Abstract
As the novel coronavirus (SARS-CoV-2) continues to spread rapidly across the globe, we aimed to identify and summarize the existing evidence on epidemiological characteristics of SARS-CoV-2 and the effectiveness of control measures to inform policymakers and leaders in formulating management guidelines, and to provide directions for future research. We conducted a systematic review of the published literature and preprints on the coronavirus disease (COVID-19) outbreak following predefined eligibility criteria. Of 317 research articles generated from our initial search on PubMed and preprint archives on 21 February 2020, 41 met our inclusion criteria and were included in the review. Current evidence suggests that it takes about 3-7 days for the epidemic to double in size. Of 21 estimates for the basic reproduction number ranging from 1.9 to 6.5, 13 were between 2.0 and 3.0. The incubation period was estimated to be 4-6 days, whereas the serial interval was estimated to be 4-8 days. Though the true case fatality risk is yet unknown, current model-based estimates ranged from 0.3% to 1.4% for outside China. There is an urgent need for rigorous research focusing on the mitigation efforts to minimize the impact on society.
Keywords: COVID-19, SARS-CoV-2, epidemiology, basic reproduction number, incubation period, serial interval, severity
1. Introduction
The coronavirus disease 2019 (COVID-19) outbreak, which originated in Wuhan, China, has now spread to 192 countries and administrative regions infecting nearly 800,000 individuals of all ages as of 31 March 2020 [1]. Though most infected individuals exhibit mild symptoms including fever, upper respiratory tract symptoms, shortness of breath, and diarrhea [2], or are asymptomatic altogether [3], severe cases of infection can lead to pneumonia, multiple organ failure, and death [4]. Globally, at least 7900 deaths have been directly attributed to COVID-19 [1], and this number is expected to rise with the ongoing epidemic.
Posing a significant global health threat, COVID-19 has drawn unprecedented attention from public health researchers around the globe, with more than 200 research articles published in academic journals in two months. There are also nearly 150 epidemiological and modeling preprints on COVID-19. Many of these articles seek to investigate the epidemiological parameters of the disease at different locations to disseminate critical information among both modelers and policymakers for a timely control response to be implemented. This is particularly crucial as the current outbreak involves a new pathogen (SARS-CoV-2), on which limited knowledge exists of its infectivity and clinical profile.
The current estimates from this research vary widely, partly due to the differences in analytical methods and assumptions. This variance is also reflected in the estimates on the effectiveness of public health interventions implemented worldwide. As the COVID-19 outbreak is at a decisive point—as pointed out by the Director-General of the WHO on 27 February 2020—it is imperative to synthesize all existing evidence available to date and summarize the key findings to identify research gaps and to assist policymakers in evidence-based decision making for better pandemic preparedness.
Based on a framework of parameter requirements and necessary modeling efforts outlined by Cowling and Leung [5], we performed a systematic review of the literature available on transmission dynamics, severity, susceptibility and control measures to inform policymakers and leaders in formulating management guidelines and to provide directions for future research.
2. Materials and Methods
In this systematic review, we identified studies that describe or assess the transmission dynamics, severity, and susceptibility of COVID-19 and that examine the impact of early control measures.
2.1. Inclusion Criteria
We selected research articles that contain estimates for at least one of the following epidemiological parameters: (i) the size of the epidemic, (ii) the epidemic doubling time, which is defined as the time it takes for the number of cases to double in the epidemic, (iii) the basic reproduction number (), which is defined as an average number of secondary cases generated by an index case in a totally susceptible population, (iv) the incubation period, which is defined as the time between exposure and symptom onset, (v) the serial interval, which is defined as the time between symptom onset of successive infections, (vi) the susceptibility, in demographical and clinical profile, and (vii) the severity, in terms of symptom profile and case fatality risk. We also included articles that estimated the effectiveness of (viii) control measures, such as travel restrictions, quarantine, or airport screening.
2.2. Literature Search
We searched PubMed and preprint archives for research articles published up to 21 February 2020 using the following terms: “COVID-19”, “SARS-CoV-2”, “2019-nCoV”, “n-CoV”, and “coronavirus.”
At the initial stage of screening, any articles that were published before 1 December 2019 were excluded. Research titles were independently reviewed by two authors to eliminate studies that did not meet our inclusion criteria before the full review of abstracts and full-text of selected studies.
2.3. Additional Analysis
In addition, we calculated the final attack rate F based on the reproduction number estimates extracted from the published and preprint studies using the implicit formula [6]:
| (1) |
3. Results
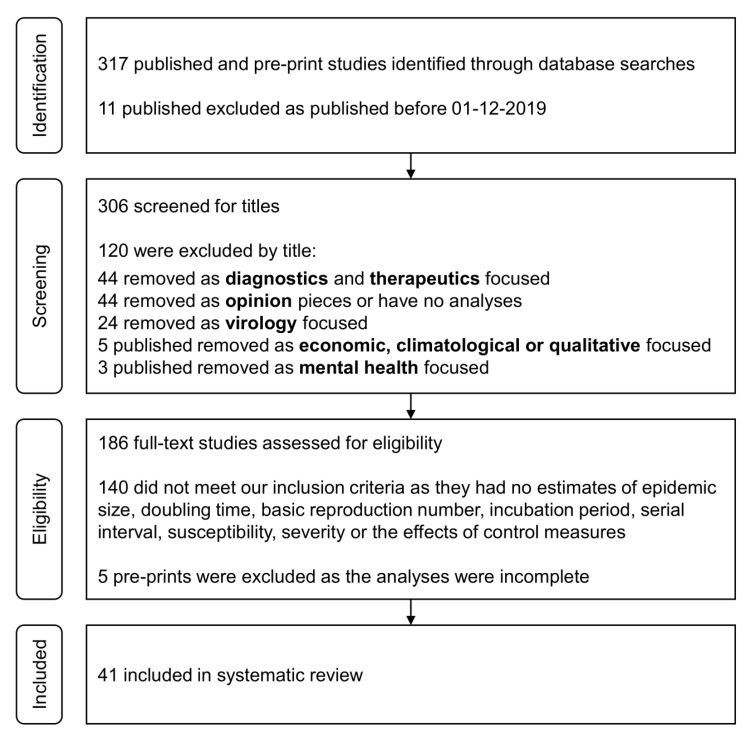
Of 317 articles generated from the initial search on PubMed and preprint archives, we identified 41 epidemiological and modeling studies on the COVID-19 outbreak in 2019–20 through title and abstract screening. The detailed selection process is illustrated in Figure 1.
Figure 1.
PRISMA flow diagram.
3.1. Size of the Outbreak at Epicentre
Current estimates for the epidemic size in Wuhan largely varies, ranging from 12,400 (95% CrI 3112–58,465) to 75,815 (37,304–130,330) for published articles and from 18,566 (14,134–22,978) to 58,956 (90% CI: 40,759–87,471) for preprints, by the 4th week of January 2020 (Table 1). The epidemic doubling time is estimated to be between 6.4 and 7.4 days based on three studies that have been published to date, and between 2.9 and 4.6 days according to two preprint studies.
Table 1.
Estimated size of the outbreak and epidemic doubling time from selected studies.
| Author | Data | Estimates | Estimation Period | Doubling Time |
|---|---|---|---|---|
| Published (2020) | ||||
| Du et al. [7] | Number of confirmed cases outside China and travel data | 12,400 in Wuhan | By 22 Jan 2020 | 7.31 days |
| Wu et al. [8] | Number of confirmed cases outside China and travel data | 75,815 in Wuhan | By 25 Jan 2020 | 6.4 days |
| Nishiura et al. [9] | Proportion of asymptomatic cases among Japanese evacuated from Wuhan | 20,767 in Wuhan | By 29 Jan 2020 | - |
| Li et al. [10] | Case reports from Wuhan | - | By 22 Jan 2020 | 7.4 days |
| Preprint | ||||
| Cao et al. [11] | Number of confirmed cases in China and travel data | 18,556 in Wuhan | By 23 Jan 2020 | - |
| Chinazzi et al. [12] | Number of confirmed cases outside China and travel data | 58,956 in Wuhan | By 23 Jan 2020 | 4.6 days |
| Xiong et al. [13] | Number of confirmed cases in China | 49,093 in China | By 16 Feb 2020 | - |
| Q. Zhao et al. [14] | Number of confirmed cases outside China and travel data | − | By 23 Jan 2020 | 2.9 days |
Three published studies have estimated the final epidemic size in Wuhan. An early analysis based on a mathematical model and recent travel data estimated that more than 75,815 (95% CrI 37,304–130,330) individuals had been infected in Wuhan by 25 Jan 2020, as the epidemic doubled in size every 6.4 days [8]. Using a similar approach, an analysis of mobility data including the number of passengers traveling between Wuhan and other cities in China estimated 12,400 infections (3112–58,465) in Wuhan by 22 Jan 2020 with a case detection rate of 8.95% (2.22–28.72%) [7]. Another study, which used case reports of Japanese citizens evacuated from Wuhan, reported that there might be 20,767 (9528–38,421) infected individuals in Wuhan [9] with a 9.2% ascertainment rate.
We also reviewed three preprints that included the final epidemic size in Wuhan and China. A preprint study found that the median estimate for the total number of infections in Wuhan was 58,956 (90% CI: 40,759–87,471) before the travel ban was implemented on 23 January 2020 [12]. The article also reported an epidemic doubling time of 4.6 days (4.2–5.1). The same study estimated the median ascertainment rate to be 19.59% (IQR: 14.36% and 35.58%). Accounting for the travel ban that was implemented in 23 Jan 2020, a more recent analysis found that the disease was spreading quicker, which was based on cases confirmed in six countries and administrative regions across Asia by 5 February 2020 with the estimated epidemic doubling time of 2.9 days (95% CrI: 2.0–4.1) [14].
Overall, these estimates support an ascertainment rate of 2.22–35.58%, providing evidence of a large number of non-severe cases being undocumented by healthcare systems.
3.2. Transmissibility of SARS-CoV-2
3.2.1. Basic Reproduction Number ()
Current estimates of the mean range from 1.9 to 6.5 based on eight published and eight preprint papers. Of 20 estimates, 13 are within the range of 2.0 and 3.0 (Table 2). The estimates are comparable to that of SARS-CoV, which was estimated when excluding superspreading events in the early phase of the outbreak in Hong Kong (2.7) and Singapore (2.2–3.6).
Table 2.
Estimated basic reproduction number () from selected studies.
| Author | Method | Estimates | Uncertainty | Estimation Period | |||
|---|---|---|---|---|---|---|---|
| Published (2020) | |||||||
| S. Zhao et al. [15] | Exponential growth model | 2.56 | 2.49 | – | 2.63 | 1–15 Jan 2020 | 95%CI |
| S. Zhao et al. [16] # | Exponential growth model | 2.24 | 1.96 | – | 2.55 | 10–24 Jan 2020 | 95%CI |
| S. Zhao et al. [16] ^ | Exponential growth model | 3.58 | 2.89 | – | 4.39 | 10–24 Jan 2020 | 95%CI |
| Riou et al. [17] | Stochastic simulations of outbreak trajectories | 2.2 | 1.4 | – | 3.8 | By 18 Jan 2020 | 90%HDI * |
| Li et al. [10] | Analysis of epidemiological data | 2.2 | 1.4 | – | 3.9 | By 22 Jan 2020 | 95%CI |
| Tang et al. [18] | SEIR model § | 6.47 | 5.71 | – | 7.23 | By 22 Jan 2020 | 95%CI |
| Du et al. [7] | Hierarchical model | 1.90 | 1.47 | – | 2.59 | By 22 Jan 2020 | 95%CI |
| Jung et al. [19] † | Epidemic growth model | 2.1 | 2 | – | 2.2 | By 24 Jan 2020 | 95%CI |
| Jung et al. [19] ‡ | Epidemic growth model | 3.2 | 2.7 | – | 3.7 | By 24 Jan 2020 | 95%CI |
| Wu et al. [8] | SEIR model | 2.68 | 2.47 | – | 2.86 | By 25 Jan 2020 | 95%CI |
| Preprint | |||||||
| Shen et al. [20] | SEIJR model §§ | 4.71 | 4.5 | – | 4.92 | On 12 Dec 2019 | 95%CI |
| Shen et al. [20] | SEIJR model | 2.08 | 1.99 | – | 2.18 | On 22 Jan 2020 | 95%CI |
| Read et al. [21] | SEIR model | 3.8 | 3.6 | – | 4.0 | By 22 Jan 2020 | 95%CI |
| Liu et al. [22] | Exponential growth model | 2.90 | 2.32 | – | 3.63 | By 23 Jan 2020 | 95%CI |
| Liu et al. [22] | MLE ¶ | 2.92 | 2.28 | – | 3.67 | By 23 Jan 2020 | 95%CI |
| Chinazziet al. [12] | GLEAM ** and SLIR ## | 2.4 | 2.2 | – | 2.6 | By 23 Jan 2020 | 90%CI |
| Q. Zhao et al. [14] | Exponential growth model | 5.7 | 3.4 | – | 9.2 | By 23 Jan 2020 | 95%CI |
| Cao et al. [11] | Geo-stratified debiasing estimation framework | 3.24 | By 23 Jan 2020 | ||||
| Majumder et al. [23] | Incidence Decay and Exponential Adjustment | 2.5 | 2.0 | – | 3.1 | By 26 Jan 2020 | Range |
| Xiong et al. [13] | EIR model (I = Identified) | 2.7 | By 16 Feb 2020 | ||||
| Comparison with SARS-CoV and MERS-CoV | |||||||
| SARS-CoV [24] | Hong Kong (2003) | 2.7 | 2.2 | – | 3.7 | Early phase | 95%CI |
| SARS-CoV [25] | Singapore (2003) | - | 2.2 | – | 3.6 | Early phase | Range |
| MERS-CoV [26] | South Korea (2012-2013) | 0.91 | 0.36 | – | 1.44 | 95%CI | |
#: assuming 8-fold increase in the reporting rate; ^: assuming 2-fold increase in the reporting rate; * HDI: high density interval; §: SEIR= susceptible-exposed-infectious-recovered; †: start date for exponential growth fixed at 8 Dec 2019; ‡: start date for exponential growth varying between 1–10 Dec 2019; §§: SEIJR = SEIR with J = isolated with treatment; ¶: MLE = maximum likelihood estimation; **: GLEAM = global epidemic and mobility model; ## SLIR = susceptible-latent-infectious-recovered
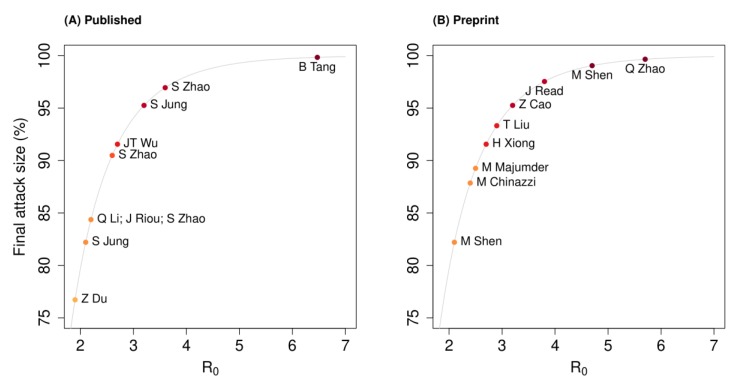
The highest estimate of 6.47 is from an early analysis using the number of cases reported in China until 22 Jan 2020, where contact rates were assumed to be higher during the Lunar New Year holiday period [18]. Assuming no interventions, the final attack rate would lie between 75% and 100% in a completely susceptible population, as presented in Figure 2.
Figure 2.
Projected final attack rate based on the basic reproduction number (Ro) estimates from: (A) published and (B) preprint articles, assuming no interventions are implemented.
3.2.2. Incubation Period
We identified six published articles and two preprints that reported the estimated incubation period distribution based on the epidemiological data collected from China and several other countries (Table 3).
Table 3.
Estimated incubation period from selected studies.
| Author | Country/Region | Sample Size | Estimate | Uncertainty | |||
|---|---|---|---|---|---|---|---|
| Published (2020) | |||||||
| Li et al. [10] | Wuhan | 10 cases | 5.2 | 4.1 | – | 7.0 | 95% CI |
| Backer et al. [27] | Outside Wuhan | 88 cases | 6.4 | 5.6 | – | 7.7 | 95% CI |
| Linton et al. [28] | Wuhan | 158 cases | 5.6 | 5.0 | – | 6.3 | 95% CI |
| Linton et al. [28] | Outside Wuhan | 52 cases | 5.0 | 4.2 | – | 6.0 | 95% CI |
| Ki [29] | South Korea | 22 cases | 3.6 | 1.0 | – | 9.0 | Range |
| Jiang et al. [30] | Global | 50 cases | 4.9 | 4.4 | – | 5.5 | 95% CI |
| Guan et al. * [2] | China | 291 cases | 4.0 | 2.0 | – | 7.0 | IQR |
| Preprint | |||||||
| Lauer et al. [31] | Global (excl. Hubei) | 101 cases | 5.2 | 4.4 | – | 6.0 | 95% CI |
| Zhang et al. [32] | China (excl. Hubei) | 49 cases | 5.2 | 1.8 | – | 12.4 | 95% CI |
| Comparison with SARS-CoV and MERS-CoV | |||||||
| SARS-CoV (2003) [33] | Hong Kong | 4.4 | |||||
| MERS-CoV (2012-3) [26] | Global | 5.5 | 3.6 | – | 10.2 | 95% CI | |
| MERS-CoV (2015) [34] | South Korea | 6.7 | 6.1 | – | 7.3 | 95% CI | |
*: The study was initially included as a preprint but has been moved to ‘Published’ as it was published on 28 February 2020.
All nine estimates extracted from eight published literature and preprints have the mean (or median) and uncertainty of shorter than 13 days (Table 3).
Current estimates for the mean or median incubation period range from 4 to 6 days, comparable to SARS-CoV (4.4 days) [33] and MERS-CoV (5.5 days worldwide) [26]. Two published studies, including an early analysis of 291 patients confirmed in China, reported the median incubation period of 4 days [2]. Five out of nine estimates indicated a mean incubation period of 5 days. A case analysis of 10 confirmed patients in Wuhan reported a mean incubation period of 5.2 days (95% CI: 4.1–7.0) with a 95th percentile of the distribution at 12.5 days (9.2–18) [10]. Another analysis estimated the mean incubation period to be 5.0 days (4.2–6.0) and the time from symptom onset to hospital admission to be 3.3 days (2.7–4.0) [28]. A recently published article reported a mean incubation period of 4.9 days (4.4–5.5) based on an analysis of confirmed patients with well-defined exposure dates. This study also measured the incubation period for other coronaviruses and concluded that there is no significant difference in the incubation period among SARS-CoV-2, SARS-CoV (4.7 days), and MERS-CoV (5.8 days) [30].
Similarly, a preprint using cases diagnosed in China (excluding Wuhan) and other countries reported a median incubation period of 5.2 days (4.4–6.0) [31], which is the same as that estimated based on the cases from Wuhan [10]. The study also noted, however, that 64 out of 10,000 cases may develop symptoms after the 14-day quarantine period. Another preprint also estimated the mean incubation period to be 5.2 days but with a larger confidence interval (1.8–12.4) compared to the aforementioned study [32].
3.2.3. Serial Interval
Current estimates of the mean serial interval for COVID-19 ranges from 5 to 8 days from two published studies, and ~4 to 5 days based on four preprints (Table 4).
Table 4.
Estimated serial interval from selected studies.
| Author | Country | Sample Size | Estimate | 95% CI | ||
|---|---|---|---|---|---|---|
| Published (2020) | ||||||
| Li et al. [10] | Wuhan | 6 pairs | 7.5 | 5.3 | – | 19.0 |
| Ki [29] | Korea | 7 pairs | 4.6 | 3.0 | – | 9.0 |
| Preprint | ||||||
| Du et al. [35] | China (excl. Hubei) | 468 pairs | 3.96 | 3.53 | – | 4.39 |
| Zhang et al. [32] | China (excl. Hubei) | 35 pairs | 5.1 | 1.3 | – | 11.6 |
| Nishiura et al. [36] | Global | 28 pairs | 4.0 | 3.1 | – | 4.9 |
| Nishiura et al. [36] | Global | 18 pairs | 4.6 | 3.5 | – | 5.9 |
| S. Zhao et al. [37] | Hong Kong | 21 pairs | 4.4 | 2.9 | – | 6.7 |
| Comparison with SARS-CoV and MERS-CoV | ||||||
| SARS-CoV (2003) [25] | Singapore | 8.4 | - | - | ||
| MERS-CoV (2013) [38] | Saudi Arabia | 7.6 | 2.5 | – | 23.1 | |
| MERS-CoV (2015) [34] | South Korea | 12.6 | 12.1 | – | 13.1 | |
A relatively short serial interval of ~4 to 5 days was estimated from recent preprint studies. An analysis of 468 infector–infectee pairs confirmed in China reported a mean serial interval of 3.96 days (3.53–4.39) [35]. The study also noted that 59 of 468 pairs (12.6%) had negative-valued serial intervals, suggesting pre-symptomatic transmission. Two more studies estimated the median serial interval to be around 4 days based on data collected from several countries (4.0, 95% CrI: 3.1–4.9) [36] and Hong Kong (4.4, 95% CI: 2.9–6.7) [37]. Both studies highlighted the high possibility of pre-symptomatic infections given that the estimated serial interval is shorter than the incubation period, which is currently estimated to be around 5 days on average. Two published articles estimated a longer mean serial interval of 4.6 days (range: 3.0–9.0) [29] and 7.5 days (95% CI: 5.3–19.0) [10].
3.3. Susceptibility
There is limited information from published literature on susceptibility regarding neutralizing immunity. Existing evidence suggests that everyone (regardless of age, sex, or race) who has had close contact with an infected individual is susceptible to COVID-19. According to recently published literature, which described demographic and clinical characteristics of 44,672 laboratory-confirmed patients in China [39], COVID-19 has infected both men and women (male to female ratio = 1.06:1) and individuals of all ages (range = 0 to 90+). While there is no evidence of vertical transmission, two published studies reported adverse health outcomes (including death) on infants born to mothers infected with COVID-19 in China [40,41].
3.4. Severity
Current evidence suggests that older individuals and those with compromised immune systems from pre-existing conditions are more likely to develop severe forms of COVID-19. To date, few modeling studies have examined the case fatality risk (CFR) mostly because the outbreak is in the early phases in most countries, and considerable uncertainty exists in regards to the ascertainment rate and asymptomatic rate.
3.4.1. Descriptive Analysis
The crude CFR of COVID-19 from confirmed cases in China largely varied from 2% to 15%. Earlier analyses using a relatively small number of laboratory-confirmed patients in Wuhan in January reported an overall CFR of 11% [4] and 15% [42]. More recently, the analysis of 44,672 patients reported an overall CFR of 2.3% [39], which is much lower than that from earlier analyses and that for other coronaviruses (9.6% for SARS-CoV [43] and 34.5% for MERS-CoV [44]). The study showed that most cases (81%) had mild symptoms, while 14% developed severe conditions and the remaining 5% fell critically ill. Severity increased with age, and the CFR was highest (14.8%) in individuals aged 80 and above. The CFR was higher among individuals with underlying chronic conditions, such as cardiovascular diseases (10.5%), diabetes (7.3%), or chronic respiratory diseases (6.3%) than among individuals with no pre-existing conditions (0.9%). Another study on pediatric patients reported that most children with COVID-19 had developed mild symptoms, such as fever and cough, with good prognoses for recovery [45].
3.4.2. Modeling Studies: Estimates for China
We identified one published study and four preprints that estimated the CFR in China. In an early analysis, an estimated confirmed case fatality rate (cCFR) of 5.3% (3.5–7.5%) to 8.4% in China (5.3–12.3%) was provided using an exponential growth spread model with case reports collected up to 24 January 2020 [19]. A CFR of 7.24% (95% CI: 6.61–8.01) was reported for Hubei province in a preprint, which used a survival analysis in a competing risk model, with a much lower CFR of 1.00% (0.87–1.18%) in other Chinese provinces [46]. Using Bayesian methods, another estimated a crude CFR of 4.5% (95% CrI: 4.02–5.31) in Wuhan and a time-delay adjusted CFR of 15.93% (14.6–17.28%) [47]. However, the crude risk of death among all infected individuals (IFR) in Wuhan city and time-delay adjusted IFR were estimated to be much lower at 0.07% (0.05–0.09%) and 0.23% (0.17–0.3%), respectively [47].
3.4.3. Modeling Studies: Estimates for Outside China
One published study and one preprint reported modeling-based estimates of the CFR outside China ranging from 0.3% [9] to 1.4% [48], while one published study estimated the asymptomatic rate of around 18% [49]. The IFR was estimated to be 0.3% to 0.6% based on an estimated ascertainment rate of 10% in a study of 565 Japanese citizens who were evacuated from Wuhan on 29–31 January 2020 [9]. The study noted that their estimate is similar to the CFR of the Asian pandemic flu (1957–8). A CFR of 1.37% (95% CI: 0.57–3.22) was estimated in another preprint using the proportion of confirmed cases in ICU outside China [48]. In the meantime, a recently published study [49] estimated the asymptomatic rate of COVID-19 among all infected cases to be 17.9% (95% CrI: 15.5–20.2) based on a report of 634 infected individuals who were on a Princess Cruises’ Ship. To note, this study was previously included in our review as a preprint but has been published while preparing the manuscript.
3.5. Control Measures
We identified three published studies and four preprints of modeling interventions for COVID-19. These measures include non-pharmaceutical interventions, quarantine, and mobility reductions on the population level, and airport screening.
3.5.1. Travel Restrictions
We found a published study and two preprints that examined the impacts of the travel restriction policies, which were implemented in Wuhan on 23 January 2020 as part of the efforts to contain the further spread of COVID-19. Using a deterministic SEIR model, a published study [18] estimated that travel restrictions might lead to a 91% reduction in the number of cases in seven days in Beijing, compared to the baseline scenario with no restrictions. Similarly, two more preprint studies found that the travel restriction in Wuhan delayed the epidemic peak by 2.91 days [50], and 3–5 days [12].
3.5.2. Non-Pharmaceutical Interventions and Quarantine
We found three published studies and one preprint that examined the impacts of social distancing and other non-pharmaceutical interventions (e.g., face masks) on the epidemic trajectories. A mathematical modeling study [8] showed that a 25% reduction in transmissibility from the nationwide implementation of control measures would lead to a 50% reduction in the magnitude of the epidemic and a one month delay in the epidemic peak. The study, however, found that citywide quarantine in Wuhan in which all inbound and outbound mobility were to be eliminated would have little effects because local epidemics may have already occurred in other provinces across China by then [8]. Similarly, another published study concluded that the quarantine of exposed individuals identified through contact tracing in Wuhan had little effect in reducing the number of infections and slowing down the epidemic across China [18]. Incorporating the possibility of pre-symptomatic transmission in simulating the spread of COVID19 across China with different levels of quarantining, a recent preprint highlighted that the effectiveness of quarantine largely depends on when it is implemented and the proportion in quarantine. The study suggested that the quarantine rate should be at least 63% (threshold) for the epidemic to be averted [13] and that such strong control measures should be kept in place over the course of the outbreak.
More recently, a modeling study in Singapore which simulated the impact of different control measures found that workplace distancing was more effective in reducing the spread of COVID-19 than school closure [51]. According to the study, a combined strategy of case isolation and close contact quarantining, school closure and workplace distancing was the most effective in reducing the outbreak size, with the estimated median number of infections reduced by 99.3% (IQR 92.6–99.9), 93.0% (81.5–99.7), and 78.2% (59.0 −94.4) when R0 was 1.5, 2.0, or 2.5, respectively.
3.5.3. Airport Screening
One published study and one preprint examined the effectiveness of airport screening in containing the spread of COVID-19. A simulation study reported that ~46% (95% CI: 36–58) of infected travelers would not be detected through airport screening [52]. The study also noted that exit screening is more effective for longer flights compared to entry screening with a higher probability of developing symptoms on the flight. Using probabilistic methods to model the efficacy of travel screening, a study estimated that current practices will detect 34% (median; 95% CrI: 20–50%) of all infected travelers in the best case scenario where only 5% of cases are asymptomatic [53].
4. Discussion
The rapid spread of COVID-19 with pandemic potential poses one of the most significant global challenges in recent years. With more than 300 scientific reports and articles on this topic published in the past two months, it is crucial to disseminate the main findings through a comprehensive review of the existing evidence. In this review, we extracted and synthesized key epidemiological, demographic, and clinical features of COVID-19 from the published literature and preprints available to date.
Our findings suggest that the true size of the epidemic is much larger than what has been reported worldwide, though these figures largely depend on the effectiveness of the control measures. The outbreak is growing fast with an infected individual infecting two to three other persons on average and doubling in size every 3 to 7 days. While the incubation period ranges from 3 to 6 days based on eight published literature and preprints, its mean (or median) is most likely to be around 5 days on average, which is similar to that of other coronaviruses, such as SARS-CoV (4.4 days) [33] and MERS-CoV (5.5 to 6.7 days) [26,34].
Current estimates for the mean serial interval for COVID-19 range from 4 to 8 days from published articles and prints. However, it should be noted that the sample size is relatively small (six and seven pairs) for both of the published articles in which the mean serial interval was estimated to be between 5 and 8 days [10,29], respectively. More recent analyses in preprint used a much larger sample that includes up to 468 pairs [35], making their estimates of between 4 to 5 days more statistically reliable (given that all methodologies are sound and valid) with a smaller margin of errors. As highlighted in the literature, a serial interval shorter than the incubation period could imply pre-symptomatic transmission and thereby should be considered in formulating intervention strategies, as it may impede containment efforts. The serial interval estimates for SARS-CoV-2 is also shorter than that of SARS-CoV (8.4 days) [25] and MERS-CoV (8-13 days) [34,38], which suggests that it may be more challenging to contain the spread compared to other coronaviruses.
While the true CFR of SARS-CoV-2 still remains uncertain with modeling-based estimates largely varying between studies, it is found to be less severe than other coronaviruses, such as SARS-CoV (9.6%) [42] and MERS-CoV (34.5%) [43]. According to the latest statistics, the observed mortality rate of COVID-19 is estimated to be around 4.8% worldwide [1]. The mortality rate in the five most affected countries largely varies across countries, from as low as 1.0% for Germany to 11.4% for Italy [1]. However, it is possible that estimates for countries with relatively recent local establishment may be overestimated and vary between them. It is because during the initial stage of an outbreak, severe cases are more likely to be picked up by health authorities, while the vast majority of cases with no or mild symptoms are left undetected.
Control measures such as quarantine, travel restrictions, and airport screening for travelers have been widely implemented to contain the spread of infections. The effectiveness of these containment measures in controlling the outbreak, however, remains inconclusive. Current evidence from modeling studies on COVID-19 suggests that travel restrictions leading to reduced transmissibility can be highly effective in containing the spread. While school closure is less effective than workplace distancing or quarantine of exposed individuals, a combined strategy which implements all three measures together was found to be most effective in reducing the spread. Airport screening is shown to be not as effective either, detecting only 34% [52] to 54% [51] of infected travelers through thermal scanning. An important implication to this is that the effectiveness of such control measures could be further hampered by a significant portion of asymptomatic patients (17.9%, 95% CrI: 15.5–20.2%) [49] and pre-symptomatic transmission (12.1%), as evidenced from early analyses [35,36].
Some important limitations should be noted. In this review, we included preprints that are “in-press” or have not yet been peer-reviewed, which might impair the overall quality of the review to a certain extent. At the time of writing, however, there is still very limited information on some of the key epidemiological parameters of COVID-19. For example, we found only two published studies with an estimate for the serial interval using a small number of samples. Including five more estimates from preprints gave greater confidence of the means and distributions for the epidemiological parameters as they had bigger sample sizes with more data becoming available. It should also be noted that of preprints, we included only those that provided sound rationales for the methodology they used in the form of a complete manuscript.
Another potential limitation is that most of the studies included in this review are based on data collected during the early phase of the outbreak in China. As COVID-19 is rapidly evolving, these early estimates may change as more information is collected. We believe that additional research using case reports from other countries would be extremely useful since different demographic and cultural characteristics of the population may play an important role in determining the outbreak trajectories and clinical outcomes at the population level.
More rigorous research to estimate the effects of other control measures currently implemented in many affected countries, such as social distancing or school closure, could also provide important evidence for countries with sustained human-to-human transmission. As few suspected cases of reinfection have been reported in China, Japan, and South Korea, serologic studies, which examine the possibility of reinfection (or reactivation of the virus) among recovered individuals, should also be made an urgent research priority.
5. Conclusions
Understanding the epidemiology and transmission dynamics of an emerging infectious disease is a key for successful outbreak control. As the COVID-19 pandemic continues to rapidly spread across continents, there is an urgent need for more rigorous research focusing on mitigation strategies (shift from containment). Here we disseminate key findings of epidemiological parameters from the literature at this time point, which can be used by modelers and policymakers for epidemic planning purposes.
Author Contributions
Conceptualization, M.P. and A.R.C.; methodology, M.P. and B.L.D.; data collection and analysis, M.P., J.T.L., and Y.S.; writing—original draft preparation, M.P.; writing—review and editing, M.P., B.L.D., and A.R.C.; visualization, M.P. and B.L.D.; supervision, A.R.C. All authors have read and agreed to the published version of the manuscript.
Funding
B.L.D. and A.R.C. have received study funding from the Singapore Population Health Improvement Centre (NMRC/CG/C026/2017_NUHS).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.BNO Tracking Coronavirus: Map, Data and Timeline. [(accessed on 18 March 2020)]; Available online: https://bnonews.com/index.php/2020/02/the-latest-coronavirus-cases/
- 2.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowling B.J., Leung G.M. Epidemiological research priorities for public health control of the ongoing global novel coronavirus (2019-nCoV) outbreak. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.6.2000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kretzschmar M. International Encyclopedia of Public Health. Academic Press; Cambridge, MA, USA: 2017. Measurement and Modeling: Infectious Disease Modeling; pp. 579–585. [Google Scholar]
- 7.Du Z., Wang L., Cauchemez S., Xu X., Wang X., Cowling B.J., Meyers L.A. Risk for Transportation of 2019 Novel Coronavirus Disease from Wuhan to Other Cities in China. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2605.200146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishiura H., Kobayashi T., Yang Y., Hayashi K., Miyama T., Kinoshita R., Linton N.M., Jung S.M., Yuan B., Suzuki A., et al. The Rate of Underascertainment of Novel Coronavirus (2019-nCoV) Infection: Estimation Using Japanese Passengers Data on Evacuation Flights. J. Clin. Med. 2020;9:419. doi: 10.3390/jcm9020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Z., Zhang Q., Lu X., Pfeiffer D., Wang L., Song H., Pei T., Jia Z., Zeng D.D. Incorporating Human Movement Data to Improve Epidemiological Estimates for 2019-nCoV. medRxiv. 2020 doi: 10.1101/2020.02.07.20021071. [DOI] [Google Scholar]
- 12.Chinazzi M., Davis J.T., Ajelli M., Gioannini C., Litvinova M., Merler S., Pastore y Piontti A., Rossi L., Sun K., Viboud C., et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (2019-nCoV) outbreak. medRxiv. 2020 doi: 10.1126/science.aba9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong H., Yan H. Simulating the infected population and spread trend of 2019-nCov under different policy by EIR model. medRxiv. 2020 doi: 10.2139/ssrn.3537083. [DOI] [Google Scholar]
- 14.Zhao Q., Chen Y., Small D.S. Analysis of the epidemic growth of the early 2019-nCoV outbreak using internationally confirmed cases. medRxiv. 2020 doi: 10.1101/2020.02.06.20020941. [DOI] [Google Scholar]
- 15.Zhao S., Musa S.S., Lin Q., Ran J., Yang G., Wang W., Lou Y., Yang L., Gao D., He D., et al. Estimating the Unreported Number of Novel Coronavirus (2019-nCoV) Cases in China in the First Half of January 2020: A Data-Driven Modelling Analysis of the Early Outbreak. J. Clin. Med. 2020;9:388. doi: 10.3390/jcm9020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao S., Lin Q., Ran J., Musa S.S., Yang G., Wang W., Lou Y., Gao D., Yang L., He D., et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int. J. Infect. Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riou J., Althaus C.L. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang B., Wang X., Li Q., Bragazzi N.L., Tang S., Xiao Y., Wu J. Estimation of the Transmission Risk of the 2019-nCoV and Its Implication for Public Health Interventions. J. Clin. Med. 2020;9:462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung S.M., Akhmetzhanov A.R., Hayashi K., Linton N.M., Yang Y., Yuan B., Kobayashi T., Kinoshita R., Nishiura H. Real-Time Estimation of the Risk of Death from Novel Coronavirus (COVID-19) Infection: Inference Using Exported Cases. J. Clin. Med. 2020;9:523. doi: 10.3390/jcm9020523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen M., Peng Z., Xiao Y., Zhang L. Modelling the epidemic trend of the 2019 novel coronavirus outbreak in China. bioRxiv. 2020 doi: 10.1101/2020.01.23.916726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Read J.M., Bridgen J.R.E., Cummings D.A.T., Ho A., Jewell C.P. Novel coronavirus 2019-nCoV: Early estimation of epidemiological parameters and epidemic predictions. medRxiv. 2020 doi: 10.1101/2020.01.23.20018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu T., Hu J., Kang M., Lin L., Zhong H., Xiao J., He G., Song T., Huang Q., Rong Z., et al. Transmission dynamics of 2019 novel coronavirus (2019-nCoV) bioRxiv. 2020 doi: 10.2139/ssrn.3526307. [DOI] [Google Scholar]
- 23.Majumder M., Mandl K.D. Early Transmissibility Assessment of a Novel Coronavirus in Wuhan, China (January 26, 2020) SSRN. 2020 doi: 10.2139/ssrn.3524675. [DOI] [Google Scholar]
- 24.Riley S., Fraser C., Donnelly C.A., Ghani A.C., Abu-Raddad L.J., Hedley A.J., Leung G.M., Ho L.M., Lam T.H., Thach T.Q., et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: Impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 25.Lipsitch M., Cohen T., Cooper B., Robins J.M., Ma S., James L., Gopalakrishna G., Chew S.K., Tan C.C., Samore M.H., et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cauchemez S., Fraser C., Van Kerkhove M.D., Donnelly C.A., Riley S., Rambaut A., Enouf V., van der Werf S., Ferguson N.M. Middle East respiratory syndrome coronavirus: Quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect. Dis. 2014;14:50–56. doi: 10.1016/S1473-3099(13)70304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linton N.M., Kobayashi T., Yang Y., Hayashi K., Akhmetzhanov A.R., Jung S.M., Yuan B., Kinoshita R., Nishiura H. Incubation Period and Other Epidemiological Characteristics of 2019 Novel Coronavirus Infections with Right Truncation: A Statistical Analysis of Publicly Available Case Data. J. Clin. Med. 2020;9:538. doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ki M., Task Force for 2019-nCoV Epidemiologic characteristics of early cases with 2019 novel coronavirus (2019-nCoV) disease in Republic of Korea. Epidemiol. Health. 2020:e2020007. doi: 10.4178/epih.e2020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X., Rayner S., Luo M.H. Does SARS-CoV-2 has a longer incubation period than SARS and MERS? J. Med. Virol. 2020 doi: 10.1002/jmv.25708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H., Azman A.S., Reich N.G., Lessler J. The incubation period of 2019-nCoV from publicly reported confirmed cases: Estimation and application. medRxiv. 2020 doi: 10.1101/2020.02.02.20020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J., Litvinova M., Wang W., Wang Y., Deng X., Chen X., Li M., Zheng W., Yi L., Chen X., et al. Evolving epidemiology of novel coronavirus diseases 2019 and possible interruption of local transmission outside Hubei Province in China: A descriptive and modeling study. medRxiv. 2020 doi: 10.1101/2020.02.21.20026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau E.H., Hsiung C.A., Cowling B.J., Chen C.H., Ho L.M., Tsang T., Chang C.W., Donnelly C.A., Leung G.M. A comparative epidemiologic analysis of SARS in Hong Kong, Beijing and Taiwan. BMC Infect. Dis. 2010;10:50. doi: 10.1186/1471-2334-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowling B.J., Park M., Fang V.J., Wu P., Leung G.M., Wu J.T. Preliminary epidemiological assessment of MERS-CoV outbreak in South Korea, May to June 2015. Euro Surveill. 2015;20:7–13. doi: 10.2807/1560-7917.ES2015.20.25.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Z., Wang L., Xu X., Wu Y., Cowling B.J., Meyers L.A. The serial interval of COVID-19 from publicly reported confirmed cases. medRxiv. 2020 doi: 10.1101/2020.02.19.20025452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishiura H., Linton N.M., Akhmetzhanov A.R. Serial interval of novel coronavirus (2019-nCoV) infections. medRxiv. 2020 doi: 10.1101/2020.02.03.20019497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao S., Gao D., Zhuang Z., Chong M., Cai Y., Ran J., Cao P., Wang K., Lou Y., Wang W., et al. Estimating the serial interval of the novel coronavirus disease (COVID-19): A statistical analysis using the public data in Hong Kong from January 16 to February 15, 2020. medRxiv. 2020 doi: 10.1101/2020.02.21.20026559. [DOI] [Google Scholar]
- 38.Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Z., McGoogan J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 40.Chen H., Guo J., Wang C., Luo F., Yu X., Zhang W., Li J., Zhao D., Xu D., Gong Q., et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. Lancet. 2020 doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu H., Wang L., Fang Z., Peng S., Zhang L., Chang G., Xia S., Zhou W. Clinical analysis of 10 neonates born to mothers with 2019-nCoV pneumonia. Transl. Pediatr. 2020;9:51–60. doi: 10.21037/tp.2020.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WHO Summary of Probable SARS Cases with Onset of Illness from 1 November 2002 to 31 July 2003. [(accessed on 1 March 2020)]; Available online: https://www.who.int/csr/sars/country/table2004_04_21/en/
- 44.WHO Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [(accessed on 1 March 2020)]; Available online: https://www.who.int/emergencies/mers-cov/en/
- 45.Chen Z.M., Fu J.F., Shu Q., Chen Y.H., Hua C.Z., Li F.B., Lin R., Tang L.F., Wang T.L., Wang W., et al. Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World J. Pediatr. 2020 doi: 10.1007/s12519-020-00345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X., Ma Z., Ning Y., Chen C., Chen R., Chen Q., Zhang H., Li C., He Y., Wang T., et al. Estimating the case fatality ratio of the COVID-19 epidemic in China. medRxiv. 2020 doi: 10.1101/2020.02.17.20023630. [DOI] [Google Scholar]
- 47.Mizumoto K., Kagaya K., Chowell G. Early epidemiological assessment of the transmission potential and virulence of 2019 Novel Coronavirus in Wuhan City: China, 2019-2020. medRxiv. 2020 doi: 10.1101/2020.02.12.20022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson N., Kvalsvig A., Telfar Barnard L., Baker M.G. Estimating the Case Fatality Risk of COVID-19 using Cases from Outside China. medRxiv. 2020 doi: 10.1101/2020.02.15.20023499. [DOI] [Google Scholar]
- 49.Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian H., Liu Y., Li Y., Kraemer M.U.G., Chen B., Wu C.-H., Cai J., Li B., Xu B., Yang Q., et al. Early evaluation of transmission control measures in response to the 2019 novel coronavirus outbreak in China. medRxiv. 2020 doi: 10.1101/2020.01.30.20019844. [DOI] [Google Scholar]
- 51.Koo J.R., Cook A.R., Park M., Sun Y., Sun H., Lim J.T., Tam C., Dickens B.L. Interventions to mitigate early spread of SARS-CoV-2 in Singapore: A modelling study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quilty B.J., Clifford S., Flasche S., Eggo R.M. Effectiveness of airport screening at detecting travellers infected with novel coronavirus (2019-nCoV) Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.5.2000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gostic K., Gomez A.C.R., Mummah R.O., Kucharski A.J., Lloyd-Smith J.O. Estimated effectiveness of traveller screening to prevent international spread of 2019 novel coronavirus (2019-nCoV) medRxiv. 2020 doi: 10.1101/2020.01.28.20019224. [DOI] [Google Scholar]