Abstract
Targeting epigenetic regulators such as histone modifying enzymes provides novel strategies for cancer therapy. The GCN5 lysine acetyltransferase (KAT) functions together with MYC both during normal development and in oncogenesis. As transcription factors, MYC family members are difficult to target with small molecule inhibitors, but the acetyltransferase domain and the bromodomain in GCN5 might provide alternative targets for disruption of MYC-driven functions. GCN5 is part of two distinct multiprotein histone modifying complexes, SAGA and ATAC. This review summarizes key findings on the roles of SAGA and ATAC in embryo development and in cancer, to better understand the functional relationships of these complexes with MYC family members, as well as their future potential as therapeutic targets.
Introduction
GCN5 was defined in the late-1990s as a component of the Spt-Ada-Gcn5-Acetyl transferase (SAGA) complex in yeast (1) and related TFTC/STAGA complexes in mammals (2) [1, 2]. These multiprotein assemblies post-translationally modify histones as well as other proteins [1]. SAGA is composed of lysine acetyltransferase (KAT), deubiquitinase (DUB), SPT and TAF modules. These modules maintain SAGA architecture (SPT), regulate histone acetylation and deubiquitination (KAT and DUB) and interact with general transcriptional machinery (TAF), as well as specific transcription factors (TRRAP; TF binding module) [3]. GCN5 (KAT2A) was later identified as a component of the ADA Two A Containing (ATAC) complex, which also regulates gene transcription and chromatin organization [4]. GCN5 KAT activity and specificity is augmented by association with the Alteration/Deficiency in Activation (ADA) proteins [1, 5], and independent ADA-GCN5 complexes have been identified in yeast and metazoans [6]. GCN5 requires association with both ADA2 and ADA3 for robust acetylation of histone and nonhistone targets [7]. GCN5 is associated with two forms of ADA2 in metazoans, termed ADA2A and ADA2B [8]. ADA2B is exclusively found in SAGA, while ADA2A is found in ATAC [8]. Outside of the KAT modules, SAGA and ATAC are distinct in composition, likely reflecting unique functions [3, 4].
Both SAGA and ATAC function as transcriptional co-activators. These complexes are targeted to genomic loci through interactions with sequence specific transcription factors, and may be stabilized at certain regions through interactions of “reader” domains with specific histone modifications. Some studies indicate that SAGA acts at select genes, whereas others indicate that it functions as a general transcription factor [9–11]. Recent studies may resolve these differences, as chromatin immunoprecipitation (ChIP) experiments indicate that the SAGA subunit SUS1 is recruited to yeast genes regulated by the general transcription factor IID (TFIID), as well as to known SAGA-regulated genes and ribosomal protein genes [12, 13]. However, under conditions of heat shock, SUS1 binding shifts away from SAGA-dominated and ribosomal protein genes to stress-responsive genes [12], and SAGA colocalizes with heat shock factor 1 (Hsf1) at these genes [14]. These findings indicate that SAGA distributions across the genome can be dynamic.
SAGA regulates both transcription initiation and elongation through its KAT and DUB activities. SAGA also has non-histone substrates, such as c-MYC, which is acetylated by GCN5 and its ortholog PCAF [15] and TRF1, which is deubiquitylated by USP22 [16]. As such, SAGA influences gene expression at many levels, from transcription to protein stability [17]. GCN5 functions extend beyond the nucleus, independent of the SAGA and ATAC complexes, including mitochondrial functions in S. cerevisiae [18]. Genetic studies in mice indicate GCN5 [19, 20] and USP22 [21] are required for normal embryo development. SAGA also impacts signaling pathways, genome integrity and metabolic control in mammalian cells [17]. Although less studied, components of ATAC are also required for normal embryo development, modulating ribosome biogenesis, cell-cycle and DNA repair [22]. This review will further explore the functions of SAGA and ATAC in development and in cancer, especially in regards to MYC functions.
Links between SAGA and MYC family
GCN5 was the first identified transcription-related KAT [23], and it acetylates lysine residues in histones H3 and H2B [17]. Mammals also express PCAF (P300/CBP-Associated Factor, or KAT2B), which is highly homologous to GCN5 [24]. GCN5 and PCAF are incorporated into SAGA and ATAC complexes in a mutually exclusive way. In contrast to Gcn5, no abnormal phenotypes are associated with Pcaf deletion in mice [19, 20]. Interestingly, combined knockout of Gcn5 and Pcaf results in more severe developmental phenotypes than Gcn5 deletion alone [19], suggesting both redundant and synergistic roles for these two KATs.
Conditional deletion of Gcn5 in neural progenitor cell populations reduces brain mass in mice, with a phenotype similar to conditional loss of c-Myc or N-myc [25, 26]. Interestingly, MYC regulates chromatin structures in neural progenitors at least in part through upregulation of GCN5 [26]. Regulatory connections between GCN5 and MYC family members have been observed in numerous settings. The TRRAP protein in SAGA directly interacts with MYC in cancer cells, recruiting GCN5 for activation of MYC target genes [27–29]. Moreover, acetylation of MYC by GCN5 or PCAF extends its half-life in vivo, augmenting MYC functions [15]. In embryonic stem cells (ESCs), GCN5 [30] and other SAGA components [31] colocalize with MYC for upregulation of cell-cycle genes, promoting ESC self-renewal. Moreover, functions of MYC as a “Yamanaka factor” in somatic cell reprogramming involve upregulation of GCN5 expression followed by recruitment of GCN5 protein to MYC target genes [30]. Loss of Gcn5 also hinders transcription of c-MYC target genes associated with FGF signaling in embryoid bodies and early differentiation [32]. Altogether, these studies indicate that GCN5 and MYC work together in a feed forward loop. These functions are likely to involve SAGA, as the TAD domain of MYC interacts with SAGA but not ATAC components [33], and MYC upregulates expression of several components of SAGA in addition to GCN5 [30].
SAGA in Cancer
GCN5 likely plays a role in the oncogenic functions of MYC, although GCN5 and other components of SAGA may contribute to cancer formation or progression independently of MYC as well. GCN5 over expression is observed in multiple cancer types (Figure 1A), and GCN5 depletion inhibits cell proliferation or induces apoptosis, consistent with a role in oncogenesis. For example, inhibition of GCN5 activity reduces viability of lung cancer stem-like cells and reduces non-small cell lung cancer growth [34, 35]. GCN5 has also been linked to regulation of cell proliferation and invasion in glioma [36]. In breast cancer, GCN5 regulates the epithelial-to-mesenchymal (EMT) transition by enhancing STAT3, AKT and E2F1 signaling pathways [37]. GCN5 over expression in hepatocellular carcinoma is linked to increased AIB1 expression, which drives progression of these cancers [38]. In melanoma cells, AND-1, a protein required for the stability of GCN5, is upregulated [39]. A CRISPR screen for factors required for AML identified GCN5 (KAT2A) as a “top-hit” [40]. In lymphoma cells, GCN5 inhibition reduces cell-survival through reduction of MYC target gene expression [41]. Many of the cancer types that overexpress GCN5 also harbor MYC amplifications or increased expression of MYC family proteins.
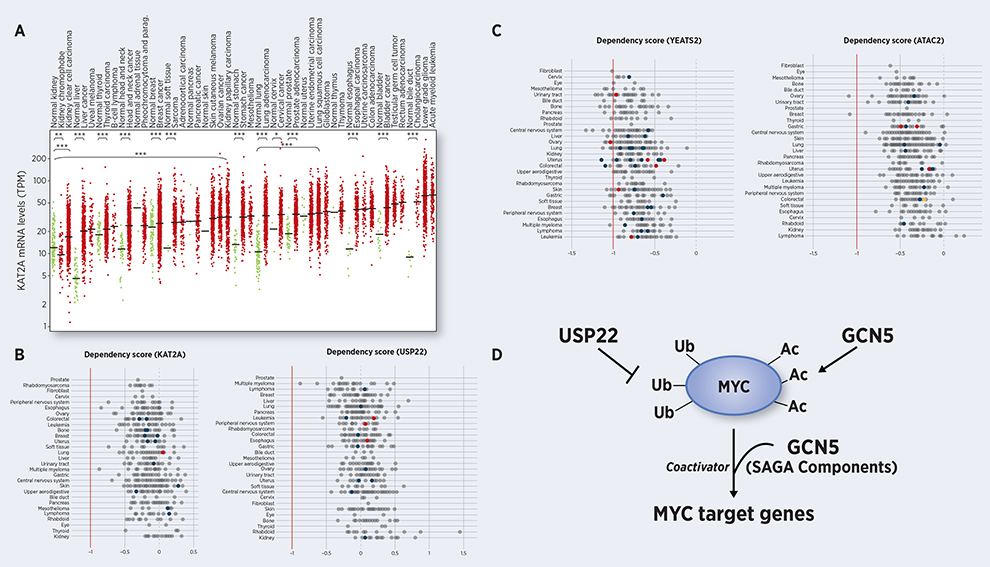
Figure 1. Targeting SAGA and ATAC in cancer.
A. GCN5 mRNA levels are significantly upregulated in most human cancers (red) relative to adjacent normal tissues (green), as indicated by analysis of The Cancer Genome Atlas (*, P<0.05; **, P<0.01; ***, P<0.001). Multiple cancer types show dependencies on B. GCN5 and USP22 as well as C. YEATS2 and ATAC2, as shown by DepMap, where a lower CERES score indicates that a gene is essential to a cell line, where a score of 0 indicates that the gene is not critical. Cell lines containing damaging, hotspot or other non-conserving mutations in YEATS2 or ATAC2 are highlighted in red, orange and blue, respectively. D. Model summarizing how SAGA components promote MYC functions, through acting as a coactivator for MYC target genes and by increasing the stability of MYC protein through acetylation (GCN5) and deubiquitination (USP22).
Although highly related to GCN5, PCAF may play different roles in cancer, acting as either an oncogene or tumor suppressor based on the tissue of origin. In hepatocellular carcinoma, PCAF promotes autophagy [42]. PCAF acts as a suppressor of gastric cancer by inhibiting cell growth and through increased immunity [43]. In contrast, upon p53 loss of function and perturbation of DNA damage response, PCAF may promote Hedgehog-dependent cell-survival and proliferation [44, 45]. Double knockdown of both PCAF and GCN5 significantly affects c-MYC levels in urothelial carcinoma cells [46]. Given the differential, but overlapping, expression patterns of GCN5 and PCAF in specific tissues, and their different functions during development, these KATS also likely have shared and unique roles in different cancers that are important to define for therapy development.
ADA proteins
Functions of the ADA proteins (ADA2 A and B isoforms and ADA3) in cancer have not yet been defined, but they are likely to be important as these proteins are critical regulators of GCN5 catalysis [6, 17]. A recent study to define gene dependencies in N-MYC-driven neuroblastomas identified ADA2B and several other SAGA components as top hits [47]. Importantly, loss of ADA2A or ADA2B inactivates both GCN5- and PCAF-containing versions of ATAC and SAGA, respectively, providing an efficient way to eliminate redundant functions of these KATs in cancer cells.
USP22
USP22 is a member of the ubiquitin-specific processing proteases (USPs) family, and it targets both histone and non-histone substrates. USP22 is particularly known for targeting mono-ubiquitinated H2B to facilitate gene regulation [45, 48, 49]. USP22 has been described as a member of an 11 gene ‘death from cancer’ signature [50] and like GCN5, genetic studies indicate that USP22 has important functions in both normal development and oncogenesis.
Deletion of Usp22 in mice leads to vascular defects in the placental labyrinth linked to decreased activity of several signaling pathways including those driven by TGFβ, VEGF and other receptor tyrosine kinases [21]. Similarly, USP22 is also critical for angiogenesis of non-small cell lung cancer where its functions mirror those observed in placenta development [51]. Early embryonic lethality of USP22 deficient mice is also linked to increased p53 expression [52]. USP22 affects the stability and expression of non-histone proteins such as TRF1, which impacts telomere structure and genome integrity, and FBP1, which regulates MYC expression [16, 53]. Other studies indicate USP22 is important for counteracting silencing in heterochromatin and serves as a positive cofactor for nuclear receptor activation [53, 54]. The SAGA DUB module is also known to impact early stages of the DNA damage response (DDR) and is required for the DNA repair phase of class switch recombination (CSR) [55].
USP22 is overexpressed in highly aggressive tumors [56], and knockdown of USP22 in cancer cells often leads to cell-cycle arrest and decreased tumor growth [57]. Here again, USP22 functions include both gene regulation, likely due to changes in H2B ubiquitination, and regulation of non-histone protein stability. USP22 deubiquitinates c-MYC in breast cancer cells, increasing MYC stability [58]. USP22 also regulates androgen receptor levels and coordinates with MYC signaling to drive prostate adenocarcinoma [59]. USP22 deubiquitinates and stabilizes the PU.1 transcription factor. Loss of USP22 in a mouse model of KRAS-driven leukemia exacerbated disease due to decreased PU.1 stability and subsequent blocks to myeloid differentiation associated with increased expression of MYC target genes in progenitor cells [60]. Multiple cancer types are dependent on GCN5 and USP22, as shown by DepMap [61], analyses (Figure 1B).
Further work is needed to determine the relationship between the KAT and DUB modules in cancer, and whether both enzymes might be simultaneously targeted for a synergistic response. Loss of GCN5 results in the depletion of USP22 from SAGA, leading to telomere fusions in human and mouse cells [16]. The DUB module may stimulate the KAT activity in SAGA [62] and USP22 is acetylated at multiple lysines [63]. Crosstalk between these enzymatic modules is consistent with functional relationships between other DUBs and KATs in oncogenesis [64].
Overview of ATAC and its role in Cancer
The YEATS2 component of ATAC is emerging as potentially important in cancer. YEATS2 contains a histone fold domain as well as a YEATS domain, which selectively binds to (“reads”) acetyl-lysine residues in histones and likely other non-histone proteins [65]. In particular, YEATS2 binds to acetylated H3K27, recruiting the ATAC complex to chromatin [66]. This recruitment allows for the maintenance of H3K9ac and H3K14ac facilitating gene transcription. YEATS2 is amplified in lung, ovarian, cervical and uterine cancers and is necessary for survival of non-small cell lung cancer cells [66]. Whether ATAC plays a role in MYC functions is currently unknown. YEATS2 physically interacts with MYC, but at much lower levels than SAGA components [33]. Cancers, such as lung, liver, breast and prostate, are dependent on YEATS2 and ATAC2, as shown by DepMap analyses (Figure 1C). The YEATS domain and the ATAC2 KAT domain might provide unique targets for cancer therapies.
Distributions of the SAGA and ATAC complexes
Identification of specific oncogenic or tumor-suppressive pathways affected by SAGA or ATAC is needed to fully understand functions of these complexes in cancers. Chromatin immunoprecipitation studies in GM12878 and HeLa cell lines indicate that SAGA and ATAC are targeted to different genomic loci by distinct regulatory elements [67]. SAGA and ATAC also are involved in distinct signaling pathways [68]. Unlike ADA2A (ATAC), ADA2B (SAGA) interacts with the tumor suppressor protein p53 and p53-response genes [68]. ATAC interacts with several MAPK pathway components during osmotic stress induction and may play a more prominent role than SAGA in this process [68].
Targetable domains within the SAGA and ATAC complexes
SAGA and ATAC subunits provide unique targets for potential therapy development. Both complexes contain multiple enzymatic activities, the HAT and DUB modules in SAGA and the two KATs in ATAC. The acetyltransferase domains of the KATs, the reader domains in GCN5 (bromodomain) and YEATS2 (the YEATS domain) and ubiquitin specific protease domain of USP22 all provide potential targets for inhibitor development.
Targeting KAT activities
KAT activity can be repressed by directly inhibiting enzymatic activity or disrupting interactions between a KAT and recruitment proteins necessary for its functions. KAT inhibitors include natural products, carboxylic acids, protein-protein interaction inhibitors, bi-substrate inhibitors and synthetic compounds [69]. Oridonin, a lysine acetyltransferase inhibitor that targets multiple KATs including GCN5/PCAF, inhibits proliferation of breast cancer both in vitro and in vivo [70] and induces apoptosis in gastric cancer cell lines [71]. In the same study, Butyrolactone 3 (MB-3), a more specific GCN5/PCAF KAT inhibitor, was also shown to inhibit gastric cancer cell proliferation, but less potently than Oridonin. Similarly, Garcinol, a PCAF and P300 KAT probe, inhibits non-homologous end joining and radio sensitizes lung and cervical cancer cells in vitro [72]. PU139, a GCN5, PCAF, CREB and P300 KAT inhibitor, triggers caspase-independent cell-death in neuroblastoma cells and blocks the growth of neuroblastoma xenografts in mice [73]. Lastly, the GCN5/PCAF KAT inhibitor CPTH6 affects growth and viability of lung cancer stem-like cells and affects the viability of other cancer cell lines [74]. Key GCN5/PCAF KAT activity probes having therapeutic potential in cancer are summarized in Table 1. Future work should identify whether ATAC2, the second KAT within the ATAC complex, might also be targeted for therapy.
Unfortunately, available KAT inhibitors have limited utility for clinical trials due to issues with selectivity, toxicity and potency [69]. Not only is it difficult to develop inhibitors specific to particular KATs, these inhibitors also affect other acetyl coenzyme A (Ac-CoA)-specific functions [69]. Since KATs are bi-substrate enzymes, multiple factors influence potency of inhibitors including the catalytic mechanism and concentration of substrates [69]. Lastly, since KATs function in multiple cellular processes beyond gene transcription, their inhibition may increase toxicity.
All KATs have non-histone substrates that may also be directly involved in tumorigenesis, such as p53, Rb and NF-κB [75]. The interplay between HATs and HDACs, which have also been targeted for therapeutic benefit, must also be considered. Use of an HDAC inhibitor on a HAT-dependent cancer, for example, might have more detrimental than beneficial effects.
Targeting Bromodomains in KATs
Bromodomains are found in KATs, methyltransferases and transcriptional coactivators, and some proteins, such as Brd4, help organize super enhancers that drive expression of oncogenes such as MYC [76]. Bromodomain and Extra-Terminal motif (BET) inhibitors provide an alternate way to target MYC and have shown therapeutic potential in hematological malignancies [77]. The BET family of proteins are characterized by two tandem bromodomains and an extra-terminal domain and regulate gene transcription through epigenetic interactions between bromodomains and acetylated histones [78]. Although BET inhibitors have efficacy as single agents, resistance to these is common and they may work best in combination with agents such as checkpoint inhibitors [76].
BET domains are just one subfamily of the more than 60 different bromodomain-containing proteins in mammals [79]. The bromodomains of CBP/P300 are essential for proliferation of leukemia and lymphoma cell lines [80], suggesting inhibitors targeting these may have therapeutic potential. Only a few small molecules targeting GCN5/PCAF bromodomains have been developed. L-Moses was the first selective bromodomain inhibitor reported for GCN5/PCAF [81]. L-Moses shows no cytotoxicity in normal blood cells and is metabolically stable in the human liver, suggesting it might be a good candidate for testing as an anticancer agent. The GSK4027 chemical probe is a more selective and potent GCN5/PCAF bromodomain inhibitor in vitro [82]. Additional studies are needed to determine the effects of L-Moses and the GSK probes on cancer cell lines and xenograft models. Development of PROTAC degraders of BET domains has proven useful in avoiding resistance mechanisms in cancer [83]. Such degraders targeting GCN5/PCAF bromodomains might also provide effective means of blocking functions of these KATs in cancer [84].
The discovery that YEATS domains, which are structurally distinct from bromodomains, also read acetyl-lysine moieties raises the possibility that chemical probes against these domains, including the YEATS2 protein in ATAC, may have therapeutic potential [85, 86].
Targeting DUBs
SAGA functions can also be targeted through USP22 inhibition. DUB catalytic centers are structurally homologous making development of selective inhibitors difficult [87]. In addition, USP27X and USP51 are not only structurally similar to USP22, they also compete with USP22 for activation by ATXN7L3 and ENY2 [88]. In some cases, a DUB may function as an oncoprotein in one cancer and a tumor suppressor in another, such as USP18, which plays an oncogenic role in lung cancer [89], but is a tumor suppressor in sarcomas. In addition, the DUB USP7 plays various roles depending on substrate abundance and physiological state [90]. Whether targeting USP22 will be effective in conferring antineoplastic effects is not clear. Proteasome inhibitors have been approved for therapies against myelomas and lymphomas [87]. Even though many DUB inhibitors show antitumor effects in vitro, they are not in clinical trials.
Conclusions and future perspectives
The proper balance between acetylation/deacetylation or ubiquitination/deubiquitination is essential in normal physiology. This balance makes targeting enzymes controlling these states a double-edged sword. However, selective inhibition of KATs in cancers that are ‘addicted’ to high expression of oncogenes may allow definition of therapeutic windows. By simultaneously repressing pathways that cancer cells depend on, there is a greater chance in reducing severity and reoccurrence.
The SAGA complex appears to play a key role in promoting MYC-driven cancers (Figure 1D). Both GCN5 and USP22 affect MYC protein stability, and SAGA serves as a co-activator of MYC target genes, which include genes encoding SAGA components. Inhibitors of SAGA and ATAC activities may be useful in targeting the functions of difficult to drug proteins, like c-MYC. Identifying whether cancers driven by “undruggable” oncogenes are sensitive to GCN5, PCAF and ADA2B inhibition is an area of active investigation. How SAGA components affect the functions of different MYC family members is also an important question. Studies on GCN5 may also provide insight into targeting other epigenetic regulators that interact with c-MYC, such as TIP60 [91]. Understanding the division of labor between transcriptional coactivator complexes in mediating oncogenic pathways will hopefully help identify new strategies to tackle cancers.
Acknowledgements
This work was supported by the Postdoctoral Fellowship 131779-PF-18–034-01-DMC from the American Cancer Society (LM Mustachio), the Postdoctoral Fellowship Award from the Center for Cancer Epigenetics at MD Anderson Cancer Center (LM Mustachio) and a Tobacco Pilot Grant from MD Anderson Cancer Center (SYR Dent) and in part from NIH grants R01 HD094400 and R35 GM131678 to SYR Dent.
We thank Drs. Michael Johnson McAndrew and Evangelia Koutelou for their helpful consultation.
Footnotes
Disclosure of Conflicts of Interest
The authors have declared that no competing interest exists.
References
- 1.Grant PA, et al. , Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev, 1997. 11(13): p. 1640–50. [DOI] [PubMed] [Google Scholar]
- 2.Martinez E, Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol, 2002. 50(6): p. 925–47. [DOI] [PubMed] [Google Scholar]
- 3.Wang L and Dent SY, Functions of SAGA in development and disease. Epigenomics, 2014. 6(3): p. 329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riss A, et al. , Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5. J Biol Chem, 2015. 290(48): p. 28997–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sterner DE, et al. , Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol, 1999. 19(1): p. 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soffers JHM, et al. , Characterization of a metazoan ADA acetyltransferase complex. Nucleic Acids Res, 2019. 47(7): p. 3383–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossl A, et al. , A synthetic non-histone substrate to study substrate targeting by the Gcn5 HAT and sirtuin HDACs. J Biol Chem, 2019. 294(16): p. 6227–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres-Zelada EF, et al. , The Drosophila Dbf4 ortholog Chiffon forms a complex with Gcn5 that is necessary for histone acetylation and viability. J Cell Sci, 2019. 132(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huisinga KL and Pugh BF, A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell, 2004. 13(4): p. 573–85. [DOI] [PubMed] [Google Scholar]
- 10.Lee TI, et al. , Redundant roles for the TFIID and SAGA complexes in global transcription. Nature, 2000. 405(6787): p. 701–4. [DOI] [PubMed] [Google Scholar]
- 11.Baptista T, et al. , SAGA Is a General Cofactor for RNA Polymerase II Transcription. Mol Cell, 2017. 68(1): p. 130–143 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Molinero V, et al. , The SAGA/TREX-2 subunit Sus1 binds widely to transcribed genes and affects mRNA turnover globally. Epigenetics Chromatin, 2018. 11(1): p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonova SV, et al. , Epigenetics and transcription regulation during eukaryotic diversification: the saga of TFIID. Genes Dev, 2019. 33(15–16): p. 888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinayachandran V, et al. , Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock. Genome Res, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel JH, et al. , The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol, 2004. 24(24): p. 10826–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atanassov BS, et al. , Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell, 2009. 35(3): p. 352–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker SP and Grant PA, The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene, 2007. 26(37): p. 5329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montanari A, et al. , Gcn5 histone acetyltransferase is present in the mitoplasts. Biol Open, 2019. 8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, et al. , Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet, 2000. 26(2): p. 229–32. [DOI] [PubMed] [Google Scholar]
- 20.Yamauchi T, et al. , Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci U S A, 2000. 97(21): p. 11303–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutelou E, et al. , USP22 controls multiple signaling pathways that are essential for vasculature formation in the mouse placenta. Development, 2019. 146(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guelman S, et al. , The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol Cell Biol, 2009. 29(5): p. 1176–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brownell JE, et al. , Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 1996. 84(6): p. 843–51. [DOI] [PubMed] [Google Scholar]
- 24.Yang XJ, et al. , A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 1996. 382(6589): p. 319–24. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Cerdeno V, et al. , N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS One, 2012. 7(6): p. e39456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knoepfler PS, et al. , Myc influences global chromatin structure. EMBO J, 2006. 25(12): p. 2723–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon SB, Wood MA, and Cole MD, The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol, 2000. 20(2): p. 556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, et al. , c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem, 2003. 278(22): p. 20405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon SB, et al. , The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell, 1998. 94(3): p. 363–74. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch CL, et al. , Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev, 2015. 29(8): p. 803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seruggia D, et al. , TAF5L and TAF6L Maintain Self-Renewal of Embryonic Stem Cells via the MYC Regulatory Network. Mol Cell, 2019. 74(6): p. 1148–1163 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, et al. , GCN5 Regulates FGF Signaling and Activates Selective MYC Target Genes during Early Embryoid Body Differentiation. Stem Cell Reports, 2018. 10(1): p. 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang N, et al. , MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim Biophys Acta, 2014. 1839(5): p. 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, et al. , Lysine acetyltransferase GCN5 potentiates the growth of non-small cell lung cancer via promotion of E2F1, cyclin D1, and cyclin E1 expression. J Biol Chem, 2013. 288(20): p. 14510–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mustachio LM, et al. , Repression of GCN5 expression or activity attenuates c-MYC expression in non-small cell lung cancer. Am J Cancer Res, 2019. 9(8): p. 1830–1845. [PMC free article] [PubMed] [Google Scholar]
- 36.Liu K, et al. , GCN5 Potentiates Glioma Proliferation and Invasion via STAT3 and AKT Signaling Pathways. Int J Mol Sci, 2015. 16(9): p. 21897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao L, Pang A, and Li Y, Function of GCN5 in the TGF-beta1-induced epithelial-to-mesenchymal transition in breast cancer. Oncol Lett, 2018. 16(3): p. 3955–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majaz S, et al. , Histone acetyl transferase GCN5 promotes human hepatocellular carcinoma progression by enhancing AIB1 expression. Cell Biosci, 2016. 6: p. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, et al. , And-1 is required for the stability of histone acetyltransferase Gcn5. Oncogene, 2012. 31(5): p. 643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzelepis K, et al. , A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep, 2016. 17(4): p. 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farria AT, et al. , GCN5 HAT inhibition reduces human Burkitt lymphoma cell survival through reduction of MYC target gene expression and impeding BCR signaling pathways. Oncotarget, 2019. 10(56): p. 5847–5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia YL, et al. , P300/CBP-associated factor (PCAF) inhibits the growth of hepatocellular carcinoma by promoting cell autophagy. Cell Death Dis, 2016. 7(10): p. e2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fei HJ, et al. , PCAF acts as a gastric cancer suppressor through a novel PCAF-p16-CDK4 axis. Am J Cancer Res, 2016. 6(12): p. 2772–2786. [PMC free article] [PubMed] [Google Scholar]
- 44.Infante P, et al. , Yin-Yang strands of PCAF/Hedgehog axis in cancer control. Trends Mol Med, 2014. 20(8): p. 416–8. [DOI] [PubMed] [Google Scholar]
- 45.Malatesta M, et al. , Histone acetyltransferase PCAF is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res, 2013. 73(20): p. 6323–33. [DOI] [PubMed] [Google Scholar]
- 46.Koutsogiannouli EA, et al. , Differential Effects of Histone Acetyltransferase GCN5 or PCAF Knockdown on Urothelial Carcinoma Cells. Int J Mol Sci, 2017. 18(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durbin AD, et al. , Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet, 2018. 50(9): p. 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durand A, et al. , Mapping the deubiquitination module within the SAGA complex. Structure, 2014. 22(11): p. 1553–9. [DOI] [PubMed] [Google Scholar]
- 49.Zhang XY, et al. , The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell, 2008. 29(1): p. 102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glinsky GV, Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle, 2006. 5(11): p. 1208–16. [DOI] [PubMed] [Google Scholar]
- 51.Zhang K, et al. , Ubiquitin-specific protease 22 is critical to in vivo angiogenesis, growth and metastasis of non-small cell lung cancer. Cell Commun Signal, 2019. 17(1): p. 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Z, et al. , USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell, 2012. 46(4): p. 484–94. [DOI] [PubMed] [Google Scholar]
- 53.Atanassov BS and Dent SY, USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep, 2011. 12(9): p. 924–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Y, et al. , A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell, 2008. 29(1): p. 92–101. [DOI] [PubMed] [Google Scholar]
- 55.Ramachandran S, et al. , The SAGA Deubiquitination Module Promotes DNA Repair and Class Switch Recombination through ATM and DNAPK-Mediated gammaH2AX Formation. Cell Rep, 2016. 15(7): p. 1554–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X, et al. , High expression of USP22 predicts poor prognosis and advanced clinicopathological features in solid tumors: a meta-analysis. Onco Targets Ther, 2018. 11: p. 3035–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melo-Cardenas J, et al. , Ubiquitin-specific peptidase 22 functions and its involvement in disease. Oncotarget, 2016. 7(28): p. 44848–44856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D, et al. , Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J Cell Physiol, 2017. 232(12): p. 3664–3676. [DOI] [PubMed] [Google Scholar]
- 59.Schrecengost RS, et al. , USP22 regulates oncogenic signaling pathways to drive lethal cancer progression. Cancer Res, 2014. 74(1): p. 272–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melo-Cardenas J, et al. , USP22 deficiency leads to myeloid leukemia upon oncogenic Kras activation through a PU.1-dependent mechanism. Blood, 2018. 132(4): p. 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyers RM, et al. , Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet, 2017. 49(12): p. 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han Y, et al. , Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J, 2014. 33(21): p. 2534–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Armour SM, et al. , A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol, 2013. 33(8): p. 1487–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pinto-Fernandez A and Kessler BM, DUBbing Cancer: Deubiquitylating Enzymes Involved in Epigenetics, DNA Damage and the Cell Cycle As Therapeutic Targets. Front Genet, 2016. 7: p. 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao D, et al. , YEATS2 is a selective histone crotonylation reader. Cell Res, 2016. 26(5): p. 629–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mi W, et al. , YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat Commun, 2017. 8(1): p. 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krebs AR, et al. , SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell, 2011. 44(3): p. 410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spedale G, Timmers HT, and Pijnappel WW, ATAC-king the complexity of SAGA during evolution. Genes Dev, 2012. 26(6): p. 527–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wapenaar H and Dekker FJ, Histone acetyltransferases: challenges in targeting bi-substrate enzymes. Clin Epigenetics, 2016. 8: p. 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia S, et al. , Oridonin inhibits breast cancer growth and metastasis through blocking the Notch signaling. Saudi Pharm J, 2017. 25(4): p. 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi M, et al. , Oridonin, a novel lysine acetyltransferases inhibitor, inhibits proliferation and induces apoptosis in gastric cancer cells through p53- and caspase-3-mediated mechanisms. Oncotarget, 2016. 7(16): p. 22623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oike T, et al. , Garcinol, a histone acetyltransferase inhibitor, radiosensitizes cancer cells by inhibiting non-homologous end joining. Int J Radiat Oncol Biol Phys, 2012. 84(3): p. 815–21. [DOI] [PubMed] [Google Scholar]
- 73.Gajer JM, et al. , Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo. Oncogenesis, 2015. 4: p. e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Di Martile M, et al. , Histone acetyltransferase inhibitor CPTH6 preferentially targets lung cancer stem-like cells. Oncotarget, 2016. 7(10): p. 11332–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh BN, et al. , Nonhistone protein acetylation as cancer therapy targets. Expert Rev Anticancer Ther, 2010. 10(6): p. 935–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doroshow DB, Eder JP, and LoRusso PM, BET inhibitors: a novel epigenetic approach. Ann Oncol, 2017. 28(8): p. 1776–1787. [DOI] [PubMed] [Google Scholar]
- 77.Garcia-Carpizo V, et al. , CREBBP/EP300 bromodomains are critical to sustain the GATA1/MYC regulatory axis in proliferation. Epigenetics Chromatin, 2018. 11(1): p. 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taniguchi Y, The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins. Int J Mol Sci, 2016. 17(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stathis A and Bertoni F, BET Proteins as Targets for Anticancer Treatment. Cancer Discov, 2018. 8(1): p. 24–36. [DOI] [PubMed] [Google Scholar]
- 80.Sunami Y, et al. , Histone Acetyltransferase p300/CREB-binding Protein-associated Factor (PCAF) Is Required for All-trans-retinoic Acid-induced Granulocytic Differentiation in Leukemia Cells. J Biol Chem, 2017. 292(7): p. 2815–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moustakim M, et al. , Discovery of a PCAF Bromodomain Chemical Probe. Angew Chem Int Ed Engl, 2017. 56(3): p. 827–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Humphreys PG, et al. , Discovery of a Potent, Cell Penetrant, and Selective p300/CBP-Associated Factor (PCAF)/General Control Nonderepressible 5 (GCN5) Bromodomain Chemical Probe. J Med Chem, 2017. 60(2): p. 695–709. [DOI] [PubMed] [Google Scholar]
- 83.An S and Fu L, Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. EBioMedicine, 2018. 36: p. 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bassi ZI, et al. , Modulating PCAF/GCN5 Immune Cell Function through a PROTAC Approach. ACS Chem Biol, 2018. 13(10): p. 2862–2867. [DOI] [PubMed] [Google Scholar]
- 85.Moustakim M, et al. , Discovery of an MLLT1/3 YEATS Domain Chemical Probe. Angew Chem Int Ed Engl, 2018. 57(50): p. 16302–16307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li X, et al. , Structure-guided development of YEATS domain inhibitors by targeting pi-pi-pi stacking. Nat Chem Biol, 2018. 14(12): p. 1140–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang X and Dixit VM, Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res, 2016. 26(4): p. 484–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Atanassov BS, et al. , ATXN7L3 and ENY2 Coordinate Activity of Multiple H2B Deubiquitinases Important for Cellular Proliferation and Tumor Growth. Mol Cell, 2016. 62(4): p. 558–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mustachio LM, et al. , The ISG15-specific protease USP18 regulates stability of PTEN. Oncotarget, 2017. 8(1): p. 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang X, et al. , Clinical Significance of Ubiquitin Specific Protease 7 (USP7) in Predicting Prognosis of Hepatocellular Carcinoma and its Functional Mechanisms. Med Sci Monit, 2018. 24: p. 1742–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frank SR, et al. , MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep, 2003. 4(6): p. 575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]