Abstract
The protozoan parasite Toxoplasma gondii lives inside a vacuole in the host cytosol where it is protected from host cytoplasmic innate immune responses. However, IFNγ‐dependent cell‐autonomous immunity can destroy the vacuole and the parasite inside. Toxoplasma strain differences in susceptibility to human IFNγ exist, but the Toxoplasma effector(s) that determine these differences are unknown. We show that in human primary fibroblasts, the polymorphic Toxoplasma‐secreted effector GRA15 mediates the recruitment of ubiquitin ligases, including TRAF2 and TRAF6, to the vacuole membrane, which enhances recruitment of ubiquitin receptors (p62/NDP52) and ubiquitin‐like molecules (LC3B, GABARAP). This ultimately leads to lysosomal degradation of the vacuole. In murine fibroblasts, GRA15‐mediated TRAF6 recruitment mediates the recruitment of immunity‐related GTPases and destruction of the vacuole. Thus, we have identified how the Toxoplasma effector GRA15 affects cell‐autonomous immunity in human and murine cells.
Keywords: GRA15, IFNγ, p62, Toxoplasma, TRAF6
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction
Inter‐strain differences in susceptibility to interferon‐induced lysosomal degradation of parasite‐containing vacuoles is determined by secretion of a protozoan effector interfering with host ubiquitin signaling.

Introduction
Toxoplasma is a highly successful obligate intracellular parasite that can establish lifelong chronic infections in a wide range of warm‐blooded animals. In humans, it causes opportunistic infections in immunosuppressed patients, congenital infections (Hill & Dubey, 2002), and blindness (Pleyer et al, 2014). Many different Toxoplasma strains exist, but in Europe and North America, human infections are dominated by the type I and type II clonal lineages (Howe & Sibley, 1995; Saeij et al, 2005). Upon host cell invasion, Toxoplasma wraps itself with the host cell plasma membrane, which becomes the nascent parasitophorous vacuole membrane. The vacuole membrane does not fuse with the endo‐lysosome system, and without immune pressure, the vacuole does not acidify thus providing Toxoplasma with a niche for replication (Jones et al, 1972; Mordue & Sibley, 1997).
Like Toxoplasma, many intracellular pathogens reside within a vacuole (pathogen‐containing vacuole or PV) in the host cytoplasm (Liehl et al, 2015). The PV membrane (PVM) protects these pathogens from detection by host cytosolic pathogen recognition receptor (PRR). However, the host has developed mechanisms to destroy the PV thereby exposing the pathogen (Liehl et al, 2015; Saeij & Frickel, 2017). For example, in mice interferons upregulate the expression of two families of large dynamin‐like GTPases: the immunity‐related GTPases (IRGs) and the guanylate‐binding proteins (GBPs) (Howard et al, 2011), which mediate the destruction of the PV of Toxoplasma and of many gram‐negative bacteria (Liehl et al, 2015). Once the pathogen is in the cytoplasm, GBPs and IRGs can also mediate the vesiculation of the pathogen itself thereby exposing pathogen‐associated molecular patterns (PAMPs) to cytosolic PRRs, which can lead to the activation of the inflammasome (Man et al, 2017) and the induction of a form of cell death called pyroptosis (Broz & Dixit, 2016). Because host cell death removes the replication niche of intracellular pathogens, this is an efficient way of inhibiting pathogen growth (Krishnamurthy et al, 2017).
The mechanism of IRG and GBP recruitment to the Toxoplasma PVM and the identity of Toxoplasma effectors influencing IRG/GBP recruitment, or their activity, are intense areas of research. These GTPases are normally held inactive on host endomembranes by regulatory “GMS motif”‐type IRGs (Haldar et al, 2013). Initially, “pioneer” effector “GKS” motif‐type IRGs are recruited to the Toxoplasma PVM (Hunn et al, 2008), which is largely devoid of regulatory IRGs, where they oligomerize and become activated. What exact signal initiates the recruitment of these pioneer IRGs to the PVM is unclear. In murine cells, the initial conjugation of a ubiquitin‐like protein (e.g., microtubule‐associated protein light chain 3 [LC3] or γ‐aminobutyric acid receptor‐associated proteins [GABARAPs]) to the PVM was proposed to be the signal that initiates recruitment of “pioneer” IRGs (Choi et al, 2014; Sasai et al, 2017). PVM recruitment of pioneer IRGs somehow promotes the ubiquitination of the PVM which subsequently leads to the recruitment of the ubiquitin‐binding protein p62 (also called sequestosome or SQSTM1) and E3 ubiquitin ligases (e.g., TNF receptor‐associated factor 6 or TRAF6 and tripartite motif‐containing 21 or TRIM21) in a co‐dependent manner (Haldar et al, 2015; Lee et al, 2015; Foltz et al, 2017). PVM ubiquitination by these ubiquitin ligases can lead to further recruitment of p62 and TRAF6, thereby creating an amplification loop ensuring the full ubiquitination of the PV. GBPs also get recruited to the PVM and eventually vesiculation of the PVM by GBPs and IRGs exposes Toxoplasma, which can lead to its destruction by GBPs (Degrandi et al, 2013; Kravets et al, 2016) and pyronecrosis of host cells (Zhao et al, 2009). To counter the host defense mechanisms, Toxoplasma secretes ROP and GRA effector proteins into the host cell from organelles called rhoptries and dense granules, respectively. In both murine and human cells, type II strains are more susceptible to IFNγ‐mediated growth inhibition than type I strains (Haldar et al, 2015; Selleck et al, 2015; Clough et al, 2016; Qin et al, 2017). Resistance of type I strains in murine cells is determined primarily by polymorphic ROP5 and ROP18, which, together with ROP17 and GRA7, cooperatively block IRG and GBP loading on the PVM and subsequent events (Khaminets et al, 2010; Steinfeldt et al, 2010; Niedelman et al, 2012; Etheridge et al, 2014; Behnke et al, 2015; Haldar et al, 2015). ROP16 and GRA15 also affect GBP loading on the PVM in murine cells through an unknown mechanism (Virreira Winter et al, 2011).
IFNγ‐stimulated human cells control Toxoplasma using diverse mechanisms dependent on the cell type (Krishnamurthy et al, 2017). For example, IFNγ‐mediated induction of indoleamine 2,3 dioxygenase (IDO) causes breakdown of L‐tryptophan, for which Toxoplasma is auxotrophic, which mediates inhibition of parasite growth in HeLa, HAP1, and fibroblast cells (Pfefferkorn, 1984; Pfefferkorn et al, 1986; Niedelman et al, 2013; Qin et al, 2017; Bando et al, 2018). In some human cell lines, another mechanism for parasite control is ubiquitination of the vacuole, which leads to lysosomal fusion in HUVEC cells, while in HeLa cells, an autophagic double membrane forms around the vacuole and parasite growth is stunted without lysosomal fusion (Selleck et al, 2015; Clough et al, 2016). In certain human cells, GBP1 also seems important for restriction of Toxoplasma growth but the mechanism of growth restriction is unclear. In a lung epithelial cell line (A549), GBP1 restricts parasite growth without its recruitment to the PVM (Johnston et al, 2016), while in human mesenchymal stem cells (MSCs), growth restriction was associated with GBP1 PVM recruitment (Qin et al, 2017). Much less is known about what initiates targeting of human immune effectors to the PVM and how this leads to parasite elimination. In contrast to mice, humans lack IFNγ‐inducible IRGs, likely explaining why ROP5, ROP18, and ROP17 do not seem to play an important role in conferring protection against IFNγ‐mediated growth inhibition in human cells (Niedelman et al, 2012; Selleck et al, 2015; Clough et al, 2016). Currently, no parasite proteins that determine strain differences in susceptibility to IFNγ‐mediated cell‐autonomous immunity in human cells have been identified. A secreted parasite effector, Toxoplasma inhibitor of STAT1‐dependent transcription (TgIST), which blocks the STAT1 transcriptional response, was recently described, but this effector functions upstream of the upregulation of IFNγ‐induced toxoplasmacidal mechanisms in both type I and type II strains (Gay et al, 2016; Olias et al, 2016).
Herein, we report that the PVM‐localized Toxoplasma GRA15 effector (Rosowski et al, 2011) enhances parasite susceptibility to IFNγ in primary human foreskin fibroblasts (HFFs) and murine embryonic fibroblasts (MEFs). GRA15 binds several ubiquitin ligases, including TRAF2 and TRAF6, which in HFFs is associated with enhanced recruitment of p62, LC3, and GABARAP to the PVM, enhanced endo‐lysosomal fusion, and parasite destruction. In MEFs, GRA15 also interacts with TRAF2 and TRAF6, and TRAF6 recruitment leads to enhanced PVM loading with IRGs and GBPs and parasite destruction. Thus, we determined that the Toxoplasma effector GRA15 mediates strain differences in susceptibility to cell‐autonomous immunity in human cells and determined the mechanism by which GRA15 enhances parasite susceptibility to IFNγ in both human and murine fibroblasts.
Results
The polymorphic effector GRA15 enhances Toxoplasma susceptibility to IFNγ‐mediated growth inhibition in HFFs
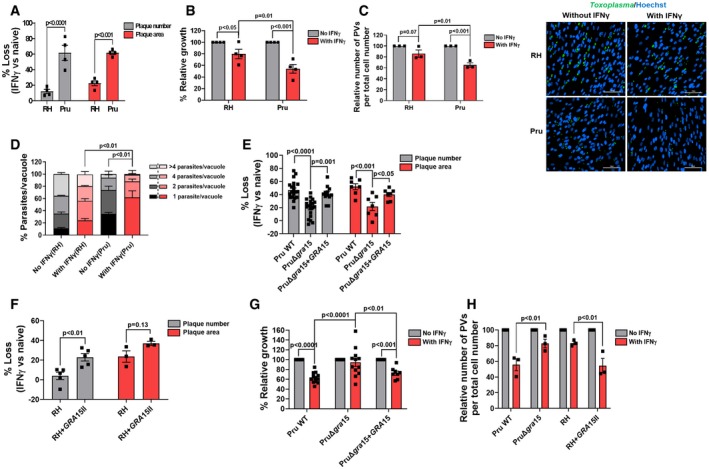
To determine whether the type I RH and the type II Pru strain differ in susceptibility to IFNγ‐mediated growth inhibition in HFFs, we measured the IFNγ‐mediated reduction in plaque numbers and plaque area. The relative reduction in the number of plaques formed in IFNγ‐stimulated vs. unstimulated cells reflects killing of Toxoplasma, while the relative reduction in the area of the plaques is a sensitive assay that reflects overall inhibition of parasite growth over multiple lysis cycles (Niedelman et al, 2012, 2013). Compared to RH, Pru had a larger IFNγ‐induced loss in plaque numbers and plaque area (Figs 1A and EV1A). We used parasites expressing luciferase to determine IFNγ‐mediated growth inhibition 24 h post‐infection (p.i.) and observed that Pru growth was more inhibited by IFNγ compared to RH growth (Fig 1B). IFNγ stimulation resulted in a decrease in the relative number of vacuoles (Fig 1C) as well as a decrease in the number of parasites per vacuole (Fig 1D), both of which were more pronounced for the Pru strain.
Figure 1. The type II (Pru) Toxoplasma strain is more susceptible to IFNγ‐mediated growth inhibition in primary human foreskin fibroblasts (HFFs) than the type I RH strain due to the presence of the polymorphic effector GRA15.
-
AHFFs were pre‐stimulated with IFNγ (10 U/ml) for 24 h. Plaque assays were performed for each strain and each condition. Plaque number and area loss were calculated 4 days p.i. for RH and 6 days p.i. for Pru. Assays were performed with RH (n = 4) and Pru (n = 4).
-
BRelative parasite growth was measured 24 h p.i. in IFNγ‐stimulated and unstimulated HFFs by luciferase assay. Growth of each strain in IFNγ‐stimulated HFFs is expressed relative to growth in unstimulated HFFs. Experiments were performed with RH (n = 4) and Pru (n = 4).
-
CNumber of PVs was calculated using HFFs grown on coverslips and stimulated with 10 U/ml IFNγ for 24 h. Following stimulation, HFFs were infected with either RH (MOI = 1) or Pru (MOI = 3) for another 24 h. Coverslips were fixed and stained with GRA7 for parasite PVM and Hoechst 33258 for nuclei. The number of PVs in 5–6 fields of the coverslips was counted for each condition and normalized with the number of host cells in each field. Experiments were performed with RH (n = 3) and Pru (n = 3). Representative images of percentage infected cells for each strain and condition are provided. Scale bar is 100 μm.
-
DParasites per vacuole were determined 24 h p.i. with similar conditions and staining as described in (C) except MOIs used were 0.5 and 1 for RH (n = 3) and Pru (n = 3), respectively.
-
EPlaque assays were performed similarly as described for (a) with Pru WT (n = 20 for plaque number and n = 7 for plaque area), PruΔgra15 (n = 20 for plaque number and n = 7 for plaque area), and PruΔgra15 + GRA15‐complemented (n = 12 for plaque number and n = 7 for plaque area).
-
FPlaque assays were performed similarly as described for (A) with RH (n = 5 for plaque number and n = 3 for plaque area) and RH + GRA15II (n = 5 for plaque number and n = 3 for plaque area).
-
GRelative parasite growth was measured as described in (B) with Pru WT (n = 12), PruΔgra15 (n = 12), and PruΔgra15 + GRA15‐complemented (n = 8).
-
HNumber of PVs was calculated as described in (C) with Pru WT (n = 3), PruΔgra15 (n = 3), RH (n = 3), and RH + GRA15II (n = 3) and normalized with the number of host cells in each field.
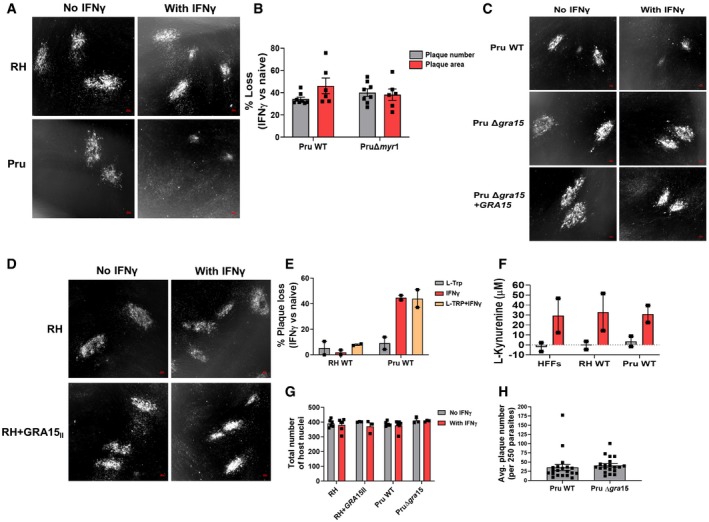
Figure EV1. IFNγ‐mediated susceptibility of the type II Pru strain is neither dependent on MYR1‐dependent parasite secreted factors nor on L‐Trp breakdown by primary human fibroblasts.
-
ARepresentative images of the plaque assay described for Fig 1A. Left panel is for unstimulated HFFs infected with either RH (top) or Pru (bottom), and right panel is for IFNγ‐stimulated HFFs infected with RH (top) and Pru (bottom). Scale bar is 100 μm.
-
BPlaque assays were performed with PruΔku80Δhpt (Pru) and PruΔku80ΔhptΔmyr1 (PruΔmyr1) strains, and plaque numbers were counted and plaque areas were measured after 6 days. Plaque numbers and plaque areas are averages from eight and six biological replicates, respectively.
-
CRepresentative images of the plaque assay described for Fig 1E. Left panel is for unstimulated HFFs infected with Pru WT (top), Pru Δgra15 (middle), or Pru Δgra15 + GRA15 (bottom) and right panel is for IFNγ‐stimulated HFFs infected with Pru WT (top), Pru Δgra15 (middle), or Pru Δgra15 + GRA15 (bottom). Scale bar is 100 μm.
-
DRepresentative images of the plaque assay described for Fig 1F. Left panel is for unstimulated HFFs infected with RH (top) or RH + GRA15II (bottom), and right panel is for IFNγ‐stimulated HFFs infected with RH (top) and RH + GRA15II (bottom). Scale bar is 100 μm.
-
ECells were supplemented with media containing 0.6 mM L‐Trp, stimulated with IFNγ for 24 h, and infected with RH or Pru parasites. Plaque number was measured 4 (RH) or 6 days (Pru) p.i. Experiments were performed two times.
-
FL‐Kynurenine was measured from HFF culture supernatant as a marker of IDO activity. Experiment was done two times.
-
GNumber of total host nuclei counted per experiment for Fig 1C and H for each indicated strains and conditions. Experiments were performed six times with RH and Pru WT and three times with RH + GRA15II and Pru Δgra15
-
HMeasurement of infectivity of Pru WT and Pru Δgra15 by plaque assay in unstimulated HFFs using equal number of parasites (250).
After invasion, Toxoplasma resides within the host cytosol in a PV and starts secreting GRAs into the PV lumen where they stay or get transported to the PVM or beyond the PVM into the host cell (Hakimi et al, 2017). The transport of GRAs beyond the PVM, but not onto the PVM, is mediated by a putative translocon containing the proteins MYR1/2/3 (Franco et al, 2016; Naor et al, 2018). We wanted to determine whether GRAs secreted beyond PVM affect susceptibility of Pru parasites to IFNγ. However, Pru and PruΔmyr1 parasites showed similar IFNγ‐mediated reductions in plaque number and area (Fig EV1B), indicating that these phenotypes are not influenced by GRAs secreted beyond the PVM. We therefore hypothesized that maybe a GRA in the PVM facing the host cytosol might be involved as these are not affected by MYR1. GRA15 is a polymorphic Toxoplasma effector protein present in the PVM that activates the NF‐κB transcription factor (Rosowski et al, 2011), a master regulator of cell signaling and cell death (Dutta et al, 2006), independent of MYR1 (Franco et al, 2016). The type I RH strain has an early stop codon in GRA15 leading to a nonfunctional GRA15 (Rosowski et al, 2011). To determine whether GRA15 plays a role in the susceptibility of Pru to IFNγ‐mediated growth inhibition, we infected IFNγ‐stimulated or naïve HFFs with Pru, PruΔgra15, and the PruΔgra15 strain complemented with an HA‐tagged copy of GRA15 (PruΔgra15 + GRA15) and measured plaque number and area after 6 days. PruΔgra15 parasites showed significantly less plaque number and area loss compared to wild‐type parasites in IFNγ‐stimulated HFFs (Figs 1E and EV1C). Complementation of Δgra15 parasites with GRA15 restored growth inhibition to wild‐type levels (Figs 1E and EV1C). RH parasites expressing type II GRA15 (RH + GRA15II) (Rosowski et al, 2011) showed significantly more plaque loss compared to the wild‐type RH strain (Figs 1F and EV1D). Furthermore, we observed that the enhanced GRA15‐mediated susceptibility of Pru to IFNγ was already apparent 24 h p.i. (Fig 1G). GRA15 also enhanced the IFNγ‐mediated elimination of vacuoles (Fig 1H and EV1E) but did not affect parasite infection of host cells (Fig EV1F).
It was recently shown that in immortalized HFFs, IFNγ‐induced IDO1 expression determines the IFNγ‐mediated growth inhibition of Toxoplasma. The MYR1‐dependent secreted Toxoplasma effector TgIST was shown to protect Toxoplasma from IDO1‐mediated growth inhibition in cells stimulated after infection by inhibiting STAT1‐mediated IDO1 expression but not in cells pre‐stimulated with IFNγ (Bando et al, 2018). In contrast, we previously showed that IDO‐mediated L‐Trp degradation only plays a minor role in inhibition of parasite growth in IFNγ‐stimulated primary HFFs (Niedelman et al, 2013). To rule out the role of IDO in the increased susceptibility of Pru parasites to IFNγ, we show that L‐Trp supplementation did not restore the reduction of plaque loss in RH and Pru strains (Fig EV1G). Furthermore, there is no difference in IDO activity in RH‐ vs. Pru‐infected IFNγ‐stimulated HFFs (Fig EV1H). Thus, in IFNγ‐stimulated HFFs parasite expression of GRA15 leads to reduced parasite growth and enhanced disappearance of vacuoles.
GRA15 enhances IFNγ‐induced endo‐lysosomal fusion with the vacuole
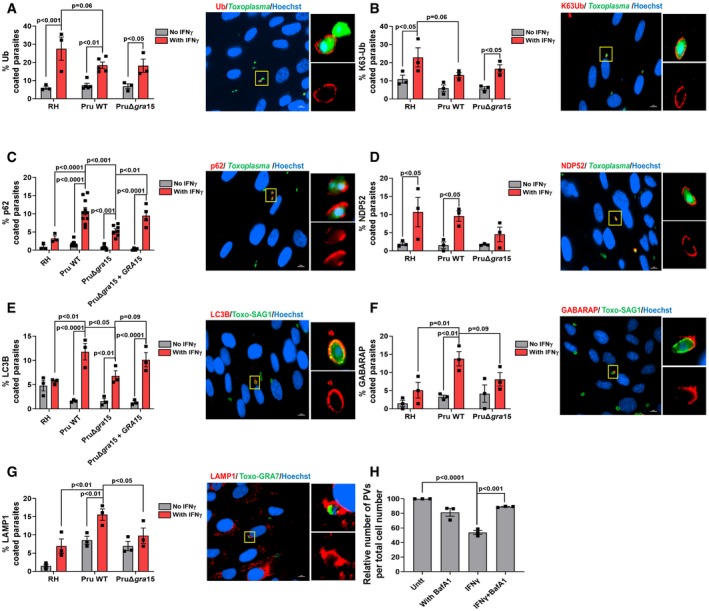
In HUVEC cells, ubiquitin, ubiquitin‐like proteins (LC3/GABARAP), and ubiquitin receptors (p62/NDP52) are recruited to the Toxoplasma PVM, eventually leading to its destruction by fusion with endo‐lysosomes (Clough et al, 2016). To determine whether this occurs in HFFs and the potential role of GRA15 in this process, we pre‐stimulated HFFs with IFNγ, infected cells with RH, Pru, or PruΔgra15 parasites, and measured accumulation of ubiquitin, p62, NDP52, LC3B, GABARAP, and LAMP1 around the PVM 3 h p.i. Surprisingly, and unlike what has been observed in HeLa, HUVEC, and murine cells (Haldar et al, 2015; Lee et al, 2015; Selleck et al, 2015; Clough et al, 2016), we observed that although PVMs of both RH and Pru strains were coated with ubiquitin in IFNγ‐stimulated HFFs, a larger fraction of RH vacuoles was coated (Fig 2A). We did not observe any difference in the ubiquitin coating intensity among the different parasite strains (Fig EV2A). Deletion of GRA15 had no effect on ubiquitination of Pru vacuoles (Fig 2A). The type of ubiquitin linkage recruited to the PVM can influence the subsequent outcome (Swatek & Komander, 2016). We observed K63‐linked, and no K48‐linked (Fig EV2B), ubiquitin localized to the PVM (Fig 2B). We observed a significantly larger fraction of Pru PVMs coated with p62 compared to RH (Fig 2C). Deletion of GRA15 from Pru resulted in significantly fewer PVMs coated with p62, while a similar fraction of PVMs of the GRA15‐complemented strains as wild‐type Pru were coated with p62 (Fig 2C). Unlike the type I RH strain, the type I GT1 strain contains a functional GRA15, which we previously showed determines RH vs. GT1 differences in activation of NF‐κB (Yang et al, 2013). Consistent with a role for GRA15 in mediating p62 PVM recruitment, a significantly larger fraction of the vacuoles from the RH + GRA15II and the GT1 strain stained positive for p62 compared to RH vacuoles (Fig EV2C). Additionally, we observed that GT1 was significantly more susceptible to IFNγ‐mediated parasite elimination compared to RH (Fig EV2D). A similar fraction of PVMs of RH and Pru vacuoles contained NDP52 in IFNγ‐stimulated HFFs (Fig 2D). However, deletion of GRA15 in Pru resulted in reduction of NDP52 recruitment to the PVM (Fig 2D). Like p62, both LC3B and GABARAP were recruited to a larger fraction of the PVs of Pru compared to RH (Fig 2E and F) in IFNγ‐stimulated HFFs. The PruΔgra15 strain had ~2‐fold less vacuoles that were coated with LC3B and GABARAP compared to wild‐type Pru (Fig 2E and F). The GRA15‐complemented strain had a larger fraction of PVs coated with LC3B compared to the GRA15‐deleted strain (Fig 2E). To determine whether the recruitment of LC3B, GABARAP, and p62 is associated with lysosomal destruction of the vacuole, we infected IFNγ‐stimulated HFFs and counted LAMP1‐positive vacuoles 3 h p.i. IFNγ enhanced the recruitment of LAMP1 to vacuoles of all strains, but significantly more LAMP1‐positive vacuoles were seen in Pru‐infected, compared to RH‐infected, cells. Deletion of GRA15 significantly reduced the number of LAMP1‐positive vacuoles (Fig 2G). In many LAMP1‐positive vacuoles, the parasites were distorted, and they often did not stain positive for GRA7, used as parasite PV marker. However, by using the DNA‐binding dye Hoechst, these vacuoles still clearly contained parasite DNA but were in advanced stages of parasite degradation (Fig EV3). The lysosomal inhibitor BafA1 significantly inhibited the disappearance of vacuoles in IFNγ‐stimulated cells (Fig 2H).
Figure 2. GRA15 enhances IFNγ‐induced PVM decoration with autophagy‐related proteins and endo‐lysosomal‐mediated vacuole destruction in HFFs.
-
A–GHFFs were stimulated with for 24 h with 10 U/ml IFNγ or left unstimulated and subsequently infected with RH, Pru, or PruΔgra15 parasites for 3 h. The percentage of vacuoles that stained positive for (A) total ubiquitin (n = 3 for RH, n = 5 for Pru, and n = 3 for PruΔgra15), (B) K63‐linked ubiquitin (n = 3 for RH, n = 3 for Pru, and n = 3 for PruΔgra15), (C) p62 (n = 3 for RH, n = 10 for Pru, n = 7 for PruΔgra15, and n = 4 for Pru Δgra15 + GRA15), (D) NDP52 (n = 3 for RH, n = 3 for Pru, and n = 3 for PruΔgra15), (E) LC3B (n = 3 for RH, n = 3 for Pru, n = 3 for PruΔgra15, and n = 3 for Pru Δgra15 + GRA15), (F) GABARAP (n = 3 for RH, n = 3 for Pru, and n = 3 for PruΔgra15), and (G) LAMP1 (n = 3 for RH, n = 3 for Pru, and n = 3 for PruΔgra15) is shown in the left bar diagram. On the right‐hand side, a representative fluorescent image is shown for the Toxoplasma Pru strain, which expresses GFP. DNA was stained with Hoechst 33258. Scale bar is 10 μm. The yellow box inside each representative image is shown as inset pictures with magnification.
-
HThe number of PVs per 20× objective field was counted and compared between IFNγ‐stimulated and IFNγ + bafilomycin A1 (100 nM)‐treated HFFs 24 h p.i. with Pru strain. Images from at least six fields were taken for each condition (n = 3).
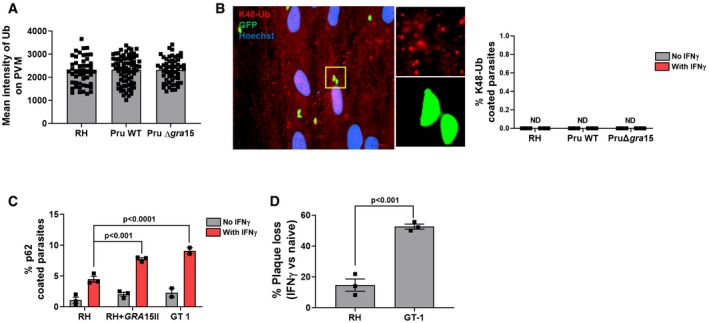
Figure EV2. PVM ubiquitination is K48‐linked ubiquitination‐independent, and the type I GT1 strain, which endogenously expresses GRA15, is more susceptible to IFNγ‐mediated parasite killing.
-
APVM ubiquitin coating intensity is similar between RH, Pru, and PruΔgra15. The measurement of fluorescence intensity of ubiquitin staining on the PVM was performed using NIS‐Elements software version 4 (Nikon) from the experiments described in Fig 2. For each strain type, intensities of at least 50 vacuoles were measured.
-
BHFFs were stimulated with for 24 h with 10 U/ml IFNγ or left unstimulated and subsequently infected with RH, Pru, or PruΔgra15 parasites for 3 h. The percentage of vacuoles that stained positive for K63‐linked ubiquitin (n = 3 for RH, n = 3 for Pru, and n = 3 for PruΔgra15) is shown in the right bar diagram. On the left‐hand side, a representative fluorescent image is shown for the Toxoplasma Pru strain, which expresses GFP. DNA was stained with Hoechst 33258. Scale bar is 10 μm.
-
Cp62 coating was performed as described in Fig 2, except here it is performed with RH, GT‐1, and RH + GRA15II. Experiment was performed three times with RH and RH + GRA15II and two times with GT‐1.
-
DPlaque assay was performed as described in Fig 1A. Plaques were counted 4 and 6 days p.i. for RH and GT‐1, respectively. Experiment was performed three times.
Figure EV3. IFNγ mediates parasite degradation through endo‐lysosomal fusion.
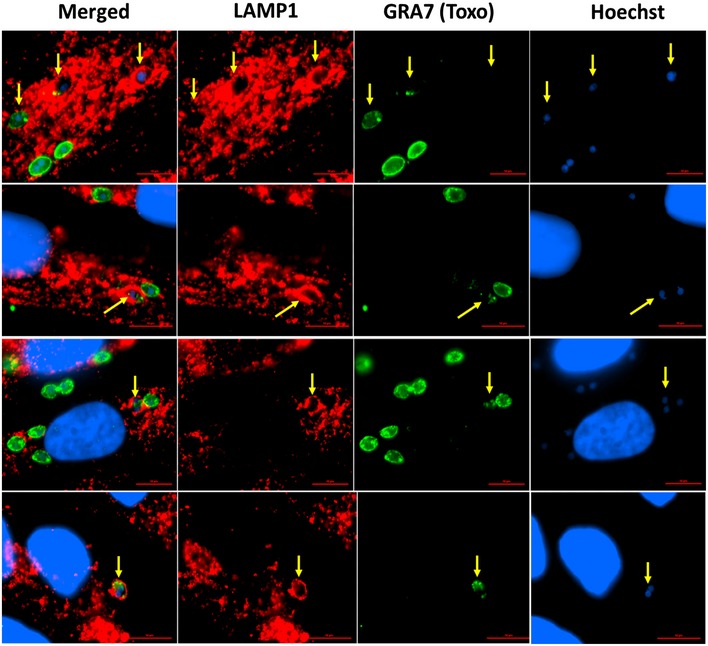
Panel of Pru‐infected HFFs that were previously stimulated with 10 U/ml of IFNγ shows different LAMP1 staining patterns characteristic of endo‐lysosomal fusion and parasite degradation. Cells were stimulated, infected, fixed, and stained as mentioned in Fig 3. Yellow arrows indicate the LAMP1‐coated parasites. Scale bar is 10 μm.
Overall, these results indicate that GRA15 enhances vacuole destruction via endo‐lysosomal fusion in IFNγ‐stimulated HFFs.
GRA15‐mediated enhancement of destruction of Pru vacuoles through endo‐lysosomal fusion in IFNγ‐stimulated HFFs correlates with PVM recruitment of p62, LC3B, GABARAP, and LAMP1 but not ubiquitin
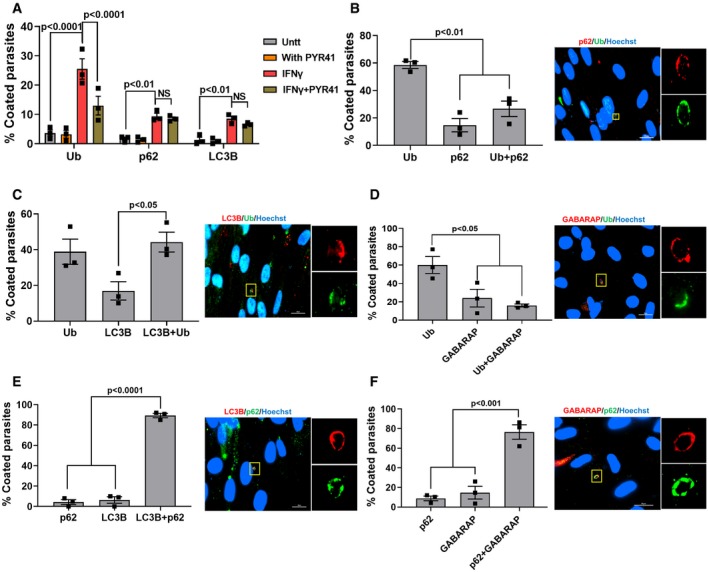
In IFNγ‐stimulated HUVEC cells, only type II strain PVMs are ubiquitinated and this ubiquitination is indispensable for subsequent endo‐lysosomal fusion and parasite elimination (Clough et al, 2016). However, in HFFs, strain differences in ubiquitination of the PVM (Fig 2A and B) did not correlate with recruitment of p62, LC3B, GABARAP, and LAMP1 to the PVM (Fig 2C–G). Furthermore, when we inhibited ubiquitination using a specific inhibitor of E1 ubiquitin‐activating enzymes (PYR 41), we observed a significant reduction in the fraction of PVMs coated with ubiquitin but no effect on the fraction of PVMs coated with p62 or LC3B (Fig 3A). Thus, the fraction of PVMs containing p62 or LC3B does not correlate with the fraction of PVMs containing ubiquitin. Consistent with this, only 25% of the vacuoles were coated with both ubiquitin and p62, only 40% of the vacuoles were coated with both ubiquitin and LC3B, and only 20% of the vacuoles were coated with ubiquitin and GABARAP (Fig 3B–D). In contrast, 90% of the vacuoles were coated with p62 and LC3B (Fig 3E) and 78% with p62 and GABARAP (Fig 3F). Given that ~20% of the vacuoles are coated with only p62 and ~60% are only Ub‐positive (Fig 3B), these data suggest that recruitment of p62 to the PVM is not dependent on PVM ubiquitination.
Figure 3. PVM decoration with autophagy adaptors correlates with p62 but not ubiquitination.
-
AHFFs were stimulated with IFNγ (10 U/ml for 24 h) or left unstimulated and subsequently treated with PYR41 (1 μM) for 2 h prior to infection. Cells were washed and subsequently infected with Pru parasites (MOI = 3) for 3 h. The percentage of vacuoles that stained positive for total ubiquitin, p62, or LC3B was determined (n = 3).
-
B–FFor all the co‐staining experiments, IFNγ‐stimulated HFFs infected for 3 h with the Pru strain were used. For each staining, at least 50 vacuoles were scored (n = 3). On the right‐hand side, a representative fluorescent image is shown for the Toxoplasma Pru strain. Scale bar is 20 μm. The yellow box inside each representative image is shown as inset pictures with higher magnification. The total number of coated vacuoles was set at 100%, and the percentage of vacuoles positive for Ub, p62, and/or LC3B/GABARAP was calculated.
GRA15 binds TRAFs and recruits TRAF6 and ubiquitin‐like molecules and receptors to the parasitophorous vacuole to mediate parasite elimination
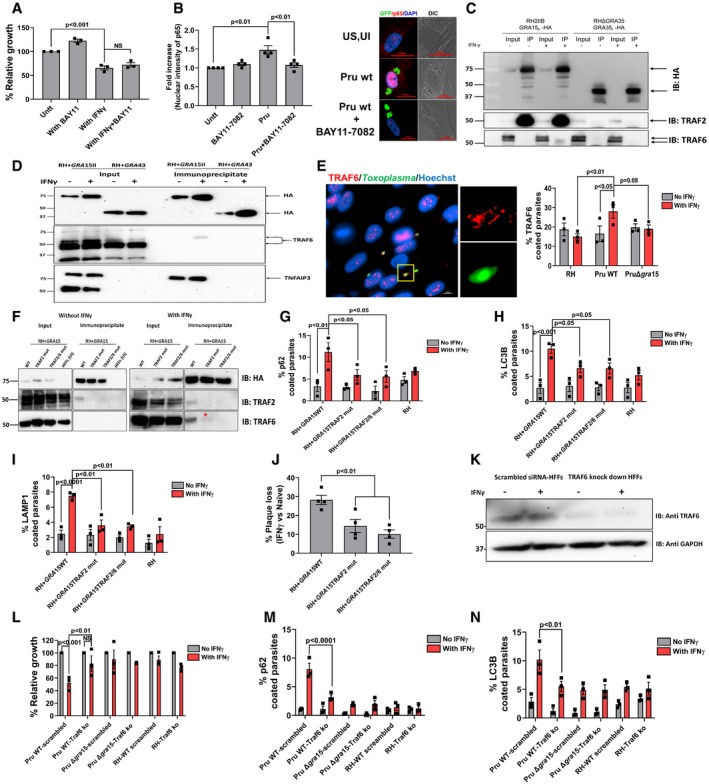
We wanted to determine whether GRA15 inhibits parasite growth in IFNγ‐stimulated HFFs through its ability to activate the NF‐κB transcription factor (Rosowski et al, 2011). To test this, we treated HFFs with BAY11‐7082, a known inhibitor of NF‐κB activation (García et al, 2005), 2 h before infection and 24 h p.i. measured parasite growth. Addition of BAY11‐7082 (1 μM) did not restore parasite growth in IFNγ‐stimulated HFFs (Fig 4A). At this concentration, BAY11‐7082 can successfully inhibit the activation of NF‐κB triggered by Pru parasites (Fig 4B). BAY11‐7082 was reported to inhibit NF‐κB activation via inhibition of E2 ubiquitin‐conjugating enzymes that mediate K63‐linked and linear polyubiquitin chains (Strickson et al, 2013). We therefore also counted the percentage of PVs coated with ubiquitin or p62 in IFNγ‐stimulated HFFs and observed that although treatment with BAY11‐7082 decreased the fraction of PVs that were coated with ubiquitin, it had no effect on the fraction of PVs coated with p62 (Fig EV4A).
Figure 4. GRA15‐mediated parasite growth reduction in IFNγ‐stimulated HFFs is independent of its ability to activate NF‐κB but dependent on GRA15's interaction with the E3 ubiquitin ligases TRAF2 and TRAF6.
-
AHFFs were stimulated with IFNγ for 24 h (10 U/ml) or left unstimulated. The NF‐κB inhibitor BAY11‐7082 (1 μM) was added 2 h pre‐infection, and HFFs were subsequently infected with Pru parasites. Parasite growth (using luciferase assay) was measured 24 h p.i. Means from unstimulated cells were set at 100%. Experiments were performed three times.
-
BNuclear translocation of the NF‐κB p65 subunit was quantified in HFFs or HFFs treated with BAY11‐7082 (1 μM, added 2 h pre‐infection) 24 h p.i. with Pru parasites. Experiments were done four times where each dot represents one experimental mean of at least 15 nuclei. In the right panel, representative images are shown. Parasites were expressing GFP, and nuclei are stained with Hoechst 33258. Scale bar is 20 μm.
-
CImmunoprecipitation and Western blot were performed in HFFs with and without IFNγ (10 U/ml) using an RH strain expressing type II GRA15‐HA and as a control RH expressing GRA35‐HA. The blots using antibodies against TRAF2 and TRAF6 were made after stripping the first blot. The inputs loaded represent 1% of total lysate prepared for immunoblotting and mass spectrometry (Table EV1). The antibodies against TRAF2 and TRAF6 were obtained from Santa Cruz Biotechnology (Appendix Table S2). Full‐length blots for this figure can be observed in the source data for this figure.
-
DImmunoprecipitation and Western blot were performed in HFFs with and without IFNγ (10 U/ml) using an RH strain expressing type II GRA15‐HA and as a control RH expressing GRA43‐HA. The blots using antibodies against TRAF6 were made after stripping the first blot that is used for HA blotting. The inputs loaded represent 10% of total lysate prepared for immunoblotting. Following TRAF6 blotting, the membrane was stripped and blotted for TNFAIP3. Full‐length blots for this figure can be observed in the source data for this figure.
-
EImmunofluorescence analysis of TRAF6 recruitment to the PVM of HFFs infected for 3 h with RH, Pru, and PruΔgra15 strains. On the right‐hand side, a representative fluorescent image is shown of TRAF6 recruitment to the PVM where Toxoplasma Pru strain expresses GFP, and DNA was stained with Hoechst 33258. Scale bar is 10 μm. n = 3 for all the strains. The antibody against TRAF6 was purchased from Abnova (Appendix Table S2).
-
FImmunoprecipitation and Western blot were performed in HFFs with and without IFNγ (10 U/ml) using a RH + GRA15WT, RH + GRA15TRAF2mut, or RH + GRA15TRAF2/6mut. For the unstimulated cells, uninfected HFFs were used an additional negative control (left panel). Left panel and right panel were run on a single gel; vertical white lines indicate excision of irrelevant lanes. Full‐length blots are in the source data for this figure. The antibodies used against TRAF2 and TRAF6 were purchased from Cell Signaling Technology and Abnova, respectively. The asterisks (*) in the lower right panel indicates the faint band of TRAF6 in the RH + GRA15TRAF2mut immunoprecipitate.
-
G–IImmunofluorescence analysis of p62, LC3B, and LAMP1 with and without IFNγ in RH and RH + GRA15WT. All the experiments were done three times with each of the strains.
-
JPlaque assays were performed with RH + GRA15WT, RH + GRA15TRAF2mut, or RH + GRA15TRAF2/6mut (n = 3).
-
KExpression of TRAF6 was detected by Western blotting of lysates from scrambled siRNA‐transfected and TRAF6‐specific siRNA‐transfected HFFs with and without IFNγ.
-
LRelative parasite growth was measured in scrambled siRNA‐transfected HFFs and TRAF6 knockdown HFFs using luciferase‐based assay with indicated strains with and without IFNγ (n = 3). The antibody against TRAF6 used here was from Abcam (Appendix Table S2). Full‐length blots are in source data available online for this figure.
-
M, NImmunofluorescence analysis of p62 and LC3B was done in scrambled siRNA‐transfected HFFs and TRAF6 knockdown HFFs with and without IFNγ using indicated strains (n = 3).
Figure EV4. PVM coating with ubiquitin does not correlate with p62 recruitment.

-
AHFFs in 24‐well plates were stimulated with IFNγ (10 U/ml for 24 h) and subsequently treated with E2 ubiquitin‐conjugating enzyme inhibitor, BAY11‐7082 (1 μM) for 2 h, and subsequently infected with Pru parasites for 3 h. The percentage of vacuoles that stained positive for ubiquitin and p62 was scored. Experiment was performed three times.
-
BSimilar to (A), 24 h post‐infection cells were treated with a TRAF6 (E3) ubiquitin ligase inhibitor, C25–140 (50 μM) for 2 h, and subsequently infected with Pru parasites for 3 h. The percentage of vacuoles that stained positive for K‐63‐linked Ub and p62 was determined. Experiment was performed three times.
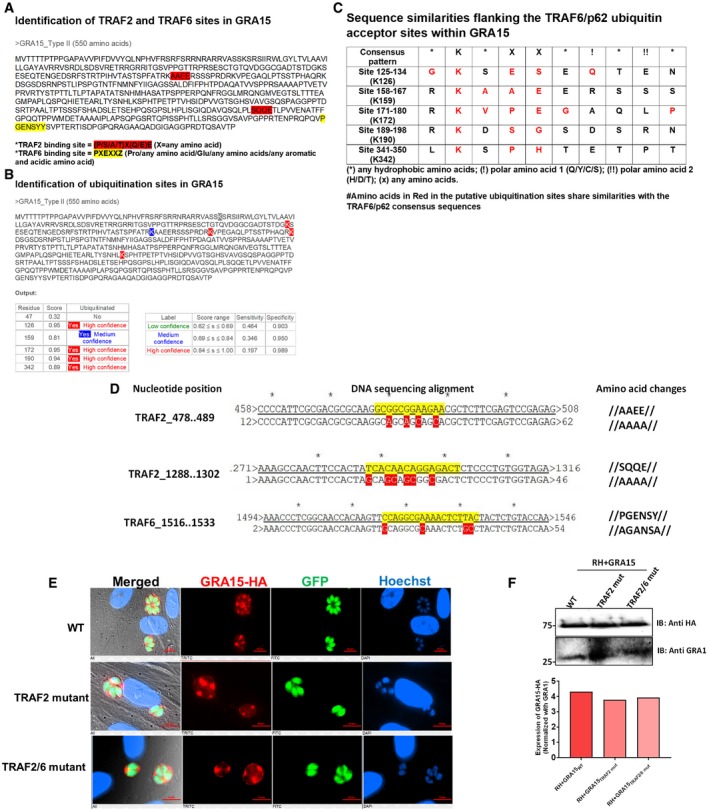
To identify the mechanism by which GRA15 enhanced the recruitment of ubiquitin‐like molecules and ubiquitin receptors to the PVM, we immunoprecipitated GRA15 from naive and IFNγ‐stimulated HFFs 8 h p.i. As a control, we immunoprecipitated GRA35, which we and others have recently shown to be a PVM‐localized GRA (Nadipuram et al, 2016; Wang et al, 2019b). GRA15 specifically immunoprecipitated multiple ubiquitin ligases, TRAF1, TRAF2, BIRC2, BIRC3, and TNFAIP3 (also named A20), while in IFNγ‐stimulated cells, also polyubiquitin and TRAF6 were immunoprecipitated (Table EV1). To confirm some of these results, we immunoprecipitated GRA15 and blotted for TRAF2 and TRAF6. Indeed, GRA15, but not GRA35, immunoprecipitated TRAF2 in both stimulated and unstimulated HFFs, while a small amount of TRAF6 was only immunoprecipitated in IFNγ‐stimulated HFFs (Fig 4C). We repeated the GRA15 immunoprecipitation using GRA43 as a control (Fig 4D) and blotted for TRAF6 and TNFAIP3 (A20). GRA15, but not GRA43, immunoprecipitated TRAF6 only in IFNγ‐stimulated HFFs, whereas TNFAIP3 (A20) was immunoprecipitated with GRA15 in both stimulated and unstimulated HFFs (Fig 4D). Interestingly, TNFAIP3 was only detected in GRA15‐infected cells, likely because it is known to be upregulated by NF‐κB. We also observed a significantly larger fraction of Pru PVs coated with TRAF6 compared to RH PVs in IFNγ‐stimulated HFFs (Fig 4E). PruΔgra15 PVMs had less recruitment of TRAF6 compared to the wild‐type Pru strain (Fig 4E). However, this antibody seemed to have quite some background staining. TRAF6 is a ring‐type E3 ubiquitin ligase and catalyzes the formation of K63‐linked ubiquitination (Deng et al, 2000), and was reported to be associated with ubiquitination of PVs in murine cells (Haldar et al, 2015). Furthermore, TRAF6, but not TRAF2, has a p62‐binding domain (Jadhav et al, 2008, 2011). We observed that although inhibition of TRAF6 ubiquitin ligase activity significantly lowered K63‐linked ubiquitination on the PVM, this had no effect on recruitment of p62 to the PVM (Fig EV4B). Because inhibiting ubiquitin‐activating (E1), ubiquitin‐conjugating (E2), and ubiquitin ligase (E3) activity did not affect p62 recruitment, if TRAF6 has a role in vacuole destruction, it is unlikely to be the ubiquitination of the PVM but more likely the recruitment of p62 via its p62‐binding domain. Recently, we have reported that activation of NF‐κB by GRA15 depends on in its interaction with TRAF2 as well as TRAF6 in HEK cells (Sangaré et al, 2019). GRA15 has two binding motifs for TRAF2 and one for TRAF6 (Fig EV5A) and contains at least four high‐confidence ubiquitination sites (Fig EV5B). However, none of the ubiquitination sites have the consensus downstream p62‐TRAF6‐binding sites (Fig EV5C). To directly determine the relevance of the GRA15 interaction with TRAF2 and TRAF6 at the PVM, we generated RH parasites that expressed either HA‐tagged wild‐type GRA15 (RH + GRA15WT) or GRA15 with mutated TRAF2‐ (RH + GRA15TRAF2mut) or TRAF2‐ and TRAF6‐binding sites (RH + GRA15TRAF2/6mut). Immunofluorescence and Western blot analysis showed that GRA15 had a similar PV/PVM localization in RH + GRA15WT, RH + GRA15TRAF2mut, and RH + GRA15TRAF2/6mut and had similar expression levels in these parasites (Fig EV5E and F). We immunoprecipitated GRA15 from these different parasites from lysates generated from naive or IFNγ‐stimulated infected HFFs 8 h p. i. Immunoprecipitated GRA15WT pulled down TRAF2 from infected HFF lysates, while no TRAF2 was pulled down after immunoprecipitation of GRA15TRAF2mut or RH + GRA15TRAF2/6mut. In the lysate from IFNγ‐stimulated infected HFFs, GRA15WT pulled down TRAF2 and TRAF6, but no TRAF2 or TRAF6 was pulled down after immunoprecipitation of GRA15TRAF2/6mut and no TRAF2 and a very small amount of TRAF6 was pulled down after immunoprecipitation of GRA15TRAF2mut. These data confirmed that mutation of the respective TRAF2/6‐binding sites indeed abrogated the binding to TRAF2 or to TRAF2 and TRAF6 (Figs 4F and EV5D). The fact that less TRAF6 was immunoprecipitated with the GRA15TRAF2mut compared to GRA15WT could indicate that TRAF6 was coming down via binding to TRAF2 (Davies et al, 2005). We observed that significantly more PVs of RH + GRA15WT contained p62 (Fig 4G), LC3B (Fig 4H), and LAMP1 (Fig 4I) compared to the PVs of RH + GRA15TRAF2mut parasites. Recruitment of p62, LC3B, and LAMP1 to the PVM of RH + GRA15TRAF2mut and RH + GRA15TRAF2/6mut parasites was similar, indicating that this recruitment was mainly mediated via the GRA15 TRAF2‐binding sites (Fig 4G–I). Furthermore, IFNγ‐mediated killing of RH + GRA15WT was significantly higher compared to RH + GRA15TRAF2mut or RH + GRA15TRAF2/6mut parasites (Fig 4J).
Figure EV5. GRA15 possesses TRAF2‐ and TRAF6‐binding motifs but not ubiquitinated TRAF6‐p62 acceptor sites.
-
AIdentification of putative TRAF2‐ and TRAF6‐binding sites in type II GRA15 protein. The amino acid sequence of type II GRA15 was derived from ToxoDB (https://toxodb.org/toxo/). TRAF2‐binding sites are highlighted in red, while TRAF6‐binding site is highlighted in yellow (Sangaré et al, 2019).
-
BUbiquitination sites within type II GRA15 sequence were identified using http://www.ubpred.org/, and the score provided was also derived from the same online tool.
- C
-
DResults from the DNA sequencing of the RH expressing GRA15 TRAF2‐binding mutants or TRAF2/6‐binding mutants. In the right column, the nucleotide positions of the displayed sequence read are mentioned; in the middle column, the exact sequence was shown; and in the right column, the corresponding changes in the amino acids are also mentioned.
-
EThe localization of HA‐tagged GRA15 in RH expressing either GRA15 wild‐type, GRA15 TRAF2‐binding mutant, or GRA15 TRAF2/6‐binding mutants, shown using immunofluorescence using anti‐HA antibody. Scale bar is 10 μm.
-
FThe expression of GRA15 among the RH strains expressing either GRA15 wild‐type, GRA15 TRAF2‐binding mutant, or GRA15 TRAF2/6‐binding mutants (upper panel). Expression was normalized to the GRA1 expression of the parasites and plotted (lower panel).
Because TRAF2 does not contain a p62‐binding domain, we hypothesized that its main role was the recruitment of TRAF6. To determine the role of TRAF6, we knocked down TRAF6 in HFFs, using a stable lentivirus‐mediated siRNA system (Fig 4K). The growth of Pru parasites in the presence of IFNγ in these TRAF6 knockdown HFFs was significantly restored compared to scrambled siRNA‐transfected HFFs (Fig 4L), whereas both RH and PruΔgra15 were resistant in both cell types (Fig 4L). Furthermore, recruitment of p62 and LC3B in IFNγ‐stimulated cells was significantly less in TRAF6 knockdown HFFs compared to scrambled siRNA‐transfected HFFs (Fig 4M and N). These results indicate that GRA15 by binding with ubiquitin ligases TRAF2 and TRAF6 mediates the susceptibility of type II Pru strains in IFNγ‐stimulated primary human fibroblasts.
GRA15 mediates susceptibility of type II parasites to IFNγ‐mediated killing in murine fibroblasts by binding to TRAF6
Our results show that GRA15 mediates Pru susceptibility to IFNγ in HFFs by enhancing lysosomal destruction of the vacuole. In IFNγ‐stimulated MEFs, type II GRA15 plays a role in recruitment of GBPs to the PVM via an unknown mechanism (Virreira Winter et al, 2011; Fisch et al, 2019). In IFNγ‐stimulated MEFs, GBP recruitment is initiated by PVM recruitment of so‐called “pioneer” IRGs such as IRGB6 (Haldar et al, 2015). In MEF recruitment of TRAF6 and TRIM21, two ubiquitin ligases, to the PVM enhances subsequent recruitment of IRGs and GBPs that target the vacuole for degradation (Haldar et al, 2015; Foltz et al, 2017). We hypothesized that TRAF6 recruitment by GRA15 might mediate the recruitment of GBPs and IRGs as TRAF6 has a p62‐binding motif and p62 has an LC3‐interacting region (LIR) motif that can mediate recruitment of LC3B and GABARAP (Lamark et al, 2017). To determine the role of GRA15 in MEFs, we infected IFNγ‐stimulated MEFs and measured PV recruitment of TRAF6, IRGB6, ubiquitin (K63 and K48), GBPs, and LC3B. As demonstrated by others (Haldar et al, 2015; Foltz et al, 2017), Pru parasites had a significantly larger fraction of its vacuoles coated with these markers compared to RH parasites. This enhanced PVM recruitment was partially mediated by GRA15 as we observed significantly fewer PVs coated with these markers in PruΔgra15 parasites (Fig 5A–F). In contrast to what we observed in HFFs, significantly more Pru vacuoles were coated with ubiquitin, predominantly K63‐linked, in IFNγ‐stimulated MEFs compared to RH vacuoles, which is in agreement with previous studies (Haldar et al, 2015; Foltz et al, 2017). We determined parasite growth in IFNγ‐stimulated MEFs 24 h p.i. and observed that PruΔgra15 had significantly less IFNγ‐mediated growth reduction compared to either wild‐type or GRA15‐complemented parasites (Fig 5G). PruΔgra15 also showed significantly less plaque loss compared to its parental or complemented strains (Fig 5H). RH expressing type II GRA15 formed less and smaller plaques compared to wild‐type RH in IFNγ‐stimulated MEFs (Fig 5I–J).
Figure 5. GRA15‐enhanced susceptibility of type II strains to IFNγ‐mediated killing in MEFs correlates with enhanced recruitment of IRGB6, GBPs, Ubiquitin, and LC3B.

-
A–FIFNγ‐stimulated MEFs were infected with Pru, PruΔgra15, or RH for 3 h and subsequently fixed, permeabilized, and stained for (A) IRGB6, (B) GBPs, (C) total ubiquitin, (D) K63‐linked ubiquitin, (E) K48‐linked ubiquitin, and (F) LC3B. For analysis, at least 100 vacuoles were scored. All experiments were performed three times. On the right‐hand side, a representative fluorescent image is shown for the Toxoplasma Pru strain, which expresses GFP. DNA was stained with Hoechst 33258. Scale bar is 10 μm. The yellow box inside each representative image is shown as an inset picture with magnification. All experiments were performed three independent times.
-
GMEFs were stimulated with IFNγ for 24 h (100 U/ml) or left unstimulated and subsequently infected with Pru, PruΔgra15, and GRA15‐complemented parasites for 24 h for measuring parasite growth by luciferase assay. All experiments were performed three independent times.
-
HPlaque numbers were measured for Pru (n = 12), PruΔgra15 (n = 12), and GRA15‐complemented parasites (n = 6).
-
I, JMEFs were stimulated with IFNγ for 24 h (100 U/ml) or left unstimulated and subsequently infected with RH or RH expressing type II GRA15. Plaque numbers were counted (n = 3), and areas were measured 4 days p.i. (n = 3).
To establish that the GRA15‐enhanced recruitment of these markers to the vacuole could be mediated by its interaction with TRAF6, we immunoprecipitated GRA15 from naïve and IFNγ‐stimulated MEFs and performed Western blot for TRAF2 and TRAF6. We observed that akin to HFFs, GRA15 in MEFs also binds TRAF2 and TRAF6 (Fig 6A). We compared recruitment of TRAF6 to PVs of RH, Pru, and PruΔgra15 in MEFs. Akin to HFFs, we observed that in IFNγ‐stimulated MEFs, significantly more Pru PVs contained TRAF6 (threefold increase) compared to either RH or PruΔgra15 PVs (Fig 6B). Significantly fewer Pru PVs were coated (threefold decrease) with IRGB6 in Traf6 ‐/‐ MEFs compared to wild‐type MEFs (Fig 6C). Furthermore, the difference in IRGB6 coating between Pru and PruΔgra15 PVMs in wild‐type MEFs disappeared in Traf6 −/− MEFs (Fig 6C). We also measured the growth of Pru, PruΔgra15, and GRA15‐complemented parasites in wild‐type, Traf6 −/−, and NF‐κB p65 −/− MEFs 24 h p.i. In wild‐type and NF‐κB p65 −/− MEFs, both Pru and GRA15‐complemented parasites showed a significant growth reduction upon IFNγ‐stimulation (Fig 6D). In contrast, in Traf6 −/− MEFs, the action of IFNγ was abolished and GRA15‐expressing parasites no longer showed reduced growth compared to the PruΔgra15 strain, which was resistant to IFNγ in all MEFs (Fig 6D). To further establish the role of GRA15‐mediated TRAF6 binding in parasite growth inhibition in IFNγ‐stimulated MEFs, we infected MEFs with RH, RH + GRA15WT, RH + GRA15TRAF2mut, or RH + GRA15TRAF2/6mut and enumerated the vacuoles with recruitment of IRGB6, GBP1‐5, p62, TRAF6, and ubiquitin. By immunoprecipitation of GRA15 and Western blotting for TRAF2 and TRAF6, we demonstrated that these mutants are indeed unable to bind TRAF2 or TRAF2 and TRAF6 (Fig 6E). Furthermore, as expected, the RH + GRA15TRAF2/6mut strain no longer recruited TRAF6 to the vacuole while TRAF6 recruitment to the PVM of RH + GRA15TRAF2mut was similar to RH + GRA15WT (Fig 6F). The reduced recruitment of TRAF6 to the PVM of the RH + GRA15TRAF2/6mut strain was associated with a reduced recruitment of IRGB6, GBP1‐5, and p62 (Fig 6G–I). In contrast, mutating the TRAF2‐ or the TRAF2‐ and TRAF6‐binding site on GRA15 did not affect ubiquitin coating of the PVM (Fig 6J), suggesting that although GRA15 expression is associated with increased ubiquitin coating (Fig 5C and D), this is not mediated by binding of TRAF6 or TRAF2 to GRA15. The RH + GRA15TRAF2/6mut strain was also less susceptible to IFNγ compared to either the RH + GRA15WT or RH + GRA15TRAF2mut strain as indicated by reduced plaque loss upon IFNγ‐stimulation (Fig 6K).
Figure 6. GRA15‐enhanced susceptibility of the type II Pru strain in IFNγ‐stimulated MEFs is dependent on TRAF6.

-
AImmunoprecipitations and Western blots were performed on MEFs stimulated or not with IFNγ (100 U/ml) and infected with an RH strain expressing type II GRA15‐HA and as a control RH expressing GRA45‐HA. The blots using antibodies against TRAF6 and TRAF2 were made after stripping the first blot. Left panel and right panel were run on a single gel; vertical white lines indicate excision of irrelevant lanes. Full‐length blots are in the source data for this figure. The antibodies used against TRAF2 and TRAF6 were purchased from Cell Signaling Technology and Abnova, respectively.
-
BIFNγ‐stimulated MEFs were infected with Pru, PruΔgra15, and RH for 3 h and subsequently stained for TRAF6. On the right‐hand side, a representative fluorescent image is shown for the Toxoplasma Pru strain, which expresses GFP. DNA was stained with Hoechst 33258. Scale bar is 10 μm. The yellow box inside each representative image is shown as an inset picture with magnification. Experiments were performed three times.
-
CIRGB6 staining on PVM after infection of wild‐type or Traf6 −/− MEFs for 3 h with Pru, PruΔgra15, and RH. On the right‐hand side, a representative fluorescent image is shown of IRGB6 coating on the PVM of both wild‐type and Traf6 −/− MEF. Scale bar is 10 μm. At least 100 different vacuoles were observed and analyzed for each experiment (n = 3).
-
DParasite growth was measured 24 h p.i. using luciferase readout from unstimulated and IFNγ‐stimulated wild‐type, NF‐κB p65 −/−, and Traf6 −/− MEFs. Growth was compared between Pru, PruΔgra15, and PruΔgra15 + GRA15 (complemented) strains. Reading from unstimulated cells was considered as 100%, and percentage growth in IFNγ‐stimulated cells was expressed relative to unstimulated cells. Experiments were performed three times with each MEF type.
-
EImmunoprecipitation and Western blot were performed in MEFs infected with RH + GRA15WT, RH + GRA15TRAF2mut, or RH + GRA15TRAF2/6mut and as a control RH expressing GRA45‐HA. Left panel and right panel were run on a single gel; vertical white lines indicate excision of irrelevant lanes. Full‐length blots are in the source data for this figure. The antibodies used against TRAF2 and TRAF6 were purchased from Cell Signaling Technology and Abnova, respectively.
-
F–JImmunofluorescence analysis of TRAF6, IRGB6, GBP1‐5, p62, and ubiquitin was done in IFNγ‐stimulated MEFs infected with RH + GRA15WT, RH + GRA15TRAF2mut, or RH + GRA15TRAF2/6mut (n = 3).
-
KPlaque assays were performed with RH + GRA15WT, RH + GRA15TRAF2mut, or RH + GRA15TRAF2/6mut (n = 4).
Thus, GRA15, by recruiting TRAF6 in IFNγ‐stimulated cells, and not by activating NF‐κB p65, enhanced parasite susceptibility to IRG/GBP‐dependent elimination in MEFs.
Discussion
Toxoplasma strain‐dependent susceptibility to IFNγ in different human and murine cells in vitro is well established (Niedelman et al, 2012, 2013; Selleck et al, 2013, 2015; Haldar et al, 2015; Clough et al, 2016; Qin et al, 2017; Bando et al, 2018). However, Toxoplasma effectors that affect strain differences in susceptibility to IFNγ‐induced cell‐autonomous immunity in human cells have not been described. We show that in IFNγ‐stimulated primary HFFs, the Toxoplasma effector GRA15 (Jensen et al, 2011; Rosowski et al, 2011) mediates the recruitment of the E3 ubiquitin ligase TRAF6, its binding partner p62 and LC3B and GABARAPs, eventually leading to endo‐lysosomal fusion with the vacuole and parasite elimination.
It was previously shown that in murine cells, Toxoplasma GRA15 enhances the recruitment of GBP1‐5 to the vacuole (Virreira Winter et al, 2011; Fisch et al, 2019). However, it was a mystery how GRA15 mediated this recruitment of GBP1‐5 as its only known function was the activation of the NF‐κB transcription factor, which does not take place until four hours after infection, while its effect on GBP recruitment can be seen within 1 h (Rosowski et al, 2011; Virreira Winter et al, 2011). Here, we solve this mystery by showing that in both HFFs and MEFs, GRA15 enhances the recruitment of TRAF6 to the PVM. TRAF6 was previously shown to be required for subsequent recruitment of p62, further ubiquitination of the vacuole, and its eventual destruction by IRGs and GBPs (Haldar et al, 2015). We confirmed those data and additionally showed that in Traf6 −/− MEFs, the difference in susceptibility between Pru and PruΔgra15 disappears further confirming that the effect of GRA15 is mediated through TRAF6. Thus, GRA15 enhances IFNγ‐mediated parasite elimination in both HFFs and MEFs although the exact mechanism of vacuole elimination is different.
Previously, it was shown that ubiquitination of the PVM is a strictly strain‐dependent phenomenon (Haldar et al, 2015; Lee et al, 2015; Selleck et al, 2015; Clough et al, 2016) where initial ubiquitination recruits p62 which further recruits the E3 ubiquitin ligases TRAF6 and TRIM21 to generate an amplification loop that recruits further p62 and LC3 eventually controlling parasite growth (Haldar et al, 2015; Foltz et al, 2017). However, our data show that in contrast to what has been observed in MEFs, PVM ubiquitination is not strain‐specific in HFFs, inhibition of PVM ubiquitination has no effect on p62 or LC3B recruitment, and a significant fraction of p62‐coated PVMs do not contain ubiquitin. The type I RH strain is much less susceptible to IFNγ‐mediated elimination despite having robust PVM ubiquitination, probably because it does not express a functional GRA15 or maybe it has unknown effectors that block the events downstream of ubiquitination. Also, in MEFs infected with RH or RH parasites expressing wild‐type GRA15 or GRA15 TRAF‐binding mutants, the recruitment of ubiquitin to the PVM did not correlate with recruitment of TRAF6, p62, IRGB6, or GBPs. Our data suggest that the PVM‐localized GRA15 parasite effector recruits TRAF6, which then likely further recruits adaptor proteins that in other cell types appear to be recruited by PVM ubiquitination (Selleck et al, 2015; Clough et al, 2016). In HeLa cells, the recruitment of p62 and other markers caused parasite growth stunting but no vacuole destruction (Selleck et al, 2015), whereas in HUVEC cells, vacuole destruction by endo‐lysosomal fusion was described to be ubiquitin‐ and p62‐dependent (Clough et al, 2016). In HFFs, we showed that p62 is recruited to the PVM, possibly via its TRAF6‐binding domain, and parasite vacuole destruction occurs through endo‐lysosomal fusion. What determines the differences between these different cell types is currently unknown. It is also still unclear why the TRAF‐dependent p62 recruitment to PVs is IFNγ‐inducible as neither the TRAFs nor p62 are induced by IFNγ.
Although in MEFs GRA15 significantly increased the percentage of vacuoles targeted by IRGB6, vacuoles of the PruΔgra15 strain were still significantly more targeted compared to the RH strain. This is likely due to strain differences in ROP5, as we and others have previously shown that type II strains have ROP5 alleles that are less effective at inhibiting IRG loading onto the PVM and that expression of type I or type III ROP5 alleles significantly reduces coating of type II vacuoles with IRGB6 (Virreira Winter et al, 2011; Fleckenstein et al, 2012; Niedelman et al, 2012; Etheridge et al, 2014). Similarly, ROP16 also plays an important role in IFNγ‐mediated parasite control in murine cells (Virreira Winter et al, 2011; Haldar et al, 2015). It is likely that in human cells, other parasite virulence factors, besides GRA15, likely determine susceptibility to IFNγ‐mediated elimination. A systematic identification of such factors could be possible through genetic screens using CRISPR‐Cas9 approaches (Wang et al, 2019c).
GRA15 also likely explains the difference in IRG coating of vacuoles of different type I strains. For example, the GT1 type I strain has a functional GRA15, while the RH type I strain does not. We have previously shown that GT1 GRA15 can activate the NF‐κB pathway and that vacuoles of the GT1 strain are significantly more coated with IRGs compared to RH vacuoles (Yang et al, 2013). Although both RH and GT1 have a lethal dose of just a single parasite in most laboratory mouse strains, deletion of ROP18 makes GT1 avirulent after low‐dose infection (Shen et al, 2014) while deletion of ROP18 in RH only delays death of the mice (Alaganan et al, 2014; Shen et al, 2014). It is likely that this difference is mediated by GRA15 as the presence of GRA15 in GT1 would make it more susceptible to IRGB6 PVM coating and subsequent parasite elimination.
Other studies have shown that IFNγ‐dependent induction of IDO expression plays a role in inhibiting Toxoplasma growth in human fibroblasts (Pfefferkorn, 1984; Pfefferkorn et al, 1986; Bando et al, 2018). However, we find that inhibition of parasite growth is minimally dependent on IFNγ‐dependent L‐Trp breakdown (this study and Niedelman et al, 2013). These differences might be due to fibroblasts derived from different tissues and/or the use of primary vs. transformed fibroblast (Bando et al, 2018), as the origin of the fibroblasts in other studies is unclear (Pfefferkorn, 1984; Pfefferkorn et al, 1986).
Thus, although we previously thought that the main role of GRA15 was the activation of the NF‐κB pathway, it is possible that its primary role is reducing parasite virulence by enhancing IFNγ‐mediated vacuole destruction. Furthermore, it was recently shown that GRA15 promotes the type I interferon response in mice by mediating STING polyubiquitination and enhanced cGAS/STING signaling through its ability to bind TRAF molecules (Wang et al, 2019a). Taken together, by observing the detrimental effect of GRA15 on the parasite, it might seem disadvantageous for Toxoplasma to have GRA15. However, the rapidly replicating tachyzoite Toxoplasma stages present during acute infection are not orally infectious, in contrast to the slowly replicating encysted bradyzoite stages. Therefore, to enhance its chances of transmission, Toxoplasma needs to balance immune evasion, to enable replication and dissemination, and immune activation, to prevent killing its host before orally infectious tissue cysts are formed. The contrasting goals of immune evasion and immune activation are reflected in Toxoplasma's arsenal of secreted effectors. GRA15 is clearly an effector that makes the parasite less virulent and helps the host survive. In contrast to GRA15, multiple Toxoplasma effectors mediating resistance against IFNγ‐mediated toxoplasmacidal mechanisms in murine cells have been identified (Hunter & Sibley, 2012; Hakimi et al, 2017). For example, ROP5 and ROP18, together with ROP17 and GRA7, cooperatively inactivate the IRGs and thereby enhance Toxoplasma virulence in mice (Alaganan et al, 2014; Etheridge et al, 2014). However, type III strains and certain atypical strains (P89 and CASTELLS) do not express ROP18 because of a large insertion in their ROP18 promoter region, while the BOF strain has very low expression of ROP5 (Niedelman et al, 2012). Similarly, Toxoplasma secretes the effector IST beyond the vacuole and IST inhibits STAT1 transcriptional activity thereby enhancing Toxoplasma virulence (Gay et al, 2016; Olias et al, 2016). However, it also secretes the GRA24 effector beyond the vacuole which activates P38 MAPK thereby activating host immune responses (Braun et al, 2013). The ultimate outcome of infection is therefore determined by the exact combination of these, often polymorphic, effectors and the species or exact genetic background of the host. A combination of effectors that is optimal in one host might kill another host or lead to complete elimination of parasites in yet another host. It is these evolutionary forces that have likely led to differences in GRA15 expression level and/or sequence in different Toxoplasma strains.
In this manuscript, we show how the polymorphic effector GRA15 determines the differential susceptibility of type I RH and type II Pru strains to IFNγ‐mediated growth inhibition in human and murine cells. This will help to understand the molecular basis of pathogenicity of different Toxoplasma strains in humans.
Materials and Methods
Reagents and antibodies
All the reagents, antibodies, primers, gDNAs, and siRNAs used in this study are described in detail in Appendix Tables S1–S3.
Culture of cells and parasites
Human foreskin fibroblasts (HFFs) were routinely maintained in DMEM with high glucose (Gibco, Invitrogen) supplemented with 10% FBS, l‐glutamine (2 mM), 100 U/ml penicillin, 100 μg/ml streptomycin, and 20 μg/ml gentamicin (complete medium). HFFs were passed using 0.25% trypsin. For all the experiments, HFFs were used at passage 5–10, but for serial passage of the parasites, higher passage number of the HFFs was used. Mouse embryonic fibroblasts (MEFs) were maintained in the complete medium supplemented with 10 mM HEPES, 1 mM sodium pyruvate, and 1× MEM nonessential amino acids. MEFs were passed using 0.05% trypsin‐EDTA. NF‐κB p65 −/− MEFs were a gift from A. Sinai (University of Kentucky College of Medicine, Lexington, KY), and Traf6 −/− MEFs were provided by K. Fitzgerald (University of Massachusetts Medical School, Worcester, MA). All parasite lines were maintained in vitro by serial passage on monolayers of HFFs and cultured in DMEM with high‐glucose medium supplemented with 1% FBS, l‐glutamine (2 mM), 100 U/ml penicillin, 100 μg/ml streptomycin, and 20 μg/ml gentamicin. A Toxoplasma gondii RH (RH 1‐1) strain expressing click beetle luciferase and green fluorescence protein (GFP) and a Pru strain expressing firefly luciferase and GFP (PruΔhpt, PruA7) were used as representative of type I and type II, respectively (Boyle et al, 2007). A genetically engineered RH strain expressing type II GRA15, PruΔgra15, and PruΔgra15 + GRA15‐HA (GRA15‐complemented strain) was described previously (Rosowski et al, 2011). Toxoplasma gondii RH strains endotagged with GRA43‐HA and GRA45‐HA were made as described previously (Wang et al, 2019b; Wang et al, 2019c).
Generation of TRAF6 knockdown human primary foreskin fibroblasts
Human foreskin fibroblasts were seeded in 12‐well plates at a concentration of 2 × 105 cells in 2 ml of complete media mentioned above for culturing HFFs, the night before the transduction. The next day, when cells were at ~70% confluency media were replaced with fresh complete media supplemented with polybrene (8 μg/ml, EMD Millipore). Ready‐to‐use lentivirus particles containing four different siRNAs targeting TRAF6 (Appendix Table S3) or containing scrambled siRNA (Appendix Table S3) were then used for transfection using three different MOIs (2/5/10) in three different wells for each virus particle (ABM Inc, BC, Canada). Cells were then kept at 37°C, 5%CO2 inside an incubator overnight. The next day, cells were checked under an inverted fluorescence microscope for expression of GFP as the lentivirus expresses GFP as a fusion protein with the puromycin resistance gene and kept for an additional 24 h in the incubator. Following this, media was replaced with fresh media with puromycin (1.5 μg/ml) for 72 h for selection of the stably transduced cells; at this time, puromycin was able to kill all the cells in untransduced cells, seeded in parallel in the same plate. Repeating the selection one more time for another 48 h, cells were divided in 6‐well plates to check by immunoblot for TRAF6 expression and knockdown of TRAF6 was confirmed.
Generation of MYR1 ko parasite
To generate the MYR1 insertional mutant in the PruΔku80 strain, the parasites were co‐transfected with a mixture of the pTOXO_Cas9CRISPR:sgMYR1 vector with purified amplicons containing the DHFR cassette flanked by sequences homologous to the sequence targeted by sgMYR1 (5:1 mass ratio). These amplicons were generated by PCR amplification of the DHFR cassette using the primers MYR1‐DHFR‐F and MYR1‐DHFR‐R, and a vector carrying the DHFR cassette as template (Donald & Roos, 1993). Stable recombinants were selected with 1 μM pyrimethamine, single‐cell‐cloned by limiting dilution, and verified by PCR analysis.
GRA15 TRAF‐binding site mutant generation
GRA15 TRAF2/6‐binding site mutation constructs were amplified from pTKO‐att‐GRA15IIHA vector (Rosowski et al, 2011) using specific primers (see Appendix Table S3) and confirmed by sequencing. 50 μg of circular vectors (pTKO‐att‐GRA15IIHA, pTKO‐att‐GRA15II‐TRA2Fmut‐HA, and pTKO‐att‐GRA15II‐TRAF2/6mut‐HA) was transfected into 1 × 107 RHΔhxgprt parasites by electroporation. Stable integrants were selected in media with 50 μg/ml mycophenolic acid and 50 μg/ml xanthine and cloned by limiting dilution. Expression and correct localization GRA15II to the PVM were confirmed by IFA for HA and Western blotting.
In vitro Toxoplasma infection
Parasites for in vitro infection were obtained from sequential syringe lysis using 27‐G and 30‐G needles of heavily infected HFF monolayers followed with a spin at 570 g for 7 min. For the infection with RH strains, MOIs of 1–3 were used, and for Pru, MOIs of 3–7 were used. Because RH and Pru strains often differ in viability and infectivity, equivalent “real” MOIs were matched from plaque assay results performed for each experiment to be able to make strain comparisons. Following infection with Toxoplasma, each time plates were centrifuged at 160 g for 3 min to synchronize the infection, prior to incubation for the required time.
IFNγ stimulation of cells
In most of the experiments, HFFs were stimulated for 18–24 h in complete medium at 37°C with 10 U/ml of human IFNγ (AbD Serotec, stock concentration is 10,000 U/ml). For some experiments, human IFNγ was used at concentration of 5–100 U/ml. MEFs were also stimulated for 18–24 h in complete medium with HEPES at 37°C with 100 U/ml murine IFNγ (PeproTech, stock concentration is 100,000 U/ml).
Use of inhibitors
For experiments using BAY11‐7082 (1 μM) or PYR41 (1 μM) or C25‐140 (50 μM), those compounds were added 24 h post‐stimulation with IFNγ but 2 h prior to infection and were kept throughout the infection for BAY11‐7082 and C25‐140 but washed away just prior to infection in the case of PYR41, as it is toxic to cells and parasites after longer incubation times. Bafilomycin A1 (100 nM) was added 1 h post‐infection as it affected the parasite invasion process.
Luciferase assay for parasite growth
Luciferase assays were performed from the 96‐well plates, and for each strain and condition, triplicate wells were used. To the confluent monolayers of HFFs/MEFs (2 × 105 cells/well), IFNγ was added (10 U/ml of human IFNγ and 100 U/ml of murine IFNγ) for 24 h prior to infection. Next day, infection with indicated parasites strains was done. For each strain, three different MOIs were used for matching of results from similar parasite infectivity between the strains later with the plaque assay. For some experiments, inhibitors were added either 2 h prior to infection or 1 h post‐infection as indicated. Following another 24‐h incubation, culture supernatants were removed and 1× lysis buffer was added (Luciferase Assay System, Promega) to the cells in the wells, followed by three freeze–thaw cycles. After that, 1× assay buffer containing luciferin was added to each well and transferred to clear centrifuge tubes for measurement of luciferase activity from the lysate using a single‐channel luminometer (Turner BioSystems). Luciferase reading of wells not treated with IFNγ was considered as 100%, and relative growth was calculated for IFNγ‐ and inhibitor‐treated wells.
Immunofluorescence assays for recruitment of host markers to the PVM and percentage of infection
Human foreskin fibroblasts or MEFs were plated on coverslips in 24‐well plates (1 × 105 cells/well) and cultured, stimulated with IFNγ for 18–24 h, and subsequently infected with Toxoplasma for 24 h (to determine the percentage of infected cells and nuclear translocation of NF‐κB), 3 h (to assess the recruitment of ubiquitin, p62, LC3B, GABARAP, GBP2, LAMP1, and TRAF6 in HFFs and MEFs), or 1 h (to determine IRGB6 coating in MEFs). Following incubation, cells were fixed with either 3% formaldehyde or 100% methanol depending on host marker (see Appendix Table S2) and then permeabilized and blocked with either buffer containing 0.2% Triton X‐100 along with 3% BSA and 5% goat serum (see Appendix Table S2) or buffer containing 0.2% freshly prepared saponin instead of Triton X‐100 (see Appendix Table S2). Cells were then treated with primary antibodies (Appendix Table S2 for overnight incubation at 4°C). Following that, each well was washed three times with 1× PBS and then secondary antibodies were added with Hoechst 33258 for 1 h. Finally, coverslips were washed five times with 1× PBS and were mounted with VECTASHIELD Antifade Mounting Medium. Imaging was done as described previously (Niedelman et al, 2013). For determination of nuclear translocation of NF‐κB, nuclear intensity of at least 15 infected cells was taken into consideration, whereas to assess percentage of infection after 24 h, cells were counted in at least six independent fields, the values observed in untreated infected cells were taken as 100%, and calculation for the rest was done relative to untreated infected cells. To measure the recruitment of host markers on the PVM, at least 100 infected cells were counted.
Plaque assay
For the plaque assay, freshly confluent 24‐well plates of HFFs or MEFs were used. The day before infection, fresh media was added replacing the media from the plates and stimulated with 10 U/ml human IFNγ or 100 U/ml mouse IFNγ or left unstimulated for 24 h. For infection, freshly harvested parasites, 100 parasites of RH, and 250 parasites of Pru strains were added to the 24‐well plates. Infected plates were incubated for 4 days at 37°C for RH strains and for 6 days in the case of Pru strains. For calculating the percentage of plaque loss, the following formula was used as described previously (Niedelman et al, 2012, 2013): [(Number of plaques in unstimulated condition − Number of plaques in stimulated condition)/Number of plaques in unstimulated condition] × 100. Plaque areas were captured and analyzed using a Nikon TE2000 inverted microscope equipped with Hamamatsu ORCA‐ER Digital Camera and NIS‐Elements Imaging Software, respectively. Plaque area loss was calculated using the same formula for plaque loss except using the plaque areas in place of plaque numbers. For all experiments, at least 20–25 plaques from technical duplicate wells were imaged.
IDO activity assay
The IDO activity upon IFNγ stimulation and Toxoplasma infection was evaluated by measuring L‐kynurenine from the culture supernatant of HFFs. Cells were cultured in 96‐well plates as mentioned in earlier assays with the complete DMEM containing total 0.6 mM L‐tryptophan (0.52 mM L‐tryptophan was added to existing 0.08 mM L‐tryptophan in the media). HFFs were either stimulated with 10 U/ml IFNγ for 18–24 h or left untreated and subsequently infected with parasite strains at different MOIs for 24 h before harvesting the culture supernatant. The concentration of L‐kynurenine was measured using 1.2% p‐dimethylaminobenzaldehyde in glacial acetic acid solution (Ehrlich's reagent). Briefly, 150 μl of culture supernatant was mixed with 20 μl of 30% trichloroacetic acid in a V‐bottom 96‐well plate followed with incubation at 50°C for 30 min. Subsequently, the plate was centrifuged for 10 min at 600 g, 100 μl of culture supernatant was mixed with Ehrlich's reagent and incubated for 10 min, and absorbance was recorded at 490 nm using a plate reader (Molecular Devices SpectraMax M2e). A standard curve of L‐kynurenine (0–1,500 μM) was used to calculate the concentrations in the samples.
Co‐Immunoprecipitation
40× T175 of human foreskin fibroblast (HFF) were stimulated or not with 10 U/ml of human IFN‐γ (AbD Serotec) for 24 h. Then, 2 h before infection (MOI: 5–10) with RH + GRA15II‐HA or RH + GRA35‐HA (for each parasite, 10× T175 containing IFN‐γ‐stimulated HFFs and 10 unstimulated), the cells were incubated with 50 μM of VX7655 (Selleckchem). Sixteen hours after infection, the cells were washed once and scraped with cold PBS. The cells were centrifuged and resuspended in 6 ml of lysis buffer (HEPES 10 mM, pH 7.9, MgCl2 1.5 mM, KCl 10 mM, EDTA 0.1 mM, 0.5 mM, NP40 0.65%, cocktail of protease inhibitor (Roche), phenylmethylsulfonyl fluoride (PMSF) 0.5 mM.) for 45 min at 4°C. 1% of these lysates were kept as input for the immunoblot. The lysate was centrifuged 30 min at 18,000 g, 4°C. Each sample was incubated with 100 μl of HA magnetic beads (Thermo Scientific) rotating overnight at 4°C. The beads were washed three times with Tris–HCl 10mM, pH 7.5, NaCl 150 mM, Triton X‐100 0.2%, PMSF 0.5 mM, and cocktail of protease inhibitor (Roche) and once with Tris–HCl 62.5 mM, pH 6.8, and subsequently resuspended in 100 μl of this buffer, performed immunoblotting (30 μl), and did the mass spectrometry analysis from the rest of the samples.
To perform immunoprecipitation with GRA15 mutants in HFFs or in MEFs, one T175 for each condition (untreated or IFNγ‐treated) was infected with RH + GRA15WT‐HA, RH + GRA15TRAF2mut‐HA, RH + GRA15TRAF2/6mut‐HA, and RH + GRA43‐HA or RH + GRA45‐HA expressing parasites for 24 h in MEF and 8 h in HFFs. Following infection, cells were washed once with ice‐cold 1× PBS and then scraped with ice‐cold PBS, centrifuged, and resuspended in 1 ml of lysis buffer mentioned above for 30 min at 4°C. After that, 10% of lysate was put aside for use as input during immunoblotting. The remaining lysates were then processed as mentioned above and incubated with 20 μl of HA magnetic beads overnight, in rotating condition at 4°C. Next day, beads were washed with the lysis buffer three times and subsequently resuspended in 60 μl of 1× loading dye to perform immunoblotting.
Immunoblotting:
20 μl of the HA magnetic beads of each sample was used to run on a 12% SDS–PAGE. The proteins were transferred to a PVDF membrane, blocked 30 min with TBST, 5% nonfat dry milk. The membrane was blotted overnight at 4°C with rat antibody against the HA‐tag (Roche, 1/500 dilution), and TRAF2 and TRAF6 rabbit antibodies (Appendix Table S2) followed by respective secondary HRP antibodies (Appendix Table S2).
Mass spectrometry‐based proteomics:
The HA magnetic beads were sent to the Proteomics Core Facility of the University of California, Davis, for mass spectrometry analysis. Briefly, the proteins were digested using Promega modified trypsin overnight at room temperature on a gently shaking device. Resulting peptides were analyzed by online LC‐MS/MS Q Exactive. All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org; version X! Tandem Alanine (2017.2.1.4)). X! Tandem was set up to search the uniprotHSTG_crap database assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 20 PPM and a parent ion tolerance of 20 PPM. Glu‐>pyro‐Glu of the n‐terminus, ammonia loss of the n‐terminus, gln‐>pyro‐Glu of the n‐terminus, deamidation of asparagine and glutamine, oxidation of methionine and tryptophan, dioxidation of methionine and tryptophan, and dicarbamidomethyl of lysine were specified in X! Tandem as variable modifications. Scaffold (version Scaffold_4.8.6, Proteome Software Inc., Portland, OR) was used to validate MS/MS‐based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 50.0% probability by the Scaffold Local FDR algorithm. Peptide identifications were also required to exceed specific database search engine thresholds, and X! Tandem identifications were also required. Protein identifications were accepted if they could be established at > 9.0% probability to achieve an FDR < 5.0% and contained at least one identified peptide. Protein probabilities were assigned by the ProteinProphet algorithm (Nesvizhskii et al, 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 7.0. All the data presented are mean ± standard error of mean (SEM), and the exact n values are mentioned in each of the figure legend. For all the calculations, P‐values of < 0.05 are considered as significant. Parameters with two different variables and groups were compared by two‐way ANOVA followed with either Bonferroni or Tukey's multiple comparison test. Parameters with one variable but three or more groups were compared by one‐way ANOVA followed with Tukey's multiple comparison test. For one variable test with two groups, two‐tailed unpaired t‐test was used. Specific statistical test performed for each figure was stated in each of the figure legend.
Author contributions
DM and JPJS designed the study. DM performed and interpreted the experimental work. LOS generated the GRA15 TRAF mutants and performed the mass spectrometry experiments and immunoprecipitation for Fig 4C. LB generated MYR1 knockout parasite strains. M‐AH provided insightful discussions and constructive suggestions and supervised the generation of knockout parasite strains. JPJS supervised the research. DM and JPJS wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank all members of the Saeij laboratory for productive discussions and Dr. Kevin Woolard for providing the instrumental facilities for assistance in the project. JPJS was supported by the National Institutes of Health NIH‐2R01AI080621‐06A1. DM was supported by American Heart Association Post‐doctoral Fellowship (18POST34030036).
The EMBO Journal (2020) 39: e103758
Data availability
The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information files are available from the authors upon request. All unique materials (e.g., the diversity of parasite lines described in this manuscript) are available upon request (contact: jsaeij@ucdavis.edu).
References
- Alaganan A, Fentress SJ, Tang K, Wang Q, Sibley LD (2014) Toxoplasma GRA7 effector increases turnover of immunity‐related GTPases and contributes to acute virulence in the mouse. Proc Natl Acad Sci USA 111: 1126–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando H, Sakaguchi N, Lee Y, Pradipta A, Ma JS, Tanaka S, Lai D‐H, Liu J, Lun Z‐R, Nishikawa Y et al (2018) Toxoplasma effector TgIST targets host IDO1 to antagonize the IFN‐gamma‐induced anti‐parasitic response in human cells. Front Immunol 9: 2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke MS, Khan A, Lauron EJ, Jimah JR, Wang Q, Tolia NH, Sibley LD (2015) Rhoptry proteins ROP5 and ROP18 are major murine virulence factors in genetically divergent South American strains of Toxoplasma gondii . PLoS Genet 11: e1005434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle JP, Saeij JPJ, Boothroyd JC (2007) Toxoplasma gondii: inconsistent dissemination patterns following oral infection in mice. Exp Parasitol 116: 302–305 [DOI] [PubMed] [Google Scholar]
- Braun L, Brenier‐Pinchart M‐P, Yogavel M, Curt‐Varesano A, Curt‐Bertini R‐L, Hussain T, Kieffer‐Jaquinod S, Coute Y, Pelloux H, Tardieux I et al (2013) A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation. J Exp Med 210: 2071–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Dixit VM (2016) Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16: 407–420 [DOI] [PubMed] [Google Scholar]
- Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, Fujita N, Saitoh T, Akira S, Yoshimori T et al (2014) The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin‐like conjugation systems of autophagy. Immunity 40: 924–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough B, Wright JD, Pereira PM, Hirst EM, Johnston AC, Henriques R, Frickel E‐M (2016) K63‐linked ubiquitination targets Toxoplasma gondii for endo‐lysosomal destruction in IFNγ‐stimulated human cells. PLoS Pathog 12: e1006027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies CC, Mak TW, Young LS, Eliopoulos AG (2005) TRAF6 is required for TRAF2‐dependent CD40 signal transduction in nonhemopoietic cells. Mol Cell Biol 25: 9806–9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrandi D, Kravets E, Konermann C, Beuter‐Gunia C, Klumpers V, Lahme S, Wischmann E, Mausberg AK, Beer‐Hammer S, Pfeffer K (2013) Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc Natl Acad Sci 110: 294–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin‐conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Donald RG, Roos DS (1993) Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase‐thymidylate synthase marker based on drug‐resistance mutations in malaria. Proc Natl Acad Sci USA 90: 11703–11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta J, Fan Y, Gupta N, Fan G, Gélinas C (2006) Current insights into the regulation of programmed cell death by NF‐κB. Oncogene 25: 6800–6816 [DOI] [PubMed] [Google Scholar]
- Etheridge RD, Alaganan A, Tang K, Lou HJ, Turk BE, Sibley LD (2014) file:///C:/Users/debmi/Downloads/citations (18).nbib the toxoplasma pseudokinase ROP5 forms complexes with ROP18 and ROP17 kinases that synergize to control acute virulence in mice. Cell Host Microbe 15: 537–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch D, Yakimovich A, Clough B, Wright J, Bunyan M, Howell M, Mercer J, Frickel E (2019) Defining host–pathogen interactions employing an artificial intelligence workflow. Elife 8: e40560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein MC, Reese ML, Konen‐Waisman S, Boothroyd JC, Howard JC, Steinfeldt T (2012) A Toxoplasma gondii pseudokinase inhibits host IRG resistance proteins. PLoS Biol 10: e1001358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz C, Napolitano A, Khan R, Clough B, Hirst EM, Frickel E‐M (2017) TRIM21 is critical for survival of Toxoplasma gondii infection and localises to GBP‐positive parasite vacuoles. Sci Rep 7: 5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M, Panas MW, Marino ND, Lee MCW, Buchholz KR, Kelly FD, Bednarski JJ, Sleckman BP, Pourmand N, Boothroyd JC (2016) A novel secreted protein, MYR1, is central to Toxoplasma's manipulation of host cells. MBio 7: e02231‐15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García MG, Alaniz L, Lopes EC, Blanco G, Hajos SE, Alvarez E (2005) Inhibition of NF‐κB activity by BAY 11‐7082 increases apoptosis in multidrug resistant leukemic T‐cell lines. Leuk Res 29: 1425–1434 [DOI] [PubMed] [Google Scholar]
- Gay G, Braun L, Brenier‐Pinchart M‐P, Vollaire J, Josserand V, Bertini R‐L, Varesano A, Touquet B, De Bock P‐J, Coute Y et al (2016) Toxoplasma gondii TgIST co‐opts host chromatin repressors dampening STAT1‐dependent gene regulation and IFN‐γ–mediated host defenses. J Exp Med 213: 1779–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi M‐A, Olias P, Sibley LD (2017) Toxoplasma effectors targeting host signaling and transcription. Clin Microbiol Rev 30: 615–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, Frickel EM, Valdivia RH, Coers J (2013) IRG and GBP host resistance factors target aberrant, “Non‐self” vacuoles characterized by the missing of “Self” IRGM proteins. PLoS Pathog 9: e1003414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla‐Moffett DM, Komatsu M, Frickel E‐M, Coers J (2015) Ubiquitin systems mark pathogen‐containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci 112: E5628–E5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D, Dubey JP (2002) Toxoplasma gondii: transmission, diagnosis and prevention. Clin Microbiol Infect 8: 634–640 [DOI] [PubMed] [Google Scholar]
- Howard JC, Hunn JP, Steinfeldt T (2011) The IRG protein‐based resistance mechanism in mice and its relation to virulence in Toxoplasma gondii . Curr Opin Microbiol 14: 414–421 [DOI] [PubMed] [Google Scholar]
- Howe DK, Sibley LD (1995) Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis 172: 1561–1566 [DOI] [PubMed] [Google Scholar]
- Hunn JP, Koenen‐Waisman S, Papic N, Schroeder N, Pawlowski N, Lange R, Kaiser F, Zerrahn J, Martens S, Howard JC (2008) Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii . EMBO J 27: 2495–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CA, Sibley LD (2012) Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat Rev Microbiol 10: 766–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav T, Geetha T, Jiang J, Wooten MW (2008) Identification of a consensus site for TRAF6/p62 polyubiquitination. Biochem Biophys Res Commun 371: 521–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav TS, Wooten MW, Wooten MC (2011) Mining the TRAF6/p62 interactome for a selective ubiquitination motif. BMC Proc 5(Suppl 2): S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KDC, Wang Y, Wojno EDT, Shastri AJ, Hu K, Cornel L, Boedec E, Ong YC, Chien YH, Hunter CA et al (2011) Toxoplasma polymorphic effectors determine macrophage polarization and intestinal inflammation. Cell Host Microbe 9: 472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AC, Piro A, Clough B, Siew M, Virreira Winter S, Coers J, Frickel EM (2016) Human GBP1 does not localize to pathogen vacuoles but restricts Toxoplasma gondii . Cell Microbiol 18: 1056–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TC, Yeh S, Hirsch JG (1972) The interaction between Toxoplasma gondii and mammalian cells. I. Mechanism of entry and intracellular fate of the parasite. J Exp Med 136: 1157–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Hunn JP, Konen‐Waisman S, Zhao YO, Preukschat D, Coers J, Boyle JP, Ong Y‐C, Boothroyd JC, Reichmann G et al (2010) Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell Microbiol 12: 939–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravets E, Degrandi D, Ma Q, Peulen T‐O, Klümpers V, Felekyan S, Kühnemuth R, Weidtkamp‐Peters S, Seidel CA, Pfeffer K (2016) Guanylate binding proteins directly attack Toxoplasma gondii via supramolecular complexes. Elife 5: e11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, Konstantinou EK, Young LH, Gold DA, Saeij JPJ (2017) The human immune response to Toxoplasma: autophagy versus cell death. PLoS Pathog 13: e1006176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Svenning S, Johansen T (2017) Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem 61: 609–624 [DOI] [PubMed] [Google Scholar]
- Lee Y, Sasai M, Ma JS, Sakaguchi N, Ohshima J, Bando H, Saitoh T, Akira S, Yamamoto M (2015) P62 plays a specific role in interferon‐γ‐induced presentation of a toxoplasma vacuolar antigen. Cell Rep 13: 223–233 [DOI] [PubMed] [Google Scholar]
- Liehl P, Zuzarte‐Luis V, Mota MM (2015) Unveiling the pathogen behind the vacuole. Nat Rev Microbiol 13: 589–598 [DOI] [PubMed] [Google Scholar]
- Man SM, Karki R, Kanneganti TD (2017) Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev 277: 61–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordue DG, Sibley LD (1997) Intracellular fate of vacuoles containing Toxoplasma gondii is determined at the time of formation and depends on the mechanism of entry. J Immunol 159: 4452–4459 [PubMed] [Google Scholar]
- Nadipuram SM, Kim EW, Vashisht AA, Lin AH, Bell HN, Coppens I, Wohlschlegel JA, Bradley PJ (2016) In vivo Biotinylation of the Toxoplasma parasitophorous vacuole reveals novel dense granule proteins important for parasite growth and pathogenesis. MBio 7: e00808–e00816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naor A, Panas MW, Marino N, Coffey MJ, Tonkin CJ, Boothroyd JC (2018) MYR1‐dependent effectors are the major drivers of a host cells early response to Toxoplasma, including counteracting MYR1‐independent effects. MBio 9: e02401–e02417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Niedelman W, Gold DA, Rosowski EE, Sprokholt JK, Lim D, Farid Arenas A, Melo MB, Spooner E, Yaffe MB, Saeij JPJ (2012) The rhoptry proteins ROP18 and ROP5 mediate Toxoplasma gondii evasion of the murine, but not the human, interferon‐gamma response. PLoS Pathog 8: e1002784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedelman W, Sprokholt JK, Clough B, Frickel EM, Saeij JPJ (2013) Cell death of gamma interferon‐stimulated human fibroblasts upon Toxoplasma gondii infection induces early parasite egress and limits parasite replication. Infect Immun 81: 4341–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olias P, Etheridge RD, Zhang Y, Holtzman MJ, Sibley LD (2016) Toxoplasma effector recruits the Mi‐2/NuRD complex to repress STAT1 transcription and block IFN‐γ‐dependent gene expression. Cell Host Microbe 20: 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn ER (1984) Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc Natl Acad Sci USA 81: 908–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn ER, Eckel M, Rebhun S (1986) Interferon‐γ suppresses the growth of Toxoplasma gondii in human fibroblasts through starvation for tryptophan. Mol Biochem Parasitol 20: 215–224 [DOI] [PubMed] [Google Scholar]
- Pleyer U, Schlüter D, Mänz M (2014) Ocular Toxoplasmosis: recent aspects of Pathophysiology and clinical implications. Ophthalmic Res 52: 116–123 [DOI] [PubMed] [Google Scholar]
- Qin A, Lai D‐H, Liu Q, Huang W, Wu Y‐P, Chen X, Yan S, Xia H, Hide G, Lun Z‐R et al (2017) Guanylate‐binding protein 1 (GBP1) contributes to the immunity of human mesenchymal stromal cells against Toxoplasma gondii . Proc Natl Acad Sci 114: 1365–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosowski EE, Lu D, Julien L, Rodda L, Gaiser RA, Jensen KDC, Saeij JPJ (2011) Strain‐specific activation of the NF‐κB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J Exp Med 208: 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij JPJ, Boyle JP, Boothroyd JC (2005) Differences among the three major strains of Toxoplasma gondii and their specific interactions with the infected host. Trends Parasitol 21: 476–481 [DOI] [PubMed] [Google Scholar]
- Saeij JP, Frickel EM (2017) Exposing Toxoplasma gondii hiding inside the vacuole: a role for GBPs, autophagy and host cell death. Curr Opin Microbiol 40: 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaré LO, Yang N, Konstantinou EK, Lu D, Mukhopadhyay D, Young LH, Saeij JPJ (2019) GRA15 activates the NF‐κB pathway through interactions with TNF receptor‐associated factors. MBio 10: e00808–e00819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M, Sakaguchi N, Ma JS, Nakamura S, Kawabata T, Bando H, Lee Y, Saitoh T, Akira S, Iwasaki A et al (2017) Essential role for GABARAP autophagy proteins in interferon‐inducible GTPase‐mediated host defense. Nat Immunol 18: 899–910 [DOI] [PubMed] [Google Scholar]
- Selleck EM, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HW IV, MacMicking JD, Sibley LD (2013) Guanylate‐binding Protein 1 (Gbp1) contributes to cell‐autonomous immunity against Toxoplasma gondii . PLoS Pathog 9: e1003320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck EM, Orchard RC, Lassen KG, Beatty WL, Xavier RJ, Levine B, Virgin HW, Sibley LD (2015) A noncanonical autophagy pathway restricts Toxoplasma gondii growth in a strain‐specific manner in IFN‐gamma‐activated human cells. MBio 6: e01157‐15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Brown KM, Lee TD, Sibley LD (2014) Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio 5: e01114‐14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfeldt T, Konen‐Waisman S, Tong L, Pawlowski N, Lamkemeyer T, Sibley LD, Hunn JP, Howard JC (2010) Phosphorylation of mouse immunity‐related GTPase (IRG) resistance proteins is an evasion strategy for virulent Toxoplasma gondii . PLoS Biol 8: e1000576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickson S, Campbell DG, Emmerich CH, Knebel A, Plater L, Ritorto MS, Shpiro N, Cohen P (2013) The anti‐inflammatory drug BAY 11‐7082 suppresses the MyD88‐dependent signalling network by targeting the ubiquitin system. Biochem J 451: 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Komander D (2016) Ubiquitin modifications. Cell Res 26: 399–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virreira Winter S, Niedelman W, Jensen KD, Rosowski EE, Julien L, Spooner E, Caradonna K, Burleigh BA, Saeij JPJ, Ploegh HL et al (2011) Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PLoS ONE 6: e24434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Li S, Zhao Y, Zhang B, Li Y, Liu S, Du H, Cao L, Ou M, Ye X et al (2019a) The GRA15 protein from Toxoplasma gondii enhances host defense responses by activating the interferon stimulator STING. J Biol Chem 294: 16494–16508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cirelli KM, Barros PDC, Sangaré LO, Butty V, Hassan MA, Pesavento P, Mete A, Saeij JPJ (2019b) Three Toxoplasma gondii dense granule proteins are required for induction of Lewis rat macrophage pyroptosis. MBio 10: e02388‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sangaré LO, Paredes‐Santos TC, Krishnamurthy S, Hassan MA, Furuta AM, Markus BM, Lourido S, Saeij JPJ (2019c) A genome‐wide loss‐of‐function screen identifies genes that determine fitness in interferon gamma‐activated murine macrophages. bioRxiv 10.1101/867705 [DOI] [Google Scholar]
- Yang N, Farrell A, Niedelman W, Melo M, Lu D, Julien L, Marth GT, Gubbels MJ, Saeij JPJ (2013) Genetic basis for phenotypic differences between different Toxoplasma gondii type I strains. BMC Genom 14: 467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YO, Khaminets A, Hunn JP, Howard JC (2009) Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNγ‐inducible immunity‐related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog 5: e1000288 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information files are available from the authors upon request. All unique materials (e.g., the diversity of parasite lines described in this manuscript) are available upon request (contact: jsaeij@ucdavis.edu).


