Abstract
Background
Teosinte branched1/Cycloidea/proliferating cell factors (TCPs) are plant-specific transcription factors widely involved in leaf development, flowering, shoot branching, the circadian rhythm, hormone signaling, and stress responses. However, the TCP function in Brassica juncea var. tumida, the tumorous stem mustard, has not yet been reported. This study identified and characterized the entire TCP family members in B. juncea var. tumida.
Methods
We identified 62 BjTCP genes from the B. juncea var. tumida genome and analyzed their phylogenetic relationship, gene structure, protein motifs, chromosome location, and expression profile in different tissues.
Results
Of the 62 BjTCP genes we identified in B. juncea var. tumida, containing 34 class I and 28 class II subfamily members, 61 were distributed on 18 chromosomes. Gene structure and conserved motif analysis showed that the same clade genes displayed a similar exon/intron gene structure and conserved motifs. Cis-acting element results showed that the same clade genes also had a similar cis-acting element; however, subtle differences implied a different regulatory pathway. The BjTCP18s members were low-expressed in Dayejie strains and the unswelling stage of Yonganxiaoye strains. Treatment with gibberellin (GA) and salicylic acid (SA) showed that GA and SA affect the expression levels of multiple TCP genes.
Conclusion
We performed the first genome-wide analysis of the TCP gene family of B. juncea var. tumida. Our results have provided valuable information for understanding the classification and functions of TCP genes in B. juncea var. tumida.
Keywords: Tumorous stem mustard, TCP transcription factors, Gene expression, Swelling, Brassica juncea var. tumida
Introduction
The teosinte branched1/Cycloidea/proliferating cell factor (TCP) family is a group of plant-specific transcription factors (TFs) reportedly involved in embryonic growth (Takeda et al., 2003), leaf development (Bresso et al., 2018; Danisman et al., 2012; Du et al., 2017; Kieffer et al., 2011; Liu et al., 2018b; Ma et al., 2016; Uberti-Manassero et al., 2012; Wang et al., 2018), branching (Aguilar-Martinez, Poza-Carrion & Cubas, 2007; Brewer, 2015; Dixon et al., 2018; Gonzalez-Grandio et al., 2013; Gonzalez-Grandio et al., 2017; Martin-Trillo et al., 2011; Maurya et al., 2020; Niwa et al., 2013; Seale, Bennett & Leyser, 2017; Shen et al., 2019; Wang et al., 2019a), flowering (Aguilar-Martinez, Poza-Carrion & Cubas, 2007; Damerval et al., 2007; Finlayson, 2007; Madrigal, Alzate & Pabon-Mora, 2017; Navarro, Cruz-Oro & Prat, 2015; Yang et al., 2015), the circadian rhythm (Beveridge et al., 2003; Giraud et al., 2010), hormone signaling (Wang et al., 2019a), and stress responses (Danisman, 2016; Guan et al., 2017; Martin-Trillo & Cubas, 2010). The TCP domain is highly conserved in the TCP family and is formed by an N-terminal region enriched in basic amino acids, followed by two amphipathic α-helices connected by a disordered loop (Cubas, Vincent & Coen, 1999; Doebley, Stec & Hubbard, 1997).
On the basis of this conserved domain, TCP proteins are divided into two subfamilies, class I and class II (Li, 2015; Martin-Trillo & Cubas, 2010). The difference between the two classes is the deletion of four amino acids in the TCP domain in class I. In Arabidopsis, TCP2–5, TCP10, TCP13, TCP17, and TCP24 are related to lateral organ organogenesis and control leaf development (Efroni et al., 2008; Hay, Barkoulas & Tsiantis, 2004; Koyama et al., 2007; Qin et al., 2005). Belonging to the same subfamily, branched1 (TCP18) and branched2 (TCP12) play an important role in controlling branch outgrowth (Aguilar-Martinez, Poza-Carrion & Cubas, 2007; Gonzalez-Grandio et al., 2013; Muhr et al., 2016; Wang et al., 2019a). By interacting with florigen proteins FLOWRING LOCUS T (FT), TCP18 inhibits floral transition of axillary meristems in Arabidopsis (Niwa et al., 2013). The BRC1 homolog gene in the hybrid aspen also mediates photoperiodic control of seasonal growth (Maurya et al., 2020), and TIG1, encoding a TCP TF, contributes to plant architecture domestication in rice (Zhang et al., 2019). TCP21 participates in the circadian rhythm by binding to TIMING OF CAB EXPRESSION 1 (TOC1) and the CIRCADIAN AND CLOCK ASSOCIATED1 (CCA1) promoter (Pruneda-Paz et al., 2009). In addition, TCP proteins, such as brassinosteroids (BRs), jasmonic acid, indole-3-acetic acid (IAA), and strigolactone (SL), involved in plant growth and development, are usually regulated by phytohormone synthesis and metabolism (Braun et al., 2012; Danisman et al., 2012; Li, 2015; Liu et al., 2017; Muhr et al., 2016; Qin et al., 2005; Schommer et al., 2008). Studies have also reported on TCP genes regulated by sugars (Wang et al., 2019b) and light (Kebrom, Burson & Finlayson, 2006).
Recently, TCP proteins have been shown to be related to defense responses. For example, TCP13, TCP14, and TCP19 are directly targeted by effectors from Pseudomonas syringae and Hyaloperonospora arabidopsidis (Mukhtar et al., 2011). Kim et al. (2014) reported that TCP8, TCP13, TCP15, TCP20, TCP22, and TCP23 can interact with the Arabidopsis immune adaptor SUPPRESSOR OF rps4-RLD1 (SRFR1), which is a negative regulator of effector-triggered immunity (Kim et al., 2014). TCP genes are also regulated by microRNA 319 (miR319) and are involved in leaf development in Arabidopsis (Bresso et al., 2018; Palatnik et al., 2003; Schommer et al., 2008; Wang et al., 2018).
The TCP family has been identified in many different plant species, such as 24 TCP genes in Arabidopsis (Martin-Trillo & Cubas, 2010), 28 in Oryza sativa, 30 in Lycopersicon esculentum (Parapunova et al., 2014), 33 in Populus euphratica (Ma et al., 2016), 27 in Citrullus lanatus s (Shi et al., 2016), 66 in Triticum aestivum (Zhao et al., 2018), 75 in Gossypium barbadense (Zheng et al., 2018), 31 in Solanum tuberosum (Wang et al., 2019c), 39 in Brassica rapa L. ssp. Pekinensis (Liu et al., 2018b), and 39 in B. rapa ssp. rapa (Du et al., 2017). Liu et al. (2019) performed a genome-wide systematic identification of the TCP proteins in the major plant lineages (47 species).
The tumorous stem mustard (B. juncea var. tumida) is an important crop of great economic value in China, so improving its yield is key issue for the Chinese pickle industry. The growth of B. juncea var. tumida involves four stages: germination, seedling, stem swelling, and flowering. Stem swelling is essential for tumorous stem formation, and the stem swelling–flowering balance is directly related to the quality and yield of tumorous mustards. B. juncea var. tumida is an annual plant, and for stem swelling, it is essential that the seeds be sown between mid-September and mid-October in Chongqing and other valleys of the Yangtze River, China. Therefore, the production period of edible stems is limited.
TCP proteins are extensively involved in branching, flowering, development, and plant morphology (Aguilar-Martinez, Poza-Carrion & Cubas, 2007; Bai et al., 2012; Braun et al., 2012; Danisman et al., 2012; Dixon et al., 2018; Feng et al., 2018; Finlayson et al., 2010; Gonzalez-Grandio et al., 2013; Gonzalez-Grandio et al., 2017; Ho & Weigel, 2014; Li, 2015; Martin-Trillo et al., 2011; Nicolas et al., 2015; Niwa et al., 2013; Prusinkiewicz et al., 2009; Rameau et al., 2014; Seale, Bennett & Leyser, 2017; Teichmann & Muhr, 2015). However, there are few reports on the TCP family in B. juncea var. tumida, and whether TCP proteins control stem swelling and flowering in B. juncea var. tumida is still unknown.
Since the entire genome of B. juncea var. tumida was sequenced (Yang et al., 2016), this study performed a genome-wide analysis of TCP genes for the first time. Of the 62 BjTCP genes identified, we analyzed their phylogenetic relationship, gene structure, protein motifs, chromosome location, and expression profile in different tissues. The results can provide valuable information for the classification of BjTCP genes and lay the foundation for exploring the molecular mechanism underlying stem swelling and flowering orchestrated by TCP genes in B. juncea var. tumida.
Material and Methods
Plant materials, growth conditions, and treatment
B. juncea var. tumida cultivar YA (with swollen tumorous stems) was used to analyze gene expression patterns. Seeds were sowed into 2:1 vermiculite:turfy soil and cultured at a constant temperature of 22 °C in a 16/8 h light/dark cycle in a culture room. Next, 3-week-old seedlings were used for exogenous hormone treatment; the seedlings were sprayed with 100 µM salicylic acid (SA) (Feng et al., 2018) and 100 µM gibberellin (GA) (Rosa et al., 2017). The second true leaf on each seedling was sampled at 0 (control), 2, 4, 6, 8, and 24 h after spraying. All treatments were repeated thrice, and each treatment was given to at least 20 seedlings. All materials were frozen immediately in liquid nitrogen and stored at −70 °C until RNA isolation.
Identification of TCP proteins in B. juncea var. tumida
The genome sequences of B. juncea var. tumida (version 1.5), B. nigra (version 1.1), and B. rapa (version 3.0) were downloaded from the Brassica database (BRAD; http://brassicadb.org/brad/datasets/pub/Genomes/) (Cheng et al., 2011; Yang et al., 2016). In addition, the TCP domain in the Pfam database (accession no. PF03634) was downloaded (Finn et al., 2010), and the domain was searched in the BRAD using HMMER 3.0 with an E-value of <1e−6 (Finn, Clements & Eddy, 2011). To confirm the results obtained by the HMMER algorithm, the TCP domain was further verified with Pfam and Smart databases (Finn et al., 2010; Letunic & Bork, 2018; Letunic, Doerks & Bork, 2015). The TCP protein sequences of A. thaliana were downloaded from the Arabidopsis information resource website (https://www.arabidopsis.org).
Sequence and phylogenetic analysis
We used the ClustalW program to perform multiple alignments of TCP protein sequences from B. juncea var. tumida and A. thaliana (Thompson et al., 1997). A phylogenetic tree was constructed using MEGA 7.0 software and the maximum likelihood method based on the Poisson correction model and a bootstrap test replicated 1,000 times (Tamura et al., 2013). A gene structure diagram was drawn using the online software of the GSDS2.0 server (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). The physical location data of BjTCP genes were retrieved from the B. juncea var. tumida genome. We subsequently mapped these TCP genes using MapInspect software. Conserved protein motifs were identified by using default parameters for the Multiple Em for Motif Elicitation (MEME; http://meme-suite.org/) program, and maximum 12 motifs were set. Subcellular localization of BjTCPs was predicted using ProtComp9.0 (http://www.softberry.com), and the identified protein motifs were further annotated using Weblogo (http://weblogo.berkeley.edu/). Finally, 2,000 bp of the 5′ sequence were used as the promoter region of each TCP gene to analyze the cis-acting elements using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Chromosomal location and prediction of miR319 target genes
The physical location data of BjTCP genes were retrieved from the B. juncea var. tumida genomes. The mapping of these TCP genes was subsequently performed using MapInspect software. To predict miR target genes, we analyzed the full lengths of candidate TCP coding sequences using the psRNATarget website (Dai & Zhao, 2011).
Expression profile of TCP genes
RNA-sequencing (RNA-seq) data from our previous research were downloaded from the National Center for Biotechnology Information Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/sra/) with the following accession numbers: SRX108496 (Dayejie [DY] stems, a mutant variety without inflated stems, were collected 22 weeks after seeding), SRX108498 (YA1; Yonganxiaoye [YA] stems were collected 18 weeks after seeding), SRX108499 (YA2; YA stems were collected 20 weeks after seeding), SRX108500 (YA3; YA stems were collected 22 weeks after seeding), SRX108501 (YA4; YA stems were collected 25 weeks after seeding), and SRX108502 (YAr; YA mix roots were collected 20 and 22 weeks after seeding) (Sun et al., 2012). Clean reads filtered from raw reads were mapped onto B. juncea genome version 1.5 (http://brassicadb.org/brad/datasets/pub/Genomes/Brassica_juncea/V1.5/) (Yang et al., 2016) using Tophat2 with default parameters (Trapnell, Pachter & Salzberg, 2009; Trapnell et al., 2012). Gene expression levels of individual genes were quantified using reads per kilobase of transcript per million (RPKM) values using Cufflinks 2.2.1 with default parameters (Trapnell et al., 2012).
RNA extraction and real-time quantitative PCR analysis
Total RNA was extracted from different plant materials using RNA plant plus reagent (Tiangen Biotech Co., Ltd., Beijing, China) and treated with DNase I (Takara, Qingdao, China) to remove genomic DNA. Reverse transcription was performed using the Hiscript II 1st strand complementary DNA (cDNA) synthesis kit (Vazyme, Nanjing, China). Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed with 20 µL volume using TB Green™ Premix Ex Taq™ II (Tli RNaseH Plus) (Takara). BjActin was used as the internal reference gene for qRT-PCR; Table S1 lists gene-specific primers.
Three replicate samples of each period were subjected to three biological replicates using the BioRad IQ5 Real-Time PCR instrument (BioRad Laboratories, Hercules, CA, USA). Amplification parameters were as follows: activation at 50 °C for 2 min, predenaturation at 95 °C for 2 min, denaturation at 95 °C for 15 s, and annealing at 60 °C for 1 min for 40 cycles. Finally, the relative gene expression level was calculated using the 2−ΔΔCt method (Livak & Schmittgen, 2001).
Results
Identification of TCP family members in B. juncea var. tumida
To identify TCP proteins in B. juncea var. tumida, we screened out 63 genes and confirmed the domain using Pfam and Smart databases. Finally, we identified 62 BjTCP genes in B. juncea var. tumida. On the basis of similarity with A. thaliana homology genes, the 62 BjTCP genes were named with BjTCP1a-BjTCP24d (Table 1). The coding amino acids were from 171 to 639, with a molecular weight of 18.6–71.87 kDa and an isoelectric point (pI) of 5.5–10.18. Of the 62 genes, 61 were located on 18 chromosomes, except BjTCP17b anchored in contig6125. There was one TCP gene each on chromosomes A04, A08, A10, B01, and B06; two TCP genes each on chromosomes A01 and A05; and three to seven genes on other chromosomes (Figs. 1A–1R). We also found that most of the BjTCP proteins were localized in the nucleus, except BjTCP13a-c, whose location information was not found (Table 1), indicating that BjTCPs are TFs. These 62 TCP proteins may have multiple functions, and they mainly enriched in multiple GO terms, such as biological regulation, response to stimulus, rhythmic process and so on (Table S2).
Table 1. The TCP protein family members in B. juncea var. tumida.
| ID | pfam domin (star-end) | Name | chr | star | end | sence+/ antisence- | Subcellular locallzation | Homolog | PI | MW (kD) | protein (aa) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BjuA007230 | 84 | 242 | BjTCP1a | A02 | 10613454 | 10614491 | − | Nuclear | AtTCP1 | 5.5 | 39.28 | 346 |
| BjuA027377 | 84 | 232 | BjTCP1b | A07 | 29105069 | 29106097 | − | Nuclear | AtTCP1 | 6.68 | 38.96 | 343 |
| BjuB030534 | 84 | 233 | BjTCP1c | B03 | 29932727 | 29933758 | − | Nuclear | AtTCP1 | 5.98 | 39.1 | 344 |
| BjuB043984 | 85 | 235 | BjTCP1d | B03 | 27732982 | 27734025 | − | Nuclear | AtTCP1 | 5.96 | 39.57 | 348 |
| BjuB045720 | 81 | 300 | BjTCP1e | B05 | 38301640 | 38302668 | − | Nuclear | AtTCP1 | 5.88 | 38.91 | 343 |
| BjuB013551 | 72 | 251 | BjTCP2a | B05 | 13742319 | 13743080 | + | Nuclear | AtTCP2 | 6.64 | 27.45 | 254 |
| BjuB044682 | 161 | 261 | BjTCP2b | B02 | 58451263 | 58452048 | + | Nuclear | AtTCP2 | 6.71 | 28.53 | 262 |
| BjuA013153 | 162 | 259 | BjTCP2c | A01 | 16646189 | 16646968 | + | Nuclear | AtTCP2 | 6.69 | 28.34 | 260 |
| BjuA003953 | 81 | 236 | BjTCP2d | A01 | 5519388 | 5520104 | − | Nuclear | AtTCP2 | 6.48 | 25.82 | 259 |
| BjuB045012 | 1 | 158 | BjTCP3 | B07 | 27416073 | 27416864 | + | Nuclear | AtTCP3 | 6.55 | 28.64 | 264 |
| BjuB027204 | 34 | 231 | BjTCP5a | B04 | 184613 | 186371 | − | Nuclear | AtTCP5 | 9.33 | 30.31 | 275 |
| BjuB037760 | 63 | 364 | BjTCP5b | B02 | 59342015 | 59343112 | + | Nuclear | AtTCP5 | 6.21 | 40.73 | 365 |
| BjuB039698 | 62 | 262 | BjTCP5c | B07 | 12846872 | 12847951 | − | Nuclear | AtTCP5 | 6.95 | 34.36 | 304 |
| BjuA016046 | 36 | 211 | BjTCP6 | A04 | 13029721 | 13033965 | − | Nuclear | AtTCP6 | 7.98 | 71.87 | 639 |
| BjuA023481 | 42 | 223 | BjTCP7a | A06 | 20165661 | 20166410 | − | Nuclear | AtTCP7 | 9.69 | 27 | 250 |
| BjuA032507 | 14 | 189 | BjTCP7b | A09 | 5290320 | 5290967 | + | Nuclear | AtTCP7 | 9.38 | 23.12 | 216 |
| BjuA045586 | 1 | 151 | BjTCP7c | A02 | 34582679 | 34583206 | − | Nuclear | AtTCP7 | 7.92 | 18.6 | 176 |
| BjuB039713 | 14 | 190 | BjTCP7d | B07 | 12703979 | 12704632 | − | Nuclear | AtTCP7 | 9.82 | 23.3 | 218 |
| BjuB044801 | 42 | 235 | BjTCP7e | B02 | 60858548 | 60859336 | + | Nuclear | AtTCP7 | 9.51 | 28.37 | 263 |
| BjuA033567 | 53 | 218 | BjTCP8a | A09 | 15197090 | 15198274 | + | Nuclear | AtTCP8 | 6.09 | 41.43 | 395 |
| BjuB028163 | 54 | 232 | BjTCP8b | B04 | 9742480 | 9743682 | − | Nuclear | AtTCP8 | 6 | 42.26 | 401 |
| BjuA010736 | 71 | 197 | BjTCP9a | A03 | 13517323 | 13518273 | − | Nuclear | AtTCP9 | 9.86 | 33.84 | 317 |
| BjuA018039 | 62 | 178 | BjTCP9b | A05 | 3386569 | 3387543 | − | Nuclear | AtTCP9 | 9.41 | 33.43 | 325 |
| BjuB016827 | 72 | 201 | BjTCP9c | B08 | 26586920 | 26587915 | − | Nuclear | AtTCP9 | 9.67 | 35.15 | 332 |
| BjuB020003 | 67 | 184 | BjTCP9d | B06 | 3171113 | 3172111 | − | Nuclear | AtTCP9 | 9.58 | 35.15 | 333 |
| BjuA041558 | 100 | 193 | BjTCP12a | A02 | 11715044 | 11716146 | − | Nuclear | AtTCP12 | 8.78 | 37.88 | 322 |
| BjuB010789 | 103 | 231 | BjTCP12b | B05 | 55023718 | 55024870 | − | Nuclear | AtTCP12 | 8.45 | 39.61 | 352 |
| BjuA011472 | 59 | 220 | BjTCP13a | A03 | 18170796 | 18171755 | + | − | AtTCP13 | 6.73 | 35.69 | 320 |
| BjuA021096 | 86 | 243 | BjTCP13b | A05 | 31265695 | 31266872 | − | – | AtTCP13 | 6.63 | 35.72 | 321 |
| BjuB026804 | 58 | 214 | BjTCP13c | B07 | 14926766 | 14927692 | − | – | AtTCP13 | 7.94 | 34.47 | 309 |
| BjuA022523 | 1 | 178 | BjTCP14a | A06 | 12060176 | 12061564 | − | Nuclear | AtTCP14 | 5.98 | 36.45 | 344 |
| BjuB019669 | 103 | 292 | BjTCP14b | B08 | 54134488 | 54135858 | − | Nuclear | AtTCP14 | 6.48 | 49.33 | 457 |
| BjuA007311 | 58 | 201 | BjTCP15a | A02 | 12233735 | 12234697 | + | Nuclear | AtTCP15 | 6.91 | 33.84 | 321 |
| BjuA016487 | 55 | 195 | BjTCP15b | A07 | 30212223 | 30213185 | + | Nuclear | AtTCP15 | 6.91 | 34.06 | 321 |
| BjuA027170 | 56 | 197 | BjTCP15c | A07 | 27540133 | 27541098 | − | Nuclear | AtTCP15 | 7.43 | 33.96 | 322 |
| BjuB000709 | 50 | 186 | BjTCP15d | B05 | 51941921 | 51942820 | − | Nuclear | AtTCP15 | 8.05 | 32.02 | 300 |
| BjuB003932 | 57 | 194 | BjTCP15e | B03 | 29008395 | 29009351 | + | Nuclear | AtTCP15 | 7.43 | 33.74 | 319 |
| BjuB030482 | 55 | 196 | BjTCP15f | B03 | 32626814 | 32627776 | + | Nuclear | AtTCP15 | 7.15 | 33.49 | 321 |
| BjuA009092 | 32 | 164 | BjTCP17a | A03 | 1624898 | 1625620 | + | Nuclear | AtTCP17 | 8.53 | 19.12 | 171 |
| BjuO008355 | 37 | 254 | BjTCP17b | Contig6125 | 66534 | 67304 | + | Nuclear | AtTCP17 | 6.7 | 28.54 | 257 |
| BjuA012606 | 154 | 319 | BjTCP18a | A03 | 22282211 | 22283992 | + | Nuclear | AtTCP18 | 8.56 | 48.45 | 425 |
| BjuB007175 | 150 | 311 | BjTCP18b | B07 | 5163145 | 5165165 | + | Nuclear | AtTCP18 | 7.32 | 40.12 | 350 |
| BjuB007177 | 150 | 311 | BjTCP18c | B07 | 5138717 | 5140787 | + | Nuclear | AtTCP18 | 6.66 | 50.08 | 437 |
| BjuB025473 | 159 | 292 | BjTCP18d | B01 | 38022785 | 38024462 | − | Nuclear | AtTCP18 | 8.4 | 46.6 | 404 |
| BjuA007026 | 52 | 174 | BjTCP19 | A02 | 9082363 | 9083205 | + | Nuclear | AtTCP19 | 5.5 | 30.18 | 281 |
| BjuA024339 | 67 | 304 | BjTCP20a | A06 | 25596111 | 25597025 | + | Nuclear | AtTCP20 | 7.97 | 32.22 | 305 |
| BjuA031722 | 53 | 239 | BjTCP20b | A09 | 3333673 | 3334532 | − | Nuclear | AtTCP20 | 5.17 | 25.25 | 241 |
| BjuB037176 | 65 | 309 | BjTCP20c | B02 | 56079378 | 56080310 | − | Nuclear | AtTCP20 | 7.3 | 32.61 | 311 |
| BjuA009108 | 31 | 206 | BjTCP21a | A03 | 1689417 | 1690118 | + | Nuclear | AtTCP21 | 10.18 | 24.26 | 234 |
| BjuA041017 | 32 | 207 | BjTCP21b | A02 | 1466529 | 1467236 | + | Nuclear | AtTCP21 | 9.29 | 24.54 | 236 |
| BjuA047338 | 31 | 208 | BjTCP21c | A10 | 17491637 | 17492344 | + | Nuclear | AtTCP21 | 7.99 | 24.73 | 236 |
| BjuB012430 | 31 | 209 | BjTCP21d | B05 | 17001527 | 17002243 | − | Nuclear | AtTCP21 | 9.57 | 25.05 | 239 |
| BjuB040955 | 33 | 209 | BjTCP21e | B08 | 2452006 | 2452713 | + | Nuclear | AtTCP21 | 10.18 | 24.46 | 236 |
| BjuB048495 | 28 | 209 | BjTCP21f | B02 | 50550592 | 50551305 | − | Nuclear | AtTCP21 | 7.99 | 24.94 | 238 |
| BjuA007449 | 57 | 190 | BjTCP22a | A02 | 13365484 | 13366605 | + | Nuclear | AtTCP22,AtTCP23 | 8.63 | 39.06 | 374 |
| BjuA043373 | 40 | 202 | BjTCP22b | A07 | 31378278 | 31379348 | + | Nuclear | AtTCP22,AtTCP23 | 6.87 | 37.34 | 357 |
| BjuB010697 | 56 | 186 | BjTCP22c | B05 | 59790473 | 59791573 | − | Nuclear | AtTCP22,AtTCP23 | 8.63 | 38.41 | 367 |
| BjuB044035 | 45 | 168 | BjTCP22d | B03 | 38473563 | 38474609 | + | Nuclear | AtTCP22,AtTCP23 | 6.31 | 36.59 | 349 |
| BjuA034777 | 46 | 138 | BjTCP24a | A09 | 31311472 | 31312425 | + | Nuclear | AtTCP24 | 7.8 | 35.29 | 318 |
| BjuB029526 | 47 | 139 | BjTCP24b | B04 | 35033858 | 35034823 | − | Nuclear | AtTCP24 | 6.90 | 35.7 | 322 |
| BjuB032913 | 66 | 306 | BjTCP24c | B03 | 5929399 | 5930376 | + | Nuclear | AtTCP24 | 7.16 | 36.57 | 326 |
| BjuA029872 | 55 | 284 | BjTCP24d | A08 | 20433785 | 20434732 | + | Nuclear | AtTCP24 | 6.81 | 35.46 | 316 |
Figure 1. The gene locations of BjTCP gene family.
The chromosome name is at the top of each bar. The scale of the chromosome is in millions of bases (Mb).
The phylogenetic tree of BjTCP genes and AtTCPs
Multiple-sequence alignment of TCP proteins showed that the conserved region was mainly focused on the TCP domain (Fig. S1).
To assess the phylogenetic relationships of the TCP family, we used the predicted TCP protein sequences from B. juncea var. tumida and A. thaliana to construct a phylogenetic tree. Results indicated that all TCP proteins are divided into two groups, class I and class II (Fig. 2A). In class II, the TCP proteins were further subdivided into CYC, TB1, and CIN groups. The CYC group was mainly clustered by AtTCP1 and AtTCP12, containing four AtTCP1 homologous proteins BjTCP1a-d, BjTCP2a-d, and two AtTCP12 homologous proteins BjTCP12a-b. The TB1 group comprised AtTCP18 and four homologous TCP proteins BjTCP18a-d. In the CIN group, we found no proteins to be homologous with AtTCP4 and AtTCP10, while the other TCP proteins had at least one homologous protein, such as AtTCP24 (four homologous proteins BjTCP24a–d), AtTCP13 (three homologous proteins BjTCP13a–c), AtTCP17 (two homology proteins BjTCP17a and BjTCP17b), and AtTCP5 (three homologous proteins BjTCP5a–c). In class I, we found no homologous proteins in B. juncea var. tumida, except AtTCP11, AtTCP16, and AtTCP23, but the other TCP proteins had multiple homologous proteins, such as AtTCP15, and AtTCP21 even had six homologous proteins.
Figure 2. Evolutionary relationships of taxa.
(A) The evolutionary history was inferred by using the Maximum Likelihood method based on the Poisson correction model. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The analysis involved 86 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 66 positions in the final dataset. Evolutionary analyses were conducted in MEGA7. (B) A conserved motif in the Class I subfamily of the BjTCP gene family. (C) A conserved motif in the Class II subfamily of the BjTCP gene family. The consensus sequences were displayed using Weblogo (http://weblogo.berkeley.edu).
Interestingly, a series of genes, such as BjTCP15b, BjTCP15c, BjTCP1b, and BjTCP22b, were located on the same chromosome A07 (Fig. 1G). Their homologous genes (BjTCP15e, BjTCP15f, BjTCP1c, and BjTCP22d) showed the same order on chromosome B03 (Figs. 1M and 2). The eight genes were searched in B. rapa (AA) and B. nigra (BB) using the BLASTP program, and four highly similar genes were screened out in B. rapa (AA) and B. nigra (BB). The evolutionary relationships suggested that the four genes located on chromosome A07 were clustered with the homologous genes of a subgenome ancestor B. rapa (AA) in one branch (Fig. S2). The four genes on chromosome B03 also corresponded to the B subgenome ancestor B. nigra (BB) (Fig. S2). These results indicated that the fragments between the four genes of chromosomes A07 and B03 might be formed from B. rapa (AA) and B. nigra (BB), respectively.
BjTCP proteins had a typical bHLH motif in all identified TCP proteins (Figs. 2B, 2C). In A. thaliana, the main difference between classes I and II was the identity of the residue at positions 10–15 of the TCP domain. Most class I BjTCP proteins lost four amino acids at positions 9–13 and had Gly at position 15, while class II BjTCP proteins had Asp at position 15 in the TCP domain (Figs. 2B, 2C).
Gene structures and conserved motif analysis of BjTCP genes
To further analyze the characteristic of BiTCP genes, we explored the exon/intron gene structure. Results indicated that most BjTCP genes only have one exon, except BjTCP18s, BjTCP12s, BjTCP20b, and BjTCP13b, which contain two or more exons. We also found that the genetic structure and evolutionary relationships of all TCP family members of B. juncea var. tumida are closely related. Genes within the same subfamily often showed similar gene structures. BjTCP12a and BjTCP12b comprised two exons, while BjTCP18a-d comprised more than three exons (Figs. 3A, 3B). In Chinese cabbage, the homologous BrTCP1a and BrTCP1b genes have two exons, and this exon number was highly similar to B. juncea var. tumida’s homology genes; for example, BrTCP6–13 and BrTCP15 had only one exon. Although BjTCP18a and BjTCP18b have two exons, BjTCP18c and BjTCP18d have four exons, which is similar with their homologous B. rapa BrTCP18a and BrTCP18b genes (Liu et al., 2018b). The conserved motifs in these BjTCP genes also showed similar characters within the same subgroup, such as three similar motifs in all BjTCP15 homologous proteins and five similar motifs in all BjTCP21 homologous proteins (Fig. 3C).
Figure 3. Genomic structure and motif composition of BjTCPs.
(A) The phylogenetic tree of BjTCP proteins. (B) Genomic structure of BjTCPs family members in tumorous stem mustard. Exons and introns are represented with blank boxes and blank lines. (C) The conserved motifs in tumorous stem mustard TCP proteins were identified using MEME. Each motif is represented with a specific color and the characters sequence were showed below.
BjTCP genes with miR319 target sites
In Arabidopsis, AtTCP2-4, AtTCP10, and AtTCP24 are post-transcriptionally regulated by miR319 (Bresso et al., 2018; Palatnik et al., 2003). In B. juncea var. tumida, the evolutionarily closest homologs of these genes are BjTCP2a–d, and BjTCP24a–d, which contain sequences well matched with miR319 and might be the targets of miRs (Fig. 4). BjTCP3 did not contain the putative miR319 recognition site, but mismatches of other genes mainly existed at 3′ of miR319 and 5′ of the targeted BjTCP mRNA, and core target sequences were conserved.
Figure 4. Alignment of putative target areas for miR319.
Mismatches and G-U wobbles were represented by yellow and green, respectively.
Promoter cis-acting element analysis of BjTCP genes
The cis-acting elements in the promoter of a gene usually regulate gene expression and function. In this study, we found multiple cis-acting elements in TCP gene promoters, such as plant hormone response elements, light response elements, stress response elements, meristem expression, circadian control, and low-temperature and wound response elements (Fig. 5 and Table S3).
Figure 5. Cis-acting elements on promoters of BjTCP genes.
The colour bar shows the number of cis-acting elements.
For hormone-related cis-acting elements, we identified abscisic acid (ABA) response elements (ABREs) and found at least two or more ABRE cis-acting elements in B. juncea var. tumida TCP gene promoters, expect BjTCP5s, BjTCP18s, BjTCP19, and BjTCP24. Auxin response elements included AuxRP and TGA elements, and AuxRP had a relatively small number of components, mainly in the BjTCP5, BjTCP19, and BjTCP20 promoters, while the TGA element was relatively more extensively distributed. The MeJA response elements CGTCA and TGACG were found on most promoters, except for BjTCP12, BjTCP19, and BjTCP20. We also found a number of other hormone-related cis-elements, such as ethylene (ET) response element (ERE), GA response elements (GAREs) P-box and TATC-box, and the SA response element TCA element in some BjTCP promoters.
In addition, we found a large number of cis-acting elements related to light response in these promoters, including the 3-AF1 binding site, ACE, AE-box, TCT-motif, ATC-motif, Box 4, GATA-motif, G-Box, GT1-motif, and I-box. We also found other elements, including WUN-motif (related to wounds), meristem element (related to the meristem), circadian element (related to circadian control), LTR element (related to low-temperature induction), and defense and stress responsiveness elements (including MBS, Myb, Myc, STRE, TC-rich, W box, and ARE elements). In particular, we identified MYB and MYC-motif elements in almost all TCP promoters.
Tissue-specific expression profiles of BjTCP genes
In A. thaliana, TCP proteins were found to be mainly involved in development and defense. We analyzed the expression patterns of all TCP genes in different development periods and tissues on the basis of previous RNA-seq data (Sun et al., 2012). BjTCP1a and BjTCP5c expression could not be detected in all samples. However, 26 TCP genes were highly expressed in at least two tissues (log2 (fragments per kilobase of transcript per million [FPKM]) ≥ 3; subgroup half-bottom in Fig. 6). BjTCP13b and BjTCP5a were weakly expressed in no-swelling strain (DA) samples. We did not detect BjTCP1b, BjTCP1c, BjTCP1d, BjTCP5c, BjTCP7c, BjTCP13a, BjTCP13c, BjTCP18d, and BjTCP19 expression in DY stem tissue. BjTCP12a, BjTCP12b and BjTCP18c were weakly expressed in DY, YA1, and/or YA2 strains (Fig. 6).
Figure 6. Expression patterns of TCP genes in different tissues and development stages of B. juncea var. tumida.

DY, Dayejie stems were collected 22 weeks after seeding (daye3bianzhong); YA1-4, The stems of Yong’an were collected 18, 20, 22, and 25 weeks after seeding; YAr, The mix roots samples of 18 and 22 weeks after seeding. The expression levels are represented by the color bar (log2-transformed).
In addition, we analyzed the expression profiles of B. juncea var. tumida seedlings and tumorous stems. The expression levels of these four genes (BjTCP18a-d) were low in these tissues; the expression levels gradually decreased along with the swelling of tumorous stems (Fig. S3).
Expression analysis of BjTCP genes in response to exogenous hormones
To predict the possible functions of TCP genes in environmental adaptation, we investigated their transcriptional profile after SA and GA treatment. Multiple gene family members in the same branch often have highly similar sequence characteristics and contain similar cis-acting elements (Fig. 5), so we selected one corresponding homologous gene from each A. thaliana TCP for qRT-PCR analysis.
After SA treatment for 2–8 h, almost all the detected BjTCP genes were upregulated, except BjTCP1a, BjTCP12a, and BjTCP17a, while after 24 h, the BjTCP genes were downregulated to a low level. BjTCP9a, BjTCP13a, BjTCP15a, BjTCP18a, BjTCP19a, BjTCP22a, and BjTCP24a were induced at early stages and maintained at a relatively high level until 8 h. BjTCP17a was induced slowly and was highly expressed 24 h after SA treatment (Fig. 7).
Figure 7. Expression levels of BjTCPs under SA and GA treatment by qRT-PCR.
The number represented the treatment times (hours). The colour scales represent relative expression data.
After GA treatment, we did not detect BjTCP12a, BjTCP17a, and BjTCP20a expression. In contrast, the expression of other genes was induced at early stages but decreased to a low level mainly after 8 h (Fig. 7).
Discussion
Plant-specific TCP TFs play various roles in plant growth and development. In many plants, the general organization of the TCP family is conserved, and there are more members in class I compared to class II (Du et al., 2017; Li et al., 2017; Liu et al., 2018b; Ma et al., 2014; Ma et al., 2016; Shi et al., 2016; Wang et al., 2018; Wang et al., 2019c; Zhao et al., 2018; Zheng et al., 2018). In B. juncea var. tumida, we found 62 BjTCP genes. As a tetraploid plant, B. juncea var. tumida contains twice as many TCP proteins as Arabidopsis (24 TCP proteins), indicating that some genes are duplicated during evolution. However, recent studies have reported 36 and 38 TCP genes in B. rapa (AA) and B. oleracea (CC), respectively (Liu et al., 2019), indicating that the B. juncea genome might contain >70 TCP genes, although we identified only 62. We used HMMER 3.0 to search for TCP domain proteins and further removed members that did not contain the TCP domain using the Pfam database. We did not find the homologous genes of AtTCP4, AtTCP10, AtTCP11, and AtTCP16, which is similar to the study by Liu et al. (2018b), who did not find the homologous genes of ATCP11 and AtTCP16 during the identification of the TCP family in the Chinese cabbage using the BLASTP program (Liu et al., 2018b). In this study, BjuA003953, BjuA026354, BjuB013551, and BjuB044682 (named BjTCP2a–d) were found to be similar to AtTCP2, but the confidence level was low when analyzed using the Pfam database, and MEME analysis showed that no motif included the TCP domain. In addition, the BjTCP2a-d amino acid sequence was not clustered with the homolog protein AtTCP2 in the phylogenetic tree, which might be because of the incompletely predicted amino acid sequences during assembling of the genome sequence.
The exon/intron gene structure, conserved motif distribution patterns, and BjTCP homologous gene domain often show high similarity, such as BjTCP21a-f, BjTCP12a/b, and so on, and we believe that these similarities within the cluster of homologous genes members suggest that they might have a similar function during B. juncea var. tumida growth and development.
We located two genes clusters (BjTCP15b, BjTCP15c, BjTCP1b, and BjTCP22b) and (BjTCP15f, BjTCP15e, BjTCP1c, and BjTCP22d) on chromosomes A07 and B03, respectively. Evolutionary relationship results showed the allopolyploid B. juncea var. tumida (B.juncea, AABB) might form by hybridization between the diploid ancestors of B. rapa (AA) and B. nigra (BB), followed by spontaneous chromosome doubling. These results also indicated that the division of BjTCP15b and BjTCP15c might occur earlier than tetraploid formation.
As mentioned before, there are 24 TCP genes in Arabidopsis. Some corresponding homologous of TCP genes were not found in B. juncea var. tumida, such as TCP4, TCP10, TCP11, and TCP16, probably due to gene loss events during evolution. B. juncea var. tumida is a tetraploid plant that belongs to the cruciferous near-source species of Arabidopsis. In B. juncea var. tumida, some TCP genes have more than two homologous genes, such as BjTCP1 (five homologous genes), BjTCP18 (four homologous genes), BjTCP21 (six homologous genes), and BjTCP15 (six homologous genes). These genes may be formed by multiple gene duplication events, and the functions of these paralogous genes gradually differentiated during evolution. Most paralogous genes had similar cis-acting elements, but there were a few differences. For example, the four paralogues of BjTCP18 had no ABA and auxin cis-acting elements, but BjTCP18b was the only member with circadian regulatory elements, suggesting that BjTCP18b might be involved in the circadian rhythm. Correspondingly, the expression patterns of these paralogous genes were also different.
In addition, six homologous genes of AtTCP15 and AtTCP21 in B. juncea var. tumida are interesting. AtTCP15 plays an important role in regulating endoreduplication during development in Arabidopsis (Li, Li & Dong, 2012; Uberti-Manassero et al., 2012). In different developmental stages, the six BjTCP genes are highly expressed in DY, but there are chronological differences among the swollen tuber cultivars, suggesting that several genes might be involved in developmental regulation at different stages.
Interestingly, as mentioned before, in Arabidopsis, AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24 are post-transcriptionally regulated by miR319, and these genes mainly regulate leaf morphogenesis and senescence (Bresso et al., 2018; Palatnik et al., 2003). In B. juncea var. tumida, no AtTCP4 and AtTCP10 homolog genes have been identified, and BjTCP3 does not contain the miR319 regulation site. Only BjTCP2a–d and BjTCP24a–d have the putative miR319 recognition site, and their expression levels in stem development of the swelling strains (YA1–YA4) are relatively low compared to the no-swelling strain (DA). These results indicate that miR319 might not be involved in stem-swelling regulation in B. juncea var. tumida.
The Arabidopsis BRANCHED1 (BRC1), the rice TB1, and the maize TB1 function as negative regulators of the growth of axillary buds and branching (Aguilar-Martinez, Poza-Carrion & Cubas, 2007; Dixon et al., 2018; Finlayson, 2007; Takeda et al., 2003). BRC1/TB1 orthologues play a similar role in the development of the primary shoot architecture and negatively regulate lateral branching (Aguilar-Martinez, Poza-Carrion & Cubas, 2007; Dixon et al., 2018; Finlayson, 2007; Gonzalez-Grandio et al., 2013; Muhr et al., 2016; Wang et al., 2019a; Yang et al., 2015). In addition, OsTB1 can be regulated by IPA1 to suppress tillering in rice, and TB1 can interact with FT1 to regulate inflorescence architecture in bread wheat (Dixon et al., 2018; Guo et al., 2013; Takeda et al., 2003). In B. juncea var. tumida, which has a close phylogenetic relationship with Arabidopsis, BjTCP18 s might play a similar function in branching. There are four BjTCP18 homologous genes in B. juncea var. tumida, which might have been formed by gene duplication. Functional differentiation might occur between the four TCP18 genes, given their differential expression patterns during tissue development. The flowering stage in B. juncea var. tumida is mainly characterized by swelling of the tumorous stem. At this time, the plant shows a bolting and flowering phenomenon similar to Arabidopsis. Since BRC1 inhibits branching and flowering, gradual downregulation of its messenger RNA (mRNA) levels might reflect a gradual decrease in the ability to inhibit branching and flowering. These events also indicate that B. juncea var. tumida is about to enter the period of reproductive growth.
There are 16 varieties of mustard species identified and used for food consumption, in which the main difference is the tissue shape, including the root, stem, leaf, and branch (Qiao, Liu & Lei, 1998). The BRC1 gene controls plant branching and interacts with the flowering time–relate gene FT, and four identified BjBRC1 genes might imply further functional differentiation of branch development and floral transition. Among these BjTCP genes, multiple BjTCP15 and BjTCP21 genes are highly expressed in the DY and/or the early stage of seedling and tumorous stem per-swelling stage as compared to the swelling stage. DY is a mutant line with no swelling, and YA1 and YA2 also have not started to swell. These findings indicate that these genes are involved in the process of stem swelling in B. juncea var. tumida.
Increasing evidence verifies that TCP proteins are involved in responses to plant hormones (Braun et al., 2012; Danisman et al., 2012; Dun et al., 2012; Feng et al., 2018; Gonzalez-Grandio et al., 2017; Hay, Barkoulas & Tsiantis, 2004; He et al., 2016; Liu et al., 2018a; Lopez et al., 2015; Nicolas & Cubas, 2016; Qin et al., 2005; Schommer et al., 2008; Shen et al., 2019; Wang et al., 2019a; Wang et al., 2013). In this study, most of B. juncea var. tumida TCP genes appeared to be regulated by SA and GA. In A. thaliana, several TCPs interact with the SA biosynthetic enzyme ISOCHORISMATE SYNTHASE 1 gene and enhance its expression by binding to the TCP-binding motif in its promoter region (Wang et al., 2015). Our results showed that there are many SA-related cis-elements in the promoter regions of BjTCP genes, and the expression levels of several BjTCP genes significantly increase after SA treatment, indicating that BjTCP genes might be involved in SA signal transduction. However, SA treatment does not seem to directly affect the expression of these genes. For example, the promoter regions of BjTCP1a and BjTCP12a contain two TCA elements, but there was almost no expression of these genes. In contrast, although the promoter regions of BjTCP7a, BjTCP8a, BjTCP17a, and BjTCP20a contain no TCA elements, the expression levels of these four genes changed after SA treatment at different times. The other ten genes containing the TCA element were upregulated almost 2 h after SA treatment. These results suggest that SA might not only directly regulate the TCA element but also link other pathways to indirectly regulate the expression of some TCP genes.
Most BjTCP genes have the polytype GA response elements GARE, P-box, and TATC-box in their promoter regions, which might lead to more complex regulation of their expression and more diverse expression patterns. Analysis of the GA response elements of these genes showed no GA-related elements in the promoter regions of BjTCP1a, BjTCP7a, BjTCP8a, BjTCP12a, BjTCP14a, BjTCP20a, and BjTCP22a. After GA treatment, no expression of BjTCP12a and BjTCP20a was detected, while BjTCP1a, BjTCP7a, BjTCP8a, BjTCP14a, and BjTCP22a expression first increased and then decreased. In addition, although there was a P-box element in BjTCP17a, the expression level did not change after GA treatment. The remaining genes including one or more GA response elements in their promoter regions were upregulated after GA treatment. These results suggest that GA affects the expression levels of most of the TCP genes.
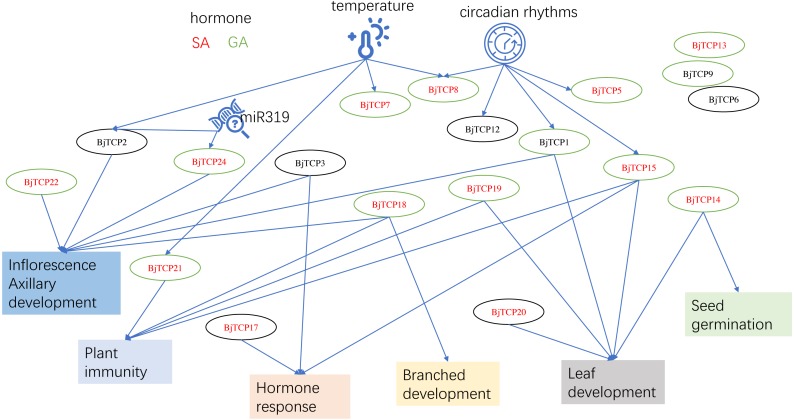
This study was the first to identify 62 BjTCP genes in B. juncea var. tumida and to investigate their roles in stem development. On the basis of our results and reports on A. thaliana, we believe that BjTCP is also regulated by many factors and is involved in hormone response, plant architecture, inflorescence development, and immune regulation in B. juncea var. tumida (Fig. 8). Our results will provide the foundation for further determining the molecular mechanism underlying stem swelling and flowering orchestrated by TCP genes in B. juncea var. tumida.
Figure 8. The putative mechanism diagram on the basis of current results and the reports of proximal species A. thaliana.
Red letter shows the genes may regulated by SA. A green circle means the genes may induced by GA. Arrows indicate possible regulatory relationships.
Conclusions
We performed a genome-wide analysis and identified 62 BjTCP genes in B. juncea var. tumida. These genes are divided into two 34 class I and 28 class II subfamilies. Of these 62 BjTCP genes, 61 are heterogeneously distributed on 18 chromosomes, 51 have no introns, and most of the BjTCP genes in the same cluster have similar patterns of exon length, intron number, and conserved motifs. Several genes are highly expressed in the development of B. juncea var. tumida, and branching-related genes have low expression in the swelling stage of vegetative growth.
Supplemental Information
The yellow background colour showed the GA response cis-acting elements, Gray background showed the SA response cis-acting elements. The Red colour gene name mean detected expression by qRT-PCR.
Acknowledgments
We acknowledge Dr Yinghong Li for his assistance in RNA-seq data processing and related bioinformatics analysis.
Funding Statement
This work was supported by the Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN201800609) and Science and Technology Research Program of Chongqing University of Posts and Telecommunications (Grant No. A2018-114) and Chongqing Natural Science Foundation (Grant No. cstc2015jcyjA0752). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Daping Gong, Email: gongdaping@caas.cn.
Quan Sun, Email: sunquan@cqupt.edu.cn.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Jing He performed the experiments, prepared figures and/or tables, and approved the final draft.
Xiaohong He and Huaizhong Jiang performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Pingan Chang and Daping Gong analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Quan Sun conceived and designed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Data Availability
References
- Aguilar-Martinez, Poza-Carrion & Cubas (2007).Aguilar-Martinez JA, Poza-Carrion C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. The Plant Cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai et al. (2012).Bai F, Reinheimer R, Durantini D, Kellogg EA, Schmidt RJ. TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12225–12230. doi: 10.1073/pnas.1202439109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge et al. (2003).Beveridge CA, Weller JL, Singer SR, Hofer JM. Axillary meristem development. Budding relationships between networks controlling flowering, branching, and photoperiod responsiveness. Plant Physiology. 2003;131:927–934. doi: 10.1104/pp.102.017525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun et al. (2012).Braun N, De Saint Germain A, Pillot JP, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, Luo D, Bendahmane A, Turnbull C, Rameau C. The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching. Plant Physiology. 2012;158:225–238. doi: 10.1104/pp.111.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresso et al. (2018).Bresso EG, Chorostecki U, Rodriguez RE, Palatnik JF, Schommer C. Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiology. 2018;176:1694–1708. doi: 10.1104/pp.17.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer (2015).Brewer PB. Plant architecture: the long and the short of branching in potato. Current Biology. 2015;25:R724–R725. doi: 10.1016/j.cub.2015.06.066. [DOI] [PubMed] [Google Scholar]
- Cheng et al. (2011).Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biology. 2011;11:136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas, Vincent & Coen (1999).Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- Dai & Zhao (2011).Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval et al. (2007).Damerval C, Le Guilloux M, Jager M, Charon C. Diversity and evolution of CYCLOIDEA-like TCP genes in relation to flower development in Papaveraceae. Plant Physiology. 2007;143:759–772. doi: 10.1104/pp.106.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman (2016).Danisman S. TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Frontiers of Plant Science. 2016;7:1930. doi: 10.3389/fpls.2016.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman et al. (2012).Danisman S, Van der Wal F, Dhondt S, Waites R, De Folter S, Bimbo A, Van Dijk AD, Muino JM, Cutri L, Dornelas MC, Angenent GC, Immink RG. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiology. 2012;159:1511–1523. doi: 10.1104/pp.112.200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon et al. (2018).Dixon LE, Greenwood JR, Bencivenga S, Zhang P, Cockram J, Mellers G, Ramm K, Cavanagh C, Swain SM, Boden SA. TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum) The Plant Cell. 2018;30:563–581. doi: 10.1105/tpc.17.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, Stec & Hubbard (1997).Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Du et al. (2017).Du J, Hu S, Yu Q, Wang C, Yang Y, Sun H, Yang Y, Sun X. Genome-wide identification and characterization of brrtcp transcription factors in brassica rapa ssp. rapa. Frontiers of Plant Science. 2017;8:1588. doi: 10.3389/fpls.2017.01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun et al. (2012).Dun EA, De Saint Germain A, Rameau C, Beveridge CA. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiology. 2012;158:487–498. doi: 10.1104/pp.111.186783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni et al. (2008).Efroni I, Blum E, Goldshmidt A, Eshed Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. The Plant Cell. 2008;20:2293–2306. doi: 10.1105/tpc.107.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng et al. (2018).Feng ZJ, Xu SC, Liu N, Zhang GW, Hu QZ, Gong YM. Soybean TCP transcription factors: evolution, classification, protein interaction and stress and hormone responsiveness. Plant Physiology and Biochemistry. 2018;127:129–142. doi: 10.1016/j.plaphy.2018.03.020. [DOI] [PubMed] [Google Scholar]
- Finlayson (2007).Finlayson SA. Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant and Cell Physiology. 2007;48:667–677. doi: 10.1093/pcp/pcm044. [DOI] [PubMed] [Google Scholar]
- Finlayson et al. (2010).Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ. Phytochrome regulation of branching in Arabidopsis. Plant Physiology. 2010;152:1914–1927. doi: 10.1104/pp.109.148833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, Clements & Eddy (2011).Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Research. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn et al. (2010).Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Research. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud et al. (2010).Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Van Aken O, Millar AH, Murcha M, Whelan J. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. The Plant Cell. 2010;22:3921–3934. doi: 10.1105/tpc.110.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Grandio et al. (2017).Gonzalez-Grandio E, Pajoro A, Franco-Zorrilla JM, Tarancon C, Immink RG, Cubas P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E245–E254. doi: 10.1073/pnas.1613199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Grandio et al. (2013).Gonzalez-Grandio E, Poza-Carrion C, Sorzano CO, Cubas P. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. The Plant Cell. 2013;25:834–850. doi: 10.1105/tpc.112.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan et al. (2017).Guan P, Ripoll JJ, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:2419–2424. doi: 10.1073/pnas.1615676114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2013).Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nature Communications. 2013;4:1566. doi: 10.1038/ncomms2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, Barkoulas & Tsiantis (2004).Hay A, Barkoulas M, Tsiantis M. PINning down the connections: transcription factors and hormones in leaf morphogenesis. Current Opinion in Plant Biology. 2004;7:575–581. doi: 10.1016/j.pbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- He et al. (2016).He Y, Liu X, Ye L, Pan C, Chen L, Zou T, Lu G. Genome-wide identification and expression analysis of two-component system genes in tomato. International Journal of Molecular Sciences. 2016;17:1204. doi: 10.3390/ijms17081204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho & Weigel (2014).Ho WW, Weigel D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. The Plant Cell. 2014;26:552–564. doi: 10.1105/tpc.113.115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2015).Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom, Burson & Finlayson (2006).Kebrom TH, Burson BL, Finlayson SA. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiology. 2006;140:1109–1117. doi: 10.1104/pp.105.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer et al. (2011).Kieffer M, Master V, Waites R, Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant Journal. 2011;68:147–158. doi: 10.1111/j.1365-313X.2011.04674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim et al. (2014).Kim SH, Son GH, Bhattacharjee S, Kim HJ, Nam JC, Nguyen PD, Hong JC, Gassmann W. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant Journal. 2014;78:978–989. doi: 10.1111/tpj.12527. [DOI] [PubMed] [Google Scholar]
- Koyama et al. (2007).Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. The Plant Cell. 2007;19:473–484. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot et al. (2002).Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic & Bork (2018).Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Research. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, Doerks & Bork (2015).Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Research. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li (2015).Li S. The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signaling & Behavior. 2015;10:e1044192. doi: 10.1080/15592324.2015.1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Li & Dong (2012).Li ZY, Li B, Dong AW. The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Molecular Plant. 2012;5:270–280. doi: 10.1093/mp/ssr086. [DOI] [PubMed] [Google Scholar]
- Li et al. (2017).Li W, Li DD, Han LH, Tao M, Hu QQ, Wu WY, Zhang JB, Li XB, Huang GQ. Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum) Scientific Reports. 2017;7:10118. doi: 10.1038/s41598-017-10609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2017).Liu J, Cheng X, Liu P, Sun J. miR156-Targeted SBP-Box Transcription Factors Interact with DWARF53 to Regulate TEOSINTE BRANCHED1 and BARREN STALK1 Expression in Bread Wheat. Plant Physiology. 2017;174:1931–1948. doi: 10.1104/pp.17.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2018b).Liu Y, Guan X, Liu S, Yang M, Ren J, Guo M, Huang Z, Zhang Y. Genome-wide identification and analysis of TCP transcription factors involved in the formation of leafy head in Chinese cabbage. International Journal of Molecular Sciences. 2018b;19:847. doi: 10.3390/ijms19030847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2019).Liu MM, Wang MM, Yang J, Wen J, Guo PC, Wu YW, Ke YZ, Li PF, Li JN, Du H. Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. International Journal of Molecular Sciences. 2019;20:3591. doi: 10.3390/ijms20143591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2018a).Liu HL, Wu M, Li F, Gao YM, Chen F, Xiang Y. TCP transcription factors in moso bamboo (Phyllostachys edulis): genome-wide identification and expression analysis. Frontiers of Plant Science. 2018a;9:1263. doi: 10.3389/fpls.2018.01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak & Schmittgen (2001).Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez et al. (2015).Lopez JA, Sun Y, Blair PB, Mukhtar MS. TCP three-way handshake: linking developmental processes with plant immunity. Trends in Plant Science. 2015;20:238–245. doi: 10.1016/j.tplants.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Ma et al. (2016).Ma X, Ma J, Fan D, Li C, Jiang Y, Luo K. Genome-wide identification of TCP family transcription factors from populus euphratica and their involvement in leaf shape regulation. Scientific Reports. 2016;6:32795. doi: 10.1038/srep32795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2014).Ma J, Wang Q, Sun R, Xie F, Jones DC, Zhang B. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Scientific Reports. 2014;4:6645. doi: 10.1038/srep06645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigal, Alzate & Pabon-Mora (2017).Madrigal Y, Alzate JF, Pabon-Mora N. Evolution and expression patterns of TCP genes in asparagales. Frontiers of Plant Science. 2017;8:9. doi: 10.3389/fpls.2017.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Trillo & Cubas (2010).Martin-Trillo M, Cubas P. TCP genes: a family snapshot ten years later. Trends in Plant Science. 2010;15:31–39. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Martin-Trillo et al. (2011).Martin-Trillo M, Grandio EG, Serra F, Marcel F, Rodriguez-Buey ML, Schmitz G, Theres K, Bendahmane A, Dopazo H, Cubas P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant Journal. 2011;67:701–714. doi: 10.1111/j.1365-313X.2011.04629.x. [DOI] [PubMed] [Google Scholar]
- Maurya et al. (2020).Maurya JP, Singh RK, Miskolczi PC, Prasad AN, Jonsson K, Wu F, Bhalerao RP. Branching regulator BRC1 mediates photoperiodic control of seasonal growth in hybrid aspen. Current Biology. 2020;30:122–126. doi: 10.1016/j.cub.2019.11.001. [DOI] [PubMed] [Google Scholar]
- Muhr et al. (2016).Muhr M, Prufer N, Paulat M, Teichmann T. Knockdown of strigolactone biosynthesis genes in Populus affects BRANCHED1 expression and shoot architecture. New Phytologist. 2016;212:613–626. doi: 10.1111/nph.14076. [DOI] [PubMed] [Google Scholar]
- Mukhtar et al. (2011).Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, Pevzner SJ, Donovan SE, Ghamsari L, Santhanam B, Romero V, Poulin MM, Gebreab F, Gutierrez BJ, Tam S, Monachello D, Boxem M, Harbort CJ, McDonald N, Gai L, Chen H, He Y, European Union Effectoromics Consortium. Vandenhaute J, Roth FP, Hill DE, Ecker JR, Vidal M, Beynon J, Braun P, Dangl JL. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, Cruz-Oro & Prat (2015).Navarro C, Cruz-Oro E, Prat S. Conserved function of FLOWERING LOCUS T (FT) homologues as signals for storage organ differentiation. Current Opinion in Plant Biology. 2015;23:45–53. doi: 10.1016/j.pbi.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Nicolas & Cubas (2016).Nicolas M, Cubas P. TCP factors: new kids on the signaling block. Current Opinion in Plant Biology. 2016;33:33–41. doi: 10.1016/j.pbi.2016.05.006. [DOI] [PubMed] [Google Scholar]
- Nicolas et al. (2015).Nicolas M, Rodriguez-Buey ML, Franco-Zorrilla JM, Cubas P. A Recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Current Biology. 2015;25:1799–1809. doi: 10.1016/j.cub.2015.05.053. [DOI] [PubMed] [Google Scholar]
- Niwa et al. (2013).Niwa M, Daimon Y, Kurotani K, Higo A, Pruneda-Paz JL, Breton G, Mitsuda N, Kay SA, Ohme-Takagi M, Endo M, Araki T. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. The Plant Cell. 2013;25:1228–1242. doi: 10.1105/tpc.112.109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik et al. (2003).Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Parapunova et al. (2014).Parapunova V, Busscher M, Busscher-Lange J, Lammers M, Karlova R, Bovy AG, Angenent GC, De Maagd RA. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biology. 2014;14:157. doi: 10.1186/1471-2229-14-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz et al. (2009).Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz et al. (2009).Prusinkiewicz P, Crawford S, Smith RS, Ljung K, Bennett T, Ongaro V, Leyser O. Control of bud activation by an auxin transport switch. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17431–17436. doi: 10.1073/pnas.0906696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Liu & Lei (1998).Qiao AM, Liu PY, Lei JJ. RAPD analysis of sixteen varieties of mustard. Acta Botanica Sinica. 1998;40:915–921. (Chinese) [Google Scholar]
- Qin et al. (2005).Qin G, Gu H, Zhao Y, Ma Z, Shi G, Yang Y, Pichersky E, Chen H, Liu M, Chen Z, Qu LJ. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. The Plant Cell. 2005;17:2693–2704. doi: 10.1105/tpc.105.034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameau et al. (2014).Rameau C, Bertheloot J, Leduc N, Andrieu B, Foucher F, Sakr S. Multiple pathways regulate shoot branching. Frontiers of Plant Science. 2014;5:741. doi: 10.3389/fpls.2014.00741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa et al. (2017).Rosa M, Abraham-Juarez MJ, Lewis MW, Fonseca JP, Tian W, Ramirez V, Luan S, Pauly M, Hake S. The maize MID-COMPLEMENTING ACTIVITY homolog CELL NUMBER REGULATOR13/NARROW ODD DWARF coordinates organ growth and tissue patterning. The Plant Cell. 2017;29:474–490. doi: 10.1105/tpc.16.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer et al. (2008).Schommer C, Palatnik JF, Aggarwal P, Chetelat A, Cubas P, Farmer EE, Nath U, Weigel D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLOS Biology. 2008;6:e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale, Bennett & Leyser (2017).Seale M, Bennett T, Leyser O. BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development. 2017;144:1661–1673. doi: 10.1242/dev.145649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen et al. (2019).Shen J, Zhang Y, Ge D, Wang Z, Song W, Gu R, Che G, Cheng Z, Liu R, Zhang X. CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:17105–17114. doi: 10.1073/pnas.1907968116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2016).Shi P, Guy KM, Wu W, Fang B, Yang J, Zhang M, Hu Z. Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biology. 2016;16:85. doi: 10.1186/s12870-016-0765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun et al. (2012).Sun Q, Zhou G, Cai Y, Fan Y, Zhu X, Liu Y, He X, Shen J, Jiang H, Hu D, Pan Z, Xiang L, He G, Dong D, Yang J. Transcriptome analysis of stem development in the tumourous stem mustard Brassica juncea var. tumida Tsen et Lee by RNA sequencing. BMC Plant Biology. 2012;12:53. doi: 10.1186/1471-2229-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda et al. (2003).Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C. The OsTB1 gene negatively regulates lateral branching in rice. Plant Journal. 2003;33:513–520. doi: 10.1046/j.1365-313X.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- Tamura et al. (2013).Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann & Muhr (2015).Teichmann T, Muhr M. Shaping plant architecture. Frontiers of Plant Science. 2015;6:233. doi: 10.3389/fpls.2015.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson et al. (1997).Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, Pachter & Salzberg (2009).Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell et al. (2012).Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberti-Manassero et al. (2012).Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH. The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. Journal of Experimental Botany. 2012;63:809–823. doi: 10.1093/jxb/err305. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2015).Wang X, Gao J, Zhu Z, Dong X, Wang X, Ren G, Zhou X, Kuai B. TCP transcription factors are critical for the coordinated regulation of isochorismate synthase 1 expression in Arabidopsis thaliana. Plant Journal. 2015;82:151–162. doi: 10.1111/tpj.12803. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2019a).Wang M, Le Moigne MA, Bertheloot J, Crespel L, Perez-Garcia MD, Oge L, Demotes-Mainard S, Hamama L, Daviere JM, Sakr S. BRANCHED1: a key hub of shoot branching. Frontiers of Plant Science. 2019a;10:76. doi: 10.3389/fpls.2019.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2019b).Wang M, Oge L, Voisine L, Perez-Garcia MD, Jeauffre J, Saint-Oyant LH, Grappin P, Hamama L, Sakr S. Posttranscriptional regulation of RhBRC1 (Rosa hybrida BRANCHED1) in response to sugars is mediated via its own 3 ’ untranslated region, with a potential role of RhPUF4 (Pumilio RNA-Binding Protein Family) International Journal of Molecular Sciences. 2019b;20:3808. doi: 10.3390/Ijms20153808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2018).Wang H, Wang H, Liu R, Xu Y, Lu Z, Zhou C. Genome-wide identification of TCP family transcription factors in medicago truncatula reveals significant roles of miR319-targeted TCPs in nodule development. Frontiers of Plant Science. 2018;9:774. doi: 10.3389/fpls.2018.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2019c).Wang Y, Zhang N, Li T, Yang J, Zhu X, Fang C, Li S, Si H. Genome-wide identification and expression analysis of StTCP transcription factors of potato (Solanum tuberosum L.) Computational Biology and Chemistry. 2019c;78:53–63. doi: 10.1016/j.compbiolchem.2018.11.009. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2013).Wang MY, Zhao PM, Cheng HQ, Han LB, Wu XM, Gao P, Wang HY, Yang CL, Zhong NQ, Zuo JR, Xia GX. The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiology. 2013;162:1669–1680. doi: 10.1104/pp.113.215673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2016).Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B, Hu Z, Chen S, Pental D, Ju Y, Yao P, Li X, Xie K, Zhang J, Wang J, Liu F, Ma W, Shopan J, Zheng H, Mackenzie SA, Zhang M. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nature Genetics. 2016;48:1225–1232. doi: 10.1038/ng.3657. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2015).Yang X, Zhao XG, Li CQ, Liu J, Qiu ZJ, Dong Y, Wang YZ. Distinct regulatory changes underlying differential expression of TEOSINTE BRANCHED1-CYCLOIDEA-PROLIFERATING CELL FACTOR genes associated with petal variations in zygomorphic flowers of petrocosmea spp. of the family gesneriaceae. Plant Physiology. 2015;169:2138–2151. doi: 10.1104/pp.15.01181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2019).Zhang W, Tan L, Sun H, Zhao X, Liu F, Cai H, Fu Y, Sun X, Gu P, Zhu Z, Sun C. Natural variations at TIG1 encoding a TCP transcription factor contribute to plant architecture domestication in rice. Molecular Plant. 2019;12:1075–1089. doi: 10.1016/j.molp.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Zhao et al. (2018).Zhao J, Zhai Z, Li Y, Geng S, Song G, Guan J, Jia M, Wang F, Sun G, Feng N, Kong X, Chen L, Mao L, Li A. Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.) Frontiers of Plant Science. 2018;9:1282. doi: 10.3389/fpls.2018.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng et al. (2018).Zheng K, Ni Z, Qu Y, Cai Y, Yang Z, Sun G, Chen Q. Genome-wide identification and expression analyses of TCP transcription factor genes in Gossypium barbadense. Scientific Reports. 2018;8:14526. doi: 10.1038/s41598-018-32626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The yellow background colour showed the GA response cis-acting elements, Gray background showed the SA response cis-acting elements. The Red colour gene name mean detected expression by qRT-PCR.
Data Availability Statement
The following information was supplied regarding data availability:
Data is available at NCBI SRA: SRX108496, SRX108498–SRX108502.