SUMMARY
The regulatory mechanisms by which neurons coordinate their physiology and connectivity are not well understood. The Drosophila olfactory receptor neurons (ORNs) provide an excellent system to investigate this question. Each ORN type expresses a unique olfactory receptor or a combination thereof, and sends their axons to a stereotyped glomerulus. Using single-cell RNA-sequencing, we identified 33 transcriptomic clusters for ORNs and mapped 20 to their glomerular types, demonstrating that transcriptomic clusters correspond well with anatomically and physiologically defined ORN types. Each ORN type expresses hundreds of transcription factors. Transcriptome-instructed genetic analyses revealed that 1) one broadly expressed transcription factor (Acj6) only regulates olfactory receptor expression in one ORN type and only wiring specificity in another type; 2) one type-restricted transcription factor (Forkhead) only regulates receptor expression; and 3) another type-restricted transcription factor (Unplugged) regulates both events. Thus, ORNs utilize diverse strategies and complex regulatory networks to coordinate their physiology and connectivity.
Graphical Abstract

eTOC
In this study, Li et al. perform single-cell RNA-sequencing of developing olfactory receptor neurons (ORNs) in Drosophila, and reveal that ORNs utilize diverse transcriptional strategies to coordinate their olfactory receptor expression and axon targeting.
INTRODUCTION
The ultimate function of a neuron is determined by both its physiology and connectivity, but the transcriptional regulatory mechanisms that coordinate these two features are not well understood [1–4]. In the Drosophila adult olfactory system, there are 50 types of olfactory receptor neurons (ORNs). Each ORN type expresses a single olfactory receptor or a unique combination thereof, which determines its physiological reponses to olfactory stimuli. Each ORN type also sends axons to a stereotyped glomerulus, which determines how the olfactory signal is represented in the brain [5, 6–8, 9]. The mammalian olfactory system shares these two features [10]. In mice, the coordination of olfactory receptor expression and wiring specificity is accomplished in part by olfactory receptors themselves regulating ORN wiring specificity [11, 12, 13]. However, Drosophila olfactory receptors do not regulate axon targeting [5, 14], raising the question of how receptor expression and wiring specificity are coordinated.
Single-cell RNA-sequencing (scRNA-seq) has been used to classify neurons and identify markers in diverse organisms and brain regions [e.g., 15–21]. Determining single cell transcriptomes should also facilitate the investigation of mechanisms that regulate olfactory receptor expression and axon targeting. Here, we performed single-cell RNA-sequencing for fly antennal ORNs, and identified 33 distinct transcriptomic clusters. We unambiguously mapped 20 clusters to their glomerular types, demonstrating that transcriptomic clusters correspond well with anatomically and physiologically defined ORN types. To understand transcriptional regulation of receptor expression and wiring, we comprehensively analyzed the expression patterns of all detected transcription factors from our sequenced cells, and found that each ORN cluster expresses hundreds of transcription factors. We then performed a targeted genetic screen focusing on transcription factors that are restricted to only a few clusters, and identified the transcription factor unpg that regulates both olfactory receptor expression and wiring, two fundamental features of sensory neurons. Together with genetic analysis of two other transcription factors, our findings revealed that ORNs utilize diverse transcriptional regulatory strategies to coordinate olfactory receptor expression and axon targeting.
RESULTS
Single-cell RNA-sequencing of Developing Drosophila ORNs
To identify molecular mechanisms underlying olfactory receptor expression and axon targeting, we first profiled transcriptomes of single ORNs from the Drosophila antenna at 42–48 hours after puparium formation (48hAPF hereafter). Transcriptomic analysis at this stage will allow us to capture molecules regulating both of these two events, because ORNs are completing their axon targeting [22] and olfactory receptors begin to express at 48hAPF [5, 7, 14]. Using plate-based single-cell RNA-sequencing with SMART-seq2 [19, 23], we obtained 1016 high-quality cells from about 44 types of antennal ORNs, with each cell sequenced to a depth of ~1 million reads resulting in ~1500 detected genes (Figure 1A; Figure S1A–C; STAR*Methods). Using an unsupervised machine-learning algorithm [19] and hierarchical density-based unbiased clustering [24], we identified 33 distinct transcriptomic clusters (Figure 1B; Figure S1D–F).
Figure 1. Single-cell RNA-seq of Drosophila Olfactory Receptor Neurons (ORNs) at 48hAPF.

(A) Schematic of the fly olfactory system and single-cell RNA-seq workflow. 50 ORN types (44 from the third segment of the antenna and 6 from the maxillary palp) send axons to the antennal lobe to form stereotypical one-to-one connections with 50 types of projection neurons (PNs). The third segments of pupal antennae were manually dissected, GFP+ ORNs were sorted into 96-well plates using fluorescence-activated cell sorting (FACS), and the Smart-seq2 protocol was used for library preparation and sequencing [23].
(B) Visualization of ORN transcriptomic clusters using t-distributed Stochastic Neighbor Embedding (tSNE) plot based on 408 genes identified by ICIM. Each dot is a cell. 1016 ORNs at 48hAPF (908 from pan-ORN nSyb-GAL4, 63 from 85A10-GAL4, and 45 from AM29-GAL4) form 33 distinct clusters. Black dots are cells that could not be assigned to any cluster.
(C) Hierarchical heat map showing clear separation of 1016 ORNs (red) and 946 PNs (green) using top 500 overdispersed genes identified across all cells. Each column is one cell and each row is one gene. ORNs are from 48hAPF. PNs are from 24hAPF [19]. Expression levels are indicated by the color bar (CPM, counts per million sequencing reads). Cells (columns) and genes (rows) are ordered using hierarchical clustering.
(D) Visualization of ORNs (red) and PNs (green) using principal component analysis followed by tSNE plot using top 500 overdispersed genes.
See also Figure S1.
Drosophila ORNs and projection neurons (PNs) are synaptic partners in the antennal lobe. We compared their transcriptomic differences using the 1016 ORNs here with 946 PNs sequenced previously [19]. Using either highly variable genes across all ORNs and PNs, or differentially expressed genes between them, ORNs and PNs were readily separated into two distinct groups (Figure 1C, D; Figure S1G). We further identified several ORN- and PN-specific genes, including widely used ORN and PN markers (Figure S1H), and validated the expression of a newly-identified ORN-specific gene [25] (Figure S1I).
Matching Transcriptomic Clusters with Glomerular Types
We next employed three strategies to map transcriptomic clusters to anatomically- and functionally-defined ORN types. First, we used AM29-GAL4 [26] and 85A10-GAL4 [27] driven GFP expression to label two and five distinct ORN types, respectively, and sequenced these cells at 48hAPF (Figure 2A). As expected, AM29+ and 85A10+ ORNs mapped to two and five distinct ORN clusters, respectively (Figure 2B; Figure S2C, I). Second, we used expression of fruitless (fru) in three ORN types [28] to define three more clusters (Figure S2M).
Figure 2. Mapping Transcriptomic Clusters to Glomerular Types.

(A) Three drivers were used to label ORNs for scRNA-seq: nSyb-GAL4 for all ORNs, AM29-GAL4 and 85A10-GAL4 for two and five specific ORN types, respectively. Confocal images showing expression patterns of these drivers in both antenna and antennal lobe at 48hAPF. All drivers were crossed with ey-Flp;UAS-FRT-STOP-FRT-mCD8:GFP to restrict the GFP expression to cells in the antenna. Elav staining labels neuronal nuclei, and N-cadherin (NCad) staining labels neuropil.
(B) Visualization of nSyb+, AM29+, and 85A10+ ORNs using tSNE plot as in Figure 1B. Cells are colored according to drivers. AM29+ cells (magenta) map to two clusters, and 85A10+ cells (green) map to five clusters (circled). Note that several individual cells from both drivers fall into other clusters, likely due to stochastic sparse labeling of these two drivers in other ORNs beyond the 7 ORN types.
(C) Heat maps showing top 19 detected olfactory receptor genes in 1016 ORNs. Each column is an individual ORN from 48hAPF. Cells are ordered using hierarchical clustering.
(D) tSNE plots showing Gr21a expression. Gr21a+ ORNs target axons to glomerulus V.
(E) Validation of the decoded cluster. Intersecting ey-Flp with DIP-epsilon-T2A-GAL4 specifically labels V-ORNs (GFP, green) at 48hAPF. N-cadherin (NCad) staining labels neuropil.
(F) Spatial distribution of ORNs from four sensillar groups in the antenna [adapted from 29].
(G) Hierarchical clustering of ORNs based on the heat map in Figure S3C. Sensillar groups are colored according to Figure 2F.
(H–J) Summary of 20 transcriptomic clusters (H) that have been mapped to 21 glomerular types (I and J). Note that cluster 9 was mapped to two ORN types, VM5d and VM5v. D, dorsal; L, lateral. Scale bar, 20 μm.
Third, we used olfactory receptor expression to map transcriptomic clusters to glomerular types based on previously established correspondences between olfactory receptor expression and glomerular targets [8, 29, 30, 31]. We first systematically assessed the expression of olfactory receptors, including all chemosensory receptors in the olfactory system belonging to the Or (odorant receptor), Gr (gustatory receptor), and Ir (ionotropic receptor) families [5, 7, 32, 33]. Excluding co-receptors expressed in multiple adult ORN types (Orco and Ir25a, which act in concert with type-specific receptors for odor detection) [31, 34], we found that 37% of ORNs expressed olfactory receptors at 48hAPF (Figure 2C), consistent with the previous finding that olfactory receptors are gradually turned on during pupal development [7]. We then used detected receptors to futher assign positive clusters to specific glomerular types (Figure S2A, B, D–H, J–L, N–U).
The three strategies gave congruent results when mapping the same clusters, and allowed us to decode the glomerular identity of 18 clusters in total. We further validated our cluster identity assignments using GAL4 drivers from specific genes. In all cases, the GAL4 expression patterns matched their cluster identities (Figure 2D, E; Figure S2V–Y), suggesting robust mapping between transcriptomic clusters and glomerular types. The only exception to the one-toone matching between transcriptomic clusters and glomerular types is that Cluster 9 corresponds to two ORN types, VM5d and VM5v (Figure S2J, K).
Relationship between Transcriptomes, Lineage, and Spatial Patterning
Each antennal ORN is housed in one of four types of sensory organs called sensilla. Each sensillum contains 1–4 ORNs derived from a common progenitor, with stereotyped combination of ORN type, morphology, and spatial distribution [8, 29] (Figure 2F; Figure S3A). To examine the relationship between transcriptomes, lineage, and spatial patterning of antennal ORNs, we compared transcriptomes that we had mapped to the glomerular types. We found that ORNs that belong to the same sensillar type (large basiconics, small basiconics, tricoids, or coeloconics) share more similar transcriptomes compared to ORNs that belong to different sensillar types (Figure 2G; Figure S3B, C), based on our decoded clusters. We also note two exceptions: two clusters (DL3 and DA4l) from antennal tricoides are more similar to thin and small basiconics ORNs from our hierarchical clustering analysis (Figure 2G, S3C). ORNs within the same sensillum did not exhibit additional similarities when compared to ORNs from the same sensillar types yet from different sensilla (Figure S3D). We further identified differentially expression genes between four sensillar types, and noticed that transcription factors (TFs) and cell-surface molecules (CSMs) account for a large proportion (Figure S4A–C), suggesting a critical role for these two sets of genes during this developmental stage.
These analyses suggest that spatial distributions across the antenna, which separate different sensillar types (Figure 2F), are an important determinant for transcriptomes of individual ORN types. We predicted the sensillar types of two previously unidentified transcriptomic clusters based on hierarchical clustering (Figure 2G), and validated the identity of their corresponding glomeruli with experimental data (Figure S3E–H).
In summary, we mapped a total of 20 transcriptomic clusters to 21 specific glomerular types (Figure 2H–J), establishing a nearly one-to-one correspondence between transcriptomic clusters and anatomically and physiologically defined ORN types. Our annotated dataset provides a valuable resource for studying development and function of individual ORN types.
Transcription Factor Expression Analyses in ORNs
We next investigated the mechanisms by which transcription factors (TFs) regulate olfactory receptor expression and wiring specificity. In principle, three types of TFs may exist: those that regulate wiring specificity only, receptor expression only, or both (Figure 3A). We first compared expression levels between TFs and other genes in the genome. Contrary to the notion that transcription factors are usually expressed at relatively low levels [35], we found no significant differences between expression levels of TFs and non-TFs in the genome (Figure 3B).
Figure 3. Transcription Factor Expression in ORNs.

(A) Schematic showing three kinds of transcription factors (TFs) that regulate olfactory receptor expression and wiring specificity.
(B) Scatter plots showing number of cells in which a gene can be detected versus the mean expression level of the gene in all cells. TFs are highlighted. A positive cell is defined as the cell expressing a gene at the level of Log2(CPM+1) ≥3. CPM, counts per million. This analysis shows that TFs have a wide range of expression patterns, from a few cells or all cells, and that the overall TF expression level is comparable with other types of genes.
(C) Distributions of the number of TFs detected per ORN at the level of Log2(CPM+1) ≥ 3.
(D) Sparsity and expression level of the TFs among ORN transcriptomic clusters. Each dot is one TF. A positive cluster is defined as more than 30% cells in the cluster expressing the TF at the level of Log2(CPM+1) ≥ 3. Highlighted in red are three example genes, unpg, eyg, and acj6, with high mean expression levels but different sparsity.
(E) tSNE plots showing expression of unpg, eyg, and acj6. Expression levels are indicated by the color bar. In the acj6 tSNE plot, two clusters corresponding to DL4 and DM6 are indicated.
Feature maps like those shown in (E) can be produced for any of the transcription factors listed in (D) using transcriptomic data we have deposited in Github.
We next characterized TF expression at the level of cells and clusters with the following criteria: a positive cluster is defined as more than 30% cells in the cluster expressing the TF at the level of Log2(CPM+1) ≥ 3, where CPM is transcript counts per million. Of the 1045 TFs in the fly genome [36], 899 were expressed in at least one ORN, and on average about 150 TFs were detected in individual ORNs (Figure 3C). At the cluster level, 423 TFs were detected in one or more clusters, exhibiting a wide range of expression patterns (Figure 3D, E). Next we sought to identify TFs that regulate olfactory receptor expression and wiring specificity based on our transcriptomic data.
POU-domain Transcription Factor Acj6 Regulates Olfactory Receptor Expression in One ORN Type, but Axon Targeting in Another
Previous studies have identified several TFs that regulate Drosophila olfactory receptor expression or wiring specificity [37, 5, 38–40, 41]. However, with the exception of one study on ORN fate diversification [26], these two processes have been investigated separately within the same ORN type because of a lack of appropriate genetic tools. Transgenes utilizing olfactory receptor promoter driven GFP were used to visualize ORN axon targeting in most cases. If a TF were required for olfactory receptor expression, deleting the TF would lead to the loss of olfactory receptor expression, such that ORN axon targeting could not be visualized. We overcame this obstacle by using a GAL4 reporter whose expression is independent of olfactory receptors, as well as additional methods (see below).
We first analyzed a POU-domain transcription factor, abnormal chemosensory jump 6 (acj6), which is expressed in most antennal ORNs and regulates receptor expression in some ORN types and axon targeting in other ORN types [37, 38, 41]. However, it is unclear if acj6 regulates both wiring specificity and receptor expression in the same ORN type. Our sequencing data showed that acj6 was expressed in all ORN clusters, including those that express AM29-GAL4 and target axons to the DL4 and DM6 glomeruli (hereafter DL4-ORNs and DM6-ORNs) (Figure 3E, 4A). Using an acj6 null allele [37], we found that acj6 was required for the expression of Or85f in DL4-ORNs but not required for the expression of Or67a in DM6-ORNs (Figure 4A), consistent with previous results [41]. Using AM29-GAL4 to independently label the axons of DL4- and DM6-ORNs, we found that DM6-ORNs showed highly penetrant mistargeting phenotypes, while DL4-ORNs still targeted to the correct glomerulus (Figure 4A). To test whether acj6 regulates the axon targeting cell-autonomously, we performed mosaic analysis with a repressible cell marker (MARCM), where homozygous mutant cells are labeled within an otherwise mostly heterozygous genetic background [42]. Sparse MARCM clones visualized by AM29-GAL4 revealed similar mistargeting of DM6-ORNs, but normal targeting of DL4-ORNs (Figure 4B). These experiments suggest that acj6 acts cell-autonomously to regulate axon targeting of DM6- but not DL4-ORNs. In summary, acj6 is required for wiring but not receptor expression in DM6-ORNs, and is required for receptor expression but not wiring in DL4-ORNs (Figure 4C).
Figure 4. Role of acj6 in Olfactory Receptor Expression and Axon Targeting.
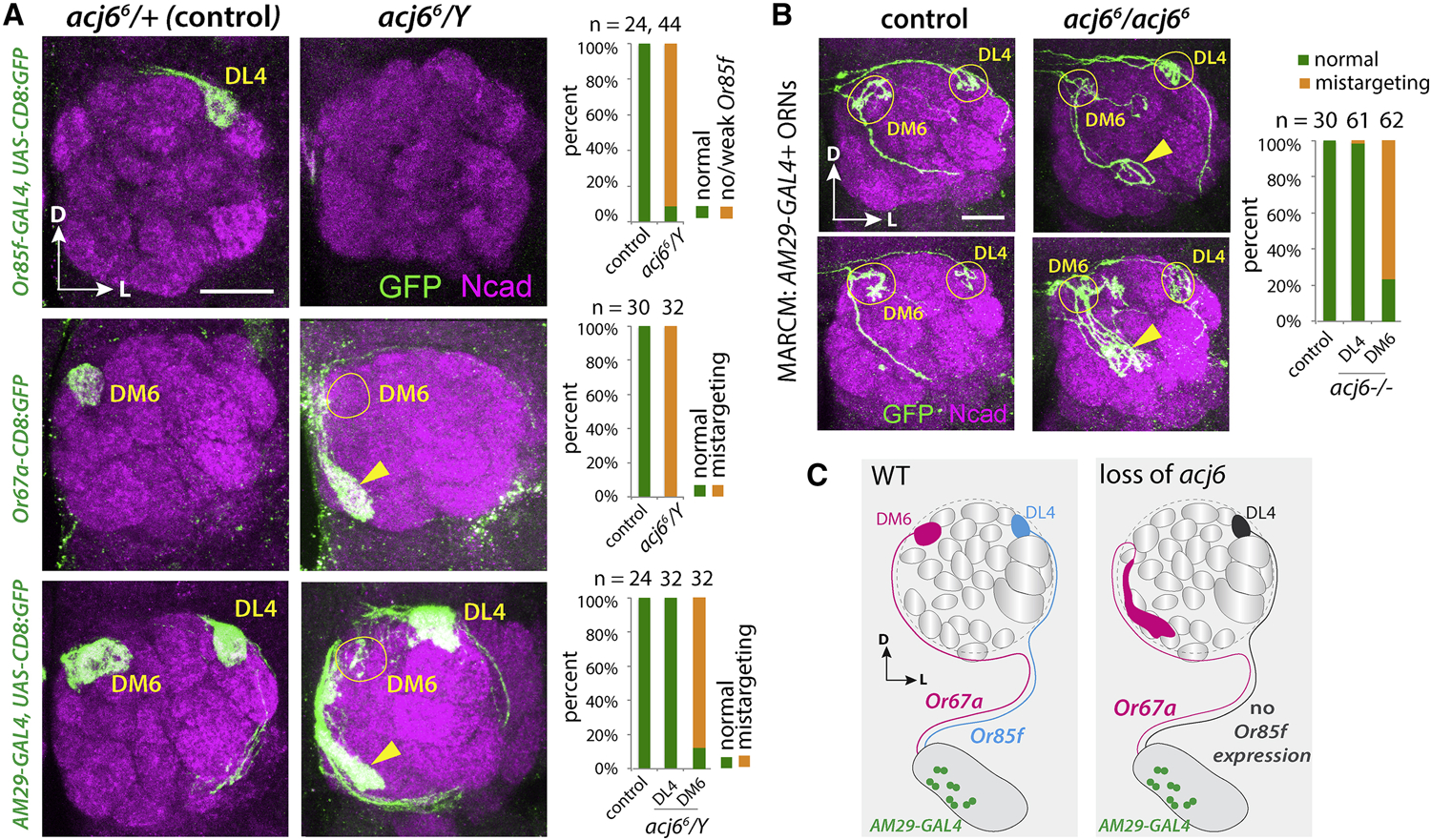
(A) Confocal images of adult antennal lobes showing ORN axon targeting in heterozygous control (left) and hemizygous mutant (right). In acj6 mutants, Or85f expression is lost, but DL4 ORN axons still target normally as shown by AM29-GAL4; Or67a expression is normal, but DM6 ORN axons mistarget (arrow in the middle panel), as confirmed by AM29-GAL4 (arrows in the bottom panel).
(B) Sparse MARCM clones visualized by AM29-GAL4 revealed mistargeting of ORN axons that took the medial trajectory, with a mistargeting phenotype (arrows) highly similar to mistargeting of DM6-ORN axons labeled by Or67a in acj6 mutant (Figure 3F). ORN axons that took the lateral trajectory targeted normally to the DL4 glomerulus. Quantifications are shown on the right, with antennal lobe numbers (n) on top.
(C) Schematic summary: acj6 is required for receptor expression but not for wiring specificity in DL4-ORNs, and is required for wiring specificity but not for receptor expression in DM6-ORNs.
All confocal images are z-stacks covering the region of target glomeruli. N-cadherin (NCad) staining labels neuropil. D, dorsal; L, lateral. Scale, 20 μm.
Homeodomain Transcription Factor Unplugged Regulates Both Olfactory Receptor Expression and ORN Axon Targeting
To test whether type-restricted transcription factor (TF) expression also contributes to olfactory receptor expression and wiring specificity, we focused on ORNs targeting to glomerulus V (hereafter V-ORNs), which co-express two olfactory receptors Gr21a and Gr63a [43, 44]. We performed an RNAi screen on 25 candidate TFs expressed in only a few transcriptomic clusters including V-ORNs. This screen identified two TFs. The first TF is the founding member of the FOX family of transcription factors, Forkhead (Fkh) [45], which is expressed in two clusters including V-ORNs (Figure 5A). Knockdown of fkh drastically reduced the reporter expression for both Gr21a and Gr63a (Figure 5B). However, the size and shape of the V glomerulus and axon targeting reflected by residual reporter expression appeared normal (Figure 5B), suggesting that fkh is required for normal receptor expression but not for wiring of V-ORNs.
Figure 5. Transcriptional Control of Olfactory Receptor Expression and Wiring Specificity in V-ORNs.
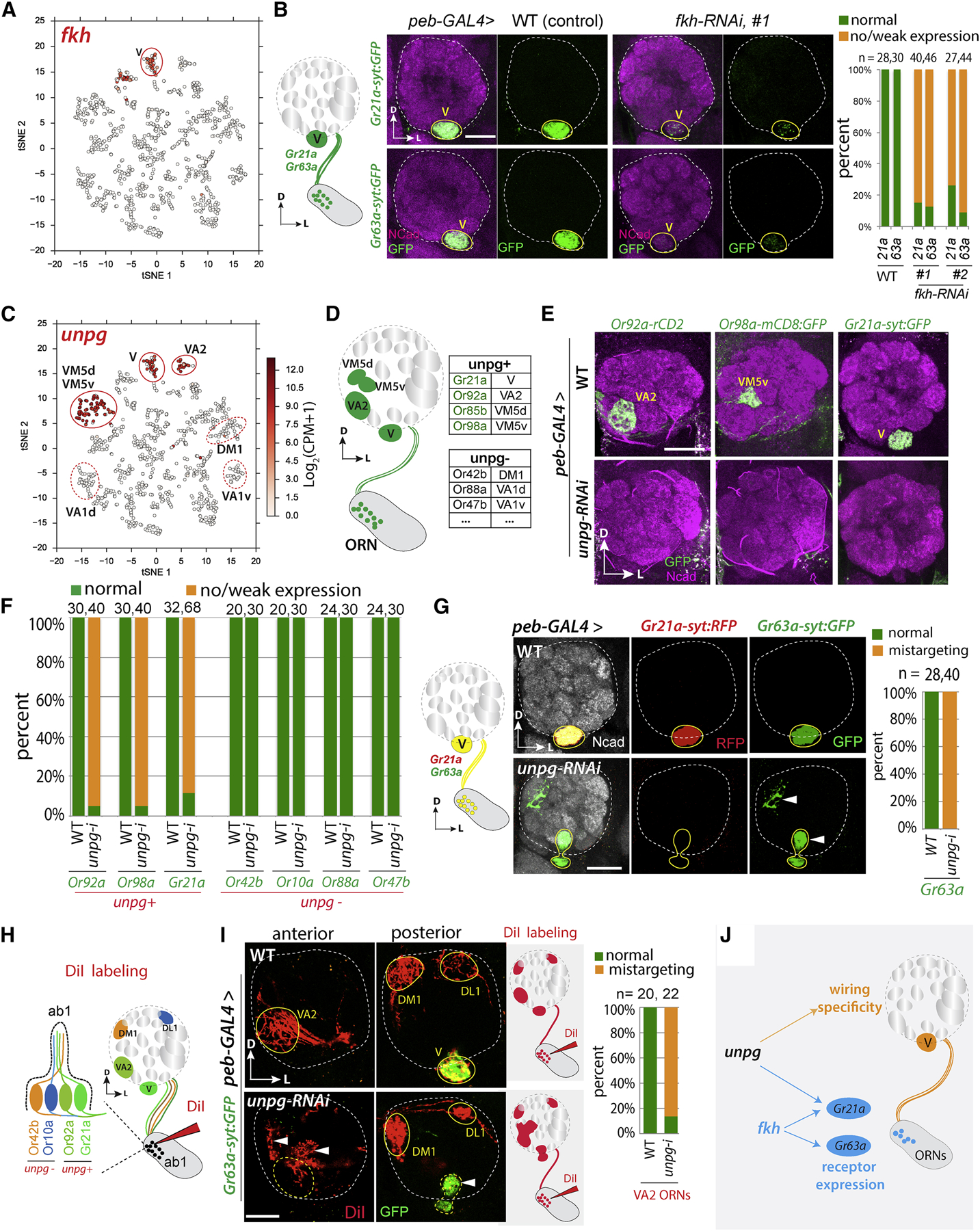
(A) tSNE plot showing that fkh is expressed in 2 clusters, one of which corresponds to V-ORNs.
(B) Olfactory receptors Gr63a and Gr21a are co-expressed in V-ORNs. Confocal images of adult antennal lobes showing that ORN knockdown of fkh using peb-GAL4;UAS-dcr2 causes markedly reduced expression of markers driven from transgenes utilizing the promoters of Gr21 and Gr63. Quantified on the right with antennal lobe number (n) on top. Residual marker expression indicates that Gr21a/Gr63a+ axons still target to V.
(C) tSNE plot showing that unpg is expressed in three clusters mapped to V, VA2, and VM5d/VM5v (solid outline). Three unpg-negative clusters are indicated (dashed outline).
(D) Schematic summarizing receptor expression and glomerular targets of all unpg-positive and some unpg-negative ORN types.
(E) Confocal images of adult antennal lobes showing olfactory receptor expression in control (WT) and unpg-RNAi flies. peb-GAL4;UAS-dcr2 was crossed with either w1118 (WT) or unpg-RNAi, and olfactory receptor promoter-driven reporters were used to monitor receptor expression. In WT flies, Or92a, Or98a, and Gr21a are expressed normally. In unpg-RNAi flies, the expression of all three receptors is lost.
(F) Quantification for the receptor expression in three unpg-positive ORN types and four unpg-negative ones.
(G) In unpg-RNAi flies, Gr21a expression is lost consistently, and Gr63a+ axons show stereotyped mistargeting (arrowheads). Quantifications are shown on the right.
(H) Schematic illustrating the strategy of DiI labeling for four ORN types from the same ab1 sensilla in the antenna: Or42b and Or10a (unpg-negative), as well as Gr21a and Or92a (unpg-positive).
(I) For DiI labeling, Gr63a-syt:GFP is used to monitor V-ORNs. In WT flies, DiI labels VA2 in an anterior section of the antennal lobe, and DM1, DL1 and V in a posterior section. In unpg-RNAi flies, DiI labels a diffuse area (arrowheads) around VA2 in the anterior section, labels DL1 and DM1 normally, and co-labels Gr63a-posistive V-ORNs, which show mistargeting phenotypes. Schematic on the right summarizes DiI labeling in WT and unpg-RNAi flies. Quantifications are shown on the right; antennal lobe numbers (n) are indicated.
(J) Summary of the regulation of olfactory receptor expression and wiring in V-ORNs by transcription factors fkh and unpg.
See also Figure S5.
The second TF is the homeobox transcription factor Unplugged (Unpg), previously identified as a regulator of tracheal branch formation [46] and as a marker for a specific neuroblast sub-lineage [47]. We found unpg expression in three transcriptomic clusters mapped to four ORN types: V (Gr21a/Gr63a+), VA2 (Or92a+), VM5d (Or85b+), and VM5v (Or98a+) (Figure 5C, D). Because the Or85b reporter was barely detectable in control flies, we focused our subsequent analysis on the other three ORN types. We found RNAi knockdown of unpg in ORNs caused loss of receptor expression in all three unpg+ ORN types, but not in four unpg– ORN types (Figure 5E, F; Figure S5A–C), suggesting that unpg is specifically required for olfactory receptor expression in unpg+ ORNs. Since two unpg− ORNs were from the same sensilla as unpg+ ORNs that express Or92a and Gr21a (see Figure 5H), these data suggest that the loss of Or92a and Gr21a in unpg-RNAi flies was not due to gross developmental defects of the sensilla.
Does unpg also regulate axon targeting? Interestingly, unlike Gr21a in V-ORNs, the co-expressed receptor Gr63a was not downregulated by unpg RNAi knockdown, and Gr63a+ axons showed stereotyped mistargeting in all unpg-RNAi flies (Figure 5G). The V glomerulus was also smaller and misshapen, likely reflecting a lack of proper V-ORN axon targeting. Thus, unpg regulates both Gr21a expression and axon targeting of V-ORNs. We note that some V-ORN axons in unpg-RNAi flies still targeted to the correct glomerulus. This is consistent with previous studies where deleting a single gene usually leads to partial mis-wiring phenotypes, presumably due to partial redundancy of regulators of wiring specificity [19, 48].
Two additional approaches using PN dendrite targeting and DiI labeling of ORN axons revealed that unpg also controls axon targeting in three other unpg-positive ORN types: VM5d, VM5v, and VA2 (Figure 5H, I; Figure S5D). We identified a Janelia-GAL4 driver [27], GMR86C10-GAL4, that specifically labels VM5d and VM5v PNs, and generated a GMR86C10-LexA driver to label these PNs independent of the GAL4/UAS system. Since ORN axon mistargeting during development can lead to dendrite mistargeting of partner PNs [49, 50], we reasoned that we might observe mistargeting of VM5d and VM5v PNs if knocking down unpg in VM5d and VM5v ORNs causes targeting defects. This was indeed the case (Figure S5D), as we observed that VM5d and VM5v PN dendrites showed mistargeting phenotypes when unpg was only knocked down in ORNs using pan-ORN peb-GAL4. This result suggests that unpg also regulates VM5d and VM5v ORN axon targeting.
Finally, we utilized the fluorescent lipophilic DiI-mediated anterograde tracing to examine axon targeting of Or92+ VA2-ORNs. Each ab1 sensillum houses four ORNs: two are unpg-negative, and two are unpg-positive (V- and VA2-ORNs; Figure 5H) [29]. By applying DiI to a small subset of ab1 sensilla to initiate anterograde tracing, we found that two unpg-negative ORN types targeted their axons normally as expected, while the two unpg-positive ones showed stereotyped mistargeting (Figure 5I), suggesting that unpg also regulates axon targeting of VA2 ORNs. Taken together, our data suggest that unpg controls both olfactory receptor expression and wiring specificity within the same ORN types (Figure 5J; S5E).
DISCUSSION
Using plate-based single-cell RNA-sequencing, we analyzed high-quality transcriptomes of 1016 antennal ORNs at a mid-pupal stage, when ORNs are completing their axon targeting to their cognate glomeruli and a subset of ORNs start to express olfactory receptors. The smaller number of transcriptomic clusters (33) compared to glomerular types (44 for antennal ORNs) may result from: 1) for some ORN types, not enough cells were captured to reach the minimal requirement of forming a cluster (see STAR*Methods for criteria for transcriptomic clusters); 2) closely-related ORN types may form one transcriptomic cluster (e.g., Cluster 9 correspond to two ORN types, VM5d and VM5v; Figure S2J, K). Besides olfactory receptor neurons, there are also other sensory cells in the third segment of the antenna, for example hygro- and thermo-sensory neurons in the sacculus and arista. It has been shown that all those neurons express Ir25a and Ir93a, and different subsets express Ir21a, Ir40a, Ir68a, and Gr28b in adult flies [33, 51–54]. Our scRNAseq data shows that Ir25a is broadly expressed in many ORN types, but all other above genes are not expressed at 48hAPF. Due to the lack of specific markers, we could not define these cells. Compared to the large number of ORNs, these other cells likely constitute a minority of cells.
Our understanding of how developing neurons coordinately regulate physiological properties and connectivity is limited to only a few examples [11–13, 55, 56]. Here, we found that even in the same group of neurons (Drosophila ORNs), the coordination of these two features uses diverse transcriptional strategies. On one hand, the broadly expressed acj6 regulates receptor expression but not wiring in one ORN type, and wiring but not receptor expression in a second type. On the other hand, the type-restricted unpg regulates both receptor expression and wiring specificity in all ORN types that express unpg (Figure S5E). However, within the V-ORNs, the type-restricted fkh regulates the expression of both co-receptors but not wiring, whereas unpg regulates only one of the two co-receptors, arguing against a simple regulatory relationship (Figure 5J). The complexity of the regulatory network inferred from our study is perhaps a result of evolution of different ORN types in a piece-meal fashion, as reflected by their utilizing three distinct families of chemoreceptors as olfactory receptors [5–7, 33, 34]. Untangling this complexity requires future studies to systematically identify transcriptional targets of these transcription factors and investigate their regulatory relationship.
In conclusion, scRNA-seq in developing Drosophila ORNs enabled us to map 20 transcriptomic clusters to glomerular types. This reinforces the idea that neuronal transcriptomic identity corresponds well with anatomical and physiological identity defined by connectivity and function in well-defined neuronal types [17, 19]. Our genetic analyses further suggest that ORNs utilize diverse regulatory strategies to coordinate their physiology and connectivity. Given that each ORN type expresses hundreds of transcription factors, it is remarkable that loss of a single transcription factor, unpg, can result in profound disruption of receptor expression and wiring specificity, two most fundamental properties of sensory neurons.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Liqun Luo (lluo@stanford.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly stocks
The following fly lines were used in this study. nSyb-GAL4 (Bloomington Drosophila Stock Center, BDSC #51635); act5C-GAL4 (BDSC #3954); AM29-GAL4 [26]; GMR86C10-GAL4 [57] and GMR85A10-GAL4 [27]; NompB-T2A-GAL4 (BDSC #76632); DIP-epsilon-T2A-GAL4 (BDSC #67502); 5-HT1B-T2A-GAL4 (BDSC #76668); tsh-T2A-GAL4 [25]; acj66 [37]; Mz19-QF [49]; fkh-RNAi (BDSC #33760, #58059); UAS-Robo2 (BDSC #66886); Robo2-RNAi (BDSC #9286), unpg-RNAi (VDRC #107638); peb-GAL4 [58]; GH146-Flp [59]; ey-Flp [60]; UAS-STOP-mCD8:GFP [49]; Or-GAL4 and Or-mCD8:GFP [29]; Or85f-GAL4, Or42b-mCD8:GFP, Or67a-mCD8:GFP, Or98a-mCD8:GFP; Or92a-rCD2 [61]; Or47b-rCD2 [62]; Or88a-mtdT [50]; Or10a-LexA [30]; Gr21a-syt:GFP and Gr63a-syt:GFP [43].
METHOD DETAILS
Immunostaining
Tissue dissection and immunostaining were performed following previously described methods [63]. Briefly, fly pupal and adult brains were dissected in 1x PBS and then fixed in 4% paraformaldehyde (20% paraformaldehyde diluted in PBS with 0.015% Triton X-100) for 20 min at room temperature. Fixed brains were washed three times with PBST (PBS with 0.3% Triton X-100) and incubated in PBST twice for 20 min. The samples were incubated in blocking buffer (5% normal goat serum in PBST) for 30 min at room temperature or overnight at 4°C. Then, primary antibodies diluted in blocking buffer were applied and samples were incubated for 24–48 h at 4°C. Then, samples were washed using PBST for 20 min twice, and secondary antibodies diluted in blocking buffer were applied and samples were incubated in dark for more than 24 h at 4°C. Samples were washed in PBST for 20 min twice and mounting solution (Slow Fade Gold) was added. Samples were left in mounting solution for at least 1 h before mounting them onto glass slides. All wash steps were performed at room temperature. Primary antibodies used in this study include rat anti-DNcad (DN-Ex #8; 1:40; DSHB), chicken anti-GFP (1:1000; Aves Labs), rabbit anti-DsRed (1:250; Clontech), mouse anti-ratCD2 (OX-34; 1:200; AbD Serotec). Secondary antibodies were raised in goat or donkey against rabbit, mouse, rat, and chicken antisera (Jackson Immunoresearch), conjugated to Alexa 405, 488, FITC, Cy3, Cy5, or Alexa 647.
MARCM analysis
For ORN MARCM clonal analysis, Tub-GAL80,hsFlp,FRT19A;AM29-GAL4 flies were crossed either with UAS-mCD8:GFP,FRT19A (control) or with acj66,UAS- mCD8:GFP,FRT19A. Newly formed pupae (0–2hAPF) were heat-shocked for 1 hour at 37°C and adult flies were dissected for axon projection analysis. While acj6 is required for dendrite targeting of a subset of PNs [64] and PN targeting can affect ORN axon targeting [50], the observed ORN axon targeting phenotype is not due to loss of acj6 in unlabeled PN clones because all PNs are born before the pupal stage and therefore could not produce clones under our heat-shock condition.
Confocal imaging
All confocal images were taken through a Z-stack scan from most anterior to the most posterior of the antenna or antennal lobe using the Zeiss LSM 780 system. Then images were processed with ImageJ and Adobe Illustrator. For quantification in Figs. 4 and 5, at least 10 flies (20 antennal lobes) were used.
DiI labeling
DiI (Sigma 468495) in saturated DMSO solution was applied to the medioproximal corner of the third antennal segment of adult flies, where the ab1 sensilla are located, through glass micropipette using a micromanipulator. Flies were recovered for 24h after labeling before their brains were dissected, mounted in 30% sucrose, and imaged using a confocal microscope. Mistargeting phenotypes of both antennal lobes from each fly were quantified in WT control and unpg-RNAi expressing flies. Since 4 types of ORNs are localized in ab1 sensilla, we only focused on flies that are labeled for 4 glomeruli. Out of 11 WT flies, 1 was labeled for many glomeruli, which is likely due to labeling of additional sensilla near ab1 during DiI application. 3 out of 14 unpg-RNAi expressing flies showed similar additional labeling and were excluded from quantification. In total, 10 WT and 11 unpg-RNAi flies (20 and 22 antennal lobes) were quantified for VA2 ORN axon targeting.
Quantitative PCR (qPCR)
Total RNA was extracted using MiniPrep kit (Zymo Research, R1054) from either actin5CGAL4, w1118 (control) or actin5C-GAL4; unpg-RNAi flies at middle pupal stage (N = 3 replicates for each condition; 5 pupae per replicate). cDNA was synthesized using an oligo-dT primer. qPCR was performed on a Bio-Rad CFX96 detection system. p-value was calculated using Student’s t test. Relative expression was normalized to actin5C. Primer sequences used for qPCR were:
actin5C (F): 5’-CTCGCCACTTGCGTTTACAGT-3’
actin5C (R): 5’-TCCATATCGTCCCAGTTGGTC-3’
unpg pair 1 (F): 5’- CTACAACGGCGAGATGGACA-3’
unpg pair 1 (R): 5’- TTGGAGTTTGAGCTGGAGCC-3’
unpg pair 2 (F): 5’- GGAACTACAACGGCGAGATG-3’
unpg pair 2 (R): 5’- GATACTTCTTGGCGTGGAACT C-3’
Single-cell RNA-seq
Single-cell RNA-seq was performed following the protocol that we developed recently [19]. Briefly, Drosophila third antennal segments with mCD8:GFP-labeled cells using specific GAL4 drivers were manually dissected. Single-cell suspensions were then prepared. Single labeled cells were sorted via Fluorescence Activated Cell Sorting (FACS) into individual wells of 96-well plates containing lysis buffer using an SH800 instrument (Sony Biotechnology). Full-length poly(A)-tailed RNA was reverse-transcribed and amplified by PCR following the SMART-seq2 protocol47 with several modifications as below. To increase cDNA yield and detection efficiency, we increased the number of PCR cycles to 25. To reduce the amount of primer dimer PCR artifacts, we digested the reverse-transcribed first-strand cDNA using lambda exonuclease (New England Biolabs) (37°C for 30 min) prior to PCR amplification. Sequencing libraries were prepared from amplified cDNA using tagmentation (Nextera XT). Sequencing was performed using the Illumina Nextseq 500 platform with paired-end 75 bp reads.
Sequence alignment and preprocessing
Reads were aligned to the Drosophila melanogaster genome (r6.10) using STAR (2.4.2a) [65] with the ENCODE standard options, except “--outFilterScoreMinOverLread 0.4 --outFilterMatchNminOverLread 0.4 --outFilterMismatchNmax 999 --outFilterMismatchNoverLmax 0.04”. Uniquely mapped reads that overlap with genes were counted using HTSeq-count (0.7.1) [66] with default settings except “-m intersection-strict”. Cells having fewer than 100,000 uniquely mapped reads were removed. To normalize for differences in sequencing depth across individual cells, we rescaled gene counts to counts per million (CPM). All analyses were performed after converting gene counts to logarithmic space via the transformation Log2(CPM+1). Sequenced cells were filtered for expression of canonical neuronal genes (elav, brp, Syt1, nSyb, CadN, and mCD8GFP), retaining only those cells that expressed at least 4/6 genes at Log2 (CPM+1) ≥ 3.
PCA and tSNE
Principal component analysis (PCA) and t-distributed Stochastic Neighbor Embedding (tSNE) were used for visualizing Drosophila scRNA-seq data as detailed previously [19]. Briefly, to visualize and interpret high dimensional gene expression data, we obtained two-dimensional projections of the cell population by first reducing the dimensionality of the gene expression matrix using PCA, then further reducing the dimensionality of these components using tSNE [67]. We performed PCA for Figure 1D on a reduced gene expression matrix composed of the top 500 overdispersed genes. The top 8 principal components (8 PCs) were used. We further reduced these components using tSNE to project them into a two-dimensional space.
Iterative Clustering for Identifying Markers (ICIM)
We previously developed an unsupervised machine learning algorithm called ICIM to identify genes that distinguish transcriptome clusters for different fly olfactory projection neuron (PN) subtypes [19] (available at https://github.com/felixhorns/FlyPN). We performed similar analysis for ORNs with several modifications of the adjustable parameters, including pearson correlation and dropout threshold, and identified 408 ICIM genes, which we used for further dimensionality reduction by PCA. The top 18 PCs were further reduced to two dimensions using tSNE. HDBSCAN, a hierarchical density-based unbiased clustering algorithm [24], was used to identify clusters.
We observed that standard dimensionality reduction and clustering methods using PCA and tSNE failed to discriminate subpopulations that corresponded to known lineages and molecular features of PNs [19] or ORNs (current study). We attributed the failure of these methods to the high degree of similarity of transcriptional states among olfactory neuron types, which represent closely-related neurons having similar functions. Thus, olfactory neuron types may be distinguished by a small number of genes. We developed ICIM as a strategy to identify the most informative genes for distinguishing subpopulations within a population of closely-related cells in an unbiased way.
Briefly, starting with a population of cells, we first identify the top 100 overdispersed genes. Next, we expand this set of genes by finding genes whose expression profiles are strongly correlated. We also filter this set of genes by (1) removing those having <2 correlated partners to remove noisy genes, and (2) those that are expressed in >80% of cells to remove housekeeping genes. Cells are then clustered. We cut the dendrogram at the deepest branch and partition the population into two subpopulations. The same steps are then performed iteratively on each subpopulation. Iteration continues until a population cannot be split. The termination condition is defined as the minimum terminal branch length being larger than 0.2. The result of this analysis is a set of genes that discriminate subpopulations within a population, which can be used for dimensionality reduction.
Overdispersion analysis and differential expression analysis
Overdispersion analysis and differential expression analysis were previously described [19]. Briefly, genes that are highly variable within a population often carry important information for distinguishing cell types. Variability of gene expression depends strongly on the mean expression level of a gene. This motivates the use of a metric called dispersion, which measures the variability of a gene’s expression level in comparison with other genes that are expressed at a similar level. Overdispersed genes are those that display higher variability than expected based on their mean expression level. To identify overdispersed genes, we binned genes into 20 bins based on their mean expression across all cells. We then calculated a log-transformed Fano factor D(x) of each gene x
where σ2(x) is the variance and μ(x) is the mean of the expression level of the gene across cells. Finally, we calculated the dispersion d(x) as the Z-score of the Fano factor within its bin
where Mean[D(x)] is the mean log-transformed Fano factor within the bin and Std[D(x)] is the standard deviation of the log-transformed Fano factor within the bin. We then rank genes by their dispersion and select the top genes for downstream analysis.
To find differentially expressed genes, we used the Mann-Whitney U test, a non-parametric test that detects differences in the level of gene expression between two populations. The Mann-Whitney U test is advantageous for this application because it makes very general assumptions: (1) observations from both groups are independent and (2) the gene expression levels are ordinal (i.e., can be ranked). Thus the test applies to distributions of gene expression levels across cells, which rarely follow a normal distribution. Using the Mann-Whitney U test, we compared the distributions of expression levels of every gene separately. P values were adjusted using the Bonferroni correction for multiple testing. Different significance thresholds for determining whether a gene is differentially expressed were used for various analyses in this work.
Cluster map in Figure 1
In Figure 1B, we used all ORNs for the tSNE plot, including AM29-GAL4+ and 85A10-GAL4+, 2 and 5 ORN types, that are used to decode cluster identities later. This is because for tSNE plots, each time when we add new cells, the overall cluster layout (e.g., the relative positions of clusters) is changed, which may cause confusion to the general readership. To avoid such confusion, we decided to plot all cells (including 2 and 5 types) in Figure 1B, so that the cluster map will remain the same as in Figure 2, which we used to decode cluster identities.
Cell type coverage in the antenna
We have analyzed 1016 ORNs, which are from 44 ORN types. We classified cells into clusters in an unbiased manner using HDBSCAN with min_cluster_size=6 and min_samples=6 on coordinates after tSNE projection; under these condtions, each cluster requires a minimum of 6 cells. This resulted in 33 clusters. It is likely that, for certain ORN types, we covered fewer than 6 cells, which were not enough to form a cluster. This is also consistent with our observation that when we project our AM29-GAL4 cells into the pan-neuronal nSyb-GAL4-based reference dataset, the accuracy is very low (data not shown). This can explain why cluster #3 (Figure 2H) contains mostly AM29-GAL4 cells, with very few nSyb-GAL4 cells.
Transcription factor (TF) gene lists
To identify genes that are transcription factors (TFs), we used manually curated lists. We obtained a list of Drosophila TFs from the FlyTF v1 database (http://www.mrclmb.cam.ac.uk/genomes/FlyTF). These lists were manually curated to remove spurious annotations and redundancies according to Flybase annotation, resulting in 1045 TFs (see https://github.com/felixhorns/FlyPN), which were used for analysis in Figure 3 and S4A. For Figure 3, we tested several levels of thresholds that define a positive cell and obtained similar results. For example, similar total numbers (915, 899, and 890) of detected TFs in at least one cell were obtained when using three different thresholds, Log2(CPM+1) ≥ 2, 3, or 4.
QUANTIFICATION AND STATISTICAL ANALYSIS
All RNA-seq data analysis was performed in Python using Numpy, Scipy, Pandas, scikit-learn, and a custom single-cell RNA-seq module. We used the Mann-Whitney U test to find differentialy expressed genes between two cell populations. For phenotype quantification in Figure 4 and 5, student’s t-test was used.
DATA AND CODE AVAILABILITY
Sequencing reads and preprocessed data are available from NCBI Gene Expression Omnibus (GSE143038). Python codes for figures are available from Github (https://github.com/Hongjie-Li/FlyORN).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-DNcad | Developmental Studies Hybridoma Bank | DN-Ex #8 |
| Chicken anti-GFP | Aves Labs | GFP-1020 |
| Rabbit anti-DsRed | Clontech | 632496 |
| Rat anti-ratCD2 | AbD Serotec | OX-34 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Critical Commercial Assays | ||
| Deposited Data | ||
| Sequencing reads | This paper | GSE143038 |
| Preprocessed sequence data | This paper | GSE143038 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: nSyb-GAL4 | Bloomington Drosophila Stock Center | BDSC: 51635 |
| D. melanogaster: act5C-GAL4 | Bloomington Drosophila Stock Center | BDSC: 3954 |
| D. melanogaster: NompB-T2A-GAL4 | Bloomington Drosophila Stock Center | BDSC: 76632 |
| D. melanogaster: 5-HT1B-T2A-GAL4 | Bloomington Drosophila Stock Center | BDSC: 76668 |
| D. melanogaster: UAS-STOP-mCD8GFP | [49] | BDSC: 41573 |
| D. melanogaster: DIP-epsilon-T2A-GAL4 | [27] | BDSC: 67502 |
| D. melanogaster: unpg-RNAi | Vienna Drosophila Resource Center | VDRC #107638 |
| D. melanogaster: AM29-GAL4 | [26] | N/A |
| D. melanogaster: GMR86C10-GAL4 | [57] | N/A |
| D. melanogaster: GMR85A10-GAL4 | [27] | N/A |
| D. melanogaster: peb-GAL4 | [58] | N/A |
| D. melanogaster: GH146-Flp | [59] | N/A |
| D. melanogaster: ey-Flp | [60] | N/A |
| D. melanogaster: Or85f-GAL4 | [29] | N/A |
| D. melanogaster: Or42b-mCD8:GFP | [29] | N/A |
| D. melanogaster: Or67a-mCD8:GFP | [29] | N/A |
| D. melanogaster: Or98a-mCD8:GFP | [29] | N/A |
| D. melanogaster: Or92a-rCD2 | [61] | N/A |
| D. melanogaster: Or47b-rCD2 | [62] | N/A |
| D. melanogaster: Or88a-mtdT | [50] | N/A |
| D. melanogaster: Or10a-LexA | [30] | N/A |
| D. melanogaster: Gr21a-syt:GFP | [43] | N/A |
| D. melanogaster: Gr63a-syt:GFP | [43] | N/A |
| D. melanogaster: GMR86C10-LexA | This paper | N/A |
| Oligonucleotides | ||
|
Actin5C (qPCR forward primer): 5’-CTCGCCACTTGCGTTTACAGT-3’ |
This paper | N/A |
|
Actin5C (qPCR reverse primer): 5’-TCCATATCGTCCCAGTTGGTC-3’ |
This paper | N/A |
|
unpg (qPCR forward primer 1F): 5’- CTACAACGGCGAGATGGACA −3’ |
This paper | N/A |
|
unpg (qPCR reverse primer 1R): 5’- TTGGAGTTTGAGCTGGAGCC −3’ |
This paper | N/A |
|
unpg (qPCR forward primer 2F): 5’- GGAACTACAACGGCGAGATG −3’ |
This paper | N/A |
|
unpg (qPCR reverse primer 2R): 5’- GATACTTCTTGGCGTGGAACT −3’ |
This paper | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Iterative Clustering for Identifying Markers (ICIM) | [19] | https://github.com/felixhorns/FlyPN |
| Other | ||
Highlights.
Single-cell RNA-seq analysis of developing Drosophila olfactory receptor neurons
20 of the 33 transcriptomic clusters of ORNs are mapped to glomerular types
Each ORN type expresses hundreds of transcription factors (TFs)
Homeodomain TF Unpg regulates both olfactory receptor expression and axon targeting
ACKNOWLEDGEMENTS
We thank H. Bellen, L. Zipursky, I. Grunwald Kadow, J. Simpson, Bloomington and Vienna Stock Centers for reagents; J. Lui and E. Richman for discussions; N. Neff, J. Okamoto, and Chan-Zuckerberg Biohub for assistance with sequencing; Y. Ge for assistance on fly work; and A. Shuster, J. Lui, and D. Pederick for comments on the manuscript. H.L. was a Stanford Neuroscience Institute Interdisciplinary Postdoctoral Scholar, F.H. acknowledges support from the National Science Foundation Graduate Research Fellowship, J.L. thanks Genentech Foundation Predoctoral and Vanessa Kong Kerzner Graduate Fellowships. S.R.Q. is a Chan Zuckerberg Investigator, and L.L. is an HHMI Investigator. This work was supported by NIH grant 1K99AG062746-01 (to H.L.) and R01-DC005982 (to L.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- 1.Sanes JR, and Zipursky SL (2010). Design principles of insect and vertebrate visual systems. Neuron 66, 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolodkin AL, and Tessier-Lavigne M (2011). Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb. Perspect. Biol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalton RP, and Lomvardas S (2015). Chemosensory receptor specificity and regulation. Annu. Rev. Neurosci 38, 331–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Shuster SA, Li J, and Luo L (2018). Linking neuronal lineage and wiring specificity. Neural Dev. 13, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, and Carlson JR (1999). A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22, 327–338. [DOI] [PubMed] [Google Scholar]
- 6.Gao Q, and Chess A (1999). Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60, 31–39. [DOI] [PubMed] [Google Scholar]
- 7.Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, and Axel R (1999). A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96, 725–736. [DOI] [PubMed] [Google Scholar]
- 8.Vosshall LB, and Stocker RF (2007). Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci 30, 505–533. [DOI] [PubMed] [Google Scholar]
- 9.Wilson RI (2013). Early olfactory processing in Drosophila: mechanisms and principles. Annu. Rev. Neurosci 36, 217–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axel R (1995). The molecular logic of smell. Sci. Am 273, 154–159. [DOI] [PubMed] [Google Scholar]
- 11.Feinstein P, Bozza T, Rodriguez I, Vassalli A, and Mombaerts P (2004). Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell 117, 833–846. [DOI] [PubMed] [Google Scholar]
- 12.Wang F, Nemes A, Mendelsohn M, and Axel R (1998). Odorant receptors govern the formation of a precise topographic map. Cell 93, 47–60. [DOI] [PubMed] [Google Scholar]
- 13.Imai T, Suzuki M, and Sakano H (2006). Odorant receptor-derived cAMP signals direct axonal targeting. Science 314, 657–661. [DOI] [PubMed] [Google Scholar]
- 14.Elmore T, and Smith DP (2001). Putative Drosophila odor receptor OR43b localizes to dendrites of olfactory neurons. Insect Biochem. Mol. Biol 31, 791–798. [DOI] [PubMed] [Google Scholar]
- 15.Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, and Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanchate NK, Kondoh K, Lu Z, Kuang D, Ye X, Qiu X, Pachter L, Trapnell C, and Buck LB (2015). Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science 350, 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci 19, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Horns F, Wu B, Xie Q, Li J, Li T, Luginbuhl DJ, Quake SR, and Luo L (2017). Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 171, 1206–1220.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jefferis GSXE, Vyas RM, Berdnik D, Ramaekers A, Stocker RF, Tanaka NK, Ito K, and Luo L (2004). Developmental origin of wiring specificity in the olfactory system of Drosophila. Development 131, 117–130. [DOI] [PubMed] [Google Scholar]
- 23.Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9, 171–181. [DOI] [PubMed] [Google Scholar]
- 24.Campello RJGB, Moulavi D, Zimek A, and Sander J (2013). A framework for semi-supervised and unsupervised optimal extraction of clusters from hierarchies. Data Min Knowl Discov 27, 344–371. [Google Scholar]
- 25.Lee P-T, Zirin J, Kanca O, Lin W-W, Schulze KL, Li-Kroeger D, Tao R, Devereaux C, Hu Y, Chung V, et al. (2018). A gene-specific T2A-GAL4 library for Drosophila. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endo K, Aoki T, Yoda Y, Kimura K, and Hama C (2007). Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nat. Neurosci 10, 153–160. [DOI] [PubMed] [Google Scholar]
- 27.Jenett A, Rubin GM, Ngo T-TB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, and Dickson BJ (2005). Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807. [DOI] [PubMed] [Google Scholar]
- 29.Couto A, Alenius M, and Dickson BJ (2005). Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol 15, 1535–1547. [DOI] [PubMed] [Google Scholar]
- 30.Fishilevich E, and Vosshall LB (2005). Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol 15, 1548–1553. [DOI] [PubMed] [Google Scholar]
- 31.Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GSXE, and Benton R (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci 31, 13357–13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott K, Brady R, Cravchik A, Morozov P, Rzhetsky A, Zuker C, and Axel R (2001). A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104, 661–673. [DOI] [PubMed] [Google Scholar]
- 33.Benton R, Vannice KS, Gomez-Diaz C, and Vosshall LB (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, and Vosshall LB (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. [DOI] [PubMed] [Google Scholar]
- 35.Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, and Weissman JS (2003). Global analysis of protein expression in yeast. Nature 425, 737–741. [DOI] [PubMed] [Google Scholar]
- 36.Pfreundt U, James DP, Tweedie S, Wilson D, Teichmann SA, and Adryan B (2010). FlyTF: improved annotation and enhanced functionality of the Drosophila transcription factor database. Nucleic Acids Res. 38, D443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clyne PJ, Certel SJ, de Bruyne M, Zaslavsky L, Johnson WA, and Carlson JR (1999). The odor specificities of a subset of olfactory receptor neurons are governed by Acj6, a POU-domain transcription factor. Neuron 22, 339–347. [DOI] [PubMed] [Google Scholar]
- 38.Komiyama T, Carlson JR, and Luo L (2004). Olfactory receptor neuron axon targeting: intrinsic transcriptional control and hierarchical interactions. Nat. Neurosci 7, 819–825. [DOI] [PubMed] [Google Scholar]
- 39.Tichy AL, Ray A, and Carlson JR (2008). A new Drosophila POU gene, pdm3, acts in odor receptor expression and axon targeting of olfactory neurons. J. Neurosci 28, 7121–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai L, and Carlson JR (2010). Distinct functions of acj6 splice forms in odor receptor gene choice. J. Neurosci 30, 5028–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jafari S, Alkhori L, Schleiffer A, Brochtrup A, Hummel T, and Alenius M (2012). Combinatorial activation and repression by seven transcription factors specify Drosophila odorant receptor expression. PLoS Biol. 10, e1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee T, and Luo L (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461. [DOI] [PubMed] [Google Scholar]
- 43.Jones WD, Cayirlioglu P, Kadow IG, and Vosshall LB (2007). Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90. [DOI] [PubMed] [Google Scholar]
- 44.Kwon JY, Dahanukar A, Weiss LA, and Carlson JR (2007). The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. USA 104, 3574–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weigel D, Jürgens G, Küttner F, Seifert E, and Jäckle H (1989). The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57, 645–658. [DOI] [PubMed] [Google Scholar]
- 46.Chiang C, Young KE, and Beachy PA (1995). Control of Drosophila tracheal branching by the novel homeodomain gene unplugged, a regulatory target for genes of the bithorax complex. Development 121, 3901–3912. [DOI] [PubMed] [Google Scholar]
- 47.Cui X, and Doe CQ (1995). The role of the cell cycle and cytokinesis in regulating neuroblast sublineage gene expression in the Drosophila CNS. Development 121, 3233–3243. [DOI] [PubMed] [Google Scholar]
- 48.Hong W, and Luo L (2014). Genetic control of wiring specificity in the fly olfactory system. Genetics 196, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong W, Mosca TJ, and Luo L (2012). Teneurins instruct synaptic partner matching in an olfactory map. Nature 484, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward A, Hong W, Favaloro V, and Luo L (2015). Toll receptors instruct axon and dendrite targeting and participate in synaptic partner matching in a Drosophila olfactory circuit. Neuron 85, 1013–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ni L, Bronk P, Chang EC, Lowell AM, Flam JO, Panzano VC, Theobald DL, Griffith LC, and Garrity PA (2013). A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature 500, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enjin A, Zaharieva EE, Frank DD, Mansourian S, Suh GSB, Gallio M, and Stensmyr MC (2016). Humidity sensing in drosophila. Curr. Biol 26, 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Budelli G, Ni L, Berciu C, van Giesen L, Knecht ZA, Chang EC, Kaminski B, Silbering AF, Samuel A, Klein M, et al. (2019). Ionotropic receptors specify the morphogenesis of phasic sensors controlling rapid thermal preference in drosophila. Neuron 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knecht ZA, Silbering AF, Cruz J, Yang L, Croset V, Benton R, and Garrity PA (2017). Ionotropic Receptor-dependent moist and dry cells control hygrosensation in Drosophila. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morey M, Yee SK, Herman T, Nern A, Blanco E, and Zipursky SL (2008). Coordinate control of synaptic-layer specificity and rhodopsins in photoreceptor neurons. Nature 456, 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Courgeon M, and Desplan C (2019). Coordination between stochastic and deterministic specification in the Drosophila visual system. Science 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Q, Wu B, Li J, Xu C, Li H, Luginbuhl DJ, Wang X, Ward A, and Luo L (2019). Transsynaptic Fish-lips signaling prevents misconnections between nonsynaptic partner olfactory neurons. Proc. Natl. Acad. Sci. USA 116, 16068–16073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sweeney LB, Couto A, Chou Y-H, Berdnik D, Dickson BJ, Luo L, and Komiyama T (2007). Temporal target restriction of olfactory receptor neurons by Semaphorin-1a/PlexinA-mediated axon-axon interactions. Neuron 53, 185–200. [DOI] [PubMed] [Google Scholar]
- 59.Potter CJ, Tasic B, Russler EV, Liang L, and Luo L (2010). The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141, 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chotard C, Leung W, and Salecker I (2005). glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron 48, 237–251. [DOI] [PubMed] [Google Scholar]
- 61.Li J, Guajardo R, Xu C, Wu B, Li H, Li T, Luginbuhl DJ, Xie X, and Luo L (2018). Stepwise wiring of the Drosophila olfactory map requires specific Plexin B levels. Elife 7:e39088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu H, and Luo L (2004). Diverse functions of N-cadherin in dendritic and axonal terminal arborization of olfactory projection neurons. Neuron 42, 63–75. [DOI] [PubMed] [Google Scholar]
- 63.Wu JS, and Luo L (2006). A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protoc 1, 2110–2115. [DOI] [PubMed] [Google Scholar]
- 64.Komiyama T, Johnson WA, Luo L, and Jefferis GSXE (2003). From lineage to wiring specificity. POU domain transcription factors control precise connections of Drosophila olfactory projection neurons. Cell 112, 157–167. [DOI] [PubMed] [Google Scholar]
- 65.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anders S, Pyl PT, and Huber W (2015). HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maaten L, and Hinton G (2008). Visualizing data using t-SNE. Journal of machine learning research.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads and preprocessed data are available from NCBI Gene Expression Omnibus (GSE143038). Python codes for figures are available from Github (https://github.com/Hongjie-Li/FlyORN).


