Abstract
The ongoing pandemic of coronavirus disease 2019 (COVID-19), caused by infection with human coronavirus 2019 (HCoV-19 / SARS-CoV-2 / 2019-nCoV), is a global threat to the human population. Here, we briefly summarize the available data for the zoonotic origins of HCoV-19, with reference to the other two epidemics of highly virulent coronaviruses, SARS-CoV and MERS-CoV, which cause severe pneumonia in humans. We propose to intensify future efforts for tracing the origins of HCoV-19, which is a very important scientific question for the control and prevention of the pandemic.
Keywords: HCoV-19 / SARS-CoV-2, Zoonotic origins, Natural Reservoir, Susceptibility, Bat, Pangolin
The world is currently experiencing its third outbreak with a highly virulent coronavirus causing severe pneumonia in humans, with all three taking place in the 21st century. The first epidemic had origins in Guangdong Province, China, during late 2002 and lasted until 2004 (Ksiazek et al., 2003; Qin et al., 2003; Rota et al., 2003; The Chinese SARS Molecular Epidemiology Consortium, 2004). The outbreak, caused by Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), killed at least 700 people and infected over 8 000, and cases were reported in all six permanently inhabited continents with 29 countries affected (de Wit et al., 2016; Stadler et al., 2003). Aside from a few instances of laboratory-linked infections, SARS-CoV has not been reported in the human population ever since (Lim et al., 2004; Orellana, 2004). The second epidemic was first characterized in a man with pneumonia in Saudi Arabia in 2012 (Zaki et al., 2012), in which sporadic cases, small clusters, and large outbreaks have been reported over subsequent years in 24 affected countries, mostly located in the Middle East, but with imported infections to North America, Africa, Europe and Asia (Assiri et al., 2013; Guery et al., 2013; Hijawi et al., 2013; Shehata et al., 2016; Zumla et al., 2015). The aetiological agent was named Middle East Respiratory Syndrome Coronavirus (MERS-CoV), in which over 800 deaths and close to 2 500 cases were reported to date (Shehata et al., 2016; Zumla et al., 2015). The third pandemic was first reported in Hubei Province, China, in late 2019 and is still ongoing as of April 2020 (Li et al., 2020; Lu et al., 2020a; Wu et al., 2020; Zhou et al., 2020a; Zhu et al., 2020). Caused by another novel coronavirus, known as the human coronavirus 2019 (HCoV-19, also named by various bodies as SARS-CoV-2 or 2019-nCoV) (Jiang et al., 2020), intense and widespread transmission was observed worldwide, with over 80 000 deaths and 1 400 000 cases reported from over 200 countries, areas or territories as of 08 April 2020 (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). From sequencing results, it was determined that HCoV-19 had over 75% genomic similarity to SARS-CoV (Wu et al., 2020; Zhou et al., 2020a; Zhu et al., 2020). To date, there are no licensed prophylactics or therapeutics available to prevent or treat Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), or Coronavirus disease 2019 (COVID-19), the diseases caused by infection with SARS-CoV, MERS-CoV or HCoV-19, respectively.
Tracking the origins of these novel coronaviruses has important implications for the control and prevention of related infectious diseases in humans. While the scientific world is focused on developing efficacious drugs, vaccines and therapeutics specific to HCoV-19 to save the maximum number of lives, elucidating the source of these viruses can lead to evidence-based policy changes that can prevent infections, and thus outbreaks, from occurring in the first place. Extensive work performed on the origins of SARS-CoV found that this virus had zoonotic origins (de Wit et al., 2016; Guan et al., 2003; Hu et al., 2017; Li et al., 2005a; Wang et al., 2005). In particular, samples taken from apparently healthy palm civets (Pagumaspp.) sold as food in local wet markets in Guangdong Province, China, during the SARS epidemic resulted in the detection and isolation of a SARS-CoV strain (SARS-CoV strain (Guan et al., 2003; Wang et al., 2005). Combined with molecular studies showing that the SARS-CoV receptor (Angiotensin-converting enzyme 2, ACE2) (Li et al., 2003) between humans and palm civets were sufficiently similar to allow for virus entry and presumptively infection, it was deduced that SARS-CoV may have crossed the species barrier from palm civets to humans (Li et al., 2005b; Peiris et al., 2004; Song et al., 2005). SARS-CoV was later detected in raccoon dogs (Nyctereuteussp.), ferret badgers (Melogalesp.) and domestic cats (Guan et al., 2003; Martina et al., 2003). In 2005, a number of SARS-like coronaviruses were discovered in Chinese horseshoe bats (Rhinolophus spp.) (Lau et al., 2005; Li et al., 2005a; Poon et al., 2005). Phylogenetic analysis of these SARS-like coronaviruses strongly suggested bat-origins for these pathogens, and horseshoe bats (as the presumptive natural reservoir) likely infected humans directly in the wild or indirectly through intermediate animal species (such as palm civets) in Chinese live animal markets (Lau et al., 2005; Li et al., 2005a; Poon et al., 2005; Shi & Hu, 2008; Wang et al., 2006).
The investigative work performed on the origins of MERS-CoV also suggests that bats are likely the natural reservoirs of this pathogen (Ithete et al., 2013; Memish et al., 2013). Studies have shown the detection of sequences phylogenetically related to MERS-CoV in bats (Nycteris sp.) in Ghana and Europe (Pipistrellus sp.) (Annan et al., 2013). In addition, camels were also implicated in the interspecies transmission of MERS-CoV to humans (Hemida et al., 2014). Multiple studies have shown that there is high seroprevalence of MERS-CoV specific antibodies in dromedary camels (Camelus dromedarius) from Egypt, Oman and other locations in the Middle East, but that sheep, goats, cattle, and other camelids were seronegative (Omrani et al., 2015). Epidemiological work also showed that a majority of confirmed MERS patients had close contact history with dromedary camels, or products derived from these animals, such as camel meat and milk (Hemida et al., 2017). In addition, the MERS-CoV sequences isolated from patients and close-contact dromedary camels were highly similar, or in some cases, 100% identical (Azhar et al., 2014). Thus, the evidence supports that while camels serve as the principal source of MERS-CoV transmission to humans, bats may be again the natural reservoir of this pathogen. Additionally, some alpacas (Vicugna pacos) in the endemic region were also seropositive for MERS-CoV, suggesting this animal may be another source for MERS-CoVs (Reusken et al., 2016). Based on the available evidence, it seems that the animal-to-human transmissions of MERS-CoV and SARS-CoV are clear, and sets a good paradigm for tracing the origins of HCoV-19.
Since HCoV-19 emerged recently in the human population, we do not know for certain the origins of this virus and much work remains to be done. There are several websites and servers, such as the 2019 Novel Coronavirus Resource (2019nCoVR: https://bigd.big.ac.cn/ncov/) and GISAID (https://www.gisaid.org/), showing the genomic epidemiology and clustering of HCoV-19 genomes in the human population, and the information there helped scientists to understand mutations in HCoV-19, designing proper diagnostic kits, and tracing ongoing outbreaks across the world. Based on a distorted elucidation and/or improper analysis of the initially released, limited numbers of HCoV-19 genome data, there are several speculations or conspiracy theories that HCoV-19 was artificially generated in the laboratory (Andersen et al., 2020; Liu et al., 2020), especially the insertion of a poly-basic (furin) cleavage site in the spike protein, which was found for the first time in beta-coronaviruses (Andersen et al., 2020). Based on the information and knowledge gained from past SARS-CoV and MERS-CoV epidemics, combined with the successful detection and isolation of SARS-like coronaviruses (Bat-CoV-RaTG13) in bats (R. affinis) with over 95% similarity to HCoV-19, it can be postulated with a degree of confidence that this novel coronavirus likely also originated from bats (Zhou et al., 2020a). In addition, the insertion of “PAA” on the cleavage site between S1 and S2 has now been found in a coronavirus genome identified in another bat (R. malaynus) (Zhou et al., 2020b). These results further indicate at least two bat species, R. affinis and R. malaynus, may be the putative natural hosts of HCoV-19, and confirmed that bats are likely the animal reservoir for the SARS-like (Shi & Hu, 2008; Wang et al., 2006) and HCoV-19-like coronaviruses. So far, there is no conclusive evidence showing that bats can directly transmit the HCoV-19-like coronaviruses to humans.
There are several recent studies showing that Malayan pangolin (Manis javanica) as a potential natural reservoir or intermediate host of HCoV-19 (Lam et al., 2020; Xiao et al., 2020; Zhang et al., 2020a). In one study, Lam et al. (2020) performed metagenomic sequencing of lung, intestine and blood samples from the Malayan pangolins and identified viral sequences that belong to two sub-lineages of HCoV-19 related coronaviruses. In particular, five critical residues on the receptor binding domain (RBD) of the pangolin virus, playing important roles for human receptor binding, are identical to HCoV-19 (Lam et al., 2020). Re-analysis of a previously reported viral metagenomics dataset of Malayan pangolins (Liu et al., 2019) also suggested a probable pangolin origin of HCoV-19 (Zhang et al., 2020a). The isolation and comprehensive characterization of a HCoV-19 related coronavirus (Pangolin-CoV) in the pangolins has further implicated the Malayan pangolin as a possible intermediate host for HCoV-19 (Xiao et al., 2020), and HCoV-19 might have originated from possible recombination between the reported Bat-CoV-RaTG13-like virus (Zhou et al., 2020a) and this Pangolin-CoV-like virus (Xiao et al., 2020). Malayan pangolins are found in nature throughout Southeast Asia (including Laos PDR, Cambodia, Thailand, Myanmar, Vietnam, and Indonesia), but not known to be native in China, where HCoV-19 was first reported (Li et al., 2020; Lu et al., 2020a; Wu et al., 2020; Zhou et al., 2020a; Zhu et al., 2020). Due to its importance as a food source and a traditional Chinese medicine, Malayan pangolins are illegally smuggled in live from Southeast Asia to China to be sold in live animal markets or as dried scales to be sold in Chinese Medicine store. Furthermore, R. affinis and R. malaynus, as suspected natural reservoir of HCoV-19 (Zhou et al., 2020a, 2020b), are also distributed widely in South and Southeast Asia. It remains to be seen whether bats and Malayan pangolins (or any other unknown animal species at the moment) played a role in the generation of HCoV-19 via recombination (Figure 1A). Most recently, Andersen et al. (2020) put forward several theories regarding the origins of the HCoV-19 and highlighted the potential role of natural selection in animal host before the zoonotic transfer, natural selection in humans after the animal-to-human transfer, and adaptation during sequential passages. The theories will help uncover the origins of HCoV-19, but direct scientific evidence is needed.
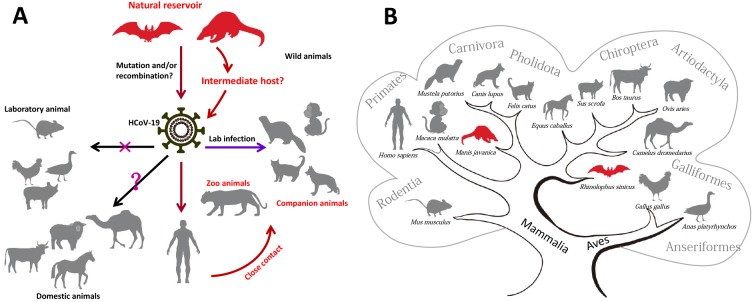
1. Natural reservoir and susceptibility of various animal species to HCoV-19.
A: Putative zoonotic origin, transmission and infection of HCoV-19 in human and animals. Domestic animals (pigs, chickens and ducks) and laboratory mice cannot be infected (as marked by an X). Other domestic animals, including the MERS-CoV intermediate host dromedary camel, have unknown susceptibility to HCoV-19 and are marked with a question mark. Ferrets, monkeys, tigers, cats and dogs can be infected with HCoV-19. B: Schematic relationship of different mammals and birds based on the phylogenetic tree of the ACE2 protein sequences.
At present, it remains to be concretely demonstrated that bats and pangolins are the possible natural reservoirs and intermediate animal species for HCoV-19 related viruses. However, whether there exist any other animal species that may also play an intermediate role in the interspecies spread of HCoV-19 remains unknown. The recent report for detection of SARS-CoV-2 neutralizing serum antibodies in cats from Wuhan, Hubei Province, indicated that cats were infected during the COVID-19 outbreak (Zhang et al., 2020b). Direct inoculation of HCoV-19 to a variety of animals showed that ferrets and cats are highly susceptible to this virus, whereas dogs have a low genetic susceptibility, and pigs, chickens and ducks could not be infected (Shi et al., 2020). In particular, cats could transmit the virus via respiratory droplets (Shi et al., 2020), which would pose difficulties for the control of the virus. Other studies showed that Syrian hamsters (Chan et al., 2020) and monkeys (Bao et al., 2020; Lu et al., 2020b; Munster et al., 2020; Rockx et al., 2020; Shan et al., 2020) are susceptible to HCoV-19 infection and may be promising as potential animal models for COVID-19, indicating a possibility of interspecies spread of HCoV-19 (Figure 1A). Anecdotal reports in a variety of news media reported that tigers in a New York Zoo tested positive for HCoV-19, in which these animals exhibited symptoms of the illness (https://www.nationalgeographic.com/animals/2020/04/tiger-coronavirus-covid19-positive-test-bronx-zoo/). These reports suggested that there may be more animals susceptible to HCoV-19 than previously thought (Figure 1A).
The wild animals (bats and pangolins) suspected to be carrying the HCoV-19-like coronaviruses are known to be abundant in Southeast Asia, which is one of the world’s most biodiverse regions, with over 2 000 new species described between 1997 and 2014 alone, and with likely many new species yet to be discovered (Hughes, 2017). There is a need to prioritize sampling of animals that may carry SARS-like or HCoV-19-like coronaviruses in this region. Regular and routine investigations into wet markets selling wild exotic animals for consumption is urgently needed, as these animals are often kept in crowded, unsanitary conditions with other species, which may promote viral reassortment and interspecies transmission. On the other hand, to conduct a comprehensive virome screening for wild animals and to establish an archive dataset for all potential viruses across the world may be helpful for future rapid tracing the origins of unknown virus outbreak.
One prerequisite for inter-species transmission of a virus is its interaction with the receptor of different hosts. The identification of human ACE2 as the cellular receptor for HCoV-19 sheds light on the viral inter-species transmission route (Hoffmann et al., 2020; Zhou et al., 2020a). Characterizing the interaction between spike (S) protein and ACE2 of different species (Figure 1B) would give clues for the origins/intermediate and susceptible hosts, as well as for drug and vaccine development (Lan et al., 2020; Wang et al., 2020; Yan et al., 2020). For instance, the ACE2 from civets, bats (R. sinicus) and pigs confer the ability to support viral entry (Zhou et al., 2020a), and in vivo challenging with HCoV-19 virus did indicate that ferrets and cats can be more efficiently infected than dogs (Shi et al., 2020). Given the importance of ACE2 and other unknown co-receptor(s) in the infection process, it is also crucial to analyze the expression patterns and levels of ACE2 and other co-receptor(s) in mediating the HCoV-19 infection, and by extension, determining the mediating host animals. This kind of molecular characterization of ACE2 and other potential receptors of HCoV-19 in different animals at the lab would provide helpful information regarding the prediction of genetic susceptibility to HCoV-19 and direct the reservoir tracing of the virus in nature.
It is very important to elucidate the potential sources of HCoV-19 in wild animals and prevent viral resurgence in countries with excellent management of the pandemic. In addition, tracing the susceptible animals and surveillance of HCoV-19-related viruses would be of important significance for preventing similar emerging viruses in the future. Findings from past outbreaks and available studies did have direct impact on subsequent public health policy. After reports on the zoonotic origins of SARS-CoV surfaced (Peiris et al., 2004; Stadler et al., 2003), Chinese authorities ordered the slaughter of over 10 000 animals due to be sold in live animal markets, including civets, raccoon dogs, and ferret badgers, and the USA instituted a trade embargo on civet cats (https://www.cdc.gov/sars/about/civet-embargo.html). After dromedary camels were implicated in the spread of MERS-CoV, the Office International des Epizooties (OIE) announced that positive real-time PCR results for MERS-CoV or isolation of the pathogen from dromedary camels is an OIE-notifiable event to mitigate the health risk of the virus to humans, and to prevent international spread. Cooking of camel meat and pasteurization of camel milk before consumption was promoted (Hemida et al., 2017). After the discovery that pangolins harbour HCoV-19 related coronaviruses, Chinese authorities put a ban on the trade and sales for wild animals. At present, we are still uncertain of the final consequence of HCoV-19 on wild animal management, and how this will shape our future life and animal conservation and usage.
In short, accumulating evidence on the discovery of SARS-, MERS- and HCoV-19-like viruses in animals makes it clear that there are likely more unknown, highly virulent coronaviruses with epidemic/pandemic potential circulating amongst wild animal species. Increased interactions between human populations and these zoonotic hosts due to deforestation, economical development and climate change will increase the probability for close contact, and thus higher chances for the next outbreak. In light of this scenario, it is ever more important for authorities to provide sustainable support not only for the discovery of novel coronaviruses, but also other unknown or neglected tropical pathogens with potential for high public health impact, such as bunyaviruses, filoviruses, arenaviruses and so on. As evidenced by the increasing frequency of viral epidemics/pandemics with novel pathogens in the 21st century (i.e., SARS-CoV, pandemic H1N1, MERS-CoV, Zika virus and now HCoV-19), it remains difficult to predict the location and timing of the next outbreak. Pathogen surveillance and discovery, facilitated by cutting-edge new sequencing technology, comparative genomic analyses and elegant molecular characterization, remains one of our best ways for pandemic preparedness, and the origins of novel pathogens, along with specific vaccine and drug development, need to be highlighted as research priorities, such that evidence-based policy for the prevention of future infections with novel pathogens can be established.ished.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Y.G.Y., G.W., and Y.H.B. conceived the review. G.W. and Y.G.Y. prepared the draft. Z.G.Z., Y.H.B. and Y.G.Y. designed the figure. All authors contributed to the discussions. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We thank Mrs. Li-Bin Wu for help with the preparation of the figure.
Biographies
garyckwong@ips.ac.cn (G.W.)
beeyh@im.ac.cn (Y.H.B.)
wangqihui@im.ac.cn (Q.H.W.)
chenxw@wh.iov.cn (X.W.C.)
zhangzhigang@ynu.edu.cn (Z.G.Z.)
yaoyg@mail.kiz.ac.cn (Y.G.Y.)
Contributor Information
Gary Wong, Email: garyckwong@ips.ac.cn.
Yu-Hai Bi, Email: beeyh@im.ac.cn.
Qi-Hui Wang, Email: wangqihui@im.ac.cn.
Xin-Wen Chen, Email: chenxw@wh.iov.cn.
Zhi-Gang Zhang, Email: zhangzhigang@ynu.edu.cn.
Yong-Gang Yao, Email: yaoyg@mail.kiz.ac.cn.
References
- 1.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF The proximal origin of SARS-CoV-2. Nature Medicine. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annan A, Baldwin HJ, Corman VM, Klose SM, Owusu M, Nkrumah EE, Badu EK, Anti P, Agbenyega O, Meyer B, Oppong S, Sarkodie YA, Kalko EKV, Lina PHC, Godlevska EV, Reusken C, Seebens A, Gloza-Rausch F, Vallo P, Tschapka M, Drosten C, Drexler JF Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerging Infectious Diseases. 2013;19(3):456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeem RF, Makhdoom HQ, Zumla AI, Memish ZA Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. The Lancet Infectious Diseases. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM, Madani TA Evidence for camel-to-human transmission of MERS coronavirus. The New England Journal of Medicine. 2014;370(26):2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 5.Bao LL, Deng W, Gao H, Xiao C, Liu JY, Xue J, Lv Q, Liu JN, Yu PF, Xu YF, Qi FF, Qu YJ, Li FD, Xiang ZG, Yu HS, Gong SR, Liu MY, Wang GP, Wang SY, Song ZQ, Zhao WJ, Han YL, Zhao LN, Liu X, Wei Q, Qin C Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.03.13.990226. [DOI] [Google Scholar]
- 6.Chan JFW, Zhang AJ, Yuan SF, Poon VKM, Chan CCS, Lee ACY, Chan WM, Fan ZM, Tsoi HW, Wen L, Liang RH, Cao JL, Chen YX, Tang KM, Luo CT, Cai JP, Kok KH, Chu H, Chan KH, Sridhar S, Chen ZW, Chen HL, To KKW, Yuen KY Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019(COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clinical Infectious Diseases. 2020 doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wit E, van Doremalen N, Falzarano D, Munster VJ SARS and MERS: recent insights into emerging coronaviruses. Nature Reviews Microbiology. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JSM, Poon LLM Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 9.Guery B, Poissy J, el Mansouf L, Séjourné C, Ettahar N, Lemaire X, Vuotto F, Goffard A, Behillil S, Enouf V, Caro V, Mailles A, Che D, Manuguerra JC, Mathieu D, Fontanet A, van der Werf S; MERS-CoV study group Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. The Lancet. 2013;381(9885):2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemida MG, Chu DKW, Poon LLM, Perera RAPM, Alhammadi MA, Ng HY, Siu LY, Guan Y, Alnaeem A, Peiris M MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerging Infectious Diseases. 2014;20(7):1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemida MG, Elmoslemany A, Al-Hizab F, Alnaeem A, Almathen F, Faye B, Chu DKW, Perera RAPM, Peiris M Dromedary camels and the transmission of Middle East respiratory syndrome coronavirus (MERS-CoV) Transboundary and Emerging Diseases. 2017;64(2):344–353. doi: 10.1111/tbed.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hijawi B, Abdallat M, Sayaydeh A, Alqasrawi S, Haddadin A, Jaarour N, Alsheikh S, Alsanouri T Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation. Eastern Mediterranean Health Journal. 2013;19(Suppl 1):S12–S18. [PubMed] [Google Scholar]
- 13.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, Xie JZ, Shen XR, Zhang YZ, Wang N, Luo DS, Zheng XS, Wang MN, Daszak P, Wang LF, Cui J, Shi ZL Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathogens. 2017;13(11):e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes AC Understanding the drivers of Southeast Asian biodiversity loss. Ecosphere. 2017;8(1):e01624. doi: 10.1002/ecs2.1624. [DOI] [Google Scholar]
- 16.Ithete NL, Stoffberg S, Corman VM, Cottontail VM, Richards LR, Schoeman MC, Drosten C, Drexler JF, Preiser W Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerging Infectious Diseases. 2013;19(10):1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang SB, Shi ZL, Shu YL, Song JD, Gao GF, Tan WJ, Guo DY A distinct name is needed for the new coronavirus. The Lancet. 2020;395(10228):949. doi: 10.1016/S0140-6736(20)30419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong SX, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ; SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. The New England Journal of Medicine. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 19.Lam TTY, Shum MHH, Zhu HC, Tong YG, Ni XB, Liao YS, Wei W, Cheung WY, Li WJ, Li LF, Leung GM, Holmes EC, Hu YL, Guan Y Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 20.Lan J, Ge JW, Yu JF, Shan SS, Zhou H, Fan SL, Zhang Q, Shi XL, Wang QS, Zhang LQ, Wang XQ Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 21.Lau SKP, Woo PCY, Li KSM, Huang Y, Tsoi HW, Wong BHL, Wong SSY, Leung SY, Chan KH, Yuen KY Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Guan XH, Wu P, Wang XY, Zhou L, Tong YQ, Ren RQ, Leung KSM, Lau EHY, Wong JY, Xing XS, Xiang NJ, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu WX, Chen CD, Jin LM, Yang R, Wang Q, Zhou SH, Wang R, Liu H, Luo YB, Liu Y, Shao G, Li H, Tao ZF, Yang Y, Deng ZQ, Liu BX, Ma ZT, Zhang YP, Shi GQ, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng ZJ Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. The New England Journal of Medicine. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li WD, Shi ZL, Yu M, Ren WZ, Smith C, Epstein JH, Wang HZ, Crameri G, Hu ZH, Zhang HJ, Zhang JH, McEachern J, Field H, Daszak P, Eaton BT, Zhang SY, Wang LF Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005a;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 24.Li WH, Moore MJ, Vasilieva N, Sui JH, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li WH, Zhang CS, Sui JH, Kuhn JH, Moore MJ, Luo SW, Wong SK, Huang IC, Xu KM, Vasilieva N, Murakami A, He YQ, Marasco WA, Guan Y, Choe H, Farzan M Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. The EMBO Journal. 2005b;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim PL, Kurup A, Gopalakrishna G, Chan KP, Wong CW, Ng LC, Se-Thoe SY, Oon L, Bai XL, Stanton LW, Ruan YJ, Miller LD, Vega VB, James L, Ooi PL, Kai CS, Olsen SJ, Ang B, Leo YS Laboratory-acquired severe acute respiratory syndrome. The New England Journal of Medicine. 2004;350(17):1740–1745. doi: 10.1056/NEJMoa032565. [DOI] [PubMed] [Google Scholar]
- 27.Liu P, Chen W, Chen JP Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins (Manis javanica) . Viruses. 2019;11(11):979. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu SL, Saif LJ, Weiss SR, Su LS No credible evidence supporting claims of the laboratory engineering of SARS-CoV-2. Emerging Microbes & Infections. 2020;9(1):505–507. doi: 10.1080/22221751.2020.1733440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu RJ, Zhao X, Li J, Niu PH, Yang B, Wu HL, Wang WL, Song H, Huang BY, Zhu N, Bi YH, Ma XJ, Zhan FX, Wang L, Hu T, Zhou H, Hu ZH, Zhou WM, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan JY, Xie ZH, Ma JM, Liu WJ, Wang DY, Xu WB, Holmes EC, Gao GF, Wu GZ, Chen WJ, Shi WF, Tan WJ Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020a;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu SY, Zhao Y, Yu WH, Yang Y, Gao JH, Wang JB, Kuang DX, Yang ML, Yang J, Ma CX, Xu JW, Qian XL, Li HY, Zhao SW, Li JM, Wang HX, Long HT, Zhou JX, Luo FY, Ding KY, Wu DJ, Zhang Y, Dong YL, Liu YQ, Zheng YQ, Lin XC, Jiao L, Zheng HY, Dai Q, Sun QM, Hu YZ, Ke CW, Liu HQ, Peng XZ Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv. 2020b doi: 10.1101/2020.04.08.031807. [DOI] [Google Scholar]
- 31.Martina BEE, Haagmans BL, Kuiken T, Fouchier RAM, Rimmelzwaan GF, van Amerongen G, Peiris JSM, Lim W, Osterhaus ADME SARS virus infection of cats and ferrets. Nature. 2003;425(6961):915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, Alhakeem R, Durosinloun A, Al Asmari M, Islam A, Kapoor A, Briese T, Daszak P, Al Rabeeah AA, Lipkin WI Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerging Infectious Diseases. 2013;19(11):1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, Avanzato VA, Rosenke R, Hanley PW, Saturday G, Scott D, Fischer ER, de Wit E Respiratory disease and virus shedding in rhesus macaques inoculated with SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.03.21.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omrani AS, Al-Tawfiq JA, Memish ZA Middle East respiratory syndrome coronavirus (MERS-Cov): animal to human interaction. Pathogens and Global Health. 2015;109(8):354–362. doi: 10.1080/20477724.2015.1122852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orellana C Laboratory-acquired SARS raises worries on biosafety. The Lancet Infectious Diseases. 2004;4(2):64. doi: 10.1016/S1473-3099(04)00911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peiris JSM, Guan Y, Yuen KY Severe acute respiratory syndrome. Nature Medicine. 2004;10(12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon LLM, Chu DKW, Chan KH, Wong OK, Ellis TM, Leung YHC, Lau SKP, Woo PCY, Suen KY, Yuen KY, Guan Y, Peiris JSM Identification of a novel coronavirus in bats. Journal of Virology. 2005;79(4):2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin ED, Zhu QY, Yu M, Fan BC, Chang GH, Si BY, Yang BA, Peng WM, Jiang T, Liu BH, Deng YQ, Liu H, Zhang Y, Wang CE, Li YQ, Gan YH, Li XY, Lü FS, Tan G, Cao WC, Yang RF, Wang J, Li W, Xu ZY, Li Y, Wu QF, Lin W, Chen WJ, Tang L, Deng YJ, Han YJ, Li CF, Lei M, Li GQ, Li WJ, Lü H, Shi JP, Tong ZZ, Zhang F, Li SG, Liu B, Liu SQ, Dong W, Wang J, Wong GKS, Yu J, Yang HM A complete sequence and comparative analysis of a SARS-associated virus (Isolate BJ01) Chinese Science Bulletin. 2003;48(10):941–948. doi: 10.1007/BF03184203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reusken CBEM, Schilp C, Raj VS, De Bruin E, Kohl RHG, Farag EABA, Haagmans BL, Al-Romaihi H, Le Grange F, Bosch BJ, Koopmans MPG MERS-CoV infection of alpaca in a region where MERS-CoV is endemic. Emerging Infectious Diseases. 2016;22(6):1129–1131. doi: 10.3201/eid2206.152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Munnink BBO, de Meulder D, van Amerongen G, van den Brand J, Okba NMA, Schipper D, van Run P, Leijten L, Sikkema R, Verschoor E, Verstrepen B, Bogers W, Langermans J, Drosten C, van Vlissingen MF, Fouchier R, de Swart R, Koopmans M, Haagmans BL Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020 doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TCT, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Günther S, Osterhaus ADME, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 42.Shan C, Yao YF, Yang XL, Zhou YW, Wu J, Gao G, Peng Y, Yang L, Hu X, Xiong J, Jiang RD, Zhang HJ, Gao XX, Peng C, Min J, Chen Y, Si HR, Zhou P, Wang YY, Wei HP, Pang W, Hu ZF, Lv LB, Zheng YT, Shi ZL, Yuan ZM. 2020. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in the rhesus macaques. Nature, under review, doi: 10.21203/rs.2.25200/v1.
- 43.Shehata MM, Gomaa MR, Ali MA, Kayali G Middle East respiratory syndrome coronavirus: a comprehensive review. Frontiers of Medicine. 2016;10(2):120–136. doi: 10.1007/s11684-016-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi ZL, Hu ZH A review of studies on animal reservoirs of the SARS coronavirus. Virus Research. 2008;133(1):74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi JZ, Wen ZY, Zhong GX, Yang HL, Wang C, Huang BY, Liu RQ, He XJ, Shuai L, Sun ZR, Zhao YB, Liu PP, Liang LB, Cui PF, Wang JL, Zhang XF, Guan YT, Tan WJ, Wu GZ, Chen HL, Bu ZG Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020 doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, Lei LC, Chen QX, Gao YW, Zhou HQ, Xiang H, Zheng HJ, Chern SWW, Cheng F, Pan CM, Xuan H, Chen SJ, Luo HM, Zhou DH, Liu YF, He JF, Qin PZ, Li LH, Ren YQ, Liang WJ, Yu YD, Anderson L, Wang M, Xu RH, Wu XW, Zheng HY, Chen JD, Liang GD, Gao Y, Liao M, Fang L, Jiang LY, Li H, Chen F, Di B, He LJ, Lin JY, Tong SX, Kong XG, Du L, Hao P, Tang H, Bernini A, Yu XJ, Spiga O, Guo ZM, Pan HY, He WZ, Manuguerra JC, Fontanet A, Danchin A, Niccolai N, Li YX, Wu CI, Zhao GP Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(7):2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadler K, Masignani V, Eickmann M, Becker S, Abrignani S, Klenk HD, Rappuoli R SARS-beginning to understand a new virus. Nature Reviews Microbiology. 2003;1(3):209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303(5664):1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 49.Wang M, Yan MY, Xu HF, Liang WL, Kan B, Zheng BJ, Chen HL, Zheng H, Xu YM, Zhang EM, Wang HX, Ye JR, Li GC, Li MC, Cui ZG, Liu YF, Guo RT, Liu XN, Zhan LH, Zhou DH, Zhao AL, Hai R, Yu DZ, Guan Y, Xu JG SARS-CoV infection in a restaurant from palm civet. Emerging Infectious Diseases. 2005;11(12):1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang LF, Shi ZL, Zhang SY, Field HE, Daszak P, Eaton BT Review of bats and SARS. Emerging Infectious Diseases. 2006;12(12):1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang QH, Zhang YF, Wu LL, Niu S, Song CL, Zhang ZY, Lu GW, Qiao CP, Hu Y, Yuen KY, Wang QS, Zhou H, Yan JH, Qi JX Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao KP, Zhai JQ, Feng YY, Zhou N, Zhang X, Zou JJ, Li N, Guo YQ, Li XB, Shen XJ, Zhang ZP, Shu FF, Huang WY, Li Y, Zhang ZD, Chen RA, Wu YJ, Peng SM, Huang M, Xie WJ, Cai QH, Hou FH, Liu YH, Chen W, Xiao LH, Shen YY Isolation and characterization of 2019-nCoV-like coronavirus from Malayan pangolins. bioRxiv. 2020 doi: 10.1101/2020.02.17.951335. [DOI] [Google Scholar]
- 54.Yan RH, Zhang YY, Li YN, Xia L, Guo YY, Zhou Q Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. The New England Journal of Medicine. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 56.Zhang T, Wu QF, Zhang ZG Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Current Biology. 2020a;30(7):1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q, Zhang HJ, Huang K, Yang Y, Hui XF, Gao JD, He XL, Li CF, Gong WX, Zhang YF, Peng C, Gao XX, Chen HC, Zou Z, Shi ZL, Jin ML SARS-CoV-2 neutralizing serum antibodies in cats: a serological investigation. bioRxiv. 2020b doi: 10.1101/2020.04.01.021196. [DOI] [Google Scholar]
- 58.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020a;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou H, Chen X, Hu T, Li J, Song H, Liu YR, Wang PH, Liu D, Yang J, Holmes EC, Hughes AC, Bi YH, Shi WF A novel bat coronavirus reveals natural insertions at the S1/S2 cleavage site of the Spike protein and a possible recombinant origin of HCoV-19. bioRxiv. 2020b doi: 10.1101/2020.03.02.974139. [DOI] [Google Scholar]
- 60.Zhu N, Zhang DY, Wang WL, Li XW, Yang B, Song JD, Zhao X, Huang BY, Shi WF, Lu RJ, Niu PH, Zhan FX, Ma XJ, Wang DY, Xu WB, Wu GZ, Gao GF, Tan WJ, The China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zumla A, Hui DS, Perlman S Middle East respiratory syndrome. The Lancet. 2015;386(9997):995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]