Abstract
Rare genetic diseases affect a limited number of patients, but their etiology is often known, facilitating the development of reliable animal models and giving the opportunity to investigate physiopathology. Lysosomal storage disorders are a group of rare diseases due to primary alteration of lysosome function. These diseases are often associated with neurological symptoms, which highlighted the importance of lysosome in neurodegeneration. Likewise, other groups of rare neurodegenerative diseases also present lysosomal alteration. Lysosomes fuse with autophagosomes and endosomes to allow the degradation of their content thanks to hydrolytic enzymes. It has emerged that alteration of the autophagy–lysosome pathway could play a critical role in neuronal death in many neurodegenerative diseases. Using a repertoire of selected rare neurodegenerative diseases, we highlight that a variety of alterations of the autophagy–lysosome pathway are associated with neuronal death. Yet, in most cases, it is still unclear why alteration of this pathway can lead to neurodegeneration.
Keywords: lysosomal storage diseases, autophagy, signaling, trafficking, neuronal death
Abbreviations: LSD, lysosomal storage disorder; CLN, neuronal ceroid lipofuscinose; NPC, Niemann–Pick type C; FTLD, frontotemporal lobar degeneration; ALS, amyotrophic lateral sclerosis; HSP, hereditary spastic paraplegia; SCA7, spinocerebellar ataxia type 7; mTOR, mammalian target of rapamycin
Graphical abstract
Highlights
-
•
Lysosome function is impaired in many rare neurodegenerative diseases, making it a convergent point for these diseases.
-
•
Impaired lysosome function is associated with alteration of the autophagy pathway.
-
•
Autophagy–lysosome pathway can be impaired at various steps in different rare neurodegenerative diseases.
-
•
The mechanisms linking impaired autophagy–lysosome pathway to neurodegeneration are still not fully elucidated.
Introduction
Macroautophagy, termed here autophagy, is a conserved pathway allowing the bulk degradation of macromolecules or organelles through their delivery to lysosomes. Cellular substrates that need to be degraded are engulfed by a double-membrane compartment, forming an autophagosome. The content of the latter is then degraded by hydrolytic enzymes upon fusion with the degradative organelles, lysosomes (Figure 1). This cellular process has been implicated in many neurodegenerative diseases [1] making it of central interest for the development of new therapies. Lysosomes play a crucial role in this pathway, but the mechanisms underlying the alterations of autophagy in many neurodegenerative diseases are still elusive.
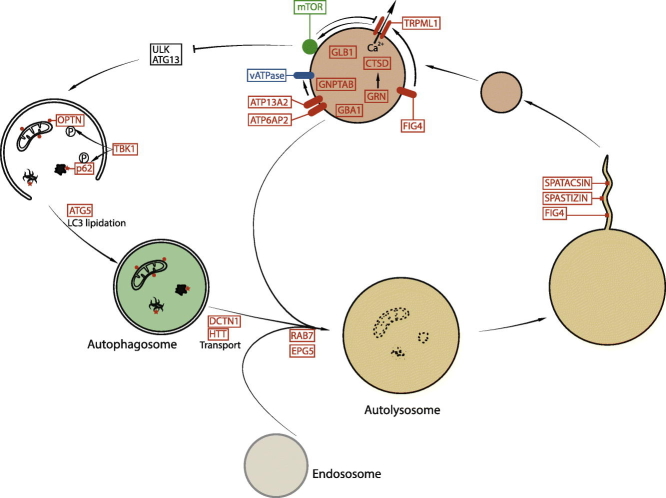
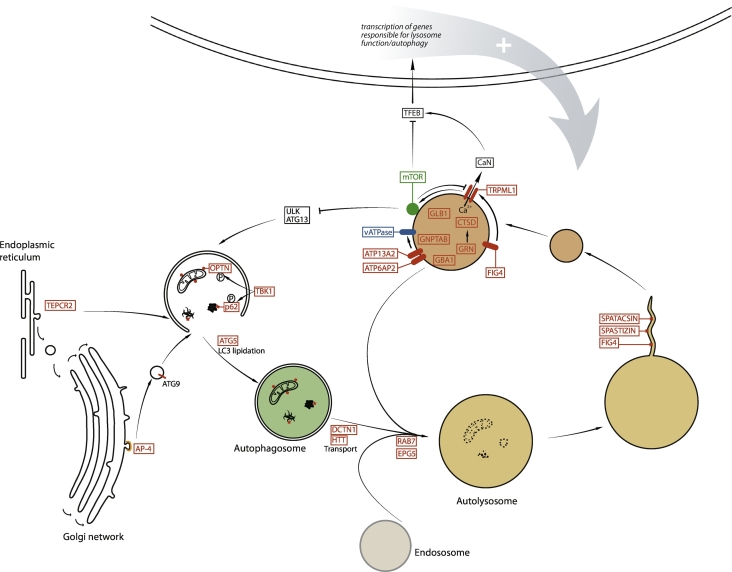
Figure 1.
Schematic view of the role of lysosomes in the autophagy pathway. Cellular substrates that need to be degraded are engulfed by a double-membrane compartment, the autophagosome, whichis formed by integrating various signals. The autophagosome fuses with lysosome, forming an autolysosome allowing the degradation of cellular substrates by hydrolytic enzymes. Upon degradation step, recycling of lysosome membrane leads to formation of new proto-lysosomes that can maturate into functional lysosomes. The latter are at the crossroad of signaling pathways regulating the autophagy–lysosome pathway. The proteins indicated in red are encoded by genes mutated in rare neurodegenerative diseases, highlighting the critical role of this pathway for these disorders. AP4, adaptor complex 4 subunits; CaN, calcineurin; CTSD, cathepsin D; GBA1, glucocerebrosidase; GNPTAB, N-acetylglucosamine-1-phosphotransferase; GLB1, β-galactosidase; GRN, granulin; HTT, huntingtin; OPTN, optineurin.
Among the diseases presenting impairment of the lysosome function, many are rare neurodegenerative diseases that are caused by defined genetic mutations. The variety of diseases and causative mutations represents a challenge for the development of therapies for each pathology. However, the knowledge of the genetic etiology of these diseases represents an opportunity to investigate the mechanisms leading to impairment of the autophagy–lysosome pathway. In that respect, lysosomal storage disorders (LSDs) are a group of diseases presenting primary alteration of lysosomal function, often associated with impaired autophagy [2]. Patients affected by LSDs often present with neurological symptoms, highlighting the crucial role of lysosomes for neuronal survival. However, other rare neurodegenerative diseases also present lysosomal dysfunction and impaired autophagy, either as a primary dysfunction or as a consequence of the alteration of other related pathways. In this review, we do not aim to present an exhaustive list of rare diseases with lysosomal dysfunction, but rather select few examples to highlight the variety of altered functions and the various aspects of the lysosome–autophagy pathway that can be altered.
Lysosomal Dysfunction in Rare Neurodegenerative Diseases
Lysosomes are involved in the degradation of most macromolecules, such as proteins, glycosaminoglycans, or lipids, into their basic building blocks. This function relies on the presence in the lysosome lumen of more than 50 different hydrolases. The latter are synthesized in the endoplasmic reticulum, trafficked through the Golgi apparatus where they are tagged by mannose-6-phosphate residues that allows their targeting to the endolysosomal compartment by the mannose-6-phosphate receptors [3]. The proper function of these enzymes requires the acidic intraluminal pH of lysosomes that is maintained by vATPase. Due to the central function of lysosomes for cell physiology, it is not surprising that lysosomal dysfunction is implicated in a variety of diseases.
LSDs and neurodegeneration
The critical role of lysosomes for neuronal survival was first evidenced by the investigation of LSDs (Table 1). They are a group of rare diseases in which the function of lysosomes is primarily impaired. The concept of LSD was first defined in the case of Pompe disease, where the absence of acidic α-glucosidase was associated with the accumulation of the substrate of this enzyme [97]. Loss of function of others hydrolytic enzymes or their cofactors, which are required for catabolism of various macromolecules, were later associated with other LSD such as gangliosidoses, sphingolipidoses (Fabry disease, Gaucher disease, …), and mucopolysaccharidoses [98]. However, defects in lysosomal transporters or proteins required for the trafficking of lysosomal hydrolases can also lead to accumulation of undigested substrates in lysosomes. Such alterations are, for example, observed in neuronal ceroid lipofuscinoses (CLNs), also known as Batten disease, a group of LSDs that share physiopathological features such as accumulation of ceroid lipofuscin consisting of lysosomal autofluorescent material. These diseases are caused by mutations in at least 14 different genes encoding proteins of different functions [99]. Some of the gene products encode lysosomal hydrolases such as cathepsin D (CLN10) or cathepsin F (CLN13). CLN8 encodes an endoplasmic reticulum cargo implicated in the trafficking of lysosomal hydrolases [17], explaining how mutations can lead to lysosomal accumulation of material in patients. Yet, the function of other CLN gene products are still debated, and further physiopathological studies will allow the dissection of the basic molecular mechanisms regulating lysosomal function. In another example, physiopathological investigations demonstrated that the Niemann–Pick type C (NPC)-associated proteins NPC1 and NPC2 play a role in cholesterol egress from lysosomes [[100], [101], [102]]. Consistently, NPC patients present lysosomal accumulation of cholesterol that is used for diagnosis in clinical practice [32]. In NPC neurons, lysosomes also present secondary accumulation of other lipids such as sphingomyelin or gangliosides that could contribute to pathogenic mechanisms [33].
Table 1.
Main alterations in lysosome function and autophagy pathways observed in a subset of rare neurodegenerative diseases
| Disease | Mutated gene | Function | Accumulation of material | Autophagy step altered | Secondary cellular dysfunctions, possibly associated to neurodegeneration | References |
|---|---|---|---|---|---|---|
| Primary lysosomal dysfonction | ||||||
| GM1 gangliosidosis | GLB1 | β-Galactosidase | GM1 | Increased autophagy activation | Impaired mitochondrial function | [4] |
| Mucolipidosis type II (and type III) | GNPTAB | N-acetylglucosamine-1-phosphotransferase, required for trafficking of lysosomal hydrolases | Mutilamellar bodies, lipofusin, glycans, gangliosides | Accumulation of autolysosomes | [5] | |
| Mucolipidosis type IV | TRPML1 | Lysosomal calcium channel | Accumulation of enlarged autolysosomes, accumulation and impaired degradation of autophagosomes | Accumulation of dysfunctional mitochondria | [6,7] | |
| Gaucher disease | GBA1 | Glucocerebrosidase | Glucosylceramide, glucosylsphingosine, | Impaired autophagosome–lysosome fusion, decreased autophagic flux, impaired ALR | Accumulation of dysfunctional mitochondria, loss of synapses | [[8], [9], [10]] |
| Fabry disease | GLA | α-Galactosidase | Globotriaosylceramide (Gb3), lipopigment aggregates, α-synculein | Impaired autophagic flux; impaired ALR | [[11], [12], [13]] | |
| CLN3 | CLN3 | Unknown | Lipofuscin, subunit c of mitochondrial F0-ATPase | Impaired autophagic flux; impaired autolysosome degradation; impaired autophagosome–lysosome fusion | [14,15] | |
| CLN7 | CLN7 | Unknown, putative transporter | Lipofuscin, saposinD | Impairment of constitutive macroautophagy, accumulation of p62 | [16] | |
| CLN8 | CLN8 | Endoplasmic protein required for trafficking of lysosomal hydrolases | Lipofuscin, ATP synthase subunit c | ER stress; deficient mitochondrial calcium buffering | [[17], [18], [19], [20], [21]] | |
| CLN10 | CTSD | Lysosomal hydrolase cathepsin D | Lipofuscin, accumulation of autolysosomes | Impaired degradation of lysosome content | [22,23] | |
| CLN11 |
PGRN (loss of function) |
Protein processed into granulin peptides | Lipofuscin deposit, TDP43 | Impaired clearance of autophagosomes | Decreased levels of saposin and decreased activity of cathepsin D | [[24], [25], [26], [27]] |
| FTLD-GRN |
PGRN (haploinsufficiency) |
Protein processed into granulin peptides | TDP43, lipofuscin | Impaired clearance of autophagosomes | Decreased levels of saposin and decreased activity of cathepsin D | [[24], [25], [26], [27]] |
| CLN12 Parkinson's disease (PARK9) HSP (SPG78) |
ATP13A2 | P5ATPase | Lipofuscin | Impaired lysosomal degradative activity, impaired autophagosome–lysosome fusion, accumulation of autolysosomes | Impaired lysosome acidification, accumulation of impaired mitochondria | [28,29] |
| X-linked parkinsonism with spasticity | ATP6AP2 | Accessory protein of vATPase, required for lysosome acidification | Accumulation of electron dense material detected by electron microscopy | Accumulation of autophagy substrates | Abnormal synaptic function, impaired myelination | [30,31] |
| NPC |
NPC1 NPC2 |
Cholesterol transport | Cholesterol, glycosphingolipids | Impaired autophagosome–lysosome fusion; impaired clearance of autophagosomes | Accumulation of mitochondria, oxidative stress | [[32], [33], [34], [35], [36], [37]] |
| Cargo recognition | ||||||
| FTDALS3 | SQSTM1 | Autophagy receptor p62 | Ubiquitin, p62, TDP-43 | Decreased clearance of protein aggregates | Impaired mitochondrial respiration | [[38], [39], [40]] |
| ALS15 FTLD |
Ubiquilin-2 | Autophagy receptor | Poly-ubiquitinated proteins, TDP43 | Decreased clearance of poly-ubiquitinated proteins | Toxic gain of function? Impaired acidification of lysosomes? | [[41], [42], [43], [44]] |
| ALS12 FTLD |
OPTN | Autophagy receptor | TDP43 | Decreased clearance of protein aggregates, pathogens, and mitochondria | Accumulation of defective mitochondria | [45,46] |
| FTDALS4 | TBK1 | Kinase regulating autophagy receptors | TDP-43 | Impaired phosphorylation of autophagy receptors, decreasing their activities | Impaired mitophagy | [[45], [46], [47], [48]] |
| Huntington disease | HTT | Huntingtin | Aggregation of mutant huntingtin | impaired cargo recognition mediated by mutant huntingtin | [49,50] | |
| Autophagosome formation | ||||||
| SCA25 (autosomal recessive ataxia) |
ATG5 (missense) |
Conjugation of LC3 to PE | Decreased interaction with ATG12, decreased autophagosome formation | [51] | ||
| Neurodegeneration with brain iron accumulation | WDR45 | Interacts with ATG2 and ATG9 | Accumulation of autophagosomes and immature autophagic vesicles | Impaired autophagosome formation and elongation | Impaired iron homeostasis ? | [52,53] |
| HSP (SPG49) | TECPR2 | Maintenance of ER exit sites, interaction with LC3 | Impaired autophagosome formation | [54,55] | ||
| HSP (SPG47, SPG50, SPG51, SPG52); AP4 syndrome | AP4S1, AP4M1, AP4B1, AP4E1 | Adaptor protein complex-4 | Brain iron accumulation detected by MRI | Accumulation of ATG9 in trans-Golgi, impaired autophagosome formation? | [[56], [57], [58], [59]] | |
| FTDALS1 | C9ORF72 | Guanine nucleotide exchange factor? | TDP-43, Lipid accumulation in lysosomes |
Decreased autophagosome formation; Increased autophagic flux | [[60], [61], [62]] | |
| ALS10 FTLD |
TARDBP | RNA regulation | TDP-43 | Decreased or increased autophagosome formation? | [63,64] | |
| ALS6 FTLD |
FUS | RNA regulation | FUS | Decreased omegasome formation | [65] | |
| ALS1 | SOD1 | Superoxide dismutase | SOD1 | Increased induction of autophagy | [66,67] | |
| Spinocerebellar ataxia SCA3 | SCA3 | Ataxin3 | Ataxin3 with poly-glutamine expansions, positive for p62 and ubiquitin | Impaired autophagosome formation | [68,69] | |
| Spinocerebellar ataxia SCA7 | SCA7 | Ataxin7 | Ataxin7 with poly-glutamine expansions; aggregates positive for mTOR, Beclin, p62, and ubiquitin | Impaired autophagosome formation? Impaired autophagic flux | [70] | |
| Huntington disease | HTT | Huntingtin | Aggregation of mutant huntingtin | Impaired activation of autophagy or autophagosome formation | [49,50,68] | |
| Autophagosome–lysosome fusion and autolysosome clearance | ||||||
| Huntington disease | HTT | Huntingtin | Aggregation of mutant huntingtin | Impaired autophagosome trafficking leading to impaired fusion with lysosomes | [71] | |
| ALS1 | SOD1 | Superoxide dismutase | SOD1 | Impaired retrograde transport of autophagosomes, preventing fusion with lysosomes | Accumulation of dysfunctional mitochondria | [72] |
| ALS2 | ALS2 | Alsin, guanine nucleotide exchange factor for the small GTPase Rab5 | Impaired autophagosome clearance | [73] | ||
| ALS10 FTLD |
TARDBP | RNA regulation | TDP-43 | Impaired autophagosome–lysosome fusion | [64] | |
| ALS14 FTLD Inclusion body myopathy |
VCP | Valosin containing protein; AAA-ATPase | TDP43, ubiquitin positive inclusions | Accumulation of damaged lysosomes, impaired autophagosome–lysosome fusion | [[74], [75], [76]] | |
| ALS17 FTLD |
CHMP2B | Subunit of the endosomal sorting complex required for transport (ESCRT-III) | P62-positive inclusions; Lipofuscin-like autofluorescent aggregates | Impaired maturation of phagophore into autophagosome; impaired endosome-lysosome fusion | [77,78] | |
| ALS | DCTN1 | Subunit of dynein–dynactin complex | Ubiquitin-, p150Glued-positive inclusions | Impaired autophagosome trafficking, impaired autophagosome–lysosome fusion | [79,80] | |
| Charcot–Marie–Tooth 2B | RAB7 point mutations | Rab GTPase | Reduced fusion of autophagosomes with lysosomes | Impaired signaling, axon growth defects | [81,82] | |
| Vici syndrome | EPG5 | Rab7 effector | Impaired autophagosome–lysosome fusion; impaired degradation of autolysosomes | [83,84] | ||
| Autosomal recessive ataxia | SNX14 | Intracellular membrane trafficking | Lysosomal accumulation of cholesterol | Impaired autophagosome clearance; impaired autophagosome–lysosome fusion? | Impaired lipid metabolism | [85,86] |
| Dentatorubral-pallidoluysian Atrophy | ATN1 | Atrophin | Blockade of autolysosome egradation | Disrupted nuclear organization | [87,88] | |
| Lysosome recycling | ||||||
| HSP (SPG11) ALS5 |
SPG11 | Initiation of ALR | Lipids (cholesterol, gangliosides), lipofucsin-like, autolysosomes | Impaired ALR, accumulation of autolysosomes | Impaired cellular calcium homeostasis | [[89], [90], [91], [92]] |
| HSP (SPG15) | SGP15 | Initiation of ALR | Fingerprint bodies, lipofuscin-like deposits, autolysosomes | Impaired ALR, accumulation of autolysosomes | [92,93] | |
| HSP (SPG48) | SPG48 | Adaptor protein complex 5 | Membrane swirls, lipofuscin-like deposits, autolysosomes | Accumulation of autophagosomes and autolysosomes, impaired ALR | Alteration of Golgi network | [94] |
| ALS11 Charcot–Marie–Tooth 4J |
FIG4 | PI(3,5)P2 phosphatase, subunit if PIKFyve | Accumulation of large lysosomes containing electron dense material in neurons and glia | Impaired lysosomal fission | Impaired lysosomal calcium homeostasis, impaired synapse morphology | [95,96] |
ALR, autophagic lysosome reformation.
Most patients affected by LSDs present multisystemic disorders, with prominent neurological symptoms [98], connecting impairment of lysosomal function to neurodegeneration. Consistently, mutations in genes affecting lysosomal function also lead to rare neurodegenerative diseases that are not classified as LSDs, although they share similarities with LSD.
Frontotemporal lobar degeneration and amyotrophic lateral sclerosis
Frontotemporal lobar degeneration (FTLD) is the second most frequent form of dementia, characterized by behavior and language impairments [103]. Clinical and genetic data have shown that some forms of FTLD share genetic risk factors and pathological hallmarks with amyotrophic lateral sclerosis (ALS) characterized by the degeneration of cortical and spinal motor neurons leading to muscle weakness.
Heterozygous mutations in the GRN gene leading to haploinsufficiency in Progranulin (PGRN) are responsible for some forms of FTLD [104]. Identification of homozygous mutations in GRN as a cause of the LSD neuronal ceroid lipofuscinosis CLN11 highlighted the role of PGRN in lysosomes [105]. PGRN deficiency in mouse models and induced pluripotent stem cells derived from FTLD-GRN patients showed the presence of enlarged vesicles and lipofuscin deposits [24,25]. PGRN can be trafficked to lysosomes [106,107]. Actually, it can co-traffic with prosaposin, the precursor of saposin peptides that are essential for lysosomal glycosphingolipid degradation, leading to reduced levels of prosaposin in neurons both in mice deficient in PGRN and in human samples from FTLD patients due to GRN mutations [26]. Furthermore, PGRN can be processed in lysosomes into granulin peptides [27] that regulate cathepsin D activity [25], highlighting the importance of PGRN for lysosomal function.
C9ORF72 and CHMP2B are also implicated in the endolysosomal trafficking and autophagy–lysosome pathway. Heterozygous mutations in CHMP2B are responsible for rare cases of FTLD [108]. A mouse model of this form of the pathology showed that CHMP2B mutation causes a decrease in neuronal endolysosomal motility and leads to lysosomal storage pathology [109,110]. In the case of C9ORF72 pathology, expansion of hexanucleotides in intron 1 leads to haploinsufficiency of the gene product as well as production of non ATG-mediated expression of dipeptide repeats [111]. The product of C9ORF72 was reported colocalized with endosomes, lysosomes, and autophagosomes [112,113]. Consistently, an ortholog in Caenorhabditis elegans leads to impaired degradation of endolysosomal content and subsequent lysosome reformation [114] that occurs after lysosomal degradation of vesicular cargos [11]. Importantly, in mouse models, C9orf72 deficiency was associated with increased levels of lysosomal and autophagy markers in the spleen and liver, but not in the brain [115], suggesting different roles for the protein, or different regulations of the lysosomal function in peripheral tissues and in the central nervous system.
One strong argument linking the physiopathology of FTLD/ALS to lysosome dysfunction was the identification of the lysosomal protein TMEM106B as a modulator of the function of PGRN, C9ORF72, or CHMP2B. Indeed, TMEM106B is a risk factor for FTLD [116], with higher levels of TMEM106B being associated with higher risk of FTLD with GRN mutations [117,118]. TMEM106B regulates the morphology of lysosome compartments, the degradation of endocytic cargoes, and lysosomal trafficking in neurons [119,120]. The overexpression of TMEM106B notably promotes the formation of enlarged lysosomes and impairs lysosome acidification and degradative function. These consequences of TMEM106B overexpression are, however, abolished in the absence of C9ORF72 [121], suggesting a role for the two proteins in the same cellular pathways. Furthermore, overexpression of TMEM106B regulates progranulin levels [118,119]. Deletion of Tmem106b in Pgrn knockout mice was shown to restore the levels of lysosomal enzymes, but did not improve lipofuscin accumulation [122]. Together, these data highlight the importance of the lysosomal function for the FTLD–ALS-associated gene products [123].
Hereditary spastic paraplegias
Hereditary spastic paraplegias (HSPs) are a group of neurodegenerative diseases characterized by spasticity in lower limbs that is caused by the degeneration of cortical motor neuron axons. This group of diseases is clinically highly heterogenous, notably because more 60 genes have been shown to cause HSP. Despite this large heterogeneity, a subgroup of HSP appears to be caused by abnormal function of the endolysosomal pathway.
Patients with SPG78 form of HSP present loss of function mutation in ATP13A2, which is also mutated in patients diagnosed as neuronal ceroid lipofuscinosis (CLN12) [124,125]. ATP13A2 is a P5ATPase that is mainly localized in the membrane of lysosomes [126], and dysfunction of this protein impairs lysosomal degradation [28]. Consistently, homozygous mutations in fibroblasts of SPG78 patients increased the number of lysosomes and impaired lysosomal degradative activity, and electron microscopy analysis revealed that lysosomes accumulated abnormal material consisting of whirls and stacks of membranes [125]. Mutation of ATP6AP2, encoding an accessory protein of the lysosomal vATPase, has been found in patients with X-linked parkinsonism with spasticity [127]. The mutation led to skipping of exon 4 in 50% of transcript and resulted in the brain of these patients in reduced expression of ATP6AP2 [127]. Recently, a de novo intronic mutation of ATP6AP2 has been found in a patient with fulminant neurodegeneration presenting with epilepsy and spasticity [30]. The mutation also led to skipping of exon 4 in 80% of ATP6AP2 transcript. Loss of ATP6AP2 was proposed to impair vesicular acidification [30,128], although contradictory results have been published [129]. Furthermore, postnatal deletion of Atp6ap2 in a mouse model led to accumulation of electron dense material in neurons [30], consistent with impaired lysosomal function.
Alteration of lysosomal function in HSP has also been associated with loss of function of non-lysosomal proteins. In the brains of Spg15−/−, Spg11−/−, and Spg48−/− mice, autofluorescent material accumulates in lysosomes and ultrastructural analysis showed the presence of electron dense deposits in neurons [89,93,94,130]. Accumulation of membranes in lysosomes was also observed in fibroblasts of SPG48 patients with loss of function mutation in the subunit ζ of the AP-5 complex [131]. Electron microscopy analysis of fibroblasts of SPG15 patients and neurons of Spg15−/− mice showed the accumulation of zebra or fingerprint bodies [93,132], which are lysosomes with accumulation of membranes similar to those found in some LSD [133]. Analysis of SPG11 patient fibroblasts, in contrast, did not reveal the accumulation of lamellar structures by electron microscopy [132]. A lipidomic analysis performed in the brain of Spg11−/− mice showed the progressive accumulation in lysosomes of simple gangliosides [90], a process observed in a wide range of LSD [33]. Accumulation of membrane structures in endolysosomes has also been observed in cells derived from Spg4−/− or Spg31−/− mice. These defects resulted from impaired ER-mediated endosomal tubule fission [134]. Yet, it is not clear whether these structures share similarities in composition with the lysosomes accumulating membranes observed in fibroblasts of SPG11, SPG15, SPG48, or ATP13A2 patients. The accumulation of material in lysosomes in some forms of HSP led to the proposition that they may be considered as a new form of LSD [131], which is supported by the fact that mutation in ATP13A2 are responsible for either SPG78 form of HSP or CLN12 [124,125].
Ataxias
Lysosomal accumulation of biological material has also been observed in models of spinocerebellar ataxias. For example, a knock-in mouse model of spinocerebellar ataxia type 7 (SCA7) expressing ataxin 7 with 266 glutamines presents autolysosomes containing amorphous electron-dense material and lipids [70]. Similar accumulations were also observed in a mouse model of the polyglutamine disease dentatorubral–pallidoluysian atrophy [87]. It is important to note that the proteins responsible for SCA7 or dentatorubral–pallidoluysian atrophy have no known function in lysosomes, but their aggregative properties probably alter the degradation pathways [135,136]. Other forms of ataxias also have impaired lysosomal function. For example, the impairment of lysosome function due to loss of SNX14 that binds the lysosome-enriched phosphoinositide PI(3,5)P2 is responsible for recessive ataxia [85]. Enlarged lysosomes accumulating cholesterol were observed in cells derived from patients with SNX14 loss of function mutation [85,86].
The investigations of the physiopathology of LSDs, FTLD/ALS, HSPs, and ataxias highlight the importance of maintaining a proper lysosomal function for the neuronal survival. The genetic overlap observed between some LSD and some form of FTLD (PGRN) or HSP (SPG78) highlights the existence of putative common mechanisms underlying neurodegeneration in the various families of diseases. Yet, the age of onset and the evolution of diseases are quite different, showing that different lysosomal dysfunctions may occur in the various diseases, or other pathways may also contribute to the pathology.
Alteration of Lysosomal Regulation in Rare Neurodegenerative Diseases
Lysosomes play a key catabolic role for the degradation of the content of endosomes and autophagosomes, as we will explore later. However, it has emerged that they also play a crucial role in the regulation of cellular functions and represent a signaling hub, allowing the cells to integrate environmental cues, including nutrient availability and response to growth factors [137]. Two main, inter-related lysosome signaling pathways have been investigated: the pathway related to mammalian target of rapamycin (mTOR) and calcium, both converging on the transcription factor TFEB.
mTORC1 signaling pathway
mTORC1 is a key kinase for the response to metabolic state of the cells. Activation of mTOR requires its recruitment to the lysosome surface bringing it in close proximity of its activator, the GTPase Rheb [138]. The localization of mTORC1 to the surface of lysosomes is critical to sense and respond to the variations in the levels of nutrients, including amino acids, glucose, and cholesterol. The recruitment of mTOR to lysosome is regulated by a complex of the Ras-regulated GTPases (RagA/B and RagC/D) that are themselves regulated by a set of transmembrane lysosomal and cytosolic proteins recruited to the lysosomes [139]. Recruitment of mTORC1 to lysosomes by amino acids relies on the vATPase [140] and on a set of sensors such a SLC38A9 that signals arginine sufficiency to mTORC1 [141]. In the presence of nutrients, mTORC1 is activated and promotes the phosphorylation of downstream targets. One of the mTOR targets is the transcription factor TFEB that is maintained inactive in a phosphorylated state [142].
Deprivation of any nutrients promoting mTORC1 activation signals prevents the phosphorylation of downstream targets and leads to activation of autophagy [139]. For example, autophagy is induced in case of nutrient stress, allowing recycling of intracellular components to restore or compensate the deficiency. Alteration of autophagy has been associated with many LSDs [2]. The activation of mTOR is, however, rarely impaired in these diseases. A recent study demonstrated that the lysosomal membrane transporter SLC38A9 allows activation of mTOR by cholesterol. Knocking-out NPC1 in HEK293 cells abolished mTOR activation by cholesterol [143]. Yet, in a mouse model of NPC where high amounts of cholesterol accumulate in lysosomes, no change in the activation of mTOR was observed [34], suggesting that mTOR is not directly implicated in the pathology. Increased levels of mTOR phosphorylation were observed upon downregulation of ATP13A2 in SH-SY5Y neuroblastoma cells [144] or in the brain of a mouse model of GM1 gangliosidosis [4]. Yet, in the GM1 gangliosidosis model, no increase in the phosphorylation state of the mTOR substrate S6 was observed [4]. In contrast in the twitcher mouse model of Krabbe disease, the accumulation of psychosin was associated with a slight decrease in mTOR phosphorylation [145]. Upon long-term deprivation, mTOR can be reactivated to promote the reformation of new lysosomes and promote the lysosomal homeostasis [11]. Fibroblasts derived from Cln7 knockout mice present a defect in the ability to adapt to long-term starvation conditions as shown by impaired mTORC1 reactivation [146].
In contrast to LSD, decreased activity of mTOR has been observed in many models of rare neurodegenerative diseases where lysosomal function was not primarily impaired. For example, loss of C9orf72 function decreases phosphorylation of the mTOR substrate S6K [60]. C9orf72 forms a stable complex with SMCR8 (Smith–Magenis syndrome chromosome region, candidate 8) [60,61,147], and loss of SMCR8 also increased mTOR activity [147]. In models of Huntington's disease or cerebellar ataxia SCA7, mTOR is trapped in polyglutamine aggregates [49,70], which could prevent its kinase activity.
Overactivation of the mTOR has been associated with neurodevelopmental defects and focal epilepsies [148]. Loss of function mutations in the GATOR1 or in the tuberous sclerosis complex that both inhibit mTOR or gain of function in mTOR have been found in patients affected by focal cortical dysplasia or focal epilepsies. These mutations lead to activation of both mTORC1 and mTORC2 complexes that regulate autophagy, but also other metabolic pathways. It is therefore not clear yet which pathway contributes to the pathology in this group of neurodevelopmental diseases.
Lysosomal calcium signaling and activation of the transcription factor TFEB
The role of calcium in the regulation of lysosomal function benefited from the investigation of the late endosome or lysosome cation-permeant channel TRPML1 whose function is lost in mucolipidosis type IV [149]. The best-characterized action of TRPML1 is the transport of calcium ions from lysosomal lumen to cytosol [149], which allowed to investigate the importance of calcium for the regulation of lysosomal function. The release of calcium by TRPML1 is regulated by the levels of PI(3,5)P2, a phosphoinositide predominantly found in late endosomes and lysosomes [150]. It was suggested that PI(3,5)P2 could activate TRPML1 by direct binding to the N terminus of the channel [151]. PI(3,5)P2 is generated by a protein complex that includes the lipid kinase Pikfyve, the scaffolding protein Vac14, and the lipid phosphatase Fig4 [152]. Inhibition of PI(3,5)P2 synthesis by the YM201636 inhibitor results in the accumulation of large late endosomes and lysosomes and a defect in endocytic trafficking similar that the one observed in cells lacking TRPML1 [151,153].
The local release of calcium by TRPML1 can regulate several functions of the lysosomes. Local calcium release by lysosomes was proposed to stimulate the kinase activity of mTORC1 complex [154]. It can also activate the calcium-dependent phosphatase calcineurin that dephosphorylates the transcription factor TFEB, allowing its nuclear translocation [155]. As a result, TFEB promotes the transcription of target genes that supports lysosome biogenesis and catabolism. It is therefore possible that calcium released by TRPML1 could simultaneously activate, via the calcineurin, and inhibit, via mTOR phosphorylation, the nuclear translocation of TFEB (Figure 1). The equilibrium between the two functions of TRPML1 is unclear, but it could contribute to a fine-tuning of TFEB nuclear translocation. An indirect control of lysosome on calcium homeostasis has also recently been proposed, where the impaired trafficking of cholesterol out of lysosome leads to a decrease in the concentration of cholesterol in the plasma membrane. This change in plasma membrane composition leads to the entry of calcium by store-operated calcium entry leading to a slight increase in cytosolic calcium levels [91]. Accumulation of cholesterol in lysosome was proposed to inhibit calcium release by TRPML1 [156]. Unlike the local release of calcium by TRPML1 in proximity of lysosome, the second mechanism does not allow a change in local calcium concentration. The two mechanisms may differently contribute to the regulation of TFEB translocation into the nucleus. It should also be noted that late endosomes and lysosomes have another type of calcium channel, Two Pore Channels, that are activated by the NAADP signaling molecules or by sphingosine [157,158]. Calcium release by Two Pore Channels can also promote the activation of TFEB [157,159], highlighting the complexity of the regulation of TFEB.
Beside its crucial role for the activation of TFEB, local release of calcium was also proposed to regulate membrane fusion/fission events in late endosome and lysosomes [151], although fusion of lysosomes and autophagosomes was not impaired in Drosophila model of mucolipidosis type IV [160]. Finally, calcium release by TRPML1 also promotes the traffic of lysosomes toward the perinuclear region by allowing the recruitment of dynein–dynactin motors [6].
The function of the TRPML1 channel is altered in several models of rare neurodegenerative diseases, highlighting the importance of lysosomal calcium signaling for the physiopathology of many diseases and not only mucolipidosis type IV. The function of TRPML1 is modulated by kinase and phosphatase allowing synthesis and degradation of the channel agonist PI(3,5)P2. Inhibiting PI(3,5)P2 synthesis in yeast leads to accumulation of large vacuoles [161]. In human, mutations of FIG4, a PI(3,5)P2 phosphatase, have been found in patients with the Charcot–Marie–Tooth type 4J neuropathy [162] as well as in patients affected by Yunis–Varon syndrome, a severe syndrome associated with brain abnormalities, facial dysmorphisms, and skeletal abnormalities [163]. It was proposed that the complete loss of function of FIG4 activity leads to Yunis–Varon syndrome, whereas hypomorphic mutations are responsible for Charcot–Marie–Tooth disease [163]. FIG4 belongs to a complex with the PI3P kinase PIK-FYVE and VAC 14, and its loss of function leads to a depletion in PI(3,5)P2 [162]. Loss of FIG4 function leads to higher levels of calcium in lysosomes and impaired lysosomal fission. These phenotypes can be rescued by application of the TRPML1 agonist ML-SA1 [95]. The activity of the TRPML1 channel is also inhibited by accumulation of sphingomyelin in lysosomes in models of NPC cells. Increasing TRPML1 expression or activity was sufficient to correct the trafficking defects and reduce lysosome storage and cholesterol accumulation in NPC cells [156]. This suggests that abnormal accumulation of luminal lipids causes secondary lysosome storage by blocking TRPML1- and calcium-dependent lysosomal trafficking.
Together, these observations highlight that the signaling pathways controlled by lysosomes are altered in a variety of rare neurodegenerative diseases, highlighting some physiopathological pathways contributing to neurodegeneration. It is also important to note that the activation of mTOR and TFEB pathways can also be used to monitor the alteration of the autophagy pathway.
Impact of Lysosome Dysfunction on the Autophagy Pathway
Lysosomes play a critical role in the autophagy pathway, as they degrade the autophagic material following fusion with autophagosomes. Again, LSD helped deciphering the mechanisms linking lysosome dysfunction to impairment of the autophagy pathway. In most LSD, impairment of lysosomal function leads to accumulation of autophagosomes, accumulation of the autophagic substrate p62, and a decrease in the autophagic flux that represents the rate at which the material is degraded through autophagy [2,164]. Similar alterations are also observed in most rare neurodegenerative diseases. However, despite these similarities, the defects in autophagy can be due to impairment at different steps of the process, including the formation of autophagosomes, fusion of autophagosomes with lysosomes, degradation of the autophagosomes, or recovery of lysosome membrane after the end of autophagy (Table 1).
Impaired autophagosome formation and autophagy cargo recognition
It is striking to note that several forms of FTLD or ALS are caused by mutations in genes encoding autophagy receptors, such as p62/SQSTM1, Optineurin, or Ubiquilin-2, or by the kinase Tank-binding kinase 1 (TBK1) that phosphorylates and regulates the autophagy receptors [165]. These proteins contribute to selectively recognize cargos that will be degraded by autophagy. P62/SQSTM1 and Optineurin play notably key role in the clearance of protein aggregates or damaged mitochondria, respectively [38,45]. Cargo recognition is also impaired by mutant huntingtin. The latter preferentially affects organelle sequestration, which could explain the higher presence of altered mitochondria observed in Huntington's disease patient cells [50]. However, huntingtin can also impair other critical steps in the initiation of autophagy. Indeed, expansion of polyglutamines was also shown to impair autophagosome formation. Sequestration of the key initiator of autophagy into mutant huntingtin or mutant ataxin-7 polyglutamine aggregates impairs the formation of autophagosomes [70,166]. Furthermore, the polyglutamine expansion of ataxin-3 impairs interaction of wild-type ataxin-3 with beclin1, promoting its degradation and thus decreasing the formation of autophagosomes [68].
Impaired autophagosome formation is also observed in genetic forms of FTLD or ALS. Downregulation of C9ORF72 in N2a cells or neurons derived from C9ORF72-induced pluripotent stem cells showed a decrease in the number of autophagosomes that was interpreted as impaired formation of autophagosomes [61,62]. Yet, downregulation or knockout of C9ORF72 decreased the activation of mTOR [60], which is supposed to promote activation of autophagy. This discrepancy could be due to the increase in autophagic flux observed in the absence of C9ORF72 [60,61]. C9ORF72 forms a complex with SMCR8, and both proteins are important to stabilize each other [147]. However, downregulation of C9ORF72 and SMCR8 has an opposite effect on autophagy induction and autophagic flux [61]. It was proposed that the difference in the action of the two proteins could be due to interaction with different Rab GTPases [61], but this must be clarified. These studies are important to reveal the role of C9ORF72 in the regulation of autophagy. However, patients with C9ORF72 mutations express reduced levels of the protein [167]. Thus, knockout models are not the best systems to investigate the alteration of autophagy pathway in the FTLD or ALS. Instead, investigation in cells or tissues derived from patients is more likely to reveal the role of autophagy due to mutations in C9ORF72.
Progranulin is also required for the formation of autophagosomes around Listeria in infected macrophages to clear cells from bacteria [168]. Progranulin deficiency causes impairment of autophagy, with contradictory results. While PGRN knockout neurons showed a decrease in autophagic flux, an acute ~ 50% reduction of PGRN levels using siRNA increased the autophagic flux [168,169]. These discrepancies are still unexplained but could result from differential regulation of autophagy in the models, as PGRN was proposed to stimulate autophagy induction [168]. Impaired autophagosome formation has been observed in other cases of rare neurodegenerative diseases. Loss of WDR45 in patients with a form of neurodegeneration with brain accumulation leads to impaired autophagy [52]. A study in C. elegans showed that its ortholog plays a role in the formation of autophagosomes [53]. The loss of tectonin-beta-propeller containing protein 2 (TECPR2), mutated in SPG49 form of HSP, is also required for the formation of autophagosomes [54,55].
These examples illustrate that rare neurodegenerative diseases can be caused by alteration in the early steps of autophagy, either autophagic cargo recognition or autophagosome formation. However, a recent study showed that loss of the presumed autophagy receptor ubiquilin in Drosophila was associated with accumulation of autophagosomes and autolysosomes as well as impaired acidification of lysosomes [41]. This highlights the need for a thorough characterization of the protein functions to elucidate their implication in neuronal death.
Impaired autophagosome–lysosome fusion and digestion of autolysosomes
Many models of rare neurodegenerative diseases present the accumulation of autophagosomes and/or autolysosomes. These accumulations can result from impaired degradation of autolysosomes or impaired fusion between lysosomes and autophagosomes. Yet, the frontier between the two types of alterations is not clear, as impaired autolysosome degradation can secondarily lead to impaired fusion. In both cases, this dysfunction results in a decrease in the autophagic flux.
Fusion of autophagosomes with lysosomes relies on proper retrograde transport of these vesicles toward the center of the cell to increase their probability of meeting. Consistently, mutant huntingtin, or some variants of dynactin observed in the forms of familial ALS impair the vesicular trafficking of autophagosomes [71,79], decreasing their fusion with lysosomes. Models of other rare neurodegenerative diseases have also pointed the importance of lysosomes motility for cell survival. For example, in a cellular model of CLN7, anterograde movement of lysosomes or endolysosomes was impaired and associated with cell survival while the cells were apparently autophagy competent [170]. Impaired traffic of endolysosomes was also observed in a mouse model with Chmp2b mutation that presents lysosomal pathology [109]. Although mutations in CHMP2B were shown to impair autophagosome degradation [171], the aberrant lysosomes observed in the animal model were not positive for the autophagy marker p62 [110], suggesting that aberrant lysosomes and accumulation of autophagososomes could be distinct pathologies. These observations highlight the importance of regulated lysosome trafficking to prevent neurodegeneration. However, in the latter examples, it is not clear whether the alteration of lysosome trafficking would affect the autophagy pathway.
The fusion of autophagososomes with lysosomes is also a critical step that is altered in several rare neurodegenerative diseases. For example, in cellular model of NPC, fusion of autophagosomes with late endosomes is impaired, due to perturbations of the formation of specific SNARE complexes [172], while no overt defects in the proteolytic activity of lysosomes were detected. This example highlights the importance of fusion machinery for the processing of autophagosomes. In other cases, the fusion of autophagosomes with lysosomes is impaired as a consequence of impaired degrative capacity of lysosomes. Retinal pigment epithelium of Cln3 mouse model presented the accumulation of autophagosomes fused with lysosomes containing undigested material, as well as accumulation of autophagosomes docked to lysosomes, without mixing of the content of the two compartments [14]. This suggests that impaired lysosomal processing could secondarily affect the process of fusion between autophagosomes and lysosomes. Recently, CLN3 was reported to be required for the interaction between Rab7A and PLEKHM1, which contributes to autophagosome–lysosome fusion [173].
Impaired digestion of autolysosomes has also been observed in models of cathepsin D or saposin C deficiency [174,175]. The impaired degradation of autolysosomes in Saposin C-deficient cells was restored by overexpression of the lysosomal proteases Cathepsin B or D [175], highlighting the importance of hydrolases for the degradation of the autolysosome content. The degradation of the content of autolysosomes can also be impeded by the loss of non-hydrolytic enzymes. For example, loss of ATP13A2 impaired lysosomal acidification, decreased proteolytic processing of lysosomal enzymes, reduced degradation of lysosomal substrates, and diminished lysosomal-mediated clearance of autophagosomes [28]. Clearance of autophagosomes was also defective in models of mucolipidosis type IV, where the loss of lysosomal calcium channel TRPML1 affects lysosomal function [160,176]. Some studies claimed that loss of TRPML1 affected only the degradative activity of lysosomes, but not the fusion between lysosomes and autophagosomes [160], whereas others showed a delay in the fusion between autophagosomes and late endosomes or lysosomes [177]. This highlights the interdependency of autophagosome–lysosome fusion and lysosomes hydrolytic activity.
In many diseases, the precise alterations in the autophagy process still need to be defined. For example, loss of EPG5 observed in patients with Vici syndrome was proposed to block the degradation of autolysosomes [83]. Another study suggested that it could block the degradative activities of autolysosomes or block the fusion of autophagosomes with lysosomes [84]. This highlights the need for thorough investigation of the autophagy pathway in the different models of rare diseases.
Impaired lysosome reformation from autolysosomes
mTOR is a key regulator of the autophagy pathway, as its inhibition following amino acid starvation promotes autophagy. However, upon long starvation, mTOR is reactivated, which attenuates autophagy and generates proto-lysosomal tubules and vesicles that extrude from autolysosomes [11]. This mechanism is thought to contribute to the maintenance of a pool of functional lysosomes. This process requires the degradation of the content of the autolysosomes, and was thus impaired in cells derived from patients affected by LSD such as Fabry disease or Gaucher disease [8,11]. The proteins spatacsin and spastizin, associated with HSPs SPG11 and SPG15, play a role in the initiation of the formation of tubules in autolysosomes [92]. Consequently, their absence leads in mouse models to the accumulation in neurons of autolysosomes together with the autophagy receptor p62 [89,93,130]. The formation of tubules on autolysosomes also requires the TRPML1 channel and its activation by the phosphoinositide PI(3,5)P2 [6]. Consistently, loss of FIG4 impairs the production of PI(3,5)P2 and impairs the fission of lysosomes [95]. Loss of FIG4 also downregulated the expression and activity of dynamin1, which interacts with spatacsin [90,95]. Importantly, loss of FIG4 and loss of spatacsin are both associated with the storage of material in lysosomes [90,96,130]. These data therefore suggest that the machinery required for lysosome reformation is also implicated in the clearance of lysosomal material. However, further investigations are required to define the molecular mechanisms leading to lysosome reformation and to understand its link with storage of material in lysosomes.
Claiming that a specific step of the autophagy pathway is altered in each neurodegenerative disease is, however, over-simplistic. For example, mutant huntingtin has been shown to alter various steps of the autophagy pathway. Indeed, mutant huntingtin was proposed to contribute to activation of the autophagy pathway [49], whereas another study claimed it could prevent autophagosome formation [68]. Furthermore, it impairs cargo recognition [50] and disrupts autophagosome trafficking leading to impaired fusion of autophagosomes with lysosomes [71], highlighting the diversity of alterations of the autophagy–lysosome pathway that can take place in neurodegenerative diseases.
Mechanisms Linking Impaired Autophagy/Lysosome Pathway to Neurodegeneration
One of the difficulties in investigating the role of the lysosome–autophagy pathway in neurodegeneration is to determine whether the detrimental consequences are solely due to the impairment of this cellular pathway, or whether they result from secondary impairments of its dysfunction. The difficulty to identify the cause of neuronal death following impaired autophagy lysosome pathway is particularly evident if we consider some LSD, where alterations of biochemical function due to the pathogenic mutations are well characterized. Yet, it is not known why the primary lysosomal defect leads to neurodegeneration and it has been proposed that engorgement of autophagy–lysosome pathway may not fully explain the clinical manifestations of some LSD [2,178]. Indeed, dysfunction of the autophagy–lysosome pathway can lead to accumulation of toxic proteins or dysfunctional organelles or can promote inflammation that may contribute to neurodegeneration.
Accumulation of impaired mitochondria and oxidative stress
The accumulation of impaired mitochondria has been reported in many forms of LSD [179], but also in other forms of rare neurodegenerative diseases. FTLD/ALS causing mutations in optineurin inhibit its ability to promote mitophagy [180], linking FTLD/ALS to mitochondrial dysfunction. P62 mutations associated with FTLD and ALS were also shown to inhibit complex I mitochondrial respiration and promote reactive oxygen species production [39]. Autophagy is critical for quality control of mitochondria by selectively degrading dysfunctional mitochondria [181]. Impairment of the autophagy–lysosome pathway has been proposed to promote the persistence of dysfunctional mitochondria and inducing oxidative stress in models of mucolipidosis type IV or in models of CLN [160,182]. This hypothesis is appealing, as impairment of mitophagy is implicated in some genetic forms of Parkinson's disease [183]. Yet, this hypothesis is challenged by some observations. For example, inhibition of autophagy using 3-methyladenine restored mitochondrial function in mucolipidosis II and III skin fibroblasts [184]. It was also recently proposed that mitochondrial biogenesis is impaired in models of NPC and acid sphingomyelinase deficiency [35], although it is not clear yet whether this dysfunction is a direct consequence of the impaired autophagy–lysosome pathway.
Accumulation of toxic proteins
Many neurodegenerative diseases are associated with the aggregation of proteins, which can be cleared by autophagy [1]. Autophagy process plays a role in removal of impaired organelles, but also in the clearance of protein aggregates. In most cases of FTLD and ALS, aggregation of TAR DNA-binding protein 43 (TDP43) is observed, although the mechanisms leading to such aggregation are poorly understood. It has been hypothesized that impairment of autophagy could contribute to such accumulation [165]. Impairment of autophagy by loss of progranulin or C9ORF72 promoted the neuronal aggregation of TDP-43 [168,185]. Expansion of polyglutamines in models of cerebellar ataxias or Huntington disease leads to the formation of insoluble protein aggregates that can trap some proteins important for activation of autophagy such as mTOR or Beclin 1 [49,70,166]. Stimulation of autophagy by rapamycin prevented huntingtin accumulation and prevented neurodegeneration in models of Huntington disease [49], highlighting the importance of aggregated huntingtin in the neurodegenerative process. However, it is still not clear why and how the so-called toxic proteins induce neuronal death.
Metabolic stress
Lysosomes hydrolyze the content of endocytic and autophagic vesicles, allowing the recycling of basic building blocks that can be used by the cells for the biosynthesis of new macromolecules. It is therefore conceivable that impairment of the autophagy–lysosome pathway could lead to metabolic stress, where some key metabolites are limiting for the synthesis of macromolecules [186]. Similarly, blockade of protein degradation by proteasome leads to a deleterious shortage of intracellular amino acids, which can be compensated by amino acid supplementation [187]. The activation of mTOR at the lysosome surface is under the control of amino acid sensors, including the lysosomal membrane SLC38A9 [141,188], supporting the idea that recycling of amino acids by lysosomes is important for cell physiology. Other metabolites also need to be recycled by lysosomes to ensure proper cell function and survival. For example, the recycling of sugars and lipids coming from the lysosomal degradation of glycosphingolipids is reused and represents the majority of glycosphingolipids that are synthesized in several cell lines [189].
Inflammation
The molecular deficits responsible for neurodegeneration in rare diseases are not restricted to neurons and can affect other cell types. Accordingly, it has emerged that microglia and astrocyte activation contributes to neurodegenerative phenotype in LSD or other neurodegenerative diseases [190,191]. Microglia are the main immune cells in the brain parenchyma, and they contribute to tissue homeostasis, notably by remodeling synapses and secreting neurotrophic factors. During neurodegeneration, microglia have been proposed to play both protective or deleterious roles. Indeed, primary neuronal damages can lead to activation of microglia that phagocytose cellular debris [192]. For example, in a mouse model of Sandhoff disease, the progression of symptoms was slowed down by deletion of tumor necrosis factor alpha, a pro-inflammatory cytokine [193]. In contrast, in a model of Niemann–Pick type A, disease progression was accelerated by chemical ablation of microglia [194]. These examples illustrate the complexity of the mechanisms underlying neuronal death in rare diseases caused by lysosomal dysfunction and call for thorough characterization and elucidation of these mechanisms.
Conclusions and Perspectives
Rare neurodegenerative diseases, by definition, affect a small number of patients, and it is elusive to develop a targeted therapeutic approach for each disease. Instead, it would be more rational to develop few therapeutic strategies that can apply to defined subgroups of rare diseases that share at least partially common physiopathology. Lysosome dysfunction is observed in a wide variety of rare neurodegenerative diseases, and often leads to impairment of autophagy. As reviewed here, a few alterations are observed in a large number of rare diseases, as, for example, the accumulation of autophagosomes or autolysosomes. However, the mechanisms underlying these cellular dysfunctions are often elusive. Furthermore, lysosomes are not only required for the degradation of autophagosomes. They also allow the degradation of macromolecules internalized by endocytosis, and lysosome dysfunction can impair the endocytosis pathway. For example, dominant mutations in Rab7 that are responsible for the Charcot–Marie–Tooth type 2B neuropathy [195] alter both endosomal trafficking and autophagy, highlighting the interdependence of late endosome and autophagosome degradation [81].
It will thus be important to elucidate in the future how and why the alterations of the autophagy–lysosome pathway lead to neurodegeneration. In most cases, the consequences of the impairment of lysosome on cellular functions that could contribute to neurodegeneration are not known (Table 1). We proposed and highlighted several potential pathways that are not mutually exclusive and that could be simultaneously altered in some diseases.
Based on the observation that autophagy–lysosome pathway is impaired in many forms of rare neurodegenerative diseases, several strategies have been tested to enhance autophagy as a therapeutic strategy [123]. The definition of subgroups of rare neurodegenerative diseases sharing similar physiopathology should support the use of similar strategies to a larger number of disorders. Yet, we must be aware that enhancement of autophagy may not be the panacea for all neurodegenerative diseases. This calls for further investigations to thoroughly decipher the mechanisms linking lysosomal dysfunction to impairment of the autophagy pathway and to neuronal death. These investigations should allow the clustering of rare neurodegenerative diseases into subgroups sharing similar pathological mechanisms and that could respond to similar therapy.
Acknowledgments
We thank B. Gurchenkov for his help with illustration. This work was supported by the “Investissements d'avenir” program grants (ANR-10-IAIHU-06) and (ANR-11-INBS-0011) and received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No. 779257-SOLVE-RD (to G.S.), the GIS-Maladies Rares Foundation (to G.S.), the E-Rare program (Neurolipid and Prepare consortia, to G.S.),and the European Research Council [European Research Council Starting Grant (No. 311149) to F.D.].
Edited by Manolis Fanto.
References
- 1.Menzies F.M., Fleming A., Rubinsztein D.C. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman A.P., Puertollano R., Raben N., Slaugenhaupt S., Walkley S.U., Ballabio A. Autophagy in lysosomal storage disorders. Autophagy. 2012;8:719–730. doi: 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braulke T., Bonifacino J.S. Sorting of lysosomal proteins. Biochim. Biophys. Acta (BBA) Molec Cell Res. 2009;1793:605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 4.Takamura A., Higaki K., Kajimaki K., Otsuka S., Ninomiya H., Matsuda J., Ohno K., Suzuki Y. Enhanced autophagy and mitochondrial aberrations in murine GM1-gangliosidosis. Biochem. Biophys. Res. Commun. 2008;367:616–622. doi: 10.1016/j.bbrc.2007.12.187. [DOI] [PubMed] [Google Scholar]
- 5.Kollmann K., Damme M., Markmann S., Morelle W., Schweizer M., Hermans-Borgmeyer I., Röchert A.K., Pohl S. Lysosomal dysfunction causes neurodegeneration in mucolipidosis II ‘knock-in’ mice. Brain. 2012;135:2661–2675. doi: 10.1093/brain/aws209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Rydzewski N., Hider A., Zhang X., Yang J., Wang W., Gao Q., Cheng X. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 2016;18:404–417. doi: 10.1038/ncb3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jennings J.J., Zhu J., Rbaibi Y., Luo X., Chu C.T., Kiselyov K. Mitochondrial aberrations in mucolipidosis type IV. J. Biol. Chem. 2006;281:39041–39050. doi: 10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- 8.Magalhaes J., Gegg M.E., Migdalska-Richards A., Doherty M.K., Whitfield P.D., Schapira A.H.V. Autophagic lysosome reformation dysfunction in glucocerebrosidase deficient cells: relevance to Parkinson disease. Hum. Mol. Genet. 2016;25:3432–3445. doi: 10.1093/hmg/ddw185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y., Liou B., Ran H., Skelton M.R., Williams M.T., Vorhees C.V., Kitatani K., Hannun Y.A. Neuronopathic Gaucher disease in the mouse: viable combined selective saposin C deficiency and mutant glucocerebrosidase (V394L) mice with glucosylsphingosine and glucosylceramide accumulation and progressive neurological deficits. Hum. Mol. Genet. 2010;19:1088–1097. doi: 10.1093/hmg/ddp580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osellame L.D., Rahim A.A., Hargreaves I.P., Gegg M.E., Richard-Londt A., Brandner S., Waddington S.N., Schapira A.H.V. Mitochondria and quality control defects in a mouse model of Gaucher disease—links to Parkinson’s disease. Cell Metab. 2013;17:941–953. doi: 10.1016/j.cmet.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liebau M.C., Braun F., Höpker K., Weitbrecht C., Bartels V., Müller R.-U., Brodesser S., Saleem M.A. Dysregulated autophagy contributes to podocyte damage in Fabry’s disease. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson M.P., Tse T.E., O’Quinn D.B., Percival S.M., Jaimes E.A., Warnock D.G., Shacka J.J. Autophagy–lysosome pathway associated neuropathology and axonal degeneration in the brains of alpha-galactosidase A-deficient mice. Acta Neuropathol Commun. 2014;2:20. doi: 10.1186/2051-5960-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wavre-Shapton S.T., Calvi A.A., Turmaine M., Seabra M.C., Cutler D.F., Futter C.E., Mitchison H.M. Photoreceptor phagosome processing defects and disturbed autophagy in retinal pigment epithelium of Cln3Δex1-6 mice modelling juvenile neuronal ceroid lipofuscinosis (Batten disease) Hum. Mol. Genet. 2015 doi: 10.1093/hmg/ddv406. ddv406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goebel H.H. Topical review: the neuronal ceroid-lipofuscinoses. J. Child Neurol. 1995;10:424–437. doi: 10.1177/088307389501000602. [DOI] [PubMed] [Google Scholar]
- 16.Brandenstein L., Schweizer M., Sedlacik J., Fiehler J., Storch S. Lysosomal dysfunction and impaired autophagy in a novel mouse model deficient for the lysosomal membrane protein Cln7. Hum. Mol. Genet. 2016;25:777–791. doi: 10.1093/hmg/ddv615. [DOI] [PubMed] [Google Scholar]
- 17.di Ronza A., Bajaj L., Sharma J., Sanagasetti D., Lotfi P., Adamski C.J., Collette J., Palmieri M. CLN8 is an endoplasmic reticulum cargo receptor that regulates lysosome biogenesis. Nat. Cell Biol. 2018;20:1370–1377. doi: 10.1038/s41556-018-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolikova J., Afzalov R., Surin A., Lehesjoki A.-E., Khiroug L. Deficient mitochondrial Ca2+ buffering in the Cln8mnd mouse model of neuronal ceroid lipofuscinosis. Cell Calcium. 2011;50:491–501. doi: 10.1016/j.ceca.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Galizzi G., Russo D., Deidda I., Cascio C., Passantino R., Guarneri R., Bigini P., Mennini T. Different early ER-stress responses in the CLN8mnd mouse model of neuronal ceroid lipofuscinosis. Neurosci. Lett. 2011;488:258–262. doi: 10.1016/j.neulet.2010.11.041. [DOI] [PubMed] [Google Scholar]
- 20.Bronson R.T., Lake B.D., Cook S., Taylor S., Davisson M.T. Motor neuron degeneration of mice is a model of neuronal ceroid lipofuscinosis (Batten’s disease) Ann. Neurol. 1993;33:381–385. doi: 10.1002/ana.410330408. [DOI] [PubMed] [Google Scholar]
- 21.C.A. Pardo, B.A. Rabin, D.N. Palmer, D.L. Price,Accumulation of the adenosine triphosphate synthase subunit C in the mnd mutant mouse. A model for neuronal ceroid lipofuscinosis, Am. J. Pathol. 144 (1994) 829–835. [PMC free article] [PubMed]
- 22.Shacka J.J., Klocke B.J., Young C., Shibata M., Olney J.W., Uchiyama Y., Saftig P., Roth K.A. Cathepsin D deficiency induces persistent neurodegeneration in the absence of Bax-dependent apoptosis. J. Neurosci. 2007;27:2081–2090. doi: 10.1523/JNEUROSCI.5577-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marques A.R.A., Di Spiezio A., Thießen N., Schmidt L., Grötzinger J., Lüllmann-Rauch R., Damme M., Storck S.E. Enzyme replacement therapy with recombinant pro-CTSD (cathepsin D) corrects defective proteolysis and autophagy in neuronal ceroid lipofuscinosis. Autophagy. 2019:1–15. doi: 10.1080/15548627.2019.1637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed Z., Sheng H., Xu Y., Lin W.-L., Innes A.E., Gass J., Yu X., Hou H. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am. J. Pathol. 2010;177:311–324. doi: 10.2353/ajpath.2010.090915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valdez C., Wong Y.C., Schwake M., Bu G., Wszolek Z.K., Krainc D. Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum. Mol. Genet. 2017;26:4861–4872. doi: 10.1093/hmg/ddx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X., Sun L., Bracko O., Choi J.W., Jia Y., Nana A.L., Brady O.A., Hernandez J.C.C. Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat. Commun. 2017;8:15277. doi: 10.1038/ncomms15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holler C.J., Taylor G., Deng Q., Kukar T. Intracellular proteolysis of progranulin generates stable, lysosomal granulins that are haploinsufficient in patients with frontotemporal dementia caused by GRN mutations, ENeuro. ENEURO. 2017;4(2017):0100–0117. doi: 10.1523/ENEURO.0100-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dehay B., Ramirez A., Martinez-Vicente M., Perier C., Canron M.-H., Doudnikoff E., Vital A., Vila M. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc. Natl. Acad. Sci. 2012;109:9611–9616. doi: 10.1073/pnas.1112368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R., Tan J., Chen T., Han H., Tian R., Tan Y., Wu Y., Cui J. ATP13A2 facilitates HDAC6 recruitment to lysosome to promote autophagosome–lysosome fusion. J. Cell Biol. 2019;218:267–284. doi: 10.1083/jcb.201804165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirose T., Cabrera-Socorro A., Chitayat D., Lemonnier T., Féraud O., Cifuentes-Diaz C., Gervasi N., Mombereau C. ATP6AP2 variant impairs CNS development and neuronal survival to cause fulminant neurodegeneration. J. Clin. Investig. 2019;129:2145–2162. doi: 10.1172/JCI79990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. Dubos, A. Castells-Nobau, H. Meziane, M.A.W. Oortveld, X. Houbaert, G. Iacono, C. Martin, C. Mittelhaeuser, V. Lalanne, J.M. Kramer, A. Bhukel, C. Quentin, J. Slabbert, P. Verstreken, S.J. Sigrist, N. Messaddeq, M.-C. Birling, M. Selloum, H.G. Stunnenberg, Y. Humeau, A. Schenck, Y. Herault, Conditional depletion of intellectual disability and Parkinsonism candidate gene ATP6AP2 in fly and mouse induces cognitive impairment and neurodegeneration, Hum. Mol. Genet. 24 (2015) 6736–6755. 10.1093/hmg/ddv380. [DOI] [PMC free article] [PubMed]
- 32.Patterson M. Niemann–Pick Disease Type C. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J., Stephens K., Amemiya A., editors. GeneReviews®. University of Washington; Seattle, Seattle (WA): 2000. http://www.ncbi.nlm.nih.gov/books/NBK1296/ [PubMed] [Google Scholar]
- 33.Walkley S.U. Secondary accumulation of gangliosides in lysosomal storage disorders. Semin. Cell Dev. Biol. 2004;15:433–444. doi: 10.1016/j.semcdb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Pacheco C.D., Kunkel R., Lieberman A.P. Autophagy in Niemann–Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum. Mol. Genet. 2007;16:1495–1503. doi: 10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- 35.Yambire K.F., Fernandez-Mosquera L., Steinfeld R., Mühle C., Ikonen E., Milosevic I., Raimundo N. Mitochondrial biogenesis is transcriptionally repressed in lysosomal lipid storage diseases. ELife. 2019;8 doi: 10.7554/eLife.39598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao G., Yao Y., Liu J., Yu Z., Cheung S., Xie A., Liang X., Bi X. Cholesterol accumulation is associated with lysosomal dysfunction and autophagic stress in Npc1 −/− mouse brain. Am. J. Pathol. 2007;171:962–975. doi: 10.2353/ajpath.2007.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meske V., Erz J., Priesnitz T., Ohm T.-G. The autophagic defect in Niemann–Pick disease type C neurons differs from somatic cells and reduces neuronal viability. Neurobiol. Dis. 2014;64:88–97. doi: 10.1016/j.nbd.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 38.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.-A., Outzen H., Øvervatn A., Bjørkøy G. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 39.Bartolome F., Esteras N., Martin-Requero A., Boutoleau-Bretonniere C., Vercelletto M., Gabelle A., Le Ber I., Honda T. Pathogenic p62/SQSTM1 mutations impair energy metabolism through limitation of mitochondrial substrates. Sci. Rep. 2017;7:1666. doi: 10.1038/s41598-017-01678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Øvervatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Şentürk M., Lin G., Zuo Z., Mao D., Watson E., Mikos A.G., Bellen H.J. Ubiquilins regulate autophagic flux through mTOR signalling and lysosomal acidification. Nat. Cell Biol. 2019;21:384–396. doi: 10.1038/s41556-019-0281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Q., Liu M., Huang C., Liu X., Huang B., Li N., Zhou H., Xia X.-G. Pathogenic Ubqln2 gains toxic properties to induce neuron death. Acta Neuropathol. 2015;129:417–428. doi: 10.1007/s00401-014-1367-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothenberg C., Srinivasan D., Mah L., Kaushik S., Peterhoff C.M., Ugolino J., Fang S., Cuervo A.M. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet. 2010;19:3219–3232. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.N’Diaye E., Kajihara K.K., Hsieh I., Morisaki H., Debnath J., Brown E.J. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–179. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richter B., Sliter D.A., Herhaus L., Stolz A., Wang C., Beli P., Zaffagnini G., Wild P. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wild P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R., Richter B., Korac J. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto G., Shimogori T., Hattori N., Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum. Mol. Genet. 2015;24:4429–4442. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- 48.Moore A.S., Holzbaur E.L.F. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E3349–E3358. doi: 10.1073/pnas.1523810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., de Vries R., Arias E. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim M., Sandford E., Gatica D., Qiu Y., Liu X., Zheng Y., Schulman B.A., Xu J. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. ELife. 2016;5 doi: 10.7554/eLife.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitsu H., Nishimura T., Muramatsu K., Kodera H., Kumada S., Sugai K., Kasai-Yoshida E., Sawaura N. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 2013;45:445–449. doi: 10.1038/ng.2562. [DOI] [PubMed] [Google Scholar]
- 53.Lu Q., Yang P., Huang X., Hu W., Guo B., Wu F., Lin L., Kovács A.L. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 54.Oz-Levi D., Ben-Zeev B., Ruzzo E.K., Hitomi Y., Gelman A., Pelak K., Anikster Y., Reznik-Wolf H. Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am. J. Hum. Genet. 2012;91:1065–1072. doi: 10.1016/j.ajhg.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stadel D., Millarte V., Tillmann K.D., Huber J., Tamin-Yecheskel B.-C., Akutsu M., Demishtein A., Ben-Zeev B. TECPR2 cooperates with LC3C to regulate COPII-dependent ER export. Mol. Cell. 2015;60:89–104. doi: 10.1016/j.molcel.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 56.Mattera R., Park S.Y., De Pace R., Guardia C.M., Bonifacino J.S. AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E10697–E10706. doi: 10.1073/pnas.1717327114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Pace R., Skirzewski M., Damme M., Mattera R., Mercurio J., Foster A.M., Cuitino L., Jarnik M. Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies A.K., Itzhak D.N., Edgar J.R., Archuleta T.L., Hirst J., Jackson L.P., Robinson M.S., Borner G.H.H. AP-4 vesicles contribute to spatial control of autophagy via RUSC-dependent peripheral delivery of ATG9A. Nat. Commun. 2018;9:3958. doi: 10.1038/s41467-018-06172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ivankovic D., Drew J., Lesept F., White I.J., López Doménech G., Tooze S.A., Kittler J.T. Axonal autophagosome maturation defect through failure of ATG9A sorting underpins pathology in AP-4 deficiency syndrome. Autophagy. 2019:1–17. doi: 10.1080/15548627.2019.1615302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ugolino J., Ji Y.J., Conchina K., Chu J., Nirujogi R.S., Pandey A., Brady N.R., Hamacher-Brady A. Loss of C9orf72 enhances autophagic activity via deregulated mTOR and TFEB signaling. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang M., Liang C., Swaminathan K., Herrlinger S., Lai F., Shiekhattar R., Chen J.-F. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi Y., Hung S.-T., Rocha G., Lin S., Linares G.R., Staats K.A., Seah C., Wang Y. Identification and therapeutic rescue of autophagosome and glutamate receptor defects in C9ORF72 and sporadic ALS neurons. JCI Insight. 2019;4 doi: 10.1172/jci.insight.127736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bose J.K., Huang C.-C., Shen C.-K.J. Regulation of autophagy by neuropathological protein TDP-43. J. Biol. Chem. 2011;286:44441–44448. doi: 10.1074/jbc.M111.237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia Q., Wang H., Hao Z., Fu C., Hu Q., Gao F., Ren H., Chen D. TDP-43 loss of function increases TFEB activity and blocks autophagosome–lysosome fusion. EMBO J. 2016;35:121–142. doi: 10.15252/embj.201591998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soo K.Y., Sultana J., King A., Atkinson R., Warraich S.T., Sundaramoorthy V., Blair I., Farg M.A. ALS-associated mutant FUS inhibits macroautophagy which is restored by overexpression of Rab1. Cell Death Dis. 2015;1:15030. doi: 10.1038/cddiscovery.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morimoto N., Nagai M., Ohta Y., Miyazaki K., Kurata T., Morimoto M., Murakami T., Takehisa Y. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 67.Bandyopadhyay U., Nagy M., Fenton W.A., Horwich A.L. Absence of lipofuscin in motor neurons of SOD1-linked ALS mice. Proc. Natl. Acad. Sci. 2014;111:11055–11060. doi: 10.1073/pnas.1409314111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ashkenazi A., Bento C.F., Ricketts T., Vicinanza M., Siddiqi F., Pavel M., Squitieri F., Hardenberg M.C. Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature. 2017;545:108–111. doi: 10.1038/nature22078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seidel K., den Dunnen W.F.A., Schultz C., Paulson H., Frank S., de Vos R.A., Brunt E.R., Deller T. Axonal inclusions in spinocerebellar ataxia type 3. Acta Neuropathol. 2010;120:449–460. doi: 10.1007/s00401-010-0717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alves S., Cormier-Dequaire F., Marinello M., Marais T., Muriel M.-P., Beaumatin F., Charbonnier-Beaupel F., Tahiri K. The autophagy/lysosome pathway is impaired in SCA7 patients and SCA7 knock-in mice. Acta Neuropathol. 2014;128:705–722. doi: 10.1007/s00401-014-1289-8. [DOI] [PubMed] [Google Scholar]
- 71.Wong Y.C., Holzbaur E.L.F. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 2014;34:1293–1305. doi: 10.1523/JNEUROSCI.1870-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie Y., Zhou B., Lin M.-Y., Wang S., Foust K.D., Sheng Z.-H. Endolysosomal deficits augment mitochondria pathology in spinal motor neurons of asymptomatic fALS mice. Neuron. 2015;87:355–370. doi: 10.1016/j.neuron.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hadano S., Otomo A., Kunita R., Suzuki-Utsunomiya K., Akatsuka A., Koike M., Aoki M., Uchiyama Y. Loss of ALS2/Alsin exacerbates motor dysfunction in a SOD1H46R-expressing mouse ALS model by disturbing endolysosomal trafficking. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Papadopoulos C., Kirchner P., Bug M., Grum D., Koerver L., Schulze N., Poehler R., Dressler A. VCP/p97 cooperates with YOD 1, UBXD 1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2017;36:135–150. doi: 10.15252/embj.201695148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arhzaouy K., Papadopoulos C., Schulze N., Pittman S.K., Meyer H., Weihl C.C. VCP maintains lysosomal homeostasis and TFEB activity in differentiated skeletal muscle. Autophagy. 2019;15:1082–1099. doi: 10.1080/15548627.2019.1569933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson A.E., Shu H., Hauswirth A.G., Tong A., Davis G.W. VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo. ELife. 2015;4 doi: 10.7554/eLife.07366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu Y., Zhang Z., Sun D., Sweeney S.T., Gao F.-B. Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol. Cell. 2013;52:264–271. doi: 10.1016/j.molcel.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Urwin H., Authier A., Nielsen J.E., Metcalf D., Powell C., Froud K., Malcolm D.S., Holm I. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum. Mol. Genet. 2010;19:2228–2238. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ikenaka K., Kawai K., Katsuno M., Huang Z., Jiang Y.-M., Iguchi Y., Kobayashi K., Kimata T. dnc-1/dynactin 1 knockdown disrupts transport of autophagosomes and induces motor neuron degeneration. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Laird F.M., Farah M.H., Ackerley S., Hoke A., Maragakis N., Rothstein J.D., Griffin J., Price D.L. Motor neuron disease occurring in a mutant dynactin mouse model is characterized by defects in vesicular trafficking. J. Neurosci. 2008;28:1997–2005. doi: 10.1523/JNEUROSCI.4231-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Colecchia D., Stasi M., Leonardi M., Manganelli F., Nolano M., Veneziani B.M., Santoro L., Eskelinen E.-L. Alterations of autophagy in the peripheral neuropathy Charcot–Marie–Tooth type 2B. Autophagy. 2018;14:930–941. doi: 10.1080/15548627.2017.1388475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ponomareva O.Y., Eliceiri K.W., Halloran M.C. Charcot–Marie–Tooth 2b associated Rab7 mutations cause axon growth and guidance defects during vertebrate sensory neuron development. Neural Dev. 2016;11:2. doi: 10.1186/s13064-016-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 84.Zhao H., Zhao Y.G., Wang X., Xu L., Miao L., Feng D., Chen Q., Kovács A.L. Mice deficient in Epg5 exhibit selective neuronal vulnerability to degeneration. J. Cell Biol. 2013;200:731–741. doi: 10.1083/jcb.201211014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akizu N., Cantagrel V., Zaki M.S., Al-Gazali L., Wang X., Rosti R.O., Dikoglu E., Gelot A.B. Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome–autophagosome dysfunction. Nat. Genet. 2015;47:528–534. doi: 10.1038/ng.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bryant D., Liu Y., Datta S., Hariri H., Seda M., Anderson G., Peskett E., Demetriou C. SNX14 mutations affect endoplasmic reticulum-associated neutral lipid metabolism in autosomal recessive spinocerebellar ataxia 20. Hum. Mol. Genet. 2018;27:1927–1940. doi: 10.1093/hmg/ddy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baron O., Boudi A., Dias C., Schilling M., Nölle A., Vizcay-Barrena G., Rattray I., Jungbluth H. Stall in canonical autophagy-lysosome pathways prompts nucleophagy-based nuclear breakdown in neurodegeneration. Curr. Biol. 2017;27:3626–3642. doi: 10.1016/j.cub.2017.10.054. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nisoli I., Chauvin J.P., Napoletano F., Calamita P., Zanin V., Fanto M., Charroux B. Neurodegeneration by polyglutamine Atrophin is not rescued by induction of autophagy. Cell Death Differ. 2010;17:1577–1587. doi: 10.1038/cdd.2010.31. [DOI] [PubMed] [Google Scholar]
- 89.Varga R.-E., Khundadze M., Damme M., Nietzsche S., Hoffmann B., Stauber T., Koch N., Hennings J.C. In vivo evidence for lysosome depletion and impaired autophagic clearance in hereditary spastic paraplegia type SPG11. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boutry M., Branchu J., Lustremant C., Pujol C., Pernelle J., Matusiak R., Seyer A., Poirel M. Inhibition of lysosome membrane recycling causes accumulation of gangliosides that contribute to neurodegeneration. Cell Rep. 2018;23:3813–3826. doi: 10.1016/j.celrep.2018.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boutry M., Pierga A., Matusiak R., Branchu J., Houllegatte M., Ibrahim Y., Balse E., El Hachimi K.-H. Loss of spatacsin impairs cholesterol trafficking and calcium homeostasis. Cell Biology. 2019 doi: 10.1101/605964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang J., Lee S., Blackstone C. Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J. Clin. Invest. 2014;124:5249–5262. doi: 10.1172/JCI77598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khundadze M., Kollmann K., Koch N., Biskup C., Nietzsche S., Zimmer G., Hennings J.C., Huebner A.K. A hereditary spastic paraplegia mouse model supports a role of ZFYVE26/SPASTIZIN for the endolysosomal system. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khundadze M., Ribaudo F., Hussain A., Rosentreter J., Nietzsche S., Thelen M., Winter D., Hoffmann B. A mouse model for SPG48 reveals a block of autophagic flux upon disruption of adaptor protein complex five. Neurobiol. Dis. 2019;127:419–431. doi: 10.1016/j.nbd.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 95.Zou J., Hu B., Arpag S., Yan Q., Hamilton A., Zeng Y.-S., Vanoye C.G., Li J. Reactivation of lysosomal Ca2+ efflux rescues abnormal lysosomal storage in FIG4-deficient cells. J. Neurosci. 2015;35:6801–6812. doi: 10.1523/JNEUROSCI.4442-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martyn C., Li J. Fig4 deficiency: a newly emerged lysosomal storage disorder? Prog. Neurobiol. 2013;101–102:35–45. doi: 10.1016/j.pneurobio.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.H. Hers, α-Glucosidase deficiency in generalized glycogen-storage disease (Pompe's disease), Biochem. J. 86 (1963) 11–16. 10.1042/bj0860011. [DOI] [PMC free article] [PubMed]
- 98.Jeyakumar M., Dwek R.A., Butters T.D., Platt F.M. Storage solutions: treating lysosomal disorders of the brain. Nat. Rev. Neurosci. 2005;6:713–725. doi: 10.1038/nrn1725. [DOI] [PubMed] [Google Scholar]
- 99.Butz E.S., Chandrachud U., Mole S.E., Cotman S.L. Moving towards a new era of genomics in the neuronal ceroid lipofuscinoses. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2019 doi: 10.1016/j.bbadis.2019.165571. [DOI] [PubMed] [Google Scholar]
- 100.Carstea E.D. Niemann–Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 101.Loftus S.K. Murine model of Niemann–Pick C disease: mutation in a cholesterol homeostasis gene. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- 102.Naureckiene S. Identification of HE1 as the second gene of Niemann–Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 103.Strong M.J., Abrahams S., Goldstein L.H., Woolley S., Mclaughlin P., Snowden J., Mioshi E., Roberts-South A. Amyotrophic lateral sclerosis—frontotemporal spectrum disorder (ALS-FTSD): revised diagnostic criteria. Amyotroph. Lateral Scler. Frontotemporal Degener. 2017;18:153–174. doi: 10.1080/21678421.2016.1267768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 105.Smith K.R., Damiano J., Franceschetti S., Carpenter S., Canafoglia L., Morbin M., Rossi G., Pareyson D. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet. 2012;90:1102–1107. doi: 10.1016/j.ajhg.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hu F., Padukkavidana T., Vægter C.B., Brady O.A., Zheng Y., Mackenzie I.R., Feldman H.H., Nykjaer A. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68:654–667. doi: 10.1016/j.neuron.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou X., Sun L., Bastos de Oliveira F., Qi X., Brown W.J., Smolka M.B., Sun Y., Hu F. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J. Cell Biol. 2015;210:991–1002. doi: 10.1083/jcb.201502029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 109.Clayton E.L., Milioto C., Muralidharan B., Norona F.E., Edgar J.R., Soriano A., Jafar-nejad P., Rigo F. Frontotemporal dementia causative CHMP2B impairs neuronal endolysosomal traffic-rescue by TMEM106B knockdown. Brain. 2018;141:3428–3442. doi: 10.1093/brain/awy284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.The FReJA consortium, Clayton E.L., Mizielinska S., Edgar J.R., Nielsen T.T., Marshall S., Norona F.E., Robbins M. Frontotemporal dementia caused by CHMP2B mutation is characterised by neuronal lysosomal storage pathology. Acta Neuropathol. 2015;130:511–523. doi: 10.1007/s00401-015-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mori K., Weng S.-M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 112.Amick J., Roczniak-Ferguson A., Ferguson S.M. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. MBoC. 2016;27:3040–3051. doi: 10.1091/mbc.e16-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Farg M.A., Sundaramoorthy V., Sultana J.M., Yang S., Atkinson R.A.K., Levina V., Halloran M.A., Gleeson P.A. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 2014;23:3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Corrionero A., Horvitz H.R. A C9orf72 ALS/FTD ortholog acts in endolysosomal degradation and lysosomal homeostasis. Curr. Biol. 2018;28:1522–1535. doi: 10.1016/j.cub.2018.03.063. e5. [DOI] [PubMed] [Google Scholar]
- 115.Sullivan P.M., Zhou X., Robins A.M., Paushter D.H., Kim D., Smolka M.B., Hu F. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy–lysosome pathway. Acta Neuropathol Commun. 2016;4:51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Van Deerlin V.M., Sleiman P.M.A., Martinez-Lage M., Chen-Plotkin A., Wang L.-S., Graff-Radford N.R., Dickson D.W., Rademakers R. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen-Plotkin A.S., Unger T.L., Gallagher M.D., Bill E., Kwong L.K., Volpicelli-Daley L., Busch J.I., Akle S. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J. Neurosci. 2012;32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Finch N., Carrasquillo M.M., Baker M., Rutherford N.J., Coppola G., DeJesus-Hernandez M., Crook R., Hunter T. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76:467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brady O.A., Zheng Y., Murphy K., Huang M., Hu F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum. Mol. Genet. 2013;22:685–695. doi: 10.1093/hmg/dds475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schwenk B.M., Lang C.M., Hogl S., Tahirovic S., Orozco D., Rentzsch K., Lichtenthaler S.F., Hoogenraad C.C. The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J. 2013 doi: 10.1002/embj.201385857. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Busch J.I., Unger T.L., Jain N., Skrinak R.T., Charan R.A., Chen-Plotkin A.S. Increased expression of the frontotemporal dementia risk factor TMEM106B causes C9orf72-dependent alterations in lysosomes. Hum. Mol. Genet. 2016 doi: 10.1093/hmg/ddw127. ddw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Klein Z.A., Takahashi H., Ma M., Stagi M., Zhou M., Lam T.T., Strittmatter S.M. Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron. 2017;95:281–296. doi: 10.1016/j.neuron.2017.06.026. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sharma J., di Ronza A., Lotfi P., Sardiello M. Lysosomes and brain health. Annu. Rev. Neurosci. 2018;41:255–276. doi: 10.1146/annurev-neuro-080317-061804. [DOI] [PubMed] [Google Scholar]
- 124.Bras J., Verloes A., Schneider S.A., Mole S.E., Guerreiro R.J. Mutation of the parkinsonism gene ATP13A2 causes neuronal ceroid-lipofuscinosis. Hum. Mol. Genet. 2012;21:2646–2650. doi: 10.1093/hmg/dds089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Estrada-Cuzcano A., Martin S., Chamova T., Synofzik M., Timmann D., Holemans T., Andreeva A., Reichbauer J. Loss-of-function mutations in the ATP13A2/PARK9 gene cause complicated hereditary spastic paraplegia (SPG78) Brain. 2017;140:287–305. doi: 10.1093/brain/aww307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ramirez A., Heimbach A., Gründemann J., Stiller B., Hampshire D., Cid L.P., Goebel I., Mubaidin A.F. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 127.Korvatska O., Strand N.S., Berndt J.D., Strovas T., Chen D.-H., Leverenz J.B., Kiianitsa K., Mata I.F. Altered splicing of ATP6AP2 causes X-linked parkinsonism with spasticity (XPDS) Hum. Mol. Genet. 2013;22:3259–3268. doi: 10.1093/hmg/ddt180. https://doi.org/10.1093/hmg/ddt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kinouchi K., Ichihara A., Sano M., Sun-Wada G.-H., Wada Y., Kurauchi-Mito A., Bokuda K., Narita T. The (Pro)renin receptor/ATP6AP2 is essential for vacuolar H+-ATPase assembly in murine cardiomyocytes. Circ. Res. 2010;107:30–34. doi: 10.1161/CIRCRESAHA.110.224667. [DOI] [PubMed] [Google Scholar]
- 129.Kissing S., Rudnik S., Damme M., Lüllmann-Rauch R., Ichihara A., Kornak U., Eskelinen E.-L., Jabs S. Disruption of the vacuolar-type H +-ATPase complex in liver causes MTORC1-independent accumulation of autophagic vacuoles and lysosomes. Autophagy. 2017;13:670–685. doi: 10.1080/15548627.2017.1280216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Branchu J., Boutry M., Sourd L., Depp M., Leone C., Corriger A., Vallucci M., Esteves T. Loss of spatacsin function alters lysosomal lipid clearance leading to upper and lower motor neuron degeneration. Neurobiol. Dis. 2017;102:21–37. doi: 10.1016/j.nbd.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hirst J., Edgar J.R., Esteves T., Darios F., Madeo M., Chang J., Roda R.H., Dürr A. Loss of AP-5 results in accumulation of aberrant endolysosomes: defining a new type of lysosomal storage disease. Hum. Mol. Genet. 2015;24:4984–4996. doi: 10.1093/hmg/ddv220. https://doi.org/10.1093/hmg/ddv220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Renvoisé B., Chang J., Singh R., Yonekawa S., FitzGibbon E.J., Mankodi A., Vanderver A., Schindler A.B. Lysosomal abnormalities in hereditary spastic paraplegia types SPG15 and SPG11. Ann Clin Transl Neurol. 2014;1:379–389. doi: 10.1002/acn3.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Parkinson-Lawrence E.J., Shandala T., Prodoehl M., Plew R., Borlace G.N., Brooks D.A. Lysosomal storage disease: revealing lysosomal function and physiology. Physiology. 2010;25:102–115. doi: 10.1152/physiol.00041.2009. [DOI] [PubMed] [Google Scholar]
- 134.Allison R., Edgar J.R., Pearson G., Rizo T., Newton T., Günther S., Berner F., Hague J. Defects in ER–endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol. 2017;216:1337–1355. doi: 10.1083/jcb.201609033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yvert G. SCA7 mouse models show selective stabilization of mutant ataxin-7 and similar cellular responses in different neuronal cell types. Hum. Mol. Genet. 2001;10:1679–1692. doi: 10.1093/hmg/10.16.1679. [DOI] [PubMed] [Google Scholar]
- 136.Miyashita T., Nagao K., Ohmi K., Yanagisawa H., Okamura-Oho Y., Yamada M. Intracellular aggregate formation of dentatorubral-pallidoluysian atrophy (DRPLA) protein with the extended polyglutamine. Biochem. Biophys. Res. Commun. 1998;249:96–102. doi: 10.1006/bbrc.1998.9096. [DOI] [PubMed] [Google Scholar]
- 137.Lie P.P.Y., Nixon R.A. Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol. Dis. 2019;122:94–105. doi: 10.1016/j.nbd.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bai X., Ma D., Liu A., Shen X., Wang Q.J., Liu Y., Jiang Y. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318:977–980. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- 139.Kim J., Guan K.-L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019;21:63–71. doi: 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- 140.Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H +-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang S., Tsun Z.-Y., Wolfson R.L., Shen K., Wyant G.A., Plovanich M.E., Yuan E.D., Jones T.D. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB: self-regulation of the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Castellano B.M., Thelen A.M., Moldavski O., Feltes M., van der Welle R.E.N., Mydock-McGrane L., Jiang X., van Eijkeren R.J. Lysosomal cholesterol activates mTORC1 via an SLC38A9–Niemann–Pick C1 signaling complex. Science. 2017;355:1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gusdon A.M., Zhu J., Van Houten B., Chu C.T. ATP13A2 regulates mitochondrial bioenergetics through macroautophagy. Neurobiol. Dis. 2012;45:962–972. doi: 10.1016/j.nbd.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.T. Sural-Fehr, H. Singh, L. Cantuti-Catelvetri, H. Zhu, M.S. Marshall, R. Rebiai, M.J. Jastrzebski, M.I. Givogri, M.M. Rasenick, E.R. Bongarzone, Inhibition of the IGF-1–PI3K–Akt–mTORC2 pathway in lipid rafts increases neuronal vulnerability in a genetic lysosomal glycosphingolipidosis, Dis. Model. Mech. 12 (2019) dmm036590. 10.1242/dmm.036590. [DOI] [PMC free article] [PubMed]
- 146.Danyukova T., Ariunbat K., Thelen M., Brocke-Ahmadinejad N., Mole S.E., Storch S. Loss of CLN7 results in depletion of soluble lysosomal proteins and impaired mTOR reactivation. Hum. Mol. Genet. 2018;27:1711–1722. doi: 10.1093/hmg/ddy076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lan Y., Sullivan P.M., Hu F. SMCR8 negatively regulates AKT and MTORC1 signaling to modulate lysosome biogenesis and tissue homeostasis. Autophagy. 2019;15:871–885. doi: 10.1080/15548627.2019.1569914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Marsan E., Baulac S. Review: mechanistic target of rapamycin (mTOR) pathway, focal cortical dysplasia and epilepsy. Neuropathol. Appl. Neurobiol. 2018;44:6–17. doi: 10.1111/nan.12463. [DOI] [PubMed] [Google Scholar]
- 149.Waller-Evans H., Lloyd-Evans E. Regulation of TRPML1 function. Biochm. Soc. Trans. 2015;43:442–446. doi: 10.1042/BST20140311. [DOI] [PubMed] [Google Scholar]
- 150.Ho C.Y., Alghamdi T.A., Botelho R.J. Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic. 2012;13:1–8. doi: 10.1111/j.1600-0854.2011.01246.x. [DOI] [PubMed] [Google Scholar]
- 151.Dong X., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y. PI(3,5)P2 controls membrane trafficking by direct activation of mucolipin Ca2 + release channels in the endolysosome. Nat. Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zolov S.N., Bridges D., Zhang Y., Lee W.-W., Riehle E., Verma R., Lenk G.M., Converso-Baran K. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl. Acad. Sci. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.H.B.J. Jefferies, F.T. Cooke, P. Jat, C. Boucheron, T. Koizumi, M. Hayakawa, H. Kaizawa, T. Ohishi, P. Workman, M.D. Waterfield, P.J. Parker, A selective PIKfyve inhibitor blocks PtdIns(3,5)P2 production and disrupts endomembrane transport and retroviral budding, EMBO Rep. 9 (2008) 164–170. 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed]
- 154.Li R.-J., Xu J., Fu C., Zhang J., Zheng Y.G., Jia H., Liu J.O. Regulation of mTORC1 by lysosomal calcium and calmodulin. ELife. 2016;5 doi: 10.7554/eLife.19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Medina D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shen D., Wang X., Li X., Zhang X., Yao Z., Dibble S., Dong X., Yu T. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Höglinger D., Haberkant P., Aguilera-Romero A., Riezman H., Porter F.D., Platt F.M., Galione A., Schultz C. Intracellular sphingosine releases calcium from lysosomes. ELife. 2015;4 doi: 10.7554/eLife.10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Morgan A.J., Platt F.M., Lloyd-Evans E., Galione A. Molecular mechanisms of endolysosomal Ca 2+ signalling in health and disease. Biochem. J. 2011;439:349–378. doi: 10.1042/BJ20110949. [DOI] [PubMed] [Google Scholar]
- 159.Rah S.-Y., Lee Y.-H., Kim U.-H. NAADP-mediated Ca2+ signaling promotes autophagy and protects against LPS-induced liver injury. FASEB J. 2017;31:3126–3137. doi: 10.1096/fj.201601290R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Venkatachalam K., Long A.A., Elsaesser R., Nikolaeva D., Broadie K., Montell C. Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell. 2008;135:838–851. doi: 10.1016/j.cell.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rudge S.A., Anderson D.M., Emr S.D. Vacuole size control: regulation of PtdIns(3,5)P 2 levels by the vacuole-associated Vac14–Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. MBoC. 2004;15:24–36. doi: 10.1091/mbc.e03-05-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chow C.Y., Zhang Y., Dowling J.J., Jin N., Adamska M., Shiga K., Szigeti K., Shy M.E. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Campeau P.M., Lenk G.M., Lu J.T., Bae Y., Burrage L., Turnpenny P., Román Corona-Rivera J., Morandi L. Yunis–Varón syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am. J. Hum. Genet. 2013;92:781–791. doi: 10.1016/j.ajhg.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lumkwana D., du Toit A., Kinnear C., Loos B. Autophagic flux control in neurodegeneration: progress and precision targeting—where do we stand? Prog. Neurobiol. 2017;153:64–85. doi: 10.1016/j.pneurobio.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 165.Deng Z., Sheehan P., Chen S., Yue Z. Is amyotrophic lateral sclerosis/frontotemporal dementia an autophagy disease? Mol. Neurodegener. 2017;12:90. doi: 10.1186/s13024-017-0232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B. Regulation of intracellular accumulation of mutant huntingtin by Beclin 1. J. Biol. Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 167.Viodé A., Fournier C., Camuzat A., Fenaille F., NeuroCEB Brain Bank, Latouche M., Elahi F., Ber I.L. New antibody-free mass spectrometry-based quantification reveals that C9ORF72 long protein isoform is reduced in the frontal cortex of hexanucleotide-repeat expansion carriers. Front. Neurosci. 2018;12:589. doi: 10.3389/fnins.2018.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chang M.C., Srinivasan K., Friedman B.A., Suto E., Modrusan Z., Lee W.P., Kaminker J.S., Hansen D.V. Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J. Exp. Med. 2017;214:2611–2628. doi: 10.1084/jem.20160999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Elia L.P., Mason A.R., Alijagic A., Finkbeiner S. Genetic regulation of neuronal progranulin reveals a critical role for the autophagy–lysosome pathway. J. Neurosci. 2019;39:3332–3344. doi: 10.1523/JNEUROSCI.3498-17.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.von Kleist L., Ariunbat K., Braren I., Stauber T., Storch S., Danyukova T. A newly generated neuronal cell model of CLN7 disease reveals aberrant lysosome motility and impaired cell survival. Mol. Genet. Metab. 2019;126:196–205. doi: 10.1016/j.ymgme.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 171.Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M.C., Isaacs A., Brech A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sarkar S., Carroll B., Buganim Y., Maetzel D., Ng A.H.M., Cassady J.P., Cohen M.A., Chakraborty S. Impaired autophagy in the lipid-storage disorder Niemann–Pick type C1 disease. Cell Rep. 2013;5:1302–1315. doi: 10.1016/j.celrep.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yasa S., Modica G., Sauvageau E., Kaleem A., Hermey G., Lefrancois S. CLN3 regulates endosomal function by modulating Rab7A effector interactions. J. Cell Sci. 2020 doi: 10.1242/jcs.234047. [DOI] [PubMed] [Google Scholar]
- 174.Koike M., Nakanishi H., Saftig P., Ezaki J., Isahara K., Ohsawa Y., Schulz-Schaeffer W., Watanabe T. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J. Neurosci. 2000;20:6898–6906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tatti M., Motta M., Di Bartolomeo S., Scarpa S., Cianfanelli V., Cecconi F., Salvioli R. Reduced cathepsins B and D cause impaired autophagic degradation that can be almost completely restored by overexpression of these two proteases in Sap C-deficient fibroblasts. Hum. Mol. Genet. 2012;21:5159–5173. doi: 10.1093/hmg/dds367. [DOI] [PubMed] [Google Scholar]
- 176.Curcio-Morelli C., Charles F.A., Micsenyi M.C., Cao Y., Venugopal B., Browning M.F., Dobrenis K., Cotman S.L. Macroautophagy is defective in mucolipin-1-deficient mouse neurons. Neurobiol. Dis. 2010;40:370–377. doi: 10.1016/j.nbd.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vergarajauregui S., Connelly P.S., Daniels M.P., Puertollano R. Autophagic dysfunction in mucolipidosis type IV patients. Hum. Mol. Genet. 2008;17:2723–2737. doi: 10.1093/hmg/ddn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fiorenza M.T., Moro E., Erickson R.P. The pathogenesis of lysosomal storage disorders: beyond the engorgement of lysosomes to abnormal development and neuroinflammation. Hum. Mol. Genet. 2018;27:R119–R129. doi: 10.1093/hmg/ddy155. [DOI] [PubMed] [Google Scholar]
- 179.Platt F.M., Boland B., van der Spoel A.C. Lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J. Cell Biol. 2012;199:723–734. doi: 10.1083/jcb.201208152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wong Y.C., Holzbaur E.L.F. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kim I., Rodriguez-Enriquez S., Lemasters J.J. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fossale E., Wolf P., Espinola J.A., Lubicz-Nawrocka T., Teed A.M., Gao H., Rigamonti D., Cattaneo E. Membrane trafficking and mitochondrial abnormalities precede subunit c deposition in a cerebellar cell model of juvenile neuronal ceroid lipofuscinosis. BMC Neurosci. 2004;5:57. doi: 10.1186/1471-2202-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Corti O. Neuronal mitophagy: lessons from a pathway linked to Parkinson’s disease. Neurotox. Res. 2019;36:292–305. doi: 10.1007/s12640-019-00060-8. [DOI] [PubMed] [Google Scholar]
- 184.Otomo T., Higaki K., Nanba E., Ozono K., Sakai N. Inhibition of autophagosome formation restores mitochondrial function in mucolipidosis II and III skin fibroblasts. Mol. Genet. Metab. 2009;98:393–399. doi: 10.1016/j.ymgme.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 185.Sellier C., Campanari M., Julie Corbier C., Gaucherot A., Kolb-Cheynel I., Oulad-Abdelghani M., Ruffenach F., Page A. Loss of C9 ORF 72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 2016;35:1276–1297. doi: 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Walkley S.U. Pathogenic mechanisms in lysosomal disease: a reappraisal of the role of the lysosome: pathogenic mechanisms in lysosomal disease. Acta Paediatr. 2007;96:26–32. doi: 10.1111/j.1651-2227.2007.00202.x. [DOI] [PubMed] [Google Scholar]
- 187.Suraweera A., Münch C., Hanssum A., Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol. Cell. 2012;48:242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rebsamen M., Pochini L., Stasyk T., de Araújo M.E.G., Galluccio M., Kandasamy R.K., Snijder B., Fauster A. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519:477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tettamanti G. Ganglioside/glycosphingolipid turnover: new concepts. Glycoconj. J. 2003;20:301–317. doi: 10.1023/B:GLYC.0000033627.02765.cc. [DOI] [PubMed] [Google Scholar]
- 190.M.E. Bosch, T. Kielian, Neuroinflammatory paradigms in lysosomal storage diseases, Front. Neurosci. 9 (2015). 10.3389/fnins.2015.00417. [DOI] [PMC free article] [PubMed]
- 191.Skaper S.D., Facci L., Zusso M., Giusti P. An inflammation-centric view of neurological disease: beyond the neuron. Front. Cell. Neurosci. 2018;12:72. doi: 10.3389/fncel.2018.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Heneka M.T., McManus R.M., Latz E. Inflammasome signalling in brain function and neurodegenerative disease. Nat. Rev. Neurosci. 2018;19:610–621. doi: 10.1038/s41583-018-0055-7. [DOI] [PubMed] [Google Scholar]
- 193.Abo-ouf H., Hooper A.W., White E.J., Janse van Rensburg H.J., Trigatti B.L., Igdoura S.A. Deletion of tumor necrosis factor-α ameliorates neurodegeneration in Sandhoff disease mice. Hum. Mol. Genet. 2013;22:3960–3975. doi: 10.1093/hmg/ddt250. [DOI] [PubMed] [Google Scholar]
- 194.Gabandé-Rodríguez E., Pérez-Cañamás A., Soto-Huelin B., Mitroi D.N., Sánchez-Redondo S., Martínez-Sáez E., Venero C., Peinado H. Lipid-induced lysosomal damage after demyelination corrupts microglia protective function in lysosomal storage disorders. EMBO J. 2019;38 doi: 10.15252/embj.201899553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Verhoeven K., De Jonghe P., Coen K., Verpoorten N., Auer-Grumbach M., Kwon J.M., FitzPatrick D., Schmedding E. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot–Marie–Tooth type 2B neuropathy. Am. J. Hum. Genet. 2003;72:722–727. doi: 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]