Abstract
The YABBY family are a group of seed plant-specific transcription factors, which are involved in the specification of abaxial polarity in lateral organs. In Arabidopsis thaliana, YABBY3 (YAB3) plays a critical role in regulating abaxial patterning, growth of lateral organs, and inflorescence phyllotaxy. In this study, the BcYAB3 gene was isolated from Pak-choi (Brassica rapa subsp. chinensis). The tissue-specific expression analysis indicated that the BcYAB3 gene has significantly high transcript levels in stem, leaf, and flower. We investigated the subcellular localization of BcYAB3 and found the protein to be expressed in the nucleus. In the transgenic Arabidopsis thaliana plants expressing the BcYAB3 gene, the leaves were curling downward with the plant growth, and the bolting and flowering stages were delayed. These results not only validate the function of BcYAB3 in the leaf and flower development in Arabidopsis, but also contribute to unravel the molecular regulatory mechanism of YAB3 gene in the establishment of adaxial–abaxial polarity of the lateral organs in Pak-choi.
Keywords: BcYAB3, leaf development, adaxial–abaxial polarity, Pak-choi, Arabidopsis
1. Introduction
The leaf is a major organ of plants, responsible for photosynthesis, respiration, and transpiration. Therefore, the flattening of the leaf is essential for maximizing photosynthesis. Leaf shape generally depends on the development of three axes of polarity, namely the proximal–distal axis, adaxial–abaxial axis, and medial–lateral axis. Among them, the adaxial–abaxial axis serve as the basis for establishment of the medial–lateral axis and proximal–distal axis [1]. Moreover, the juxtaposition of adaxial and abaxial cells is essential for normal lamina outgrowth [2].
The adaxial–abaxial polarity is established in the development of lateral organs and controlled by several transcriptional factors and small non-coding RNAs, such as the HD-Zip III family, AS1/AS2 (ASYMMETRIC LEAVES1/2), KANADI family, ARF3/4 (AUXIN RESPONSE FACTOR3/4), YABBY family, and miR165/166 (microRNA165/166) [3,4,5,6,7,8]. YABBY genes belong exclusively to the seed plants [9], have a conserved C2C2 zinc finger like domain at the N-terminus, and helix–loop–helix motif analogous to a high mobility group (HMG) box (termed as YABBY domain) at the C-terminus [10,11,12]. In angiosperms, the YABBY family can be divided into five subgroups according to evolutionary analysis and expression profiles of YABBY genes [12,13], namely FILAMENTOUS FLOWER (FIL)/YABBY3 (YAB3), YABBY2 (YAB2), INNER NO OUTER (INO), CRABS CLAW (CRC), and YABBY5 (YAB5). In the previous works, a large number of YABBY genes were identified from different species; they participate in SAM development, establishment of leaf adaxial–abaxial polarity, and lamina and floral organ growth [14,15,16,17,18].
In Arabidopsis, FIL/YAB3/YAB5 are expressed in the abaxial regions of leaves and floral tissues [14,19]. The fil yab3 double mutant results in ectopic SAM structures, partial loss of abaxial cell identity, and abnormal vasculature, while the ectopic expression of FIL and YAB3 causes partial abaxialization of leaves [14]. In addition, fil yab3 yab5 triple and fil yab2 yab3 yab5 quadruple mutants exhibit dramatic polarity defects of lateral organs and loss of lamina expansion [11]. In Antirrhinum majus, the mutation of GRAMINIFOLIA (GRAN) gene, ortholog of FIL in Arabidopsis, leads to significant adaxialization along the abaxial margin of leaves [15]. In Solanum lycopersicum, LeYAB2 (a YABBY2-like gene) is involved in the development of pericarp cells in the abaxial region [20,21,22]. These results illustrate that YABBY genes play a pivotal role in defining the abaxial identity of lateral organs in eudicots. However, YABBY genes have different expression patterns in monocots. For example, the OsYAB1 gene, ortholog of YAB2 in Arabidopsis, has no polar characteristics when expressed in lateral organs [23]. In maize, YABBY genes are expressed in the adaxial sides of the leaves [24]. TaYAB1, a wheat YABBY gene, was overexpressed in Arabidopsis, the adaxial surface characteristic trend, and the formation of leaf adaxial polarity were affected [25]. These results demonstrated that functional divergence of YABBY genes occurred in monocots and dicots [26].
Pak-choi (Brassica rapa subsp. chinensis), which belongs to the Brassica species of the Cruciferae family, is an economically important vegetable and widely cultivated in Asia. Leaves are the main edible organs of Pak-choi. Therefore, it is significant to understand the regulatory mechanism of leaf polarity in Pak-choi. In Arabidopsis, FIL and YAB3 proteins are involved in the regulation of inflorescence phyllotaxy, abaxial patterning, and development of lateral organs. The function of the FIL homologous gene has been extensively studied in many species [19,27,28,29,30], while a functional role of the YAB3 gene has not been fully documented. In this study, the BcYAB3 gene was isolated from Pak-choi, and the transgenic Arabidopsis overexpressed BcYAB3 was acquired using the floral dip method. The ectopic expression of BcYAB3 induced strong leaf curling and flowering stage delay in Arabidopsis.
2. Materials and Methods
2.1. Plant Materials and Cultivation Management
In this study, the Pak-choi cultivar “suzhouqing” was used. The seeds were scattered in a plastic petri dish with wet filter paper and grown in pots containing an organic matrix, namely a vermiculite (2:1) mixture in phytotron under controlled environmental conditions (16 h light/8 h dark photoperiod at 23 °C/17 °C). The different tissue (leaf, petiole, stem, root, hypocotyl, flower bud, flower bud, pod) samples were collected at seedling, rosette, flowering, and podding stages. Nicotiana benthamiana and Arabidopsis thaliana wild type (ecotype Col-0) were used in this study and grown in illumination incubators under the same conditions.
2.2. Isolation and Characterization Analysis
Total RNAs were isolated from different tissues using an RNAeasy Mini Kit (Tiangen, Beijing, China), and the first strands of cDNA were synthesized via reverse transcription using a PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). The CDS of BcYAB3 was amplified using gene-specific primers based on the sequence of Chinese cabbage BrYAB3 (Bra037320, http://brassicadb.org/brad/index.php) via homology cloning according to a previous report [31]. The PCR product was cloned into the pMD18-T vector and sequenced by GenScript Company (Nanjing, China). The physicochemical characteristics of BcYAB3 were predicted using the Expasy website (https://web.expasy.org/protparam/). The secondary structure of BcYAB3 was predicted by PSIPRED 4.0 (http://bioinf.cs.ucl.ac.uk/psipred/).
2.3. Sequence Alignment and Phylogenetic Analysis
The sequences of Arabidopsis YABBY proteins were retrieved from the TAIR database (https://www.arabidopsis.org/). The protein sequence data of Solanum lycopersicum, Vitis vinifera, Zea mays, and Brassica oleracea were downloaded from Plant-TFDB (http://planttfdb.cbi.pku.edu.cn/index.php). The protein sequences for Oryza sativa, Antirrhinum majus, Triticum aestivum, Vitis pseudoreticulata, and Pak-choi were obtained from previous reports [29,32]. Sequencing alignment was obtained using ClustalW software. The phylogenetic analysis was performed using MEGA 7.0 software using the neighbor-joining (NJ) method, and the bootstrap value was set at 1000 replications. All protein sequences used in this study are listed in Table S1.
2.4. Subcellular Localization Analysis
The protein-coding region of BcYAB3 was introduced into the pRI101-GFP vector digested with BamHI and NdeI restriction enzymes to generate the construct 35S:BcYAB3-GFP. Then, the plasmid of fusion construct and empty vector were transformed into Agrobacterium GV3101. The epidermal cell transformation of tobacco leaves was carried out with injection of Agrobacterium. After dark culture for 24 h, the seedlings were transferred to normal growth for 24–36 h, and then fluorescent pictures were taken using a confocal laser scanning microscope (Zeiss, LSM780, Jena, Germany).
2.5. Transformation and Screening of BcYAB3 Transgenic Plants
The BcYAB3 CDS was subcloned into the binary vector pCAMBIA1301 to generate the 35S:BcYAB3–GUS plant expression vector. The recombinant construct was introduced into Agrobacterium GV3101 and then transformed into Arabidopsis thaliana (Col) using the floral dip method [33]. The seeds of the T0 generation were harvested and screened on solid Murashige and Skoog (MS) medium containing 30 mg/L hygromycin. Resistant plants were subjected to further verification using PCR amplification and qRT-PCR experiments. The T3 transgenic lines were used for subsequent phenotype observation and functional analysis. The leaf length and leaf width of transgenic and wild type Arabidopsis thaliana were measured using a Vernier caliper for 25-day-old seedlings, and 25 plants of each line were analyzed. ANOVA (analysis of variance) was used for statistical analysis.
2.6. Real-Time PCR
Total RNA of Pak-choi and Arabidopsis plants was extracted using an RNAeasy Mini Kit (Tiangen, Beijing, China), and the cDNA for real time PCR was synthesized using a PrimeScript™RT reagent Kit with gDNA Eraser (Takara, Dalian, China). qRT-PCR was carried out with SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (Takara, Dalian, China) using the ABI StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The PCR procedure was carried out with the following parameters: 95 °C for 30 s, 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. Furthermore, a melting curve was employed to verify the specificity of all reactions. The Actin gene (referred to as Bra028615) was used as a housekeeping gene to normalize the transcript levels of BcYAB3 genes among different tissues [34]. Actin of Arabidopsis was used as an internal control gene, and the transcript levels were calculated using the 2−ΔΔCt method [35]. Three biological and technical replicates were implemented for each reaction. All primers used in this study are listed in Table S2.
3. Results
3.1. Phylogenetic Analysis of BcYAB3
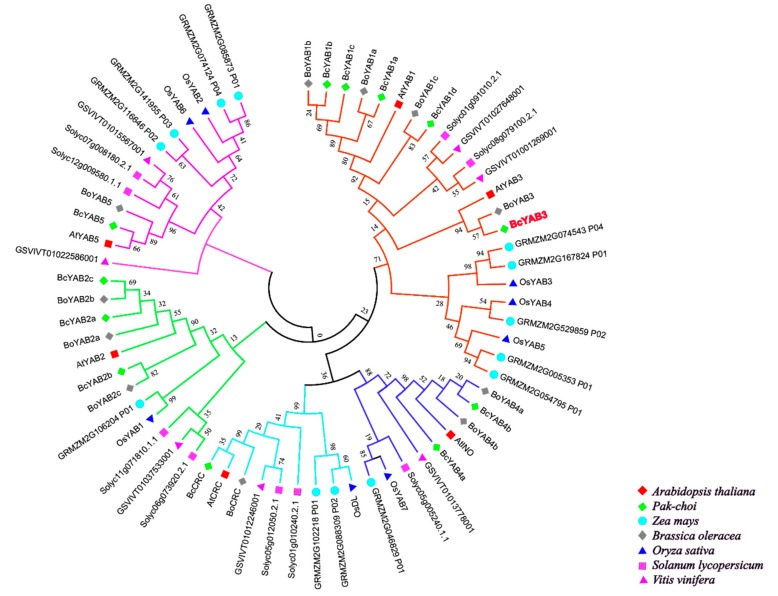
Until now, hundreds of YABBY genes have been identified in plants. For example, a total of 6 YABBY genes were isolated in Arabidopsis, 8 members in rice [36], 9 members in tomato [37], 12 members in Pak-choi [32], 13 members in maize [38], 17 members in soybean [39], et al. In this study, an unrooted phylogenetic tree was constructed based on protein sequences from different species, including two monocots (Oryza sativa, Zea mays) and five dicots (Arabidopsis, Solanum lycopersicum, Vitis vinifera, Brassica oleracea, Pak-choi) (Figure 1, Table S1). Several lines of evidence have shown that YABBY family members can be divided into five groups [9,13,36,40]. As shown in the tree, Pak-choi YABBY genes are distributed in five subclasses, and most YABBY genes from dicots are clustered together and separated from monocots, suggesting that they have functional differentiation. Therefore, the functions of Pak-choi YABBY genes could be deduced from previous studied YABBYs via phylogenetic relationships.
Figure 1.
Phylogenetic tree analysis of BcYAB3. Phylogenetic relationships of YABBY genes involving with Arabidopsis thaliana, Oryza sativa, Zea mays, Solanum lycopersicum, Vitis vinifera, Brassica oleracea, and Pak-choi. The phylogenetic tree was constructed using the neighbor-joining method, and the bootstrap value was set at 1000.
3.2. Characterization and Expression Profile of BcYAB3 Gene in Pak-choi
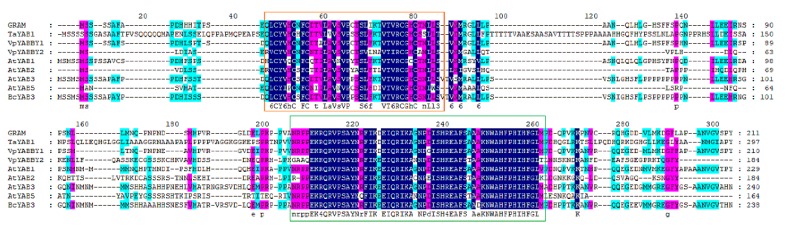
The BcYAB3 gene was isolated from Pak-choi using the homology cloning method. The full-length fragment of BcYAB3 contained a 717 bp coding sequence encoding 238 amino acid residues. Expasy online prediction indicated that the BcYAB3 protein is an unstable hydrophilic protein (GRAVY = −0.432); the molecular weight is 26.21 KDa, and theoretical isoelectric point (pI) is 8.66. The BcYAB3 protein has a highly conserved domain of the YABBY family at the N-terminal (20–198 amino acid sites), containing C2C2 zinc finger and HMG domains. In addition, secondary structure analysis demonstrated that the BcYAB3 protein is composed of a strand, helix, and coil (Figure 2, Figure S1).
Figure 2.
Multiple sequence alignment of BcYAB3. The BcYAB3 gene was aligned with YABBY genes and have been reported to be in Antirrhinum majus, Triticum aestivum, Vitis pseudoreticulata, and Arabidopsis thaliana. The conserved C2C2 zinc finger domain and YABBY domain are presented using an orange box and green box, respectively.
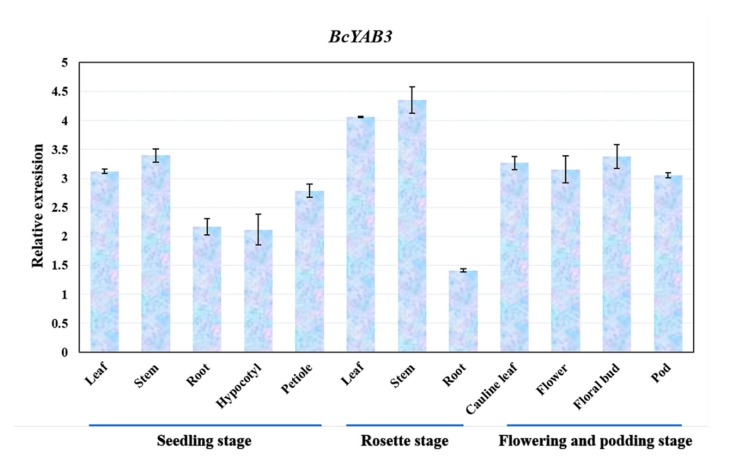
To understand the expression patterns of BcYAB3, samples of different tissues in three developmental growth stages of Pak-choi were collected and identified using a qRT-PCR assay. The result showed that BcYAB3 was expressed in diverse tissues and had a high expression in stems and leaves, followed by floral organs, but low expression in roots and the hypocotyl (Figure 3). This result is consistent with the function of YABBY genes in SAM and leaf development in previous studies.
Figure 3.
Expression patterns of BcYAB3 in different tissues. The expression levels of BcYAB3 with qRT-PCR amplification in leaf, stem, root, petiole, hypocotyl, flower bud, flower, and pod. The data represent the means of three replicates, and error bars represent the standard deviations of means.
3.3. BcYAB3 Shows Nuclear Localization
To verify whether the expression of BcYAB3, as a putative transcription factor, is localized in the cell nucleus, the 35S:BcYAB3-GFP recombinant construct was generated and transformed into tobacco epidermal cells through the Agrobacterium injection method. Under a laser scanning confocal microscope, the green fluorescence of the 35S:BcYAB3-GFP fusion protein was observed and localized specifically in the nuclei, whereas the protein of 35S:GFP was found to be expressed in both the nucleus and the cytoplasm (Figure 4). The results demonstrated that BcYAB3 is a nuclear-localized protein.
Figure 4.
Subcellular localization of BcYAB3. (A) The construct of 35S:GFP and 35S:BcYAB3-GFP. GFP: green fluorescent protein; NOS: nopaline synthase gene. (B) Transient expression of 35S:BcYAB3-GFP fusion protein in tobacco. 35S:GFP was used as a control. Fluorescence images of mCherry (a nuclear marker), GFP, and merged were captured with confocal laser scanning microscopy and are displayed in red, green, and yellow, respectively. Scale bars = 50 um.
3.4. Ectopic Expression of BcYAB3 Resulted in Pleiotropic Phenotypes in Arabidopsis
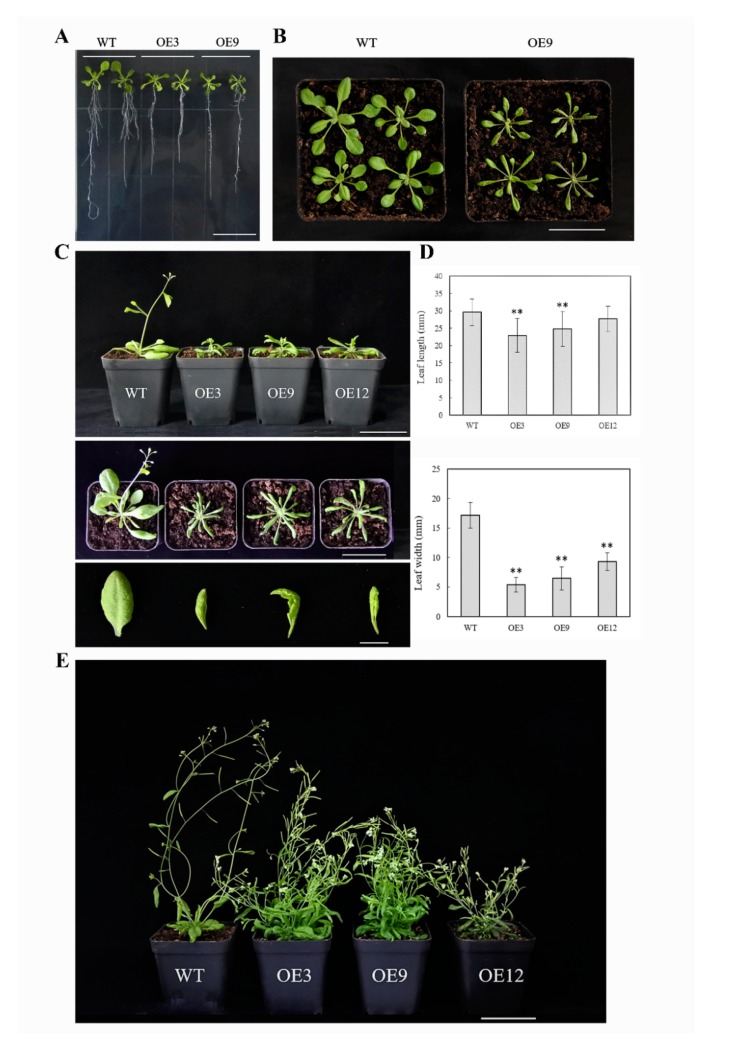
To investigate the function of BcYAB3, we firstly transformed BcYAB3 into Arabidopsis for overexpression. In this study, a total of thirteen transgenic lines (named as OE1–13) were acquired after screening by PCR amplification and qRT-PCR assay (Figure S2). Among them, seven transgenic lines with relatively higher expression levels were selected for further analysis. The overexpression lines of BcYAB3 could be classified into two types according to the phenotype observation.
In type I, which had a normal growth and development, the transgenic plants (OE3, OE9, OE12) were relatively dwarf compared to the wild type (Figure 5). In the seedling stage, the root system with fewer lateral roots was weaker than the wild type (Figure 5A), the leaves were mildly curled, and then became long–narrow and significantly curled downwards with plant growth (Figure 5B,C). The bolting and flowering stages were delayed; the height of the bolt was shorter than the wild type, while the number of floral axes and leaves increased tremendously. There was no obvious main stem in transgenic plants, and the plants were capable of bearing fruit.
Figure 5.
Type I phenotype of 35S:BcYAB3–GUS transgenic plants in Arabidopsis thaliana. (A) Fourteen days of BcYAB3 transgenic plants grown on Murashige and Skoog (MS) medium with hygromycin. Scale bar = 2 cm. (B) The leaves of 22-day-old seedlings. Scale bar = 5 cm. (C) Phenotypes of the whole plant and leaves during bolting stage. Scale bars indicate 5 cm and 1 cm, respectively. (D) Leaf length and width statistics of the rosette leaves of 25-day-old plants. Error bars represent standard deviation of the mean number of 25 plants for each line. ** means significant differences compared to wild type (P < 0.01). (E) Phenotype of BcYAB3 and wild type plants during flowering and fruiting stage. WT: wild-type Arabidopsis thaliana; OE3/9/12: transgenic lines of BcYAB3. Scale bar = 5 cm.
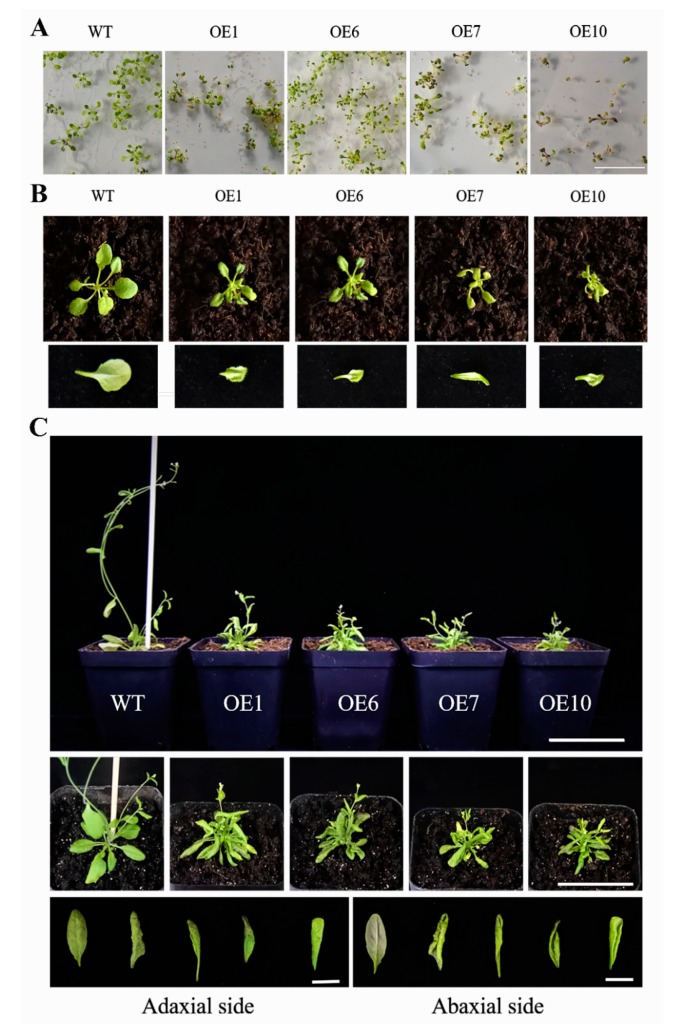
Type II, which had a severe phenotype, exhibited a diminutive and bushy plant phenotype (Figure 6). The leaf blades were clearly smaller than the wild type; the degree of downward curling of leaves gradually increased with the development of plants. The rosette leaves clustered together, and the flower stalks were shorter than the wild type at bolting and flowering stages; the height of transgenic plants was markedly decreased, and the ability to bear fruit was largely suppressed. In addition, a small number of purple seedlings were found in transgenic plants (Figure 6A, OE1/OE7/OE10), which may be caused by the accumulation of anthocyanins [14]. These results revealed a pivotal contribution of BcYAB3 in leaf development; moreover, the overexpression of BcYAB3 in Arabidopsis prolonged the vegetative period and delayed the flowering stage.
Figure 6.
Type II phenotype of transgenic Arabidopsis with BcYAB3 gene. (A) Phenotype of 10-old-day seedlings. Scale bar = 2 cm. (B) Phenotype of 18-old-day seedlings and characteristic of abaxial side of leaves in wild type and transgenic plants. Scale bar = 1 cm. (C) Phenotype of transgenic plants and wild type during flowering and fruiting stages. The phenotype was observed in a front view and top view, respectively. Scale bars = 5 cm. Comparison of the adaxial and abaxial sides of the leaves of wild type and transgenic plants. Scale bar = 1 cm. WT: wild-type Arabidopsis thaliana; OE1/6/7/10: transgenic lines of BcYAB3.
4. Discussion
Several lines of evidence show that YABBY genes have a general role in the development of the meristem [28,41], leaves [30], floral organs [42], and fruits [43]. In Arabidopsis, the YABBY gene family plays a pivotal role in the development of abaxial cell fate in lateral organs. Numerous YABBY genes involved in diverse processes of growth and development were also identified and studied in other plant species [14,16,44,45]. Nevertheless, the functions of Pak-choi YABBY genes in plant development have not yet been revealed. BcYAB3, belonging to the FIL/YAB3 subgroup, is homologous to YAB3 in Arabidopsis, which is related to leaf polarity, implying that BcYAB3 possibly has a similar function in the regulation of leaf development. We profiled the transcript level of BcYAB3 in various tissues at three developmental stages, including leaves, stems, roots, petioles, hypocotyls, floral organs, and pods. The qRT-PCR results indicated that BcYAB3 was highly expressed in stems and leaves, especially in the rosette stage, followed by floral organs, suggesting that BcYAB3 possibly participates in the regulation of leaf and floral development.
Previous reports showed that FIL, YAB2, YAB3, and YAB5 genes were associated with the development of vegetative tissues in Arabidopsis, while INO and CRC were involved in the ovules and carpels [10,19,46]. Moreover, the function of YABBY genes in tomato, Antirrhinum, and cabbage is similar to the Arabidopsis YABBY genes [15,37], indicating that YABBY genes have a conserved function in dicotyledonous plants. However, YABBY genes have different roles in monocotyledons, such as maize, rice and wheat [23,24,25,36,47], shown by the functional divergent phenomena YABBY genes produced in monocotyledons and dicotyledons. In this study, an unrooted phylogenetic tree was constructed and exhibited an evident separation between monocotyledons and dicotyledons, which was consistent with previous studies. Thus, the roles of Pak-choi YABBY genes can be speculated based on the results of previous research.
To further investigate the function of BcYAB3 gene, we firstly transformed the BcYAB3 gene into Arabidopsis, and thirteen transgenic lines were obtained via screening and verification of PCR amplification and GUS staining. In comparison with wild-type plants, the leaves of transgenic plants with 35S:BcYAB3–GUS exhibited outward curled cotyledons, and then became long–narrow and dramatically curled towards the abaxial side with increases in leaf age. Measurement of leaf traits with 25-day-old seedlings showed that the width of transgenic Arabidopsis plants was significantly shorter than that of the wild type. This phenotype has a similarity with Arabidopsis overexpressed AtYAB3 to some extent, which produced epinastic and narrow leaves. However, in most cases, the SAM of transgenic seedlings expressing ectopic AtYAB3 was disrupted and only produced cotyledons [14]. In this study, the transgenic plants overexpressed BcYAB3 produced cotyledons and leaves in both type I and type II, suggesting the functional difference of YAB3 gene existed in different species. Furthermore, the bolting and flowering stages were delayed in transgenic plants with 35S:BcYAB3–GUS, the inflorescence rachis increased, and leaves clustered together during the flowering stage. Conclusively, these data confirmed that BcYAB3 plays a crucial role in specifying abaxial cell fate in lateral organs originated from apical and flower meristems.
5. Conclusions
In summary, this study provides first insights into the function of the Pak-choi BcYAB3 gene and showed its implications in the development of leaves. This study paves the way for future research on YABBY genes to understand the regulatory mechanism of leaf polarity in Brassica species. Furthermore, the function of BcYAB3 in delaying the flowering stage will contribute to vegetable crops to prolong the harvest period for improving crop yield and achieving the objective of long-term market supply.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/4/370/s1, Table S1: Sequences of YABBY proteins used in this study. Table S2: Primers used in this study. Figure S1: The secondary structure of BcYAB3. Figure S2: The verification of T3 positive transgenic plants.
Author Contributions
Conceptualization, X.H. and H.H.; Formal Analysis, H.H.; Investigation, H.H. and Y.L.; H.H. wrote the manuscript; Writing—Original Draft Preparation, H.H.; Writing—Review and Editing, X.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by The Key Projects of National Key Research and Development Plan (2017YFD0101803), the China Agriculture Research System (CARS-23-A-06), the National Key R&D Program of China (2016YFD0101701).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Reinhardt D., Mandel T., Kuhlemeier C. Auxin Regulates the Initiation and Radial Position of Plant Lateral Organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waites R., Hudson A. phantastica: A gene required for dorsiventrality of leaves in Antirrhinum majus. Development. 1995;121:2143–2154. [Google Scholar]
- 3.Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C.P. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- 4.Chitwood D.H., Guo M., Nogueira F.T.S., Timmermans M.C.P. Establishing leaf polarity: The role of small RNAs and positional signals in the shoot apex. Development. 2007;134:813–823. doi: 10.1242/dev.000497. [DOI] [PubMed] [Google Scholar]
- 5.Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 6.Iwakawa H., Iwasaki M., Kojima S., Ueno Y., Soma T., Tanaka H., Semiarti E., Machida Y., Machida C. Expression of the ASYMMETRIC LEAVES2 Gene in the Adaxial Domain of Arabidopsis Leaves Represses Cell Proliferation in This Domain and Is Critical for the Development of Properly Expanded Leaves. Plant J. 2007;51:173–184. doi: 10.1111/j.1365-313X.2007.03132.x. [DOI] [PubMed] [Google Scholar]
- 7.Eckardt N.A. YABBY Genes and the Development and Origin of Seed Plant Leaves. Plant Cell. 2010;22:2103. doi: 10.1105/tpc.110.220710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pekker I., Alvarez J.P., Eshed Y. Auxin Response Factors Mediate Arabidopsis Organ Asymmetry via Modulation of KANADI Activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finet C., Floyd S.K., Conway S.J., Zhong B., Scutt C.P., Bowman J.L. Evolution of the YABBY gene family in seed plants. Evol. Dev. 2016;18:116–126. doi: 10.1111/ede.12173. [DOI] [PubMed] [Google Scholar]
- 10.Bowman J.L., Smyth D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development. 1999;126:2387–2396. doi: 10.1242/dev.126.11.2387. [DOI] [PubMed] [Google Scholar]
- 11.Sarojam R., Sappl P.G., Goldshmidt A., Efroni I., Floyd S.K., Eshed Y., Bowman J.L. Differentiating Arabidopsis Shoots from Leaves by Combined YABBY Activities. Plant Cell. 2010;22:2113–2130. doi: 10.1105/tpc.110.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowman J.L. The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 2000;3:17–22. doi: 10.1016/S1369-5266(99)00035-7. [DOI] [PubMed] [Google Scholar]
- 13.Bartholmes C., Hidalgo O., Gleissberg S. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae) Plant Biol. 2012;14:11–23. doi: 10.1111/j.1438-8677.2011.00486.x. [DOI] [PubMed] [Google Scholar]
- 14.Siegfried K.R., Eshed Y., Baum S.F., Otsuga D., Drews G.N., Bowman J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- 15.Golz J.F., Roccaro M., Kuzoff R., Hudson A. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development. 2004;131:3661–3670. doi: 10.1242/dev.01221. [DOI] [PubMed] [Google Scholar]
- 16.Han H.Q., Liu Y., Jiang M.M., Ge H.Y., Chen H.Y. Identification and expression analysis of YABBY family genes associated with fruit shape in tomato (Solanum lycopersicum L.) Genet. Mol. Res. 2015;14:7079–7091. doi: 10.4238/2015.June.29.1. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X.L., Zhang L.G. Molecular cloning and expression of the male sterility-related CtYABBY1 gene in flowering Chinese cabbage (Brassica campestris L. ssp. chinensis var. parachinensis) Genet. Mol. Res. 2014;13:4336–4347. doi: 10.4238/2014.June.10.1. [DOI] [PubMed] [Google Scholar]
- 18.Yang C.J., Kursel L.E., Studer A.J., Bartlett M.E., Whipple C.J. A Gene for Genetic Background in Zea mays: Fine-Mapping enhancer of teosinte branched1.2 to a YABBY Class Transcription Factor. Genetics. 2016;204:1573. doi: 10.1534/genetics.116.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawa S., Watanabe K., Goto K., Liu Y.G., Shibata D., Kanaya E., Morita E.H., Okada K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999;13:1079–1088. doi: 10.1101/gad.13.9.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartley G.E., Ishida B.K. Digital fruit ripening: Data mining in the TIGR tomato gene index. Plant Mol. Biol. Report. 2002;20:115–130. doi: 10.1007/BF02799427. [DOI] [Google Scholar]
- 21.Bartley G.E., Ishida B.K. Developmental gene regulation during tomato fruit ripening andin-vitrosepal morphogenesis. BMC Plant Biol. 2003;3:4. doi: 10.1186/1471-2229-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartley G.E., Ishida B.K. Ethylene-sensitive and insensitive regulation of transcription factor expression during in vitro tomato sepal ripening. J. Exp. Bot. 2007;58:2043–2051. doi: 10.1093/jxb/erm075. [DOI] [PubMed] [Google Scholar]
- 23.Jang S., Hur J., Kim S.-J., Han M.-J., Kim S.-R., An G. Ectopic expression ofOsYAB1causes extra stamens and carpels in rice. Plant Mol. Biol. 2004;56:133–143. doi: 10.1007/s11103-004-2648-y. [DOI] [PubMed] [Google Scholar]
- 24.Juarez M.T., Twigg R.W. Specification of adaxial cell fate during maize leaf development. Development. 2004;131:4533–4544. doi: 10.1242/dev.01328. [DOI] [PubMed] [Google Scholar]
- 25.Zhao W., Su H.Y., Song J., Zhao X.Y., Zhang X.S. Ectopic expression of TaYAB1, a member of YABBY gene family in wheat, causes the partial abaxialization of the adaxial epidermises of leaves and arrests the development of shoot apical meristem in Arabidopsis. Plant Sci. 2006;170:364–371. doi: 10.1016/j.plantsci.2005.09.008. [DOI] [Google Scholar]
- 26.Zhao X.Y., Xie H.T., Chen X.B., Wang S.S., Zhang X.S. Ectopic Expression of TaYAB2, a Member of YABBY Gene Family in Wheat, Causes Partial Abaxialization of Adaxial Epidermises of Leaves in Arabidopsis. Acta Agron. Sin. 2012;38:2042–2051. doi: 10.3724/SP.J.1006.2012.02042. [DOI] [Google Scholar]
- 27.Filyushin M.A., Slugina M.A., Shchennikova A.V., Kochieva E.Z. Identification and Expression Analysis of the YABBY1 Gene in Wild Tomato Species. Russ. J. Genet. 2018;54:536–547. doi: 10.1134/S1022795418050022. [DOI] [Google Scholar]
- 28.Zhang X.L., Yang Z.-P., Zhang J., Zhang L.-G. Ectopic Expression of BraYAB1-702, a Member of YABBY Gene Family in Chinese Cabbage, Causes Leaf Curling, Inhibition of Development of Shoot Apical Meristem and Flowering Stage Delaying in Arabidopsis thaliana. Int. J. Mol. Sci. 2013;14:14872–14891. doi: 10.3390/ijms140714872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H., Shi G., Li X., Hu D., Cui Y., Hou J., Yu D., Huang F. Overexpression of a soybean YABBY gene, GmFILa, causes leaf curling in Arabidopsis thaliana. BMC Plant Biol. 2019;19:234. doi: 10.1186/s12870-019-1810-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vosnakis N., Maiden A., Kourmpetli S., Hands P., Drea S. A FILAMENTOUS FLOWER orthologue plays a key role in leaf patterning in opium poppy. Plant J. 2012;72:662–673. doi: 10.1111/j.1365-313X.2012.05112.x. [DOI] [PubMed] [Google Scholar]
- 31.Song X., Li Y., Liu T., Duan W., Huang Z., Wang L., Tan H., Hou X. Genes associated with agronomic traits in non-heading Chinese cabbage identified by expression profiling. BMC Plant Biol. 2014;14:71. doi: 10.1186/1471-2229-14-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou H., Wu P., Gao L., Zhang C., Hou X. Characterization and expression profile analysis of YABBY family genes in Pak-choi (Brassica rapa ssp. chinensis) under abiotic stresses and hormone treatments. Plant Growth Regul. 2019;87:421–432. doi: 10.1007/s10725-019-00475-5. [DOI] [Google Scholar]
- 33.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thal-iana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C., Lyu S., Gao L., Song X., Li Y., Hou X. Genome-Wide Identification, Classification, and Expression Analysis of SNARE Genes in Chinese Cabbage (Brassica rapa ssp. pekinensis) Infected by Turnip mosaic virus. Plant Mol. Biol. Report. 2018;36:210–224. doi: 10.1007/s11105-017-1060-0. [DOI] [Google Scholar]
- 35.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toriba T., Harada K., Takamura A., Nakamura H., Ichikawa H., Suzaki T., Hirano H.Y. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genom. 2007;277:457–468. doi: 10.1007/s00438-006-0202-0. [DOI] [PubMed] [Google Scholar]
- 37.Huang Z., Van Houten J., Gonzalez G., Xiao H., van der Knaap E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genom. 2013;288:111–129. doi: 10.1007/s00438-013-0733-0. [DOI] [PubMed] [Google Scholar]
- 38.Min G.E., Yuan-Da L., Zhang T.F., Tan L.I., Zhang X.L., Zhao H. Genome-wide identification and analysis of YABBY gene family in maize. Jiangsu J. Agric. Sci. 2014;30:1267–1272. [Google Scholar]
- 39.Zhao S.P., Lu D., Yu T.-F., Ji Y.-J., Zheng W.-J., Zhang S.-X., Chai S.-C., Chen Z.-Y., Cui X.-Y. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol. Biochem. 2017;119:132–146. doi: 10.1016/j.plaphy.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 40.Yamada T., Ito M., Kato M. YABBY2-Homologue Expression in Lateral Organs ofAmborella trichopoda (Amborellaceae) Int. J. Plant Sci. 2004;165:917–924. doi: 10.1086/423793. [DOI] [Google Scholar]
- 41.Tanaka W., Toriba T., Hirano H.Y. Three TOB1-related YABBY genes are required to maintain proper function of the spikelet and branch meristems in rice. New Phytol. 2017;215:825–839. doi: 10.1111/nph.14617. [DOI] [PubMed] [Google Scholar]
- 42.Alvarez J., Smyth D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development. 1999;126:2377–2386. doi: 10.1242/dev.126.11.2377. [DOI] [PubMed] [Google Scholar]
- 43.Cong B., Barrero L.S., Tanksley S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008;40:800–804. doi: 10.1038/ng.144. [DOI] [PubMed] [Google Scholar]
- 44.Yang C., Ma Y.M., Li J.X. The rice YABBY4 gene regulates plant growth and development through modulating the gibberellin pathway. J. Exp. Bot. 2016;18:5545–5556. doi: 10.1093/jxb/erw319. [DOI] [PubMed] [Google Scholar]
- 45.Sun X., Guan Y., Hu X. Isolation and Characterization of IaYABBY2 Gene from Incarvillea arguta. Plant Mol. Biol. Report. 2014;32:54144–54148. doi: 10.1007/s11105-014-0725-1. [DOI] [Google Scholar]
- 46.Villanueva J.M., Broadhvest J., Hauser B.A., Meister R.J., Schneitz K., Gasser C.S. INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev. 1999;13:3160–3169. doi: 10.1101/gad.13.23.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H.Y. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell. 2004;16:500–509. doi: 10.1105/tpc.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.