Abstract
Mutations in WASHC5 (also known as KIAA0196) cause autosomal dominant hereditary spastic paraplegia (HSP) type SPG8. WASHC5, commonly called strumpellin, is a core component of the Wiskott-Aldrich syndrome protein and SCAR homologue (WASH) complex that activates actin nucleation at endosomes. Although various other cellular roles for strumpellin have also been described, none account for how SPG8-associated mutations lead to HSP. Here, we identified protein interactors of the WASH complex by immunoprecipitation and mass spectrometry and assessed the functions of strumpellin in cultured cells using both overexpression and RNA interference plus cell spreading assays to investigate cell adhesion. We uncovered a decrease in CAV1 protein abundance as well as endosomal fission defects resulting from pathogenic SPG8 mutations. Caveolin-1 (CAV1), a key component of caveolae, interacted with strumpellin in cells, and strumpellin inhibited lysosomal degradation of CAV1. SPG8-associated missense mutations in strumpellin did not rescue endosomal tubulation defects, reduction of CAV1 protein, or integrin-mediated cell adhesion in strumpellin-deficient cells. Mechanistically, we demonstrated that the WASH complex maintained CAV1 and integrin protein levels by inhibiting their lysosomal degradation through its endosomal actin nucleation activity. In addition, the interaction of strumpellin with CAV1 stimulated integrin recycling, thereby promoting cell adhesion. These findings provide a clear molecular link between WASHC5 mutations and impairment of CAV1- and integrin-mediated cell adhesion, providing potentially important insights into the cellular pathogenesis of SPG8.
One-Sentence Summary:
Hereditary spastic paraplegia–associated mutations in a WASH complex component impair integrin-mediated cell adhesion.
Editor’s Summary:
Disease-associated mutations impair integrin recycling
Hereditary spastic paraplegia type SPG8 is an inherited neurologic disorder associated with mutations in WASHC5, which encodes strumpellin, a component of the Wiskott-Aldrich syndrome protein and SCAR homolog (WASH) complex. The WASH complex localizes to endosomes and plays a role in endosomal sorting. Lee et al. found that strumpellin interacted with and inhibited the degradation of the caveolar protein CAV1. Cultured cells lacking strumpellin or expressing SPG8-associated strumpellin mutant proteins exhibited impaired endosomal tubulation, reduced CAV1 at intracellular vesicles, and reduced integrin-mediated cell adhesion. The actin nucleation activity of the WASH complex was required for inhibiting the degradation of CAV1 and integrin α5, and the interaction between strumpellin and CAV1 was required for proper recycling of integrin α5 and for integrin-mediated cell adhesion. These findings suggest that the axon defects that characterize HSP type SPG8 may be due to disruption of strumpellin- and CAV1-dependent integrin-mediated cell adhesion.
Introduction
Hereditary spastic paraplegias (HSPs) are a large and diverse group of inherited neurologic disorders with a defining and unifying feature of prominent lower extremity spasticity due to axonopathy of the longest corticospinal neurons (1). Historically, HSPs have been bisected into “pure” and “complicated” (or “complex”) forms. Pure forms are characterized by lower limb spasticity without prominent additional clinical findings other than mild urinary symptoms and impaired distal vibratory sensation. Complicated forms are associated with additional, often prominent, clinical features such as neuropathy, seizures, parkinsonism, cognitive impairment, amyotrophy, short stature, and visual abnormalities, among others (1).
HSPs are among the most genetically-diverse Mendelian disorders, with >80 distinct spastic gait genetic loci (SPG1-80, plus others) and over 60 genes already identified (1-4). Numerous studies elucidating the molecular pathogenesis underlying HSPs have highlighted the importance of a relatively small number of converging basic cellular functions -- especially membrane trafficking, mitochondrial function, organelle shaping and biogenesis, axon transport, and lipid metabolism -- in disease pathogenesis (1, 5).
Autosomal dominant mutations in the WASHC5 (also known as KIAA1096) gene, which encodes a protein known as strumpellin or WASHC5, cause an often severe, mostly pure HSP termed SPG8 (MIM #603563) (6), though ataxia and axonal sensory neuropathy have also been observed in SPG8 patients (7, 8). Strumpellin is a 1159-residue protein with an N-terminal domain (residues 1-240), a central spectrin-repeat domain (five repeats, each comprising three α-helices; residues 241-791), and a C-terminal domain (residues 792-1159). Strumpellin is a core component of the hetero-pentameric Wiskott-Aldrich syndrome protein and SCAR homolog (WASH) complex that activates Arp2- and Arp3-mediated actin polymerization on endosomes (9, 10). The other core components are WASH1 (also called WASHC1), FAM21 (also called WASHC2), CCDC53 (also called WASHC3), and SWIP (also called WASHC4). The WASH complex functions in the endocytic recycling of various types of membrane proteins, maintenance of endolysosomal network integrity, exocytosis, cell adhesion, and cell migration (11-23).
Although the WASH complex has been extensively studied, little is known about how SPG8-associated mutations in WASHC5 cause HSP. To date, 11 missense mutations, a large deletion, and a frame-shift nucleotide deletion in WASHC5 have been reported to underlie SPG8 (6, 8, 24, 25). Previous studies investigating the molecular mechanisms of SPG8-associated strumpellin mutations have found that expression of missense mutants in a wild-type background does not exert a dominant-negative effect (6, 26). The expressed mutant proteins do not induce defects in endosomal tubulation and mis-localization of β2-adrenergic receptors, which are known to be triggered by strumpellin depletion (26). Therefore, it is necessary to assess other cellular roles for strumpellin that may be related to HSP pathogenesis.
Here we report that strumpellin interacts with caveolin-1 (CAV1), a major component of caveolae (27). Strumpellin was required for maintenance of CAV1 abundance, integrin localization to focal adhesions, and fibronectin-mediated cell adhesion. Strumpellin-depleted cells expressing SPG8-associated mutant forms of strumpellin were deficient in maintaining CAV1 and integrin abundance as well as in integrin-mediated cell adhesion, suggesting that aberrant CAV1- and integrin–mediated cell adhesion might play a role in SPG8 pathogenesis. Furthermore, the actin-nucleating activity of WASH1 at endosomes was essential to promote a CAV1- and integrin–mediated cell adhesion pathway.
Results
Strumpellin interacts with CAV1
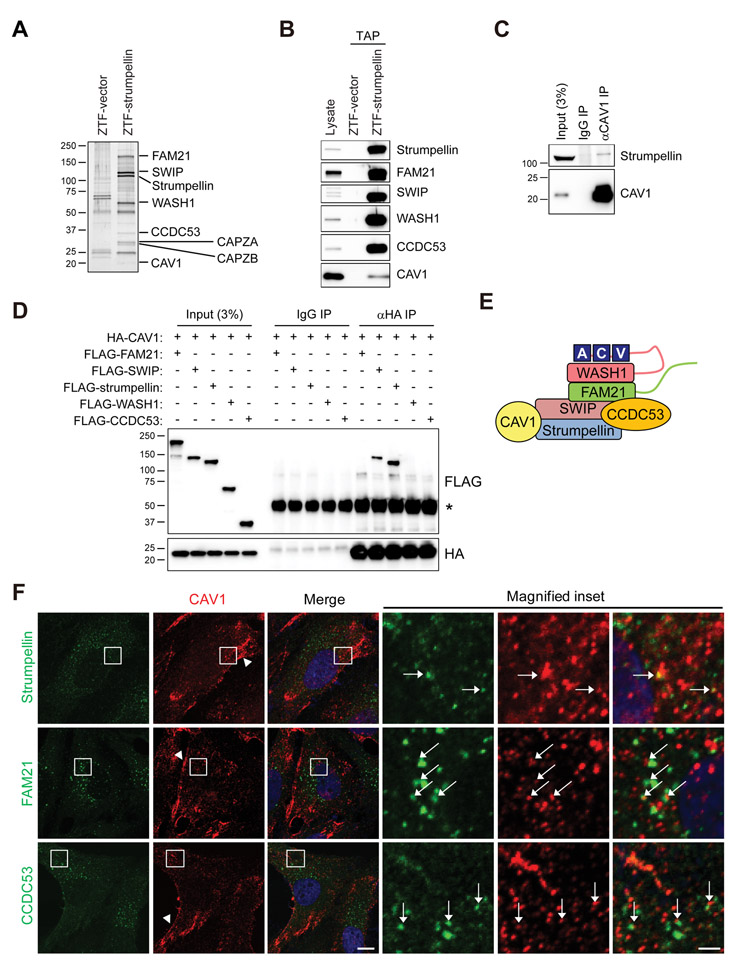
To identify strumpellin-interacting proteins, we generated human hTERT-RPE1 cells stably expressing full-length strumpellin fused with ZZ protein (an Fc region-binding domain originating from the B domain of protein A), a cleavage site for TEV protease, and a FLAG epitope (ZTF). We purified proteins that associated with strumpellin-ZTF using tandem affinity purification (TAP) (Fig. 1, A and B). All other core proteins of the WASH complex (FAM21, SWIP, WASH1, and CCDC53) as well as two known peripheral components of the complex (CAPZA and CAPZB) (9) co-precipitated robustly with strumpellin, as expected. Additionally, we identified CAV1, a major membrane protein component of caveolae – flask-shaped, lipid-rich pits enriched in the plasma membrane but also present in some intracellular membranes – as a previously unknown strumpellin-interacting protein (Fig. 1A and table S1). The interaction of CAV1 with the WASH complex was confirmed by co-immunoprecipitation (Fig. 1, C and D). Ectopically-expressed, HA-tagged CAV1 interacted only with strumpellin and SWIP (Fig. 1D). This interaction pattern for CAV1 fits well with the proposed model of the WASH regulatory complex, wherein SWIP and strumpellin constitute a sub-complex (Fig. 1E) (9). As reported previously, CAV1 localized to caveolae at both the cell membrane and intracellular vesicles (Fig. 1F) (28, 29). Fluorescence signals for WASH complex components strumpellin, FAM21, and CCDC53, all of which are known to localize to endosomes, partially overlapped with CAV1-immunoreactive signals on intracellular vesicles (Fig. 1F), suggesting that CAV1 and WASH components interacted at endosomes.
Fig. 1. Strumpellin interacts with CAV1.
(A) Strumpellin-associated proteins were immunoaffinity purified from hTERT-RPE1 cells stably expressing ZTF-strumpellin, with ZTF only (ZTF-vector) as a control. Eluted proteins were separated by SDS-PAGE and silver stained, then specific proteins were identified by mass spectrometry (table S1). Proteins identified by mass spectrometry are noted, along with the positions of standards (in kDa). Gel is representative of three independent experiments. (B) A small amount of tandem-affinity purified proteins was subjected to immunoblotting for the indicated proteins. Blot is representative of three independent experiments. (C) HEK293T cell lysates were immunoprecipitated (IP) with antibodies specific for CAV1 or control IgG, then immunoblotted for strumpellin and CAV1. Blot is representative of three independent experiments. (D) HEK293T cells were co-transfected with HA-CAV1 and individual 3×FLAG-tagged-WASH complex proteins as indicated. Lysates were immunoprecipitated (IP) with antibodies against HA or control IgG, then immunoblotted for FLAG and HA. An asterisk (*) denotes the IgG heavy chain. The IgG light chain band is visible in the IgG IP in the panel probed for HA. Blots are representative of five independent experiments. (E) Schematic model of possible interacting configuration of CAV1 with the WASH complex. VCA, verprolin, cofilin acidic domain of WASH1. (F) hTERT-RPE1 cells were immunostained for endogenous CAV1 (red) along with the indicated WASH complex proteins (green). Magnified images show boxed areas. Arrowheads indicate CAV1 staining at cell periphery; arrows indicate puncta exhibiting co-localization of CAV1 with WASH components. Images are representative of three independent experiments. Scale bars, 10 μm, 2 μm (magnified inset).
Maintenance of CAV1 protein levels depends on the WASH complex
To further investigate the relationship between CAV1 and the WASH complex, we depleted WASH complex proteins from hTERT-RPE1 cells using small interfering RNAs (siRNAs). Although treatment with an siRNA targeting strumpellin efficiently decreased the amounts of strumpellin, WASH1, and CCDC53 in cells, depletion of WASH1 did not affect the abundance of strumpellin (Fig. 2A) (9). Total amounts of CAV1 were greatly decreased upon depletion of either strumpellin or WASH1 (Fig. 2, A and B). Fluorescence signals for CAV1 at intracellular vesicles were also abolished in strumpellin- or WASH1-depleted cells, whereas CAV1 signals at the plasma membrane remained robust (Fig. 2C). Overall, total fluorescence intensities for CAV1 were reduced to about half of that in control cells, as also seen by immunoblotting (Fig. 2D). We infer that less stable CAV1 at intracellular vesicles could be preferentially affected by depletion of the WASH complex as compared to more stable membrane-associated CAV1, which undergoes slower turnover (30). This decrease in CAV1 protein was not due to transcriptional changes, because CAV1 transcripts were unaffected by the depletion of WASH complex components (fig. S1). In cultured rat cerebral cortical neurons depleted of strumpellin using short hairpin RNAs (shRNAs), a similar reduction in CAV1 abundance occurred, suggesting that this reduction may be relevant for SPG8 pathogenesis (Fig. 2, E and F).
Fig. 2. Maintenance of CAV1 protein depends on the WASH complex.
(A) hTERT-RPE1 cells transfected with control (CTL), strumpellin, or WASH1 siRNAs were lysed and immunoblotted. (B) Relative intensities of CAV1 immunoreactive signals were measured and normalized to GAPDH. n = 3 independent experiments. Graph shows means ± SD. (C) hTERT-RPE1 cells transfected with the indicated siRNAs were immunostained for CAV1 (green) and Alexa 555-conjugated phalloidin A (actin, red). Merged images are shown with DAPI staining to label nuclei. Scale bar, 10 μm. Images are representative of five independent experiments. (D) Total cell intensities of CAV1 were measured from >30 cells per group in three independent experiments as shown in (C). To calculate the fold-increase across groups, the mean for each group was divided by that for control cells. Means ± SD are shown. (E) Cultured rat cerebral cortical neurons were infected with viral particles from vector only or with the indicated shRNAs for 5 d, then immunoblotted for strumpellin, CAV1, and GFP. GFP is an indicator of viral infection. (F) Relative intensities of CAV1 and strumpellin immunoreactive signals in (E) were normalized to GAPDH. Graphs show means ± SD of 4 independent experiments. *p < 0.05, **p < 0.01, ****p < 0.0001. P values were obtained using the Kruskal-Wallis test with Dunnett’s post hoc test.
CAV1 is synthesized in the endoplasmic reticulum and then transported to the Golgi apparatus (29). To identify the mechanism underlying strumpellin-dependent accumulation of CAV1, we assessed newly-synthesized CAV1 at the Golgi in hTERT-RPE1 cells by immunostaining using a selective antibody (BD Biosciences C37120) that specifically labels CAV1 at the Golgi (Fig. 3A) (31). Depletion of strumpellin by siRNA did not affect steady-state amounts of CAV1 at the Golgi (Fig. 3A, top row). Treatment of cells with cycloheximide (CHX) for 3 h abolished Golgi-associated CAV1 protein, indicating that CAV1 translation was effectively blocked (Fig. 3A, middle row). After 1 h recovery from CHX treatment, newly-synthesized CAV1 was detected at the Golgi in both control and strumpellin-depleted cells (Fig. 3A, bottom row). Taken together, these results indicate that the decrease of CAV1 protein upon WASH complex depletion most likely occurs post-translationally.
Fig. 3. Decreases in CAV1 protein upon WASH complex depletion most likely occur post-translationally.
(A) hTERT-RPE1 cells transfected with control (CTL) or strumpellin siRNAs were incubated with cycloheximide (CHX) for 3 h. Cells were fixed and immunostained before CHX treatment, after CHX treatment, and after 1 hr recovery for the Golgi marker GM130 (red) and an antibody specific for the Golgi pool of CAV1 (green). Merged images are shown with DAPI labeling (blue). Images are representative of three independent experiments. Scale bar, 10 μm. (B) hTERT-RPE1 cells transfected with the indicated siRNAs were incubated in the absence or presence of Baf A1 for 24 h, then cells were lysed and immunoblotted for the indicated proteins. Numbers across the bottom of the blot indicate the relative abundances of CAV1 normalized to GAPDH.
CAV1 is ubiquitylated and targeted to lysosomes for degradation (30). Treatment of cells with bafilomycin (Baf) A1, which inhibits protein degradation at lysosomes, restored CAV1 abundance in strumpellin-depleted cells, similarly to its effects on control cells (Fig. 3B), indicating that strumpellin depletion enhanced lysosomal degradation of CAV1. Thus, maintenance of CAV1 protein levels depended on the WASH complex.
SPG8 mutant strumpellin proteins are defective in endosomal fission and maintenance of CAV1
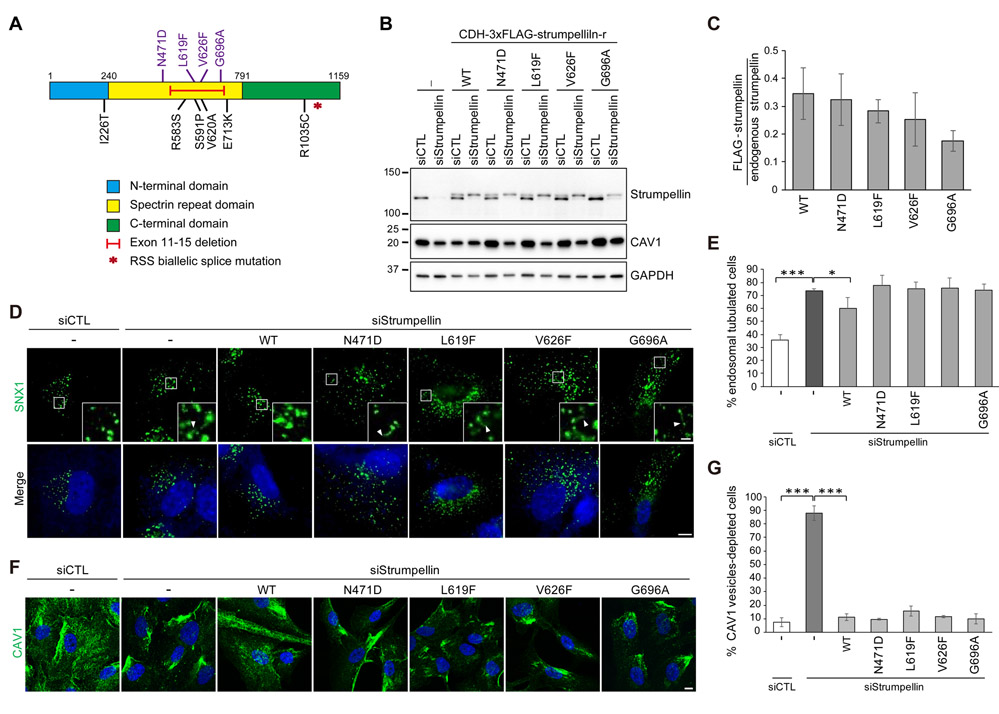
Previously published studies investigating strumpellin depletion or expression of strumpellin proteins harboring SPG8-associated missense mutations have not identified the cellular pathogenesis underlying SPG8 (6, 26). To assess the characteristics of autosomal dominant mutations in WASHC5 at the cellular level, we generated hTERT-RPE1 cells stably expressing strumpellin proteins harboring known SPG8-associated missense mutations along with silent mutations rendering them resistant to siRNA-mediated knockdown. Eight of eleven mutations identified in SPG8 patients localize to the spectrin-repeat domain, suggesting that this region could be particularly critical for SPG8 pathogenesis (8, 25, 32). Thus, we chose four missense mutations within this domain (N471D, L619F, V626F, and G696A) for further analysis (Fig. 4A). Amounts of these stably-expressed mutant strumpellin proteins were ~30% of that of the endogenous protein (Fig. 4, B and C), and the mutant proteins localized to endosomes like endogenous strumpellin (fig. S2). Stably-expressed wild-type strumpellin proteins rescued both an endosomal tubulation phenotype and aggregation of the WASH complex at collapsed endosomes, which are induced by the depletion of endogenous strumpellin (Fig. 4, D and E, and fig. S2). However, none of the SPG8 mutants tested rescued the endosomal tubulation phenotype, whereas all tested SPG8 mutants rescued the aberrant formation of collapsed endosomes. These results suggest that impaired endosomal tubulation could be linked to SPG8 pathogenesis.
Fig. 4. Pathogenic SPG8 mutations in strumpellin induce defects in endosomal fission and CAV1 maintenance.
(A) Schematic diagram of strumpellin showing locations of reported SPG8 and Ritscher-Schinzel/3C syndrome (RSS) mutations. (B) hTERT-RPE1 cells stably expressing either empty vector (−) or one of the indicated WT or SPG8 missense mutant forms of siRNA-resistant 3×FLAG-tagged strumpellin (CDH-3xFLAG-strumpellin-r) were transfected with control (CTL) or strumpellin siRNAs and immunoblotted. (C) Relative intensities of strumpellin immunoreactive signals were measured and normalized to GAPDH from four independent experiments. Graph shows means ± SD. (D) Cells from (B) were transfected with the indicated siRNAs and then immunostained for the endosomal marker sorting nexin 1 (SNX1, green) and labelled with DAPI. Arrowheads indicate ends of endosomal tubules. (E) Quantification of cells in (D) with at least one SNX1-positive tubule longer than 2 μm. n = 3 independent experiments with 100 cells per group. (F) Cells from (B) were transfected with the indicated siRNAs and then immunostained for CAV1 (green) and labelled with DAPI. (G) Quantification of cells lacking CAV1-positive vesicles in the cytoplasm. n = 3 independent experiments with 200 cells per group. Graphs show means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. P values were obtained by Kruskal-Wallis test with Dunnett’s post hoc test. Scale bars, 10 μm; 2 μm (inset).
Next, we examined whether CAV1 was affected by SPG8-associated mutations in WASHC5. Strikingly, all tested SPG8 missense mutants failed to rescue the reduction of CAV1 abundance caused by strumpellin depletion (Fig. 4B). In addition, although the expression of wild-type strumpellin rescued the loss of CAV1-positive vesicles in strumpellin-depleted cells, expression of none of the SPG8 mutants rescued this phenotype (Fig. 4, F and G). Endosomal recycling is a critical process in determining the destination of endocytosed cargoes for recycling or degradation (19, 22). Given that endosomal tubules appear temporarily during endosomal recycling, the increased lysosomal degradation of CAV1 in cells expressing SPG8-associated strumpellin mutants could be due to defects in endosomal sorting. Altogether, these results indicate that expression of SPG8-associated strumpellin mutants impaired fission of endosomal tubules and maintenance of CAV1 protein levels.
SPG8-associated mutations impair integrin-mediated cell adhesion
Caveolae are critical for the internalization of various cargoes, including integrins, monosialotetrahexosylgangliosides (GM1 gangliosides), small viruses, and cholera toxin-B (CT-B) (28, 33, 34). Among the known caveolar cargoes we selected the trafficking of integrins, the major receptor for extracellular matrix (ECM) ligands, for examination because the WASH complex also regulates cell adhesion and migration through integrin trafficking (13, 16). In control cells, α5 integrin primarily localized to the ends of actin filaments, corresponding to focal adhesions (Fig. 5A, arrowheads). However, depletion of strumpellin mostly abolished α5 integrin localization at focal adhesions, as also shown previously in WASH1-depleted cells (13, 16); the intracellular pool of α5 integrin increased (Fig. 5A, arrows), reflecting a failure in integrin recycling (35). Intracellular α5 integrin mainly colocalized with the lysosomal marker LAMP1 in strumpellin-depleted cells (Fig. 5B), suggesting that loss of strumpellin facilitated lysosomal degradation of α5 integrin. Indeed, total protein amounts of α5 integrin were reduced in strumpellin-depleted cells (Fig. 5C). As expected, treatment with Baf A1 restored α5 integrin abundance in strumpellin-depleted cells to approximately those in control cells, further indicating that depletion of strumpellin facilitated lysosomal degradation of α5 integrin. On the other hand, trafficking of CT-B, another cargo of caveolae, was not affected by strumpellin depletion (Fig. 5D). CT-B strongly accumulated at the Golgi apparatus even after strumpellin depletion, indicating that CT-B was actively internalized from the cell surface in a strumpellin-independent manner. These results demonstrate that strumpellin selectively affected specific types of caveolar-mediated trafficking. Furthermore, α5 integrin distribution to focal adhesions was rescued upon expression of wild-type strumpellin, but not by the the expression of SPG8-associated strumpellin mutants (Fig. 5A), suggesting that caveolar-mediated trafficking of α5 integrin may be linked to SPG8 pathogenesis.
Fig. 5. Pathogenic SPG8 mutations perturb integrin-mediated cell adhesion.

(A) hTERT-RPE1 cells stably expressing empty vector (−) or siRNA-resistant 3×FLAG-tagged strumpellin, wild-type strumpellin, or the indicated SPG8 missense strumpellin mutants were transfected with the control (CTL) or strumpellin siRNAs and stained for α5 integrin (green) and Alexa 555-conjugated phalloidin A (actin, red). Arrows indicate the intracellular pool of α5 integrin; arrowheads indicate focal adhesions. Images are representative of three independent experiments. (B) hTERT-RPE1 cells were transfected with the indicated siRNAs and immunostained for α5 integrin (green) and LAMP1 (red). Images are representative of three independent experiments. (C) hTERT-RPE1 cells transfected with the indicated siRNAs were incubated in the absence or presence of Baf A1 for 16 h, then cells were lysed and immunoblotted for the indicated proteins. The band intensities for α5 integrin were determined and normalized to the corresponding α-tubulin intensities. Graph shows means ± SD of 5 independent experiments. (E) hTERT-RPE1 cells transfected with the indicated siRNAs were incubated with FITC-conjugated CT-B at 4 °C for 30 min. then warmed to 37 °C for 1 h to allow CT-B internalization prior to fixation. Cells were then immunostained for the Golgi marker GM130 (red). Images are representative of three independent experiments. (F) Suspended cells were plated on coverslips coated with fibronectin and allowed to spread for 45 min before fixation and immunostaining for Alexa 555-conjugated phalloidin A (actin, red) and labelling with DAPI. (G) Spread cell area (μm2) in (F) was measured from more than 30 cells for each group from 3 independent experiments. The average for each group was divided by the average for control cells. Graphs express means ± SD. *p < 0.05. P values were calculated by the Wilcoxon rank-sum test for (D) and Kruskal-Wallis test with Dunnett’s post hoc test for (G). Scale bars, 10 μm.
Because integrins play a critical role in cell adhesion by binding to fibronectin in the ECM (36), we next performed cell spreading assays to examine the role of strumpellin in cell adhesion. Strumpellin-depleted cells spread less on an intermediate concentration of fibronectin (20 μg/ml) than did control cells (Fig. 5, E and F), similar to findings in WASH1-depleted cells (16). This spreading defect was not rescued by expression of SPG8-associated strumpellin mutants, but it was rescued by wild-type strumpellin (Fig. 5, E and F). Together, these results demonstrate that SPG8-associated mutant strumpellin proteins were defective in CAV1-dependent, integrin-mediated cell adhesion, suggesting that SPG8 could be associated with abnormalities in CAV1-dependent, integrin-mediated cell adhesion.
Integrin recycling is mediated by an interaction between strumpellin and CAV1
Because the WASH complex promotes integrin recycling (13, 16, 37), and SPG8-associated strumpellin mutant proteins cause defects in CAV1-dependent, integrin-mediated adhesion, we examined whether CAV1 mediates the role of strumpellin in promoting integrin recycling. We identified the central spectrin-repeat domain of strumpellin (residues 241-790) as a major CAV1-binding region (Fig. 6A), with residues 608-790 being critical for binding to CAV1 in cells (Fig. 6B). Whereas the expression of wild-type strumpellin rescued the reduction in α5 integrin protein abundance caused by the depletion of endogenous strumpellin, the strumpellin deletion mutant that was deficient in CAV1 binding failed to rescue (Fig. 6, C and D). These results indicate that the interaction of strumpellin with CAV1 mediates the recycling of α5 integrin. Eight of eleven pathogenic SPG8 mutations are located in the central spectrin domain of strumpellin, and five of those mutations are located between residues 608 and 790 within this domain (Fig. 4A). The fact that the regions in which pathogenic SPG8 mutations are concentrated are required for CAV1 binding strongly supports the notion that SPG8 etiology is closely linked to CAV1-dependent, integrin-mediated cell adhesion.
Fig. 6. Integrin recycling is mediated by the interaction of strumpellin with CAV1.

(A) HEK293T cells were co-transfected with HA-CAV1 and the indicated individual Myc-tagged-strumpellin constructs. Lysates were immunoprecipitated (IP) with antibodies specific for HA, then immunoblotted with antibodies against the Myc and HA tags. Blots are representative of five independent experiments. (B) HEK293T cells were co-transfected as indicated, and HA immunoprecipitates were immunoblotted for Myc and HA. Blot is representative of five independent experiments. (C) HeLa cells expressing control (CTL) or strumpellin siRNAs were transfected with empty Myc-tagging vector (−) or Myc-tagged siRNA-resistant strumpellin (Myc-strumpellin-r) expression constructs. After 24 hours, the cells were lysed and immunoblotted for the indicated proteins. (D) Band intensities for α5 integrin were determined from (C) and normalized against the corresponding α-tubulin intensities. Graph shows means ± SD of 5 independent experiments. *p < 0.05, **p < 0.01. P values were calculated by Kruskal-Wallis test with Dunnett’s post hoc test.
The actin nucleation activity of WASH1 is required for CAV1-dependent, integrin-mediated cell adhesion
Given that endosomal recycling mediated by the WASH complex depends on the actin nucleation activity of WASH1 (13, 35), we examined whether the restoration of actin nucleation activity at endosomes was sufficient to rescue the CAV1-dependent, integrin-mediated cell adhesion defects in WASH complex–depleted cells. To restore actin nucleation activity to endosomes in strumpellin- or WASH1-depleted cells, we stably expressed the WASH1 VCA motif, which can stimulate actin nucleation (Fig. 7, A and B), fused to a C-terminal fragment of FAM21 (Δ356) for endosomal targeting (35). The stably-expressed, FLAG-tagged FAM21Δ356-WASH1-VCA fusion protein reversed the reduction of CAV1 abundance caused by depletion of WASH1 or strumpellin (Fig. 7A). The FAM21Δ356-WASH1-VCA fusion protein also rescued CAV1 localization at intracellular vesicles (Fig. 7, B and C). These results indicate that the actin nucleation activity of WASH1 was required for maintenance of CAV1 at intracellular membranes.
Fig. 7. The actin nucleation activity of WASH1 is required for CAV1-dependent, integrin-mediated cell adhesion.

(A) To rescue actin nucleation activity at endosomes, hTERT-RPE1 cells stably expressing the WASH1 VCA motif fused to the C-terminal domain of FAM21 (FAM21Δ356-WASH-VCA) were generated. Cells were transfected with the control (CTL), strumpellin, or WASH1 siRNAs and immunoblotted for the indicated proteins. Blots are representative of three independent experiments. (B) Cells in (A) were transfected with the indicated siRNAs, then immunostained for FLAG epitope (green) and endogenous CAV1 (red). (C) Cells in (B) lacking CAV1-positive vesicles in the cytoplasm were counted. n = 3 independent experiments with 100 cells per group. *p < 0.05, **p < 0.01. P values were calculated using the Kruskal-Wallis test with Dunnett’s post hoc test. Scale bar, 10 μm.
Discussion
Despite numerous efforts to identify the function of strumpellin, the effects of mutations underlying SPG8 at the cellular level have remained unclear. We sought to identify proteins that interact with strumpellin through an unbiased proteomics approach and found that strumpellin interacted with CAV1, a protein that regulates trafficking of various cargoes, including integrins. Depletion of strumpellin resulted in a concomitant decrease in CAV1 protein in both heterologous cell lines and primary cortical neurons, with impairments in integrin localization and fibronectin-mediated cell adhesion. We sought to model SPG8 at the cellular level by stably expressing pathogenic mutants of strumpellin in a background in which endogenous strumpellin was depleted. Using this approach, we identified an abnormal cellular phenotype for the pathogenic SPG8 missense mutants -- defective CAV1-dependent, integrin-mediated cell adhesion. Our results indicate that aberrant CAV1-dependent, integrin-mediated cell adhesion might underlie the axon degeneration that occurs in SPG8 patients.
CAV1 is a principal component of caveolae, small invaginations of the plasma membrane that have various cellular functions in endocytosis, exocytosis, lipid regulation, and mechanosensation (29, 34). To investigate the effects of decreases in CAV1 abundance induced by strumpellin depletion, we examined internalization of the well-characterized caveolar cargoes CT-B and integrin. Only integrin localization was altered upon strumpellin depletion, suggesting that strumpellin specifically affects CAV1-mediated sorting pathways (Fig. 5, A and E). This differential regulatory mechanism of strumpellin appears to originate from its role in endosomal recycling. Expression of four different SPG8 missense mutant strumpellin proteins showed a similar endosomal tubulation defect (Fig. 4D), which is known to induce aberrant endosomal recycling. Indeed, lysosomal degradation of CAV1 and integrin was accelerated in strumpellin-depleted cells (Fig. 3B and 5C). Because the gene products implicated in several other forms of HSP, including the most common SPG4 form, are also involved in endosomal tubulation and impact lysosomal function, this pathway could represent an important converging theme in HSP pathogenesis (21).
Previous studies have identified disturbances of integrin trafficking and cell adhesion in WASH-depleted cells (13, 16), and we now demonstrate that the WASH complex can positively regulate integrin-mediated cell adhesion through CAV1. Our results further indicate that strumpellin fine-tunes CAV1 protein levels by inhibiting its lysosomal degradation (Fig. 8), perhaps akin to its role in low-density lipoprotein receptor sorting and lysosomal degradation (18). Interestingly, valosin-containing protein (VCP), which interacts with strumpellin (38, 39), targets mono-ubiquitylated CAV1 for degradation. Depletion or inhibition of VCP blocks endolysosomal sorting of CAV1, a prerequisite for its degradation (40, 41). Furthermore, the ANKRD13 family of ubiquitin-interacting motif–bearing proteins facilitates VCP-mediated lysosomal trafficking of CAV1 (42). Meanwhile, the related protein ANKRD50 has been identified as an essential component of the SNX27-retromer-WASH supercomplex, which prevents entry into the degradative lysosomal pathway (43). Perhaps strumpellin and VCP work antagonistically to fine-tune CAV1 abundance, which could be particularly important for HSP pathogenesis more generally, given that bioinformatic analyses have suggested VCP as a central player in HSP-related neurodegeneration (44).
Fig. 8. Schematic representation of the pathogenic mechanism for SPG8 mutations.

(A) The WASH complex promotes endosomal recycling of CAV1 and integrin through the actin nucleation activity of WASH1, which is mediated by the VCA domain. Lysosomal degradation of CAV1 is prevented by the physical interaction with strumpellin at the endosome. The interaction of CAV1 with the WASH complex also inhibits lysosomal degradation of integrin, a cargo of CAV1. The WASH complex cooperates with CAV1 to promote integrin trafficking to focal adhesions, inducing integrin-mediated cell adhesion. (B) Loss of strumpellin accelerates lysosomal degradation of CAV1 and integrin and inhibits the actin-nucleating activity of WASH1, thereby impairing CAV1-dependent integrin recycling and integrin-mediated cell adhesion. This defect in adhesion could ultimately lead to length-dependent axonopathy, a defining characteristic of HSP.
How might defects in CAV1-dependent, integrin-mediated cell adhesion ultimately result in the distal axonopathy that defines the HSPs? One possibility is through impairment of the pro-survival effects of CAV1 (45), which is highly abundant in both neurons and glia. For instance, CAV1 enhances pro-survival and pro-growth signaling by concentrating neurotransmitter and neurotrophin receptors at membrane microdomains and/or lipid rafts, thus modulating signaling pathways such as the formation of cyclic adenosine monophosphate (cAMP), which has been shown to regulate the growth and guidance of developing axons (46). Furthermore, mice deficient in Cav1 have neurologic motor phenotypes including clasping, abnormal spinning, muscle weakness, and gait abnormalities (47), confirming the importance of CAV1 in motor function. Finally, a study identified heterozygous 1 bp del-A (p.Ile134fsThr137) mutations in CAV1 that segregate with spastic paraparesis, spastic-ataxic gait, congenital cataracts and pigmentary retinopathy in a Brazilian family (48), further buttressing the case for a CAV1-dependent HSP mechanism.
Pathogenic missense mutations associated with SPG8 are most generally considered to exert a dominant-negative or gain-of-function effect, because SPG8 is an autosomal dominant HSP (6, 49). Furthermore, heterozygous WASHC5+/− mice do not show HSP-related phenotypes, suggesting heterozygous loss of SPG8 might not lead to haploinsufficiency (32). Moreover, expression of some SPG8 missense mutants do not cause alterations in endosomal tubulation, endosomal recycling, or WASH complex assembly, which are known to be induced by strumpellin depletion (26, 38). On the other hand, depletion of strumpellin in zebrafish impairs motility, and siRNA knockdown in human neuroblastoma SH-SY5Y cells reduces neurite length (38). A case report describing a frameshift deletion mutation in WASHC5 in a Chinese subject with spastic paraparesis (25) could be compatible with a haploinsufficiency disease mechanism, though the Genome Aggregation Database (gnomAD; https://gnomad.broadinstitute.org) lists dozens of frameshift or stop gain mutations in WASHC5, plus numerous splice site mutations, in normal subjects, which would argue strongly against haploinsufficiency as a disease mechanism for SPG8.
Even so, the range of disease phenotypes due to different types of WASHC5 mutations may also be more complex. A biallelic splice-site mutation at the C-terminus has been reported in numerous members of a First Nations community in northern Manitoba, Canada with Ritscher-Schinzel/3C syndrome (RSS), a disorder characterized by distinctive craniofacial features as well as cerebellar and cardiac abnormalities (50). This mutation, c.3335+2T>A, results in skipping of exon 27 near the C-terminus, and one patient sample showed a 60% reduction in strumpellin protein. Thus, it seems likely that different types of strumpellin mutations could be linked to distinct clinical phenotypes.
Taken together, our results suggest that defects in the CAV1-dependent, integrin-mediated adhesion pathway could account for the pathogenic mechanism of HSP caused by the most compelling SPG8 missense mutations. In the future, the various roles that different types of WASHC5 mutations play in different phenotypic presentations will be important to correlate with cellular alterations in strumpellin function.
Materials and Methods
DNA constructs
Human WASHC5/KIAA0196 (GenBank NM_014846), FAM21 (GenBank BC082258), SWIP (GenBank NM_001293640), WASH1 (GenBank NM_182905), CCDC53 (GenBank NM_016053), and CAV1 (GenBank NM_001753) cDNAs were obtained from Open Biosystems. The CAV1 cDNA was subcloned into the GW1-HA vector (51). FLAG epitope-tagged FAM21, strumpellin, WASH1, and CCDC53 were previously described (19). WASHC5/KIAA0196 was subcloned into pCDH-CMV-MCS-EF1-Puro-ZTF and pCDH-CMV-MCS-EF1-Puro-3×FLAG lentiviral vectors (SBI; the sequence for ZTF or 3×FLAG was inserted into XbaI/EcoRI sites). Strumpellin cDNA with SPG8 missense mutations and FAM21Δ356-WASH-VCA were also subcloned into the pCDH-CMV-MCS-EF1-Puro-3×FLAG lentiviral vector. siRNA-resistant strumpellin constructs were generated by introducing four silent, mismatched mutations against the siStrumpellin target sequence. All constructs were confirmed by DNA sequencing. Oligoduplex shRNAs for rat strumpellin (GenBank accession: XM_021791095) were created by annealing the paired oligomers. Strumpellin shRNA-1: 5’- TGCTGGACACTGCACAATTTTTCAAGAGAAAATTGTGCAGTGTCCAGCTTTTTTC-3’ and 5’- tcgaGAAAAAAGCTGGACACTGCACAATTTTCTCTTGAAAAATTGTGCAGTGTC CAGCA-3’; Strumpellin shRNA-2: 5’- TGCACCTGTGGATGAGTCTATTCAAGAGATAGA CTCATCCACAGGTGCTTTTTTC-3’ and 5’- tcgaGAAAAAAGCACCTGTGGATGAGTCT ATCTCTTGAATAGACTCATCCACAGGTGCA-3’. The oligoduplexes were then directly cloned into XhoI and HpaI sites of a modified pLentiLox 3.7 vector (gift from Dr. Zheng Li) (52).
Antibodies
The following antibodies were used in this study: anti-FLAG (M2, mouse, Sigma-Aldrich), anti-HA (F-7, mouse, Santa Cruz Biotechnology), anti-HA (Y-11, rabbit, Santa Cruz Biotechnology), anti-CAV1 (D46G3, rabbit, Cell Signaling), anti-CAV1 (2297, mouse, BD Biosciences), anti-strumpellin (C-14, rabbit, Santa Cruz Biotechnology), anti-FAM21 (polyclonal, rabbit, EMD Millipore), anti-CCDC53 (polyclonal, rabbit, EMD Millipore), anti-WASH1 (polyclonal, rabbit, EMD Millipore), anti-SWIP (polyclonal, rabbit, EMD Millipore), anti-GAPDH (6C5, mouse, Santa Cruz Biotechnology), Alexa 555-conjugated phalloidin A (Molecular Probes), anti-GM130 (35, mouse, BD Biosciences), anti-c-myc (polyclonal, goat, Bethyl), anti-α5 integrin (CD49e, mouse, BD Biosciences), anti-α5 integrin (C-9, mouse, Santa Cruz Biotechnology), anti-LAMP1 (H4A3, mouse, abcam), anti-α-tubulin (1E4C11, mouse, Proteintech), anti-β-tubulin (D66, mouse, Sigma-Aldrich), anti-SNX1 (51, mouse, BD Biosciences), FITC-conjugated CT-B (Molecular Probes), and anti-GFP (598, rabbit, MBL). The working dilutions for each antibody are listed in table S2.
Chemicals
Cycloheximide (C4859) and Baf A1 (19-148) were purchased from Sigma-Aldrich and Millipore, respectively.
Cell culture, transfection and RNA interference
hTERT-RPE1 and HEK293T cells (ATCC) were cultured in DMEM/F-12 and DMEM (Gibco), respectively, supplemented with 10% fetal bovine serum (Gibco) and maintained at 37 °C in a 5% CO2 humidified incubator. Fibroblasts derived from human subjects were cultured in DMEM supplemented with 10% fetal bovine serum. Cells were transfected with Avalanche-Omni (EZ Biosystems) for plasmid DNAs and Lipofectamine RNAiMax (Invitrogen) for siRNAs, as per the instructions of the manufacturers. The following siRNAs were used: siStrumpellin (Dharmacon): 5’-GGAUGAGUCUGUAACGUUU-3’; siWASH1 (Ambion): 5’-ACUACUUCUAUGUGCCAGA-3’. Control siRNAs were from Ambion.
Generation of stable cell lines
WT strumpellin, SPG8 strumpellin mutants, or FAM21Δ356-WASH1-VCA fusion proteins were stably expressed in hTERT-RPE1 or human fibroblast cells using lentiviral infection as described previously (53). Cells infected with the indicated viruses were selected using 20 μg/ml of puromycin.
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting were performed as previously described (54). Immunoreactive proteins were revealed using Amersham ECL Prime (GE Healthcare). Images were obtained and processed using a ChemiDoc XRS+ System with Image Lab software (Bio-Rad).
Tandem-affinity purification and mass spectrometry
hTERT-RPE1 cells stably expressing ZZ-TEV-FLAG or ZZ-TEV-FLAG-strumpellin were grown on eight 100-mm culture plates. Cells were washed with PBS and then lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EGTA [pH 8.0], 1.5 mM EDTA [pH 8.0], 1% Triton X-100, Complete Protease Inhibitor Cocktail [Roche], and PhosSTOP phosphatase inhibitor cocktail [Roche]) on ice for 30 min. Lysates were clarified by centrifugation (15,700 g, 4 °C) for 15 min, and supernatants were incubated with IgG Sepharose 6 Fast Flow (GE Healthcare) at 4 °C for 5 h. Beads were washed 3 times with lysis buffer, and incubated with AcTEV Protease (Thermo Fisher Scientific) in lysis buffer at 4 °C for 16 h. Supernatants collected after spinning down the beads were incubated with anti-FLAG M2 Affinity Gel (Sigma-Aldrich) at 4 °C for 5 h, and the beads were washed 3 times with lysis buffer. Beads were then incubated with lysis buffer containing 100 mg/ml of 3× FLAG Peptide (Sigma-Aldrich) at 25 °C for 1 h, and eluted proteins were collected and digested with trypsin overnight at 37 °C. Tryptic peptides were desalted and analyzed on a nano-LC/MS/MS system with an Ultimate 3000 HPLC (Thermo-Dionex) connected to an Orbitrap Elite mass spectrometer via an Easy-Spray ion source (Thermo Fisher Scientific). Peptides were separated on an ES800 Easy-Spray column (75 μm inner diameter, 15 cm length, 3 μm C18 beads; Thermo Fisher Scientific) at a flow rate of 300 nl/min with a 25-min linear gradient of 2–30% mobile phase B (98% acetonitrile, 0.1% formic acid). The Orbitrap Elite mass spectrometer was operated in a positive nano-electrospray mode. Resolution of the survey scan was set for 60k at m/z 400, with a target value of 106 ions. The m/z range for MS scans was 300–1600. MS/MS data were acquired in a data-dependent mode, and the two most abundant ions were selected for product ion analysis. Data are summarized in Table 1 and available at https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=0bd8015ecc3d4076b8878133b46888ee.
Immunofluorescence staining and confocal microscopy
hTERT-RPE1 cells and primary human fibroblasts cultured on cover slips were fixed with 4% paraformaldehyde for 15 min. Fixed cells were blocked with 3% normal donkey serum in PBSS (PBS with 0.05% saponin) for 15 min. Next, cells were incubated with primary antibodies diluted in blocking solution for 2 h, then washed four times with PBSS, incubated with Alexa Fluor-conjugated secondary antibodies (Molecular Probes) diluted in blocking solution for 45 min, and washed with PBSS four times. Actin was visualized using Alexa-conjugated phalloidin A. DAPI solution (Molecular Probes) was applied to the cells to stain the nucleus, and cover slips were mounted on glass slides using Fluoromount-G (SouthernBiotech). Cells were observed using a Zeiss LSM710 confocal microscope with a 63× 1.4 NA Plan-Apochromat lens. ZEN 2009 (Carl Zeiss Microimaging) and ImageJ (NIH) software packages were used to acquire and process all the images.
Quantitative real-time PCR
RNA and cDNA were prepared as previously described (19). Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (Invitrogen) using an Applied Biosystems StepOnePlus System (Applied Biosystems). Experiments were performed in quadruplicate using three independent cDNAs. The results were normalized to GAPDH and calculated following the 2−ΔΔCt method (55). The following primer sequences were used: GAPDH-F: 5’-GGTCGGAGTC AACGGATTTGGTCG-3'; GAPDH-R: 5’-CCTCCGACGCCTGCTTCACCAC-3’; CAV1-F: 5’-CGTAGACTCGGAGGGACATC-3’; CAV1-R: 5’-GTTGAGGTGTTTAGGGTCGC-3’.
Cell spreading assay
Cell spreading assays were performed as previously described (16). Briefly, suspended cells were seeded onto cover slips coated with fibronectin (20 μg/ml) for 45 min. Adherent cells were fixed with 4% paraformaldehyde, and then stained with Alexa-conjugated phalloidin A. Spread cell areas were measured using ImageJ software.
CT-B uptake assay
hTERT-RPE1 cells grown on cover slips were washed with ice-cold culture media, then incubated with FITC-conjugated CT-B (Molecular Probes) at 4 °C for 30 min. After washing with warm culture media, cells were placed at 37 °C for 1 h to permit CT-B internalization (56). Cells were then fixed with 4% paraformaldehyde and immunostained.
Neuronal cultures
Animal studies were approved by the NINDS/NIDCD Animal Care and Use Committee. Rat cerebral cortical neurons were prepared from E18 embryos, plated at densities of 2×104/cm2 on 18-mm cover slips or 2×106/well on 6-well plates, and maintained as described previously (57). Viral transductions were performed 1 h after cells were plated. Cultured neurons were harvested using RIPA buffer supplemented with protease inhibitors (Roche) at day 5 in culture, followed by brief sonication. Proteins (30 μg per well) were subjected to SDS-PAGE and immunoblotted.
Statistical analysis
Data were tested for normality with the Shapiro-Wilk test. Continuous variables between two groups were compared using the Wilcoxon rank-sum test. To establish differences in numerical values among multiple groups, the Kruskal-Wallis test was performed along with Dunnett’s post hoc test using ranks. P < 0.05 was considered significant.
Supplementary Material
Fig. S1. CAV1 expression in WASH complex knockdown cells.
Fig. S2. SPG8 strumpellin proteins suppress abnormal aggregation of WASH complex components induced by strumpellin depletion.
Table S1. Mass spectrometry interaction data for hTERT-RPE1 cells stably expressing ZZ-TEV-FLAG-strumpellin.
Table S2. Antibodies used in this study
Acknowledgements:
We thank Dr. Yan Li (NINDS Protein/Peptide Sequencing Facility) for assistance with mass spectrometry and Young Ju Suh (College of Medicine, Inha University) for statistical analysis consultation.
Funding: This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke (to S.L., P.-P.Z., C.B., and J.C.) and the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2016R1A6A3A04006478, NRF-2014R1A5A2009392 to S.L., NRF-2019R1F1A1051699, NRF-2019R1A5A2026045 to J.C).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials, and the mass spectrometry dataset is available at https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=0bd8015ecc3d4076b8878133b46888ee.
References and Notes
- 1.Blackstone C, Hereditary spastic paraplegia. Handb. Clin. Neurol 148, 633–652 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Fink JK, Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 126, 307–328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Souza PVS, de Rezende Pinto WBV, de Rezende Batistella GN, Bortholin T, Oliveira ASB, Hereditary spastic paraplegia: clinical and genetic hallmarks. Cerebellum 16, 525–551 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Tesson C, Koht J, Stevanin G, Delving into the complexity of hereditary spastic paraplegias: how unexpected phenotypes and inheritance modes are revolutionizing their nosology. Hum. Genet. 134, 511–538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackstone C, Converging cellular themes for the hereditary spastic paraplegias. Curr. Opin. Neurobiol 51, 139–146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valdmanis PN, Meijer IA, Reynolds A, Lei A, MacLeod P, Schlesinger D, Zatz M, Reid E, Dion PA, Drapeau P, Rouleau GA, Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am. J. Hum. Genet 80, 152–161 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettencourt C, Morris HR, Singleton AB, Hardy J, Houlden H, Exome sequencing expands the mutational spectrum of SPG8 in a family with spasticity responsive to L-DOPA treatment. J. Neurol 260, 2414–2416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichinose Y, Koh K, Fukumoto M, Yamashiro N, Kobayashi F, Miwa M, Nagasaka T, Shindo K, Ishiura H, Tsuji S, Takiyama DY, Exome sequencing reveals a novel missense mutation in the KIAA0196 gene in a Japanese patient with SPG8. Clin. Neurol. Neurosurg 144, 36–38 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD, WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl. Acad. Sci. U. S. A 107, 10442–10447 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotty JD, Wu C, Bear JE, New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol 14, 7–12 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A, The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Gomez TS, Billadeau DD, A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zech T, Calaminus SDJ, Caswell P, Spence HJ, Carnell M, Insall RH, Norman J, Machesky LM, The Arp2/3 activator WASH regulates α5β1-integrin-mediated invasive migration. J. Cell Sci 124, 3753–3759 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carnell M, Zech T, Calaminus SD, Ura S, Hagedorn M, Johnson SA, May RC, Soldati T, Machesky LM, Insall RH, Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J. Cell Biol 193, 831–839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez TS, Gorman JA, de Narvajas AA, Koenig AO, Billadeau DD, Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol. Biol. Cell 23, 3215–3228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duleh SN, Welch MD, Regulation of integrin trafficking, cell adhesion, and cell migration by WASH and the Arp2/3 complex. Cytoskeleton (Hoboken) 69, 1047–1058 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, Tavaré JM, Cullen PJ, A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol 15, 461–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartuzi P, Billadeau DD, Favier R, Rong S, Dekker D, Fedoseienko A, Fieten H, Wijers M, Levels JH, Huijkman N, Kloosterhuis N, van der Molen H, Brufau G, Groen AK, Elliott AM, Kuivenhoven JA, Plecko B, Grangl G, McGaughran J, Horton JD, Burstein E, Hofker MH, van de Sluis B, CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat. Commun 7, 10961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Chang J, Blackstone C, FAM21 directs SNX27-retromer cargoes to the plasma membrane by preventing transport to the Golgi apparatus. Nat. Commun 7, 10939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyrrell BJ, Woodham EF, Spence HJ, Strathdee D, Insall RH, Machesky LM, Loss of strumpellin in the melanocytic lineage impairs the WASH Complex but does not affect coat colour. Pigment Cell Melanoma Res. 29, 559–571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allison R, Edgar JR, Pearson G, Rizo T, Newton T, Günther S, Berner F, Hague J, Connell JW, Winkler J, Lippincott-Schwartz J, Beetz C, Winner B, Reid E, Defects in ER-endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol 216, 1337–1355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burd C, Cullen PJ, Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol 6, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeman CL, Hesketh G, Seaman MNJ, RME-8 coordinates the activity of the WASH complex with the function of the retromer SNX dimer to control endosomal tubulation. J. Cell Sci 127, 2053–2070 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chrestian N, Dupré N, Gan-Or Z, Szuto A, Chen S, Venkitachalam A, Brisson JD, Warman-Chardon J, Ahmed S, Ashtiani S, MacDonald H, Mohsin N, Mourabit-Amari K, Provencher P, Boycott KM, Stavropoulos DJ, A Dion P, Ray PN, Suchowersky O, Rouleau GA, Yoon G, Clinical and genetic study of hereditary spastic paraplegia in Canada. Neurol. Genet. 3, e122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma L, Shi Y, Chen Z, Li S, Qin W, Zhang J, A novel KIAA0196 mutation in a Chinese patient with spastic paraplegia 8: a case report. Medicine (Baltimore) 97, e10760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman C, Seaman MNJ, Reid E, The hereditary spastic paraplegia protein strumpellin: characterisation in neurons and of the effect of disease mutations on WASH complex assembly and function. Biochim. Biophys. Acta 1832, 160–173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamaze C, Tardif N, Dewulf M, Vassilopoulos S, Blouin CM, The caveolae dress code: structure and signaling. Curr. Opin. Cell Biol 47, 117–125 (2017). [DOI] [PubMed] [Google Scholar]
- 28.del Pozo MA, Balasubramanian N, Alderson NB, Kiosses WB, Grande-García A, ANderson RGW, Schwartz MA, Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat. Cell Biol 7, 901–908 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parton RG, Simons K, The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol 8, 185–194 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Hayer A, Stoeber M, Ritz D, Engel S, Meyer HH, Helenius A, Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J. Cell Biol 191, 615–629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pol A, Martin S, Fernández MA, Ingelmo-Torres M, Ferguson C, Enrich C, Parton RG, Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol. Biol. Cell 16, 2091–2105 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jahic A, Khundadze M, Jaenisch N, Schüle R, Klimpe S, Klebe S, Frahm C, Kassubek J, Stevanin G, Schöls L, Brice A, Hübner CA, Beetz C, The spectrum of KIAA0196 variants, and characterization of a murine knockout: implications for the mutational mechanism in hereditary spastic paraplegia type SPG8. Orphanet J. Rare Dis 10, 147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi F, Sottile J, Caveolin-1-dependent β1 integrin endocytosis is a critical regulator of fibronectin turnover. J. Cell Sci 121, 2360–2371 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng JPX, Nichols BJ, Caveolae: one function or many? Trends Cell Biol. 26, 177–189 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Hao Y-H, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR, Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell 152, 1051–1064 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul NR, Jacquemet G, Caswell PT, Endocytic trafficking of integrins in cell migration. Curr. Biol 25, R1092–1105 (2015). [DOI] [PubMed] [Google Scholar]
- 37.McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, Langton P, Pearson N, Danson CM, Nägele H, Morris LL, Singla A, Overlee BL, Heesom KJ, Sessions R, Banks L, Collins BM, Berger I, Billadeau DD, Burstein E, Cullen PJ, Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol 19, 1214–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clemen CS, Tangavelou K, Struckberg K-H, Just S, Gaertner L, Regus-Leidig H, Stumpf M, Reimann J, Coras R, Morgan RO, Fernandez M-P, Hofmann A, Müller S, Schoser B, Hanisch F-G, Rottbauer W, Blümcke I, von Hörsten S, Eichinger L, Schröder R, Strumpellin is a novel valosin-containing protein binding partner linking hereditary spastic paraplegia to protein aggregation diseases. Brain 133, 2920–2941 (2010). [DOI] [PubMed] [Google Scholar]
- 39.de Bot ST, Schelhaas HJ, Kamsteeg EJ, van de Warrenburg BP, Hereditary spastic paraplegia caused by a mutation in the VCP gene. Brain 135, e223; author reply e224 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Ritz D, Vuk M, Kirchner P, Bug M, Schütz S, Hayer A, Bremer S, Lusk C, Baloh RH, Lee H, Glatter T, Gstaiger M, Aebersold R, Weihl CC, Meyer H, Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol 13, 1116–1123 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirchner P, Bug M, Meyer H, Ubiquitination of the N-terminal region of caveolin-1 regulates endosomal sorting by the VCP/p97 AAA-ATPase. J. Biol. Chem 288, 7363–7372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burana D, Yoshihara H, Tanno H, Yamamoto A, Saeki Y, Tanaka K, Komada M, The Ankrd13 Family of ubiquitin-interacting motif-bearing proteins regulates valosin-containing protein/p97 protein-mediated lysosomal trafficking of caveolin 1. J. Biol. Chem 291, 6218–6231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kvainickas A, Orgaz AJ, Nägele H, Diedrich B, Heesom KJ, Dengjel J, Cullen PJ, Steinberg F, Retromer- and WASH-dependent sorting of nutrient transporters requires a multivalent interaction network with ANKRD50. J. Cell Sci 130, 382–395 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mancour L, Masri A, Kayserili H, Al-Aama JY, Abdel-Salam GMH, Karminejad A, Kara M, Kara B, Bozorgmehri B, Ben-Omran T, Mojahedi F, El Din Mahmoud IG, Bouslam N, Bouhouche A, Benomar A, Hanein S, Raymond L, Forlani S, Mascaro M, L Selim, Shehata N, Al-Allawi N, Bindu PS, Azam M, Gunel M, Caglayan A, Bilguvar K, Tolun A, Issa MY, Schroth J, Spencer EG, Rosti RO, Akizu N, Vaux KK, Johansen A, Koh AA, Megahed H, Durr A, Brice A, Stevanin G, Gabriel SB, Ideker T, Gleeson JG, Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 343, 506–511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Head BP, Hu Y, Finley JC, Saldana MD, Bonds JA, Miyanohara A, Niesman IR, Ali SS, Murray F, Insel PA, Roth DM, Patel HH, Patel PM, Neuron-targeted caveolin-1 protein enhances signaling and promotes arborization of primary neurons. J. Biol. Chem 286, 33310–33321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray AJ, Tucker SJ, Shewan DA, cAMP-dependent axon guidance is distinctly regulated by Epac and protein kinase A. J. Neurosci 29, 15434–15444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trushina E, Du Charme J, Parisi J, McMurray CT, Neurological abnormalities in caveolin-1 knock out mice. Behav. Brain Res 172, 24–32 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Souza PVS, Bortholin T, Dias RB, Chieia MAT, Burlin S, Naylor FGM, Pinto WBVR, Oliveira ASB, New genetic causes for complex hereditary spastic paraplegia. J. Neurol. Sci 379, 283–292 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Hedera P, Rainier S, Alvarado D, Zhao X, Williamson J, Otterud B, Leppert M, Fink JK, Novel locus for autosomal dominant hereditary spastic paraplegia, on chromosome 8q. Am. J. Hum. Genet 64, 563–569 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elliott AM, Simard LM, Coghlan G, Chudley AE, Chodirker BN, Greenberg CR, Burch T, Ly V, Hatch GM, Zelinski T, A novel mutation in KIAA0196: identification of a gene involved in Ritscher-Schinzel/3C syndrome in a First Nations cohort. J. Med. Genet 50, 819–822 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Zhu P-P, Patterson A, Lavoie B, Stadler J, Shoeb M, Patel R, Blackstone C, Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J. Biol. Chem 278, 49063–49071 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Hu Z, Yu D, Gu Q.-h., Yang Y, Tu K, Zhu J, Li Z, miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat. Commun 5, 3263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee S, Chang J, Renvoisé B, Tipirneni A, Yang S, Blackstone C, MITD1 is recruited to midbodies by ESCRT-III and participates in cytokinesis. Mol. Biol. Cell 23, 4347–4361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang J, Lee S, Blackstone C, Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J. Clin. Invest 124, 5249–5262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmittgen TD, Livak KJ, Analyzing real-time PCR data by the comparative C(T) method. Nat. Proto c 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Oh P, McIntosh DP, Schnitzer JE, Dynamin at the neck of caveolae mediates their budding to form transport vesicles by GTP-driven fission from the plasma membrane of endothelium. J. Cell Biol 141, 101–114 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu P-P, Soderblom C, Tao-Cheng J-H, Stadler J, Blackstone C, SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum. Mol. Genet 15, 1343–1353 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. CAV1 expression in WASH complex knockdown cells.
Fig. S2. SPG8 strumpellin proteins suppress abnormal aggregation of WASH complex components induced by strumpellin depletion.
Table S1. Mass spectrometry interaction data for hTERT-RPE1 cells stably expressing ZZ-TEV-FLAG-strumpellin.
Table S2. Antibodies used in this study