Abstract
The COVID-19 pandemic urgently needs therapeutic and prophylactic interventions. Here, we report the rapid identification of SARS-CoV-2-neutralizing antibodies by high-throughput single-cell RNA and VDJ sequencing of antigen-enriched B cells from 60 convalescent patients. From 8,558 antigen-binding IgG1+ clonotypes, 14 potent neutralizing antibodies were identified, with the most potent one, BD-368-2, exhibiting an IC50 of 1.2 and 15 ng/mL against pseudotyped and authentic SARS-CoV-2, respectively. BD-368-2 also displayed strong therapeutic and prophylactic efficacy in SARS-CoV-2-infected hACE2-transgenic mice. Additionally, the 3.8 Å cryo-EM structure of a neutralizing antibody in complex with the spike-ectodomain trimer revealed the antibody’s epitope overlaps with the ACE2 binding site. Moreover, we demonstrated that SARS-CoV-2-neutralizing antibodies could be directly selected based on similarities of their predicted CDR3H structures to those of SARS-CoV-neutralizing antibodies. Altogether, we showed that human neutralizing antibodies could be efficiently discovered by high-throughput single B cell sequencing in response to pandemic infectious diseases.
Keywords: single-cell sequencing, SARS-CoV-2, COVID-19, neutralizing antibody, convalescent patient, B cell, CDR3
Graphical Abstract
Neutralizing antibodies, which could effectively block virus entry into host cells, are urgently needed for intervention against COVID-19. Using high-throughput single-cell RNA sequencing, Cao et al. identified fourteen potent neutralizing antibodies from 60 convalescent patients’ B cells. The most potent antibody, BD-368-2, exhibits high therapeutic and prophylactic efficacy in SARS-CoV-2-infected mice.
Introduction
Coronavirus disease 2019 (COVID-19) caused by a novel coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide as a severe pandemic (Callaway et al., 2020). Both SARS-CoV-2 and SARS-CoV belong to lineage B of the betacoronavirus genus (Zhou et al., 2020, Wu et al., 2020), and their RNA genomes share around 82% identity (Chan et al., 2020). The mechanisms by which SARS-CoV-2 infects target cells have been well studied and recently reported (Hoffmann et al., 2020, Walls et al., 2020). Similar to SARS-CoV, the spike (S) glycoprotein on the surface of SARS-CoV-2 mediates membrane fusion and receptor recognition of the virus (Wrapp et al., 2020). The S1 subunit at the N-terminal region is responsible for virus attachment and contains the receptor-binding domain (RBD), which directly binds to the ACE2 receptor on the host cell. Currently, no validated therapeutics against virus-target interactions are available for COVID-19.
Convalescent patients’ plasma, which contains neutralizing antibodies produced by the adaptive immune response, has led to a clear clinical improvement of both mild and severe COVID-19 patients when used as a therapeutic modality (Chen et al., 2020, Shen et al., 2020, Cao, 2020). However, therapeutic use is limited since plasma cannot be produced on a large scale. On the other hand, neutralizing monoclonal antibodies (mAbs) isolated from convalescent patient’s memory B cells may serve as a promising intervention to SARS-CoV-2 due to their scalability and therapeutic effectiveness. Human-sourced mAbs targeting viral surface proteins have increasingly shown their therapeutic and prophylactic efficacy against infectious diseases such as HIV, Ebola, and Middle Eastern respiratory syndrome (MERS) (Corti et al., 2016, Wang et al., 2018, Scheid et al., 2009). Their safety and potency in patients have been demonstrated in multiple clinical trials (Xu et al., 2019, Caskey et al., 2017). Despite their advantages, screening for potent neutralizing mAbs from human memory B cells is often a slow and laborious process, which is not ideal when responding to a worldwide health emergency. A rapid and efficient method for screening SARS-CoV-2-neutralizing mAbs is urgently needed.
Due to VDJ recombination and somatic hypermutation, B cells exhibit diverse B cell repertoires, necessitating the analysis of one B cell at a time (Bassing et al., 2002). Techniques, such as single-cell clonal amplification of memory B cells, are usually utilized to obtain paired immunoglobulin heavy-light chain RNA sequences from the heterogeneous B cell population to produce mAbs (El Debs et al., 2012, Niu et al., 2019). Clonal amplification of Epstein-Barr virus (EBV)-immortalized memory B cells from convalescent patients has proved successful in isolating neutralizing mAbs against viral infections such as HIV, Dengue, and MERS (Burton et al., 2009, Corti et al., 2015, Scheid et al., 2009, Setthapramote et al., 2012). Yet, due to the time-consuming incubation and screening steps, the technique takes several months at least to complete a successful screen.
On the other hand, single-cell RT-PCR combined with fluorescence-activated cell sorting (FACS) or optofluidics platform, such as Beacon (Berkley Light), could obtain antibody sequences in several days by performing nested PCR on single antigen-binding memory B cells after single-cell sorting (Tiller et al., 2008, Wardemann et al., 2003, Wrammert et al., 2008, Liao et al., 2009). The method has led to efficient isolation of neutralizing mAbs in various infectious diseases, including HIV and MERS (Scheid et al., 2009, Wang et al., 2018). Nevertheless, the recent development of high-throughput single-cell RNA and VDJ sequencing of B cell receptor repertoires using 10X Chromium has outperformed single-cell RT-PCR in terms of B cell screening throughput (Goldstein et al., 2019, Horns et al., 2020). The microfluidic-based technique could obtain auto-paired heavy- and light-chain sequences from tens of thousands single B cells in one run and has successfully been used for the isolation of human neutralizing mAbs against HIV (Setliff et al., 2019).
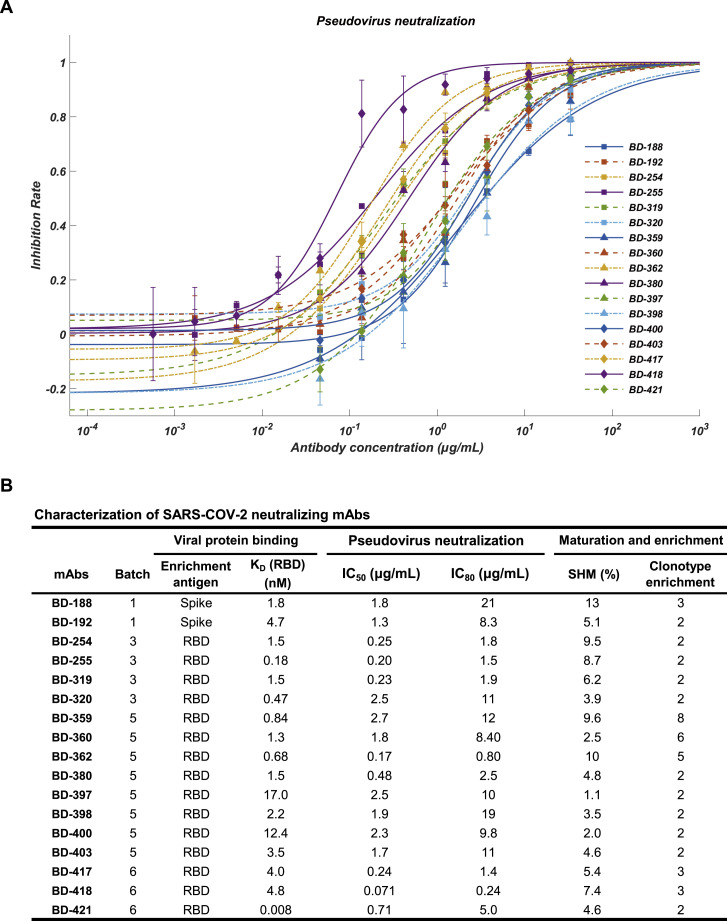
Here, we report the rapid and efficient identification of SARS-CoV-2-neutralizing antibodies achieved by high-throughput single-cell RNA and VDJ sequencing of antigen-binding B cells from convalescent COVID-19 patients. Over 8,500 antigen-binding B cell clonotypes expressing immunoglobulin G1 (IgG1) antibodies were identified from 60 convalescent patients. In total, we identified 14 potent neutralizing mAbs, among which the most potent mAb, BD-368-2, exhibited an IC50 of 1.2 and 15 ng/mL against pseudotyped and authentic SARS-CoV-2. Additionally, in vivo experiments confirmed that BD-368-2 could provide strong therapeutic efficacy and prophylactic protection against SARS-CoV-2, using the hACE2 transgenic mice model (Bao et al., 2020, Yang et al., 2007, McCray et al., 2007).
Further, we solved the cryoelectron microscopy (cryo-EM) structure of one neutralizing mAb, BD-23, in complex with the SARS-CoV-2 spike ectodomain trimer and showed that its epitope overlaps with the RBD/ACE2 binding motif. Moreover, based on the high conservation between SARS-CoV-2 and SARS-CoV, we demonstrated that potent neutralizing mAbs against SARS-CoV-2 could be directly chosen from the large antigen-binding clonotype library, utilizing the CDR3H structure’s similarity to that of SARS-CoV-neutralizing antibody m396 (Prabakaran et al., 2006, Zhu et al., 2007). Overall, we showed that high-throughput single-cell sequencing could lead to the identification of highly potent neutralizing mAbs that have strong therapeutic and prophylactic efficacy, which could greatly assist in the intervention of prevailing and emerging infectious diseases, such as COVID-19.
Results
High-Throughput Sequencing of Single B Cells from Convalescent Patients
Unlike traditional methodologies, large-scale data obtained from high-throughput scVDJ sequencing (scVDJ-seq) enabled us to examine B cell clonotype enrichment prior to in vitro antibody expression (Goldstein et al., 2019, Croote et al., 2018). B cells that share an identical CDR3 region for both heavy and light chains were grouped into the same clonotypes, and their enrichment was calculated based on the number of cells observed for the clonotype. Since antigen-activated B cells would go through clonal selection and expansion from pre-existing naive and memory B cells (Murugan et al., 2018, Seifert and Kü ppers, 2016), we hypothesized that enriched B cell clonotypes would more likely yield high-affinity SARS-CoV-2 binding and neutralizing antibodies.
To exploit this hypothesis, we first collected peripheral blood mononuclear cells (PBMCs) and isolated the B cells from 12 COVID-19 convalescent patients from Beijing Youan Hospital (Table S1). We performed small conditional RNA (scRNA) and scVDJ sequencing on the freshly isolated B cells or CD27+ memory B cell subsets using 10X Chromium 5′ mRNA and VDJ sequencing (Figures 1 A and S1 A). The scVDJ data indeed revealed enriched IgG1 clonotypes (Figures S1F and S1H), and the CD27+ memory B cell selection largely improved the number of memory B cells sequenced (Figures S1D and S1E) as well as the IgG1 clonotypes discovered (Figures S1G and S1I). However, from the 130 in vitro-expressed mAbs selected from memory B cells containing enriched IgG1+ clonotypes (Table S2), only one out of the two RBD-binding mAbs, named BD-23, showed a weak virus neutralization ability against pseudotyped and authentic SARS-CoV-2 (Figures S1B and S1C). The low efficiency in terms of RBD-binding mAbs and neutralizing mAbs identification necessitates an antigen-affinity based selection, which could greatly enrich for spike/RBD binding B cells.
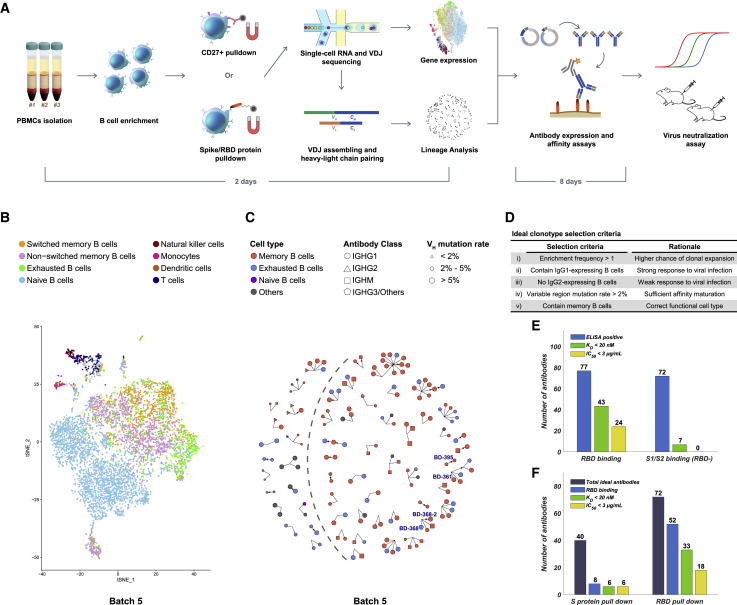
Figure 1.
Efficient Neutralizing Antibody Identification through Antigen-Enriched High-Throughput Single-Cell RNA Sequencing
(A) Schematic overview of the neutralizing antibody identification process. The sequence of the mAbs could be obtained within 2 days using 10X Genomics 5′ VDJ sequencing.
(B) Cell-type identification of the single B cells binding RBD (batch 5) based on gene expression. Only productive heavy-light chain-paired single cells are analyzed. See also Figures S1 and S2.
(C) Ideal clonotype selection for in vitro expression showing clonotype’s enrichment frequency, immunoglobulin class, cell type, and variable region mutation rate (batch 5). Ideal clonotypes are on the right side of the dashed line with four potent neutralizing mAbs selected for further characterization are labeled.
(D) Ideal clonotype selection criteria.
(E) Characteristics of RBD-binding and spike-protein binding (RBD-) antibodies. Only RBD-binding antibodies showed neutralizing ability in pseudovirus neutralization assays. An antibody was determined as ELISA positive if it showed saturated absorption at 1 μg/mL antigen and 1 μg/mL antibody concentration. KD was measured by using SPR with a 1:1 binding model.
(F) Characteristics of the antibodies selected based on different antigen enrichment methods.
See also Figure S3.
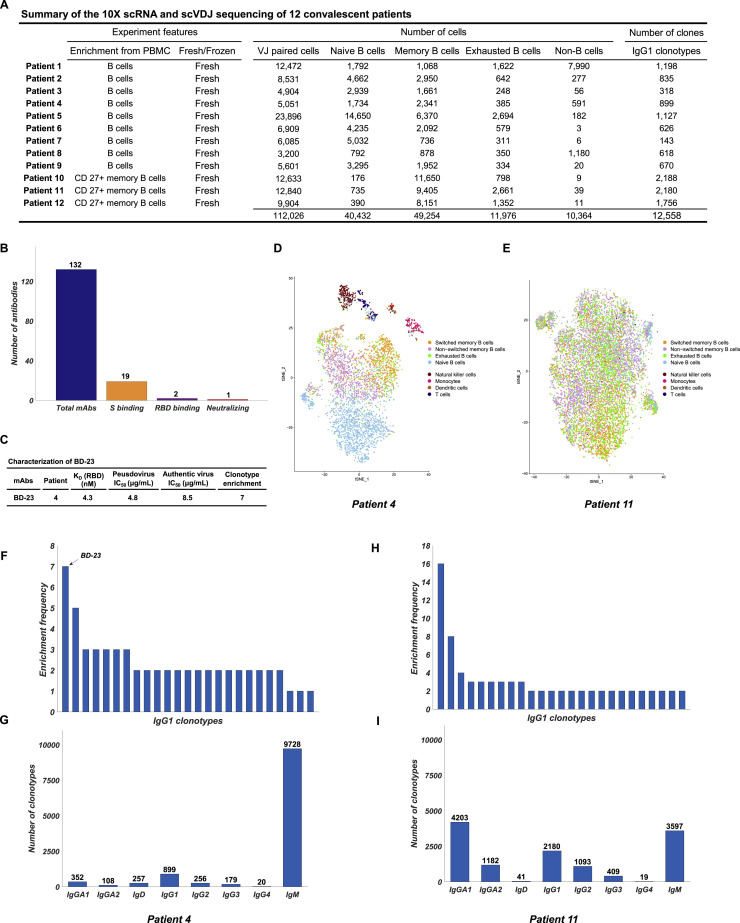
Figure S1.
Summary of the 10X scRNA and scVDJ Sequencing of 12 Convalescent Patients’ B Cells, Related to Figure 1
(A) Summary of the 10X scRNA and scVDJ sequencing of 12 convalescent patients’ B cells. Patient 1-9 used a MACS-based negative selection for B cell enrichment from PBMC. Patient 10-12 used a MACS-based CD27+ selection for memory B cell enrichment from PBMC. Cells are assigned to the same clonotype if they have identical heavy and light chain CDR3 DNA sequences. (B) Characteristics of the selected mAbs identified from the 12 patients. mAbs are selected from clonotypes that contain IgG1-presenting memory B cells. (C) Characteristics of the neutralizing mAb BD-23. (D) t-SNE plot of patient 4’s scRNA-seq result. Only productive Heavy-Light chain paired cells were shown. Cells are colored based on cell types. (E) t-SNE plot of patient 11’s scRNA-seq result. (F) Top 25 most enriched clonotypes of patient 4. The clonotype containing BD-23 is labeled. (G) Ig class distribution of patient 4’s clonotypes. (H) Top 25 most enriched clonotypes of patient 11. (I) Ig class distribution of patient 11’s clonotypes.
Efficient Identification of Neutralizing mAbs by High-Throughput Sequencing of Antigen-Binding Single B Cells
To enrich for RBD-binding B cells, we utilized both biotinylated RBD and S protein to select for antigen-binding B cells through magnetic beads separation (Figure 1A). Since the selection introduces a large reduction in the number of B cells, we combined the PBMCs from different patients for sufficient 10X loading. In total, we analyzed 60 convalescent patients’ antigen-binding B cells in 6 different batches (Table S1). Since the goal was to produce neutralizing mAbs, we filtered out all B cells that failed to have productive V-J spanning heavy-light chain pairs. A total of 8,558 distinct IgG1-presenting antigen-binding clonotypes were detected (Table 1 ). Furthermore, the scRNA-seq data enabled us to perform cell-typing based on mRNA expression for memory B cell identifications (Figures 1B and S2 ). We observed sufficient separation between naive B cells and memory B cells, as well as non-B cells versus memory B cells. However, a clear separation between switched/non-switched memory B cells and exhausted memory B cells may not be observed in some batches, mostly due to the relatively low sequencing depth and the low number of genes detected per cell of the scRNA sequencing (Table S3).
Table 1.
Summary of the 10X scRNA and scVDJ Sequencing of Antigen-Binding B Cells
| Enrichment Antigen | PBMC Status | VJ Paired Cells | Naive B Cells | Memory B Cells | Exhausted B Cells | Non-B Cells | IgG1 Clonotypes | |
|---|---|---|---|---|---|---|---|---|
| Batch 1 | S protein | fresh | 14,895 | 5,417 | 6,333 | 1,680 | 1,465 | 2,598 |
| Batch 2 | S protein | frozen | 6,962 | 1,051 | 1,161 | 362 | 4,388 | 883 |
| Batch 3 | RBD protein | fresh | 6,000 | 1,668 | 2,603 | 498 | 1,231 | 812 |
| Batch 4 | S1 (RBD-) | fresh | 11,536 | 3,202 | 6,413 | 1,710 | 211 | 3,093 |
| Batch 5 | RBD protein | fresh | 8,020 | 3,532 | 3,302 | 823 | 363 | 916 |
| Batch 6 | RBD protein | fresh | 2,925 | 1,177 | 1,353 | 260 | 135 | 256 |
| Total | 50,338 | 16,047 | 21,165 | 5,333 | 7,793 | 8,558 |
The experimental details for each sequencing batch. Enrichment antigen refers to the antigen used for the MACS pull-down. Batch 4 was performed on samples that have RBD-binding B cells filtered. PBMC status refers to whether the PBMC used were previously frozen. VJ paired cells refer to cells that both full-length heavy- and light-chain variable regions were detected. Cells are assigned to the same clonotype if they have identical heavy- and light-chain CDR3 DNA sequences. See also Figure S2.
Figure S2.
Summary of the 10X scRNA and scVDJ Sequencing of Antigen-Binding B Cells, Related to Table 1
A) – E) t-SNE plot of productive heavy-light chain paired single cells in each batch. Cells are colored based on cell types.
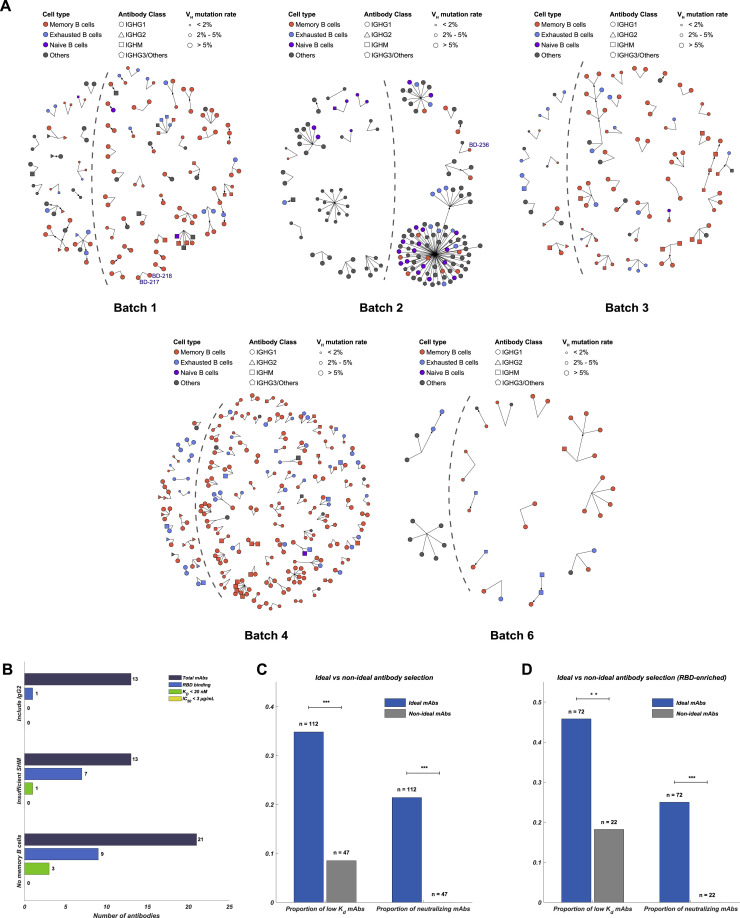
To increase the efficiency of neutralizing mAbs identification, we developed a set of criteria to filter out the clonotypes that have low chances to produce neutralizing mAbs (Figure 1D). First, only enriched clonotypes containing IgG1-expressing B cells were selected (Figure 1C and S3 A), since IgG1-expressing B cells respond strongly to viral stimuli (Vidarsson et al., 2014). Second, IgG2-expressing B cells normally respond weakly to viral pathogens, and thus IgG2-presenting clonotypes were not included as ideal candidates (Irani et al., 2015). Next, clonotypes that did not contain any B cells with a somatic hypermutation rate (SHM) higher than 2% indicate insufficient affinity maturation and were excluded (Croote et al., 2018, Methot and Di Noia, 2017). Last, as exhausted B cells and naive B cells respond less effective to antigen stimuli (Moir and Fauci, 2014), clonotypes that contained only exhausted memory B cells or naive B cells were not considered as ideal candidates (Figures 1B and S2). Together, a total of 169 ideal candidates were selected from enriched clonotypes and were expressed in HEK293 cells through transfection (Table S2). Meanwhile, 47 non-ideal candidates were also expressed in vitro as a reference to validate the effectiveness of our selection criteria (Table S2).
Figure S3.
Ideal Antibody Selection from Enriched IgG1+ clonotypes for In Vitro Expression, Related to Figure 1
(A) Ideal antibody selection based on clonotype enrichment frequency, Ig class, cell type, and VDJ region mutation rate. Ideal antibodies are on the right side of the dashed line. (B) Non-ideal candidates showed less strong binding and neutralizing mAbs. (C) The comparison of the proportion of strong binding and neutralizing mAbs identified from ideal candidates and non-ideal candidates (Fisher’s exact test, ∗∗∗p < 0.001). (D) The comparison of the proportion of strong binding and neutralizing mAbs identified from RBD-enriched ideal candidates and non-ideal candidates (Fisher’s exact test, ∗∗p < 0.01, ∗∗∗p < 0.001).
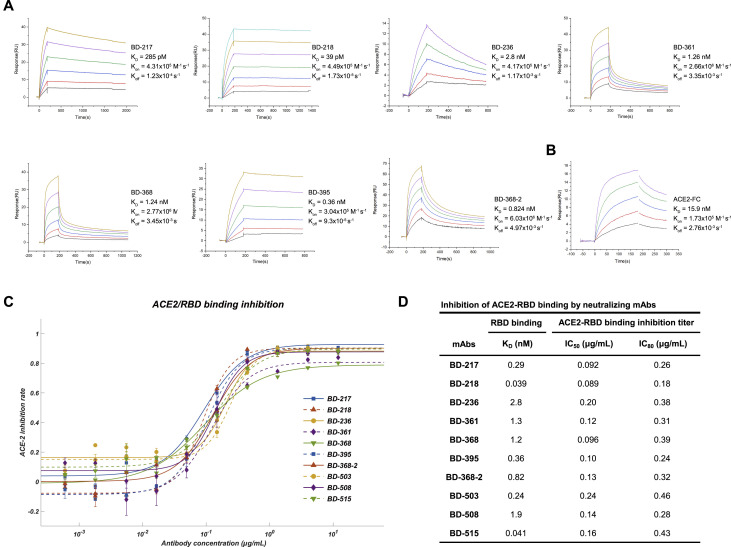
The purified mAbs were tested for SARS-CoV-2 RBD/spike reactivity by ELISA and surface plasmon resonance (SPR), and 149 S-binding mAbs were identified, among which over half of the mAbs bind to the RBD. We further screened all ELISA-positive mAbs for neutralizing ability using a SARS-CoV-2 pseudovirus system. We found that only RBD-binding mAbs showed pseudovirus neutralization effects (Figure 1E), and only mAbs bound to the RBD with a KD smaller or close to the dissociation constant of ACE2/RBD (15.9 nM) (Figure S5 B), would have significant neutralization effects (IC50 < 3 μg/mL) on SARS-CoV-2 pseudovirus (Figures S4 A and S4B). Both full-length S protein and the RBD protein could enrich for RBD-binding mAbs but with distinct efficiency (Figure 1F). Using the RBD protein as the enrichment antigen resulted in an RBD ELISA-positive rate 3-fold higher than using S protein. Most importantly, mAbs selected from ideal candidates showed an efficiency of 46% and 25% in identifying strong RBD-binding mAbs (KD < 20 nM) and SARS-CoV-2 neutralizing mAbs, respectively (Figure S3D), which is significantly higher compared to the non-ideal mAbs candidates (Figures S3B and S3C).
Figure S5.
KD Measurement and ACE2/RBD Binding Inhibition for the Potent Neutralizing mAbs, Related to Figure 2
(A) Dissociation constant measurement of the representing mAbs against RBD. KD is calculated using a 1:1 binding model. All measurements are performed by using a serial 2-fold dilution of biotinylated RBD, starting from 50 nM (Yellow) to 1.56 nM (Black). (B) Dissociation constant measurement of ACE2 against RBD. (C) ACE2/RBD binding inhibition rate determined by ACE2 competition ELISA assay. The data were obtained from a single representative experiment with three replicates. Data are represented as mean ± SD (D) IC50 and IC80 calculated from ACE2 competence ELISA assays by fitting a four-parameter logistic regression model.
Figure S4.
Binding Specificity and Neutralizing Abilities of the Non-potent Neutralizing mAbs, Related to Figure 2
(A) Neutralization potency measured by a SARS-CoV-2 spike-pseudotyped VSV neutralization assay. Data for each mAb were obtained from a representative neutralization experiment, which contains three replicates. Data are represented as mean ± SD. IC50 and IC80 were calculated by fitting a four-parameter logistic curve. (B) Characteristics of the neutralizing mAbs. KD is measured using SPR with a 1:1 binding model targeting biotinylated RBD protein. The somatic hypermutation rate (SHM) is calculated from mutated DNA sequences of the heavy- chain variable regions (V, D, and J regions) using Igblast.
High Neutralizing Potency against SARS-CoV-2 among mAbs from Enriched Clonotypes
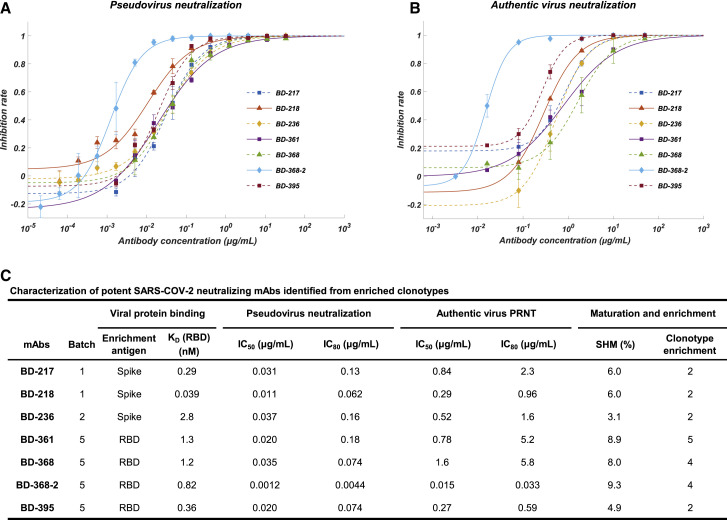
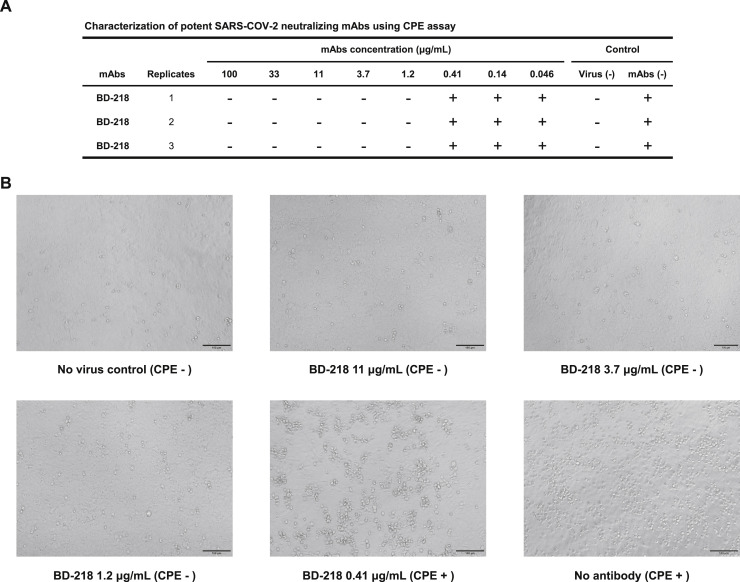
Among all the neutralizing mAbs isolated from enriched clonotypes, seven of them showed potent neutralization ability with an IC50 lower than 0.05 μg/mL against SARS-CoV-2 pseudovirus (Figures 2A and 2C). The most potent mAb BD-368-2, which was selected from the same clonotype as BD-368 (Figure 1C), exhibited an IC50 of 1.2 ng/mL. All seven mAbs bind strongly to the RBD with nM or sub-nM KD, revealed by SPR (Figure S5A), and could competitively inhibit the ACE2 binding with the RBD (Figure S5C and S5D). To evaluate their neutralization potential against the authentic virus, we performed the plaque reduction neutralization test (PRNT) using authentic SARS-CoV-2 isolated from COVID-19 patients. Among all mAbs tested, BD-368-2 displayed the highest potency against the authentic virus with an IC50 of 15 ng/mL (Figures 2B and 2C). To further validate the authentic virus neutralization shown by PRNT, we performed the cytopathic effect (CPE) inhibition assay using BD-218 on authentic SARS-CoV-2. BD-218 showed complete viral inhibition across all three replicates, at the concentration of 1.2 μg/mL (Figures S6 A and S6B), consistent with the PRNT results. Together, our data have shown that highly potent neutralizing mAbs could be identified from convalescent patients by high-throughput single-cell sequencing of antigen-binding B cells.
Figure 2.
Affinity Specificity and Neutralizing Abilities of the Potent Neutralizing mAbs
(A) Neutralization potency measured by using a pseudotyped virus neutralization assay. Data for each mAb were obtained from a representative neutralization experiment, which contains three replicates. Data are represented as mean ± SD. See also Figure S4.
(B) Neutralization potency measured by an authentic SARS-CoV-2 plaque reduction neutralizing test (PRNT) assay. Data for each mAb were obtained from a representative neutralization experiment, which contains three replicates. Data are represented as mean ± SD. See also Figure S6.
(C) Characteristics of the neutralizing mAbs. IC50 and IC80 were calculated by using a four-parameter logistic curve fitting. KD targeting RBD was measured by using SPR with a 1:1 binding model. The somatic hypermutation rate (SHM) was calculated by comparing DNA sequences of the heavy-chain variable regions (V, D, and J regions) to germline sequences using Igblast.
See also Figure S5.
Figure S6.
Authentic SARS-CoV-2 Neutralization Potency Measured by Cytopathic Effect (CPE) Assay Showed High Consistency with PRNT, Related to Figure 2
(A) A serial dilution of each mAbs is tested against the authentic SARS-CoV-2 on Vero-E6 cells examined by CPE (B) Phase-contract image of Vero-E6 cells examining CPE. BD-218 shows no CPE at 1.2 μg/mL, which is consistent with the PRNT results of BD-218.
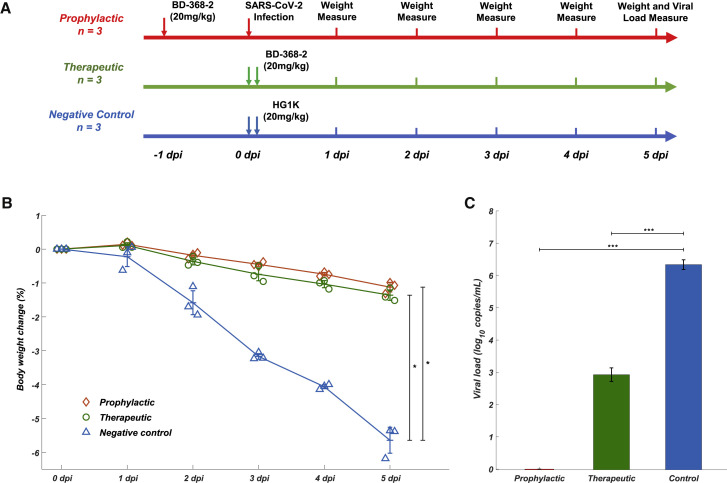
BD-368-2 Showed High Therapeutic and Prophylactic Efficacy in hACE2 Mice
To evaluate whether the identified neutralizing mAbs could serve as therapeutic interventions and prophylactic protection against SARS-COV-2 in vivo, we tested the neutralization efficacy of BD-368-2 on hACE2 transgenic mice infected with SARS-CoV-2 (Bao et al., 2020). BD-368-2 was chosen since it exhibits the highest potency against both pseudotyped and authentic SARS-CoV-2. A total of 9 hACE2 transgenic mice were used in this study and were equally split into three groups (n = 3). The prophylactic efficacy was examined by intraperitoneally injecting 20 mg/kg (of mice body weight) BD-368-2 into hACE2 transgenic mice 24 h before viral infection (Figure 3 A). For the therapeutic group, 20 mg/kg BD-368-2 was injected 2 h after infection. As a negative control, 20 mg/kg HG1K, an IgG1 antibody against the H7N9 virus, was injected 2 h after infection. We recorded the body weight for each mice daily after infection for 5 days (Figure 3A) and found that the therapeutic and prophylactic treatment groups maintained their body weight, whereas the negative control group significantly lost weight (Figure 3B). This indicates that BD-368-2 applied both before and after the infection could greatly improve the physiological condition of the SARS-CoV-2-infected mice. Moreover, we analyzed the viral load by qRT-PCR of the lungs at 5 dpi (Yang et al.,2007) and found that injections of BD-368-2 before the viral challenge could completely inhibit SARS-CoV-2 (Figure 3C). Furthermore, applying BD-368-2 2 h after infection could result in a 3–4 log decrease in virus copies in mice lung, indicating an effective reduction of SARS-CoV-2 replication (Figure 3C). Together, BD-368-2 exhibits high therapeutic and prophylactic efficacy in vivo.
Figure 3.
BD-368-2 Showed High Therapeutic and Prophylactic Efficacy in SARS-CoV-2-Infected hACE2 Transgenic Mice
(A) Experimental design for therapeutic and prophylactic testing of BD-368-2 in hACE2 transgenic mice. BD-368-2 or unrelated antibody HG1K (20 mg/kg of body weight) was intraperitoneally injected into the transgenic mice before or after SAR-CoV-2 infection.
(B) Body-weight change (%) of the hACE2 transgenic mice recorded over 5 days (one-sided permutation test, ∗p < 0.05). Each group contains 3 mice. Data are represented as mean ± SD.
(C) Virus titers of lung tissue at 5 dpi. The viral loads of the lung were determined by qRT-PCR (one-tailed t test, ∗∗∗p < 0.001). Data are represented as mean ± SD.
Cryo-EM Structure of the Neutralizing mAb Bound to the Trimeric Spike Protein
Neutralizing mAbs against SARS-CoV-2 were identified efficiently after analyzing enriched clonotypes. However, due to the shallow sampling of B cells for each patient, the majority of the clonotypes did not show any enrichment. Nevertheless, the non-enriched antigen-binding clonotypes still remain as a valuable library for future identification of neutralizing mAbs. A new antibody selection criterion for non-enriched clonotypes is needed, since arbitrary selection may lead to a high percentage of weak antigen-binding mAbs and is also prone to non-specific binding during the antigen pull-down. Previously, people have used antibody structure prediction to annotate Ig-seq data for better mAbs selection (DeKosky et al., 2016). By selecting the mAbs that share similar CDR3H structures with reference neutralizing mAbs, the structural annotation approach has proved to be successful in improving the performance of Ig-seq as well as in vitro antibody maturation (Kovaltsuk et al., 2017, Krawczyk et al., 2014, Sela-Culang et al., 2014). Inspired by these results, we hypothesized that CDR3H-based structure prediction could also help to annotate the scVDJ-seq data from convalescent patients, which may serve to improve the efficiency in identifying neutralizing mAbs against SARS-CoV-2.
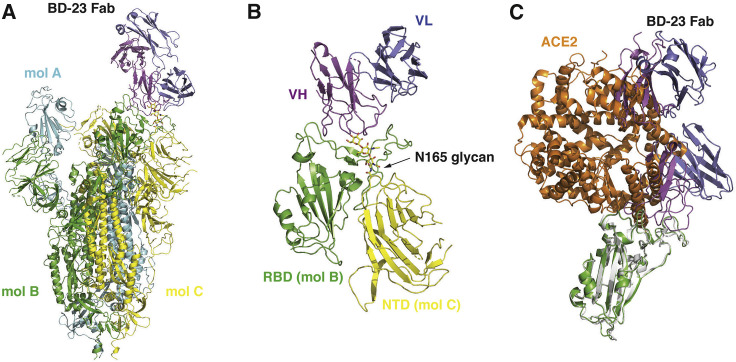
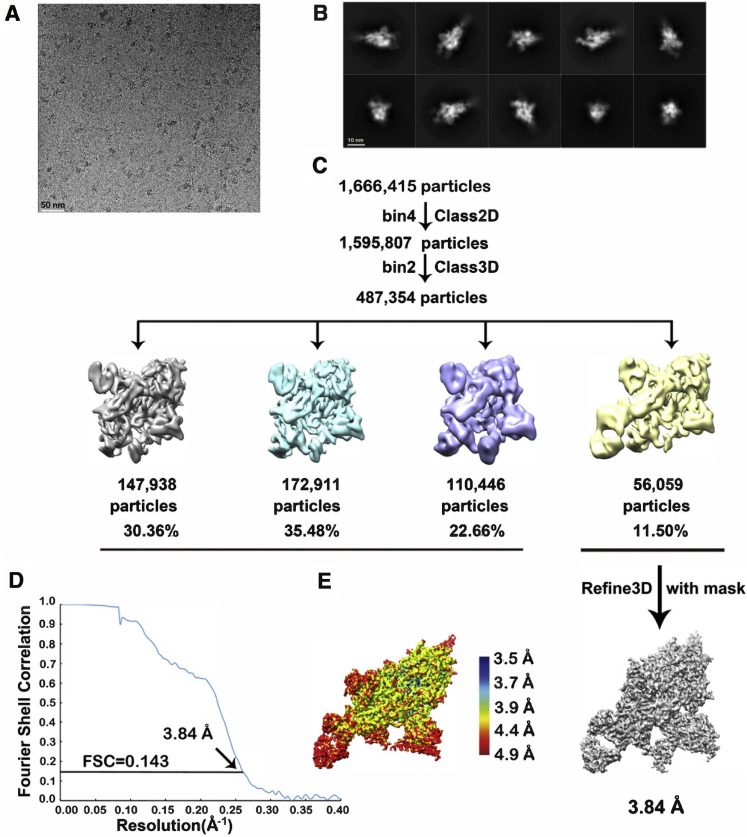
The main difficulty, however, lies in the selection of the reference antibody structure. To date, no SARS-CoV-2 neutralizing mAb structures are recorded in the Protein Database (PDB), which could serve as a reference. To address this, we solved the cryo-EM structure of BD23-Fab in complex with the S ectodomain trimer at an overall resolution of 3.8 Å (Figures 4 A and S7 ; Table S4). The S ectodomain here adopts an asymmetric conformation as previously reported (Wrapp et al., 2020), with the RBD in one protomer (mol A) adopting an “up” position, whereas the other two RBDs (mol B and C) adopt “down” positions. In this 3D reconstruction, a single BD23-Fab is observed per S trimer, and it binds the “down” RBD in protomer B. Only the heavy chain variable domain of BD-23 is involved in binding to the RBD (Figure 4B). Interestingly, the binding is also facilitated by an N-linked glycan on Asn165 of protomer C. Although the current resolution does not support further analysis of the molecular interactions between BD23-Fab and RBD, it is evident that BD23’s epitope overlaps with the motif recognized by ACE2. In fact, a comparison with the RBD-ACE2 complex structure demonstrates that BD23-Fab competes with ACE2 for binding to RBD (Figure 4C). This observation suggests that the SARS-CoV-2 neutralization ability of BD-23 comes from the disruption of the ACE2-RBD binding, which is highly similar to previously identified neutralizing mAbs against SARS-CoV (Prabakaran et al., 2006, Yuan et al., 2020). Indeed, several studies have suggested that SARS-CoV-neutralizing mAbs could cross-neutralize SARS-CoV-2 (Wang et al., 2020, Tian et al., 2020, Lv et al., 2020), which further highlights the resemblance between the RBDs of SARS-CoV and SARS-CoV-2. Altogether, the structural observations and the high conservation between SARS-CoV and SARS-COV-2 suggest that it may also be feasible to use SARS-CoV-neutralizing mAbs’ crystal structures as a reference, to screen for SARS-CoV-2 antigen-binding clonotypes that share similar CDR3H structures to identify neutralizing mAbs.
Figure 4.
Cryo-EM Structure of BD23-Fab in Complex with the Spike Trimer
(A) Cryo-EM structure of the S trimer in complex with BD23-Fab reconstructed at 3.8 Å resolution. The three protomers in the S trimer are depicted in cyan, green, and yellow, respectively. BD23-Fab is depicted in magenta (heavy chain) and blue (light chain).
(B) N165 glycan in the NTD of protomer C facilitates the interaction between BD23-Fab and the RBD of protomer B.
(C) The crystal structure of the RBD/ACE2 complex is overlaid onto the RBD/BD23-Fab structure. BD23-Fab would collide with ACE2 and therefore block the interaction between RBD and ACE2. RBD is shown in green and white, whereas ACE2 in orange.
See also Figure S7.
Figure S7.
Workflow for BD-23 Cryo-EM 3D Reconstructions, Related to Figure 4
A) A representative raw image collected using a Titan Krios 300 kV microscope with a K2 detector. B) Representative 2D classes. C) Flow chart of image processing. D) Gold standard Fourier shell correlation (FSC) curve with estimated resolution. E) Local resolution estimation of the EM map analyzed by ResMap.
Selecting Potent Neutralizing mAbs Based on the Structural Similarity of CDR3H
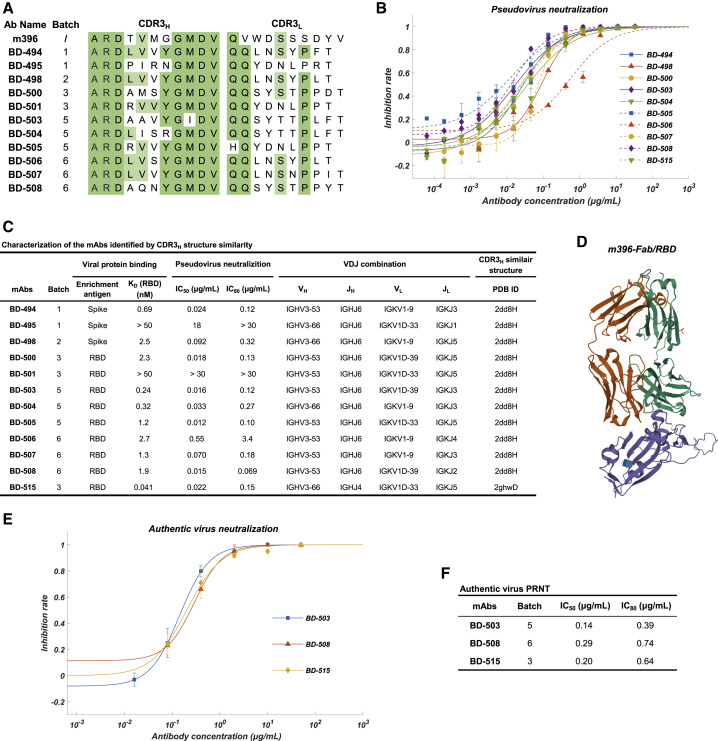
We compared every IgG1-presenting clonotype’s predicted structure based on CDR3H using FREAD to the available SARS-CoV-neutralizing mAbs’ crystal structure in the PDB database as well as the structure of BD-23. Unfortunately, no IgG1-presenting clonotypes showed high CDR3H structural similarity with BD-23. However, a total of 12 IgG1 clonotypes showed high structure similarity with PDB ID 2dd8 and 2ghw (Table S2; Figure 5C), which corresponds to previously isolated SARS-CoV-neutralizing mAbs m396 and 80R, respectively (Prabakaran et al., 2006, Hwang et al., 2006). The CDR3H sequence of the identified mAbs shared high homology with m396 (2dd8) (Figure 5A), which neutralizes SARS-CoV by disrupting ACE2/RBD binding (Figure 5D).
Figure 5.
Characteristics of the Neutralizing mAbs Identified Based on CDR3H Structural Similarity to SARS-CoV-Neutralizing mAbs
(A) The CDR3 sequence comparison between SARS-CoV-neutralizing mAb m396 and the SARS-CoV-2-neutralizing mAbs identified based on CDR3H structure similarity.
(B) Neutralization potency measured by using a pseudotyped virus neutralization assay. Data for each mAb were obtained from a representative neutralization experiment, which contains three replicates. Data are represented as mean ± SD.
(C) Characteristics of the neutralizing mAbs identified based on structure similarity. IC50 and IC80 were calculated by using a four-parameter logistic curve-fitting. KD targeting RBD was measured by using SPR with a 1:1 binding model. VDJ alignment is determined by Igblast. The CDR3H structure prediction was performed using FREAD.
(D) The crystal structure of m396-Fab/SARS-CoV-RBD. The regions corresponding to the RBD, m396-H, and m396-L domain are shown in purple, green, and orange, respectively. The structure is from PDB ID 2dd8.
(E) Authentic SARS-CoV-2 plaque reduction neutralizing test (PRNT) assay on the three selected mAbs, BD-503, BD-508, and BD-515. Data for each mAb were obtained from a representative neutralization experiment, which contains three replicates. Data are represented as mean ± SD.
(F) Neutralization potency on authentic SARS-CoV-2 of the three selected mAbs. IC50 and IC80 were calculated by fitting a four-parameter logistic curve.
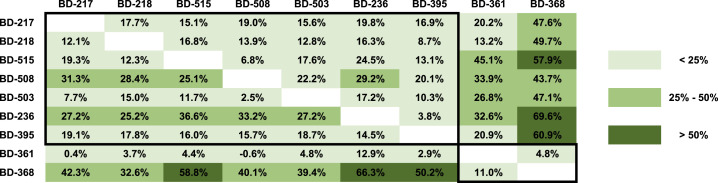
Surprisingly, we found that 7 out of the 12 mAbs exhibited a strong RBD-binding affinity and a potent neutralizing ability against pseudotyped SARS-CoV-2 (Figures 5B and 5C). The neutralization abilities of three representative mAbs, BD-503, BD-508, and BD-515, were also verified in the PRNT against authentic SARS-CoV-2 and showed high potencies (Figures 5E and 5F). Moreover, these mAbs were mostly encoded by VH3-66/JH6 or VH3-53/JH6 gene segments for the heavy chain and VLK1-9 or VLK1D-33/39 for the light chain (Figure 5C). Due to the similarity of amino acid sequences and VDJ combinations, we questioned whether these mAbs came from the same lineage. After RNA mutation analysis, we found that none of the mAbs was from the same lineage nor the same patients. Together, these observations suggest that stereotypic B cell receptors that bind to a particular SARS-CoV-2 epitope may exist in different individuals, similar to what has been observed in HIV, influenza, and hepatitis C (Gorny et al., 2009, Ekiert et al., 2009, Marasca et al., 2001). Indeed, epitope binning experiments using a double-antibodies sandwich ELISA has shown that BD-503, BD-508, and BD-515 likely share an overlapping epitope (Figure 6 ). However, more structural analysis is needed to obtain the exact epitopes to confirm whether they are largely conserved.
Figure 6.
Epitope Binning of the Potent Neutralizing mAbs Using Competitive ELISA
Competition tolerance was shown for each pair of neutralizing mAbs measured by double-antibody sandwich ELISA (RBD). The column indicates the primary antibody, and the row indicates the secondary antibody. Competition tolerance larger than 50% indicates a high possibility of no overlapping epitope.
Discussion
To improve the efficiency of discovering new neutralizing mAbs, we applied a systematic analysis of antibody-presenting B cells before performing in vitro expression. Large-scale, single-cell resolution data obtained from scRNA and scVDJ sequencing provided the chance to perform lineage analysis, which cannot easily be accomplished using hybridoma and single-cell RT-PCR-based techniques. The ability to calculate the enrichment frequency for each clonotype not only enables priority selection of natural clonally expanded B cell lineages but also reduces the false-positive rates of non-specific antigen-binding during the antigen-enrichment step (Figure 1A). The simultaneous measurement of mRNA expression at the single-cell level further enables cell typing of individual B cells, which is crucial for the removal of the exhausted and naive B cell subsets. However, our current shallow scRNA sequencing barely enables the accurate separation of exhausted memory B cells from non-exhausted memory B cells. Therefore, deeper sequencing of the scRNA libraries is needed to further evaluate the effectiveness of removing exhausted memory B cells to improve the identification of neutralizing mAbs.
Besides the clonally enriched B cell clonotypes, the vast majority of the sequenced B cells were not utilized in this study but remained as a valuable library of potential neutralizing mAbs. Nevertheless, due to the lack of enrichment, mAbs selection from the large pool of unenriched B cells continued to be a problem. We, therefore, utilized bioinformatic-based selection approaches, such as CDR3H structure prediction. mAbs that shared a highly similar CDR3H structure with SARS-CoV-neutralizing antibody m396 showed a surprisingly high percentage of high neutralization potency for SARS-CoV-2. The largely conserved combination of the VDJ gene segment and likely overlapping epitopes of those mAbs suggests the existence of stereotyped B cell receptors against SARS-CoV-2. Preliminary studies of mAbs selected based on VH3-66/JH6 or VH3-53/JH6 encoded heavy chains from different individuals indeed revealed a high proportion of SARS-CoV-2 neutralization mAbs with high potency (data not shown) and support the existence of stereotyped B cell receptors for SARS-CoV-2.
Although 72 S1/S2 (non-RBD) binding mAbs were identified, none of them exhibited pseudovirus neutralizing ability. This was an interesting result since non-RBD binding neutralizing mAbs were observed for MERS-CoV (Xu et al., 2019). More antibodies from the remaining pool of antigen-binding B cell clonotypes that target different regions of the antigens would be screened for neutralization ability, to further confirm the observation. Additionally, more strains of pseudoviruses could be utilized to examine potential risks of neutralization escape after virus mutation, especially at the RBD region (Wang et al., 2018). A possible solution to the virus mutation could be to combine two potent mAbs targeting two different epitopes. Epitope binning results (Figure 6) suggested that BD-368, which shares the same epitope with BD-368-2, exhibited non-overlapping epitopes with most of the potent neutralizing mAbs. An antibody cocktail combining BD-368-2 with another neutralizing mAb could greatly boost SARS-CoV-2 neutralization potency compared to any individual mAbs and could largely prevent mutation escape, since the possibility of mutations appearing on both epitopes is unlikely.
The continual dissemination of COVID-19 as a worldwide pandemic demands effective interventions against SARS-CoV-2 infections. The potent neutralizing antibodies we identified may provide an effective therapeutic and prophylactic solution, evidenced by the results on hACE2 transgenic mice. Clinical trials using BD-368-2 are underway. The application of high-throughput single-cell sequencing methodology in this study could also be expanded to infectious diseases other than COVID-19 and serve as a technical reserve for rapid neutralizing mAbs discovery during future pandemics.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human IgG (H+L)/HRP | JACKSON | Cat #109-035-088; RRID: AB_2337584 |
| Anti-His/HRP | ThermoFisher | Cat # MA1-21315; RRID: AB_557403 |
| Bacterial and Virus Strains | ||
| SARS-COV-2 VSV pseudotyped virus | NIFDC (Nie et al., 2020) | N/A |
| Authentic SARS-CoV-2 virus (2019-nCoVBetaCoV/Wuhan/AMMS01/2020) | Qin’s lab | N/A |
| Biological Samples | ||
| PBMCs from COVID-19 convalescent patients | Youan hospital | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ficoll-Paque Plus | GE Healthcare | Cat #17144002 |
| Fetal bovine serum (FBS) | ThermoFisher | Cat #10099 |
| RPMI 1640 medium | ThermoFisher | Cat #12633012 |
| SARS-COV-2 spike protein, His-tag | Sino Biology | Cat # 40589-V08B1 |
| SARS-COV-2 RBD protein, His-tag | Sino Biology | Cat # 40592-V08B |
| Biotinylated SARS-COV-2 spike protein | Sino Biology | Cat # 40589-V08B-B |
| Biotinylated SARS-COV-2 RBD protein | Sino Biology | Cat # 40592-V08B-B |
| 0.4% (w/v) Trypan blue stain | ThermoFisher | Cat # 15250061 |
| Streptavidin M-280 Dynabeads | ThermoFisher | Cat # 60210 |
| 1x PBS, pH 7.4 | ThermoFisher | Cat # 10010031 |
| EDTA 0.5M, pH8.0, RNase-free | ThermoFisher | Cat # AM9260G |
| TMB Chromogen Solution (for ELISA) | ThermoFisher | Cat # 002023 |
| NaCl (5 M), RNase-free | ThermoFisher | Cat # AM9760G |
| Surfactant P20 | GE Healthcare | Cat #BR100054 |
| ACE2 Protein, Human, Recombinant (His Tag) | Sino Biology | Cat # 10108-H08B |
| HG1K IgG1 antibody against H7N9 | Sino Biology | Cat #HG1K |
| DMEM | ThermoFisher | Cat #11965092 |
| HEPES (1 M) | ThermoFisher | Cat #15630080 |
| LMP agarose | Promega | Cat #V3841 |
| Crystal violet | SigmaAldrich | Cat # C0775 |
| Formaldehyde solution | SigmaAldrich | Cat # F8775 |
| Polyethylenimine | Polysciences | Cat # 23966-2 |
| Papain | Sangon Biotech | Cat # A501612-0025 |
| Iodoacetamide | SigmaAldrich | Cat # I1149 |
| Critical Commercial Reagents | ||
| EasySep Human B Cell Enrichment Kit | STEMCELL | Cat #19054 |
| EasySep Human Memory B Cell Isolation Kit | STEMCELL | Cat #17864 |
| Chromium Single Cell 5′ Library Construction Kit | 10X Genomics | Cat #1000020 |
| Chromium i7 Multiplex Kit | 10X Genomics | Cat #120262 |
| Chromium Single Cell V(D)J Enrichment Kit, Human B Cell | 10X Genomics | Cat #1000016 |
| Chromium Single Cell 5′ Library & Gel Bead Kit | 10X Genomics | Cat #1000014 |
| Chromium Single Cell A Chip Kit | 10X Genomics | Cat #1000009 |
| SA sensor chip | GE Healthcare | Cat #29104992 |
| Spike Protein ELISA kit | Sino Biology | Cat #KIT40591 |
| Ni Sepharose 6 Fast Flow | GE healthcare | Cat # 17531803 |
| Superose 6 Increase 10/300 GL | GE healthcare | Cat # 29091596 |
| RNeasy Mini Kit | QIAGEN | Cat # 74104 |
| PrimerScript RT Reagent Kit | TaKaRa | Cat # RR037B |
| PowerUp SYBR Green Master Mix Kit | Applied Biosystems | Cat # A25741 |
| Deposited Data | ||
| Cryo-EM structure of SARS-COV-2 spike and BD23-Fab | This paper | PDB: 7BYR |
| Electron microscopy density map of SARS-COV-2 spike and BD23-Fab | This paper | EMDB: EMD-30247 |
| Antibody amino acid sequences | This paper | European Genome-Phenome Archive: EGAS00001004412 |
| Experimental Models: Cell Lines | ||
| HEK293F cells | ThermoFisher | Cat # 11625019 |
| Huh7 cells | NIFDC (Nie et al., 2020) | N/A |
| Vero E6 cells | Qin’s lab | N/A |
| Experimental Models: Cell Lines | ||
| hACE2 transgenic mice | Qin’s lab | N/A |
| Oligonucleotides | ||
| 5′-TCGTTTCGGAAGAGACAGGT-3′ | Qin’s lab | N/A |
| 5′-GCGCAGTAAGGATGGCTAGT-3′ | Qin’s lab | N/A |
| Recombinant DNA | ||
| SARS-CoV-2 S gene, residues 1-1208, pcDNA | Sino Biology | N/A |
| SARS-CoV-2 S gene, residues 1-1208, 2P and furin cleavage mutation, T4 fibritin trimerization motif, 8xHisTag, pcDNA | This paper | N/A |
| Software and Algorithms | ||
| cutadapt (2.9) | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/installation.html |
| CellRanger (3.1.0) | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| SingleR (1.0.5) | Aran et al., 2019 | https://bioconductor.org/packages/release/bioc/html/SingleR.html |
| Seurat (3.1.3) | Satija et al., 2015 | https://satijalab.org/seurat/install.html |
| IgBlast-1.15.0 | National Center for Biotechnology Information (NCBI) | ftp://ftp.ncbi.nih.gov/blast/executables/igblast/release/1.15.0/ |
| igraph (1.2.5) | Csardi and Nepusz, 2006 | https://cran.r-project.org/web/packages/igraph/index.html |
| SAAB+ | Kovaltsuk et al., 2020 | https://github.com/oxpig/saab_plus |
| SerialEM software | Mastronarde, 2005 | https://bio3d.colorado.edu/SerialEM |
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/ucsf-motioncor2 |
| Gctf program (v1.06) | Zhang, 2016 | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf |
| RELION (v3.07) | Zivanov et al., 2018 | https://www2.mrc-lmb.cam.ac.uk/relion |
| ResMap | Kucukelbir et al., 2014 | http://resmap.sourceforge.net |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot |
| Pymol | Schrödinger | https://pymol.org/2/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, X. Sunney Xie (sunneyxie@biopic.pku.edu.cn)
Materials Availability
There are restrictions on the availability of antibodies due to limited stock and continued consumption. We are glad to share remaining antibodies with reasonable compensation for processing and shipping upon completion of a Material/Data Transfer Agreement for non- commercial usage.
Data and Code Availability
The accession number for the human antibody sequences reported in this paper is EGA: S00001004412. Material/Data Transfer Agreements, which allow the use of the antibody sequences for non-commercial purposes but not their disclosure to third parties, are needed to obtain the sequences by contacting the Data Access Committee.
Experimental Model and Subject Details
Convalescent patients
A total of 64 confirmed COVID-19 patients discharged from Beijing Youan Hospital, China, from February 01 to February 28, 2020, were enrolled in this study (Table S1). Patients 1-12 were enrolled in the prelinamery study without antigen-enrichment. Patients 13-64, as well as 8 patents from ID 1-12, enrolled in the study corresponding to sequence batch 1-6 (Table S1). Discharged COVID-19 patients were isolated and observed continually for 28 days. Follow-ups and SARS-CoV-2 RNA detection were performed weekly. The throat swabs from the upper respiratory tract, and whole blood was collected from patients at various time-points after hospitalization and during followed-up. Sample collection, processing, and laboratory testing were performed as recommended by China CDC and complied with WHO guidance. All COVID-19 patients were confirmed infected according to positive respiratory RT-PCR tests. The discharge criteria were listed as follows. First, body temperature returns to normal for more than three days. Second, absence of respiratory tract symptoms. Third, two consecutively SARS-CoV-2 RT-PCR assays of sputum with a one-day sampling interval turn negative. None of the study participants was co-infected with HIV, hepatitis B virus/hepatitis C virus, or influenza viruses. All participants did not have a comorbid condition, tuberculosis, autoimmune diseases, or related drug usage. This study and all the relevant experiments were approved by the Beijing Youan Hospital Research Ethics Committee, and written informed consent was obtained from each participant in accordance with the Declaration of Helsinki. All participants provided written informed consent for the collection of information, and their clinical samples were stored and used for research. Data generated from the research were agreed to be published.
PBMCs from blood
All human blood samples were collected for laboratory assessments according to the doctor’s instruction from Beijing Youan Hospital, China. Peripheral blood mononuclear cells (PBMCs) were isolated immediately from fresh blood. Primary human B lymphocytes were obtained by purification of human blood CD19+ B cells or memory CD19+ CD27+ B cells, by immunomagnetic bead isolation (Catalog #19054 and #17864, STEMCELL), after Ficoll-Hypaque sedimentation according to standard density-gradient centrifugation methods (GE Healthcare). For each sample, the cell viability exceeded 85%. Several patients’ PBMCs were stored frozen and thawed before use. Cryopreserved PBMCs were thawed in RPMI 1640 medium supplemented with 20% fetal bovine serum.
Recombinant RBD and full-length Spike protein from SARS-CoV-2
Spike/RBD recombinant proteins were purchased from Sino Biological Inc. with > 95% purity. Full-length Spike/RBD protein was expressed in the Baculovirus-Insect cell system using the DNA sequence encoding the SARS-CoV-2 spike protein (S1+S2 ECD) or RBD protein. The protein is expressed with a polyhistidine tag at the C terminus and purified in sterile 20 mM Tris, 300 mM NaCl, 10% glycerol, pH 8.0.
SARS-CoV-2 pseudovirus
The SARS-CoV-2 pseudovirus is a gift from the Division of HIV/AIDS and Sex-transmitted Virus Vaccines, National Institutes for Food and Drug Control (NIFDC). The pseudovirus was constructed as previously described using the spike genes from strain Wuhan-Hu-1 (GenBank: MN908947)(Nie et al., 2020). Briefly, the SARS-CoV-2 spike gene was codon-optimized and cloned into a eukaryotic expression plasmid. 293T cells were transfected by the plasmid and later infected with a VSV pseudotyped virus (G∗ΔG-VSV), which substituted the VSV-G gene with luciferase expression cassettes. The culture supernatants were harvested and filtered 24 h post-infection. The SARS-CoV-2 pseudovirus could not be neutralized by VSV-G antibodies, and no G∗ΔG-VSV was mixed with the SARS-CoV-2 pseudovirus stock (Nie et al., 2020).
Authentic SARS-CoV-2 virus
The authentic SARS-CoV-2 virus (2019-nCoV BetaCoV/Wuhan/AMMS01/2020) was isolated from a COVID-19 patient from Beijing. The RNA sequence of the isolated virus was verified by Sanger sequencing. All experiments associated with the authentic virus were conducted in Biosafety Level 3 laboratory with standard operating procedures.
hACE2 transgenic mice
Pathogen-free, male, and female human ACE2 transgenic mice (6 - 8 wo) were obtained from the Institute of Laboratory Animal Science, Peking Union Medical College, China. The hACE2 transgenic mice have been demonstrated to be a feasible model for the SARS-CoV-2 infection study (Bao et al., 2020). hACE2 mice were generated as previously described (Yang et al., 2007), by microinjecting DNA fragments of mouse ACE2 promoter driving hACE2 coding sequence into the pronuclei of ICR mice’s fertilized ova. The zygotes were then transplanted into the oviducts of pseudopregnant mice. These mice were bred to produce offspring for the experiment.
Method Details
MACS enrichment of B cells from PBMC
B cells were isolated from fresh or previously frozen PBMCs by immunomagnetic negative selection according to the manufacturer’s protocol (EasySep Human B Cell Enrichment Kit, STEMCELL). Non-B cells are labeled with magnetic beads and separated using an EasySep magnet. Purified B cells were eluted and washed in PBS containing 2% (v/v) fetal bovine serum (FBS) and 1 mM EDTA. Purified B cells were counted by using 0.4% (w/v) trypan blue stain and Countess Automated Cell Counter according to the manufacturer’s protocol (Thermofisher).
CD27+ memory B cell enrichment
CD27+ memory B cells were isolated from purified B cells by immunomagnetic positive selection according to the manufacturer’s protocol (EasySep Human Memory B Cell Isolation Kit, STEMCELL). Briefly, CD27+ B cells are labeled with magnetic beads combined with CD27 antibodies and separated using an EasySep magnet. Purified CD27+ B cells were eluted and washed in PBS containing 2% (v/v) fetal bovine serum (FBS) and 1 mM EDTA. CD27+ B cells were counted by using 0.4% (w/v) trypan blue stain and Countess Automated Cell Counter according to the manufacturer’s protocol.
Antigen-binding B cells enrichment
Biotinylated Spike/RBD recombinant proteins were purchased from Sino Biological Inc. Fresh antigen/streptavidin M-280 Dynabeads (Thermofisher) complex was made every time before B cell enrichment. Briefly, 100 μL of M-280 beads containing 6.5 × 107 beads were vortexed for 30 s and brought to room temperature before use. Beads were washed with 1 mL 1x PBS twice on a magnetic rack, and eluted in 100 μL 1x PBS. Beads (100 μL) were mixed with 20 μg of biotinylated spike/RBD protein and incubated at room temperature for 30 mins. After incubation, the complex was washed three times using 500 μL 1x PBS on a magnetic rack. The washed complex was eluted in 100 μL 1x PBS and stored on ice until use.
Before antigen-enrichment, the complex was equilibrated at room temperature. The amount of beads complex used was calculated based on a 1:1 ratio with the number of purified B cells. The spike/RBD magnetic beads complex was added directly into the B cell mixture, mixed and incubated at 4°C on a thermomixer for 30 mins. After the incubation, the mixture was put on a magnetic rack, and the supernatant was removed. The beads were washed times by mixing off the magnet four times in total. The final antigen-enrichment B cells were eluted in 1x PBS containing 2% (v/v) fetal bovine serum (FBS) and 1 mM EDTA. Enriched B cells were counted by using 0.4% (w/v) trypan blue stain and Countess Automated Cell Counter according to the manufacturer’s protocol.
Single-cell 5′ mRNA and VDJ sequencing
After CD27+ memory B cell enrichment or antigen-specific enrichment, all B cells were loaded on a 10X Chromium A Chip. If the total cell number exceeded 20,000, two individual runs would be performed. Since SARS-CoV-2 may still appear in convalescent patients’ blood (An et al., 2020, Zhang et al., 2020), we performed all the experimental processes before library preparation in a P3-level laboratory. Single-cell lysis and RNA first-strand synthesis were performed using 10X Chromium Single Cell 5′ Library & Gel Bead Kit according to the manufacturer’s protocol. After RNA first-strand synthesis, all viruses should be denatured since all samples were heated to 85°C for 5 mins. RNA samples were wiped with 80% EtOH for three times and sealed in a plastic box for sample transportation.
The following RNA and VDJ library preparation is performed according to the manufacturer’s protocol (Chromium Single Cell V(D)J Reagent Kits, 10X Genomics) in a P2 laboratory. All libraries were quantified by using Qubit 3.0 (ThermoFisher), Fragment Analyzer, and qPCR. Sequencing was performed on a Hiseq 2500 platform running Rapid SBS Kit V2 2x100bp kit (Illumina), with a 26x91 pair-end reading mode. Sequencing results were obtained within two days. Average sequencing depth aimed for the mRNA library is 10,000 read pairs per cell and 5,000 read pairs per cell for the VDJ libraries (Table S3).
In Vitro expression of mAbs
The selected paired heavy- and light-chain’s cDNA was codon-optimized and cloned into expression vectors containing the IgG1 constant regions of human. IgG mAbs were expressed by transfecting HEK293 cells with equal amounts of heavy- and light-chain plasmids. The mAbs were resuspended into PBS and analyzed by SDS-PAGE.
The numbering of mAbs is based on the order of DNA synthesis submission. BD 1-175 were mAbs produced from the experiments without using antigen enrichment (Table S2). BD 176 – 425 were mAbs produced from the Batch 1-6 with antigen enrichment, and were all selected from clonotypes that have an enrichment frequency larger than 1 (Table S2). BD 492 – 515 were mAbs selected based on CDR3H structural similarity to SARS-CoV neutralizing antibodies (Table S2). BD 426 – 491, which were not shown, were attempts trying to substitute rare DNA mutations in the FR region of the verified neutralizing mAbs, aiming for higher neutralization potency. None of the mAbs showed improvements in terms of neutralization potency, and the results were not shown. Also, the antibodies in each group may have discontinuous numbering. The excluded mAbs were the antibodies selected from clonotypes that consist of only IgM- or IgG3-presenting B cells. None of mAbs showed neutralizing ability and thus were excluded from the results.
ELISA quantification
ELISA plates were coated with SARS-CoV-2 RBD or S protein at 0.01 μg/mL and 1 μg/mL in PBS at 4°C overnight. After standard washing and blocking, 100 μL 1 μg/mL antibodies was added to each well. After a 2 h incubation at room temperature, plates were washed and incubated with 0.08 μg/mL goat anti-human IgG (H+L)/HRP (JACKSON) for 1 h incubation at room temperature. Chromogen solution was used as the substrate, and absorbance at 450nm was measured by a microplate reader. A mAb is defined as ELISA-positive when the OD450 is saturated using 1 μg/mL RBD/S protein.
mAbs binding affinity measurement
The dissociation coefficient is measured by using surface plasmon resonance (SPR). Biotinylated RBD/S (Sino Biological Inc.) was immobilized to a SA sensor chip (GE Healthcare) at a level of ∼100 response units (RUs) using a Biacore T200 (GE Healthcare). Running buffer is composed of PBS pH 7.4 and 0.005% (v/v) P20. Serial dilutions of purified antibody were injected, ranging in concentration from 50 to 0.78 nM. The resulting data were fit to a 1:1 binding model using Biacore Evaluation Software.
ACE2 competition assay
Biotinylated RBD (0.3 μg/mL, Sino Biological Inc.) was immobilized to an ELISA plate. After standard washing, 0.05 μg/mL His-tagged hACE2 protein was added to the wells, and then the diluted mAbs were added and mixed immediately. After 2 h of incubation at room temperature, plates were washed, and 0.08 μg/mL anti-His/HRP was added. After 1 h of incubation at room temperature, the chromogen solution was used as the substrate, and absorbance at 450 nm was measured by a microplate reader. The ACE2/RBD-binding inhibition rate was calculated by comparing to the mAbs negative control well.
Protein expression and purification for cryo-EM
The SARS-CoV-2 S gene was synthesized by Sino Biological Inc. The DNA fragment encoding the ectodomain of S (residues 1-1208) was cloned into a pcDNA vector. Two stabilizing proline substitutions at residues 986-987 and a “GSAS” substitution at the furin cleavage site (residues 682–685) were introduced as previously described to improve protein expression (Wrapp et al., 2020). A C-terminal T4 fibritin trimerization motif and an 8 × His tag were also engineered to stabilize trimer formation and facilitate protein purification.
HEK293F cells were cultured in SMM 293T-I medium (Sino Biological Inc.) at 37°C, with 5% CO2 and 55% humidity. The plasmid was transfected into the cells using polyethylenimine (Polysciences) when the cell density reached 1 × 106 cells per mL. Four days after transfection, the conditioned media were collected by centrifugation at 500 × g, concentrated using a Hydrosart Ultrafilter (Sartorius), and exchanged into the binding buffer (25 mM Tris-HCl, pH 8.0, 200 mM NaCl). The S protein was subsequently isolated using the Ni-NTA resin, and purified using a Superose 6 10/300 gel filtration column (GE Life Sciences). The final buffer used for the gel filtration step is 20 mM HEPES, pH 7.2, 100 mM NaCl.
To obtain BD23-Fab, BD-23 (2 mg/mL) was digested with papain (0.1 mg/mL) at 37°C for 2 h in 50 mM phosphate buffer saline, pH 7.0, 2 mM EDTA, and 5.5 mM cysteine. The reaction was terminated using 30 mM iodoacetamide at room temperature for 20 minutes. The Fc region was removed by protein A chromatography, and BD23-Fab was further purified using a Superdex 200 Increase 10/300 gel filtration column (GE Life Sciences) and eluted using the final buffer. Ab23-Fab was incubated with the purified S protein in a 1.5:1 molar ratio overnight on ice. After that, the complex was purified using the Superose 6 10/300 column and eluted with the final buffer.
Cryo-EM data collection and processing
Holy-carbon gold grids (Quantifoil, R1.2/1.3) were glow-discharged with a Solarus 950 plasma cleaner (Gatan) for 30 s. Four-microliter aliquots of the S/BD23-Fab complex (0.6 mg/mL) were applied to the grids, blotted with filter paper at 4°C and 100% humidity, and plunged into the liquid ethane using a Vitrobot Mark IV (FEI). The grids were first screened on a 200 kV Talos Arctica microscope equipped with Ceta camera (FEI). Data were collected using a Titan Krios electron microscope (FEI) operated at 300 kV. Movies were recorded with a K2 Summit direct electron detector (Gatan) in the super-resolution mode using the SerialEM software (Mastronarde, 2005). Statistics for data collection are summarized in (Table S4).
The workflow of data processing was illustrated in Figure S7. Raw movie frames were aligned and averaged into motion-corrected summed images using MotionCor2 (Zheng et al., 2017). The Gctf program (v1.06) was used to estimate the contrast transfer function (CTF) parameters of the motion-corrected images (Zhang, 2016). Relion (v3.07) was used for all the following data processing (Zivanov et al., 2018). The S trimer (PDB ID: 6VSB) was used as a reference for the 3D classifications. The local resolution map was analyzed using ResMap (Kucukelbir et al., 2014) and displayed using UCSF Chimera (Pettersen et al., 2004).
A structural model of BD23-Fab was generated using the Phyre2 server (Kelley et al., 2015). This BD23-Fab model and the S trimer structure (PDB ID: 6VSB) were docked into the cryo-EM density using UCSF Chimera. One round of rigid-body refinement was performed using the real-space refinement in Phenix (Liebschner et al., 2019). Further structure refinement of BD23-Fab was not performed due to limited resolution in this region. Nevertheless, it is evident that BD23-Fab engages an epitope on RBD that predominantly overlaps with the ACE2-binding site.
Pseudovirus neutralization assay
The pseudovirus neutralization assays were performed using Huh-7 cell lines. Huh-7 are human hepatocellular carcinoma cells that express both ACE2 and TMPRSS2 (Matsuyama et al., 2018). Various concentrations of mAbs (3-fold serial dilution using DMEM, 50 μL aliquots) were mixed with the same volume of SARS-CoV-2 pseudovirus with a TCID50 of 1.3 × 104 in a 96 well-plate. The mixture was incubated for 1 h at 37°C, supplied with 5% CO2. Negative control wells were supplied with 100 μL DMEM (1% (v/v) antibiotics, 25 nM HEPES, 10% (v/v) FBS). Positive control wells were supplied with 100 μL DMEM. Pre-mixed Huh-7 cells (100 μL, 2 × 105 in DMEM) were added to all wells, and the 96-well plates were incubated for 24 h at 37°C supplied with 5% CO2. After the incubation, 150 μL of supernatants were removed, and 100 μL D-luciferin reagent (Invitrogen) was added to each well and incubated for 2 mins. After the incubation, every well is mixed 10 times by pipetting, and 150 μL of the mixture was used to measure luciferase activity using a microplate spectrophotometer (Perkinelmer EnSight). The inhibition rate is calculated by comparing the OD value to the negative and positive control wells. IC50 and IC80 were determined by a four-parameter logistic regression using GraphPad Prism 8.0 (GraphPad Software Inc.). The raw data of a representative neutralization assay on BD-368-2 is described in Table S5.
Authentic SARS-CoV-2 PRNT
Plaque reduction neutralization test (PRNT) was performed using a clinical isolate of SARS-CoV-2 (BetaCoV/Wuhan/AMMS01/2020) (Roehrig et al., 2008). Briefly, 5-fold serial dilutions of mAbs were added to the same volume of approximately 100 PFU of SARS-CoV-2 and incubated for 1 h at 37°C. The mixture was added to a monolayer of Vero cells in a 24-well plate and incubated for 1 h at 37°C. The mixture was removed, 0.5 mL of 1.0% (w/v) LMP agarose (Promega) in 2 × DMEM supplied with 4% (v/v) FBS was added onto the infected cells. After further incubation at 37°C supplied with 5% CO2 for 2 days, the wells were stained with 1% (w/v) crystal violet dissolved in 4% (v/v) formaldehyde to visualize the plaques. IC50 values were determined using four-parameter logistic regression analysis by using GraphPad Prism 8.0 (GraphPad Software Inc.). All experiments were performed in a Biosafety Level 3 facility.
Authentic SARS-CoV-2 neutralization CPE assay
A neutralization assay of authentic SARS-CoV-2 was performed using a cytopathic effect (CPE) assay. Briefly, various concentrations (3-fold serial dilution using DMEM) of mAbs were mixed with the same volume of 100 PFU of authentic SARS-CoV-2 and incubated at 37°C for 1 h. The mixture was added to a monolayer of Vero-E6 cells (5x103 cell per well) in a 96-well plate and incubated for 1 h at 37°C. The supernatant was removed, and 200 μL of DMEM supplied with 2% (v/v) FBS was added onto the infected cells. After incubation at 37°C supplied with 5% CO2 for 5 days, all wells examine for the CPE effect. All experiments were performed in a Biosafety Level 3 facility.
hACE2 in vivo SARS-CoV-2 neutralization assay
A total of 9 hACE2 transgenic mice were used in this study. The mice were equally split into three groups for prophylactic and therapeutic study, as well as the negative control. For the prophylactic group, BD-368-2 (20 mg/kg) was intraperitoneally injected one day before infection. For the therapeutic group, BD-368-2 (20 mg/kg) was intraperitoneally injected 2 h after infection. For the negative control group, HG1K (20 mg/kg, Sino Biology), which is an anti-influenza antibody, was intraperitoneally injected 2 h after infection. The hACE2 mice were inoculated intranasally with the SARS-CoV-2 stock virus with a dosage of 105 TCID50. Body weights were recorded daily for all infected mice for 5 continuous days. The mice were sacrificed at 5 dpi, and the lungs were collected for viral load analysis.
Viral load detection was performed by qRT-PCR on RNA extracted from lung homogenate supernatants, as described previously (Bao et al., 2020). Briefly, lung homogenates were prepared by homogenizing perfused whole lung using an electric homogenizer. The supernatant was collected, and total RNA was extracted. A quantitative real-time reverse transcription-PCR (qRT-PCR) reactions were performed using the previously described primers and protocol. qRT-PCR was performed on each mouse with two replicates. All experiments were performed in a Biosafety Level 3 facility.
Epitope Binning
Double-antibody sandwich ELISA was used to categorize the epitope of the identified potent neutralizing mAbs. Briefly, SARS-CoV-2 RBD-His Recombinant Protein (Biotinylated, Sino Biological Inc.) was diluted with PBST buffer to 3 μg/mL, and was captured by streptavidin probes. The neutralizing mAbs were diluted with PBST buffer to 133.3 nM each, and the primary antibody was first incubated with the RBD proteins. After 1 h incubation, the response value was measured by OCTET RED. After the measurement, the secondary antibody was added, and the response value was again measured after 30 mins incubation. Competition tolerance was calculated by the percentage increase of response after the secondary antibody was added, as shown in Figure 6. The column indicates the primary antibody, and the row indicates secondary antibodies. Competition tolerance larger than 50% indicates a high possibility of no overlapping epitope.
Quantification and Statistical Analysis
B cell scRNA-seq data analysis
The raw fastq files were processed using the 10X Genomics CellRanger (3.1.0) pipeline. Reads produced from the 5′ gene expression profiling were aligned to the GRCh38 genome, and feature-barcode matrices were generated using “cellranger count” with default parameters. To filter out low-quality genes and cells, genes expressed in over 10 cells were retained, and cells were filtered based on the number of genes and the mitochondrial gene percentage to remove possible doublets. Cell types were identified using SingleR (Aran et al., 2019) against a human immune reference dataset (Monaco et al., 2019). Clusters of cells were visualized using T-distributed stochastic neighbor embedding (t-SNE) in Seurat (Satija et al., 2015).
B cell scVDJ data analysis
The raw fastq files were processed using 10X Genomics CellRanger (3.1.0) pipeline. 3′ ends of bases with quality scores lower than 30 were trimmed using cutadapt (Martin, 2011). Contig assembly, annotation, and clonotype analysis were performed using “cellranger vdj” with the Cell Ranger V(D)J compatible reference:
(refdata-cellranger-vdj-GRCh38-alts-ensembl-3.1.0).
Structural annotation of BCR repertoires
Structures of the three CDR regions in heavy- and light chains were annotated using the SAAB+ pipeline (Kovaltsuk et al., 2020) with default parameters. SAAB+ utilizes SCALOP (Wong et al., 2019) to identify the canonical classes of CDR1 and CDR2 sequences. Structural annotations of CDR3 sequences were made by FREAD (Choi and Deane, 2010) integrated in the SAAB+ pipeline.
Somatic hypermutation rate determination
The mapping of V(D)J sequencing reads was performed by IgBlast-1.15.0 (Ye et al., 2013) against the germline reference (202007-1 release) downloaded from the international ImMunoGeneTics information system (IMGT) (Giudicelli et al., 2006). The hypermutation rate was calculated as , in which the gaps were calculated as the number of base pairs in the insertion or deletion regions.
Ideal antibody selection
All B cells belonging to enriched clonotypes that contain IgG1 were extracted and their corresponding cell type, somatic hypermutation (SHM) rate, and Ig class were recorded. The ideal antibody selection process strictly followed the criteria listed in Figure 1D. The lineage within each clonotype was determined by the DNA mutation pattern as well as the Ig class. The lineage relationship would not interfere with ideal antibody selection, and only helps when selecting the mAbs sequences from a particular clonotype. When the B cells in the same clonotype did not share identical VDJ sequences, the DNA sequence of the most mature B cell in the lineage was usually selected for in vitro expression. The lineage plot was drawn by igraph (Csardi and Nepusz, 2006). The position of the cells on the graph has no physical meaning and was arbitrarily adjusted to group all ideal clonotypes on one side.
Acknowledgments
This work was made possible by the support from the Beijing Advanced Innovation Center for Genomics (ICG) at Peking University and the Fundamental Research Funds for the Central Universities (A20ZX00846). We thank the Beijing Municipal Commission of Science and Technology for the coordination of collaboration between institutes and financial support (Z201100005420018). We are grateful to Prof. Zhijian Chen of UT Southwestern for the stimulating discussions. We thank Prof. Yanyi Huang, Prof. Zemin Zhang, and Prof. Fuchou Tang of Peking University for their help on experimental equipment and supplies. We thank Prof. Zhanguo Li of Renmin Hospital and Prof. Chengqi Yi of Peking University for the loan of critical equipment. We thank Prof. Yingchun Wang and Dr. Weijin Huang from the Division of HIV/AIDS and Sex-transmitted Virus Vaccines, National Institutes for Food and Drug Control (NIFDC) for providing the SARS-CoV-2 pseudovirus strain and the Huh7 cell line. We thank Dr. Jie Zhang, Dr. Yutao Lin, and Dr. Wei Ren from Sino Biological Inc. for the technical assistance on mAbs expression and characterization. We thank the National Center for Protein Sciences at Peking University in Beijing, China, for the assistance with SPR measurement. We thank the National Mega projects of China for Major Infectious Diseases (2017ZX10304402), National Key Research and Development Project of China (2016YFD0500304), and CAMS initiative for Innovative Medicine of China (2016-12M-2-006) for the support on the animal model study. We thank Dr. Chuck Bailey and Prof. John Rasko from the University of Sydney and Royal Prince Alfred Hospital and Dr. Patricia Purcell from the Massachusetts Institute of Technology for critical reading and editing of the manuscript.
Author Contributions
Y.C. and X.S.X. conceptualized the project and designed and coordinated the experiments. X.S.X. obtained the funding for the project. R.J. and Y.F. organized the collection of convalescent patients’ blood samples. B.S. and X.G. prepared B cells and performed 10X experiments. Y.W. and L.Q. arranged sample transportation. Y.C. and C.G. performed scRNA and scVDJ library preparation. X.Z. and Y.C. optimized the antigen pull-down experiment. W.S. and X.C. analyzed the sequencing data. Y.C. selected the antibody sequences for synthesis. Y.Z. performed the pseudovirus neutralization assays. R.S.H. performed SPR assays. Z.D. and C.Q. coordinated the experiments on the authentic virus, which was developed by C.Q. Y.D., X.L., and Y.T. performed the PRNT on the authentic virus in the P3 laboratory. C.Q. designed and J.L. coordinated the animal experiment. L.B., H.Z., W.D., and Y.X. performed the animal experiment in the P3 laboratory. Q.L. performed the CPE assay in the P3 laboratory. D.S. and J.X. led the cryo-EM structure experiment. Q.Z., G.W., X.D., and N.G. collected and analyzed the structure data. Y.C. and X.S.X. wrote the manuscript with the help of all authors. L.S. arranged the figures.
Declaration of Interests
X.S.X, Y.C, W.S, and X.Z. are co-inventors on patent applications describing the neutralizing mAbs. X.Z. is the CEO of Singlomics (Beijing DanXu Pharmaceuticals, Co. Ltd.). The other authors declare no competing interests.
Published: May 18, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.05.025.
Supplemental Information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: A Comprehensive Python-based System for Macromolecular Structure Solution. Acta Crystallogr D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J., Liao X., Xiao T., Qian S., Yuan J., Ye H., Qi F., Shen C., Liu Y., Wang L. Clinical characteristics of the recovered COVID-19 patients with re-detectable positive RNA test. MedRxiv. 2020 doi: 10.1101/2020.03.26.20044222. Published online March 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. Published online May 7, 2020. [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Swat W., Alt F.W. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Burton D.R., Walker L.M., Phogat S.K., Chan-Hui P.Y., Wagner D., Phung P., Goss J.L., Wrin T., Simek M.D., Fling S. Broad and potent neutralizing antibodies from an african donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E., Cyranoski D., Mallapaty S., Stoye E., Tollefson J. The coronavirus pandemic in five powerful charts. Nature. 2020;579:482–483. doi: 10.1038/d41586-020-00758-2. [DOI] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M., Schoofs T., Gruell H., Settler A., Karagounis T., Kreider E.F., Murrell B., Pfeifer N., Nogueira L., Oliveira T.Y. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 2017;23:185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Deane C.M. FREAD revisited: Accurate loop structure prediction using a database search algorithm. Proteins. 2010;78:1431–1440. doi: 10.1002/prot.22658. [DOI] [PubMed] [Google Scholar]
- Corti D., Zhao J., Pedotti M., Simonelli L., Agnihothram S., Fett C., Fernandez-Rodriguez B., Foglierini M., Agatic G., Vanzetta F. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. USA. 2015;112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Misasi J., Mulangu S., Stanley D.A., Kanekiyo M., Wollen S., Ploquin A., Doria-Rose N.A., Staupe R.P., Bailey M. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- Croote D., Darmanis S., Nadeau K.C., Quake S.R. High-affinity allergen-specific human antibodies cloned from single IgE B cell transcriptomes. Science. 2018;362:1306–1309. doi: 10.1126/science.aau2599. [DOI] [PubMed] [Google Scholar]
- Csardi G., Nepusz T. 2006. igraph - The network analysis package.https://igraph.org/ [Google Scholar]
- DeKosky B.J., Lungu O.I., Park D., Johnson E.L., Charab W., Chrysostomou C., Kuroda D., Ellington A.D., Ippolito G.C., Gray J.J., Georgiou G. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. USA. 2016;113:E2636–E2645. doi: 10.1073/pnas.1525510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert D.C., Bhabha G., Elsliger M.A., Friesen R.H.E., Jongeneelen M., Throsby M., Goudsmit J., Wilson I.A. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Debs B., Utharala R., Balyasnikova I.V., Griffiths A.D., Merten C.A. Functional single-cell hybridoma screening using droplet-based microfluidics. Proc. Natl. Acad. Sci. USA. 2012;109:11570–11575. doi: 10.1073/pnas.1204514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V., Duroux P., Ginestoux C., Folch G., Jabado-Michaloud J., Chaume D., Lefranc M.-P. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L.D., Chen Y.J.J., Wu J., Chaudhuri S., Hsiao Y.C., Schneider K., Hoi K.H., Lin Z., Guerrero S., Jaiswal B.S. Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun. Biol. 2019;2:304. doi: 10.1038/s42003-019-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny M.K., Wang X.H., Williams C., Volsky B., Revesz K., Witover B., Burda S., Urbanski M., Nyambi P., Krachmarov C. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol. Immunol. 2009;46:917–926. doi: 10.1016/j.molimm.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horns F., Dekker C.L., Quake S.R. Memory B Cell Activation, Broad Anti-influenza Antibodies, and Bystander Activation Revealed by Single-Cell Transcriptomics. Cell Rep. 2020;30:905–913. doi: 10.1016/j.celrep.2019.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.C., Lin Y., Santelli E., Sui J., Jaroszewski L., Stec B., Farzan M., Marasco W.A., Liddington R.C. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J. Biol. Chem. 2006;281:34610–34616. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani V., Guy A.J., Andrew D., Beeson J.G., Ramsland P.A., Richards J.S. Molecular properties of human IgG subclasses and their implications for designing therapeutic monoclonal antibodies against infectious diseases. Mol. Immunol. 2015;67(2 Pt A):171–182. doi: 10.1016/j.molimm.2015.03.255. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaltsuk A., Krawczyk K., Galson J.D., Kelly D.F., Deane C.M., Trück J. How B-cell receptor repertoire sequencing can be enriched with structural antibody data. Front. Immunol. 2017;8:1753. doi: 10.3389/fimmu.2017.01753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaltsuk A., Raybould M.I.J., Wong W.K., Marks C., Kelm S., Snowden J., Trück J., Deane C.M. Structural diversity of B-cell receptor repertoires along the B-cell differentiation axis in humans and mice. PLoS Comput. Biol. 2020;16:e1007636. doi: 10.1371/journal.pcbi.1007636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk K., Liu X., Baker T., Shi J., Deane C.M. Improving B-cell epitope prediction and its application to global antibody-antigen docking. Bioinformatics. 2014;30:2288–2294. doi: 10.1093/bioinformatics/btu190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Levesque M.C., Nagel A., Dixon A., Zhang R., Walter E., Parks R., Whitesides J., Marshall D.J., Hwang K.K. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H., Wu N.C., Tsang O.T.-Y., Yuan M., Perera R.A.P.M., Leung W.S., So R.T.Y., Chan J.M.C., Yip G.K., Chik T.S.H. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. bioRxiv. 2020 doi: 10.1101/2020.03.15.993097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasca R., Vaccari P., Luppi M., Zucchini P., Castelli I., Barozzi P., Cuoghi A., Torelli G. Immunoglobulin gene mutations and frequent use of VH1-69 and VH4-34 segments in hepatitis C virus-positive and hepatitis C virus-negative nodal marginal zone B-cell lymphoma. Am. J. Pathol. 2001;159:253–261. doi: 10.1016/S0002-9440(10)61691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011;17:10–12. [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Matsuyama S., Shirato K., Kawase M., Terada Y., Kawachi K., Fukushi S., Kamitani W. Middle East Respiratory Syndrome Coronavirus Spike Protein Is Not Activated Directly by Cellular Furin during Viral Entry into Target Cells. J. Virol. 2018;92 doi: 10.1128/JVI.00683-18. e00683–e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot S.P., Di Noia J.M. Molecular Mechanisms of Somatic Hypermutation and Class Switch Recombination. Adv. Immunol. 2017;133:37–87. doi: 10.1016/bs.ai.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Moir S., Fauci A.S. B-cell exhaustion in HIV infection: the role of immune activation. Curr. Opin. HIV AIDS. 2014;9:472–477. doi: 10.1097/COH.0000000000000092. [DOI] [PubMed] [Google Scholar]
- Monaco G., Lee B., Xu W., Mustafah S., Hwang Y.Y., Carré C., Burdin N., Visan L., Ceccarelli M., Poidinger M. RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Reports. 2019;26:1627–1640.e7. doi: 10.1016/j.celrep.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan R., Buchauer L., Triller G., Kreschel C., Costa G., Martí G.P., Imkeller K., Busse C.E., Chakravarty S., Kim Lee Sim B. Clonal selection drives protective memory B cell responses in controlled human malaria infection. Sci. Immunol. 2018;3:eaap8029. doi: 10.1126/sciimmunol.aap8029. [DOI] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Zhao L., Qu L., Yao Z., Zhang F., Yan Q., Zhang S., Liang R., Chen P., Luo J. Convalescent patient-derived monoclonal antibodies targeting different epitopes of E protein confer protection against Zika virus in a neonatal mouse model. Emerg. Microbes Infect. 2019;8:749–759. doi: 10.1080/22221751.2019.1614885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehrig J.T., Hombach J., Barrett A.D.T. Guidelines for plaque-reduction neutralization testing of human antibodies to dengue viruses. Viral Immunol. 2008;21:123–132. doi: 10.1089/vim.2008.0007. [DOI] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Feldhahn N., Seaman M.S., Velinzon K., Pietzsch J., Ott R.G., Anthony R.M., Zebroski H., Hurley A. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Seifert M., Küppers R. Human memory B cells. Leukemia. 2016;30:2283–2292. doi: 10.1038/leu.2016.226. [DOI] [PubMed] [Google Scholar]
- Sela-Culang I., Benhnia M.R.E.I., Matho M.H., Kaever T., Maybeno M., Schlossman A., Nimrod G., Li S., Xiang Y., Zajonc D. Using a combined computational-experimental approach to predict antibody-specific B cell epitopes. Structure. 2014;22:646–657. doi: 10.1016/j.str.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Setliff I., Shiakolas A.R., Pilewski K.A., Murji A.A., Mapengo R.E., Janowska K., Richardson S., Oosthuysen C., Raju N., Ronsard L. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell. 2019;179:1636–1646.e15. doi: 10.1016/j.cell.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setthapramote C., Sasaki T., Puiprom O., Limkittikul K., Pitaksajjakul P., Pipattanaboon C., Sasayama M., Leuangwutiwong P., Phumratanaprapin W., Chamnachanan S. Human monoclonal antibodies to neutralize all dengue virus serotypes using lymphocytes from patients at acute phase of the secondary infection. Biochem. Biophys. Res. Commun. 2012;423:867–872. doi: 10.1016/j.bbrc.2012.06.057. [DOI] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidarsson G., Dekkers G., Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N. Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape. J. Virol. 2018;92:1–21. doi: 10.1128/JVI.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., Haperen R. van, Osterhaus A.D.M.E., Kuppeveld F.J.M. van, Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020 doi: 10.1038/s41467-020-16256-y. Published online May 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H., Yurasov S., Schaefer A., Young J.W., Meffre E., Nussenzweig M.C. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Wong W.K., Georges G., Ros F., Kelm S., Lewis A.P., Taddese B., Leem J., Deane C.M. SCALOP: sequence-based antibody canonical loop structure annotation. Bioinformatics. 2019;35:1774–1776. doi: 10.1093/bioinformatics/bty877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J., Smith K., Miller J., Langley W.A., Kokko K., Larsen C., Zheng N.Y., Mays I., Garman L., Helms C. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;80(361):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Jia W., Wang P., Zhang S., Shi X., Wang X., Zhang L. Antibodies and vaccines against Middle East respiratory syndrome coronavirus. Emerg. Microbes Infect. 2019;8:841–856. doi: 10.1080/22221751.2019.1624482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.H., Deng W., Tong Z., Liu Y.X., Zhang L.F., Zhu H., Gao H., Huang L., Liu Y.L., Ma C.M. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007;57:450–459. [PubMed] [Google Scholar]
- Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Cui X., Zhao X., Wang J., Zheng J., Zheng G., Guo W., Cai C., He S., Xu Y. Detectable SARS-CoV-2 viral RNA in feces of three children during recovery period of COVID-19 pneumonia. J. Med. Virol. 2020 doi: 10.1002/jmv.25795. Published online March 29, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X., Hensley L.E., Prabakaran P., Rockx B., Sidorov I.A. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. USA. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J.H., Lindahl E., Scheres S.H.W. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e4216. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the human antibody sequences reported in this paper is EGA: S00001004412. Material/Data Transfer Agreements, which allow the use of the antibody sequences for non-commercial purposes but not their disclosure to third parties, are needed to obtain the sequences by contacting the Data Access Committee.