Abstract
Systemic fungal infections pose a serious clinical problem. Treatment options are limited and antifungal drug resistance is increasing. In addition, a substantial proportion of patients do not respond to therapy despite being infected with fungi that are susceptible to the drug. The discordance between overall treatment outcome and low levels of clinical resistance may be attributable to antifungal drug tolerance. In this Review, we define and distinguish resistance and tolerance, and discuss the current understanding of the molecular, genetic and physiological mechanisms that contribute to those phenomena. Distinguishing tolerance from resistance might provide important insights into the reasons for treatment failure in some settings.
Introduction
Drug-resistant fungal infections are emerging as an important clinical problem1,2. This is mainly due to an increase in patients at-risk for invasive fungal infections from complex surgical procedures, immune-suppression or reduced immune function3. Patients who develop invasive fungal infections in the setting of other serious medical conditions frequently have increased mortality rates compared with uninfected patients. For example, the 90-day mortality rate for recipients of solid organ transplants who develop candidemia [G] is between 22%-44%, depending on the Candida species4.
The recent emergence of fungi that are resistant to more than one class of antifungal drug is a serious concern. Currently, only three primary classes of agents are used to treat invasive fungal infections5: polyenes (for example, amphotericin B); azoles (for example, fluconazole) and echinocandins (for example, caspofungin) (Table 1). Thus, one antifungal drug class becoming ineffective in treating a fungal infection owing to resistance reduces the therapeutic options by at least 33%, and frequently by 50% because azoles and echinocandins are not active against all fungi. By comparison, six distinct classes of antibiotics are currently available to treat methicillin-resistant Staphylococcus aureus (MRSA), an important and widespread drug-resistant bacterium6. Thus, the loss of one antifungal drug through resistance has a major impact on patient care.
Table 1. Mechanisms of resistance to clinically relevant classes of antifungal drugs.
| Drug class | Examples of clinically used drugs | Mechanism of action | Target | Mechanism of acquired resistance | Species with intrinsic or high rates of resistance |
|---|---|---|---|---|---|
| Azole | Fluconazole, voriconazole, posaconazole | Inhibition of ergosterol biosynthesis | Lanosterol 14α-demethylase (Erg11 or Cyp51 in other organisms) | Increased efflux pump activity, ERG11 mutations, CYP51 promoter mutations | Candida glabrata, Candida krusei, Candida auris |
| Echinocandins | Caspofungin, micafungin, anidulafungin | Inhibition of β-1,3-glucan synthase | β-1,3-glucan synthase (Fks) | Mutations in hot-spot regions of FKS mutations | Cryptococcus spp. Fusarium spp. |
| Polyene | Amphotericin B, | Ergosterol sequestration | Ergosterol | Decreased membrane ergosterol, ERG mutations | Candida auris, Aspergillus terreus |
Resistance to all classes of antifungal drugs has been characterized in most fungal species that infect humans. In general, antifungal resistance has two operational definitions. First, in the clinical microbiology laboratory, fungal strains are classified using standardized assays (Box 1) to compare the minimum inhibitory concentration (MIC) [G] to the clinical breakpoints for that drug, which take into account in vivo aspects of drug efficacy. Resistant strains have MICs above the breakpoint and are more likely to cause a therapeutic failure than susceptible strains with MICs below the breakpoint. Second, in research laboratories studying the mechanisms of antifungal activity, in vitro resistance refers to a strain that is less susceptible (that is, has a higher MIC) to drug than a control or reference strain, and will be termed resistant in this Review.
Box 1. Measuring antifungal resistance and tolerance.
Standard assays of antifungal resistance
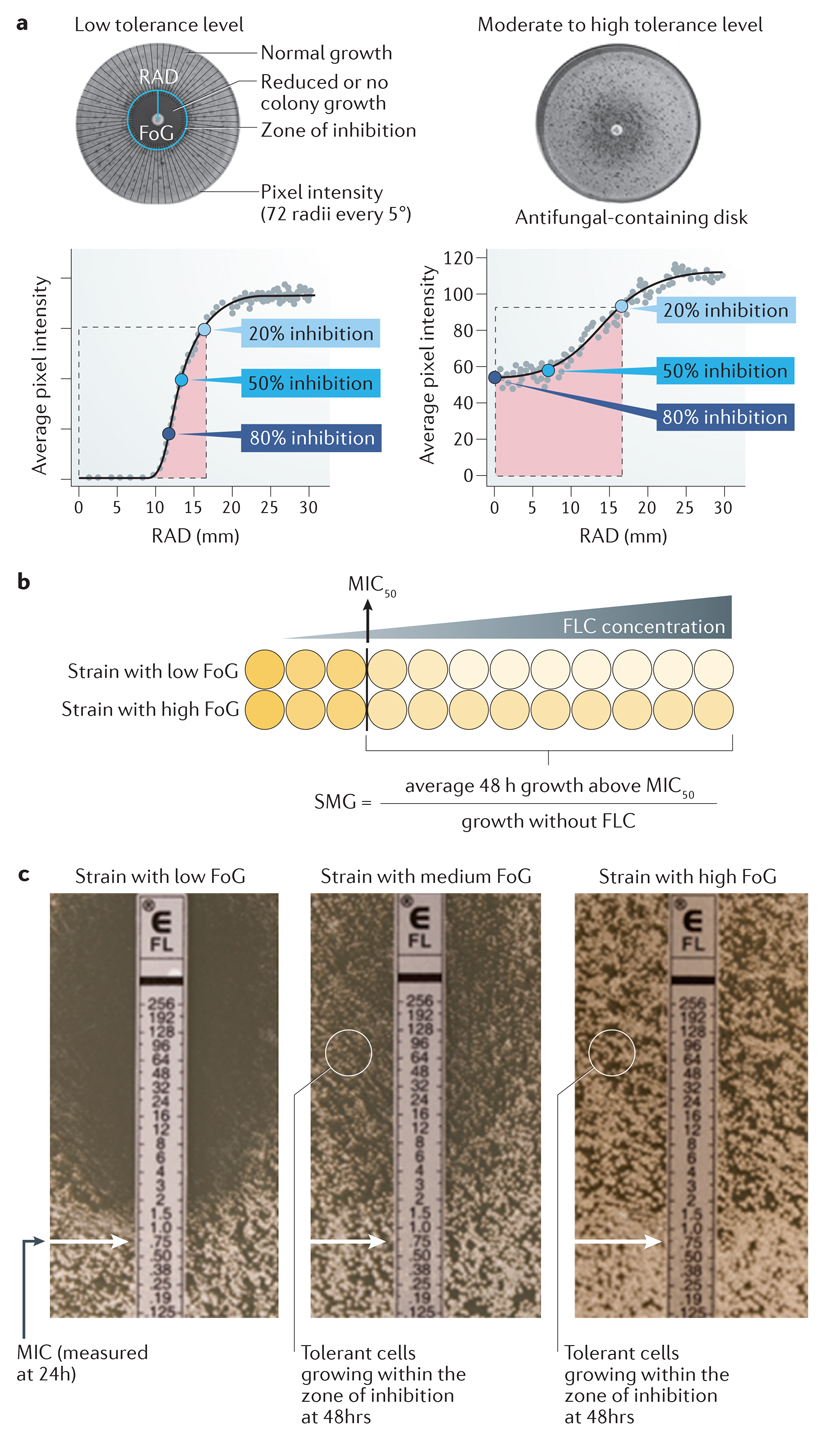
Clinical laboratory definitions of resistance are based on assays performed according to stringent criteria. Resistance is measured and quantified using three types of assays: disk diffusion assays (see the figure, part a), broth microdilution assays (see the figure, part b) or e-test® strips (see the figure, part c). Those tests are performed usually after 24h of growth. Disk diffusion assays are performed by spreading fungal cells on agar medium and then placing a filter disk containing a standard concentration of drug on the plate; fungal growth produces a lawn outside the zone of inhibition (see the figure, part a). Images of the plates are analyzed using diskImageR99, a software program that averages pixel intensity along 72 radii emanating from the disk and estimates the average pixel intensity corresponding to cell density as a function of distance from the disk. The level of susceptibility is measured as the average radius (RAD), which corresponds to the point where growth is inhibited by 20%, 50% or 80% (RAD20, RAD50 or RAD80, respectively, relative to the maximum radius, (these the bottom panels curve)). The minimum inhibitory concentration (MIC) is inversely correlated with the RAD. Shown are disk diffusion assays for two isolates with similar MIC levels and different tolerance levels, as determined from the degree of growth within the region of growth inhibition detected visually on the plates and calculated using diskImageR (as described below).
Broth microdilution assays (see the figure, part b) are performed by diluting a standard number of cells in microtiter wells containing drug in increasing concentrations (usually 2-fold increments) and propagating them. The lowest concentration that inhibits growth by 50% or more is generally considered the MIC (indicated by the yellow line and boxed wells); the Clinical and Laboratory Standards Institute (CSLI) suggests visual reading of the plates, and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommends measuring optical density. E-test strips ® (see the figure, part c) contain a gradient of drug and include markings that correspond to the drug concentration. The MIC is determined from the position of the edge of the zone of inhibition (indicated by the arrow) after 24h of growth.
Clinical breakpoints refer to MIC values above which isolates are considered to be resistant. Specific clinical breakpoints for fluconazole, voriconazole and the echinocandins have been established for Candida albicans, Candida glabrata, Candida parapsilosis, Candida krusei, Candida tropicalis and Candida guilliermondii. No amphotericin B breakpoints are available for any organisms. Based on the clinical breakpoints, interpretive guidelines are established by CLSI or EUCAST for these organisms and classify strains as sensitive, resistant or intermediate.
Measuring antifungal tolerance
Tolerance is quantified using either disk diffusion assays or broth microdilution assays, generally after 48h of growth. In disk diffusion assays, diskImageR calculates the fraction of growth (FoG) inside the zone of inhibition for a given RAD (usually at 20% or 50% inhibition) as the pixel intensity in the area under the curve (pink region, see the figure, part a)) relative to the maximum area possible (at the chosen RAD level (illustrated as the dashed line at RAD20, see the figure, part a)). Strains with similar levels of susceptibility (determined as MIC/RAD) often exhibit different levels of tolerance and are illustrated in the figure. Note that tolerant growth is often similar in density, measured as a similar pixel intensity, over some distance from the disk. Broth microdilution assays analyzed at 48h also provide a tolerance metric, termed supra-MIC growth (SMG)14. SMG is quantified as the average growth per well (measured as optical density), for all drug concentrations above the MIC, normalized to growth in the absence of the drug (see the figure, part c). E-test strip® assays provide a visual approximation of tolerance as the proportion of growth within the zone of inhibition relative to growth outside it (see the figure, part c). Solid media assays (see the figure, parts a and c) enable the visualization of subpopulation growth, evident as individual colonies, as well as the effect of drug concentration. By contrast, liquid assays do not distinguish between very slow growth of all cells in the population and slow growth of a subpopulation of cells. Box figure parts a and b ae modified from Ref. 9. Image in Box figure part c was provided by Mohamed Hajooj, Tel-Aviv University.
Although relatively low levels of resistance for most Candida species have been described, the outcomes for candidemia remain poor, as high levels of mortality (15%-50%) have been associated with candida infections7. For example, the rate of fluconazole and echinocandin resistance in Candida. albicans, the most prevalent Candida species, is generally less than 1%7, yet existing therapies often fail to treat infections caused by susceptible Candida isolates. This has been attributed to non-mycologic causes such as host factors, immune status, and/or pharmacologic issues8. However, specific interactions between the drug and the fungus, aside from the MIC, may affect therapeutic responses as well. Specifically, the discordance between overall treatment outcome and low levels of clinical resistance may be attributable to antifungal drug tolerance.
The terms antifungal drug resistance and tolerance are frequently used as synonyms in the literature. However, a more nuanced understanding of the fungal response to drugs may help explain the poor outcomes for infections caused by apparently susceptible isolates (that is, isolates that have a susceptible MIC at 24h, the standard endpoint for MIC measurements for Candida species). In this Review, we define antifungal tolerance as the ability of a drug-susceptible fungal strain to grow in the presence of an antifungal drug at concentrations above the MIC9. Subpopulations of tolerant cells grow slowly in drug concentrations above the MIC, which is usually visually evident after time periods longer than the standard 24 h for MIC measurements. In the same isogenic population, a second subpopulation of cells does not grow in the drug or its extremely slow growth is not detectable following 48 h in the drug. We posit that antifungal tolerance is a property of pathogenic fungi that can be distinguished from resistance using the same in vitro assay that can quantify both susceptibility [G] (measured as MIC at 24 h) and growth properties at 48 h (the fraction of growth [G] or supra-MIC growth [G]) (Box 1). We further suggest that the question of whether tolerance may explain some of the discordance between clinical outcome and clinical MIC measurements is worthy of larger scale clinical studies.
A recent quantitative study9 identified four characteristics of fluconazole-tolerant cells across a set of seven diverse C. albicans isolates. First, tolerance of an isolate is relatively reproducible, with differences in tolerance levels between isolates presumed to be affected by intrinsic genetic differences between those isolates. (Fig. 1A). Second, within a given isolate, tolerant cells (those that do grow slowly in the presence of the drug, while the remaining cells do not grow) represent a subpopulation (generally 5%-90% of the cells, depending on the isolate) (Fig. 1B). Third, tolerance is largely independent of drug concentration (Box 1). Fourth, the time required for cells to appear as visible colonies on a petri plate containing drug is shorter for isolates with high tolerance levels than for isolates with lower tolerance levels (Fig. 1C). Thus, drug tolerance is a manifestation of phenotypic heterogeneity intrinsic to a given fungal isolate.
Figure 1. Phenotypic characteristics and pathways required for antifungal tolerance.
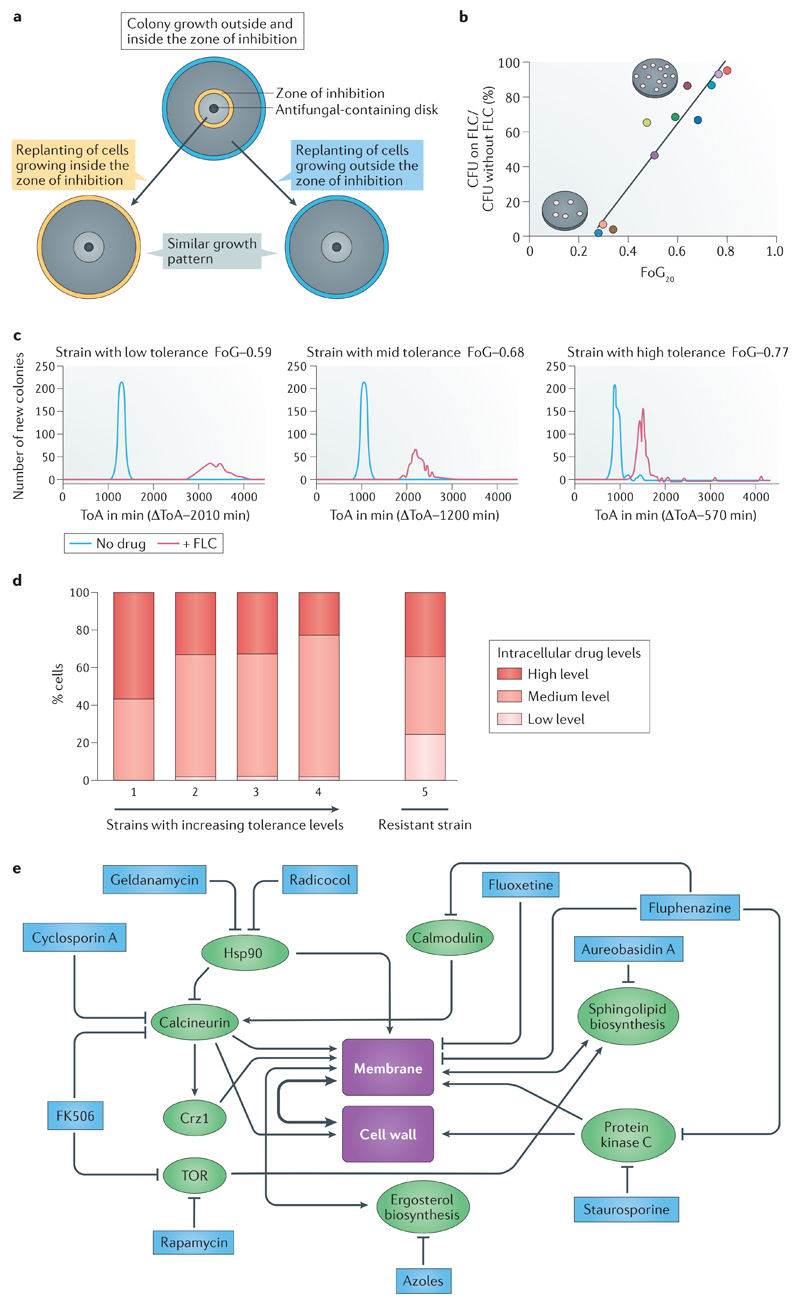
A. Tolerance is a phenotypic property of a given isolate that differs between different clinical isolates. The subpopulation of cells in the tolerant state (characterized by slow growth within the zone of inhibition in disk diffusion assays (Box 1)) is isogenic to cells in the non-tolerant state (the whole population of cells that grow outside the zone of inhibition). A disk diffusion assay with a single drug disk in the center of a petri plate is shown; the zone of inhibition border is indicated in yellow (top panel). When cells from this plate were spread on new plates with the same amount of drug, similar patterns of growth are observed for tolerant cells taken from within the zone of inhibition (bottom left) and non-tolerant cells taken from outside the zone of inhibition (between the yellow and cyan circles), indicating cells retain a similar potential to give rise to both types of subpopulations (tolerant and non-tolerant) in similar ratios. The radius (RAD) of the zone of inhibition, used to determine the minimum inhibitory concentration (MIC), and the fraction of growth (FoG) used to measure tolerance (Box 1), are similar in all three plates (not indicated). Similar results also are obtained with broth microdilution assays (not shown)9.
B. Tolerance (measured as FoG at 20% drug inhibition (FoG20) (see Box 1)) correlates with the proportion of colonies that appear at fluconazole (FLC) concentrations above the MIC (relative to total colonies growing in the absence of drug). Different strains are represented by different coloured dots.
C. The time required for cells to appear as visible colonies on a petri plate containing drug (time of colony appearance (ToA)) is shorter for isolates with high tolerance levels than for isolates with lower tolerance levels. Shown is a comparison between 3 clinical isolates that exhibit different levels of tolerance (indicated by the different FoG levels). The average difference between the ToA on plates containing an inhibitory drug concentration (red lines) and the ToA on plates without a drug (blue lines), ΔToA, inversely correlates with the level of tolerance (FoG levels). The growth rates of visible colonies and the final colony sizes are largely independent of tolerance levels.
D. Strains with low and increasing levels of tolerance are composed of cells with different amounts of intracellular azole drug. Shown are four susceptible strains, with increasing levels of tolerance (from left to right), and one resistant strain (far right). Among the susceptible strains, the proportion of cells with high amounts of intracellular drug decreases with increasing tolerance levels. The proportion of cells with high (red), intermediate (orange) and low intracellular drug concentrations of a fluorescent azole antifungal (Cy5-azole) drug differ from each other by an order of magnitude as measured by flow cytometry9. The resistant strain has a much larger proportion of cells with low intracellular drug concentration.
E. Studies using mutants and inhibitors reveal general stress pathways that affect tolerance. Proteins and pathways implicated in tolerance (green) and compounds that inhibit those pathways and also inhibit tolerance (blue) seem to converge on the cellular processes involved in membrane integrity and/or cell wall function. Some of the inhibitors affect more than one gene or pathway. For example, heat-shock protein 90 (Hsp90) inhibits calcineurin, which in turn activates Crz1, a transcription factor required for membrane integrity as well for tolerance26,25,96. Fluphenazine, an adjuvant drug that inhibits tolerance and renders fluconazole cidal9, is an inhibitor of both protein kinase C, which has been implicated in fluconazole tolerance22, and calmodulin, which is a regulator of calcineurin activity and ultimately, membrane and cell wall integrity97. This implies that tolerance relies on one or both of these central regulators for cell viability in the presence of fluconazole. Noteworthy, published studies investigating drug responses are difficult to interpret because the terms resistance and tolerance have been used interchangeably. Nonetheless, most published data is consistent with the notion that the regulation of both membrane integrity and cell wall function are important for tolerance. Figures 1a-d are modified from Ref.9. Figure 1e is based on data from Ref. 9 together with many other studies (Reviewed in98).
Although the definition of in vitro resistance is similar for bacteria and fungi, some aspects of antifungal tolerance differ from antibacterial tolerance. Tolerant bacterial and fungal isolates have MICs in the susceptible range, but tolerant bacteria are characterized by longer minimum duration to killing (MDK) than susceptible isolates10, based on responses to bactericidal drugs10–12. By contrast, antifungal tolerance is most evident with fungistatic drugs [G], and thus measuring cell death is less relevant. Furthermore, higher tolerance in bacterial isolates is associated with longer lag phase length, whereas fungal isolates with higher tolerance levels have shorter lag phase length (measured as time required for colonies to become visible13) relative to those with lower tolerance levels9.
Studies are beginning to provide evidence that tolerance is associated with multiple genetic components that differ between isolates. These genetic components, that differ between isolates, affect the ability of isogenic cells within a single isolate to respond physiologically or metabolically (to environmental or nutritional signals). The genetic make up of an individual isolate may also affect the degree of variability between the responses of different isogenic cells in the population. We know that chaperone activity, the availability of calcium and iron utilization and membrane proteins that regulate drug uptake and/or efflux, along with components of several central stress-response pathways such as protein kinase C/RAS and map kinase cascades are required for the ability to grow slowly (and to grow at all) in the drug. However, the degree to which these pathways affect differences between genetically different isolates, and the degree to which they are critical at the level of cell-to-cell differences, remains to be determined (Fig. 1E).
Growth of subpopulations of susceptible fungal isolates in the presence of drugs at concentrations above the MIC can be due to tolerance, heteroresistance [G] or persistence (see Box 2), depending on the drug, organism and the size of the subpopulation. Whereas tolerance is generally due to substantial subpopulations of cells (5-90%) that grow in drug, antifungal heteroresistance or persistence involve rare cells (generally <<1%) that transiently survive high concentrations of antifungal drugs14. The distinctions between susceptibility, resistance, tolerance and heteroresistance, all mechanisms that involve phenotypic heterogeneity [G] are discussed in Box 2.
Box 2. Antifungal heteroresistance.
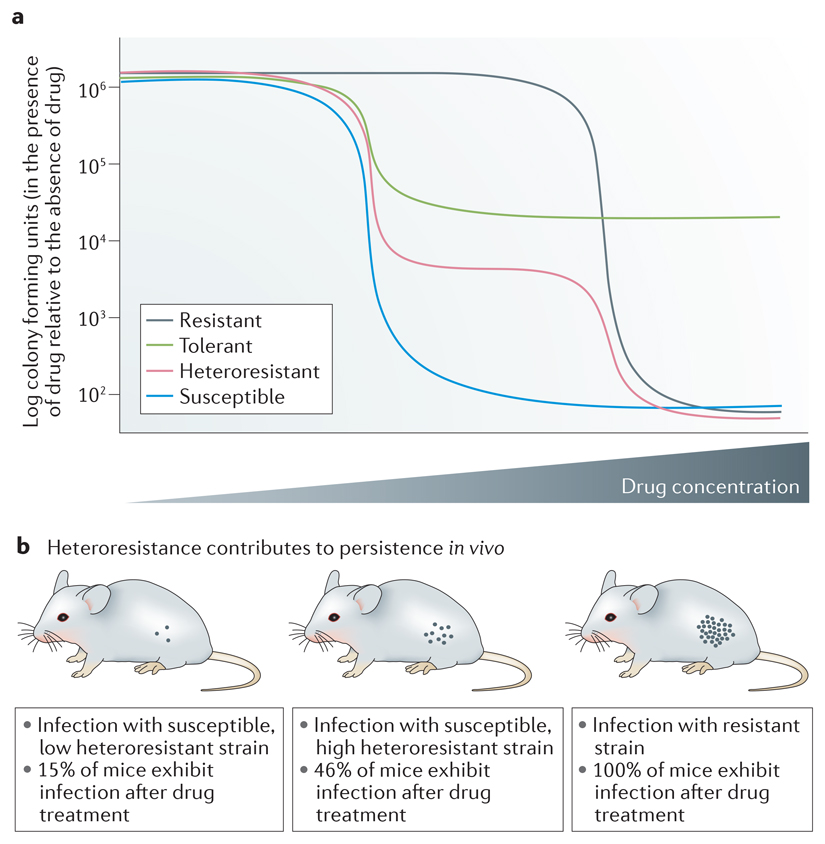
Heteroresistant clinical isolates contain a small subpopulation of cells (usually <<1%) that grow at drug concentrations ≥8-fold higher than the majority of the population11. Heteroresistance is detected, and distinguished from tolerance, by population analysis profiling (PAP) assays (see the figure, part a) that measure the proportion of cells growing at a range of drug concentrations. For Candida glabrata, upon a second exposure to drug, heteroresistant subpopulations again produce a subpopulation that grows, despite the presence of the drug; thus heteroresistance is not equivalent to genetic resistance.
Antifungal heteroresistance was first reported for Cryptococcus neoformans and Cryptococcus gattii100,101, and is an intrinsic property of these species101,102. Extra copies of an individual chromosome are associated with cryptococcal heteroresistant isolates; the degree to which heteroresistance is due to a particular aneuploidy seems to be strain-specific103. In Cryptococus, one molecular mechanism of heteroresistance seems to involve extra copies of the chromosome carrying both ERG11, the target of azoles, and AFR1, a major efflux pump. However, some Cryptococcus heteroresistant isolates do not have obvious aneuploidies104. C. glabrata isolates exhibit a range of heteroresistance levels and highly heteroresistant isolates exhibit increased levels of efflux activity100; however, the molecular mechanisms that yield heteroresistance in C. glabrata and in non-aneuploid Cryptococcus isolates are not known. Very few studies have analyzed the role of heteroresistance in in vivo persistence of a fungal pathogen in animal tissues after antifungal treatment. In vivo persistence of C. glabrata was compared using strains with low and high heteroresistance that had the same susceptible minimum inhibitory concentration (MIC). In a mouse assay that monitored CFUs in kidneys 7 days after infection and fluconazole treatment100 (see the figure, panel b), resistant isolates demonstrated a high fungal burden in the kidney in all mice, consistent with a failure to respond to the drug. Mice infected with the susceptible strain having low heteroresistance levels had a lower proportion of the mice with detectable C. glabrata in the kidneys compared to mice that were infected with an isolate that had high heteroresistance levels (numbers of mice with detectable C. glabrata are indicated). This result is consistent with the idea that high heteroresistance may underlie in vivo persistence of C. glabrata infections. Part b is modified from Ref100.
Trailing growth [G] is a term used primarily in the clinical context and has been defined as reduced but persistent visible growth of Candida spp. at fluconazole concentrations above the MIC15. Trailing growth is often estimated as the percent of growth at different drug concentrations relative to growth in the absence of drug16, a measure similar to the ‘supra-MIC growth’ (SMG) parameter used to measure tolerance (Fig. 1B). Like tolerance, trailing growth is sensitive to growth conditions17–19; the degree of sensitivity to these conditions differs for different clinical isolates9,(reviewed in 20), and leads to considerable laboratory-to-laboratory variation in the estimation of trailing growth. Noteworthy, tolerant or trailing cells, when re-tested, have a similar MIC and a similar slow/partial growth in the presence of drugs at concentrations above the MIC (Fig. 1A). Thus, we use the term ‘trailing’ for describing studies that used that term, with the assumption that tolerance and trailing growth in fluconazole probably reflect the same phenomenon19–21 in this Review.
Moreover, the ability of Candida spp. to form biofilms, and a phenomenon known as paradoxical growth [G], also affect fungal interaction with drugs. Candida spp. biofilms induce a transient, physiological alteration in drug responses22 (Box 3); that is, fungal biofilms exhibit reduced antifungal drug susceptibility via mechanisms that are dependent on cells growing within a biofilm, in which cells adhere to a substrate and form a thick mat of cells and secreted extracellular matrix that holds them together and reduces drug efficacy. The antifungal responses of biofilms and paradoxical growth are discussed in Box 3.
Box 3. Fungal biofilm and paradoxical growth responses to antifungals.
Biofilms have increased antifungal resistance
The ability of Candida species to form biofilms, where they attach to a surface and produce a community of cells with a range of filamentous morphologies surrounded by an extracellular matrix, is not only relevant for their ability to cause disease but also has an enormous effect on fungal interaction with drugs. As with bacterial biofilms, fungi within biofilms are much less susceptible to antifungal drugs 105. Assays used to characterize the susceptibility of planktonic cells are not applicable to Candida biofilms because of their physical properties. Methods available to assay antifungal responses within biofilms were recently reviewed 14;106. The most widely used method measures the effect of the drug on biofilm metabolism using dyes such as resazurin or 2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide reduction (XTT)107.
The mechanisms that contribute to the decreased susceptibility of cells in the biofilms to antifungal drugs are distinct from those in planktonic cells. Specifically, unlike with planktonic cells, efflux pumps and target gene mutations have only a limited role in Candida biofilm resistance to drugs such as fluconazole. Instead, the extracellular matrix of biofilms directly sequesters drugs such as fluconazole and amphotericin108. As a result, the major mechanism leading to the resistance of Candida biofilms is a decrease in the exposure of the cells within the biofilm to the drug itself, as evidenced by the finding that mutants that fail to generate biofilm-associated extracellular matrix remain susceptible to fluconazole108. Therefore, exposure of biofilms to a given concentration of drug results in a much lower intracellular concentration of drug. Furthermore, the resistant state is phenotypic, rather than genetic, as it relies on cells being in a biofilm and is not seen in the planktonic cells shed by the biofilm. Finally, persister cells have been proposed to explain the reduced susceptibility of Candida biofilms to fungicidal drugs such as echinocandins and amphotericin B; however, there are conflicting reports, such that the existence of persister cells in fungal biofilms, and the question of whether rare survivors are dependent on them being in a biofilm, remains controversial in the literature109–111.
Paradoxical growth
A phenomenon that shares some features of tolerance is paradoxical growth, whereby fungal strains are inhibited by low concentrations of a drug, most typically an echinocandin, yet they are able to grow at much higher drug concentrations. The mechanism or mechanisms of paradoxical growth are not yet well understood. Like tolerance, paradoxical growth involves a substantial proportion of the cell population; and it is unlikely to be due to alterations in the activity or stability of the drug target, ß-glucan synthase112–114. A potential mechanistic link between the paradoxical effect and tolerance is their common dependence on stress pathways regulated by heat-shock protein 90 (Hsp90) and calcineurin (reviewed in Ref. 115). Aspergillus nidulans cells that exhibit paradoxical growth also appear only after relatively long periods of drug exposure and appear to reverse many of the phenotypes associated with the responses of susceptible cells to echinocandins (for example, internalization of the ß-glucan synthases (reviewed in Ref. 115)).
In this Review, we explore the molecular mechanisms that contribute to antifungal drug resistance, with a focus on Candida spp., the most common cause of fungal infections in humans7. We then contrast the concepts of resistance and antifungal drug tolerance and present our current understanding of the mechanisms and phenotypes associated with tolerance. We assert that distinguishing tolerance from resistance may provide important insights into the reasons for treatment failure in some settings and the eventual development of resistance. Finally, we also discuss therapeutic approaches to reduce and/or overcome antifungal drug resistance and tolerance.
Antifungal drug resistance
Measuring and defining resistance
Resistance, the ability to grow in levels of drug that inhibit susceptible isolates, is measured clinically as an increase the MIC of a given antifungal drug when tested according to approved methods standardized by governing bodies such as The Clinical and Laboratory Standards Institute (CLSI)64 or European Committee on Antimicrobial Susceptibility Testing (EUCAST)23. Clinical breakpoints provide guidance regarding the likelihood that a given isolate will respond to therapy. Epidemiological cut-off values use MICs to identify isolates with acquired resistance, usually via mutations24.
It is important to distinguish clinical laboratory definitions of resistance from less stringent criteria frequently used in the literature. Specifically, many published reports refer to resistance as an increase in the MIC relative to a reference strain, particularly in the setting of work focused on elucidating the mechanisms of resistance. In addition, non-standard assays are frequently used to measure antifungal activity but do not generate an MIC value. For example, serial dilution agar plating assays frequently compare the susceptibility of one isolate or a mutant to its appropriate reference strain; isolates and mutants that grow better than the reference strain in these assays are often referred to as resistant. Further effects could be due to tolerance or trailing, rather than resistance9,25–28 because the MIC was not altered at 24 h. Instead, increased growth in the presence of the drug (relative to a reference strain) was usually detected at 48 h. Thus, when evaluating the literature, it is prudent to note the author’s definition of resistance and the conditions used to measure the phenomenon.
Mechanisms of drug resistance in fungi
Although resistance is emerging in non-Candida species such as Aspergillus spp.,29 the general molecular mechanisms of resistance are conserved; therefore, we will focus primarily on Candida spp., the most prevalent species and the most common cause of both life-threatening invasive disease and clinically important mucosal diseases, such as vulvovaginal candidiasis, thrush and esophagitis.
Antifungal drug resistance rates in Candida species are dynamic and variable between medical institutions and countries7. The highest rates of both azole and echinocandin resistance are observed in Candida glabrata and Candida tropicalis30 (Table 1). The most prevalent mechanism of azole resistance involves increased drug efflux pump activity, primarily due to gain-of-function mutations in transcription factors that regulate ATP-binding cassette (ABC) transporters (TAC1 in C. albicans and PDR1 in C. glabrata) and major facilitator super family (MFS) pumps (MRR1 in C. albicans)31,32. Of note, many of these transcription factors also regulate other genes that also contribute to resistance, virulence or general yeast cell fitness32. Furthermore, because of the overlapping functions of different efflux pumps, mutations such as loss-of-function of a single gene encoding an efflux pump, or gain-of-function of less efficient efflux pumps, have only modest effects on bona fide resistance and the most dramatic effects are from overexpression combined with homozygosis (loss of heterozygosity) of hyperactive transcription factor alleles33,37.
Alterations in antifungal drug targets, due to increased target expression or mutations within the target protein sequence, also mediate antifungal drug resistance (Table 1). Erg11, which is the target of azoles, is a cytochrome P450-dependent lanosterol 14α-demethylase required for ergosterol biosynthesis. In C. albicans, gain-of-function mutations in Upc2, a key transcriptional regulator of ergosterol biosynthetic genes34, lead to increased ERG11 expression and reduced azole susceptibility. Increased ERG11 expression is also seen in intrinsically resistant non-C. albicans species; however, the underlying mechanism is unknown. Mutations in ERG11 have also been observed in Candida spp. and over 100 different ERG11 alleles have been described35.
Many ERG11 mutations seem to interfere with drug binding and to decrease susceptibility in an additive manner36. Finally, in Candida auris, ERG11 polymorphisms as well as an increased number of ERG11 gene copies are thought to be responsible for its largely intrinsic azole resistance37–40 C. auris was discovered only a decade ago41 and has led to major outbreaks that have shuttered or paralyzed hospital wards. C. auris appears to rapidly acquire resistance or tolerance to echinocandins as well polyenes1,42, leaving few, if any, treatment options. Its ability to survive and adhere to surfaces over long time periods also makes C. auris contaminated areas difficult to decontaminate. How C. auris rapidly acquires drug resistance and/or tolerance to the remaining antifungal drug classes is only beginning to be explored.
Copy number variations, including whole-chromosome aneuploidy and shorter-range amplifications, are genome changes that can increase resistance32,43–49. For example, isochromosome (5L), a specific aneuploidy in C. albicans, results in the formation of two extra copies of the left arm of chromosome 543, and, consequently, extra copies of TAC1 and ERG11, genes that confer resistance to azoles in these strains50. Similarly, extra copies of the right arm of chromosome 3 may increase drug efflux because CDR1 and MRR1, which encode a major ABC transporter and a transcriptional activator of MDR1 (the MFS transporter), respectively, are found on this chromosome (Candida Genome Database [http://www.candidagenome.org/]). In agreement with this, increased copies of the right arm of chromosome 3, together with unidentified genes on trisomic chromosome 7 were associated with increased efflux51.
For heterozygous diploids such as C. albicans, loss of heterozygosity (LOH) is another frequent mechanism of resistance (reviewed in Refs. 52 and 53). For example, in a series of clinical C. albicans isolates from the same patient with HIV treated with fluconazole54,55, LOH events that were retained in all subsequent isolates arose concomitant with the acquisition of bona fide resistance. These LOH events included homozygosis of a region of the left arm of chromosome 5 that includes a point mutation in ERG1155. Homozygosis of this ERG11 allele was persistent (retained in the infecting population for more than 8 months55), consistent with the notion that mutations in ERG11 promote pathogen survival under drug selection56. Finally, some drugs enhance genome instability and thus promote aneuploidy and whole-chromosome LOH57,58. For example, exposure to fluconazole promotes whole-chromosome LOH via unusual mitotic divisions (chromosome duplications and losses)58–60. Furthermore, both LOH and aneuploidy occur at much higher frequency (2-4-fold) than point mutations59, and are readily detected following a single passage through the bloodstream or oral cavity of a mouse61–63.
Recent guidelines of the Infectious Disease Society of America recommend that an echinocandin, rather than azoles, be used as initial therapy for most invasive Candida spp. infections64. Echinocandin resistance is almost exclusively due to mutations in FKS genes (Table 1), which encode the target enzyme of the drug, 1,3-β-glucan synthase65. Echinocandin resistance is less prevalent than azole resistance (less than 1% of C. albicans infections7, but up to 10% of C. glabrata infections)66. Point mutations in one of two specific hot-spot regions of FKS genes increase the half maximal inhibition concentration (IC50) of an echinocandin in vitro by up to 3 orders of magnitude67. Of note, long periods of echinocandin therapy are associated with the emergence of echinocandin-resistant strains65. In addition, a substantial number of infections with echinocandin-susceptible C. glabrata or C. albicans isolates are not readily cleared68, and the degree to which echinocandin tolerance may play a role in this remains to be determined.
The emergence of multi-drug resistant (MDR) C. glabrata isolates is increasing at an alarming rate. A hypermutation mechanism was proposed to drive this MDR phenotype and was thought to be attributable to variant alleles of MSH2, a DNA repair gene; however, more recent work did not find a correlation between MSH2 mutations and antifungal resistance in vitro or in vivo69,70. Regardless of the mechanism, the emergence of MDR C. glabrata71. is a major concern, because it leaves the polyene amphotericin B, which is toxic8, as the only treatment option for these infections.
2. Antifungal drug tolerance
Measuring and characterizing tolerance in fungi
Antifungal tolerance is a characteristic of drug-susceptible strains that have the ability to grow slowly at inhibitory drug concentrations; usually only some of the cells in a given population exhibit this slow growth. Tolerance has been measured and characterized most extensively in C. albicans isolates treated with fluconazole, although tolerance is observed in most Candida species as well as for other drugs. The clinical importance of tolerance and/or trailing remains an open question (Box 4).
Box 4. Clinical relevance of fungal tolerance.
The clinical implications of fungal tolerance, also termed trailing growth in the clinical literature, remain to be resolved. The 24h clinical endpoint for standardized minimum inhibitory concentration (MIC) assays was established to minimize the detection of trailing growth, based largely on studies that failed to find a positive correlation between trailing and virulence or mortality116,117,118. For example, Candida albicans and Candida tropicalis trailing isolates with similar MICs showed no difference in response to fluconazole in a mouse model for up to 5 days after infection119, supporting the recommendation of a 24h MIC endpoint. One caveat is that the study involved only a single isolate with a high level of trailing growth from each species; a second issue is that longer-term clinical outcomes, such as infection recurrence, were not investigated. This is particularly important as the effect of tolerance on infection persistence may take time to be detected. We posit that the ability of the fungus to continue to divide, even slowly, in the presence of drug in the host may be an important contributor to recurrent infections as well as to the ability to acquire drug resistance.
A large clinical study analyzed 690 Candida isolates (8 species, 9 antifungals and isolates collected from 29 hospitals)116, with 83% of the ‘trailing’ isolates having <50% residual growth and not significantly affecting clinical outcomes. However, trailing of >50% was not analyzed separately. A more recent study of C. tropicalis isolates defined trailing levels as weak, moderate and heavily trailing (≤25%, 26%-50% and >50% residual growth, respectively); these isolates were tested in Galleria larvae and two mouse models15. In all three models, heavily trailing isolates (which made up ~7% of the isolates) responded poorly to fluconazole, leading the authors to suggest that they should be regarded as clinically resistant to fluconazole. They also recommended that caution should be exercised in using fluconazole to treat patients infected with isolates that have moderate trailing levels15.
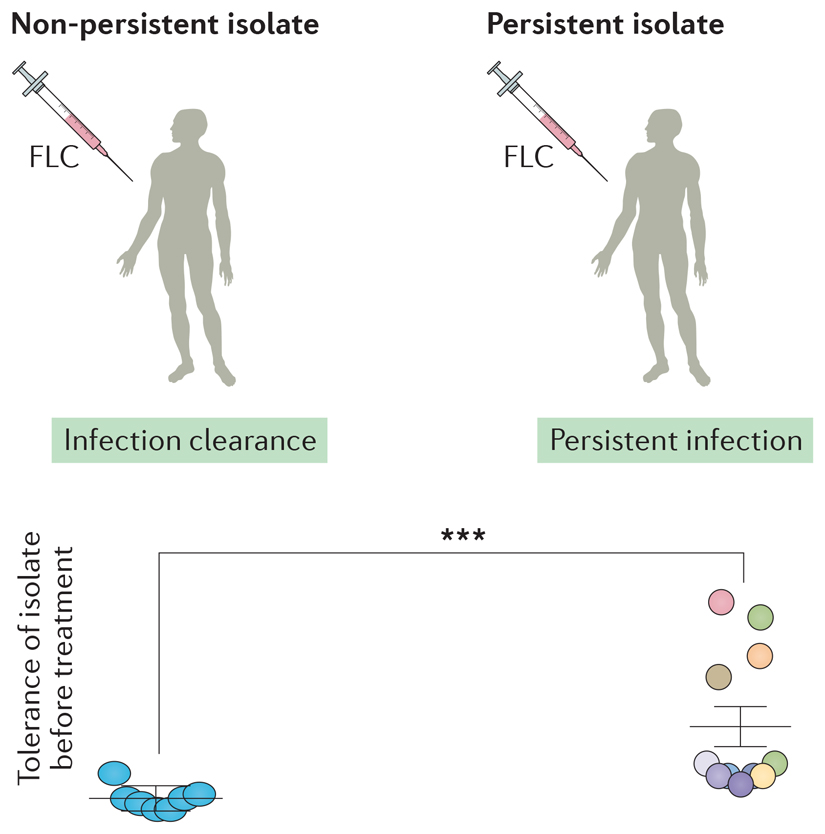
A small-scale retrospective study (19 patients) compared the tolerance levels of C. albicans strains isolated from patients treated with fluconazole whose infections were rapidly cleared by fluconazole, and strains isolated from patients in which candidemia was persistent because fluconazole failed to clear the infection120. Initial isolates (prior to antifungal therapy) from the cleared infections consistently had lower levels of tolerance (see the figure), relative to the isolates that caused persistent infections (those that are not cleared by drug treatment). This suggests that tolerance may affect the efficacy of fluconazole treatment and supports the idea that retrospective, as well as prospective studies, should be conducted to test the relationship between patient outcome and the initial tolerance level of the infecting isolates.
Clearly, it will be important to conduct additional studies that quantify tolerance in large numbers of diverse isolates and their known clinical outcomes. Moving forward, in vivo experiments also will be important. It is crucial to evaluate clinical isolates displaying a wide range of in vitro tolerance phenotypes, particularly if clinical data regarding patient outcome is available. It is well-accepted that diverse C. albicans isolates exhibit heterogeneity with respect to the expression of virulence traits and determinants of pathogenicity121,122. Although the heterogeneity of clinical responses to treatment is likely to be multifactorial, improved methods to systematically characterize tolerance can address the degree to which tolerance contributes to this heterogeneity. Figure is modified from reference9.
Several features distinguish fungal tolerance from fungal resistance: tolerant cell growth is slower and more time-dependent than resistant cell growth, and is less dependent on drug concentration9. In addition, different clinical isolates exhibit different levels of tolerance, measured as the proportion of growth above the MIC at 48 h in disk diffusion or broth microdilution assays (Box 1). The degree of tolerance correlates with the size of the subpopulation of cells that grow above the MIC, and the rate at which such cells form visible colonies at supra-MIC concentrations9. This highlights the idea that tolerance is due to the slow growth of some cells in the population and raises questions about how such cells differ from those that do not grow (or grow much more slowly).
The degree of tolerance varies across different isolates, presumably owing to the intrinsic allele diversity of each isolate. Phenotypic differences between clinical isolates involves many thousands, to tens of thousands of SNPs, and is also evident with phenotypes such as biofilm formation and its dependence upon specific transcription factors72. In this sense, the differences in tolerance levels measured as FoG or SMG (Box 1) between isolates have a genetic component. By contrast, for a single isolate, the phenotypic heterogeneity that drives some cells to grow slowly, and others to arrest growth, must be due to physiological or metabolic shifts that differ between isogenic cells in the population.
Mechanisms of antifungal tolerance
Although the mechanisms by which clinical responses to treatment vary are likely to be multifactorial, the ability to quantitatively characterize tolerance will permit the examination of these mechanisms. In addition to heterogeneity between isolates there is heterogeneity within a population in that different colonies of the same sample behave differently when exposed to fluconazole. Upon exposure to fluconazole, all cells undergo 1-3 rounds of division before growth arrest9,58; tolerant cells in the population resume growth several hours later, whereas non-tolerant cells arrest for considerably longer and only resume division in the absence of the drug. Furthermore, colonies from higher tolerance isolates appear faster than those from low tolerance isolates (Fig. 1C). Thus, tolerance is likely to be due to physiological responses that emerge heterogeneously among the cell population, where the proportion of cells exhibiting such responses is a function of the genetic potential of the specific isolate. Point mutations that affect this genetic potential remain to be identified, partly because it is difficult to screen for recessive mutations in this heterozygous diploid organism and because unrelated clinical isolates can diverge at tens of thousands of loci. Newly available haploid isolates of C. albicans73 have the potential to facilitate screens for recessive point mutations and insertion mutants74,75. For example, this approach identified sphingolipid biosynthesis as an important mechanism by which cells survive azole-induced ergosterol depletion73.
Aneuploidy and/or loss of heteroresistance are rapidly acquired genome changes that can result in increased tolerance. and are likely to affect one or several genes that alter tolerance level. Importantly, exposure to azoles promotes mitotic defects that yield tetraploid and aneuploid cells58. Aneuploidy can alter fluconazole tolerance in C. albicans drug38,51. For example, segmental trisomy of chromosome 3 conferred an increased fitness in the presence of fluconazole that was attributable, at least partially, to extra copies of NPR2, which encodes a urea transporter51. This is tolerance, rather than resistance, because the MIC remained susceptible and the increased growth was detected after incubation for more than the 24 h in standard MIC assays. Notably, a single aneuploid chromosome can confer more than one unrelated phenotype. Trisomy of chromosome 2 promotes the growth of C. albicans cells in the presence of the antifungal drug caspofungin47 without altering the MIC. Interestingly, trisomy of chromosome 2 also promotes cell survival in the presence of hydroxyurea, a cancer chemotherapy drug. The response to hydroxyurea was shown to be dependent on two of the 1087 different genes located on chromosome 2: RNR1 and RNR21; the genes responsible for survival in the presence of caspofungin are distinct from these RNR genes and have not yet been identified47. This further suggests that exposure to hydroxyurea, by selecting for chromosome 2 trisomy, has the potential to increase the subsequent survival of C. albicans cells exposed to caspofungin.
Adjuvant [G] drugs and mutations that affect several stress-responsive pathways often reduce tolerance9 (Box 5); and adjuvants reduce tolerance to similar baseline levels irrespective of the tolerance level of the isolate9. This implies that tolerance depends on a shared set of critical pathways across diverse isolates. It also suggests that drugs that interfere with tolerance may be useful in combination therapies that extend the useful lifespan of currently available antifungal drugs (Box 5).
Box 5. Synergistic drug combinations in vitro and in vivo.
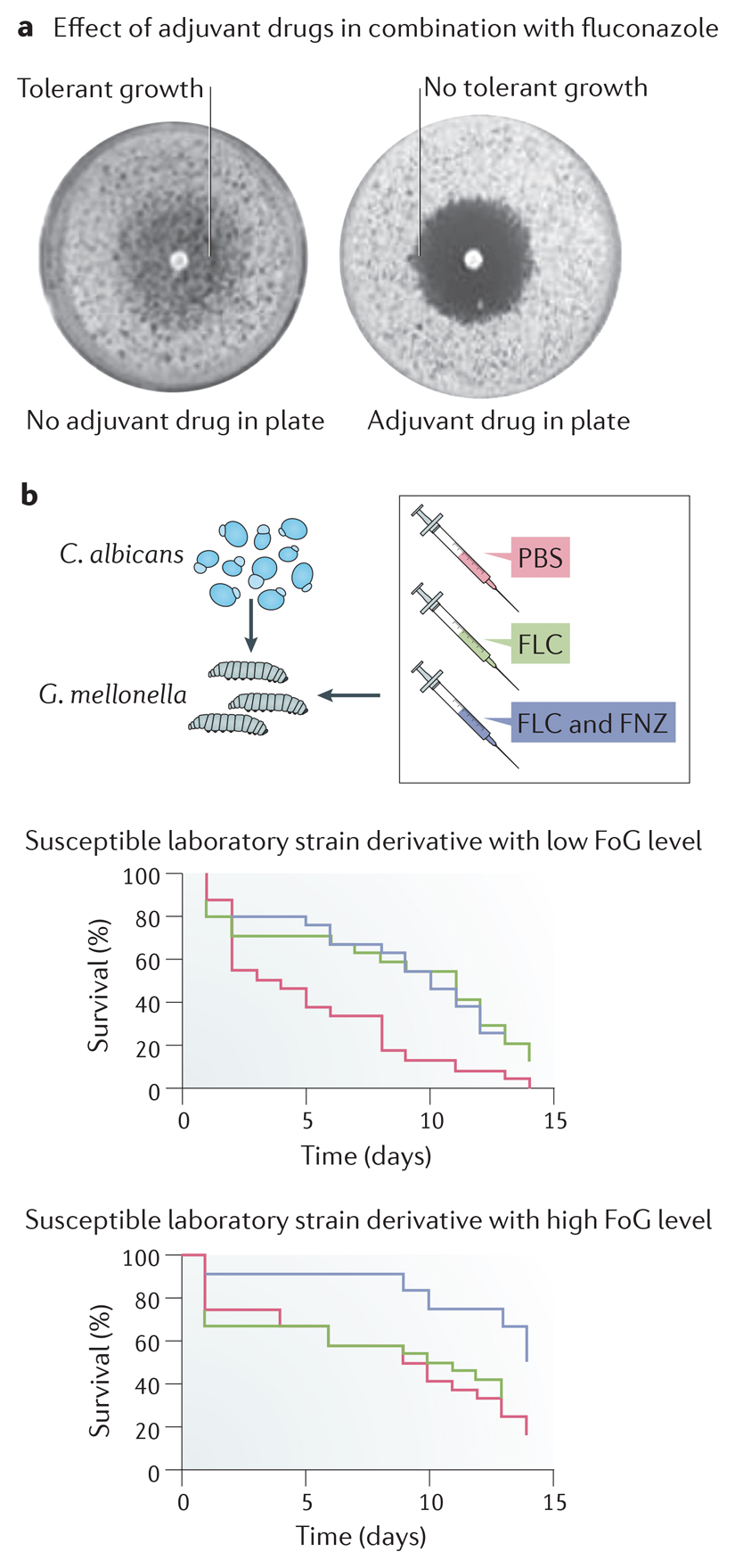
Synergistic combinations of fluconazole and an adjuvant drug are active, and often cidal, against Candida albicans in vitro. Comparison of a series of adjuvant drugs, reported to affect resistance and/or tolerance, were compared across a set of C. albicans strains. Most adjuvants, when added to the entire petri plate, dramatically reduced the fraction of growth (FoG) measured as growth within the zone of inhibition surrounding a disk containing fluconazole (see the figure, part a). This included inhibitors of heat-shock protein 90 (Hsp90) (geldanamycin and radicicol), calcineurin (cyclosporin A and FK506), TOR (rapamycin), the unfolded protein response (tunicamycin) and sphingolipid biosynthesis (aureobasidin A)9, as well as antibacterials123,124, anti-arrhythmic agents (amiodarone)125, immunosuppressive agents (FK506),126 anti-depressants (fluoxetine),127 antipsychotics (fluphenazine) and non-steroidal anti-inflammatory agents (ibuprofen)128. The minimum inhibitory concentration (MIC; radius of zone of inhibition) is largely unaffected by these adjuvants, supporting the idea that the adjuvants mainly inhibit tolerance rather than resistance. Within the zone of inhibition129, cells are not viable9,21,130, which indicates that the adjuvants in combination with fluconazole are fungicidal.
Drug combinations that reduce fluconazole tolerance have been examined in several animal studies and in one small scale retrospective analysis of patient outcomes. A study tested the combination of fluconazole and the immunosuppressant cyclosporine, which is fungicidal and affects tolerance but not resistance21, in a rat endocarditis model. The cyclosporine-fluconazole combination was more effective compared to amphotericin B and fluconazole treatment alone in clearing the infection21. Although the authors only tested a single laboratory Candida albicans isolate, and the concentrations of cyclosporine used in the study were above the corresponding therapeutic dose in humans, this early study suggested that adjuvant drugs that eliminate tolerance may provide a strategy for extending the useful lifespan of fluconazole (reviewed in Ref. 87). Similarly, although fluphenazine alone had no effect on survival, the psychotherapeutic agent fluphenazine combined with fluconazole prolonged the survival of Galleria larvae infected with highly tolerant C. albicans isolates (high fraction of growth (FoG)), but not those infected with low tolerance isolates (low fraction of growth (FoG)) (see the figure, part b). Figure parts a and b adapted from 9.
In addition to genetic determinants of tolerance level, growth conditions influence the degree of tolerance exhibited by a strain. For example, conditions that increase cell wall chitin promote cell survival in the presence of echinocandin76,77, whereas calcineurin inhibitors reduce echinocandin tolerance (measured using MIC assays analyzed after 72 h)68,78. Similarly, pH, temperature and nutrient availability affect the level of fluconazole tolerance9. Thus, tolerance has a genetic component that varies between isolates, and a non-genetic component that is influenced by growth conditions and cell physiology.
Two general concepts underlying the mechanisms of fluconazole tolerance need molecular explanations: the inter-isolate heterogeneity of tolerance levels; and cell-to-cell variation within a given isolate. The inter-isolate heterogeneity is likely due to genetic differences that contribute to several other clinically relevant phenotypes, including biofilm formation, filamentous growth responses and sensitivity to temperature52,60,79,80. Cell-to-cell phenotypic heterogeneity occurs at a frequency that is far too high (10-90% of cells) to be due to the genetic acquisition of mutations, implying that it is due to non-genetic differences between the cells.
Earlier studies identified stress response pathways important for drug responses that are most likely affecting tolerance, although at the time little distinction was made between tolerance and resistance (reviewed in Ref. 81). Inhibitors of heat shock protein 90 (Hsp90)82, calcineurin21,25,26, target of rapamycin (TOR)83 and mutants that affect Rim10184 (a transcription factor involved in the pH response pathway) effectively eliminate tolerance and greatly increase the fungicidal activity [G] of fluconazole across a diverse set of isolates with different tolerance levels (Fig. 1E; Box 5). This implies that tolerance in all strains is dependent on all of these major hubs of cell homeostasis, irrespective of both the subpopulation size and its ability to in the presence of the drug. Consistent with this, either pharmacological or genetic disruption of Hsp90, calcineurin, TOR, lysine deacetylases or Rim101 function enhance the effect of fluconazole (reviewed in Ref. 85). It appears that most, or all, of these fluconazole-associated stress response pathways affect tolerance, because inhibiting these pathways does not affect MIC levels (Box 5). Importantly, many of these same core stress response pathways in C. albicans (Hsp90, Ca2+-calmodulin-calcineurin, lysine deacetylases, protein kinase C and HOG (high osmolarity glycerol responses) pathways) t also affect echinocandin responses68,86,87. Specifically, studies of these pathways using either inhibitors or mutant strains showed they reduce fungal growth in combination with caspofungin or anidulafungin. Indeed, the involvement of lysine deacetylases and calcineurin implies a connection between tolerance and the metabolic anti-stress machinery (for example Ref. 88). Thus, variations on the function of the critical pathways listed above are candidate drivers of differences in tolerance levels between isolates. However, the complex constellation of genes responsible for differences in tolerance levels between isolates and their progeny remain to be explored.
The large number of central cellular pathways that affect tolerance also implies that many types of mutations could lead to changes in tolerance. We posit that inter-isolate heterogeneity in tolerance is a composite phenotype that emerges from variations at multiple loci; that is, different stress response pathways probably contribute to tolerance to a different degree in different isolates. Accordingly, a current challenge is to systematically study the effects of natural variation in the genes that affect tolerance. Several approaches are likely to be informative: using near-isogenic strains that differ in tolerance levels; engineering candidate mutations in different isolates; and performing unbiased screens for mutants that increase or decrease tolerance without affecting resistance.
Mutations in some pathways seem to affect resistance as well as tolerance. For example, mutations in Tac1, a positive transcriptional regulator of efflux pump genes, increases efflux activity49. In addition to affecting resistance, drug uptake and efflux appears to contribute, at least partially, to tolerance9, based on studies with fluorescent azole probes and strains with similar MIC levels but different tolerance levels (measured as FoG and SMG, see Box ##)89,90 Transcription factors can regulate multiple targets, and therefore the downstream effects of mutations in those transcription factors on resistance and tolerance could be independent of one another. Alternatively, the stress pathways important for tolerance may indirectly modify efflux activity, thereby enabling cells to grow slowly in the presence of the drug. Thus, although resistance and tolerance are different (as described above), they may share the same processes (albeit to different degrees) to promote growth in the presence of antifungal drugs.
For example, isolates that are more tolerant have lower average intracellular drug than less tolerant isolates9 (Fig 1D). Furthermore, intracellular drug concentrations vary among cells within a given isolate, with the more tolerant isolates (those that grow slowly) having a larger population of cells with moderate amounts of drug per cell. By contrast, an isolate that is resistant because of increased efflux has a proportion of cells with very low levels of intracellular drug (10X less than those with moderate drug levels compared to tolerant isolates)9 (Fig. 1D). This suggests a mechanistic distinction between resistance and tolerance. In resistant isolates, mutations (for example, gain-of-function alleles in drug efflux pumps or their positive regulators) affecting efflux would enable most (or all) cells to reduce intracellular drug concentration to very low levels. In tolerant isolates, physiological differences (for example, normal distribution in the amount of efflux gene expression) would enable some cells to reduce intracellular drug concentrations modestly relative to others, thereby facilitating slow growth in those cells with higher efflux gene expression. This idea is consistent with the results thus far9; more isolates with characterized mechanisms of resistance or tolerance will need to be tested to determine if it is generalizable.
Moreover, the molecular processes that contribute to the cell-to-cell phenotypic heterogeneity within an isogenic population are beginning to be elucidated. We do not yet know the degree to which a give daughter cell in a population inherits the phenotypic characteristics of its mother: for example, are tolerant mother cells more likely to have tolerant daughter cells, or is the degree of drug response distributed randomly amongst the progeny? The phenotypic differences between cells can be the consequence of cell-to-cell differences in: the gene expression state (for example, via chromatin modification and/or copy number variation); the metabolic state (for example, metabolite exchange interactions between cells that specialize metabolically and, as a consequence, diverge phenotypically (reviewed in Ref.91); and/or the biophysical state (via the formation of macromolecular condensates such as stress granules, prions and prion-like aggregates and/or phase transitions (reviewed in Ref. 92)). These different states are not mutually exclusive and they may require a similar set of pathways that affect azole tolerance (for example, Hsp90 and TOR).93 Once the state or states that affect tolerance are better understood, it will be possible to address critical mechanistic questions such as how the state of tolerant versus non-tolerant individual cells is maintained in a given cell and its progeny during growth in the presence of drug, and why that state is not present in other cells in the population.
Organelles such as the vacuole make important contributions to tolerance and/or trailing growth. In Saccharomyces cerevisiae several genes involved in retrograde trafficking modulate tolerance and/or trailing growth, including VPS2127,51, VPS15 and VPS5194; Rim101 may influence tolerance via its ability to regulate the transcription of key components of the vacuolar trafficking pathway95. Thus, tolerance seems to depend on intracellular processes that facilitate adaptation in response to drug stresses. Clearly, as the molecular and cellular mechanisms of tolerance, as well as the clinical implications of tolerance, become clearer, it may be possible to design therapeutic strategies that inhibit tolerance and provide new avenues to treat fungal infections.
Role of tolerance in the acquisition of resistance
Tolerance has been proposed to increase the frequency of antifungal resistance82. In part, this argument is based on the observation that inhibition of Hsp90 in the presence of fluconazole not only eliminates tolerance9 but also renders fluconazole fungicidal87. Thus, no colonies grow when fluconazole and an Hsp90 inhibitor are combined, because dead cells cannot evolve. Nonetheless, because a highly tolerant strain will divide more frequently in the presence of fluconazole than a strain with low tolerance levels, it will also have a higher population size and a higher likelihood of acquiring new mutations. In addition, drug pressure will lead to the positive selection of mutants that exhibit improved fitness. It will be important to determine the degree to which population size, mutation rate and selection pressure contribute to antifungal responses in tolerant cells and to the rate of the emergence of resistance.
Summary and future directions
In this Review, we distinguish between antifungal resistance and tolerance, clarifying terms that have been used interchangeably in the literature, to describe growth in the presence of an inhibitory drug. Resistance is due to genetic changes that directly affect either the drug target and/or intracellular drug concentrations and cause a heritable effect on the entire population of cells in a given isolate. By contrast, tolerance is a feature of susceptible (non-resistant) isolates that relies upon several central stress response pathways; it is a consequence of phenotypic heterogeneity, in which some cells in the population grow, albeit slowly, in drug. Differences in tolerance levels between isolates is assumed to have a genetic basis, and to affect the proportion of cells able to grow in drug. The mechanisms that drive cell-to-cell differences in drug responses within a single isolate remain to be determined; but, at least for azoles, they seem to be connected to differences in intracellular drug concentration between different cells.
Combination therapies with adjuvant drugs that interfere with the compensatory stress responses required for tolerance have the potential to increase the lethality of an inhibitory antifungal drug. However, the ability of many molecules to achieve exactly these purposes in vitro is only a first step; very few adjuvants have progressed to efficacy studies in animal models of infection. Furthermore, to date, most studies have not considered the role of tolerance in the efficacy of the drug combinations. Nonetheless, systematic and large-scale screens for molecules that affect tolerance provide a promising approach to overcoming antifungal drug resistance and improving the care of patients with fungal infections (see supplementary information S1).
Supplementary Material
Glossary.
| Susceptibility | Sensitivity to a drug, either arresting growth (static drugs) and/or killing cells (cidal drugs). |
|---|---|
| Fraction of Growth (FoG) | The fraction of growth (FoG) is a measure of tolerance based on assays performed on solid medium. Measured at 48 hours, the growth within the zone of inhibition (and thus above the minimum inhibitory concentration (MIC)) is estimated as a proportion of total growth possible outside the zone of inhibition. |
| Supra-MIC growth (SMG) | Supra-MIC growth (SMG) is a measure of tolerance based on assays performed in liquid medium. Growth at concentrations above the minimum inhibitory concentration (MIC) is estimated as a proportion of the total growth without drug. SMG provides a quantitative measure of growth similar to some measures of trailing growth. |
| Fungistatic drugs | Fungistatic drugs inhibit growth but do not necessarily kill a majority of the cell population at concentrations at or above the minimum inhibitory concentration (MIC). |
| Heteroresistance | Heteroresistance is a clinical term for isolates that contain small subpopulations of cells (generally <1%) that have the ability grow at drug concentrations that are at least 8-times above the minimum inhibitory concentration (MIC) for the vast majority of susceptible cells in the population. |
| Phenotypic heterogeneity | The expression of different phenotypes in different cells within an isogenic population of cells. For example, some fungal cells grow, while other sister cells do not grow (or grow too slowly to be detected) in the presence of an antifungal drug. |
| Trailing growth | Trailing growth is generally defined as reduced but persistent visible growth of Candida spp. at fluconazole concentrations above the minimum inhibitory concentration (MIC). Trailing has also been described as an increase in the MIC during growth beyond 24h (the standard endpoint for MIC measurements for Candida species). It can be measured as the residual growth in the presence of fluconazole concentrations above the MIC. Trailing was quantified in a recent study as the percentage of residual yeast growth at fluconazole concentrations above the MIC in each well and mean trailing as the geometric mean of trailing observed in all the wells above the MIC. |
| Paradoxical growth | Paradoxical growth, also referred to as the Eagle effect, is the ability of a fungal isolate to reconstitute growth in the presence of high drug concentrations, whereas being fully susceptible at lower concentrations. Paradoxical growth appears with a delay of one to several days, but resembles growth in the absence of the drug. Paradoxical growth has been reported primarily for echinocandins. |
| Adjuvants | A drug that potentiates the effect of an anti-infective, but is not an anti-infective on its own. |
| Fungicidal activity | Drugs with fungicidal activity reduce a population of cells by >99.9% or 3 log 10 units at a concentration equal to or greater than the minimum inhibitory concentration (MIC). |
Acknowledgements
The authors thank M. Ralser, D. Jarosz and members of the Berman laboratory for helpful comments, and S. Everson, Iuliana Ene, Brown University, Anton Levitan, Tel-Aviv University, Aleeza C. Gerstein, University of Manitoba and M. Hajooj for help with illustrations. Work in the author’s laboratory was supported by the European Research Council, RAPLODAPT 340087 and the Israel Science Foundation (grant number 997/18) (J.B.), and the National Institutes of Health 1R01AI125094 (D.J.K.).
Footnotes
Author contributions
Both authors contributed to all aspects of the article.
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Microbiology thanks J. Morschhauser, D. Perlin and D. Sanglard for their contribution to the peer review of this work.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arendrup MC, Patterson TF. Multidrug-Resistant Candida: Epidemiology, Molecular Mechanisms, and Treatment. The Journal of infectious diseases. 2017;216:S445–s451. doi: 10.1093/infdis/jix131. [DOI] [PubMed] [Google Scholar]
- 2.Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. The global problem of antifungal resistance: prevalence, mechanisms, and management. The Lancet Infectious diseases. 2017;17:e383–e392. doi: 10.1016/s1473-3099(17)30316-x. [DOI] [PubMed] [Google Scholar]
- 3.Alangaden GJ. Nosocomial fungal infections: epidemiology, infection control, and prevention. Infectious Disease Dlinics of North America. 2011;25:201–225. doi: 10.1016/j.idc.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Andes DR, et al. The epidemiology and outcomes of invasive Candida infections among organ transplant recipients in the United States: results of the Transplant-Associated Infection Surveillance Network (TRANSNET) Transplant infectious disease : an official journal of the Transplantation Society. 2016;18:921–931. doi: 10.1111/tid.12613. [DOI] [PubMed] [Google Scholar]
- 5.Krysan DJ. The unmet clinical need of novel antifungal drugs. Virulence. 2017;8:135–137. doi: 10.1080/21505594.2016.1276692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassetti M, Carnelutti A, Castaldo N, Peghin M. Important new therapies for methicillin-resistant Staphylococcus aureus. Expert Opin Pharmacother. 2019;20:2317–2334. doi: 10.1080/14656566.2019.1675637. [DOI] [PubMed] [Google Scholar]
- 7.Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. Twenty years of the SENTRY Antifungal Surveillance Program: Results for Candida species from 1997-2016. Open forum infectious diseases. 2019;6:S79–s94. doi: 10.1093/ofid/ofy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lepak AJ, Andes DR. Antifungal pharmacokinetics and pharmacodynamics. Cold Spring Harb Perspect Med. 2014;5 doi: 10.1101/cshperspect.a019653. a019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg A, et al. Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nature communications. 2018;9:2470. doi: 10.1038/s41467-018-04926-x. [This manuscript provides a quantitative characterization of fluconazole tolerance in C. albicans, distinguishing it from resistance and highlighting its phenotypic heterogeneity and association with subpopulation growth, and sensitivity to adjuvant drugs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nature reviews Microbiology. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 11.Balaban NQ, et al. Definitions and guidelines for research on antibiotic persistence. Nature reviews Microbiology. 2019 doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brauner A, Shoresh N, Fridman O, Balaban NQ. An experimental framework for quantifying bacterial tolerance. Biophysical journal. 2017;112:2664–2671. doi: 10.1016/j.bpj.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin-Reisman I, Fridman O, Balaban NQ. ScanLag: high-throughput quantification of colony growth and lag time. Journal of visualized experiments : JoVE. 2014 doi: 10.3791/51456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuyts J, Van Dijck P, Holtappels M. Fungal persister cells: The basis for recalcitrant infections? PLoS pathogens. 2018;14:e1007301. doi: 10.1371/journal.ppat.1007301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Astvad KMT, Sanglard D, Delarze E, Hare RK, Arendrup MC. Implications of the EUCAST Trailing Phenomenon in Candida tropicalis for the In Vivo Susceptibility in Invertebrate and Murine Models. Antimicrob Agents Ch. 2018;62 doi: 10.1128/AAC.01624-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcos-Zambrano LJ, Escribano P, Sanchez-Carrillo C, Bouza E, Guinea J. Scope and frequency of fluconazole trailing assessed using EUCAST in invasive Candida spp. isolates. Med Mycol. 2016;54:733–739. doi: 10.1093/mmy/myw033. [DOI] [PubMed] [Google Scholar]
- 17.Coenye T, De Vos M, Vandenbosch D, Nelis H. Factors influencing the trailing endpoint observed in Candida albicans susceptibility testing using the CLSI procedure. Clin Microbiol Infect. 2008;14:495–497. doi: 10.1111/j.1469-0691.2008.01956.x. [DOI] [PubMed] [Google Scholar]
- 18.Agrawal D, Patterson TF, Rinaldi MG, Revankar SG. Trailing end-point phenotype of Candida spp. in antifungal susceptibility testing to fluconazole is eliminated by altering incubation temperature. Journal of medical microbiology. 2007;56:1003–1004. doi: 10.1099/jmm.0.47168-0. [DOI] [PubMed] [Google Scholar]
- 19.Marr KA, Rustad TR, Rex JH, White TC. The trailing end point phenotype in antifungal susceptibility testing is pH dependent. Antimicrob Agents Ch. 1999;43:1383–1386. doi: 10.1128/aac.43.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delarze E, Sanglard D. Defining the frontiers between antifungal resistance, tolerance and the concept of persistence. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2015;23:12–19. doi: 10.1016/j.drup.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Marchetti O, Moreillon P, Glauser MP, Bille J, Sanglard D. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob Agents Ch. 2000;44:2373–2381. doi: 10.1128/aac.44.9.2373-2381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taff HT, Mitchell KF, Edward JA, Andes DR. Mechanisms of Candida biofilm drug resistance. Future microbiology. 2013;8:1325–1337. doi: 10.2217/fmb.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meletiadis J, Curfs-Breuker I, Meis JF, Mouton JW. In vitro antifungal susceptibility testing of Candida isolates with the EUCAST methodology, a new method for ECOFF determination. Antimicrob Agents Ch. 2017;61 doi: 10.1128/aac.02372-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockhart SR, Ghannoum MA, Alexander BD. Establishment and use of epidemiological cutoff values for molds and yeasts by use of the Clinical and Laboratory Standards Institute M57 standard. J Clin Microbiol. 2017;55:1262–1268. doi: 10.1128/jcm.02416-16. [This review provides an important and accessible over-review of the concept of epidemiological cut-offs as well as the methodology that underlies their establishment.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onyewu C, Wormley FL, Jr, Perfect JR, Heitman J. The calcineurin target, Crz1, functions in azole tolerance but is not required for virulence of Candida albicans. Infection and immunity. 2004;72:7330–7333. doi: 10.1128/IAI.72.12.7330-7333.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Molecular microbiology. 2003;48:959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- 27.Luna-Tapia A, Tournu H, Peters TL, Palmer GE. Endosomal Trafficking Defects Can Induce Calcium-Dependent Azole Tolerance in Candida albicans. Antimicrob Agents Ch. 2016;60:7170–7177. doi: 10.1128/AAC.01034-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luna-Tapia A, Butts A, Palmer GE. Loss of C-5 Sterol desaturase activity in Candida albicans: Azole resistance or merely trailing growth? Antimicrob Agents Ch. 2019;63 doi: 10.1128/aac.01337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Linden JW, et al. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2013;57:513–520. doi: 10.1093/cid/cit320. [DOI] [PubMed] [Google Scholar]
- 30.McCarty TP, Pappas PG. Invasive Candidiasis. Infectious Disease Dlinics of North America. 2016;30:103–124. doi: 10.1016/j.idc.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Prasad R, Rawal MK, Shah AH. Candida efflux ATPases and antiporters in clinical drug resistance. Advances in experimental medicine and biology. 2016;892:351–376. doi: 10.1007/978-3-319-25304-6_15. [DOI] [PubMed] [Google Scholar]
- 32.Sasse C, et al. The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans. Mol Microbiol. 2012;86:539–556. doi: 10.1111/j.1365-2958.2012.08210.x. [This manuscript showed that when a strain accumulates multiple resistance mutations that affect different transcription factors, the effects are independent an additive. In addition, these effects, generated by engineered deletion mutations, were associated with reduced fitness in the absence of drug stress. Since many resistant isolates do not have obvious fitness defects, it is assumed that they must have acquired compensatory mechanisms in addition to the characterized mechanisms of drug resistance.] [DOI] [PubMed] [Google Scholar]
- 33.Holmes AR, et al. Heterozygosity and functional allelic variation in the Candida albicans efflux pump genes CDR1 and CDR2. Mol Microbiol. 2006;62:170–186. doi: 10.1111/j.1365-2958.2006.05357.x. [DOI] [PubMed] [Google Scholar]
- 34.Flowers SA, et al. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell. 2012;11:1289–1299. doi: 10.1128/ec.00215-12. [Established that gain-of-function mutations in UPC2, the key regulator of ergosterol biosynthesis gene expression, are a source of clinically important fluconazole resistance. Previously, only transcriptional regulators of efflux pump expression were known to do so.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang MJ, et al. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS yeast research. 2013;13:386–393. doi: 10.1111/1567-1364.12042. [DOI] [PubMed] [Google Scholar]
- 36.Flowers SA, Colon B, Whaley SG, Schuler MA, Rogers PD. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Ch. 2015;59:450–460. doi: 10.1128/aac.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz JF, et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018;9:5346. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Healey KR, et al. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Ch. 2018;62 doi: 10.1128/aac.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chowdhary A, et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. The Journal of antimicrobial chemotherapy. 2018;73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 40.Lockhart SR, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satoh K, et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 42.Kordalewska M, et al. Understanding Echinocandin Resistance in the Emerging Pathogen Candida auris. Antimicrob Agents Ch. 2018;62 doi: 10.1128/aac.00238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science (New York, NY) 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson MZ, Saha A, Haseeb A, Bennett RJ. A chromosome 4 trisomy contributes to increased fluconazole resistance in a clinical isolate of Candida albicans. Microbiology. 2017;163:856–865. doi: 10.1099/mic.0.000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang F, et al. Tolerance to caspofungin in Candida albicans is associated with at least three distinctive mechanisms that govern expression of FKS genes and cell wall remodeling. Antimicrob Agents Ch. 2017;61 doi: 10.1128/aac.00071-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang F, Kravets A, Bethlendy G, Welle S, Rustchenko E. Chromosome 5 monosomy of Candida albicans controls susceptibility to various toxic agents, including major antifungals. Antimicrob Agents Ch. 2013;57:5026–5036. doi: 10.1128/aac.00516-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang F, et al. Aneuploidy enables cross-adaptation to unrelated drugs. Molecular biology and evolution. 2019 doi: 10.1093/molbev/msz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Todd RT, Wikoff TD, Forche A, Selmecki A. Genome plasticity in Candida albicans is driven by long repeat sequences. eLife. 2019;8 doi: 10.7554/eLife.45954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coste A, et al. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics. 2006;172:2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS genetics. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mount HO, et al. Global analysis of genetic circuitry and adaptive mechanisms enabling resistance to the azole antifungal drugs. PLoS genetics. 2018;14:e1007319. doi: 10.1371/journal.pgen.1007319. [This manuscript screened a library of C. albicans deletion strains and identified two genes that affect azole responses and have roles in stress response pathways and whose functions were suppressed by the acquisition of aneuploid chromosomes. Whether the mutations were affecting resistance or tolerance is not clear, but activities of the identified genes (vacuolar retrograde trafficking and a cell wall and polarity GTPase activating protein) suggest that the mechanism is involved in processes that affect tolerance rather than in directly affecting resistance.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todd RT, Forche A, Selmecki A. Ploidy variation in fungi: Polyploidy, aneuploidy, and genome evolution. Microbiology spectrum. 2017;5 doi: 10.1128/microbiolspec.FUNK-0051-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett RJ, Forche A, Berman J. Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a019604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ford CB, et al. The evolution of drug resistance in clinical isolates of Candida albicans. eLife. 2015;4:e00662. doi: 10.7554/eLife.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White TC. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Ch. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [The first detailed study of the molecular mechanisms underlying the development of fluconazole resistance over time in patients. A landmark manuscript because of its patient-based approach and because it yielded strains that are still under study today.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cowen LE, et al. Population genomics of drug resistance in Candida albicans. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9284–9289. doi: 10.1073/pnas.102291099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Forche A, et al. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS biology. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrison BD, et al. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS biology. 2014;12:e1001815. doi: 10.1371/journal.pbio.1001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forche A, et al. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio. 2011;2 doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ene IV, et al. Global analysis of mutations driving microevolution of a heterozygous diploid fungal pathogen. P Natl Acad Sci USA. 2018;115:E8688–e8697. doi: 10.1073/pnas.1806002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forche A, et al. Rapid Phenotypic and Genotypic Diversification After Exposure to the Oral Host Niche in Candida albicans. Genetics. 2018;209:725–741. doi: 10.1534/genetics.118.301019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forche A, May G, Magee PT. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans strains during infection. Eukaryot Cell. 2005;4:156–165. doi: 10.1128/ec.4.1.156-165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forche A, Magee PT, Selmecki A, Berman J, May G. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics. 2009;182:799–811. doi: 10.1534/genetics.109.103325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pappas PG, et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016;62:409–417. doi: 10.1093/cid/civ1194. [DOI] [PubMed] [Google Scholar]
- 65.Perlin DS. Echinocandin Resistance in Candida. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2015;61(Suppl 6):S612–617. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farmakiotis D, Tarrand JJ, Kontoyiannis DP. Drug-resistant Candida glabrata infection in cancer patients. Emerging infectious diseases. 2014;20:1833–1840. doi: 10.3201/eid2011.140685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-D-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob Agents Ch. 2009;53:3690–3699. doi: 10.1128/aac.00443-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. Mechanisms of Antifungal Drug Resistance. Csh Perspect Med. 2015;5 doi: 10.1101/cshperspect.a019752. doi:ARTN a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bordallo-Cardona MA, et al. MSH2 gene point mutations are not antifungal resistance markers in Candida glabrata. Antimicrob Agents Ch. 2019;63 doi: 10.1128/aac.01876-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh A, et al. Absence of Azole or Echinocandin Resistance in Candida glabrata Isolates in India despite Background Prevalence of Strains with Defects in the DNA Mismatch Repair Pathway. Antimicrobial Agents and Chemotherapy. 2018;62 doi: 10.1128/AAC.00195-18. doi:ARTN e00195-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shor E, Schuyler J, Perlin DS. A Novel, Drug Resistance-Independent, Fluorescence-Based Approach To Measure Mutation Rates in Microbial Pathogens. Mbio. 2019;10 doi: 10.1128/mBio.00120-19. doi:ARTN e00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ropars J, et al. Gene flow contributes to diversification of the major fungal pathogen Candida albicans. Nature communications. 2018;9:2253. doi: 10.1038/s41467-018-04787-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hickman MA, et al. The 'obligate diploid' Candida albicans forms mating-competent haploids. Nature. 2013;494:55–59. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao J, et al. Candida albicans gains azole resistance by altering sphingolipid composition. Nat Commun. 2018;9:4495. doi: 10.1038/s41467-018-06944-1. [This manuscript used a haploid C. albicans isolate and a transposon mutagenesis system to identify mutants that are able to improve growth in very high fluconazole levels. However, the degree to which the mutants are resistant vs tolerance is not addressed, as drug responses did not include MIC assays. The study provides convincing data showing that alterations in sphingolipid composition of membranes is associated with the ability to grow and divide in the presence of fluconazole.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Segal ES, et al. Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans. mBio. 2018;9 doi: 10.1128/mBio.02048-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walker LA, Gow NA, Munro CA. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob Agents Ch. 2013;57:146–154. doi: 10.1128/aac.01486-12. [This manuscript found that the ability of Candida species to grow in the presence of sub-MIC levels of caspofungin was associated with increased chitin levels in the cell wall. Pretreatment of cells with CaCl2 and calcofluor white induced a similar increase in chitin and ability to grow in sub-MIC caspofungin levels. Whether this effect is an alteration in tolerance or resistance levels remains to be determined; higher cell wall chitin levels are induced by reagents such as CaCl2 and calcofluor white, the effect is transient and physiological, and thus sharing some features of tolerance.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee KK, et al. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob Agents Ch. 2012;56:208–217. doi: 10.1128/aac.00683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singh-Babak SD, et al. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS pathogens. 2012;8:e1002718. doi: 10.1371/journal.ppat.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Selmecki A, Forche A, Berman J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryotic cell. 2010;9:991–1008. doi: 10.1128/ec.00060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sitterle E, et al. Within-host genomic diversity of Candida albicans in healthy carriers. Scientific reports. 2019;9:2563. doi: 10.1038/s41598-019-38768-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cowen LE. Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS pathogens. 2009;5:e1000471. doi: 10.1371/journal.ppat.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science (New York, N.Y.) 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 83.Khandelwal NK, et al. Azole resistance in a Candida albicans mutant lacking the ABC transporter CDR6/ROA1 depends on TOR signaling. Journal of Biological Chemistry. 2018;293:412–432. doi: 10.1074/jbc.M117.807032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garnaud C, et al. The Rim Pathway Mediates Antifungal Tolerance in Candida albicans through Newly Identified Rim101 Transcriptional Targets, Including Hsp90 and Ipt1. Antimicrobial agents and chemotherapy. 2018;62 doi: 10.1128/AAC.01785-17. doi:ARTN e01785-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robbins N, Caplan T, Cowen LE. Molecular evolution of antifungal drug resistance. Annual review of microbiology. 2017;71:753–775. doi: 10.1146/annurev-micro-030117-020345. [DOI] [PubMed] [Google Scholar]
- 86.Shapiro RS, Zaas AK, Betancourt-Quiroz M, Perfect JR, Cowen LE. The Hsp90 co-chaperone Sgt1 governs Candida albicans morphogenesis and drug resistance. PloS one. 2012;7:e44734. doi: 10.1371/journal.pone.0044734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cowen LE, et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2818–2823. doi: 10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Olin-Sandoval V, et al. Lysine harvesting is an antioxidant strategy and triggers underground polyamine metabolism. Nature. 2019;572:249–253. doi: 10.1038/s41586-019-1442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Benhamou RI, et al. Real-time imaging of the azole class of antifungal drugs in live Candida cells. ACS chemical biology. 2017;12:1769–1777. doi: 10.1021/acschembio.7b00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Benhamou RI, Bibi M, Berman J, Fridman M. Localizing Antifungal Drugs to the Correct Organelle Can Markedly Enhance their Efficacy. Angewandte Chemie (International ed. in English) 2018;57:6230–6235. doi: 10.1002/anie.201802509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Campbell K, Herrera-Dominguez L, Correia-Melo C, Zelezniak A, Ralser M. Biochemical principles enabling metabolic cooperativity and phenotypic heterogeneity at the single cell level. Current Opinion in Systems Biology. 2018;8:97–108. doi: 10.1016/j.coisb.2017.12.001. [DOI] [Google Scholar]
- 92.Franzmann TM, Alberti S. Protein Phase Separation as a Stress Survival Strategy. Csh Perspect Med. 2019;9 doi: 10.1101/cshperspect.a0340.58. doi:ARTN a034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Delarue M, et al. mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell. 2018;174:338–349.e320. doi: 10.1016/j.cell.2018.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roemer T, Krysan DJ. Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cornet M, et al. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infection and immunity. 2005;73:7977–7987. doi: 10.1128/Iai.73.12.7977-7987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu SJ, Chang YL, Chen YL. Calcineurin signaling: lessons from Candida species. FEMS yeast research. 2015;15 doi: 10.1093/femsyr/fov016. fov016. [DOI] [PubMed] [Google Scholar]
- 97.Hubbard M, Bradley M, Sullivan P, Shepherd M, Forrester I. Evidence for the occurrence of calmodulin in the yeasts Candida albicans and Saccharomyces cerevisiae. FEBS letters. 1982;137:85–88. doi: 10.1016/0014-5793(82)80320-7. [DOI] [PubMed] [Google Scholar]
- 98.O'Meara TR, Robbins N, Cowen LE. The Hsp90 Chaperone Network Modulates Candida Virulence Traits. Trends in microbiology. 2017;25:809–819. doi: 10.1016/j.tim.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gerstein AC, Rosenberg A, Hecht I, Berman J. diskImageR: quantification of resistance and tolerance to antimicrobial drugs using disk diffusion assays. Microbiology (Reading, England) 2016;162:1059–1068. doi: 10.1099/mic.0.000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ben-Ami R, et al. Heteroresistance to fluconazole is a continuously distributed phenotype among Candida glabrata clinical strains associated with in vivo persistence. mBio. 2016;7 doi: 10.1128/mBio.00655-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sionov E, Chang YC, Garraffo HM, Kwon-Chung KJ. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob Agents Ch. 2009;53:2804–2815. doi: 10.1128/aac.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Varma A, Kwon-Chung KJ. Heteroresistance of Cryptococcus gattii to fluconazole. Antimicrob Agents Ch. 2010;54:2303–2311. doi: 10.1128/aac.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sionov E, Chang YC, Kwon-Chung KJ. Azole heteroresistance in Cryptococcus neoformans: emergence of resistant clones with chromosomal disomy in the mouse brain during fluconazole treatment. Antimicrob Agents Ch. 2013;57:5127–5130. doi: 10.1128/aac.00694-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stone NR, et al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. The Journal of clinical investigation. 2019;129:999–1014. doi: 10.1172/JCI124516. [This manuscript tracked series of clinical C. neoformans isolates from individual patients with HIV from Tanzania and found that heteroresistance was quite common and was often, but not always, associated with specific chromosomal aneuploidies.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Desai JV, Mitchell AP, Andes DR. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb Perspect Med. 2014;4 doi: 10.1101/cshperspect.a019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lohse MB, Gulati M, Johnson AD, Nobile CJ. Development and regulation of single- and multi-species Candida albicans biofilms. Nature reviews. Microbiology. 2018;16:19–31. doi: 10.1038/nrmicro.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Ch. 2001;45:2475–2479. doi: 10.1128/aac.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taff HT, et al. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS pathogens. 2012;8:e1002848. doi: 10.1371/journal.ppat.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li P, Seneviratne CJ, Alpi E, Vizcaino JA, Jin L. Delicate metabolic control and coordinated stress response critically determine antifungal tolerance of Candida albicans biofilm persisters. Antimicrob Agents Ch. 2015;59:6101–6112. doi: 10.1128/aac.00543-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Denega I, d'Enfert C, Bachellier-Bassi S. Candida albicans biofilms are generally devoid of persister cells. Antimicrob Agents Ch. 2019;63 doi: 10.1128/aac.01979-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Al-Dhaheri RS, Douglas LJ. Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob Agents Ch. 2008;52:1884–1887. doi: 10.1128/aac.01473-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stevens DA, Espiritu M, Parmar R. Paradoxical effect of caspofungin: reduced activity against Candida albicans at high drug concentrations. Antimicrob Agents Ch. 2004;48:3407–3411. doi: 10.1128/aac.48.9.3407-3411.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stevens DA, White TC, Perlin DS, Selitrennikoff CP. Studies of the paradoxical effect of caspofungin at high drug concentrations. Diagn Micr Infec Dis. 2005;51:173–178. doi: 10.1016/j.diagmicrobio.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 114.Rueda C, Cuenca-Estrella M, Zaragoza O. Paradoxical growth of Candida albicans in the presence of caspofungin is associated with multiple cell wall rearrangements and decreased virulence. Antimicrob Agents Ch. 2014;58:1071–1083. doi: 10.1128/aac.00946-13. [A detailed assessment of the cellular changes that accompany growth beyond the MIC in caspofungin as well as the effects that those changes have on fitness in mammalian hosts.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wagener J, Loiko V. Recent insights into the paradoxical effect of echinocandins. Journal of fungi (Basel, Switzerland) 2017;4 doi: 10.3390/jof4010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rueda C, et al. Evaluation of the possible influence of trailing and paradoxical effects on the clinical outcome of patients with candidemia. Clin Microbiol Infect. 2017;23:49.e41–49.e48. doi: 10.1016/j.cmi.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 117.Lee MK, et al. Susceptibility and trailing growth of Candida albicans to fluconazole: results of a Korean multicentre study. Mycoses. 2007;50:148–149. doi: 10.1111/j.1439-0507.2006.01329.x. [DOI] [PubMed] [Google Scholar]
- 118.Arthington-Skaggs BA, et al. Comparison of visual and spectrophotometric methods of broth microdilution MIC end point determination and evaluation of a sterol quantitation method for in vitro susceptibility testing of fluconazole and itraconazole against trailing and nontrailing Candida isolates. Antimicrob Agents Ch. 2002;46:2477–2481. doi: 10.1128/aac.46.8.2477-2481.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rex JH, et al. Optimizing the correlation between results of testing in vitro and therapeutic outcome in vivo for fluconazole by testing critical isolates in a murine model of invasive candidiasis. Antimicrob Agents Ch. 1998;42:129–134. doi: 10.1128/aac.42.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Colombo AL, et al. Prognostic factors and historical trends in the epidemiology of candidemia in critically ill patients: an analysis of five multicenter studies sequentially conducted over a 9-year period. Intensive Care Med. 2014;40:1489–1498. doi: 10.1007/s00134-014-3400-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hirakawa MP, et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome research. 2015;25:413–425. doi: 10.1101/gr.174623.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang MY, Woolford CA, May G, McManus CJ, Mitchell AP. Circuit diversification in a biofilm regulatory network (vol 15, e1007787, 2019) PLoS pathogens. 2019;15 doi: 10.1371/journal.ppat.1007966. doi:ARTN e1007966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hooper RW, Ashcraft DS, Pankey GA. In vitro synergy with fluconazole plus doxycycline or tigecycline against clinical Candida glabrata isolates. Med Mycol. 2019;57:122–126. doi: 10.1093/mmy/myy008. [DOI] [PubMed] [Google Scholar]
- 124.Lu M, et al. Linezolid in combination with azoles induced synergistic effects against Candida albicans and protected Galleria mellonella against experimental Candidiasis. Front Microbiol. 2018;9:3142. doi: 10.3389/fmicb.2018.03142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.da Silva CR, et al. Synergistic effects of amiodarone and fluconazole on Candida tropicalis resistant to fluconazole. Antimicrob Agents Ch. 2013;57:1691–1700. doi: 10.1128/aac.00966-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li H, et al. Resistance reversal induced by a combination of fluconazole and tacrolimus (FK506) in Candida glabrata. Journal of medical microbiology. 2015;64:44–52. doi: 10.1099/jmm.0.081760-0. [DOI] [PubMed] [Google Scholar]
- 127.Gu W, Guo D, Zhang L, Xu D, Sun S. The Synergistic Effect of Azoles and Fluoxetine against Resistant Candida albicans Strains Is Attributed to Attenuating Fungal Virulence. Antimicrob Agents Ch. 2016;60:6179–6188. doi: 10.1128/AAC.03046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Costa-de-Oliveira S, et al. Ibuprofen potentiates the in vivo antifungal activity of fluconazole against Candida albicans murine infection. Antimicrob Agents and chemotherapy. 2015;59:4289–4292. doi: 10.1128/aac.05056-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.(European Research Council Advanced Grant, 2014–2018)
- 130.Uppuluri P, Nett J, Heitman J, Andes D. Synergistic effect of calcineurin inhibitors and fluconazole against Candida albicans biofilms. Antimicrobial agents and chemotherapy. 2008;52:1127–1132. doi: 10.1128/Aac.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.