5, CH3 |
| 3-OH | 5.20, br s | |||||
| 11-OH | 4.21, d (4.7) | |||||
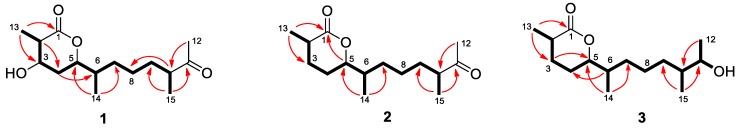
Figure 2.
Key COSY (homonuclear chemical shift correlation spectroscopy) (bold line) and HMBC (heteronuclear multiple bond correlation) (arrow) correlations of compounds 1–3.
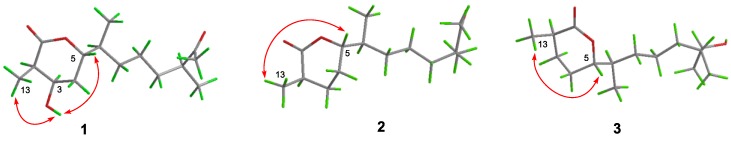
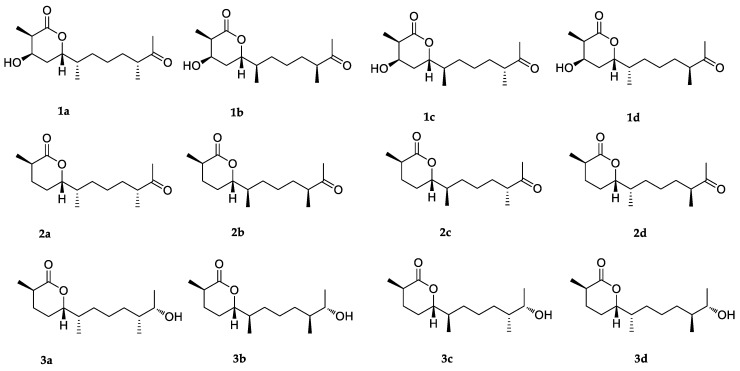
Based on the NOESY (nuclear overhauser effect spectroscopy) spectrum (Figure S6), only a part of the relative configuration in compound 1 could be determined. The NOESY correlations from 3-OH to H3-13 and H-5 confirmed that they were on the same side of 1 (Figure 3). However, NOESY data cannot be used to determine the relative configuration of the methyl groups at the alkyl chain due to the flexibility of this moiety. Thus, DP4+ probability analysis was used to indicate the relative configuration at C-6 and C-10. This method was used to assign the relative configurations of organic molecules employing GIAO (gauge-independent atomic orbital) NMR shift calculations [17,18]. Based on a DP4+ protocol [19], both proton and carbon data of four possible isomers were calculated, and the results were analyzed with the experimental values. The statistical comparison showed that the isomer 1d was the equivalent structure with a probability of 99.90% (Figure 4 and Figure S17). Thus, the results indicated that the relative configuration of 1 is 2R*, 3R*, 5S*, 6S*, and 10S*.
Figure 3.
NOESY (nuclear overhauser effect spectroscopy) correlations of compounds 1–3.
Figure 4.
Structures of possible isomers for DP4+ probability analysis of compounds 1–3.
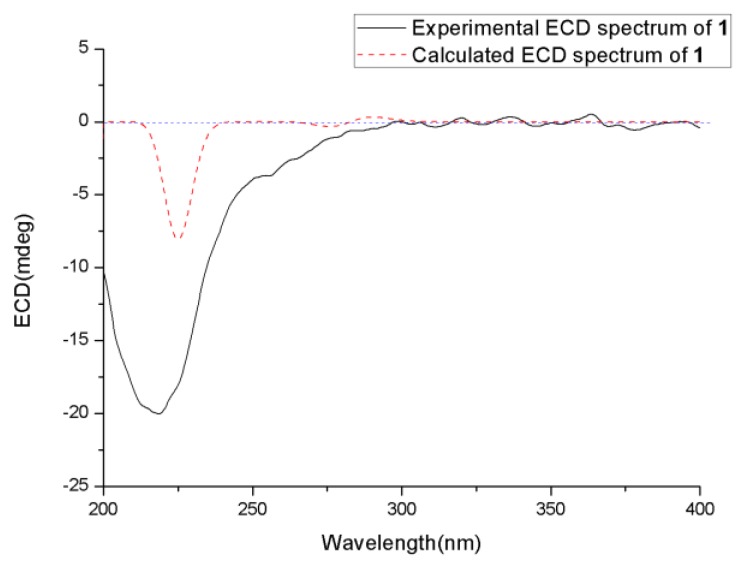
The absolute configuration of 1 was determined using ECD calculation. Gaussian 09.10 was used to perform conformational search and geometry optimization. The obtained minimum energy conformers were calculated using the time-dependent density functional theory (TDDFT) method at the B3LYP/6-31G (d) PCM (polarizable continuum model) at MeOH level for their ECD spectra. The experimental ECD spectrum of 1 matched well with that calculated for 2R, 3R, 5S, 6S, and 10S configurations, which showed negative cotton effect (CE) at approximate 218 nm (Figure 5). Thus, the structure of 1 was determined and was named chermesiterpenoid A.
Figure 5.
Experimental and calculated ECD (electronic circular dichroism) spectra of 1.
Compound 2 was obtained as a yellowish solid. Its molecular formula was determined as C15H26O3, with one O atom less than that of 1, on the basis of (+)-HRESIMS data. The 1H and 13C NMR spectra of 2 are similar to compound 1, indicating that it is also a sesquiterpenoid. However, the NMR data revealed that the oxygenated methine signal at C-3 (δC 65.8; δH 3.95) of compound 1 was replaced by a methylene signal (δC 27.9; δH 1.90/1.56) in 2 (Table 1 and Figure S7 and S8), showing that the hydroxyl group at C-3 of 1 disappeared in that of 2. The observed COSY correlations (Figure S9) from H-3 to H-2 and H-4, as well as the HMBC correlation (Figure S10) from H3-13 to C-3, provided further evidence (Figure 2).
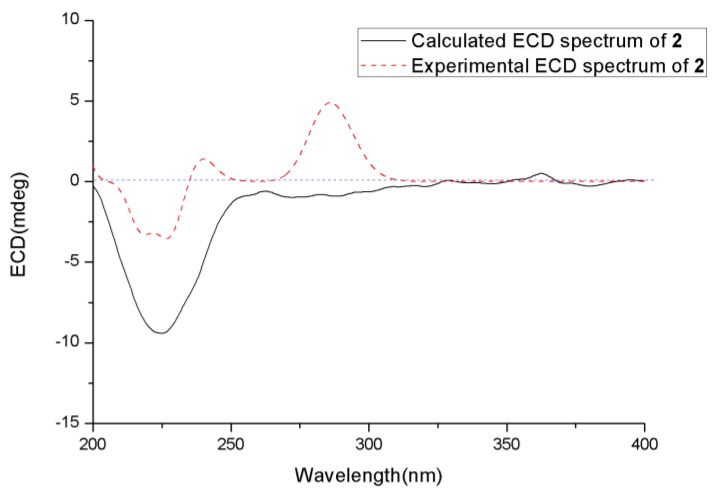
The relative configuration of 2 was also determined using NOESY spectrum (Figure S11) and DP4+ probability analysis. The NOESY correlation between H3-13 and H-5 indicated that they were on the same side. The DP4+ probability analysis of both proton and carbon data showed that the isomer 2a was the equivalent structure with the probability of 100.00% (Figure 4 and Figure S18). The absolute configuration of 2 was also determined using the TDDFT-ECD calculation, and the experimental ECD spectrum of 2 showed excellent agreement with that calculated for 2R, 5S, 6S, and 10R configurations (Figure 6). The structure of 2 was thus determined and was named chermesiterpenoid B.
Figure 6.
Experimental and calculated ECD spectra of 2.
Compound 3 was also obtained as a yellowish solid. Its molecular formula was determined as C15H28O3, with two H atoms more than that of 2 and with two degrees of unsaturation (index of hydrogen deficiency) [16], on the basis of (+)-HRESIMS data. The NMR data of 3 (Figures S12 and S13) were very similar to those of compound 2 except that the ketone carbonyl at C-11 (δC 211.8) in 2 was missing and a hydroxyl methine (δC 68.8; δH 3.48, CH-11) in 3 appeared, indicating that the ketone carbonyl at C-11 was replaced by the hydroxyl group. The chemical shift of H3-12 was shielded from δH 2.09 (3H, s) in 2 to δH 0.97 (3H, d, J = 6.3 Hz) in 3 in the 1H NMR spectrum. The planar structure was further confirmed using COSY and HMBC correlations (Figure 2, Figures S14 and S15).
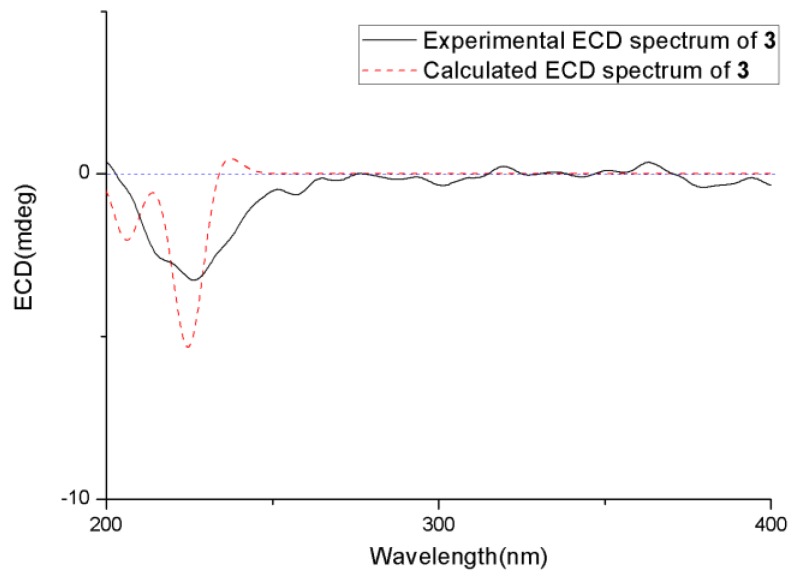
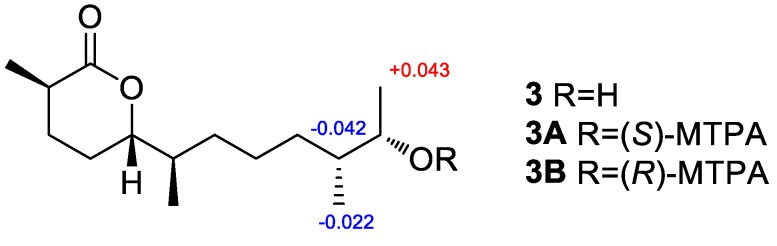
The relative and absolute configurations of 3 were determined using NOESY data, ECD calculation, the modified Mosher’s method, and DP4+ probability analysis. First, the NOESY correlation (Figure S16) from H3-13 to H-5 indicated that they were on the same side. Then, the absolute configuration of C-2 and C-5 was determined using the ECD calculation. The experimental ECD spectrum of 3 agreed well with that calculated for 2R and 5S configurations (Figure 7). After that, modified Mosher’s method was used to determine the absolute configuration of C-11, and the observed Δδ (δS-δR) values of α-methoxy-α-(trifluoromethyl) phenylacetyl (MTPA) esters suggested that the absolute configuration at C-11 was S (Figure 8). Finally, both proton and carbon data of four possible isomers with different absolute configuration at C-6 and C-10 were calculated using the DP4+ probability analysis, and the result showed that the isomer 3c was the equivalent structure with a probability of 100.00% (Figure 4 and Figure S19). The absolute configuration of 3 was determined as 2R, 5S, 6R, 10R, and 11S. Thus, the structure of 3 was determined and was named chermesiterpenoid C.
Figure 7.
Experimental and calculated ECD spectra of 3.
Figure 8.
Δδ values (Δδ (in ppm) = δS – δR) obtained for the (S)-and (R)-MTPA (α-methoxy-α-(trifluoromethyl) phenylacetyl) esters of compound 3.
2.2. Biological Activities of the Isolated Compounds
The isolated new compounds (1–3) were examined for antimicrobial activities against several human-, aqua-, and plant-pathogenic microbes. Compound 2 showed activities against the aquatic pathogens Vibrio anguillarum, Vibrio parahaemolyticus, Micrococcus luteus, and human pathogen Escherichia coli with minimum inhibitory concentration (MIC) values of 0.5, 16, 64, and 64 μg/mL, respectively, which are comparable to that of the positive control, chloromycetinum (MIC = 0.5, 1, 0.5, and 1 μg/mL, respectively). Similarly, compound 3 showed activities against the aquatic pathogens V. anguillarum, V. parahaemolyticus, and M. luteus with MIC values of 1, 32, and 64 μg/mL, respectively. Compounds 1–3 exhibited activity against the plant pathogenic fungus Colletottichum gloeosporioides with MIC values of 64, 32, and 16 μg/mL, respectively, whereas the positive control amphotericin B had a MIC value of 1.0 μg/mL.
These data indicated that compound 1, with a hydroxyl group at C-3, has weaker antimicrobial activities than compounds 2 and 3, regardless of whether against aquatic and human pathogens or plant pathogenic fungi. Compound 2 has somewhat stronger activities against aquatic and human pathogens but weaker activity against plant pathogenic fungus C. gloeosporioides than compound 3, probably due to the different degree of oxidation at C-11.
3. Materials and Methods
3.1. General
UV spectra were obtained using a PuXi TU-1810 UV-visible spectrophotometer (Shanghai Lengguang Technology Co. Ltd., Shanghai, China). Optical rotations were recorded using an Optical Activity AA-55 polarimeter (Optical Activity Ltd., Cambridgeshire, U.K.). ECD spectra were evaluated on a JASCO J-715 spectropolarimeter (JASCO, Tokyo, Japan). The 1D and 2D NMR spectra were determined using a Bruker Avance 500 spectrometer (Bruker Biospin Group, Karlsruhe, Germany). A VG Autospec 3000 (VG Instruments, London, U.K.) was used to measure mass spectra. Analytical and semi-preparative HPLC were performed on a Dionex HPLC system equipped with a P680 pump (Dionex, Sunnyvale, CA, USA), ASI-100 automated sample injector (Dionex, Sunnyvale, CA, USA), and UVD340U multiple wavelength detector (Dionex, Sunnyvale, CA, USA) controlled by Chromeleon software (version 6.80) (Dionex, Sunnyvale, CA, USA). Column chromatography (CC) was performed with silica gel (100–200 and 200–300 mesh; Qingdao Haiyang Chemical Factory, Qingdao, China), Sephadex LH-20 (18–110 μm, Merck, Darmstadt, Germany), and Lobar LiChroprep RP-18 (40–60 μm, Merck, Darmstadt, Germany).
3.2. Fungal Material
The fungus Penicillium chermesinum EN-480 was isolated from the fresh tissue of marine red algal Pterocladiella tenuis, collected from Rongcheng, Shandong province, in China in July 2014. The fungus was identified as Penicillium chermesinum using sequence analysis of the ITS (internal transcribed spacer) region of its 18S rDNA as described previously [20]. The resulting sequence data obtained were deposited in GenBank (accession no. KT119566). The strain is preserved at the key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences.
3.3. Fermentation
The fermentation was conducted dynamically in a 500 L fermentation tank (containing 1% glucose, 2% mannose, 2% maltose, 0.3% yeast extract powder, 0.1% corn flour, 1% monosodium glutamate, 0.05% K2HPO4, 0.03% MgSO4•7H2O, and 300 L naturally sourced and filtered seawater, pH 6.5−7.0) for six days at 28 °C.
3.4. Extraction and Isolation
After incubation, the fermentation broth was exhaustively extracted with EtOAc (ethyl acetate) three times. The combined EtOAc solution was concentrated under reduced pressure to yield an extract (155.3 g).
The organic extract was subjected to vacuum liquid chromatography (VLC) eluting with different solvents of increasing polarity from petroleum ether (PE) to MeOH to yield 9 fractions (Fr. 1–9). Fr. 6 (20 g), eluted with PE-EtOAc (1:1), was purified using reverse-phase column chromatography (CC) over a Lobar LiChroprep RP-18 (40–60 μm, Merck, Darmstadt, Germany) with a MeOH-H2O gradient (from 10:90 to 100:0) to yield four subfractions (Fr. 6-1–6-4). Fr. 6-1, eluted with MeOH-H2O (20:80), was further purified using CC on silica gel (eluted with CH2Cl2-MeOH, 100:1) and Sephadex LH-20 (18–110 μm, Merck, Darmstadt, Germany) (MeOH) to obtain compound 1 (17.4 mg). Fr. 6-2 (eluted with MeOH-H2O (40:60)) was further purified using prep. TLC (plate: 20 × 20 cm, developing solvents: CH2Cl2-MeOH, 50:1) and Sephadex LH-20 (18–110 μm, Merck, Darmstadt, Germany) (MeOH) to produce compounds 4 (5.0 mg), 5 (4.5 mg), and 6 (3.0 mg). Fr. 6-3 (eluted with MeOH-H2O (50:50)) was further purified using CC on silica gel (eluted with CH2Cl2-MeOH, 150:1) and prep. TLC (plate: 20 × 20 cm, developing solvents: CH2Cl2-MeOH, 100:1) to produce compound 2 (19.1 mg). Fr. 6-4 (eluted with MeOH-H2O (60:40)) was further purified using CC on silica gel (eluted with CH2Cl2-MeOH, 200:1 to 100:1) and Sephadex LH-20 (18–110 μm, Merck, Darmstadt, Germany) (MeOH) to produce compounds 3 (10.0 mg) and 7 (7.2 mg).
Chermesiterpenoid A (1): yellowish solid; +6.25 (c 0.16, MeOH); ECD λmax (Δε) 218.5 (−1.64), 1H and 13C NMR data, see Table 1; HRESIMS m/z 271.1906 [M + H]+, 288.2172 [M + NH4]+ (calcd for C15H26O4, 270.1831).
Chermesiterpenoid B (2): yellowish solid; +10.00 (c 0.10, MeOH); ECD λmax (Δε) 224.5 (−0.72) nm;1H and 13C NMR data, see Table 1; HRESIMS m/z 255.1951 [M + H]+, 277.1765 [M + Na]+ (calcd for C15H26O3, 254.1882).
Chermesiterpenoid C (3): yellowish solid; +3.70 (c 0.27, MeOH); ECD λmax (Δε) 226.5 (−0.25) nm; 1H and 13C NMR data, see Table 1; HRESIMS m/z 257.2113 [M + H]+, 279.1930 [M + Na]+ (calcd for C15H28O3, 256.2038).
3.5. Computational NMR Chemical Shift Calculation and DP4+ Analyses
All theoretical calculations were performed via the Gaussian 09 program package. Conformational searches for all possible isomers were conducted via molecular mechanics using the Merck molecular force field (MMFF) method with Macromodel software (Schrödinger, LLC, New York City, USA) and the corresponding stable conformer, from which distributions higher than 2% were collected. After this, B3LYP/6-31G (d) PCM level in DMSO (dimethyl sulfoxide) was used to optimize the conformers. Then, the NMR shielding tensors of all optimized conformers were calculated using the DFT method at mPW1PW91\6-31+G (d) PCM level on DMSO and then averaged based on Boltzmann distribution theory [19]. NMR chemical shifts were calculated using an equation described previously [21]. Finally, the NMR chemical shifts and shielding tensors (1H and 13C) were analyzed and compared with the experimental chemical shifts using DP4+ probability [19].
3.6. ECD Calculation
Conformational searches were conducted via molecular mechanics using the MMFF method in Macromodel software (Schrödinger, LLC, New York City, NY, USA), and the geometries were reoptimized at B3LYP/6-31G(d) PCM/MeOH level using Gaussian 09 software [22] to generate the energy-minimized conformers. After, the optimized conformers were used to calculate ECD spectra using TDDFT at CAM-B3LYP and BH&HLYP/TZVP; the solvent effects of the MeOH solution were estimated at the same DFT level using the SCRF/PCM method.
3.7. Modified Mosher‘s Method
Compound 3 (2.0 mg) was separated equally into two flasks, and both were dried completely. Anhydrous pyridine (400 μL), dimethylaminopyriridine (DMAP, 4 mg), and (S)-(+)-α-methoxy-α-(trifluoromethyl) phenylacetyl chloride (10 μL) or (R)-(−)-α-methoxy-α-(trifluoromethyl) phenylacetyl chloride (10 μL) were added to the two flasks. Then, the flasks were shaken carefully to mix the samples, and the reaction was performed at room temperature for 12 hours. After that, three drops of water were added to stop the reaction. The reaction mixtures were reduced pressure distillated and purified using prep. TLC (plate: 20 × 20 cm, developing solvents: CH2Cl2-MeOH, 50:1) to produce (R)- and (S)-MTPA ester derivatives, respectively, and the 1H NMR spectra and COSY were detected [23].
3.8. Antimicrobial Assay
Antimicrobial assays against human- and aqua-pathogenic microbes Aeromonas hydrophilia, Edwardsiella megi, Edwardsiella tarda, Escherichia coli, Micrococcus luteus, Pseudomonas aeruginosa, Vibrio alginolyticus, V. anguillarum, V. harveyi, V. parahaemolyticus, V. vulnificus, and plant pathogenic fungi Bipolaris sorokiniana, Colletottichum gloeosprioides, Fusarium graminearum, F. oxysporum, Phytophthora nicotiana, Physalospora piricola, and Valsa mali were conducted using the well diffusion method [24]. Chloromycetin was used as a positive control for the bacteria, whereas amphotericin B was used as a positive control for the fungi.
4. Conclusions
Three new types of sesquiterpenoids (1–3) and four known alkaloids (4–7) were isolated from the marine red algal-derived fungus Penicillium chermesinum EN-480. The structures of these new sesquiterpenoids were elucidated using detailed interpretation of the spectroscopic data. The absolute configurations were elucidated using DP4+ probability analysis, ECD calculations, and modified Mosher’s method. Compounds 2 and 3 exhibited prominent activity against aquatic pathogenic bacteria Vibrio anguillarum, V. parahaemolyticus, and Micrococcus luteus, human pathogen Escherichia coli, and plant pathogenic fungus Colletottichum gloeosporioides.
Acknowledgments
The authors appreciate the High Performance Computing Environment Qingdao Branch of Chinese Academy of Science (CAS)—High Performance Computing Center of Institute of Oceanology of CAS for CPU time.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/18/4/194/s1, Figure S1: HRESI mass spectrum of compound 1. Figure S2: 1H NMR (500 MHz, DMSO-d6) spectrum of compound 1. Figure S3: 13C NMR (125 MHz, DMSO-d6) and DEPT spectra of compound 1. Figure S4: COSY spectrum of compound 1. Figure S5: HMBC spectrum of compound 1. Figure S6: NOESY spectrum of compound 1. Figure S7: 1H NMR (500 MHz, DMSO-d6) spectrum of compound 2. Figure S8: 13C NMR (125 MHz, DMSO-d6) and DEPT spectra of compound 2. Figure S9: COSY spectrum of compound 2. Figure S10: HMBC spectrum of compound 2. Figure S11: NOESY spectrum of compound 2. Figure S12: 1H NMR (500 MHz, DMSO-d6) spectrum of compound 3. Figure S13: 13C NMR (125 MHz, DMSO-d6) and DEPT spectra of compound 3. Figure S14: COSY spectrum of compound 3. Figure S15: HMBC spectrum of compound 3. Figure S16: NOESY spectrum of compound 3. Figure S17: DP4+ probability Excel sheets of compound 1. Figure S18: DP4+ probability Excel sheets of compound 2. Figure S19: DP4+ probability Excel sheets of compound 3.
Author Contributions
X.-Y.H. performed the experiments for the isolation, structure elucidation, and antimicrobial evaluation, and prepared the manuscript; X.-M.L. performed the 1D and 2D NMR experiments; S.-Q.Y. performed the ECD calculations; H.L. isolated the fungus from algae, L.-H.M. jointly supervised the research; B.-G.W. supervised the research work and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Natural Science Foundation of China (Grant No. 31330009 and 31600267), and by the Qingdao National Laboratory for Marine Science and Technology (YQ2018NO08) and the project of Qingdao Science and Technology Bureau (18-2-2-59-jch). B.-G.W. acknowledges the support of Taishan Scholar Project from Shandong Province.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H.G., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2015;32:116–211. doi: 10.1039/C4NP00144C. [DOI] [PubMed] [Google Scholar]
- 2.Barzkar N., Tamadoni J.S., Poorsaheli H.B., Vianello F. Metabolites from marine microorganisms, micro, and macroalgae: Immense scope for pharmacology. Mar. Drugs. 2019;17:464. doi: 10.3390/md17080464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll A.R., Copp B.R., Davis R.A., Keyzers R.A., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2019;36:122–173. doi: 10.1039/C8NP00092A. [DOI] [PubMed] [Google Scholar]
- 4.Liu H., Li X.M., Liu Y., Zhang P., Wang J.N., Wang B.G. Chermesins A–D: Meroterpenoids with a drimane-type spirosesquiterpene skeleton from the marine algal-derived endophytic fungus Penicillium chermesinum EN-480. J. Nat. Prod. 2016;79:806–811. doi: 10.1021/acs.jnatprod.5b00893. [DOI] [PubMed] [Google Scholar]
- 5.Darsih C., Prachyawarakorn V., Wiyakrutta S., Mahidol C., Ruchirawat S., Kittakoop P. Cytotoxic metabolites from the endophytic fungus Penicillium chermesinum: Discovery of a cysteine-targeted Michael acceptor as a pharmacophore for fragment-based drug discovery, bioconjugation and click reactions. RSC Adv. 2015;5:70595–70603. doi: 10.1039/C5RA13735G. [DOI] [Google Scholar]
- 6.Huang H.B., Feng X.J., Xiao Z.E., Liu L., Li H.X., Ma L., Lu Y.J., Ju J.H., She Z.G., Lin Y. Azaphilones and p-terphenyls from the mangrove endophytic fungus Penicillium chermesinum (ZH4-E2) isolated from the South China Sea. J. Nat. Prod. 2011;74:997–1002. doi: 10.1021/np100889v. [DOI] [PubMed] [Google Scholar]
- 7.Singh P.D., Johnson J.H., Aklonis C.A., Bush K., Fisher S.M., O’Sullivan J. Two new inhibitors of phospholipase A2 produced by Penicillium chermesinum. J. Antibiot. 1985;38:706–712. doi: 10.7164/antibiotics.38.706. [DOI] [PubMed] [Google Scholar]
- 8.Wang B., Park E.M., King J.B., Mattes A.O., Cichewicz R.H. Transferring fungi to a deuterium-enriched medium results in assorted, conditional changes in secondary metabolite production. J. Nat. Prod. 2015;78:1415–1421. doi: 10.1021/acs.jnatprod.5b00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedras M.S.C., Taylor J.L. Metabolism of the phytoalexin brassinin by the “blackleg” fungus. J. Nat. Prod. 1993;56:731–738. [Google Scholar]
- 10.Fukuhara K.I., Murao S., Nozawa T., Hatano M. Structural elucidation of talopeptin (MK-I), a novel metallo proteinase inhibitor produced by Streptomyces mozunensis MK-23. Tetrahedron Lett. 1982;23:2319–2322. doi: 10.1016/S0040-4039(00)87331-1. [DOI] [Google Scholar]
- 11.Guo Z.K., Wang R., Chen F.X., Liu T.M., Yang M.Q. Bioactive aromatic metabolites from the sea urchin-derived actinomycete, streptomyces spectabilis, strain HDa1. Phytochem. Lett. 2018;25:132–135. doi: 10.1016/j.phytol.2018.04.014. [DOI] [Google Scholar]
- 12.Fan T.T., Zhang H.H., Tang Y.H., Zhang F.Z., Han B.N. Two new neo-debromoaplysiatoxins—A pair of stereoisomers exhibiting potent kv1.5 ion channel inhibition activities. Mar. Drugs. 2019;17:652. doi: 10.3390/md17120652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z., Fan Z., Sun Z., Liu H., Zhang W. Dechdigliotoxins A–C, three novel disulfide-bridged gliotoxin dimers from deep-sea sediment derived fungus Dichotomomyces cejpii. Mar. Drugs. 2019;17:596. doi: 10.3390/md17110596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S.R., Lee D., Eom H.J., Rischer M., Ko Y.J., Kang K.S., Kim C.S., Beemelmanns C., Kim K.H. Hybrid polyketides from a hydractinia-associated Cladosporium sphaerospermum SW67 and their putative biosynthetic origin. Mar. Drugs. 2019;17:606. doi: 10.3390/md17110606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guilet D., Guntern A., Ioset J.R., Queiroz E.F., Ndjoko K., Foggin C.M., Hostettmann K. Absolute configuration of a tetrahydrophenanthrene from Heliotropium ovalifolium by LC-NMR of its Mosher esters. J. Nat. Prod. 2003;66:17–20. doi: 10.1021/np020366k. [DOI] [PubMed] [Google Scholar]
- 16.Pellegrin V. Molecular formulas of organic compounds: The nitrogen rule and degree of unsaturation. J. Chem. Educ. 1983;60:626–633. doi: 10.1021/ed060p626. [DOI] [Google Scholar]
- 17.Park K.J., Kim C.S., Khan Z., Oh J., Kim S.Y., Choi S.U., Lee K.R. Securinega alkaloids from the twigs of Securinega suffruticosa and their biological activities. J. Nat. Prod. 2019;82:1345–1353. doi: 10.1021/acs.jnatprod.9b00142. [DOI] [PubMed] [Google Scholar]
- 18.Kim C.S., Oh J., Subedi L., Kim S.Y., Choi S.U., Lee K.R. Structural characterization of terpenoids from Abies holophylla using computational and statistical methods and their biological activities. J. Nat. Prod. 2018;81:1795–1802. doi: 10.1021/acs.jnatprod.8b00245. [DOI] [PubMed] [Google Scholar]
- 19.Grimblat N., Zanardi M.M., Sarotti A.M. Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 2015;80:12526–12534. doi: 10.1021/acs.joc.5b02396. [DOI] [PubMed] [Google Scholar]
- 20.Wang S., Li X.M., Teuscher F., Li D.L., Diesel A., Ebel R., Proksch P., Wang B.G. Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceolata. J. Nat. Prod. 2006;69:1622–1625. doi: 10.1021/np060248n. [DOI] [PubMed] [Google Scholar]
- 21.Lee S.R., Lee D., Park M., Lee J.C., Park H., Kang K.S., Kim C., Beemelmanns C., Kim K.H. Absolute configuration and corrected NMR assignment of 17-hydroxycyclooctatin, a fused 5-8-5 tricyclic diterpene. J. Nat. Prod. 2020;83:354–361. doi: 10.1021/acs.jnatprod.9b00837. [DOI] [PubMed] [Google Scholar]
- 22.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., et al. Gaussian 09, Revision D.01. Gaussian, Inc.; Wallingford, UK: 2013. [Google Scholar]
- 23.Ohtani I., Kusumi T., Kashman Y., Kakisawa H. High-field FT NMR application of Mosher’s method. The absolute configurations of marine terpenoids. J. Am. Chem. Soc. 1991;113:4092–4096. doi: 10.1021/ja00011a006. [DOI] [Google Scholar]
- 24.Al-Burtamani S.K.S., Fatope M.O., Marwah R.G., Onifade A.K., Al-Saidi S.H. Chemical composition, antibacterial and antifungal activities of the essential oil of Haplophyllum tuberculatum from Oman. J. Ethnopharmacol. 2005;96:107–112. doi: 10.1016/j.jep.2004.08.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.