Abstract
The amounts of the intracellular glycosylation, O-GlcNAc modification, are increased in essentially all tumors when compared to healthy tissue, and lowering O-GlcNAcylation levels results in reduced tumorigenesis and increased cancer cell death. Therefore, the pharmacological reduction of O-GlcNAc may represent a therapeutic vulnerability. The most direct approach to this goal is the inhibition of O-GlcNAc transferase (OGT), the enzyme that directly adds the modification to proteins. However, despite some recent success, this enzyme has proven difficult to inhibit. An alternative strategy involves starving OGT of its sugar substrate UDP-GlcNAc by targeting enzymes of the hexosamine biosynthetic pathway (HBP). Here, we explore the potential of the rate-determining enzyme of this pathway, glutamine fructose-6-phosphate amidotransferase (GFAT). We first show that CRISPR-mediated knockout of GFAT results in inhibition of cancer cell growth in vitro and a xenograft model that correlates with O-GlcNAcylation levels. We then demonstrate that pharmacological inhibition of GFAT sensitizes a small panel of cancer cells to undergo apoptosis in response to diamide-induced oxidative stress. Finally, we find that GFAT expression and O-GlcNAc levels are increased in a spontaneous mouse model of liver cancer. Together these experiments support the further development of inhibitors of the HBP as an indirect approach to lowering O-GlcNAcylation levels in cancer.
Graphical Abstract

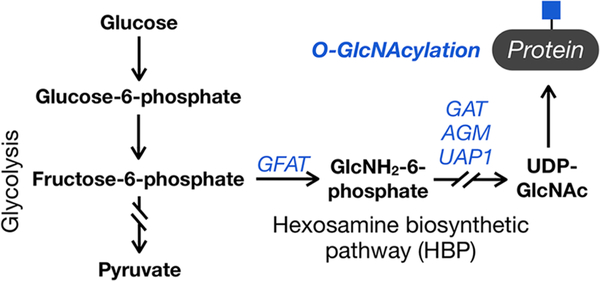
The hexosamine biosynthetic pathway (HBP) transforms fructose-6-phosphate into uridine diphosphate N-acetyl-glucosamine (UDP-GlcNAc) (Figure 1). UDP-GlcNAc can then serve as a direct substrate for various glycosyltransferases that install N-acetylglucosamine onto proteins, lipids, and proteoglycans. Of these many classes of glycans, intracellular O-GlcNAc modification (O-GlcNAcylation) is particularly sensitive to changes in UDP-GlcNAc concentrations. For example, genetic knockout of one member of the HBP biosynthetic pathway, glucosamine 6-phosphate N-acetyltransferase, results in a notable loss of O-GlcNAc, but less so in cell-surface glycosylation.1 We believe that this observation is explained by the uniquely dynamic nature of O-GlcNAcylation, which is added to intracellular proteins by O-GlcNAc transferase (OGT) and removed by the enzyme O-GlcNAcase (OGA).2,3 This creates a potential sink into lumenal and cell-surface glycans, which appear to be maintained by salvaging GlcNAc from serum,1 and away from O-GlcNAc. Additionally, O-GlcNAcylation is particularly well suited to change as a consequence of metabolism because OGT responds to different concentrations of its donor substrate UDP-GlcNAc. Specifically, OGT displays a range of apparent KM values for UDP-GlcNAc with different protein substrates, resulting in both higher and different O-GlcNAcylation events at elevated UDP-GlcNAc concentrations.4 Therefore, O-GlcNAc modification can function as a biochemical readout of the cellular UDP-GlcNAc concentration.
Figure 1.
The hexosamine biosynthetic pathway and O-GlcNAcylation. A percentage of the glucose taken into a cell is converted to the donor sugar UDP-GlcNAc by the hexosamine biosynthetic pathway (HBP), where it can be used for the O-GlcNAc modification of intracellular proteins. This type of glycosylation is particularly sensitive to changes in UDP-GlcNAc concentration.
O-GlcNAcylation is the addition of N-acetylglucosamine (GlcNAc) to serines and threonines of hundreds to thousands of intracellular proteins.5–7 Unlike most other forms of glycosylation, the core O-linked-GlcNAc residue is not further elaborated by additional monosaccharides, and it can compete for phosphorylation at identical or nearby sites. Although O-GlcNAcylation appears to have many roles in mammalian cells, one of its major functions is promoting cell survival in response to cellular stress. For example, global O-GlcNAcylation levels are rapidly increased upon exposure of cells to a variety of stresses (e.g., heat, osmotic, oxidative),8 as well as in models of heart attack and ischemia.9–11 In the majority of cases, these modest increases in O-GlcNAcylation are phenotypically protective and prevent cell death. Elevated O-GlcNAcylation is also a common feature of essentially every cancer type compared to the corresponding healthy tissue.12–15 Notably, several of these studies demonstrated that the shRNA knockdown of OGT results in cancer apoptosis in culture and prevents tumor formation in xenografts, and pharmacological inhibition of OGT has antigrowth and pro-apoptotic effects in culture.
Given these results, several laboratories have explored the development of OGT inhibitors. Until very recently,16 these inhibitors either had a documented off-target activity17 or require the metabolism of a sulfur-containing analogue of GlcNAc.18,19 Additionally, these inhibitors are competitive, and the cellular levels of UDP-GlcNAc are relatively high at double-digit μM levels.20,21 Therefore, there has been interest in lowering O-GlcNAc levels by instead targeting the biosynthesis of UDP-GlcNAc and thereby starving OGT of its substrate, by targeting the rate-determining enzyme of the HBP, glutamine fructose-6-phosphate amidotransferase (GFAT), a tetrameric enzyme that converts glutamine and fructose-6-phosphate to glutamate and glucosamine-6-phosphate.22,23 Furthermore, an increased flux through the HBP is a feature of several cancers.24–27 Most of these studies take advantage of the RNAi-mediated knockdown of GFAT and/or two naturally occurring small-molecule inhibitors: azaserine and 6-diazo-5-oxo-norleucine (DON). For example, RNAi of GFAT reduced the growth of mutant Kras-driven pancreatic cancer cells in cultures and xenografts.25 More recently, RNAi and DON were used to demonstrate that inhibition of the hexosamine biosynthetic pathway through GFAT sensitizes lung cancer cells to cisplatin in a culture.28 Unfortunately, these approaches have their drawbacks. GFAT knockdown by RNAi is generally incomplete, and even small amounts of UDP-GlcNAc can be enough to maintain glycosylation levels.1 Additionally, both azaserine and DON are reactive electrophiles that mimic glutamine and therefore inhibit a range of important enzymes beyond GFAT in cancer cells, including enzymes involved in the biosynthesis of amino acids and nucleosides.29
Here, we use CRISPR/Cas9 to generate genetic knockouts of GFAT in the lung (H1299) and breast (MCF7) cancer cell lines. Complete loss of GFAT resulted in cell death of both cell lines in the culture, which could be rescued by the addition of exogenous GlcNAc and significantly less H1299 tumor growth in a xenograft model. We then demonstrate that treatment of lung (A549 and H1299) and breast (MCF7) cancer cells with DON alone does indeed lower their O-GlcNAcylation levels; however, it does not cause a large amount of apoptosis. When we combined this inhibitor with either oxidative stress, we observed a significant increase in program cancer cell death, which could again be rescued by providing GlcNAc in the media. To confirm these results, we then synthesized a noncovalent GFAT inhibitor previously developed by Roche,30 which we term here R28. Similar to DON, treatment of the same with cell lines with R28 resulted in the dose-dependent loss of O-GlcNAcylation levels. However, the cellular potency of R28 was somewhat disappointing, demonstrating that further medicinal chemistry is needed to produce a potent and selective inhibitor of GFAT. Finally, we explored whether GFAT expression levels may be induced during tumor formation to maintain elevated O-GlcNAc levels. More specifically, we used a previously reported spontaneous model of liver cancer based on the specific deletion of lipid phosphatase and tensin homologue deleted on chromosome 10 (Pten).31 Not surprisingly, we find well-established cancer-associated changes in the transcription of the metabolic enzymes hexokinase-2 and the pyruvate kinase M2 isoform. Notably, we also find increased transcription and protein expression of GFAT that coincides with the formation of liver tumors and increased O-GlcNAcylation. This indicates that increased GFAT expression may drive flux through the HBP to support tumor O-GlcNAcylation in vivo. Collectively, our results further support the possibility of inhibiting GFAT to lower O-GlcNAcylation levels in cancer that may result in cancer cell death due to tumor environmental factors like oxidative stress. They also indicate that GFAT is indeed a key target for the anticancer activity of DON, and likely azaserine.
METHODS AND EXPERIMENTAL DETAILS
Cell Culture.
H1299 cells were cultured in RPMI 1640 (CellGro) medium enriched with 10% fetal bovine serum (FBS, Atlanta Biologicals). A549 cells were cultured in F-12K (CellGro) medium enriched with 10% FBS. MCF-7 cells were cultured in high-glucose DMEM media (CellGro) enriched with 10% FBS and 100 μg mL−1 insulin (Gibco Insulin-Transferrin-Selenium, Thermo Fisher). Hepatocytes isolated from livers of Pten and control mice were immortalized as previously described.32 The resulting hepatocyte cell line was subsequently cultured in DMEM media (Mediatech), enriched with 10% FBS (Atlas Biologicals), 5 μg mL−1 insulin (Sigma), and 10 ng mL−1 epidermal growth factor (EGF) (Invitrogen). All cell lines were grown in a humidified incubator at 37 °C in an atmosphere of 5.0% CO2.
GFAT Knockout with CRISPR.
Expression vector pSpCas9(BB)-2A-Puro (PX459) (Addgene no. 48139) targeting GFAT1 was generated as previously described.33 Briefly, annealed oligonucleotides corresponding to the first exon of human GFAT1 (5′-CACCGCTTCAGAGACTGGAGTACAG-3′ and 5′-AAACCTGTACTCCAGTCT CTGAAGC-3′) were ligated into the BbsI restriction site in the plasmid to generate SpCas9-GFAT. H1299 or MCF7 cells were then transiently transfected with SpCas9-GFAT1 using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s protocol. Then, 48 h post-transfection, 1 μg of mL−1 puromycin and GlcNAc (10 mM) was added to the media for the next 72 h. The clonal selection was then performed through trypsinization and resuspension of a confluent 10 cm plate of either cell line into 10 mL of the appropriate media. Five μL of the cell mixture was then plated into a 20 cm plate with 20 mL of complete media with GlcNAc (10 mM). The media was replaced every 3 days until colonies of cells were established throughout the plate. About 10 days after plating, individual colonies were isolated using PYREX Cloning Cylinders (Corning) and trypsinization. Each clone was then propagated until confluency in a 10 cm dish before further characterization.
Western Blotting.
Collected cells were washed three times with PBS before lysis with NP-40 lysis buffer (1% NP-40, 50 mM triethanolamine, 150 mM NaCl, pH 7.4) containing Complete Mini protease inhibitor cocktail (Roche Biosciences). After 15 min, the supernatant (soluble cell lysate) was collected by centrifugation (4 °C for 10 min at 10 000g), and the protein concentration was determined by bicinchoninic acid (BCA) assay (Pierce, Thermo Scientific). Equal amounts of lysate were then mixed with 4× SDS loading buffer and separated by SDS-PAGE using 4–20% CriterionTM Tris-HCl polyacrylamide precast gel (Biorad) before being transferred to PVDF membrane (Biorad) through standard Western blotting protocols. All Western blots besides anti-O-GlcNAc were blocked in TBST (0.05% Tween-20, 150 mM NaCl, 10 mM Tris pH 8.0) containing 5% nonfat milk for 1 h at rt. They were then incubated with the appropriate primary antibody in blocking buffer overnight at 4 °C. Anti-O-GlcNAc blots were blocked in TBST containing 5% bovine serum albumin (BSA) for 1 h at room temperature and incubated overnight at 4 °C. The antiactin (Sigma), antitubulin (Sigma), anti-O-GlcNAc (RL2, Sigma) pan-anti-GFAT (IBL-America, 663), and antiGFAT2 (Cell Signaling) were used at 1:1000 dilution. The blots were then washed three times in TBST for 10 min and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody in the appropriate blocking buffer for 1 h at room temperature. These secondary antibodies, HRP-conjugated antimouse (Jackson ImmunoResearch, no. 715–035-150) and antirabbit (Jackson ImmunoResearch, no. 711–035-152), were both used at 1:10 000 dilutions. After being washed three more times with TBST for 10 min, the blots were developed using ECL reagents (Bio-Rad) and visualized using a ChemiDoc XRS+ molecular imager (BioRad).
Lectin Blotting.
MCF7, A549, and H1299 cells were treated at 70–75% confluency with either 100 μM DON or H2O vehicle for 24 h. Cells were harvested via scraping, pelleted by centrifugation (5 min, 2000g, 4 °C), and washed with PBS (1 mL) one time. Cell pellets were then resuspended in 50 μL of 4% SDS buffer (4% SDS, 150 mM NaCl, 50 mM TEA, pH 7.4) with Complete Mini protease inhibitor cocktail (Roche Biosciences) before being lysed by tip sonication (5 s on, 5 s off, 3×) and centrifuged (10 min, 10 000g, 15 °C). The supernatant was collected, and the protein concentration was determined by a BCA assay (Pierce, ThermoScientific). Final protein samples at 2 mg mL−1 were then prepared by the addition of 2× SDS loading buffer (20% glycerol, 0.2% bromophenol blue, 1.4% β-mercaptoethanol, 200 mM Tris-HCl, pH 6.8). After boiling for 5 min at 98 °C, 10 μg of total protein was loaded per lane for SDS-PAGE (Criterion TGX 4–20%, Bio-Rad). Proteins were transferred to a PVDF membrane (Bio-Rad) following a standard protocol and were blocked for 1 h in blocking buffer at room temperature. Blots were then washed (3 × 5 min) in TBST and incubated with biotinylated-LCA (Vector Laboratories, 1:1000 in TBST) for 1 h at room temperature. The blots were then washed (3 × 10 min) in TBST and incubated HRP-streptavidin (Jackson Laboratories, 1:10 000 in TBST) for 1 h at room temperature. Following the second incubation, blots were again washed 3 × 10 min with TBST and developed using ECL reagents (Bio-Rad) and visualized on the ChemiDoc XRS+ molecular imager (Bio-Rad).
Growth Curves.
Wild-type or GFAT knockout clones of either H1299 or MCF cells were plated into 6-well plates (5 × 104 cells per well) in triplicate for each set of the GlcNAc concentrations and time points specified in Figure 2. After different lengths of time (0, 24, 48, and 72 h), the number of cells was determined using a Countess II Automated Cell Counter (Thermo Fisher).
Figure 2.
Characterization of GFAT knockout cancer cells. (A) GFAT, the rate-determining enzyme of the hexosamine biosynthetic pathway, was knocked out in H1299 and MCF7 cells using CRISPR/ Cas9. (B, D) Several populations of H1299 or MCF7 knockout cells were characterized by Western blotting, and one was confirmed by sequencing. (C, E) O-GlcNAcylation levels of GFAT knockout cells can be rescued by exogenous GlcNAc and correlate with cellular proliferation (n = 3). H1299 or MCF7 knockout cells were supplemented with the indicated amounts of GlcNAc in the media and then analyzed by Western blotting and cell counting. Data shown in panels C and E represent the mean ± s.e.m.
Mice.
Mice were housed in specific pathogen-free conditions with ad libitum access to water and a standard diet. Mice greater than 8 weeks of age were used, with an equal number of each sex represented. All animal experiments were conducted in accordance with the Office of Laboratory Animal Welfare and approved by the University of Michigan Institutional Animal Care and Use Committees.
Mouse Xenograft Experiments.
H1299 cells (1 × 106) in 200 μL of 1:1 Matrigel-RPMI + 10% FBS were injected subcutaneously into the flanks of NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratories). Tumor growth was monitored via calipers and the tumor excised to determine the final mass. Tumors were quickly segmented and snap-frozen in LN2 for downstream analysis. Frozen tissue was homogenized in RIPA buffer supplemented with the EDTA-free protease inhibitor and PhosSTOP phosphatase inhibitor using a Pro 250 Homogenizer. The lysate was then cleared by centrifugation. Protein concentrations in the lysates were quantified by the BCA assay, and equal protein amounts were then separated by SDS-PAGE. Proteins were transferred to the Immobilon-FL PVDF membrane, and Western blotting was performed as described above before development with West Pico ECL and visualized using autoradiography film.
DON Treatment and Caspase-3/7 Activity Assay.
Cells (5000 per well) were plated into each well in a 96-well plate (Costar, black plate, clear bottom with lid). After 8 h, the existing media were replaced with 100 μL of fresh media containing the H2O vehicle or 6-diazo-5-oxo-L-norleucine (DON, Bachem, 100 μM). After an additional 24 h, 100 μL of fresh media containing the H2O vehicle or DON (200 μM) or diamide (Sigma-Aldrich; A549:400 μM, H1299:200 μM, MCF7:300 μM) or both DON (200 μM) and diamide (A549:400 μM, H1299:200 μM, MCF7:300 μM) was added into each well without removing the existing media. At the conclusion of the experiment, caspase activity was measured using the Caspase-Glo 3/7 Assay (Promega) according to the manufacturer’s protocol. After incubation for 30 min, luminescence was measured using a Synergy H4 Hybrid Reader (BioTek; Gain = 135, integration time = 5 s, read height of 1 mm). The signal was then normalized to the cell number (measured by trypsinolysis and counting by hemocytometer) of an identically treated well. GlcNAc-rescue experiments were performed identically to those described above but with the addition of GlcNAc (10 mM) where appropriate.
Metabolite Sample Preparation and Metabolomics.
Aqueous metabolite fractions were collected from cells by lysing with cold 80% methanol and then clarified by centrifugation. Samples were then normalized, processed, and analyzed as previously reported.34
R28 Treatment.
Cells at approximately 40% confluency in 10 cm dishes were treated with the indicated concentrations of R28 in fresh media (10 mL). After 48 h, the cells were collected by washing three times with PBS (10 mL) followed by trypsinization. They were then subjected to the Western blotting conditions described above.
Knockout Mice.
Targeted deletion of Pten in the liver was achieved by crossing PtenloxP/loxP mice with Alb-Cre+ mice, as previously described.31 Control animals are PtenloxP/loxP; Alb-Cre− mice. Mice were genotyped from tail DNA using standard genomic PCR techniques. For tissue collection, mice fasted overnight, and livers were then perfused with cold PBS, collected in formalin for histology, and freshly frozen in liquid nitrogen for protein extraction.
DNA Microarray.
Total RNA was isolated from liver tissues from control and Pten-null mice of various ages using the Trizol reagent (Life Technologies). The quality of the RNA was verified before 1 μg of RNA was used for microarray analysis at the City of Hope microarray core using the Affymetric Gene array ST-1.0. The array data set was analyzed and further filtered. Genes that display more than 2-fold differences (P < 0.05) between the control and Pten-null groups were used for further functional and signaling pathway analysis.
Immunohistochemistry.
Liver tissues were preserved in 10% Zn-formain. Hemotoxylin and eosin (H&E) staining was performed on all liver samples for morphologic analysis. Liver sections were stained using various antibodies using the standard protocol, as we previously reported.35 Briefly, paraffin-embedded tissue sections were deparaffinized after baking by going through xylene and alcohol gradients. Antigen retrieval is performed with a pH balanced citric buffer in a pressure cooker. Following blocking with hydrogen peroxide, power block (Biogenix), and serum, primary and secondary antibodies were applied. After the washing step, the bound secondary antibody was detected using horseradish peroxidase (HRP) conjugated with streptavidin (SA-HRP, Vector Laboratories). 3,3′-Diaminobenzidine (DAB) was used as a chromagen for visualization. All immunochemically stained slides were counterstained with hematoxylin. Primary antibodies used are anti-PKM2 (Cell Signaling) and anti-O-GlcNAc (RL2, Sigma).
RESULTS
Knockout of GFAT Results in Loss of O-GlcNAc, Decreased Cancer Cell Growth, and Tumor Formation.
Mammals can express two isoforms of GFAT from two separate genes, GFPT1 and GFPT2. The resulting proteins, GFAT1 and 2, are essentially identical in their enzymatic behavior.36,37 They are both tetrameric and consist of two domains. The N-terminus of GFAT is an aminohydrolase domain that converts glutamine to glutamate and ammonia. The ammonia is then funneled to the C-terminal isomerase domain, where it reacts with fructose-6-phosphate to yield glucosamine-6-phosphate. GFAT1 contains a longer linker between these two domains compared to GFAT2, resulting in a difference of ∼2 kDa in size. GFAT1 is broadly expressed in tissues, while GFAT2 has a more limited expression profile, and GFAT2 mRNA is only found at very low levels in lung and breast cancer cell lines (The Human Protein Atlas, www.proteinatlas.org).38–40 Therefore, we chose to use CRISPR/Cas9 to target and delete GFAT1 in H1299 and MCF7 cells, lung, and breast cancer cell lines, respectively (Figure 2A). Specifically, these cells were transiently transfected with a Cas9 plasmid containing a GFPT1 targeted gRNA and a puromycin-resistance selection marker. After 48 h of puromycin treatment, we plated the cells at a very low density and collected different populations of cells using cloning cylinders. During these experiments and subsequent expansion of the cells, we supplemented the culture media with GlcNAc (10 mM) to bypass GFAT and maintain O-GlcNAcylation levels through the GlcNAc salvage pathway. Under these conditions, the KO cells grew similarly to wild-type cells (Figure S1). We then analyzed these populations by Western blotting with a pan-selective anti-GFAT antibody and sequencing (Figure 2B,D). Specifically, we used a GFAT antibody (IBL-America, 663) raised against the C-terminal sequence of GFAT that is identical in GFAT1 and GFAT2. In both cell lines, we found populations that had no detectable GFAT by Western blotting. Sequencing of one population of H1299 cells revealed two different mutations in GFPT1, while MCF7 cells contained four mutations. The average copy number of chromosome 2 in MCF7 cells is 3.09 (COSMIC cell lines project), consistent with the possibility that our parent MCF7 cell line contains four copies of chromosome 2. Notably, we did not detect any mutations at the analogous position in the GFPT2 gene, which, together with our Western blotting, confirms that GFAT1 is the major isoform of GFAT expressed in H1299 and MCF7 cells. This result contrasts with a recent report of Western blot detection of GFAT2 in H1299 cells.28 We believe that this could result from divergence in the transcript patterns between the two different populations of cells or potentially from a posttranslational modification of GFAT2 that is obscuring its detection by our antibody. In either case, our results are consistent with GFAT1 either being the only or major form of the enzyme expressed in H1299 cells.
Next, we examined the consequences of GFAT knockout on cell proliferation by growing the knockout cells in media containing different concentrations of GlcNAc. For both cell lines, we chose CRISPR population 1 (Figure 2B,D) and observed a reduction in proliferation compared to wild-type cells that correlated well with media concentrations of GlcNAc that resulted in a loss of global O-GlcNAcylation levels (Figure 2C,E). Interestingly, we also found large differences between the ability of the H1299 and MCF7 knockout cells to scavenge the exogenously added GlcNAc from the media (Figure 2C,E). H1299-knockout cells displayed a dramatic loss of O-GlcNAcylation and proliferation between 4 and 2 mM GlcNAc, which MCF7 knockout cells showed a similar sensitivity to GlcNAc concentrations between 0.75 and 0.25 mM. In both cases, GlcNAc was added directly to the media. Together, these results confirm that GFAT activity is required for the growth and propagation of cancer cells in culture and that this defect can be rescued by salvaging exogenous GlcNAc. However, the efficiency of this salvaging appears to differ wildly between different cancer cell lines.
We then tested the consequences of GFAT loss on tumorigenesis in vivo using a xenograft model using either wild-type or H1299 knockout cells. Notably, up until the injection into the mice, the GFAT knockout cells were cultured in 10 mM GlcNAc, which supports their health and normal growth. We measured tumor volumes at different times after implantation (Figure 3A), as well as the volume and mass of tumors removed from the sacrificed mice at the end of the experiment (25 days, Figure 3B). By all metrics, the GFAT knockout H1299 cells grew significantly slower than the corresponding wild-type cells. Finally, we performed Western blotting on isolated tumors (Figure 3C). Importantly, the pan-GFAT antibody detected no GFAT in the knockout cells, demonstrating that the cells have not induced the expression of GFAT2 during the course of the experiment. As predicted by this loss of GFAT, we also observed lower overall O-GlcNAcylation compared to the wild-type H1299 cells, and we believe that the remaining modification levels most likely result from the tumors scavenging GlcNAc from their environment. These data confirm the importance of GFAT activity for tumor formation in vivo and indicate that even moderate (50–75%) reduction in O-GlcNAcylation levels can significantly affect tumor growth.
Figure 3.
GFAT knockout inhibits H1299 xenograft growth and lowers tumor O-GlcNAcylation levels. (A) H1299 wild-type or GFAT knockout cells (1 × 106) were injected into the flanks of mice, and the resulting tumor volume was measured after the indicated lengths of time (n = 5). (B) At the termination of the xenograft experiment, the tumors were removed, measured, and weighed (n = 5). Data shown represent the mean ± s.e.m. Statistical significance was determined using an unpaired, two-tailed Student’s t test. (C) Tumors were lysed, and O-GlcNAcylation levels and GFAT expression were measured using Western blotting.
Pharmacologic Inhibition of GFAT Sensitizes Cancer Cells to Apoptosis under Oxidative Stress.
As mentioned above, one of the main roles of increased O-GlcNAcylation is the protection of cells from stress, including inhibition of apoptosis under oxidative stress.8 Therefore, we next asked if inhibition of GFAT would induce apoptosis in cancer cells exposed to oxidative stress. Specifically, we treated lung (A549 and H1299) or breast (MCF7) cancer cells with the H2O vehicle or the irreversible GFAT inhibitor 6-diazo-5-oxo-norleucine (DON, Figure 4A) for 24 h, followed by either the DMSO vehicle or diamide, an inducer of oxidative stress, for an additional 16 h. Treatment of these cells with DON resulted in decreased O-GlcNAcylation levels that remained low under the cumulative addition of oxidative stress (Figure 4B). UDP-GlcNAc is also a substrate for cell-surface glycosylation, in particular, all forms of N-linked glycosylation. We therefore also blotted using a biotin-conjugated LCA lectin, which recognized the core structure of N-linked glycans (Figure S2), and was previously shown to be one of the most sensitive lectins to inhibition of N-linked glycosylation by the GlcNAc analogue 5SGlcNAc.17 In H1299 and MCF7 cells, we observed essentially no loss of N-linked glycosylation. In contrast, N-linked levels in A549 cells were decreased but not to the same extent as O-GlcNAcylation. Next, in order to confirm that DON treatment lowers O-GlcNAcylation by preventing flux through the HBP, we performed a 13C glucose tracing experiment and measured the amounts of isotopically labeled UDP-GlcNAc. As expected, we found that DON eliminated essentially all 13C labeling of UDP-GlcNAc after 4 h (Figure S3, Supporting Information). To examine any downstream effect on apoptosis, we next visualized the activation of the apoptotic executioner caspases-3/7 (Promega Caspase-Glo 3/7) under a similar set of conditions (Figure 4C). We observed a significant increase in apoptosis in cells treated with both DON and diamide compared to single treatment of either compound, indicating that GFAT inhibition can indeed sensitize cancer cells to programmed cell death caused by oxidative stress. To test if our observation is dependent on GFAT activity and not another DON target, we then repeated this experiment in the absence or presence of exogenously added GlcNAc (10 mM directly dissolved in the media). We found that the addition of GlcNAc prevented the induction of apoptosis (Figure 4D) and recovered the O-GlcNAcylation levels to baseline levels (Figure 4E), supporting GFAT as the main target of DON in these assays.
Figure 4.
GFAT inhibition sensitizes cancer cells to undergo apoptosis under oxidative stress. (A) GFAT can be inhibited by the natural product 6-diazo-5-oxo-norleucine (DON). (B) Oxidative stress induces a small increase in O-GlcNAcylation that is inhibited by DON treatment. A549, H1299, or MCF7 cells were treated with diamide to induce oxidative stress and/or DON to inhibit GFAT before analysis by Western blotting. (C) DON and diamide cotreatment results in increased apoptosis. A549, H1299, or MCF7 cells were treated with the indicated concentrations of diamide and/or DON, and caspase-3/7 activity was measured using the Caspase-Glo 3/7 Assay (Promega) (n = 3). (D) Exogenous GlcNAc rescues cells from apoptosis. A549, H1299, or MCF7 cells were treated with the indicated concentrations of diamide, DON, and/or GlcNAc, and caspase-3/7 activity was measured using the Caspase-Glo 3/7 Assay (Promega) (n = 3). (E) Exogenous GlcNAc rescues O-GlcNAc levels during DON treatment. A549, H1299, or MCF7 cells were treated with DON and/or GlcNAc, and O-GlcNAcylation levels were visualized using Western blotting. Data shown in panels C and D represent the mean ± s.e.m. Statistical significance was determined using a one-way ANOVA test followed by the Dunnett test.
Characterization of the Heterocyclic GFAT Inhibitor R28.
Roche previously carried out a high-throughput screen and subsequent medicinal chemistry and published a small series of drug-like heterocyclic GFAT inhibitors.30 Their interest was in the potential role of GFAT in diabetes and characterized one compound, termed R28 here (Figure 5A), that displayed reasonable GFAT inhibition in vitro (IC50 = 1 μM) and good pharmacokinetics in rats. However, they did not explore the effects of R28 treatment on O-GlcNAcylation levels in cells or animals. Given that DON is a pan-aminotransferase inhibitor, we were hopeful that R28 could serve as a selective GFAT inhibitor that would lower O-GlcNAcylation levels for subsequent in vivo studies. We, therefore, synthesized R28 following published procedures30 (Figure S2) and treated A549, H1299, or MCF7 cells with increasing concentrations of the inhibitor before visualization of O-GlcNAcylation levels by Western blotting (Figure 5B). Unfortunately, we only observed reductions in overall O-GlcNAcylation at high concentrations of R28 (∼50 μM) that are subjectively near the solubility limit of the inhibitor in media. These results demonstrate that R28 itself is insufficient for in vivo experimentation and will also likely suffer from off-target effects in cells at the lowest effective concentrations. However, we hope they will also encourage the further development of potentially more potent inhibitors based around the R28 scaffold.
Figure 5.
The Roche inhibitor “R28” lowers O-GlcNAcylation levels at high concentrations. (A) In vitro GFAT is inhibited by the small molecule R28 with an IC50 of 1 μM. (B) R28 can lower O-GlcNAcylation levels in cells but only at high concentrations around 50 μM. A549, H1299, or MCF7 cells were treated with the indicated concentrations of R28 before analysis by Western blotting.
GFAT Expression and Increased O-GlcNAcylation Are Induced and Coassociate in a Mouse Model of Liver Tumorigenesis.
Next, to investigate whether increased GFAT expression might drive O-GlcNAcylation levels in an in vivo model of tumorigenesis, we took advantage of a previously reported Pten-null model of liver cancer (Figure 6A).31,41,42 These mice (PtenloxP/loxP; Alb-Cre+) have a liver-specific deletion in Pten, which is a major antagonist of the insulin-phosphoinositide-3 kinase (PI3K) signaling pathway. These mice develop the fatty-liver disease by 6 months of age and spontaneous liver carcinomas in mice 9 months of age and beyond. We became interested in this tumor model as hepatocytes from the Pten KO-mice are resistant to oxidative stress compared to their wild-type (PtenloxP/loxP; Alb-Cre−) counterparts.32 To gain insight into whether glycolysis or O-GlcNAcylation is altered during tumorigenesis, we first used a DNA microarray to measure the mRNA levels of genes in glycolysis, the HBP, and O-GlcNAc cycling (Figure 7A). At 3 months of age, there are no significant (P < 0.05) differences, of at least 2-fold, in the transcription of these genes between the Pten KO-mice and the wild-type controls. However, by 9 months, the Pten KO-mice had increased levels mRNA encodinghexokinase-2 and the M2 isoform of pyruvate kinase (PKM2), two markers of tumor-associated metabolic changes as the liver normally expresses PKL, and after 15 months, the KO-mice showed an increase in GFAT2 mRNA.
Figure 6.
GFAT expression and O-GlcNAcylation levels are increased in immortalized hepatocytes from the a Pten-null mouse model of liver cancer. (A) Liver-specific loss of PTEN expression leads to fatty-liver disease (3 months old), hepatocyte death and proliferation of tumor progenitor cells (9 months old), and the formation of liver tumors (12–15 months old). (B) Immortalized hepatocytes from wild-type (PtenloxP/loxP; Alb-Cre−) and knockout mice (PtenloxP/loxP; Alb-Cre+) were analyzed by Western blotting. Knockout cells had higher levels of GFAT2 that correspond with increased O-GlcNAcylation compared to the wild-type controls.
Figure 7.
GFAT expression correlated with tumor formation and elevated O-GlcNAcylation in a Pten-null mouse model of liver cancer. (A) Fold changes in liver mRNA between Pten-knockout (KO) and wild-type mice as determined by the DNA microarray. Hexokinase-2, PKM2, and GFAT2 are the only glycolytic or O-GlcNAc-associated genes that show more than a 2-fold change. (B, C) Liver lysates from wild-type and Pten-null mice were analyzed by Western blotting. Each lane represents an individual mouse. (D) Adjacent slices of Pten-null livers were stained for PKM2 and O-GlcNAc, which showed high levels and overlap in the tumor. Red line = tumor boundary; DC = ductal cells; NH = normal hepatocytes.
To determine if these changes were translated to the protein level, Western blotting was next performed on both immortalized hepatocytes and whole livers. No GFAT2 was detectable in the wild-type cells but could be readily visualized in the Pten KO cell lines, and the Pten KO cells had dramatically increased O-GlcNAcylation levels (Figure 6B). Notably, the levels of GFAT2 and O-GlcNAcylation in the different KO cell lines were also correlated (Figure 6B). Consistent with the microarray data, no GFAT2 or PKM2 as a tumor marker could be detected in whole-liver lysates from either the wild-type or Pten-null mice at 3 months of age (Figure 7B). By 9 months, GFAT2 and PKM2 are beginning to be perceptible in the Pten-null livers, and at 15 months, both enzymes are selectively overexpressed in the KO mice (Figure 7B). Furthermore, the overall O-GlcNAcylation levels are also increased in the Pten-null livers at 15 months (Figure 7C). To ascertain the location of the increased O-GlcNAcylation levels, immunohistochemical analysis was also performed from livers taken from the Pten-null animals, which showed that the modification was increased within the tumors compared to the surrounding normal hepatocytes (Figure 7D). Notably, the staining of adjacent slices of the livers with an anti-PKM2 antibody showed almost complete overlap between this tumor marker and increased O-GlcNAcylation levels (Figure 7D). Together, these results indicate that increased GFAT expression may be a hallmark of certain tumors and help drive increased O-GlcNAcylation levels.
DISCUSSION
Despite the observations that global O-GlcNAcylation is dynamically upregulated in response to cellular stress8 and that increased levels of modification inhibit apoptosis and promote cancer cell survival,12,43 there have been relatively few studies aimed at understanding the potential protein targets for lowering O-GlcNAcylation. Here, we first use a combination of CRISPR gene knockout and small-molecule inhibitors to test whether the rate-determining enzyme of the HBP, GFAT, could be one such target. We found that genetic loss of GFAT resulted in low O-GlcNAcylation levels in a culture that resulted in slower cancer cell proliferation, which could be rescued by the addition of exogenous GlcNAc to the media (Figure 2). In the case of H1299 lung cancer cells, the loss of GFAT leads to the formation of significantly smaller tumors that displayed reduced overall O-GlcNAcylation (Figure 3). We attribute the remaining O-GlcNAc modification levels to scavenging of GlcNAc from the tumor environment. This suggests that targeting the downstream enzymes of the HBP, particularly UAP1 (also known as AGX1/2), might be more efficient for lowering O-GlcNAcylation levels. However, our results also demonstrate that the O-GlcNAcylation levels in cancer cells do not necessarily need to be lowered more than ∼50% to affect tumor growth significantly in vivo.
To further explore the link between glycolysis and the biosynthesis of UDP-GlcNAc, we examined the consequences of small-molecule inhibition of GFAT. We found that either moderate oxidative stress or GFAT inhibition alone with DON did not result in large amounts of apoptosis, but the treatments in combination yielded significantly higher amounts of programmed cell death (Figure 4). Lectin blotting demonstrated that O-GlcNAcylation levels are more sensitive to reduced UDP-GlcNAc levels compared to N-linked glycosylation (Figure S2), indicated that most of the observed phenotype is due to O-GlcNAc. However, we cannot rule out the possibility that changes to cell-surface glycosylation could grow upon longer treatment times and also contribute to the sensitivity to oxidative stress, particularly in cell lines like A549 with more perturbed N-linked glycans. Disruption of GFAT and other glutaminases also prevents the conversion of glutamine to glutamate, which can play important roles in cancer from energy metabolism and biosynthesis to receptor activation.44,45 Importantly, however, the apoptosis we observe could be reversed by exogenous GlcNAc, suggesting that the major target of DON in our experiments is the production of glucosamine by GFAT and not another biosynthetic enzyme or glutamate production. Genetic interruption of UDP-GlcNAc biosynthesis or OGT expression are both embryonic lethal,1,46 and OGT simultaneously regulates many cellular pathways, potentially raising concerns about the appropriateness of these targets for the development of anticancer therapies. However, our results suggest that a reasonable therapeutic index might be found by lowering O-GlcNAcylation levels enough to sensitize stressed cancer cells to undergo apoptosis while leaving healthy tissues minimally effected. To explore this idea further, we resynthesized and characterized the heterocyclic GFAT inhibitor R28 from Roche but found that it has a fairly low potency in a cell culture (Figure 5). While this result is disappointing, we believe that our results should encourage further medicinal chemistry efforts to improve R28 or identify alternative scaffolds as GFAT inhibitors for further studies.
Finally, we found an increase in the expression of both GFAT2 and O-GlcNAcylation levels during tumorigenesis in a Pten-null mouse model of liver carcinoma and that the elevated O-GlcNAcylation colocalizes in liver tumors (Figures 6 and 7). These results are consistent with the upregulation of GFAT in a pancreatic cancer mouse model,25 indicate a potential role for GFAT in our model in the formation of tumors in vivo, and support further studies on the contribution of metabolism through the HBP to tumorigenesis in a variety of cancers.
Supplementary Material
Funding
C.J.H. was supported by the NIH/NCI (F32CA228328), a Michigan Institute of Clinical Health Research Postdoctoral Translational Scholar Award (UL1TR000433), a University of Michigan Center for Gastrointestinal Research Pilot Feasibility Award (P30DK034933), and T32CA009676. K.N.C. was a fellow of the National Science Foundation Graduate Research Fellowship Program (DGE-0937362). N.J.P. was supported by T32GM118289. B.L.S. was supported by the NIH/NCI (R01CA154986). C.A.L. was supported by a 2017 AACR NextGen Grant for Transformative Cancer Research (17–20-01-LYSS), an ACS Research Scholar Grant (RSG-18–186-01), and Cancer Center Support Grant P30 CA046592. M.R.P. was supported by an ACS Research Scholar Grant (RSG-14–225-01-CCG). Metabolomics studies performed at the University of Michigan were supported by the NIH (DK097153).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.9b00560.
Growth of GFAT knockout cells in 10 mM GlcNAc, lectin blotting, metabolomics data, and NMR characterization of R28 (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Boehmelt G, Wakeham A, Elia A, Sasaki T, Plyte S, Potter J, Yang Y, Tsang E, Ruland J, Iscove NN, Dennis JW, and Mak TW (2000) Decreased UDP-GlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO J. 19, 5092–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Vocadlo DJ (2012) O-GlcNAc processing enzymes: catalytic mechanisms, substrate specificity, and enzyme regulation. Curr. Opin. Chem. Biol 16, 488–497. [DOI] [PubMed] [Google Scholar]
- (3).Joiner CM, Li H, Jiang J, and Walker S (2019) Structural characterization of the O-GlcNAc cycling enzymes: insights into substrate recognition and catalytic mechanisms. Curr. Opin. Struct. Biol. 56, 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Shen DL, Gloster TM, Yuzwa SA, and Vocadlo DJ (2012) Insights into O-linked N-acetylglucosamine (O-GlcNAc) processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates. J. Biol. Chem. 287, 15395–15408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Bond MR, and Hanover JA (2015) A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yang X, and Qian K (2017) Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hart GW (2019) Nutrient regulation of signaling and transcription. J. Biol. Chem. 294, 2211–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Groves JA, Lee A, Yildirir G, and Zachara NE (2013) Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperon. 18, 535–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Darley-Usmar VM, Ball LE, and Chatham JC (2012) Protein O-linked β-N-acetylglucosamine: A novel effector of cardiomyocyte metabolism and function. J. Mol. Cell. Cardiol. 52, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zachara NE (2012) The roles of O-linked βN-acetylglucosamine in cardiovascular physiology and disease. AJP: Heart and Circulatory Physiology 302, H1905–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jensen RV, Andreadou I, Hausenloy DJ, and Bøtker HE (2019) The Role of O-GlcNAcylation for Protection against Ischemia-Reperfusion Injury. Int. J. Mol. Sci. 20, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ma Z, and Vosseller K (2013) O-GlcNAc in cancer biology. Amino Acids 45, 719–733. [DOI] [PubMed] [Google Scholar]
- (13).Ferrer CM, Sodi VL, and Reginato MJ (2016) O-GlcNAcylation in Cancer Biology: Linking Metabolism and Signaling. J. Mol. Biol. 428, 3282–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Vasconcelos-dos-Santos A, Queiroz RM, Rodrigues BC, Todeschini AR, and Dias WB (2018) Hyperglycemia and aberrant O-GlcNAcylation: contributions to tumor progression. J. Bioenerg. Biomembr. 50, 175–187. [DOI] [PubMed] [Google Scholar]
- (15).Hanover JA, Chen W, and Bond MR (2018) O-GlcNAc in cancer: An Oncometabolism-fueled vicious cycle. J. Bioenerg. Biomembr. 50, 155–173. [DOI] [PubMed] [Google Scholar]
- (16).Martin SES, Tan Z-W, Itkonen HM, Duveau DY, Paulo JA, Janetzko J, Boutz PL, Toörk L, Moss FA, Thomas CJ, Gygi SP, Lazarus MB, and Walker S (2018) Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors. J. Am. Chem. Soc. 140, 13542–13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ortiz-Meoz RF, Jiang J, Lazarus MB, Orman M, Janetzko J, Fan C, Duveau DY, Tan Z-W, Thomas CJ, and Walker S (2015) A small molecule that inhibits OGT activity in cells. ACS Chem. Biol. 10, 1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, and Vocadlo DJ (2011) Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat. Chem. Biol. 7, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Liu T-W, Zandberg WF, Gloster TM, Deng L, Murray KD, Shan X, and Vocadlo DJ (2018) Metabolic Inhibitors of O-GlcNAc Transferase That Act In Vivo Implicate Decreased O-GlcNAc Levels in Leptin-Mediated Nutrient Sensing. Angew. Chem., Int. Ed. 57, 7644–7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Marshall S, Nadeau O, and Yamasaki K (2004) Dynamic actions of glucose and glucosamine on hexosamine biosynthesis in isolated adipocytes: differential effects on glucosamine 6-phosphate, UDP-N-acetylglucosamine, and ATP levels. J. Biol. Chem. 279, 35313–35319. [DOI] [PubMed] [Google Scholar]
- (21).Taylor RP, Geisler TS, Chambers JH, and McClain DA (2009) Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexosamine pathway flux. J. Biol. Chem. 284, 3425–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Milewski S (2002) Glucosamine-6-phosphate synthase–the multi-facets enzyme. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1597, 173–192. [DOI] [PubMed] [Google Scholar]
- (23).Durand P, Golinelli-Pimpaneau B, Mouilleron S, Badet B, and Badet-Denisot M-A (2008) Highlights of glucosamine-6P synthase catalysis. Arch. Biochem. Biophys. 474, 302–317. [DOI] [PubMed] [Google Scholar]
- (24).Akella NM, Ciraku L, and Reginato MJ (2019) Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 17, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik J-H, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, and DePinho RA (2012) Oncogenic Kras Maintains Pancreatic Tumors through Regulationof Anabolic Glucose Metabolism. Cell 149, 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Guillaumond F, Leca J, Olivares O, Lavaut M-N, Vidal N, Berthezène P, Dusetti NJ, Loncle C, Calvo E, Turrini O, Iovanna JL, Tomasini R, and Vasseur S (2013) Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. U. S. A. 110, 3919–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lucena MC, Carvalho-cruz P, Donadio JL, Oliveira IA, de Queiroz RM, Marinho-Carvalho MM, Sola-Penna M, de Paula IF, Gondim KC, McComb ME, Costello CE, Whelan SA, Todeschini AR, and Dias WB (2016) Epithelial Mesenchymal Transition Induces Aberrant Glycosylation through Hexosamine Biosynthetic Pathway Activation. J. Biol. Chem. 291, 12917–12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chen W, Do KC, Saxton B, Leng S, Filipczak P, Tessema M, Belinsky SA, and Lin Y (2019) Inhibition of the hexosamine biosynthesis pathway potentiates cisplatin cytotoxicity by decreasing BiP expression in non-small-cell lung cancer cells. Mol. Carcinog. 58, 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Pinkus LM (1977) Glutamine binding sites. Methods Enzymol. 46, 414–427. [DOI] [PubMed] [Google Scholar]
- (30).Qian Y, Ahmad M, Chen S, Gillespie P, Le N, Mennona F, Mischke S, So S-S, Wang H, Burghardt C, Tannu S, Conde-Knape K, Kochan J, and Bolin D (2011) Discovery of 1arylcarbonyl-6,7-dimethoxyisoquinoline derivatives as glutamine fructose-6-phosphate amidotransferase (GFAT) inhibitors. Bioorg. Med. Chem. Lett. 21, 6264–6269. [DOI] [PubMed] [Google Scholar]
- (31).Stiles B, Wang Y, Stahl A, Bassilian S, Lee WP, Kim Y-J, Sherwin R, Devaskar S, Lesche R, Magnuson MA, and Wu H (2004) Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected]. Proc. Natl. Acad. Sci. U. S. A. 101, 2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zeng N, Li Y, He L, Xu X, Galicia V, Deng C, and Stiles BL (2011) Adaptive basal phosphorylation of eIF2α is responsible for resistance to cellular stress-induced cell death in Pten-null hepatocytes. Mol. Cancer Res. 9, 1708–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Darabedian N, Gao J, Chuh KN, Woo CM, and Pratt MR (2018) The Metabolic Chemical Reporter 6-Azido-6-deoxy-glucose Further Reveals the Substrate Promiscuity of O-GlcNAc Transferase and Catalyzes the Discovery of Intracellular Protein Modification by O-Glucose. J. Am. Chem. Soc. 140, 7092–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Halbrook CJ, Pontious C, Kovalenko I, Lapienyte L, Dreyer S, Lee H-J, Thurston G, Zhang Y, Lazarus J, Sajjakulnukit P, Hong HS, Kremer DM, Nelson BS, Kemp S, Zhang L, Chang D, Biankin A, Shi J, Frankel TL, Crawford HC, Morton JP, Pasca di Magliano M, and Lyssiotis CA (2019) Macrophage-Released Pyrimidines Inhibit Gemcitabine Therapy in Pancreatic Cancer. Cell Metab. 29, 1390–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).He L, Gubbins J, Peng Z, Medina V, Fei F, Asahina K, Wang J, Kahn M, Rountree CB, and Stiles BL (2016) Activation of hepatic stellate cell in Pten null liver injury model. Fibrog. Tissue Repair 9, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Broschat KO, Gorka C, Page JD, Martin-Berger CL, Davies MS, Huang H-C, Gulve EA, Salsgiver WJ, and Kasten TP (2002) Kinetic characterization of human glutamine-fructose-6-phosphate amidotransferase I: potent feedback inhibition by glucosamine 6-phosphate. J. Biol. Chem. 277, 14764–14770. [DOI] [PubMed] [Google Scholar]
- (37).Hu Y, Riesland L, Paterson AJ, and Kudlow JE (2004) Phosphorylation of mouse glutamine-fructose-6-phosphate amidotransferase 2 (GFAT2) by cAMP-dependent protein kinase increases the enzyme activity. J. Biol. Chem. 279, 29988–29993. [DOI] [PubMed] [Google Scholar]
- (38).Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CAK, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, and Ponten F (2015) Tissue-based map of the human proteome. Science 347, 1260419–1260419. [DOI] [PubMed] [Google Scholar]
- (39).Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjoörk L, Breckels LM, Bäckstroö A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson Å, Sjoöstedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Pontén F, von Feilitzen K, Lilley KS, Uhlén M, and Lundberg E (2017) A subcellular map of the human proteome. Science 356, eaal3321–14. [DOI] [PubMed] [Google Scholar]
- (40).Uhlén M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnstroö H, Glimelius B, Sjoöblom T, Edqvist P-H, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, and Ponten F. (2017) A pathology atlas of the human cancer transcriptome. Science 357, eaan2507–13. [DOI] [PubMed] [Google Scholar]
- (41).Galicia VA, He L, Dang H, Kanel G, Vendryes C, French BA, Zeng N, Bayan JA, Ding W, Wang KS, French S, Birnbaum MJ, Rountree CB, and Stiles BL (2010) Expansion of Hepatic Tumor Progenitor Cells in Pten-Null Mice Requires Liver Injury and Is Reversed by Loss of AKT2. Gastroenterology 139, 2170–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Debebe A, Medina V, Chen C-Y, Mahajan IM, Jia C, Fu D, He L, Zeng N, Stiles BW, Chen C-L, Wang M, Aggarwal K-R, Peng Z, Huang J, Chen J, Li M, Dong T, Atkins S, Borok Z, Yuan W, Machida K, Ju C, Kahn M, Johnson D, and Stiles BL (2017) Wnt/β-catenin activation and macrophage induction during liver cancer development following steatosis. Oncogene 36, 6020–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ma Z, Vocadlo DJ, and Vosseller K (2013) Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J. Biol. Chem. 288, 15121–15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hensley CT, Wasti AT, and DeBerardinis RJ (2013) Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Invest. 123, 3678–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Henderson MX, Cornblath EJ, Darwich A, Zhang B, Brown H, Gathagan RJ, Sandler RM, Bassett DS, Trojanowski JQ, and Lee VM-Y (2019) Spread of α-synuclein pathology through the brain connectome is modulated by selective vulnerability and predicted by network analysis. Nat. Neurosci. 22, 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).O’Donnell N, Zachara NE, Hart GW, and Marth JD (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.