Abstract
Purpose:
A general problem of machine-learning algorithms based on the convolutional neural network (CNN) technique is that the reason for the output judgement is unclear. The purpose of this study was to introduce a strategy that may facilitate better understanding of how and why a specific judgement was made by the algorithm. The strategy is to preprocess the input image data in different ways to highlight the most important aspects of the images for reaching the output judgement.
Materials and Methods:
T2-weighted brain image series falling into two age-ranges were used. Classifying each series into one of the two age-ranges was the given task for the CNN model. The images from each series were preprocessed in five different ways to generate five different image sets: (1) subimages from the inner area of the brain, (2) subimages from the periphery of the brain, (3–5) subimages of brain parenchyma, gray matter area, and white matter area, respectively, extracted from the subimages of (2). The CNN model was trained and tested in five different ways using one of these image sets. The network architecture and all the parameters for training and testing remained unchanged.
Results:
The judgement accuracy achieved by training was different when the image set used for training was different. Some of the differences was statistically significant. The judgement accuracy decreased significantly when either extra-parenchymal or gray matter area was removed from the periphery of the brain (P < 0.05).
Conclusion:
The proposed strategy may help visualize what features of the images were important for the algorithm to reach correct judgement, helping humans to understand how and why a particular judgement was made by a CNN.
Keywords: convolutional neural network, deep learning, diagnosis, magnetic resonance imaging
Introduction
Machine-learning approaches are increasingly becoming a topic of interest in medical diagnostic imaging. Amongst these approaches, deep-learning techniques, which are sometimes referred to as a sort of artificial intelligence, have developed very rapidly in recent years and are now attracting a great deal of attention.1,2 Before these techniques emerged, computer-aided or -assisted diagnosis usually required humans to extract specific features from the raw data (e.g. CT number, Apparent Diffusion Coefficient, and connectivity) that could be used as seed elements for diagnosis. The extracted features were input to a computer algorithm so that it could learn how to combine the information to reach a correct judgement. On the other hand, in addition to learning how to combine information to reach a judgement, deep-learning techniques can also automatically extract the features from more primitive data inputs (e.g. image data).3,4 In short, a deep-learning algorithm is a network of many simple data-processing modules that include multiple adjustable parameters within each module, and the feature extraction and learning is automatically accomplished by optimizing these parameters through a training process.
The convolutional neural network (CNN) technique, the main subject of the present research, is a form of deep learning that is specialized for data sets such as images of two or more dimensions. A number of achievements using the CNN technique have already been reported.5–9 As an example of CNN-based diagnosis,10 in a previous report, MRI images of Alzheimer’s disease patients were labeled with information about what stage of the disease the patient was at, and the image/label pairs were then used to train a CNN model. After training, the CNN model achieved a high accuracy when judging the disease stage for a new series of unlabeled MRI images.
Even though there have been some excellent results with the CNN technique, there are nevertheless some persistent issues that have not yet been solved.1 One particular example of this is the ambiguity in how and why the algorithm reached its final judgment. At present, this seems to be a ubiquitous problem in the deep-learning field, and it is an especially important problem that needs to be resolved in order to promote the application of a CNN as a useful tool for clinical diagnostic imaging. In the clinic, the importance of knowing the reasons for a judgement is never less than the importance of the accuracy of the diagnosis itself.
A method named Gradient-weighted Class Activation Mapping (Grad-CAM)11 is one approach towards solving this problem. When applied to an image classification task, this method visualizes which regions in the input image and/or which of the various parameters across the CNN model have a strong influence on the judgement. However, since Grad-CAM is completely data driven, it only specifies positions in a specific image that had a large influence on the judgement, and neither the medical rationality nor the consistency of the specified area are considered. In this research we introduce a strategy that may help to reduce the ambiguity of CNN-based judgement while considering the medical rationality and consistency. The basic idea is to extract and/or mask a part of the image information in various ways that can be medically meaningful, and then train a CNN model separately for each pattern. As it can be assumed that the performance of a trained CNN model is related to how important the masked information was to the accuracy of the judgement, the differences between the achieved accuracy of the respective training may promote human understanding of how and why a particular decision was made by the model. In short, the largest difference with Grad-CAM is that the users (clinicians) can narrow down the possible candidates responsible for the conclusion that is obtained (e.g. which tissue’s structure is most important in reaching the judgement) via image preprocessing before model training. This additional step may facilitate a link with the model prediction and one or more of the possible conclusions (e.g. diagnoses), so that the users can more easily interpret the model prediction.
The purpose of this study was to evaluate the usefulness of this strategy.
Materials and Methods
In this study, the images for training and testing were preprocessed in five different ways by extracting and/or masking different parts of the original images. Each image set was then used to train a CNN model separately to observe how the result of training and testing (i.e. judgement accuracy) changes as a function of the different preprocessings. Details are described in the following subsections.
Original image data
Four hundred and ninety-nine T2-weighted brain image series acquired from 499 healthy volunteers were downloaded from the website (http://www.humanconnectomeproject.org/) of the Human Connectome Project.12 All the image series were acquired using 3 T MRI systems developed by a single vendor (Human Connectome Scanner12) with identical scanning parameters, including TR: 3200 ms, TE: 561 ms, FOV: 224 × 224 mm2, and voxel size 0.7 mm iso.
Each image series was tagged with the sex and one of two age-ranges for the volunteers: 22–25 years (male: 111, female: 73) and 31–35 years (male: 111, female: 204). In addition, each series was bundled with the results of anatomical segmentation performed using the FSL software,13 which enabled us to extract gray matter area and white matter area from the original image slices in this study.
Image preprocessing to create five different image sets for training and testing
The goal of the training in this study was to classify the image series for each volunteer to the correct age-range. The following steps were performed to generate data sets for training and testing:
The data from 80 volunteers, consisting of 20 randomly extracted image series for each combination of age-range and sex, was selected as data for testing, and the remaining data sets were used for training.
Every second slice was selected from both the training and testing data starting from the slice that included the largest brain area to the slice 42 mm below the tip of the brain. This step was performed to generally exclude slices that may not include sufficient brain area (i.e. the slice at the tip of the brain and the slice at the skull base). An in-house software program working on MATLAB 2016a (The MathWorks, Inc., Natick, MA, USA) referencing the anatomical segmentation data was used for this step as well as for all the following image preprocessing.
Twelve subimages from around the periphery of the brain (64 × 64 pixels), and another 12 same-sized medial subimages were subsampled from each slice selected in the previous step (Fig. 1 contains a detailed description of the procedure). The initial angular location for the subimage selection was randomly selected, but after that the remaining subimages were selected at regular angular intervals (i.e. each angular interval was 30°).
The subsampling was repeated up to four times for the training images to increase the number of images for training; the number of repetitions was automatically adjusted so that the number of series included in the training data for each sex and age-range combination was approximately equal. Note that it was expected that the subimages corresponding to each repetition were different because the initial rotation of the image was updated randomly for each repetition.
Finally, the subimages from around the periphery of the brain were further processed to extract the brain parenchyma, gray matter area, and white matter area (Fig. 2). The deleted parts of an image were filled with Gaussian noise: mean and dispersion adjusted for each subimage so that mean and dispersion of all the pixel in the subimage before and after this process were the same (Fig. 2).
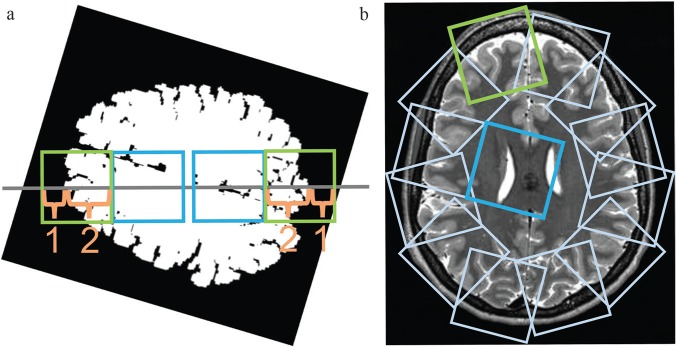
Fig. 1.
Twelve subimages from around the periphery of the brain and another 12 subimages from the inner area of the brain were subsampled automatically from each slice selected in preprocessing step 3. (a) First, each slice image was rotated through a random angle around the center of the brain. Four small subimages (64 × 64 pixels each) were then defined on a horizontal line passing through the center of the brain (gray line). The first two images were taken from the peripheral brain area (green squares), with the ratio of the brain parenchyma length to the extra-parenchyma length along the line being 2:1. Next, another two subimages (64 × 64 pixels each, blue squares) adjacent to the peripheral images on the medial sides were selected as images from the inner area of the brain. (b) The subsampling of peripheral and medial images was repeated 12 times after rotating the line in 30° increments beginning from the initial orientation. Overall, the procedure results in 12 subimages sampled at regular angular intervals for both the peripheral and the inner area of the brain.
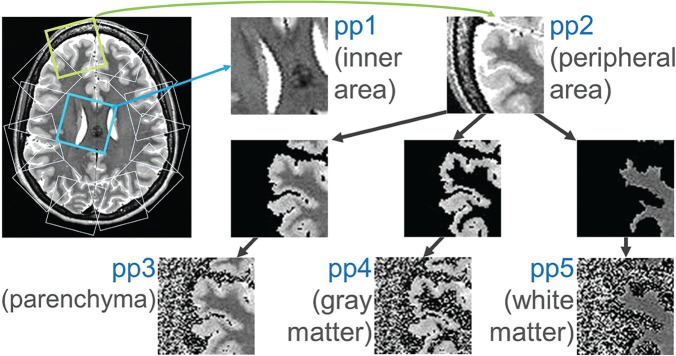
Fig. 2.
Five differently preprocessed image sets (pp1–5) were generated for training and testing the model in five different ways. pp1 and 2: the subimages from around the inner area of the brain and those from around the periphery of the brain, respectively (images generated in steps 1–4 of the section “Image preprocessing to create five different image sets for training and testing”, see Fig. 1); pp3–5: the brain parenchymal area, gray matter area, and white matter area, respectively, were further extracted from the pp2 images. The deleted parts of the images were with Gaussian noise so that the mean and dispersion of pp3–5 images was the same as for the original pp2 image.
After performing all these steps, five different image groups had been generated for training and testing (Fig. 2). The five groups were: subimages from the inner area of the brain (preprocessed image set 1: pp1); subimages from the peripheral area of the brain (pp2); subimages of the brain parenchyma area, gray matter area, and white matter area generated from the pp2 images (pp3–5). The number of subimages in each image set was approximately 200000. As described later, we aimed to focus on the importance of cortical gray matter and white matter for the classification of images from the periphery of the brain. Therefore, the method above was adopted so that both the shape and the fraction of cortical gray matter and white matter regions contained in each subimage from the periphery area are roughly the same, and that the sampling was performed evenly from the whole brain. In addition, subsampling from the inner area of the brain was performed as above in order to balance the number of images and the positional distribution of the subimages in the brain between pp1 and pp2–5.
Training the CNN in five different patterns using image sets pp1–5
A custom-made personal computer (CPU: Intel Core i7-5930K, RAM: 32 GB, GPU: NVIDIA Tesla40c, OS: Ubuntu14.04LTS) equipped with the Caffe software package14 was used to perform the training and testing. A two-dimensional neural network model bundled in this package, named CaffeNet,15 was used throughout this study. Briefly, the network model included five convolutional layers, with each accompanied by its own rectified linear unit layer, along with three max-pooling layers and three fully-connected layers. The final fully-connected layer was connected to a softmax layer for classification.
The CNN was trained in five different patterns (CNN1–5) to judge the correct age-range (i.e. 22–25 or 31–35 years) for each subimage. The trainings were performed separately using the training data of one of the sets pp1–5 for each training (CNN1 corresponded to the trained model using pp1, CNN2 to the trained model using pp2, and so on). The network structure as well as the training parameters were identical for each training. The training parameters included the number of iterations as 150000, a batch size of 128, and learning rate was fixed to 0.003 (Optimization was performed with the stochastic gradient descent [SGD] algorithm).
Testing the CNNs
Testing was performed using the test data sets of pp1 to 5 (CNN1 was tested with the test images of pp1, CNN2 with those of pp2, and so on). To statistically compare the accuracy of classification among the trained models, the fraction of slices that were accurately classified per series was defined as follows:
Each subimage for testing from ppi (i = 1–5) was passed to the corresponding trained CNNi, which outputs the probabilities of the subimage belonging to each age-range. The age-range assigned with the highest probability was considered as the classification judgement of the CNNi for that subimage. When the probabilities for each age-range were equal, the classification for that subimage was regarded as failed and this judgement was fixed automatically to the incorrect one.
The judgement for each slice was determined as the most common judgement amongst the 12 subimages corresponding to that slice. For a case of the judgement being divided evenly (i.e. 6 vs. 6), the classification was regarded as a failed judgement, similar to the previous step.
The fraction of slices that were accurately classified was recorded for each volunteer and used for statistical comparison between CNN1-5. The Steel–Dwass test was used for this multiple comparison, and P < 0.05 was considered to indicate a significant difference.
Results
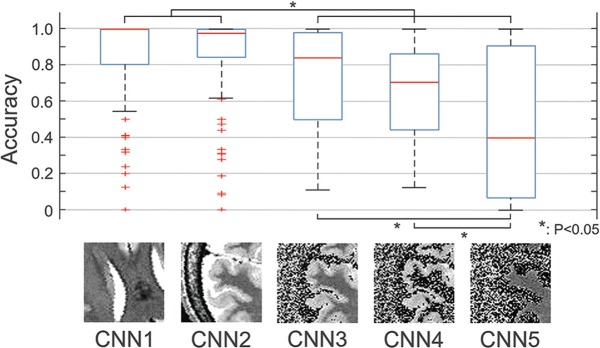
The results are summarized in Fig. 3. The fraction of accurately classified slices per series differed among the differently trained models. CNN1 and 2 achieved higher fractions than CNNs 3–5, and the differences were significant (P < 0.05). Amongst CNNs 3–5, the judgement accuracies of CNNs 3 and 4 were higher than that of CNN5, and the differences were significant (P < 0.05). The difference between CNNs 1 and 2, as well as the difference between CNNs 3 and 4, were not significant.
Fig. 3.
The fraction of accurately classified slices per series differed among the five differently trained models (CNN1–5). The differences between CNN 1, 2 and CNN 3–5, as well as the differences between CNN 3, 4 and CNN 5 were significant (P < 0.05). Apart from the image sets used for training and testing, the settings were identical for all training patterns. CNN1–5: the models trained by preprocessed image sets 1–5 (pp1–5, see Fig. 2), respectively. *P < 0.05. CNN, convolutional neural network.
Discussion
In this study, we applied differently preprocessed image sets for training and testing in order to facilitate better understanding of how and why a decision was made by a CNN. As described in the introduction, the proposed strategy does not aim to extract important features for classification as mere positions on the image, but to extract important features as medically meaningful answers. In the example shown in this study, the question “which feature is most important for classification?” was refined to the question “which tissue is most important for classification?” by narrowing down the candidate tissues through the image preprocessing. As shown in the results, differences were found in the accuracy of the trained models, but some differences were statistically significant and others were not (Fig. 3). From the differences in this statistical significance, it may be possible to estimate which tissue of the image was most important for a CNN to reach a correct judgement.
There was a significant difference in judgement accuracy between CNNs 2 and 3, with that of CNN2 being higher (Fig. 3). As the network model and all the training parameters were identical to each other, the image set used for training, which was the only difference between the two, can be regarded as the reason for this difference. Here, the image information that existed in pp2 but not in pp3 was the extra-parenchymal space and the areas of subcutaneous fat and skull. Assuming that there was relatively little difference in subcutaneous fat and skull due to age, it is most likely that the size and/or shape of the extra-parenchymal space differed between the two age-range groups. Also, as there was no difference in accuracy between CNNs 3 and 4, but the judgement accuracy of CNN5 was significantly lower (Fig. 3), it is possible to postulate that the image information included in the gray matter area was more important for a correct judgement than that included in the white matter area. On the other hand, as the difference in accuracy was not significant between CNNs 1 and 2, it might be inferred that the image information obtained from the ventricles (e.g. size and shape) is roughly equivalent to that obtained from the extra-parenchymal space. In this way, the method used in this study, which performed various image preprocessing procedures to isolate different image information, and then separately trained and tested the CNN model in multiple ways, may facilitate better understanding of how and why a judgement was made. It was not surprising that a trained model that was fed less information than another (e.g. CNN5 compared with CNN3) had an inferior accuracy. However, the important part of the strategy was to divide those differences in accuracy into those that were significant and those that were not significant. This study suggested through a binary classification example that focusing on this difference in significance may help evaluating what specific structure/tissue was important for the classification.
Solving the ambiguity for CNN-based clinical image diagnosis is important. As, in reality, it is impossible to achieve an accuracy of 100% in medical image diagnosis, it is necessary to know the limitations of the method to predict possible mistakes. On the other hand, if the conditions under which mistakes are likely to occur are understood, an imperfect diagnosis may not be such a serious problem. Software previously developed to support the image-based diagnosis of Alzheimer’s disease (Voxel-based specific regional analysis system for Alzheimer’s disease [VSRAD]) is a good example.16,17 VSRAD detects the pattern of brain atrophy of a patient from MRI images based on a voxel-based-morphometry technique and compares it to the pattern of Alzheimer’s disease patients. A physician can use the software as a useful supporting tool while keeping in mind that a patient with brain volume loss due to an old infarction or a patient with hydrocephalus, for example, is more likely to be misjudged by the software. Conversely, without any understanding of how a judgement was reached by VSRAD, a physician may have doubts about the reliability of the judgement. The proposed method might be useful to predict such limitation for a CNN.
The suggested method may be useful for highlighting image features that are important for judgement accuracy. However, it should be remembered that the appropriateness of the highlighted features is not by itself proven. This study suggests that several anatomical areas are more important for selecting the age-range correctly, but it has been only partly proven within this study. Whether the suggestions are valuable or not can be assessed by comparing them with other established evidence and also with empirical expectations. For example, the suggestion that the extra-parenchymal space is different for the two age-range groups can be tested by comparing the data with established brain atlases.18 If a suggestion proves to be reasonable, physicians will be more likely to accept a CNN as a clinical tool because they can understand its limitations. On the other hand, even if a suggestion is not in accord with established knowledge, it does not automatically mean that it is incorrect. It might in fact indicate a candidate for a new biomarker to be investigated in additional studies. The biomarkers established in this way might provide unique information compared to conventional biomarkers designed based on biological and anatomical knowledge.19–21
The proposed strategy does not completely solve the ambiguity of CNN based judgement. This is because the present method only indirectly suggests from the parallel training what the important characteristic for the classification was. It still does not explain exactly how the information was processed in each model during training. Nevertheless, the suggested method would be useful for decreasing the ambiguity of the CNN based decision in the clinic because the feature of the input image that was important to the training can be related to a medically meaningful object such as tissue type.
Detailed information about how a trained model made a judgement from the images is essentially coded and retained in the parameters of the optimized layers of the model.22–24 Analyzing that information may help understand the decision process in a more detailed and concrete way than the proposed method. However, there is as yet no general technique that can decode such information into an understandable format. The justification for a judgement needs to be simple enough to be quickly and easily understandable for clinical use by physicians. This is one of the benefits of the method proposed in this study, because the results are easily translatable as information that is familiar to physicians. The grad-CAM method11 can also displays the analysis results in an easy-to-understand heat-map. However, as mentioned above, it can only show the important positions for individual images. The strategy proposed here makes it easier to interpret the model prediction by refining the problem to a more specific set of possible candidates such as tissue types.
Another benefit of the proposed method is that it narrows down the candidate tissue to be evaluated using medical knowledge. This may be useful for suppressing clinically meaningless results, and also for accomplishing the training and testing with a relatively small sample size.
A possible limitation of this study was that the effect of the degree of image processing on the results was not considered. Particularly, the pp4 images (gray matter area) contained a long border between the extracted gray matter and the parts that were removed in the segmenting process, but those of pp1 and 2 (inner and peripheral areas of the brain, respectively) did not. Such strong artificial contrast may potentially affect the training itself. However, considering the fact that the difference in judgement accuracy between CNNs 3 and 4 was not significant, but the differences between CNNs 3 and 5, as well as between CNNs 4 and 5 were (Fig. 3), the degree of image processing probably did not strongly influence accuracy. Acquiring images with variable contrasts in one scan session may help to overcome this limitation, as it may enable masking and/or emphasizing certain information by changing the contrast between different tissue types. For example, if an additional image set were available, where the images are similar to pp3 but have no contrast between the gray and white matter areas, the comparison between the classification results using that image set and that using pp3 will enable direct assessment about the importance of the margin between the gray and white matters. The degree of image processing does not need to be considered in such a case.
Conclusion
Parallel training and testing using image sets preprocessed in different ways may represent a useful strategy to facilitate better understanding of how and why a judgement was made by a CNN.
Acknowledgment
The authors appreciate the assistance of H. Kamada, E. Mitsui, and S. Komai during the study.
Footnotes
Funding
This research was supported by a Grant-in-Aid for Scientific Research (KAKENHI #17K10385) from the Japan Society for the Promotion of Science (JSPS).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017; 42:60–88. [DOI] [PubMed] [Google Scholar]
- 2.Perone CS, Calabrese E, Cohen-Adad J. Spinal cord gray matter segmentation using deep dilated convolutions. 2017; arXiv:1710.01269v1. doi.org/ 10.1038/s41598-018-24304-3 [DOI] [PMC free article] [PubMed]
- 3.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Proceedings of the 25th International Conference on Neural Information Processing Systems. 2012; 1:1097–1105. [Google Scholar]
- 4.Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc. IEEE 1998; 86:2278–2324. [Google Scholar]
- 5.Hosseini-Asl E, Gimel’farb G, El-Baz A. Alzheimer’s disease diagnostics by a deeply supervised adaptable 3D convolutional network. 2016; arXiv:1607.00556v1. [DOI] [PubMed]
- 6.Sarraf S, Tofighi G. Classification of Alzheimer’s disease using fMRI data and deep learning convolutional neural networks. 2016; arXiv:1603.08631v1.
- 7.Yuehao P, Weimin H, Zhiping L, et al. Brain tumor grading based on neural networks and convolutional Neural Networks. Proceedings of the 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, 2015; 15585959. [DOI] [PubMed] [Google Scholar]
- 8.Dou Q, Chen H, Yu L, et al. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging 2016; 35:1182–1195. [DOI] [PubMed] [Google Scholar]
- 9.Ghafoorian M, Karssemeijer N, Heskes T, et al. Deep multi-scale location-aware 3D convolutional neural networks for automated detection of lacunes of presumed vascular origin. Neuroimage Clin 2017; 14:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam J, Zhang Y. Brain MRI analysis for Alzheimer’s disease diagnosis using an ensemble system of deep convolutional neural networks. Brain Inform 2018; 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. 2017 International Conference on Computer Vision (ICCV), Venice, 2017; 1:618–626. https://arxiv.org/abs/1610.02391 (accessed 07 February 2019). [Google Scholar]
- 12.Van Essen DC, Ugurbil K, Auerbach E, et al. The Human Connectome Project: a data acquisition perspective. Neuroimage 2012; 62:2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage 2012; 62:782–790. [DOI] [PubMed] [Google Scholar]
- 14.Jia Y, Shelhamer E, Donahue J, et al. Caffe: convolutional architecture for fast feature embedding. 2014; arXiv:1408.5093.
- 15.Donahue J. BVLC Reference CaffeNet. http://caffe.berkeleyvision.org/model_zoo.html. 2012.
- 16.Kamiyama K, Wada A, Sugihara M, et al. Potential hippocampal region atrophy in diabetes mellitus type 2: a voxel-based morphometry VSRAD study. Jpn J Radiol 2010; 28:266–272. [DOI] [PubMed] [Google Scholar]
- 17.Tomita Y, Nagashima T, Takahashi A, Tsukakoshi Y, Takao M, Mihara B. Usefulness of the VSRAD (voxel-based specific regional analysis system for Alzheimer’s disease) system for diagnosis of the early phase of Alzheimer’s disease. Eur J Neurol 2007; 14:51. [Google Scholar]
- 18.Dickie DA, Shenkin SD, Anblagan D, et al. Whole brain magnetic resonance image atlases: a systematic review of existing atlases and caveats for use in population imaging. Front Neuroinform 2017; 11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tachibana Y, Obata T, Tsuchiya H, et al. Diffusion-tensor-based method for robust and practical estimation of axial and radial diffusional kurtosis. Eur Radiol 2016; 26:2559–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tachibana Y, Obata T, Yoshida M, et al. Analysis of normal-appearing white matter of multiple sclerosis by tensor-based two-compartment model of water diffusion. Eur Radiol 2015; 25:1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hori M, Fukunaga I, Masutani Y, et al. Visualizing non-Gaussian diffusion: clinical application of q-space imaging and diffusional kurtosis imaging of the brain and spine. Magn Reson Med Sci 2012; 11:221–233. [DOI] [PubMed] [Google Scholar]
- 22.Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. 2013; arXiv:1311.2901v3.
- 23.Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M. Striving for simplicity: the all convolutional net. 2014; arXiv:1412.6806v3.
- 24.Montavon G, Bach S, Binder A, Samek W, Müller KR. Explaining nonlinear classification decisions with deep taylor decomposition. 2015; arXiv:1512.02479v1. doi: 10.1016/j.patcog.2016.11.008. [DOI]