ABSTRACT
Diet is an important, modifiable lifestyle factor of cardiometabolic disease risk, and an improved diet can delay or even prevent the onset of disease. Recent evidence suggests that individuals could benefit from diets adapted to their genotype and phenotype: that is, personalized nutrition. A novel strategy is to tailor diets for groups of individuals according to their metabolic phenotypes (metabotypes). Randomized controlled trials evaluating metabotype-specific responses and nonresponses are urgently needed to bridge the current gap of knowledge with regard to the efficacy of personalized strategies in nutrition. In this Perspective, we discuss the concept of metabotyping, review the current literature on metabotyping in the context of cardiometabolic disease prevention, and suggest potential strategies for metabotype-based nutritional advice for future work. We also discuss potential determinants of metabotypes, including gut microbiota, and highlight the use of metabolomics to define effective markers for cardiometabolic disease–related metabotypes. Moreover, we hypothesize that people at high risk for cardiometabolic diseases have distinct metabotypes and that individuals grouped into specific metabotypes may respond differently to the same diet, which is being tested in a project of the Joint Programming Initiative: A Healthy Diet for a Healthy Life.
Keywords: metabotyping, personalized nutrition, precision nutrition, targeted nutrition, cardiometabolic diseases, gut microbiota, metabolomics
Introduction
Diet is among the most important modifiable lifestyle factors contributing to cardiometabolic disease risk. Changes in a diet can delay or even prevent the onset of disease. In the current work, we define cardiometabolic disease as various disease entities that are either consequences of or late stages of the metabolic syndrome. We therefore include metabolic syndrome, Type 2 diabetes, and cardiovascular disease endpoints in this definition. Prevention is considered to be the most sustainable and cost-effective way to manage chronic diseases (1). While national and international dietary guidelines serve to promote health and prevent disease from a population perspective, data from Europe, the United States, and Australia have clearly shown that these guidelines are poorly adhered to (2–6). To combat obesity and related conditions, some individuals may need to follow personalized nutrition strategies as a complement to the general population-based advice. Such recommendations have been suggested in the most recent advisory report for the Dietary Guidelines for Americans, based on recent randomized controlled trials showing that personalized interventions could lead to greater weight loss than non-personalized strategies (7). In addition to health status being improved as a result of improved biological effects when personalizing the diet, there is also evidence that people are more open to health-promoting information when it is personalized and when the individuals recognize themselves as being highly susceptible to preventable diseases (8, 9). However, it remains to be elucidated to what extent personalized nutrition has advantages over generalized strategies and whether it is feasible at a large scale (10–12).
Personalized nutrition at the individual level requires not only the comprehensive collection of information, which is both costly and demanding, but also models that are capable of accurately generating personalized advice for the individual. A more feasible approach may be to personalize diets at the group level. Over the last 10–15 years, individuals have been grouped according to genetic set up, for example based on genetic variants that are associated with disease risk (13). However, the benefits to public health have been limited. More recent studies have suggested that individuals may be grouped according to unique metabolic responses to foods and dietary changes (14, 15). In fact, what constitutes a healthy diet may differ across individuals as a consequence of a number of factors beyond genetics, including health status, medication, and also the gut microbiota (16–18). Grouping individuals based on similarities in their metabolic phenotype—that is, metabotypes—is a novel concept, and different definitions have been used (19–23). The underlying idea behind metabotyping is to identify metabolic phenotypes based on factors such as diet, anthropometric measures, clinical parameters, metabolomics data, and the gut microbiota. An optimal diet can then be tailored to fit each metabotype specifically (Figure 1). In this perspective article, we discuss the current literature reporting on metabotypes in the context of cardiometabolic disease and how metabotyping may be used as a nutritional strategy for the improved prevention of chronic disease.
FIGURE 1.
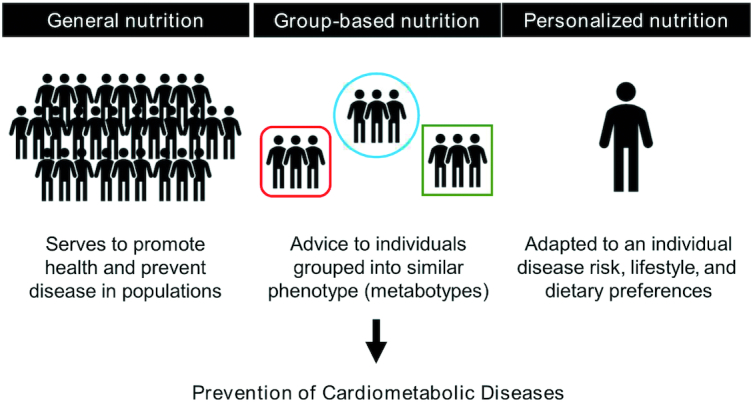
Metabotyping and group-based nutrition in the context of the conventional population-based guidelines and personalized nutrition.
Current Status of Knowledge
Definitions and strategies for personalized nutrition
There is currently no overall, generally agreed-upon definition of personalized nutrition or precision nutrition. As in other scientific fields under development, concepts have been used often without uniform definitions. In the area of personalized nutrition, concepts adapted to individuals or groups of individuals have been described: Ordovas and colleagues (10) defined “stratified and tailored nutrition” as approaches attempting to tailor diets to groups of individuals sharing similar characteristics, “personalized nutrition” as an attempt to deliver nutritional interventions or advice suited to each individual, and “precision nutrition” as the most ambitious definition, which implies a quantitative understanding of the individual, their food intake, and their phenotype (including health) to offer the nutritional interventions or advice of the greatest benefits to the individual. Furthermore, Gonzalez and Betts (24) integrated dynamics—that is, a time dimension—in their definition of precision nutrition, aiming to tailor nutrition to best meet the varying demands and requirements throughout a day, week, season, or lifespan. Most recently, the North American Branch of the International Life Sciences Institute convened a multidisciplinary panel and defined personalized nutrition as using “individual-specific information, founded in evidence-based science, to promote dietary behavior change that may result in measurable health benefits.” In addition, they suggested 10 guiding principles for personalized nutrition approaches (25). Here, we use personalized nutrition and precision nutrition interchangeably, and define it as the most appropriate diet to maximize health benefits tailored to the individual or a group of individuals.
It has been suggested that targeted nutritional advice can be personalized on 3 levels: on the basis of the current diet (Level 1); on the diet and phenotype, consisting of anthropometric and biochemical/clinical measurements (Level 2); or diet, phenotype, and genotype (Level 3) (26–29). Some evidence suggests that strategies that include genetic information have greater potential than strategies based on the background diet and health phenotype alone for improving health through personalized nutrition (28, 30–32). However, omics techniques other than genomics may also benefit personalized nutrition, such as analyses of the microbiome (metagenomics) and the metabolome (metabolomics), but these techniques have not been included in the investigation of different personalization levels. Although such omics techniques provide phenotype information, represented at Level 2, they hold immense potential compared to the traditional phenotypic measurements, as they provide a functional readout of a phenotype at a much more detailed level. Metabolomics can, to some degree, reflect food and nutrient consumption, as well as provide mechanistic evidence of the specific metabolic pathways impacted by diet (33, 34), and the gut microbiota phenotype has been shown to modulate physiological responses to diet (18).
In a landmark article from 2015, Zeevi et al. (16) showed benefits when adapting diets based on factors underlying inter-individual differences in post-prandial glucose response. In brief, the post-prandial glucose response could be accurately predicted using a tree-based machine learning algorithm, combining data on gut microbiota, blood markers, body composition, diet, and exercise. Next, the authors were able to perform personalized interventions with “good” and “bad” diets, capable of affecting post-prandial glucose levels in a favorable or non-favorable direction. Importantly, what constituted a “bad diet” for some participants resembled the “good diet” for other participants, and vice versa. This study thus showcased the potential to use phenotypic measures in combination with machine-learning algorithms on biological big data for precision nutrition at the individual level. The approach was recently validated in an American cohort of non-diabetic subjects (35). Similar phenotyping strategies can be used to identify metabotypes and, thus, personalize nutrition on a group level.
Metabotyping based on anthropometric and clinical/biochemical parameters
Metabotyping, where individuals were clustered into metabolic phenotypes based on anthropometric, biochemical, and clinical cardiometabolic measures, has been suggested as a potential nutritional strategy (21, 36–42). Riedl et al. (43) showed that 3 metabotypes, derived from BMI and 33 biochemical parameters (e.g., blood lipids, glucose, hormones, and liver enzymes) using k-means clustering, differed in cardiometabolic disease prevalence and 7-year disease incidence, thus illustrating the ability to detect high- and low-risk groups. Krishnan et al. (44) detected unique, post-prandial leptin and glucose responses for 3 distinct groups of overweight, healthy women in a cross-over study on high– versus low–glycemic index meals, using a principal component analysis. Although a firm link between post-prandial glucose response and long-term health outcomes has yet to be established among healthy subjects, differences in post-prandial metabolism are considered reflective of the metabolic health status of individuals and serve as a proxy for high-risk groups of cardiometabolic disease (45). Interestingly, when reanalyzing data from a weight-loss trial, Ritz and colleagues (46) found that metabotyping based on fasting glucose and insulin could predict weight loss from a healthy, Nordic diet rich in dietary fiber, versus a control diet (46). They also concluded that these basic measures, although seemingly of clinical relevance, likely lacked details on metabolic regulation, and that more complex measurements could provide better personalized nutrition advice. Moreover, O'Donovan and colleagues (21) demonstrated a communication strategy for large-scale, metabotype-based personalized nutrition: a decision-tree approach providing each participant with pre-defined dietary messages based on their metabotype, in combination with their BMI, waist circumference, and blood pressure. The 3 metabotypes had been identified based on 4 blood-based, clinical measures: blood glucose, triacylglycerols, total cholesterol, and high-density lipoprotein cholesterol. Compared to the personalized advice delivered by a dietitian, the decision tree generated comparable recommendations.
Gut microbiota as a potential determinant of metabotype
In recent years, the immense role of the gut microbiota in cardiometabolic disease has been highlighted, and this has been an area of intensive research (47–49). The impact of the gut microbiota on human health could partly be mediated through the ability of the gut microbiota to metabolize dietary compounds into new metabolites that impact disease risk. For example, the bacterial metabolism of dietary L-carnitine into the metabolites trimethylamine and trimethylamine N-oxide presents a mechanistic link of the relationship between red meat consumption and cardiovascular disease risk (50), and short-chain fatty acids provide a functional link between gut microbiota, dietary fiber intake, and several diet-related diseases (51–53).
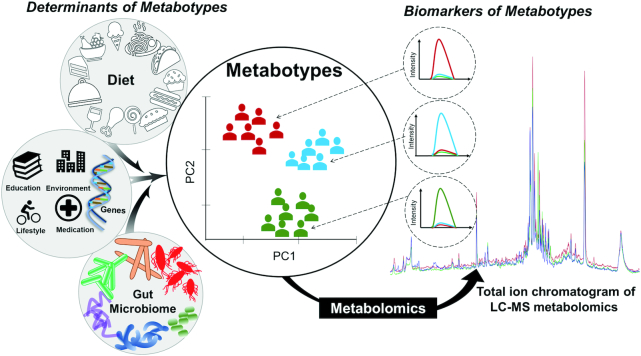
Interestingly, the diet–microbiota interactions have been shown to vary amongst individuals (54) and appear to be a determinant of the post-prandial response, as previously discussed (16). Moreover, even when a diet seemingly induces more systematic effects on the phenotype, a large part of the residual variability is associated with the microbiota in terms of composition and functionality (55). Several researchers have observed that individuals with similarities in their microbiota composition also share similarities in their phenotypic response to exposures. For instance, Romo-Vaquero et al. (56) observed that the Coriobacteriaceae could play a central role in urolithin production metabotypes (i.e., metabotypes that reflect the unique patterns of microbiota-derived metabolites from ellagitannin-rich foods), which in turn were associated with blood cholesterol levels and cardiometabolic health implications. The grouping of individuals by microbiota phenotype (enterotype) (57, 58) is a concept similar to metabotyping, with the important distinction that enterotyping is based on the microbial composition, with potential impact on diet–health interactions, whilst the metabotype partly reflects the activity of the microbiota in conjunction with dietary exposures and other phenotypic traits. The enterotype may thus be an important determinant of the metabotype, and studies investigating enterotyping in the context of diets can, therefore, provide unique and valuable insights into how to integrate metabotyping into precision nutrition (Figure 2).
FIGURE 2.
The graphic illustration of potential determinants of metabotypes and a metabolomics-based strategy for the identification of biomarkers of metabotypes by using untargeted LC-MS metabolomics. Individuals are grouped into metabotypes based on potential determinants and their interactions, using certain statistical approaches, such as PC analysis. PC, principal component.
For example, Kovatcheva-Datchary et al. (18) demonstrated the potential to predict response to diet on the enterotype level by showing that the prevalence of Prevotella determined individuals’ glycemic response to barley-based bread in healthy adults. The contribution of the enterotype was further shown by fecal transplants from study participants to germ-free mice. Improved glucose regulation and a higher capacity to digest dietary fiber were observed only in mice of the enterotype rich in Prevotella. Similarly, Korem and colleagues (17) found low– and high–glucose response groups after subjects had consumed whole-grain sourdough bread and refined wheat bread, and the groups could be accurately predicted based on the baseline gut microbial composition. Their results suggest that although there may be underlying, systematic health effects on a population level for different bread types, that effect could be overshadowed by systematic variabilities between individuals. Furthermore, enterotypes based on the relative abundance of the Prevotella and Bacteroides genera have been shown to impact the response to dietary weight loss interventions, where individuals with a high Prevotella-to-Bacteroides ratio lost more weight and body fat in response to high-fiber diets, compared to those with a low bacterial ratio (59).
Taken together, enterotypes may be a key determinant of phenotypes and be especially useful when personalizing diets in terms of dietary fiber and glycemic response. Enterotypes are also considered to influence the metabolisms of other dietary compounds, such as polyphenols, leading to the production of enterotype-specific bacterial metabolites (60–62). However, gut microbial genes, which may be important for metabolic disease phenotypes, can be present across bacterial phyla and also differ between bacterial strains that otherwise share the same genetic makeup (63). This supports the idea of metabotyping: that is, that the functionality of the gut microbiota, rather than the presence of bacteria per se, may be important for disease prevention.
Metabolites as markers for cardiometabolic disease metabotypes
Metabolomics may be particularly well suited to discover metabotypes and the biomarkers thereof, as well as to unravel how the gut microbiome contributes to variations in phenotypic responses to diets (Figure 2). Metabolites have been shown to reflect cardiometabolic disease states and risk factors (64–67), as well as host and gut microbial genetics and lifestyle factors (67–69). Moreover, recent studies have shown that microbiota composition can be effectively mirrored in both the fecal and plasma metabolomes (68).
Very few studies have been published where metabolomics has been applied for metabotyping as a means to set up improved strategies to combat cardiometabolic diseases. These studies have clustered men and women into metabolite-derived groups correlated to disease traits, risk factors, and eating patterns (15, 70–72). Men and women from a large Irish study, for example, were clustered according to their plasma fatty acid profile, creating 4 groups that differed in terms of metabolic syndrome components, anthropometric measures, dietary habits, and demographics (70). In another study, urinary metabolites associated with diabetes were used to divide diabetic and non-diabetic participants into 4 novel groups (71). Differences in plasma glucose levels led the authors to speculate on differences in disease management and the risk of future complications for the 2 metabotypes with diabetic patients. From a nutritional perspective, Fiamoncini and colleagues (15) identified 2 metabolite-based metabotypes, and evaluated their respective responses to a dietary intervention aimed at weight loss. In brief, healthy, overweight men and women were clustered into 2 distinct metabotypes based on post-prandial concentrations of metabolites related to lipid metabolism. Individuals belonging to the different metabotypes also differed in terms of amino acid and carbohydrate metabolism, post-prandial glucose and insulin levels, liver lipid contents, intra-abdominal fat mass, and eating patterns. When challenged with a 12-week weight loss trial with a reduced caloric intake, only the individuals with the more disease-associated metabotype showed improvements in glucose and insulin levels at the end of the intervention; thus, the study identified a responsive and a nonresponsive metabotype (15). In another recent study, Muniandy et al. (72) identified 2 distinct subgroups of monozygotic twin pairs associated with cardiometabolic risk factors, high-density lipoprotein cholesterol, and BMI using metabolomics.
A framework for metabotype-based nutritional advice
Given the heterogeneity in choices of variables which can be used to cluster into metabotypes, it is difficult to compare metabotypes between studies and populations (40). In fact, obtained metabotypes should be carefully assessed and validated against underlying research questions and, consequently, clustering variables. Planning the experimental study design also encompasses the choice between baseline (fasting or not fasting) and post-prandial samples, biological samples (blood, urine, or fecal samples), and continuous sampling to evaluate responses over time; these decisions are closely linked to the choice of study endpoints. Choosing statistical approaches for deciphering metabotypes from the large and complex data sets will be an instrumental part of future metabotyping studies and presents a great challenge at present. Part of the difficulty lies in how to handle and weigh the vastly different types of data. Appropriate data analytical methods will not only dictate how metabotypes are defined, but also reproducibility across research groups and, subsequently, generalizability across study populations. Although beyond the scope of this perspectives article, we note that arbitrary selection of the number of metabotypes seems to be a common practice to date; however, there are objective methods that select the ultimate number of clusters, as demonstrated by Riedl et al. (40).
Whilst the discovery of metabotypes may be complex and require careful consideration, metabotyping should preferably be simple, rapid, and affordable once metabotypes have been defined and validated. Biomarkers suitable for large-scale applications are needed, and we encourage the consideration of metabotype biomarker evaluation and validation in future metabotyping studies (Figure 2). Blood biomarkers could be useful, as they can be measured repeatedly and continuously, whilst other biological samples (e.g., fecal samples) may be more difficult to collect at defined time points and close time intervals. Moreover, work is needed to evaluate potential underlying mechanistic links and to identify traits of the metabolic phenotype undetected by measurements of a few traditional risk factors, and it may require the inclusion of other data domains, such as gut microbiota and the entire metabolome. Metabolites present as interesting candidates: they may not only be reflective of metabotypes and responses but have the advantage of being present in several biological specimens, such as blood, urine, and stool. On the other hand, future work should assess whether grouping individuals based on a small number of anthropometric, clinical, or other easily measured parameters alone might be adequate to identify those diets that would be optimal for specific metabotypes. This is relevant to make personalized nutrition more feasible and cost-effective in clinical practice.
Performing dietary intervention trials that evaluate metabotype-specific responses is a fundamental step required to bridge the current gap of knowledge with regard to the efficacy of personalized strategies in nutrition. In an ongoing Joint Programming Initiative, A Healthy Diet for a Healthy Life project, “Diet × gut microbiome-based metabotypes to determine cardio-metabolic risk and tailor intervention strategies for improved health” (73), we have hypothesized that adults at high risk of developing cardiometabolic disease will have different responses to fermentable dietary fibers, dependent on their metabotypes and specific diet–gut microbiota interactions. The idea is to define metabotypes based on the subjects' metabolomes in the context of the gut bacterial profiles, host genetics, lifestyle factors, anthropometric measures, and dietary profile. We will next test our hypothesis in a separate study population using a randomized controlled cross-over dietary intervention; we will investigate the stability of the metabotypes throughout the intervention and research whether it is possible to shift an unhealthy metabotype into a more healthy metabotype. Although the primary endpoint will be postprandial glucose, we will also evaluate other parameters that are important for cardiometabolic diseases, including blood pressure, blood lipids, and circulating C-reactive protein, as a proxy for systemic inflammation. Our study will allow us to not only investigate a new approach for personalized nutrition, but also to provide estimates of the efficacy of such interventions.
Along with short-term interventions to evaluate the effects of specific dietary interventions across metabotypes on cardiometabolic risk factors, long-term studies to evaluate adherence to metabotype-specific diets and their role for hard endpoints are warranted, but difficult to perform. However, metabotyping may also be used in an observational setting in prospective cohort studies. Such a study setting would allow for the assessment of long-term reproducibility of reported metabotypes and could be used to deduce potential biomarkers, as well as to investigate their generalizability across populations. Recent studies suggest that distinct metabotypes (and enterotypes) may be present across populations (21, 74), although the prevalence of a given metabotype may vary between countries (21). Moreover, metabolite-based metabotypes have been shown to be stable over several years (75, 76), yet some metabolites have shown higher conservation than others (76). Interestingly, metabotype instability per se has been correlated with the incidence of cardiovascular disease and all-cause mortality (75).
Practical considerations of implementing metabotype-based precision nutrition in the health-care setting
Personalized strategies rely on the premise that advice can be delivered to individuals or groups of individuals in an effective manner and that such strategies have been proven to be effective and economically feasible. Studies assessing health economic parameters for different personalized strategies, including metabotyping, are currently lacking and, therefore, needed. For the delivery of personalized nutrition advice, the ability of health-care professionals to communicate targeted nutritional advice, and their likelihood of doing so, will be instrumental if implementating metabotyping and precision nutrition in the health-care setting, together with support from policymakers, society, and other actors (8).
Several communication strategies have been employed to deliver personalized dietary advice to consumers, differing in terms of target group, means of communication, and basis of personalization (77). The previously mentioned work by O'Donovan and colleagues also investigated the delivery of metabotype-specific dietary advice on a larger scale (21, 41). Individuals’ willingness and motivation to undergo metabotyping and nutritional interventions form another practical aspect that warrants consideration. There is a need to evaluate attitudes and acceptance towards metabotyping, as well as other strategies in precision nutrition. In the meantime, studies investigating attitudes towards genetic testing as a means to prevent disease can provide some insight (78–80). For example, these studies have suggested that adults at high risk for cardiometabolic diseases may be more motivated to change their health behaviors (78); presenting with disease risk factors was associated with a higher willingness to undergo genetic testing, and perceived risks and family medical history were related to the motivation to exercise, as well as to modify lifestyle factors, such as diet and physical activity (79). Yet, the relatively low proportion of participants (14–33%) who reported improving their health behavior 3–9 months following genetic testing also raises the questions of how to achieve high intervention compliance, lasting changes in health behaviors, and improvements in health parameters. We believe it is realistic to assume that a multitude of factors will impact the outcomes and that the contribution of such factors in specific populations needs to be addressed.
Concluding Remarks
Metabotyping is a relatively new concept within the area of personalized nutrition. In the current literature, metabotypes are often defined either based on clinical and anthropometric markers (disease-associated metabotypes) or the metabolism of certain nutrients and dietary components, such as dietary fiber and polyphenols (diet-associated metabotypes). We hypothesize that the disease-associated metabotypes—that is, metabotypes present in those populations at high risk for cardiometabolic diseases—will affect the response to a specific diet. We further hypothesize that the gut microbiota is a key determinant and a modifier of metabotypes, in addition to habitual diet, genotype, anthropometric measures, and biochemical and clinical markers. In other words, the interaction between the host and exogenous exposures (e.g., diet, drugs, and gut microbiota) is an important factor regarding dietary response, and identifying such “functional” metabotypes may offer advantages over “clinical biomarker–based” metabotypes as well as enterotypes for the optimization of a diet to an individual for the prevention of cardiometabolic disease. However, much more work is needed to investigate whether responses to particular diets and dietary items are indeed metabotype-specific and whether diets tailored for metabotypes could lead to health improvements that are clinically meaningful. Ongoing and future studies will hopefully: shed light on whether metabotype clusters can be deduced in general populations, elucidate potential main determinants and, test whether the response to specific dietary interventions, such as diets high in fermentable fiber, vary across distinct metabotypes. If so, finding easily measured biomarkers of metabotypes is our highest priority, to allow the tailoring of diets for optimal prevention at a large scale.
ACKNOWLEDGEMENTS
The authors’ responsibilities were as follows – MP, CB, RL: carefully reviewed the literature and drafted the manuscript; AR-H: complemented and updated the literature search; AR-H, LS, NET, RG-D, RZ-R, YLY, JH, AT, GR, RG, GC, CV, JN, CA-L: discussed and commented on the manuscript; LS: helped with creating the figures presented in the article; and all authors: read and approved the final manuscript.
Notes
The work was developed as part of the FORMAS (Swedish Research Council for Sustainable Development) project "Diet × gut microbiome-based metabotypes to determine cardio-metabolic risk and tailor intervention strategies for improved health" (2017-02003) funded by the European Joint Programming Initiative “A Healthy Diet for a Healthy Life” (http://www.healthydietforhealthylife.eu) (to RL, GR, CA-L); and the FORMAS project “Optimal diet guided by metabotype for health & wellbeing” (2016-00314) (to RL). The work was also supported by Spanish National Grants from MINECO (the Ministry of Economy and Competitiveness) (PCIN-2017-076 and FJCI-2015-26590); the Generalitat de Catalunya's Agency AGAUR (Agency for Management of University and Research Grants) (2017SGR1546); CIBERFES (Centro de Investigacion Biomedica en Red Fragilidad y Envejecimiento Saludable) from the ISCIII (Institute of Health Carlos III) [co-funded by the FEDER (The European Regional Development Fund) Program from the European Union]; the Miguel Servet program (CP15/00100) from the ISCIII; and the European Social Fund, ICREA (The Catalan Institution for Research and Advanced Studies) Academia 2018 (to CA-L).
Author disclosures: The authors report no conflicts of interest.
Perspective articles allow authors to take a position on a topic of current major importance or controversy in the field of nutrition. As such, these articles could include statements based on author opinions or point of view. Opinions expressed in Perspective articles are those of the author and are not attributable to the funder(s) or the sponsor(s) or the publisher, Editor, or Editorial Board of Advances in Nutrition. Individuals with different positions on the topic of a Perspective are invited to submit their comments in the form of a Perspectives article or in a Letter to the Editor.
References
- 1. World Health Organization. Diet, nutrition and the prevention of chronic diseases: Report of a Joint WHO/FAO Expert Consultation, Geneva, 28 January – 1 February 2002. Geneva, Switzerland: World Health Organization; 2002. [Google Scholar]
- 2. Maiani G, Caston MJ, Catasta G, Toti E, Cambrodon IG, Bysted A, Granado-Lorencio F, Olmedilla-Alonso B, Knuthsen P, Valoti M. Carotenoids: Actual knowledge on food sources, intakes, stability and bioavailability and their protective role in humans. Mol Nutr Food Res. 2009;53(Suppl 2):S194–218. [DOI] [PubMed] [Google Scholar]
- 3. Sjors C, Hedenus F, Sjolander A, Tillander A, Balter K. Adherence to dietary recommendations for Swedish adults across categories of greenhouse gas emissions from food. Public Health Nutr. 2017;20:3381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walton J, Kehoe L, McNulty BA, Nugent AP, Flynn A. Nutrient intakes and compliance with nutrient recommendations in children aged 1–4 years in Ireland. J Hum Nutr Diet. 2017;30:665–76. [DOI] [PubMed] [Google Scholar]
- 5. Krebs-Smith SM, Guenther PM, Subar AF, Kirkpatrick SI, Dodd KW. Americans do not meet federal dietary recommendations. J Nutr. 2010;140:1832–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Australian Bureau of Statistics. Australian health survey: Consumption of food groups from the Australian Dietary Guidelines. Canberra: Australian Bureau of Statistics; 2016. [Google Scholar]
- 7. Dietary Guidelines Advisory Committee.Scientific report of the 2015 Dietary Guidelines Advisory Committee: Advisory report to the Secretary of Health and Human Services and the Secretary of Agriculture. Washington, DC: US Department of Agriculture, Agricultural Research Service; 2015. [Google Scholar]
- 8. Bouwman LI, Koelen MA.. Communication on personalised nutrition: individual-environment interaction. Genes Nutr. 2007;2:81–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Roos B. Personalised nutrition: ready for practice?. Proc Nutr Soc. 2013;72:48–52. [DOI] [PubMed] [Google Scholar]
- 10. Ordovas JM, Ferguson LR, Tai ES, Mathers JC. Personalised nutrition and health. BMJ. 2018;361:bmj.k2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang DD, Hu FB.. Precision nutrition for prevention and management of Type 2 diabetes. Lancet Diabetes Endocrinol. 2018;6:416–26. [DOI] [PubMed] [Google Scholar]
- 12. Astrup A, Hjorth MF.. Classification of obesity targeted personalized dietary weight loss management based on carbohydrate tolerance. Eur J Clin Nutr. 2018;72:1300–4. [DOI] [PubMed] [Google Scholar]
- 13. Vazquez-Vidal I, Desmarchelier C, Jones PJH. Nutrigenetics of blood cholesterol concentrations: Towards personalized nutrition. Curr Cardiol Rep. 2019;21:114–20. [DOI] [PubMed] [Google Scholar]
- 14. Hjorth MF, Zohar Y, Hill JO, Astrup A. Personalized dietary management of overweight and obesity based on measures of insulin and glucose. Annu Rev Nutr. 2018;38:245–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fiamoncini J, Rundle M, Gibbons H, Thomas EL, Geillinger-Kastle K, Bunzel D, Trezzi JP, Kiselova-Kaneva Y, Wopereis S, Wahrheit J et al.. Plasma metabolome analysis identifies distinct human metabotypes in the postprandial state with different susceptibility to weight loss-mediated metabolic improvements. FASEB J. 2018;32(10):5447–58. [DOI] [PubMed] [Google Scholar]
- 16. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M et al.. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–94. [DOI] [PubMed] [Google Scholar]
- 17. Korem T, Zeevi D, Zmora N, Weissbrod O, Bar N, Lotan-Pompan M, Avnit-Sagi T, Kosower N, Malka G, Rein M et al.. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab. 2017;25:1243–53. e5. [DOI] [PubMed] [Google Scholar]
- 18. Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Bjorck I, Backhed F. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22:971–82. [DOI] [PubMed] [Google Scholar]
- 19. Riedl A, Gieger C, Hauner H, Daniel H, Linseisen J. Metabotyping and its application in targeted nutrition: An overview. Br J Nutr. 2017;117:1631–44. [DOI] [PubMed] [Google Scholar]
- 20. Friston D, Laycock H, Nagy I, Want EJ. Microdialysis workflow for metabotyping superficial pathologies: Application to burn injury. Anal Chem. 2019;91:6541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Donovan CB, Walsh MC, Woolhead C, Forster H, Celis-Morales C, Fallaize R, Macready AL, Marsaux CFM, Navas-Carretero S, Rodrigo San-Cristobal S et al.. Metabotyping for the development of tailored dietary advice solutions in a European population: The Food4Me study. Br J Nutr. 2017;118:561–9. [DOI] [PubMed] [Google Scholar]
- 22. Brennan L. Use of metabotyping for optimal nutrition. Curr Opin Biotechnol. 2017;44:35–8. [DOI] [PubMed] [Google Scholar]
- 23. Tomas-Barberan FA, Gonzalez-Sarrias A, Garcia-Villalba R, Nunez-Sanchez MA, Selma MV, Garcia-Conesa MT, Espin JC. Urolithins, the rescue of “old” metabolites to understand a “new” concept: Metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol Nutr Food Res. 2017;61(1), doi: 10.1002/mnfr.201500901. [DOI] [PubMed] [Google Scholar]
- 24. Gonzalez JT, Betts JA.. Dietary sugars, exercise and hepatic carbohydrate metabolism. Proc Nutr Soc. 2019;78(2):246–56. [DOI] [PubMed] [Google Scholar]
- 25. Adams SH, Anthony JC, Carvajal R, Chae L, Khoo CSH, Latulippe ME, Matusheski NV, McClung HL, Rozga M, Schmid CH et al.. Perspective: Guiding principles for the implementation of personalized nutrition approaches that benefit health and function. Advances in Nutrition. 2019, doi: 10.1093/advances/nmz086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ronteltap A, van Trijp H, Berezowska A, Goossens J. Nutrigenomics-based personalised nutritional advice: In search of a business model?. Genes Nutr. 2013;8:153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gibney MJ, Walsh MC.. The future direction of personalised nutrition: My diet, my phenotype, my genes. Proc Nutr Soc. 2013;72:219–25. [DOI] [PubMed] [Google Scholar]
- 28. Livingstone KM, Celis-Morales C, Navas-Carretero S, San-Cristobal R, Macready AL, Fallaize R, Forster H, Woolhead C, O'Donovan CB, Marsaux CF et al.. Effect of an Internet-based, personalized nutrition randomized trial on dietary changes associated with the Mediterranean diet: The Food4Me Study. Am J Clin Nutr. 2016;104:288–97. [DOI] [PubMed] [Google Scholar]
- 29. Celis-Morales C, Livingstone KM, Marsaux CF, Macready AL, Fallaize R, O'Donovan CB, Woolhead C, Forster H, Walsh MC, Navas-Carretero S et al.. Effect of personalized nutrition on health-related behaviour change: Evidence from the Food4Me European randomized controlled trial. Int J Epidemiol. 2017;46:578–88. [DOI] [PubMed] [Google Scholar]
- 30. Nielsen DE, El-Sohemy A.. Disclosure of genetic information and change in dietary intake: A randomized controlled trial. PLOS One. 2014;9:e112665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hietaranta-Luoma HL, Tahvonen R, Iso-Touru T, Puolijoki H, Hopia A. An intervention study of individual, apoE genotype-based dietary and physical-activity advice: Impact on health behavior. J Nutrigenet Nutrigenomics. 2014;7:161–74. [DOI] [PubMed] [Google Scholar]
- 32. Arkadianos I, Valdes AM, Marinos E, Florou A, Gill RD, Grimaldi KA. Improved weight management using genetic information to personalize a calorie controlled diet. Nutr J. 2007;6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Toro-Martín J, Arsenault BJ, Després J-P, Vohl M-C. Precision nutrition: A review of personalized nutritional approaches for the prevention and management of metabolic syndrome. Nutrients. 2017;9:913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brennan L. Metabolomics: A powerful tool to enrich our understanding of the impact of food on health. Mol Nutr Food Res. 2019;63:1870087. [DOI] [PubMed] [Google Scholar]
- 35. Mendes-Soares H, Raveh-Sadka T, Azulay S, Edens K, Ben-Shlomo Y, Cohen Y, Ofek T, Bachrach D, Stevens J, Colibaseanu D et al.. Assessment of a personalized approach to predicting postprandial glycemic responses to food among individuals without diabetes. JAMA Netw Open. 2019;2:e188102–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Botelho PB, Fioratti CO, Abdalla DS, Bertolami MC, Castro IA. Classification of individuals with dyslipidaemia controlled by statins according to plasma biomarkers of oxidative stress using cluster analysis. Br J Nutr. 2010;103:256–65. [DOI] [PubMed] [Google Scholar]
- 37. Wilcox M, Li Q, Sun Y, Stang P, Berlin J, Wang D. Genome-wide association study for empirically derived metabolic phenotypes in the Framingham Heart Study offspring cohort. BMC Proceedings. 2009;3(Suppl 7):S53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zubair N, Kuzawa CW, McDade TW, Adair LS. Cluster analysis reveals important determinants of cardiometabolic risk patterns in Filipino women. Asia Pac J Clin Nutr. 2012;21:271–81. [PMC free article] [PubMed] [Google Scholar]
- 39. Zubair N, Kuzawa CW, Lee NR, McDade TW, Adair LS. Clustering and determinants of cardiometabolic risk factors among Filipino young adults. Asia Pac J Clin Nutr. 2014;23:148–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Riedl A, Gieger C, Hauner H, Daniel H, Linseisen J. Metabotyping and its application in targeted nutrition: An overview. Br J Nutr. 2017;117:1631–44. [DOI] [PubMed] [Google Scholar]
- 41. O'Donovan CB, Walsh MC, Nugent AP, McNulty B, Walton J, Flynn A, Gibney MJ, Gibney ER, Brennan L. Use of metabotyping for the delivery of personalised nutrition. Mol Nutr Food Res. 2015;59:377–85. [DOI] [PubMed] [Google Scholar]
- 42. van Bochove K, van Schalkwijk DB, Parnell LD, Lai CQ, Ordovas JM, de Graaf AA, van Ommen B, Arnett DK. Clustering by plasma lipoprotein profile reveals two distinct subgroups with positive lipid response to fenofibrate therapy. PLOS One. 2012;7:e38072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Riedl A, Wawro N, Gieger C, Meisinger C, Peters A, Roden M, Kronenberg F, Herder C, Rathmann W, Völzke H et al.. Identification of comprehensive metabotypes associated with cardiometabolic diseases in the population-based KORA study. Mol Nutr Food Res. 2018;62:e1800117. [DOI] [PubMed] [Google Scholar]
- 44. Krishnan S, Newman J, Hembrooke T, Keim N. Variation in metabolic responses to meal challanges differing in glycemic index in healthy women: Is it meaningful?. Nutr Metab (Lond). 2012;9(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morris C, O'Grada C, Ryan M, Roche HM, Gibney MJ, Gibney ER, Brennan L. Identification of differential responses to an oral glucose tolerance test in healthy adults. PLOS One. 2013;8:e72890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ritz C, Astrup A, Larsen TM, Hjorth MF. Weight loss at your fingertips: Personalized nutrition with fasting glucose and insulin using a novel statistical approach. Eur J Clin Nutr. 201;; 73(11):1529–35. [DOI] [PubMed] [Google Scholar]
- 47. Marchesi JR. The human microbiota and microbiome. Wallingford, UK: CABI; 2014. [Google Scholar]
- 48. Brunkwall L, Orho-Melander M.. The gut microbiome as a target for prevention and treatment of hyperglycaemia in Type 2 diabetes: From current human evidence to future possibilities. Diabetologia. 2017;60:943–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kurilshikov A, van den Munckhof ICL, Chen L, Bonder MJ, Schraa K, Rutten JHW, Riksen NP, de Graaf J, Oosting M, Sanna S et al.. Gut microbial associations to plasma metabolites linked to cardiovascular phenotypes and risk. Circ Res. 2019;124:1808–20. [DOI] [PubMed] [Google Scholar]
- 50. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L et al.. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J et al.. Gut bacteria selectively promoted by dietary fibers alleviate Type 2 diabetes. Science. 2018;359:1151–6. [DOI] [PubMed] [Google Scholar]
- 52. Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, Mujagic Z, Masclee AAM, Jonkers DMAE, Oosting M et al.. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51:600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tirosh A, Calay ES, Tuncman G, Claiborn KC, Inouye KE, Eguchi K, Alcala M, Rathaus M, Hollander KS, Ron I et al.. The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Sci Trans Med. 2019;11:eaav0120. [DOI] [PubMed] [Google Scholar]
- 54. Li F, Hullar MA, Schwarz Y, Lampe JW. Human gut bacterial communities are altered by addition of cruciferous vegetables to a controlled fruit- and vegetable-free diet. J Nutr. 2009;139:1685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loo RL, Zou X, Appel LJ, Nicholson JK, Holmes E. Characterization of metabolic responses to healthy diets and association with blood pressure: Application to the Optimal Macronutrient Intake Trial for Heart Health (OmniHeart), a randomized controlled study. Am J Clin Nutr. 2018;107:323–34. [DOI] [PubMed] [Google Scholar]
- 56. Romo-Vaquero M, Cortes-Martin A, Loria-Kohen V, Ramirez-de-Molina A, Garcia-Mantrana I, Collado MC, Espin JC, Selma MV. Deciphering the human gut microbiome of urolithin metabotypes: Association with enterotypes and potential cardiometabolic health implications. Mol Nutr Food Res. 2019;63:e1800958. [DOI] [PubMed] [Google Scholar]
- 57. Costea PI, Hildebrand F, Arumugam M, Bäckhed F, Blaser MJ, Bushman FD, de Vos WM, Ehrlich SD, Fraser CM, Hattori M et al.. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. 2018;3:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cheng M, Ning K.. Stereotypes about enterotype: The old and new ideas. Genomics Proteomics Bioinformatics. 2019;17:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hjorth MF, Roager HM, Larsen TM, Poulsen SK, Licht TR, Bahl MI, Zohar Y, Astrup A. Pre-treatment microbial Prevotella-to-Bacteroides ratio, determines body fat loss success during a 6-month randomized controlled diet intervention. Int J Obes (Lond). 2018;42:580–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tomás-Barberán FA, García-Villalba R, González-Sarrías A, Selma MV, Espín JC. Ellagic acid metabolism by human gut microbiota: Consistent observation of three urolithin phenotypes in intervention trials, independent of food source, age, and health status. J Agric Food Chem. 2014;62:6535–8. [DOI] [PubMed] [Google Scholar]
- 61. Selma MV, Gonzalez-Sarrias A, Salas-Salvado J, Andres-Lacueva C, Alasalvar C, Orem A, Tomas-Barberan FA, Espin JC. The gut microbiota metabolism of pomegranate or walnut ellagitannins yields two urolithin-metabotypes that correlate with cardiometabolic risk biomarkers: Comparison between normoweight, overweight-obesity and metabolic syndrome. Clin Nutr. 2018;37:897–905. [DOI] [PubMed] [Google Scholar]
- 62. Cortes-Martin A, Garcia-Villalba R, Gonzalez-Sarrias A, Romo-Vaquero M, Loria-Kohen V, Ramirez-de-Molina A, Tomas-Barberan FA, Selma MV, Espin JC. The gut microbiota urolithin metabotypes revisited: The human metabolism of ellagic acid is mainly determined by aging. Food Funct. 2018;9(8):4100–6. [DOI] [PubMed] [Google Scholar]
- 63. Zeevi D, Korem T, Godneva A, Bar N, Kurilshikov A, Lotan-Pompan M, Weinberger A, Fu J, Wijmenga C, Zhernakova A et al.. Structural variation in the gut microbiome associates with host health. Nature. 2019;568:43–8. [DOI] [PubMed] [Google Scholar]
- 64. Lynch CJ, Adams SH.. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10:723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C et al.. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lu Y, Wang Y, Ong C-N, Subramaniam T, Choi HW, Yuan J-M, Koh W-P, Pan A. Metabolic signatures and risk of Type 2 diabetes in a Chinese population: An untargeted metabolomics study using both LC-MS and GC-MS. Diabetologia. 2016;59:2349–59. [DOI] [PubMed] [Google Scholar]
- 67. Brial F, Le Lay A, Dumas ME, Gauguier D. Implication of gut microbiota metabolites in cardiovascular and metabolic diseases. Cell Mol Life Sci. 2018;75(21):3977–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zierer J, Jackson MA, Kastenmuller G, Mangino M, Long T, Telenti A, Mohney RP, Small KS, Bell JT, Steves CJ et al.. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. 2018;50:790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–17. [DOI] [PubMed] [Google Scholar]
- 70. Li K, Brennan L, McNulty BA, Bloomfield JF, Duff DJ, Devlin NF, Gibney MJ, Flynn A, Walton J, Nugent AP. Plasma fatty acid patterns reflect dietary habits and metabolic health: A cross-sectional study. Mol Nutr Food Res. 2016;60:2043–52. [DOI] [PubMed] [Google Scholar]
- 71. Urpi-Sarda M, Almanza-Aguilera E, Llorach R, Vazquez-Fresno R, Estruch R, Corella D, Sorli JV, Carmona F, Sanchez-Pla A, Salas-Salvadó J et al.. Non-targeted metabolomic biomarkers and metabotypes of Type 2 diabetes: A cross-sectional study of PREDIMED trial participants. Diabetes Metab. 2019;45(2):167–74. [DOI] [PubMed] [Google Scholar]
- 72. Muniandy M, Velagapudi V, Hakkarainen A, Lundbom J, Lundbom N, Rissanen A, Kaprio J, Pietiläinen KH, Ollikainen M. Plasma metabolites reveal distinct profiles associating with different metabolic risk factors in monozygotic twin pairs. Int J Obes. 2019;43:487–502. [DOI] [PubMed] [Google Scholar]
- 73.European Joint Programming Initiative, “A Healthy Diet for a Healthy Life.” JPI-HDHL INTIMIC [Internet]. Available from: https://www.healthydietforhealthylife.eu/index.php/ec-partnerships/hdhl-intimic. [Google Scholar]
- 74. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM et al.. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lacruz ME, Kluttig A, Tiller D, Medenwald D, Giegling I, Rujescu D, Prehn C, Adamski J, Greiser KH, Kastenmuller G. Instability of personal human metabotype is linked to all-cause mortality. Sci Rep. 2018;8:9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yousri NA, Kastenmuller G, Gieger C, Shin SY, Erte I, Menni C, Peters A, Meisinger C, Mohney RP, Illig T et al.. Long term conservation of human metabolic phenotypes and link to heritability. Metabolomics. 2014;10:1005–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ronteltap A, van Trijp H, Berezowska A, Goossens J. Nutrigenomics-based personalised nutritional advice: In search of a business model?. Genes Nutr. 2013;8:153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stewart-Knox BJ, Bunting BP, Gilpin S, Parr HJ, Pinhao S, Strain JJ, de Almeida MD, Gibney M. Attitudes toward genetic testing and personalised nutrition in a representative sample of European consumers. Br J Nutr. 2009;101:982–9. [DOI] [PubMed] [Google Scholar]
- 79. Nielsen DE, El-Sohemy A.. A randomized trial of genetic information for personalized nutrition. Genes Nutr. 2012;7:559–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kaufman DJ, Bollinger JM, Dvoskin RL, Scott JA. Risky business: Risk perception and the use of medical services among customers of DTC personal genetic testing. J Genet Couns. 2012;21:413–22. [DOI] [PubMed] [Google Scholar]