Abstract
Introduction
The oncogenic role of lncRNA LUADT1 has been investigated only in lung cancer. This study aimed to investigate the role of LUADT1 in oral squamous cell carcinoma (OSCC).
Patients and Methods
The expression levels of LUADT1 in paired OSCC and non-tumor tissues from OSCC patients were determined by RT-qPCR. A 5-year follow-up study was performed to analyze the prognostic value of LUADT1 for OSCC. Dual-luciferase assay and overexpression experiments were performed to assess the interactions among LUADT1, miR-34a and GASL1. Cell proliferation was analyzed by cell proliferation assay.
Results
In this study, we found that LUADT1 was upregulated in OSCC and predicted poor survival. LUADT1 was predicted to interact with miR-34a, which was confirmed by dual-luciferase activity assay. However, overexpression experiments showed that they did not affect the expression of each other. Interestingly, overexpression of LUADT1 resulted in upregulation of GAS1, a target of miR-34a. Cell proliferation assay revealed that overexpression of LUADT1 and GAS1 resulted in promoted cell proliferation. MiR-34a played an opposite role and reversed the effects of LUADT1 overexpression.
Conclusion
LUADT1 may promote OSCC proliferation by regulating miR-34a/GAS1 axis.
Keywords: oral squamous cell carcinoma, LUADT1, miR-34a, GAS1
Introduction
Oral cancer is one of the top 10 most common types of malignancies for both incidence and mortality.1 According to the latest GLOBOCAN statistics, oral cancer in 2018 only affected 354,864 new cases, accounting for 2.0% of all new cancer cases.2 During the same year, oral cancer caused 177,384 deaths, which were 1.9% of all cancer deaths in 2018.2 Occurrence of oral cancer is associated with multiple risk factors, such as tobacco smoking, alcohol consumption and human papillomavirus (HPV) infections.3–6 Increased understanding of the molecular mechanisms of oral cancer has provided novel insights into the treatment of this disease, while molecular pathways involved in this disease remain elusive.7
The development and progression of oral cancer involve the regulations of multiple molecular pathways.8,9 Understanding the interactions among molecular players involved in this disease is critical for the development of novel anticancer therapies, such as targeted therapies.10 Non-coding RNAs (ncRNAs), such as long (>200 nt) ncRNAs (lncRNAs) and miRNAs are emerging novel players in cancer biology.11 Oncology studies have shown that regulation of certain lncRNAs may suppress the development of cancer.12 However, the functionality of most lncRNAs in cancer remains elusive. LUADT1 has been characterized as an oncogenic lncRNA in lung cancer,13 while its roles in other cancers are unknown. We performed bioinformatics analysis and found that LUADT1 may interact with miR-34a, which can target GAS1 to promote cancer development.14 This study was therefore carried out to investigate the interaction between LUADT1 and miR-34a in oral squamous cell carcinoma (OSCC), which is a major subtype of oral cancer.
Patients and Methods
OSCC Patients
A total of 70 OSCC patients (43 males and 27 females, 48 to 76 years old, mean age 61.7 ± 4.9 years old) admitted at Jinan Stomatological Hospital between May 2012 and May 2014 were enrolled in this study. Before the admission of patients, this study was approved by the Ethics Committee of Jinan Stomatological Hospital (No. JSH2012OSCC02758) and was conducted in accordance with the Declaration of Helsinki. All patients had not been treated with any therapies within half year before admission. All patients were diagnosed for the first time and recurrent OSCC cases were excluded. Fine needle biopsies were performed on all 70 patients to collect OSCC tumor tissue and paired non-tumor (within 2 cm around tumors) specimens. All patients signed the written informed consent.
Therapies and 5-Year Follow-Up
The 70 patients were first staged according to AJCC criteria. The 70 patients were classified into 4 stages, including 17, 22, 17 and 14 cases at stage I, II, III, and IV, respectively. Therapies, including surgical resections, radiotherapies, chemotherapies and the combination of them, were performed on patients according to their health conditions and clinical stages. A 5-year follow-up was performed on all 70 patients and all of them completed the follow-up.
Cells and Cell Culture
Human OSCC cell line SCC090 (ATCC) was used. Cells were cultivated in a culture medium composed of 10% FBS and 90% Eagle’s Minimum Essential Medium (2mM L-glutamine) in a 5% CO2 incubator at 37 °C. Cells were collected at about 85% confluence for subsequent experiments.
Lipofectamine 2000-Mediated Transient Transfection
Constructions of LUADT1 and GAS1 were performed using pcDNA3.1 vector as backbone. Design and synthesis of LUADT1 siRNA, NC siRNA, miR-34a mimic as well as negative control (NC) miRNA were performed by Invitrogen. SCC090 cells were transfected with 45 nM miRNA or siRNA, or 10 nM vector using lipofectamine 2000 (Invitrogen). Transfection with empty pcDNA3.1 vector, NC siRNA, or NC miRNA were used as NC group. Cells with no transfections were used as the control (C) group. Overexpression effect was confirmed at 48 h post-transfection.
Dual-Luciferase Activity Assay
Basic version of pGL3 luciferase reporter vector (Promega) was used as the backbone to establish LUADT1 vector. The vector construction service was provided by Promega. SCC090 cells were co-transfected with 1) LUADT1 vector combined with NC miRNA (NC miRNA group); 2) LUADT1 vector combined with miR-34a mimic (miR-34a group) using lipofectamine 2000 (Invitrogen). The Luciferase Assay System (BPS Bioscience) was used to measure luciferase activity at 48 h after transfections.
Preparation of RNA Samples
All tissue samples were ground in liquid nitrogen. SCC090 cells were washed with PBS. Total RNAs were extracted from tissues and cells using Trizol reagent (Invitrogen, USA). RNA precipitation and washing were performed using 85% ethanol to harvest miRNA. Nanodrop 2000 (Thermo Scientific) was used to measure the RNA concentration.
RT-qPCR Assay
The qScript One-Step SYBR Green RT-qPCR (Quantabio) was used to perform RNA reverse transcriptions and qPCRs to measure the expression levels of LUADT1 and GAS1 mRNA with GAPDH as endogenous control. The All-in-One™ miRNA qRT-PCR Reagent Kit (Genecopoeia) was used to perform miRNA reverse transcriptions and qPCR reactions to measure the expression levels of mature miR-34a with U6 as endogenous control. Three replicate reactions were included in each experiment and Ct values were calculated using 2−ΔΔCT method.
Western Blot
SCC090 cells were washed with PBS and total proteins were extracted using RIPA solution (Invitrogen). Protein concentrations were measured using the BCA method. Separation of proteins was performed using 8% SDS-PAGE gel. Separated proteins were transferred to PVDF membranes and membranes were blocked with PBS containing 5% non-fat milk for 2 h. Following that, membranes were incubated with GAS1 (1:1000, ab236618, Abcam) and GAPDH (1:1000, ab8245, Abcam) primary antibodies at 4 °C for 12h. Membranes were then further incubated with anti-rabbit IgG-HRP secondary antibody (1:1000, ab97051, Abcam) at room temperature for 2 h. ECL (Sigma-Aldrich) was used to produce signals, which were then processed using Quantity One software.
CCK-8 Assay
SCC090 cells harvested at 48 h post-transfection were subjected to cell proliferation assay using the Cell Counting Kit 8 kit (CCK8, ab228554, Abcam). A 96-well cell culture plate was used to cultivate cells (6000 cells in 0.1 mL medium per well) at 37 °C. Three replicate wells were set for each experiment. Cells were harvested every 24 h after the initiation of cell culture until 96 h. CCK-8 solution was added into each well to reach a final concentration of 10% at 4 h before cell collection. OD values were measured at 450 nm to detect cell proliferation.
Statistical Analysis
All experiments were repeated 3 times and statistical analyses were performed using the mean values with SPSS19.0 (SPSS Inc., USA) software. Paired t-test was used to compare the differences between OSCC and non-tumor tissues. Unpaired t-test was used to compare the differences of luciferase activity between two groups. ANOVA Tukey’s test was used to compare differences among multiple groups. Correlations were analyzed by Pearson’s correlation coefficient. The 70 OSCC patients were grouped into high and low LUADT1 level groups (n = 35) with median expression level of LUADT1 in OSCC tissues as cutoff value. Survival curves were plotted and compared by Log rank test. P < 0.05 was considered to be statistically significant.
Results
LUADT1 Can Interact with miR-34a
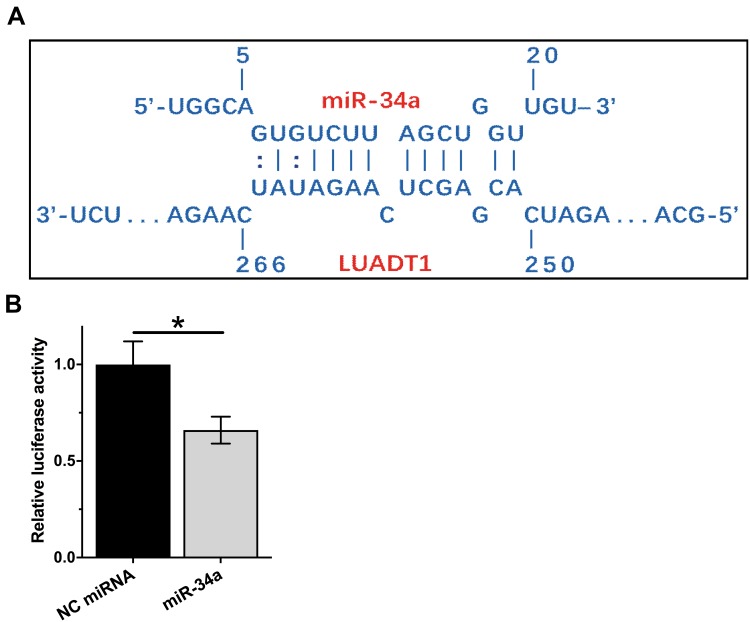
The potential binding site between LUADT1 and miR-34a was predicted using IntaRNA.15 It showed that multiple base pairing can be formed between LUADT1 and miR-34a (Figure 1A). The direct interaction was assessed by luciferase activity assay, which was performed by transfecting SCC090 cells with LUADT1 vector combined with NC miRNA (NC miRNA group) or LUADT1 vector combined with miR-34a mimic (miR-34a group). Compared to NC miRNA group, significantly lower relative luciferase activity was observed in miR-34a group (Figure 1B, p < 0.05).
Figure 1.
LUADT1 can interact with miR-34a. The potential base pairs can be formed by LUADT1 and miR-34a was predicted using IntaRNA (A). The direct interaction between them was analyzed by luciferase activity assay, which was performed by transfecting SCC090 cells with LUADT1 vector combined with NC miRNA (NC miRNA group or LUADT1 vector combined with miR-34a mimic (miR-34a group). Experiments were repeated 3 times and mean values were presented and compared by unpaired t-test (B). *p < 0.05.
The Expression of LUADT1 and miR-34a Was Altered in OSCC Tissues
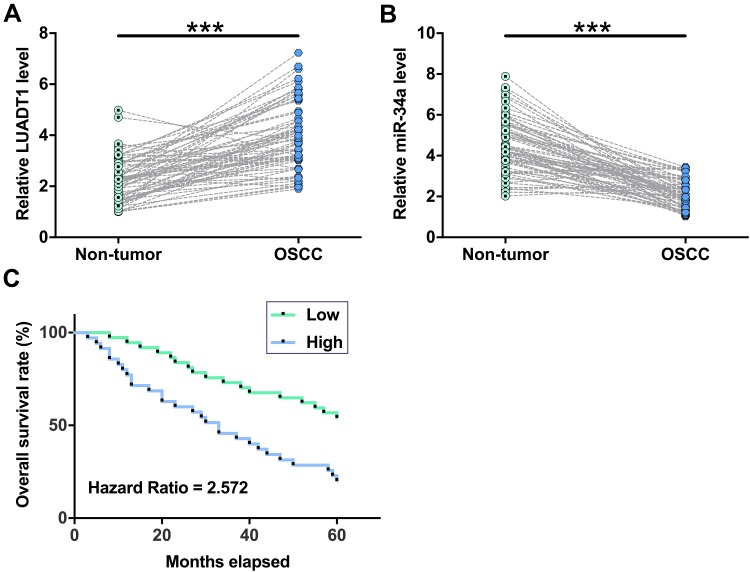
The expression levels of LUADT1 and miR-34a in both OSCC and non-tumor tissues from 70 OSCC patients were measured by qPCR. Paired t-test showed that the expression levels of LUADT1 were significantly higher in OSCC tissues in comparison to that in non-tumor tissues (Figure 2A, p < 0.001). In contrast, the expression levels of miR-34a were significantly lower in OSCC tissues in comparison to that in non-tumor tissues (Figure 2B, p < 0.001). Survival curves were plotted and compared. It was observed that patients in high LUADT1 level group experienced significantly lower overall survival rate (Figure 2C).
Figure 2.
The expression of LUADT1 and miR-34a was altered in OSCC tissues. Expression levels of LUADT1 (A) and miR-34a (B) in both OSCC and non-tumor tissues from the 70 OSCC patients were measured by performing qPCR. Paired t-test was used to compare mean values of 3 replicates. ***p < 0.001. The 70 OSCC patients were grouped into high and low LUADT1 level groups (n = 35) with median expression level of LUADT1 in OSCC tissues as cutoff value. Survival curves were plotted and compared by Log rank test (C).
LUADT1 and miR-34a Were Not Significantly Associated and They Did Not Regulate the Expression of Each Other
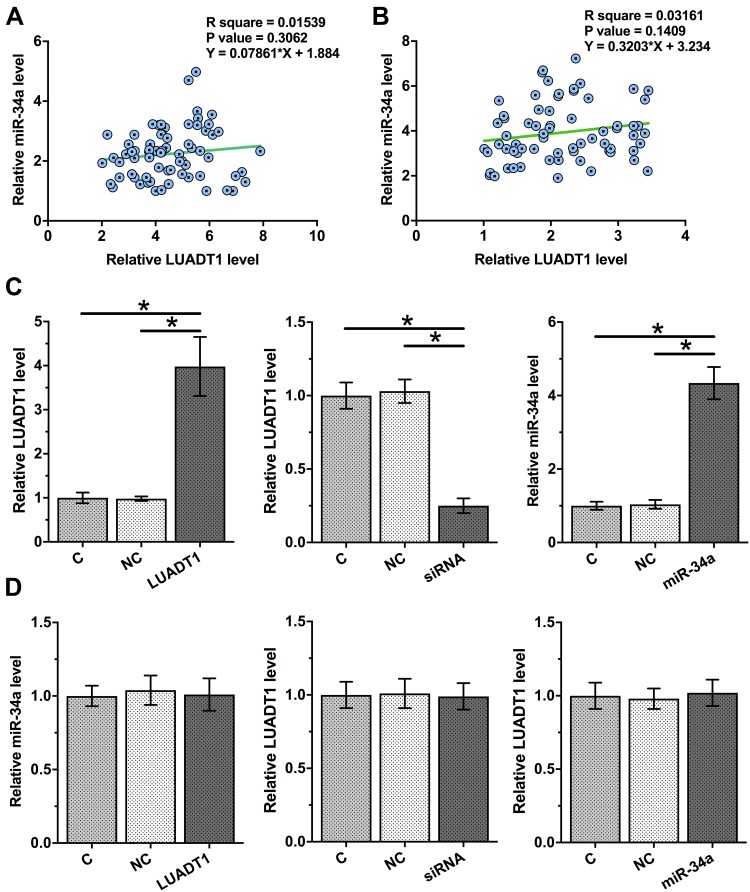
Correlations between the expression levels of LUADT1 and miR-34a were analyzed. It was observed that LUADT1 and miR-34a were not significantly correlated across OSCC tissues (Figure 3A) and non-tumor tissues (Figure 3B). To further assess the interactions between LUADT1 and miR-34a, SCC090 cells were transfected with LUADT1 expression vector, LUADT1 siRNA or miR-34a mimic. Overexpression of LUADT1 and miR-34a as well as silencing of LUADT1 was confirmed at 48 h post-transfection by qPCR (Figure 3C, p < 0.05). Compared to C and NC groups, overexpression of LUADT1 and miR-34a did not affect the expression of each other. The silencing of LUADT1 also did not affect the expression of miR-34a (Figure 3D, p < 0.05).
Figure 3.
LUADT1 and miR-34a were not significantly associated and they did not regulate the expression of each other. Correlations between expression levels of LUADT1 and miR-34a across OSCC tissues (A) and non-tumor tissues (B) were analyzed by Pearson’s correlation coefficient. SCC090 cells were transfected with LUADT1 expression vector or miR-34a mimic. Overexpression of LUADT1 and miR-34a was confirmed at 48h post-transfection by qPCR (C). The effects of overexpressing LUADT1 and miR-34a as well as silencing of LUADT1 on the expression of each other were also analyzed by qPCR (D). Experiments were performed in triplicate manner and mean values were presented and compared. *p < 0.05.
Overexpression of LUADT1 Led to Upregulated GAS1
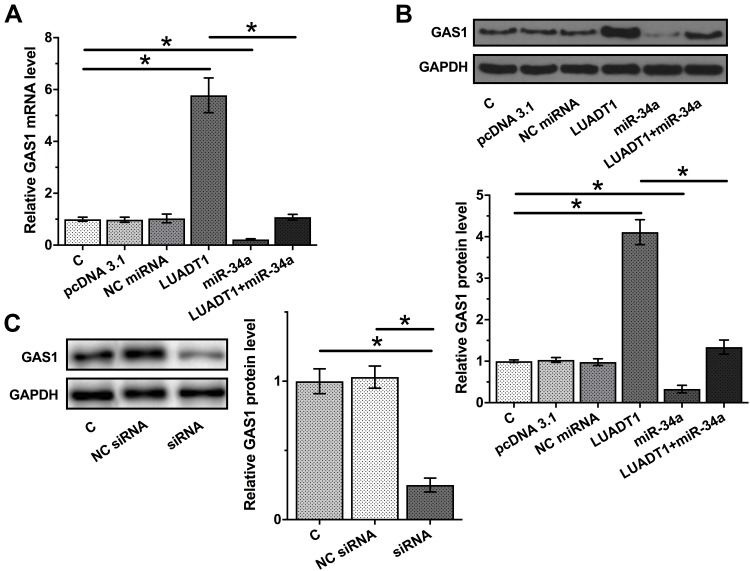
The data presented above may suggest that LUADT1 is a sponge of miR-34a. To test this possibility, the effects of LUADT1 and miR-34a on the expression of GAS1, a validated target of miR-34a, were evaluated by qPCR and Western blot at mRNA (Figure 4A) and protein (Figure 4B) levels, respectively. It was observed that overexpression of miR-34a led to downregulated GAS1 (p < 0.05). Interestingly, overexpression of LUADT1 led to upregulated GAS1 and totally reversed the role of miR-34a (p < 0.05). Moreover, downregulated expression of GAS1 was observed after the silencing of LUADT1 (Figure 4C).
Figure 4.
LUADT1 negatively regulated GAS1 through miR-34a. The effects of LUADT1 and miR-34a on the expression of GAS1, a validated target of miR-34a, were evaluated by qPCR and Western blot at mRNA (A) and protein (B) levels, respectively. The effects of silencing of LUADT1 on the expression of GAS1 were assessed by Western blot (C). Experiments were performed in triplicate manner and mean values were presented and compared. *p < 0.05.
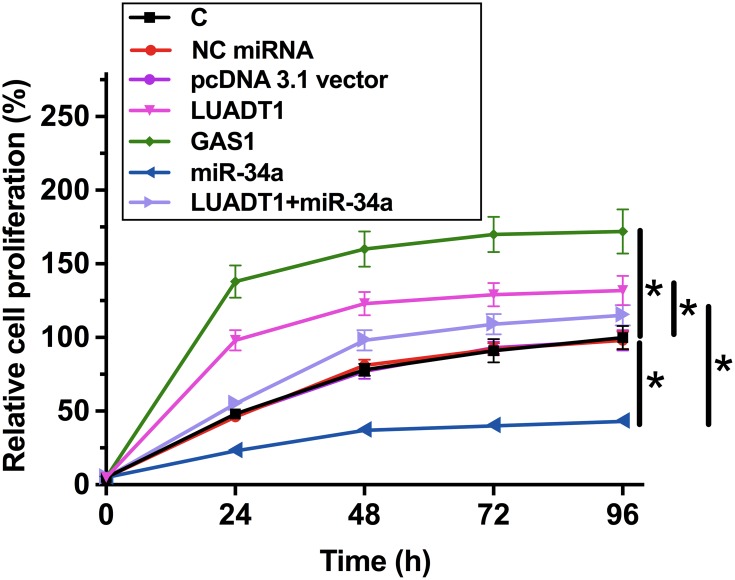
LUADT1 Regulated miR-34a/GAS1 Axis to Suppress SCC090 Cell Proliferation
The effects of LUADT1, miR-34a and GAS1 overexpression on the proliferation of SCC090 cells were assessed by CCK-8 assay. Cell proliferation assay revealed that overexpression of LUADT1 and GAS1 resulted in increased cell proliferation rate. MiR-34a plays an opposite role and reduced the effects of overexpression of LUADT1 (Figure 5, p < 0.05).
Figure 5.
LUADT1 regulated miR-34a/GAS1 axis to suppress SCC090 cell proliferation. The effects of LUADT1, miR-34a and GAS1 overexpression on the proliferation of SCC090 cells were analyzed by CCK-8 assay. Experiments were performed in triplicate manner, and mean values were presented and compared. *p < 0.05.
Discussion
This study investigated the roles of LUADT1 in OSCC, which is a common type of oral cancer. We found that the downregulation of LUADT1 in OSCC is correlated with poor survival. In addition, LUADT1 may serve as an endogenous sponge of miR-34a to upregulate GAS1, which in turn promotes cell proliferation.
The functionality of LUADT1 has only been investigated in lung adenocarcinoma.13 LUADT1 is reported to be upregulated in lung adenocarcinoma and can downregulate p27 through methylation to promote cancer cell proliferation.13 Our study also observed the upregulation of LUADT1 in OSCC. In addition, overexpression of LUADT1 led to increased proliferation rate of OSCC cells. Therefore, LUADT1 is likely an oncogenic lncRNA in OSCC. Our data and previous studies suggest that LUADT1 may play similar roles in different types of cancer, indicating the similar molecular pathogenesis shared by different types of cancer.
The survival of OSCC patients is generally poor,16,17 largely due to the low early diagnostic rate. However, this situation is unlikely to be changed due to the lack of effective early diagnostic markers. In this study, we observed that the high expression levels of LUADT1 were correlated with the poor survival of OSCC patients. Therefore, expression levels of LUADT1 measured before treatment may be used to predict the survival, which in turn provides guidance for the selection of therapies and care programs. However, clinical trials bigger sample size are needed to further investigate the prognostic values.
A recent study reported that miR-34a could target GAS1 to promote cell proliferation in papillary thyroid carcinoma.14 Consistently, our study also showed that miR-34a can downregulate GAS1 to suppress cell proliferation in OSCC. Our results showed that LUADT1 and miR-34a could interact with each other, while they did not regulate the expression of each other. Instead, overexpression of LUADT1 led to upregulation of GAS1. Therefore, the most reasonable explanation is that LUADT1 may be an endogenous control of miR-34a to suppress its oncogenic roles. However, the functions of ncRNAs may also rely on their tertiary structures. Therefore, studies on the elucidation of the structure of LUADT1 are also of great importance.
Conclusion
In conclusion, LUADT1 is upregulated in OSCC and may upregulate GAS1 by sponging miR-34a, thereby promoting cell proliferation in OSCC.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol. 2015;8(9):11884–11894. [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Kumar M, Nanavati R, Modi TG, Dobariya C. Oral cancer: etiology and risk factors: a review. J Cancer Res Ther. 2016;12(2):458–463. doi: 10.4103/0973-1482.186696 [DOI] [PubMed] [Google Scholar]
- 4.Dheilly NM, Ewald PW, Brindley PJ, Fichorova RN, Thomas F, Knoll LJ. Parasite-microbe-host interactions and cancer risk. PLoS Pathog. 2019;15(8):e1007912. doi: 10.1371/journal.ppat.1007912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drews CM, Case S, Vande Pol SB, Galloway DA. E6 proteins from high-risk HPV, low-risk HPV, and animal papillomaviruses activate the Wnt/beta-catenin pathway through E6AP-dependent degradation of NHERF1. PLoS Pathog. 2019;15(4):e1007575. doi: 10.1371/journal.ppat.1007575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang SK, Zheng R, Chen Q, Zhang S, Sun X, Chen W. Oral cancer incidence and mortality in China, 2011. Chin J Cancer Res. 2015;27(1):44–51. doi: 10.3978/j.issn.1000-9604.2015.01.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkman BM, Wong DT. Disease mechanism and biomarkers of oral squamous cell carcinoma. Curr Opin Oncol. 2006;18(3):228–233. doi: 10.1097/01.cco.0000219250.15041.f8 [DOI] [PubMed] [Google Scholar]
- 8.Ishigami T, Uzawa K, Higo M, et al. Genes and molecular pathways related to radioresistance of oral squamous cell carcinoma cells. Int J Cancer. 2007;120(10):2262–2270. doi: 10.1002/ijc.22561 [DOI] [PubMed] [Google Scholar]
- 9.Keshavarzi M, Darijani M, Momeni F, et al. Molecular imaging and oral cancer diagnosis and therapy. J Cell Biochem. 2017;118(10):3055–3060. doi: 10.1002/jcb.26042 [DOI] [PubMed] [Google Scholar]
- 10.Alhazzazi TY, Kamarajan P, Joo N, et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117(8):1670–1678. doi: 10.1002/cncr.25676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams GT, Pickard MR. Long non-coding RNAs: new opportunities and old challenges in cancer therapy. Transl Cancer Res. 2016;5(S3):S564–S566. doi: 10.21037/tcr.2016.09.04 [DOI] [Google Scholar]
- 13.Qiu M, Xu Y, Wang J, et al. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015;6:e1858. doi: 10.1038/cddis.2015.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Y, Qin H, Cui Y. MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem Biophys Res Commun. 2013;441(4):958–963. doi: 10.1016/j.bbrc.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 15.Busch A, Richter AS, Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24(24):2849–2856. doi: 10.1093/bioinformatics/btn544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auluck A, Walker BB, Hislop G, Lear SA, Schuurman N, Rosin M. Socio-economic deprivation: a significant determinant affecting stage of oral cancer diagnosis and survival. BMC Cancer. 2016;16:569. doi: 10.1186/s12885-016-2579-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Leontis NB, Zirbel CL, Bisaro DM, Ding B, Wang A. A three-dimensional RNA motif mediates directional trafficking of potato spindle tuber viroid from epidermal to palisade mesophyll cells in Nicotiana benthamiana. PLoS Pathog. 2019;15(10):e1008147. doi: 10.1371/journal.ppat.1008147 [DOI] [PMC free article] [PubMed] [Google Scholar]