Abstract
Purpose:
The role of plasma-based tumor mutation burden (pTMB) in predicting response to pembrolizumab-based first-line standard of care therapy for metastatic non-small cell lung cancer (mNSCLC) has not been explored.
Experimental Design:
A 500-gene next-generation sequencing (NGS) panel was used to assess pTMB. Sixty-six patients with newly diagnosed mNSCLC starting first-line pembrolizumab-based therapy, either alone or in combination with chemotherapy, were enrolled (Clinicaltrial.gov identifier: NCT03047616). Response was assessed using RECIST 1.1. Associations were made for patient characteristics, 6-month durable clinical benefit (DCB), progression free survival (PFS), and overall survival (OS).
Results:
Of 66 patients, 52 (78.8%) were pTMB-evaluable. Median pTMB was 16.8 mutations per megabase (mut/Mb; range 1.9–52.5) and was significantly higher for patients achieving DCB compared to no durable benefit: 21.3 mut/Mb vs. 12.4 mut/Mb, P=0.003. For patients with pTMB ≥16 mut/Mb, median PFS was 14.1 vs. 4.7 months for patients with pTMB<16 mut/Mb (HR 0.30 [0.16–0.60]) P<0.001. Median OS for patients with pTMB≥16 was not reached vs. 8.8 months for patients with pTMB<16 mut/Mb (HR 0.48 [0.22–1.03]) P=0.061. Mutations in ERBB2 exon 20, STK11, KEAP1, or PTEN were more common in patients with no DCB. A combination of pTMB≥16 and absence of negative predictor mutations was associated with PFS (HR 0.24 [0.11–0.49]) P<0.001 and OS (HR 0.31 [0.13–0.74]) P=0.009.
Conclusions:
pTMB≥16 mut/Mb is associated with improved PFS after first-line standard of care pembrolizumab-based therapy in mNSCLC. STK11/KEAP1/PTEN and ERBB2 mutations may help identify pTMB-high patients unlikely to respond. These results should be validated in larger prospective studies.
Keywords: Non-small cell lung cancer (NSCLC), liquid biopsy, pembrolizumab, next-generation sequencing (NGS), tumor mutation burden (TMB), plasma
Introduction
Immunotherapy is the current standard first-line treatment for patients with metastatic non-small cell lung cancer (mNSCLC) whose tumors lack therapeutically targetable mutations. In the US, pembrolizumab is currently approved for treatment of mNSCLC with PD-L1 Tumor Proportion Score (TPS) ≥1%, and in combination with chemotherapy regardless of PD-L1 TPS. In practice, pembrolizumab monotherapy is reserved for patients with a PD-L1 Tumor Proportion Score (TPS) ≥50%[1]; patients with PD-L1 <50% are usually treated with histology specific platinum-doublet therapy in combination with pembrolizumab[2, 3]. Nevertheless, PD-L1 TPS is an imperfect biomarker, as evidenced by a significant benefit of chemo-immunotherapy across all PD-L1 levels[4]. Therefore, there is a need to develop novel biomarkers to better identify patients likely to respond to immunotherapy. Tumor mutation burden (TMB), the number of somatic mutations per megabase (mut/Mb), is one such emerging biomarker. In retrospective studies, tissue-based TMB (tTMB) was directly related to clinical outcomes following checkpoint blockade in mNSCLC[5]. Specific negative predictor mutations in STK11, KEAP1, and other genes have also been evaluated in tissue as biomarkers for immunotherapy[6–10].
Tissue samples often provide inadequate DNA for sequencing and may under-represent tumor molecular heterogeneity[11, 12]. Circulating cell-free tumor DNA (ctDNA), shed into blood by tumor cells, is increasingly utilized to identify actionable mutations and predict response to therapy[4, 13]. Recently, ctDNA-based next-generation sequencing (NGS) was used to determine TMB in plasma; patients with pTMB ≥16 mut/Mb receiving atezolizumab on the OAK and POPLAR trials had improved overall survival (OS) compared to pTMB-low patients[14, 15]. Based on this study, using a pre-specified plasma TMB cut-off of ≥16 mut/Mb, clinical trials are underway. Preliminary analyses from this proof of concept trial reveal a numerical benefit for response rate and survival outcomes in a prospectively selected population of patients with high pTMB receiving atezolizumab for mNSCLC[16]. A similar benefit was seen with combination durvalumab and tremelimumab compared to chemotherapy on the MYSTIC trial using the 2.145 Mb GuardantOMNI assay at pTMB cutoffs of ≥16 and ≥20 mut/Mb[12]. To our knowledge, the role of plasma-based TMB and negative predictor mutations for predicting response to pembrolizumab-based therapy including in combination with chemotherapy in a real-world setting has not been explored.
Here we evaluated a plasma-based 2.145 Mb 500 gene NGS panel to measure baseline pTMB and specific negative predictor mutations for 66 patients with mNSCLC receiving first-line pembrolizumab-based treatment as standard of care.
Materials and Methods
Patients and Study Design
Patients were enrolled from 3/1/17 – 10/11/18 and included if they had pathologically confirmed mNSCLC, received pembrolizumab-based therapy as standard of care first-line treatment (Clinicaltrial.gov identifier: NCT03047616). Response Evaluation Criteria in Solid Tumors (RECIST), version 1.1 was used to perform independent radiographic response assessments. Efficacy was also defined as durable clinical benefit (DCB; complete response [CR], partial response [PR], or stable disease [SD] lasting > 6 months) or no durable benefit (NDB; PD or SD lasting ≤ 6 months)[17]. OS was calculated from the date of first pembrolizumab infusion to the time of death or censored at most recent follow-up; PFS was calculated from the date of first pembrolizumab infusion to the time of death or first documented disease progression, whichever came first, or censored at most recent follow-up. Patients were followed for a minimum of six months. We followed the REporting recommendations for tumor MARKer prognostic studies (REMARK) guidelines[18]. This study was approved by the Institutional Review Board (IRB) of the University of Pennsylvania and conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent.
Plasma-based mutation detection and statistical analysis
Plasma was obtained at baseline, prior to initiation of pembrolizumab-based therapy. Sequencing was performed using the 2.145 Mb GuardantOMNI panel[19, 20]. The mutation count included all coding somatic single nucleotide variants (SNVs; including silent SNVs) and indels. Germline alterations were filtered out[21]. Driver and resistance mutations were excluded, as well as putative clonal hematopoiesis (CH) mutations, which were identified using a curated database and in-sample context[22]. Raw mutation count was corrected for sample-specific tumor shedding and molecule coverage, with the corrected count reported as pTMB (units mut/Mb)[22]. Samples with low tumor shedding (all somatic mutations <0.3% maximum somatic allele fraction) or low unique molecule coverage were identified as pTMB-unevaluable (Supplemental Figure 1A).
Validation of plasma-based panel
Reproducibility was assessed using 11 de-identified retrospective plasma samples from multiple tumor types, including NSCLC (Supplemental Figure 1B)[23]. In silico analysis was conducted to assess concordance of pTMB with WES-determined TMB for 513 NSCLC tissue samples from The Cancer Genome Atlas (TCGA)[12, 15, 24, 25](Supplemental Figure 1C). We conducted additional in-silico analysis to simulate a comparison of TMB scores using a publicly available, retrospective cohort[17] of advanced NSCLC patients. (Supplemental Figure 1D). This builds on previous reports of a positive correlation between TMB scores for matched plasma (as measured by GuardantOMNI) and tissue, as measured by either WES[24] or FoundationOne tissue panel[12, 15] on the MYSTIC trial.
Statistics
Descriptive statistics were computed for patient, tumor, and treatment characteristics. Associations between these characteristics and pTMB were examined using Spearman’s rho correlation for continuous variables, Wilcoxon rank sum or Kruskal-Wallis tests for categorical variables due to a departure from a normal distribution for pTMB scores (Table 1). Comparisons of pTMB between 9-week response status (CR/PR versus SD/PD) and 6-month DCB were determined using a non-parametric bootstrap test of the medians. The odds ratio of binary response status with pTMB was estimated using a logistic regression. We also examined the association of a continuous pTMB with outcomes after confirming the linearity assumption using a restricted cubic spline[26]. Using the endpoint of PFS, an optimal cutoff in the range of 15 to 16 mut/Mb was identified such that the log-rank test statistic was maximized in the current data. The cutoff of 16 mut/Mb was selected for the additional analyses on this basis and also based on the existing literature[14, 15]. Kaplan-Meier curves for PFS and OS were generated and compared between patients with high pTMB (≥16 mut/Mb) and low pTMB (<16 mut/Mb) using log-rank test. Hazard ratios and the associated 95% confidence intervals (CI) were estimated using Cox proportional hazard (PH) model. PH assumption was checked using Schoenfeld residuals-based score test and no violation was identified. Potential confounders including age (≥65, <65), sex, ECOG status (≥2, <2), treatment (pembrolizumab monotherapy, pembrolizumab-chemotherapy), number of metastatic sites (≥3, <3), PD-L1 TPS (<50%, >50%) and TNM stages (M1a, M1b/c) were examined individually with the binary pTMB group using multivariate Cox or logistic regression. To determine the association of negative predictors with response to immunotherapy, mutations in ERBB2, STK11, KEAP1, PTEN, KRAS, and PIK3CA (Supplemental Table 1) were tested for association with PFS and OS using a Cox PH model. Increased chromosomal aneuploidy (fraction genome aneuploidy; FGA) has been associated with inferior outcomes[5], and was also analyzed. Two-sided p-values <0.05 were considered significant. Statistical analyses were performed using Stata, version 15 (Stata Corp, College Station, TX) and GraphPad Prism, version 7.
Table 1 –
Patient baseline characteristics
| Characteristics | All Patients N=66 | Pembrolizumab Monotherapy N=31 | Pembrolizumab + Chemotherapy N= 35 | pTMB evaluable N= 52 | Median pTMB 16.76 | Association with pTMB P-valuea |
|---|---|---|---|---|---|---|
| Age | ||||||
| Median | 67 | 68 | 66 | 66.5 | NA | 0.830 |
| Range | 47–89 | 54–89 | 47–85 | 47–83 | NA | |
| Sex | ||||||
| Male | 33 | 15 | 18 | 28 | 17.24 | 0.762 |
| Female | 33 | 16 | 17 | 24 | 15.67 | |
| Race | ||||||
| White | 48 | 20 | 28 | 36 | 15.67 | 0.679 |
| Black or African American | 15 | 9 | 6 | 13 | 21.07 | |
| Pacific Islander | 1 | 0 | 1 | 1 | 11.34 | |
| Other | 2 | 2 | 0 | 2 | 14.85 | |
| Smoking Status | ||||||
| Active | 14 | 6 | 8 | 10 | 19.76 | 0.122 |
| Former | 47 | 23 | 24 | 40 | 15.67 | |
| Never | 5 | 2 | 3 | 2 | 6.63 | |
| Histology | ||||||
| Adenocarcinoma | 54 | 22 | 32 | 42 | 17.36 | 0.069 |
| Squamous | 7 | 7 | 0 | 6 | 10.06 | |
| Poorly Differentiated | 4 | 1 | 3 | 3 | 25.80 | |
| Spindle Cell Neoplasm | 1 | 1 | 0 | 1 | 4.79 | |
| ECOG Performance Status at therapy start | ||||||
| 0 | 19 | 7 | 12 | 15 | 17.24 | 0.739 |
| 1 | 34 | 15 | 19 | 26 | 17.85 | |
| 2 | 9 | 8 | 1 | 7 | 17.24 | |
| ≥ 3 | 1 | 1 | 0 | 1 | 9.58 | |
| Unknownb | 3 | 0 | 3 | 3 | 13.24 | |
| Tissue PD-L1% | ||||||
| <1% | 19 | 0 | 19 | 16 | 21.07 | 0.325 |
| 1–49% | 12 | 0 | 12 | 7 | 11.34 | |
| ≥ 50% | 34 | 31 | 3 | 28 | 15.67 | |
| Unknownb | 1 | 0 | 1 | 1 | 4.49 | |
| Number of Metastatic Sites at blood draw | ||||||
| 1 | 6 | 3 | 3 | 3 | 16.13 | 0.283 |
| 2 | 29 | 11 | 18 | 19 | 20.11 | |
| 3 | 19 | 11 | 8 | 18 | 13.67 | |
| 4 | 7 | 3 | 4 | 7 | 13.24 | |
| ≥ 5 | 5 | 3 | 2 | 5 | 17.24 | |
| TNM Classification | ||||||
| M1a | 12 | 6 | 6 | 8 | 15.38 | 0.238 |
| M1b/c | 54 | 25 | 29 | 44 | 17.24 | |
Abbreviations: pTMB, plasma tumor mutation burden.
Footnote: Spearman’s rho rank correlation for continuous variables, Wilcoxon rank sum test or Kruskal-Wallis test for categorical variables.
Patients for whom this characteristic is unknown were excluded from analysis of the association with pTMB in right-most column.
Results
Baseline pTMB associated with response to pembrolizumab-based therapy
Sixty-six consecutive patients with mNSCLC were enrolled in this single-center prospective biomarker trial (Table 1). Thirty-one patients (47.0%) received pembrolizumab monotherapy (P; median 4.1 (0 – 29.4) months on therapy) and 35 (53.0%) received pembrolizumab with platinum-pemetrexed-based chemotherapy (PC; median 7.1 (2.0 – 21.7) months on therapy) with median OS of 22.1 months and 21.9 months respectively (Supplemental Figure 2). Fifty-two of 66 patients (78.8%) were pTMB evaluable (see Methods). pTMB could not be evaluated in 14 patients due to low tumor shedding (all somatic mutations <0.3% maximum somatic allele fraction) or low unique molecule coverage. The median pTMB was 16.8 mut/Mb (range 1.9 to 52.5; Supplemental Figure 3A). No samples were found to be Microsatellite Instability-High (MSI-high)[27]. Consistent with previous reports[14], there was no correlation between pTMB and tissue PD-L1 status (P=0.348; Supplemental Figure 3B).
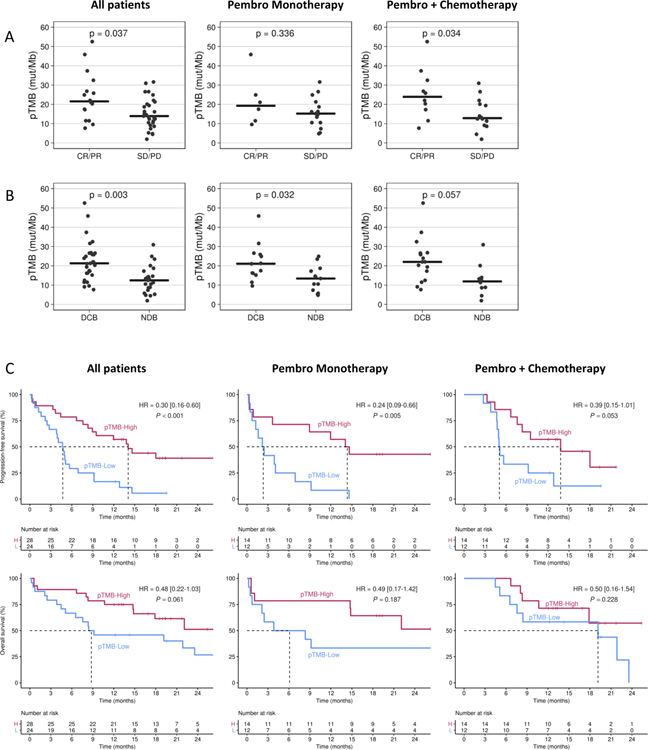
To assess outcomes following pembrolizumab-based therapy, we first analyzed whether baseline pTMB was associated with RECIST-determined response to therapy. Of all enrolled patients, 45 patients had RECIST evaluable disease at week 9. Median pTMB for patients achieving a week 9 CR/PR (responders) was 21.5 mut/MB (range 7.7 – 52.5), compared to 13.9 mut/MB (range 1.9 – 31.6) for patients with SD/PD (non-responders; P=0.037). Using a logistic regression model, pTMB score was significantly associated with week 9 response with an odds ratio (OR) of 1.09 (95%CI: 1.02–1.08, P=0.018) per one-unit increase in pTMB. Of 21 RECIST-evaluable patients at 9 weeks who received P, median pTMB for responders was 19.3 (range 9.6 – 45.9), and for non-responders, it was 15.2 (4.8 – 31.6); this difference did not reach statistical significance (P=0.336). Median pTMB for the 24 RECIST evaluable patients receiving PC who were responders was 23.9 mut/MB (7.7 – 52.5) compared to 12.8 mut/MB (1.9 – 31.0; P=0.034; Figure 1A) for non-responders. The OR per one-unit increase in pTMB was 1.07 (P=0.22) for patients treated with P and 1.11 (P=0.05) for PC.
Figure 1. pTMB and response to pembrolizumab.
A) 45 RECIST-evaluable patients (21 P and 24 PC) at week-9 on therapy were categorized as complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD), and pTMB levels assessed. B) 52 RECIST-evaluable patients (26 P and 26 PC) at month-6 on therapy were categorized as durable clinical benefit (DCB; CR, PR, or SD as of 6 months) or no durable benefit (progressive disease; NDB). Horizontal lines indicate median values. C) Kaplan-Meier survival curves using a cutoff of 16 mut/Mb for PFS (above) and OS (below) for 52 pTMB-evaluable patients (26 P and 26 PC). Left panel is all patients, middle includes patients who received P, and right panel includes patients who received PC.
Among the 52 RECIST evaluable patients at month 6, median pTMB for patients achieving a durable clinical benefit (DCB; CR/PR/SD lasting >6 months[17]) was higher than for those with no durable benefit (NDB) at 21.3 mut/Mb (range 7.7 – 52.5) vs. 12.4 mut/Mb (range 1.9 – 30.9), respectively (P=0.003). The difference in median pTMB was also significant for the 26 patients who received P, with median pTMB of 21.1 mut/MB (range 9.6 – 45.9) for those achieving DCB and a median pTMB of 13.4 mut/MB (range 4.8 – 24.9) for patients with NDB (P=0.032). Among the 26 patients who received PC, those with DCB had a higher median pTMB of 22.0 mut/MB (range 7.7 – 52.5) vs. 11.9 mut/MB (range 1.9 – 30.9) for those with NDB (P=0.057); (Figure 1B). In the multivariate logistic regression analyses, none of the covariates examined (see Methods) changed the OR of DCB for pTMB more than 15%; thus, adjusted OR was not computed.
We next assessed whether pTMB was associated with PFS and OS. When analyzed as a continuous variable, baseline pTMB was significantly associated with PFS (HR=0.93 per one-unit change, 95% CI: 0.90 – 0.97, P=0.001). To assess pTMB as a binary variable, we first identified the appropriate cutoff value. Since the optimal cutoff for measurement of TMB, whether in tissue or plasma, is a function of panel size, sequencing approach, and other factors, we utilized the endpoint of PFS in our own data set to identify the optimal cutoff (see Methods). Using this cutoff of 16 mut/Mb to assess pTMB as a binary variable, we determined a median PFS of 14.1 vs 4.7 months for pTMB ≥16 vs <16 mut/Mb; HR=0.30, 95% CI: 0.16 – 0.60, P<0.001). In our data set, 28 of 52 pTMB-evaluable patients (53.8%) had a pTMB ≥16 mut/Mb. In the multivariate Cox model analyses, the inclusion of covariates did not significantly change the estimated HRs when comparing pTMB ≥16 vs <16 mut/Mb. A similar association for PFS comparing pTMB ≥16 vs <16 mut/Mb was observed among the 26 patients who received P (median PFS of 14.1 vs. 2.2 months, HR=0.24, 95%CI: 0.09–0.66, P=0.005) and the 26 patients who received PC (median PFS of 13.8 vs. 5.0, months, HR=0.39, 95%CI: 0.15–1.01, P=0.053). There was no significant difference in median OS as a function of pTMB level overall (median not reached for pTMB-high vs 8.8 months for pTMB-low, HR 0.48 [0.22 – 1.03]; P=0.061), or among patients receiving P (median NR vs 6.1 months, HR 0.49 [0.17–1.42]; P=0.187) or PC (median NR vs 19.2 months, HR 0.50 [0.16–1.54]); P=0.228; (Figure 1C).
Exploratory analysis of association of ctDNA-detected mutations with response to pembrolizumab-based therapy
In our cohort, six pTMB-high patients did not achieve DCB (Figure 1B). Similar lack of response for some pTMB-high patients was also recently reported[28]. Primary and acquired resistance to PD1 blockade can be associated with specific mutations in JAK1 and JAK2[29, 30], although these mutations were not detected in our cohort. This led us to hypothesize that mutational profiling might improve pTMB association with response. We focused on loss of function mutations in STK11, KEAP1, and PTEN, and an activating ERBB2 exon 20 insertion, all previously shown to be associated with lack of response to checkpoint inhibitors; we also focused on KRAS and PIK3CA mutations, previously correlated with improved response to checkpoint inhibitors (Supplemental Table 1)[5–10, 31]. Increased chromosomal aneuploidy (fraction genome aneuploidy; FGA) has been associated with inferior outcomes[5], and was therefore assessed (Figure 2A). FGA was not significantly associated with response (P=0.198). While the nine putative negative predictor mutations detected for our patient cohort (Supplemental Table 1) were not significantly associated with PFS (P=0.110), we next explored the effects on PFS of a combination of these negative predictor mutations and pTMB. The median PFS for patients with pTMB ≥16 and no negative predictor mutations was 18.0 versus 4.7 months for patients with pTMB <16 or with any negative predictor mutations. This resulted in a HR of 0.24 [0.11–0.49]; P<0.001 for the combined predictors versus 0.30 [0.16–0.60] for pTMB alone. Median OS for patients with pTMB ≥16 and absence of negative predictor mutations was not reached, versus median OS of 8.4 months for pTMB <16 mut/Mb or any negative predictor mutations. This resulted in a HR of 0.31 [0.13–0.74]; P=0.009 for the combined predictors versus a HR of 0.48 [0.22–1.03] for pTMB alone (Figure 2B).
Figure 2. Mutational analysis and response to pembrolizumab.
A) pTMB scores are represented by the height of the bars (red = pTMB≥16 mut/Mb, blue = pTMB<16) and arranged in decreasing order with 29 patients who achieved a durable clinical benefit (left) and 23 patients with no durable benefit (right). Yellow horizontal line indicates pTMB=16 mut/Mb. Rows indicate: pembrolizumab cohort with P in green and PC in yellow, PD-L1 ≥50% patients in purple, patients with a negative predictor mutation in ERBB2, STK11, KEAP1, or PTEN in orange, KRAS or PIK3CA mutations in pink, and fraction genome aneuploidy (FGA; analyzed as a continuous variable) in blue (with lighter blue = lower FGA, darker blue = higher FGA). For negative predictor mutations, capital letter indicates specific mutation detected. B) Forest plots (above) and Kaplan-Meier survival curves (below) for PFS and OS. For the forest plots, black indicates the hazard ratio and 95% confidence interval for pTMB alone, and grey indicates results for pTMB and negative predictors.
Discussion
To our knowledge, this prospective study is the largest to correlate pTMB to outcomes after first-line standard of care pembrolizumab-based combination therapy in mNSCLC. Overall response rate, median PFS, and median OS were similar to those observed in large Phase III trials[1, 2]. Using a 2.145-megabase NGS panel and analyzing pTMB as a continuous variable, we determined that median pTMB was significantly higher for patients who experienced a response at 9 weeks (P=0.037) and at 6 months on therapy (P=0.003). Using a pTMB cutoff of 16 mut/Mb[14, 15], we demonstrate that patients with pTMB ≥16 mut/Mb had improved PFS (HR=0.30), and were more likely to sustain DCB (OR=8.9). We also demonstrate that combining loss of function mutations in STK11/KEAP1/PTEN and activating ERBB2 exon 20 insertion mutations with pTMB improved the ability to predict response. Similar to previous reports[5], there was no correlation between pTMB and tumor PD-L1 expression. Taken together, our results suggest pTMB is associated with response to first-line pembrolizumab-based therapy in mNSCLC.
Pembrolizumab-based immunotherapy and chemo-immunotherapy have become standard first-line therapy for mNSCLC patients without a targetable driver mutation[2]. Lack of biomarkers beyond the current standard of tissue PD-L1 has limited our ability to select patients who benefit most from immunotherapy. TMB is a promising biomarker; higher tTMB was associated with efficacy of single agent atezolizumab compared to chemotherapy[17, 32, 33]. Similarly, using a cutoff of 10 mut/Mb for tTMB, first-line treatment with nivolumab plus ipilimumab was associated with longer PFS and improved response rate compared to standard platinum-based chemotherapy[5]. However, in these trials, tissue samples were only evaluable for TMB in a subset of patients; Hellman et al. and Rizvi et al. reported 57.7% and 41.1% of tissue samples, respectively, as TMB-evaluable[5, 12, 15]. By contrast, Gandara et al. reported 77.3% and 73.1% of patients on the POPLAR and OAK studies, respectively, as TMB-evaluable from plasma[14]. In our study, 52 of 66 patients (79.8%) were pTMB-evaluable[15], suggesting that pTMB may provide a non-invasive option for predicting response in patients for whom tissue-based TMB is impossible. Although a recent report casts doubt on the association of tTMB with response to pembrolizumab plus chemotherapy[34], pTMB in our study is correlated with 9-week response (P=0.034) and 6-month durable clinical benefit (P=0.057). Just as we and others have demonstrated that plasma-based mutation analysis may provide broader sampling of the tumor mutational profile than tissue[4, 35], pTMB may associate more strongly with response than tTMB, although additional studies with matched plasma and tissue TMB measurements will be necessary.
Aside from serving as a non-invasive biomarker when tissue is lacking, pTMB may have other advantages. High spatial and intra-tumoral heterogeneity of the immune microenvironment may challenge reliance on a single tissue biopsy to predict immune signatures[36, 37]. pTMB may overcome this by more comprehensively capturing overall tumor antigenicity, including primary and metastatic sites. WES is still considered the most robust assessment of TMB, but is currently infeasible for clinical decision-making. Panel-based TMB measurements have emerged, leading to debate on panel size, variant type inclusion, interchangeability of scores from different panels, and determination of appropriate cut-points. Until consensus is reached, utility of a panel’s TMB score must be assessed against clinical outcomes.
Wang et al. reported on a NSCLC population that spanned multiple lines of therapy (first, second-line and beyond), in which a 150-gene panel with a pTMB cutoff of 6 mut/Mb could accurately predict response[28]. However, the assay required the SNV allele fraction to be >1.0%, and lacked adjustment of TMB score for low shedding. Moreover, their dataset had 15 non-responders with high pTMB, suggesting additional genomic factors that may not have been accounted for. Mutations in STK11/KEAP1 have been associated with inferior outcomes in patients treated with pembrolizumab-based chemotherapy, including among tTMB-high and PD-L1 positive patients[10]. Data from the MYSTIC trial confirmed the negative prognostic role of KEAP1 using plasma NGS in patients with mNSCLC receiving combination immunotherapy, however did not clearly confirm the predictive role for STK11, but rather showed that this may be a prognostic biomarker, with overall worse outcomes seen in patients with STK11 mutation[38]. The divergent data on mutations and their interplay with outcomes following chemotherapy, chemo-immunotherapy and immunotherapy combinations can be potentially explained by the complex molecular interactions that exist within the tumor microenvironment. While others have demonstrated an association between a subset of mutations, to our knowledge, the combination of pTMB and specific negative predictor mutations in ERBB2 exon 20, STK11, KEAP1, and PTEN from plasma has not been reported. These are small numbers and the analysis should be considered purely exploratory. Adding mutation analysis might enhance the ability of pTMB to predict outcomes from immunotherapy. These observations, if validated, suggest that including these genomic biomarkers in the predictive algorithms, may improve identification of pTMB-high patients unlikely to respond.
Our study does have certain limitations. It is a single-center, non-randomized study. Matched tissue TMB was not able to be performed as it is not yet a part of the routine clinical tissue NGS testing performed at our institution. Further study is required to validate our findings, including pTMB cutoff, in a larger dataset. Although combining tissue TMB and PD-L1 has shown improved prediction of immunotherapy response, this analysis could not be performed here, as the treatment regimens (P vs. PC) were largely dictated by tumor PD-L1 TPS. Our study also does not consider characteristics of the tumor microenvironment, immune competence, including MHC status, or the microbiome. Nevertheless, our results do argue for larger-scale validation of plasma-based TMB in the context of prospective pembrolizumab-based therapy in mNSCLC; if substantiated, this assay should be integrated into routine clinical management of patients with mNSCLC.
Supplementary Material
Statement of Translational Relevance.
Pembrolizumab based therapy is currently standard of care frontline therapy for patients with metastatic NSCLC (mNSCLC) whose tumors lack therapeutically targetable mutations. Tissue-based testing of PD-L1 Tumor Proportion Score can be used to stratify patients onto single agent pembrolizumab versus combination pembrolizumab-chemotherapy. However, it is an imperfect biomarker, and there is a need for additional predictive clinical biomarkers to aid clinical decision making. High TMB is associated with response to therapy, but testing requires sufficient tissue, which can be difficult to obtain. Here we report on the plasma TMB (pTMB) of 66 prospectively enrolled patients with mNSCLC who received frontline pembrolizumab monotherapy or in combination with chemotherapy. High baseline pTMB was associated with improved response rate (by RECIST) and PFS. Although a larger validation study is needed, our results show the potential clinical utility of a plasma based TMB test to help inform therapy selection.
Acknowledgments
Funding Sources:
This work is supported by Merck & Co., the National Cancer Institute at the National Institutes of Health [CA234225-01], and the LUNGevity Foundation.
Footnotes
Disclosures:
The authors have declared no conflicts of interest.
References
- 1.Reck M, et al. , Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med, 2016. 375(19): p. 1823–1833. [DOI] [PubMed] [Google Scholar]
- 2.Gandhi L, et al. , Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med, 2018. 378(22): p. 2078–2092. [DOI] [PubMed] [Google Scholar]
- 3.Paz-Ares L, et al. , Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N Engl J Med, 2018. 379(21): p. 2040–2051. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal C, et al. , Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol, 2018. [DOI] [PMC free article] [PubMed]
- 5.Rizvi H, et al. , Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol, 2018. 36(7): p. 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao D, et al. , Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet, 2018. 50(9): p. 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George S, et al. , Loss of PTEN Is Associated with Resistance to Anti-PD-1 Checkpoint Blockade Therapy in Metastatic Uterine Leiomyosarcoma. Immunity, 2017. 46(2): p. 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Best SA, et al. , Synergy between the KEAP1/NRF2 and PI3K Pathways Drives Non-Small-Cell Lung Cancer with an Altered Immune Microenvironment. Cell Metab, 2018. 27(4): p. 935–943 e4. [DOI] [PubMed] [Google Scholar]
- 9.Peng W, et al. , Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov, 2016. 6(2): p. 202–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skoulidis F, et al. , STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov, 2018. 8(7): p. 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellmann MD, et al. , Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med, 2018. 378(22): p. 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters S, et al. Tumor mutational burden (TMB) as a biomarker of survival in metastatic non-small cell lung cancer (mNSCLC): Blood and tissue TMB analysis from MYSTIC, a Phase III study of first-line durvalumab ± tremelimumab vs chemotherapy [abstract]. in Proceedings of the 110th Annual Meeting of the American Association for Cancer Research. 2019. Atlanta, GA: AACR. [Google Scholar]
- 13.Thompson JC, et al. , Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin Cancer Res, 2016. 22(23): p. 5772–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gandara DR, et al. , Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med, 2018. 24(9): p. 1441–1448. [DOI] [PubMed] [Google Scholar]
- 15.Rizvi NA, et al. LBA6Durvalumab with or without tremelimumab vs platinum-based chemotherapy as first-line treatment for metastatic non-small cell lung cancer: MYSTIC. in ESMO Immuno-Oncology Congress. 2018. Geneva, Switzerland: EMSO. [Google Scholar]
- 16.Kim ES, et al. , LBA55Primary efficacy results from B-F1RST, a prospective phase II trial evaluating blood-based tumour mutational burden (bTMB) as a predictive biomarker for atezolizumab (atezo) in 1L non-small cell lung cancer (NSCLC). Annals of Oncology, 2018. 29(suppl_8). [Google Scholar]
- 17.Rizvi NA, et al. , Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science, 2015. 348(6230): p. 124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McShane LM, et al. , REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer, 2005. 93(4): p. 387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odegaard JI, et al. , Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res, 2018. 24(15): p. 3539–3549. [DOI] [PubMed] [Google Scholar]
- 20.Medina Diaz I, et al. , Performance of Streck cfDNA Blood Collection Tubes for Liquid Biopsy Testing. PLoS One, 2016. 11(11): p. e0166354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nance T, et al. A novel approach to differentiation of somatic vs. germline variants in liquid biopsies using a betabinomial model. in Proceedings of the 109th Annual Meeting of the American Association for Cancer Research. 2018. Chicago, IL: AACR. [Google Scholar]
- 22.Quinn K, et al. Landscape and genomic correlates of plasma based tumor mutational burden across six solid tumor types. in Proceedings of the 110th Annual Meeting of the American Association for Cancer Research. 2019. Atlanta, GA: AACR. [Google Scholar]
- 23.Feltz CJ and Miller GE, An asymptotic test for the equality of coefficients of variation from k populations. Stat Med, 1996. 15(6): p. 646–58. [DOI] [PubMed] [Google Scholar]
- 24.Qiu P, et al. , Measuring Tumor Mutational Burden (TMB) in Plasma from mCRPC Patients Using Two Commercial NGS Assays. Sci Rep, 2019. 9(1): p. 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellrott K, et al. , Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst, 2018. 6(3): p. 271–281 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durrleman S and Simon R, Flexible regression models with cubic splines. Stat Med, 1989. 8(5): p. 551–61. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, et al. Analytical validation of MSI High detection with GuardantOMNI in Proceedings of the 110th Annual Meeting of the American Association for Cancer Research. 2019. Atlanta, GA: AACR. [Google Scholar]
- 28.Wang Z, et al. , Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol, 2019. [DOI] [PMC free article] [PubMed]
- 29.Zaretsky JM, et al. , Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med, 2016. 375(9): p. 819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin DS, et al. , Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov, 2017. 7(2): p. 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negrao MV, et al. , Association of EGFR and HER-2 exon 20 mutations with distinct patterns of response to immune checkpoint blockade in non-small cell lung cancer. Journal of Clinical Oncology, 2018. 36(15_suppl): p. 9052–9052. [Google Scholar]
- 32.Legrand FA, et al. Association of high tissue TMB and atezolizumab efficacy across multiple tumor types in ASCO 2018. Chicago, IL: J Clin Oncol. [Google Scholar]
- 33.Kowanetz M, et al. , Tumor Mutation Burden (TMB) is Associated with Improved Efficacy of Atezolizumab in 1L and 2L+NSCLC Patients. Journal of Thoracic Oncology, 2017. 12(1): p. S321–S322. [Google Scholar]
- 34.Langer CJ, et al. TMB and Outcomes for Carboplatin and Pemetrexed With or Without Pembrolizumab for Nonsquamous NSCLC in World Conference on Lung Cancer. 2019. Barcelona, Spain: IASLC. [Google Scholar]
- 35.Leighl NB, et al. , Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin Cancer Res, 2019. 25(15): p. 4691–4700. [DOI] [PubMed] [Google Scholar]
- 36.Jia Q, et al. , Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun, 2018. 9(1): p. 5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenthal R, et al. , Neoantigen-directed immune escape in lung cancer evolution. Nature, 2019. 567(7749): p. 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizvi N Mutations Associated with Sensitivity or Resistance to Immunotherapy in mNSCLC: Analysis from the MYSTIC Trial in IASLC 2019 World Conference on Lung Cancer 2019. Barcelona, Spain: Journal of Thorasic Oncology. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.