Highlights
-
•
Infection of PEDV 8aa in Vero cells leads to apoptotic cell death.
-
•
Caspase 6 or 7 can cleave PEDV 8aa N protein at the late stage of the replication.
-
•
The caspase-mediated cleavage occurs between D424 and G425 near C-terminal of N protein.
Keywords: Porcine epidemic diarrahea virus, Apoptosis, Caspase-mediated N protein cleavage, N protein cellular localication
Abstract
Porcine epidemic diarrhea virus (PEDV) infection in neonatal piglets can cause up to 100% mortality, resulting in significant economic loss in the swine industry. Like other coronaviruses, PEDV N protein is a nucleocapsid protein and abundantly presents at all stages of infection. Previously, we reported that the N protein of trypsin-independent PEDV 8aa is cleaved during virus replication. In this study, we further investigated the nature of N protein cleavage using various methods including protease cleavage assays with or without various inhibitors and mutagenesis study. We found that PEDV 8aa infection in Vero cells leads to apoptotic cell death, and caspase 6 or 7 can cleave PEDV 8aa N protein at the late stage of the replication. The caspase-mediated cleavage occurs between D424 and G425 near the C-terminal of N protein. We also report that both cleaved and uncleaved N proteins are exclusively localized in the cytoplasm of PEDV infected cells.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is an enveloped virus with a single-stranded positive-sense RNA genome of about 30 kb and a member of the genus Alphacoronavirus in the family Coronaviridae (Kocherhans et al., 2001). PEDV can cause an enteric disease (PED) with high mortality of up to 100% in neonatal piglets. There are at least two genogroups with PEDV with classical genogroup 1 and newly emerging genogroup 2 (Huang et al., 2013; Jung and Saif, 2015). Genogroup 1 PEDV has circulated among countries in Europe and Asia since the early 1980s (Huang et al., 2013). In 2010, new highly pathogenic Chinese PEDV strains emerged (Li et al., 2012) and classified into genogroup 2 later. The first US outbreak with genogroup 2a PEDV occurred in 2013 (Stevenson et al., 2013) and subsequently in Canada and Mexico (Anastasia et al., 2014; Chen et al., 2014; Pasma et al., 2016), resulting in the death of 7 million pigs during one year epidemic period (Cima, 2014). Moreover, there are several reports that the US PEDV strains (genogroup 2) caused outbreaks in European (Grasland et al., 2015; Hanke et al., 2015; Theuns et al., 2015) and Asian countries (Lee and Lee, 2014; Lin et al., 2014; Van Diep et al., 2015), which raised significant economic and public health concerns worldwide (Schulz and Tonsor, 2015).
Coronavirus genome encodes four major structural proteins including spike (S), envelope (E), membrane (M) and nucleocapsid (N) (Duarte et al., 1994). Among them, N protein is an abundant structural protein present at all stages of infection. The coronavirus N protein is composed of multiple domains including N1 (or N-terminal domain), N2 (or C-terminal domain) and N3 with spacers between them (Hurst et al., 2013). Both N1 and N2 are very basic and interact with viral RNA genome and/or N protein (Hurst et al., 2013). N3 is the carboxy-terminal part (∼45 aa) with an excess of acidic residues and known to interact with M protein (Hurst et al., 2005; Verma et al., 2006). The primary role of N protein is to act as an essential architecture component in coronavirus assembly through the interactions with N, M and viral RNA (Cavanagh and Cavanaugh, 1997). In addition to its primary role, N protein appears to perform multiple functions in the viral replication cycle and pathogenesis (McBride et al., 2014). Some previous research suggests that the N protein correlates with optimal coronavirus RNA transcription and/or replication, by acting as RNA chaperones to assist the template-switching steps (Zúñiga et al., 2007; Zuniga et al., 2010), and participating in the replicase components for efficient RNA synthesis (Hurst et al., 2013, 2010). In several studies, N protein was also shown to be involved in host cell signaling and immune responses to facilitate viral replication (Cao et al., 2015; Xu et al., 2013).
It was reported that N protein of acute respiratory syndrome coronavirus (SARS-CoV) or transmissible gastroenteritis virus (TGEV) is processed by caspases during apoptotic cell death (Diemer et al., 2008; Eléouët et al., 2000). The N protein cleavage was implied for playing roles in efficient viral replication and pathogenicity (Diemer et al., 2008; Eléouët et al., 2000). For PEDV N protein, Jaru-Ampornpan et al. demonstrated that virally-encoded 3C-like protease (3CLpro) cleaves N protein during virus replication and the cleavage is associated with viral adaptation in cell culture (Jaru-Ampornpan et al., 2017). Previously, we reported two forms of N protein from the protease independent PEDV 8aa strain, while only one form from the protease dependent PEDV KD strains (Kim et al., 2017). We hypothesized that the two forms of N protein are generated by a post-translational modification by host or cellular protease(s) during the replication of PEDV 8aa strain. Here, we demonstrated that the N protein of PEDV 8aa strain is cleaved by caspases during apoptosis of Vero cells. The PEDV N protein was cleaved by caspase 6 or 7 between D424 and G425 near the C-terminal of N protein. Furthermore, N protein was localized exclusively in the cytoplasm of PEDV infected cells regardless of N protein cleavage. Our results demonstrated characteristics during the replication of PEDV 8aa, which provides valuable information to understanding the PEDV biology.
2. Materials and methods
2.1. Viruses, cells, and reagents
The cell culture adapted PEDV US strains (8aa and KD) were propagated in Vero cells without any protease (8aa strain) or in the presence of TPCK-treated trypsin (1 μg/mL) (KD strain) as described previously (Kim et al., 2017). Vero (ATCC-CCL-81tm) cells and HEK 293 T cells were obtained from ATCC (Manassas, VA). Vero cells were maintained in Dulbecco’s minimal essential medium (DMEM) containing 5% fetal bovine serum (FBS) and antibiotics (chlortetracycline [25 μg/mL], penicillin [250 U/mL], and streptomycin [250 μg/mL]). HEK 293 T cells were maintained in minimal essential medium (MEM) containing 10% fetal bovine serum (FBS) and the antibiotics. L-1-tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin and Leupeptin was purchased from Sigma-Aldrich (St. Louis, MO). Furin inhibitor I was purchased from Cayman (Ann Arbor, MI). Pan-caspase inhibitor, Z-VAD-fmk was purchased from Enzo (Farmingdale, NY). Recombinant human Caspase 3 and 6 were purchased from Millipore (Temecula, CA) and recombinant caspase 7 were purchased from R&D Systems (Minneapolis, MIN). The synthesis of a coronavirus 3CLpro inhibitor, GC376, was previously described (Tiew et al., 2011). Anti-PEDV polyclonal antibody (Pab) was collected from a pig challenge study previously reported by us (Kim et al., 2017). A monoclonal antibody (Mab) against PEDV N protein was kindly provided by Dr. Ying Fang (Kansas State University).
2.2. Protein identification by mass spectrometry
To confirm whether the double bands from PEDV 8aa in SDS-PAGE are both N proteins, they were analyzed by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry. Briefly, concentrated 8aa strain was loaded on the SDS-PAGE gel and stained with Coomassie brilliant blue. The two bands that correspond to N protein were cut out separately and sent to Applied Biomics, Inc (Hayward, CA) for MALDI-TOF mass spectrometry study.
2.3. Western blot analysis of PEDV N protein
To study the kinetics of PEDV N protein synthesis and cleavage,confluent Vero cells were inoculated with PEDV 8aa or KD at an MOI of 0.01 and incubated with or without trypsin (1 μg/mL). Following 24, 36, or 48 h incubation, cells were lysed with Tris-glycine SDS Sample Buffer (Thermo Fisher Scientific, PA) and subjected for the Western blot analysis. To determine the effect of various protease inhibitors on N protein cleavage, Z-VAD-fmk (pan-caspase inhibitor, 100 μM), furin inhibitor I (20 μM), leupeptin (trypsin inhibitor, 20 μM), or mock-medium was added to confluent Vero cells in a 12-well plate, and the cells were then immediately inoculated with PEDV 8aa at an MOI of 0.01. Following 24, 36 or 48 h incubation at 37 °C, cells were then lysed with Tris-glycine SDS sample buffer. Cell lysates were resolved by SDS-PAGE (4–12% Tris-glycine gel) and transferred onto nitrocellulose membranes. The membranes were blocked with 5% non-fat milk in phosphate-buffered saline with Tween-20 for 1 h and probed with PEDV Pab antibody followed by horseradish peroxidase(HRP)-conjugated goat anti-swine IgG. Virus proteins were visualized by chemiluminescence reagents (Thermo Fisher Scientific, PA).
2.4. Effects of Z-VAD-fmk on PEDV 8aa replication
Confluent Vero cells in 12 well plates were inoculated with PEDV 8aa (MOI of 0.01) for 1 h at 37 °C in the presence of Z-VAD-fmk (100 μM, 10 μM), or mock (Medium). Following incubation, the plates were washed three times with PBS and replenished with fresh media containing the same concentration of Z-VAD-fmk or mock-medium. After 24 h or 48 h incubation, cells were subjected to three times of freezing and thawing and PEDV titers were determined by the 50% tissue culture infective dose (TCID50) method (Reed and Muench, 1938).
2.5. Apoptosis assays
Apoptosis in cells was identified using the TdT-mediated dUTP Nick-end labeling (TUNEL) staining according to the manufacturer’s protocol(Abcam, MA) or DNA fragmentation assay. Briefly, Vero cells were mock-infected or infected with PEDV 8aa, 8aa with Z-VAD-fmk (100 μM), or KD with trypsin(1 μg/mL) at an MOI of 1. As a positive control, Vero cells were treated with 1 μM of staurosporine. Cells were fixed at 12, 24 or 48 h and treated for 1 h at 37 °C with the reaction mixture containing terminal deoxynucleotidyl transferase and FITC-conjugated digoxigenin-11-UTP for detecting the exposed 3′hydroxyl ends of fragmented nuclear DNA. The cells were then observed using confocal microscopy. Percentage of TUNEL positive cells in images were counted using ImageJ software. To conduct DNA fragmentation assay, Vero cells were inoculated with PEDV 8aa or mock-medium and incubated for 48 h before DNA isolation as described previously (Hinshaw et al., 1994). Briefly, infected or mock-treated cells were detached with 0.5 ml of detergent buffer (10 mM Tris [pH7.4], 5 mM EDTA, 0.2%Triton) and incubated on ice for 30 min. The cell lysates were centrifuged at 10,000 × g at 4 °C for 30 min, and supernatants were used for extraction of DNA with buffered phenol, once with buffered phenol-chloroform, and once with chloroform-isoamyl alcohol (24:1). DNA was ethanol precipitated with 500 mM NaCl and resuspended in 15 μl of sterile water. The samples were run on a 2% agarose gel with ethidium bromide.
2.6. Cell-free cleavage of PEDV N protein by recombinant caspases
To examine if caspases can cleave N protein, the N gene of PEDV US 8aa strain was cloned into pIRES (Clontech, CA) plasmid. The sequences encoding HA tag was added to the N- or C-terminus of N protein using the primers, 5′-AATTCTCGAGATGTACCCATACGATGTTCCAGATTACGCTGGTGGAGCTT CTGTCAGTTTTCAG-3 and 5′-AATTCTCGAGATGGCTTCTGTCAGTTTTCA-3′ for N-terminal tag; and 5′-AATTCTCGAGATGGCTTCTGTCAGTTTTCAG-3′ and 5′-AATTCTCGAGTTAATTT CCTGTGTCGAAGATAGCGTAATCTGGAACATCGTATGGGTATCCACC-3′ for C-terminal tag. The amplified DNA was cloned into the plasmid in the downstream of the CMV promoter and the resulting plasmid was designated as pCI-N-nHA or pCI-N-cHA. The 60–80% confluent 293 T cells were transfected with pCI-N-nHA or pCI-N-cHA using Lipofectamine 3000 reagent (Thermo Fisher Scientific, PA) according to the instructions of the manufacturer and incubated for 48 h for N protein expression. Following incubation, cell lysates (5 μl) were prepared and incubated with 20 or 2 unit of recombinant human caspase 3, 6 or 7 (1:2 or 1:10, respectively) in the reaction buffer (20 μl) for 2 h at 37 °C (Stennicke and Salvesen, 1997). The samples were then subjected to SDS-PGE using 4–12% Tricine gel (Thermo Fisher Scientific, PA) and the proteins were transferred onto nitrocellulose membranes. The membranes were probed with anti-HA mouse monoclonal antibodies followed by horseradish peroxidase-conjugated anti-mouse secondary antibody.
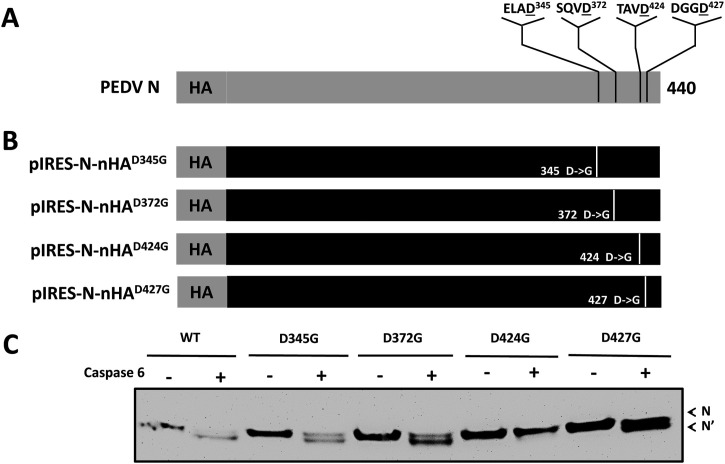
2.7. Plasmid constructions and mutagenesis assay
First, the cleavage site was determined at the C-terminus using the N protein with HA tag at N- or C-terminal.To determine the exact cleavage site in N protein, four putative cleave sites were selected using the online tool (Cascleave: http://sunflower.kuicr.kyoto-u.ac.jp/∼sjn/Cascleave/index.html) (Song et al., 2010) and aspartic acid at four different locations (345, 372, 424, and 427) at the C-terminus of pIRES-N-nHA was mutated to glycine using the QuikChange site-directed mutagenesis kit (Agilent, CA). These plasmids carrying each mutation were generated with following primers: for pIRES-N-nHAD345G, 5′ GTT CGT GAG CTA GCG GGC TCT TAC GAG ATT ACA 3′ and 5′ TGT AAT CTC GTA AGA GCC CGC TAG CTC ACG AAC 3′, for pIRES-N-nHAD372G, 5′ CTT GTT TCA CAG GTG GGT GCA TTT AAA ACT GGG 3′ and 5′ CCC AGT TTT AAA TGC ACC CAC CTG TGA AAC AAG 3′, for pIRES-N-nHAD424G, 5′ TGG GAC ACA GCT GTT GGT GGT GGT GAC ACG GCC 3′ and 5′ GGC CGT GTC ACC ACC ACC AAC AGC TGT GTC CCA 3′), and for pIRES-N-nHAD427G, 5′ GCT GTT GAT GGT GGT GGC ACG GCC GTT GAA ATT 3′ and 5′ AAT TTC AAC GGC CGT GCC ACC ACC ATC AAC AGC 3′. The underlined bold italic nucleotide in each primer is for the change in the mutagenesis analysis. The mutated sequence in each plasmid was confirmed by sequencing analysis. Each mutated N protein was expressed in 293 T cells by transfecting the cells with each mutated plasmid and they were subjected to conduct cell-free cleavage assay with the recombinant caspases 6.
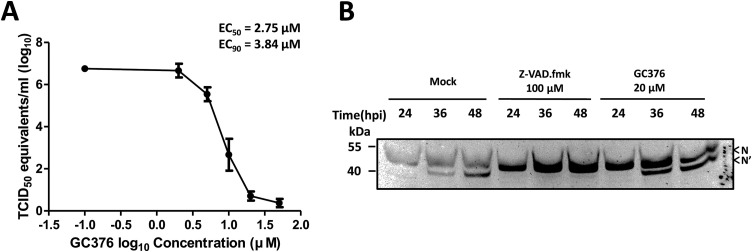
2.8. Antiviral effects of GC376 on PEDV in cell culture
Because the previous report suggested that PEDV 3CLpro can cleave N protein (Jaru-Ampornpan et al., 2017), we used a 3CLpro inhibitor(GC376) to determine its effect on N protein cleavage. First, we examined the effectiveness of GC376 against PEDV replication and determined the effective concentrations inhibiting 50% and 90% of viral replication (EC50 and EC90, respectively). Serial concentrations of GC376 were added to confluent Vero cells in 24-well plates and the cells were promptly infected with PEDV 8aa at an MOI of 0.01. The cells were then further incubated at 37 °C in the presence of GC376 until an extensive cytopathic effect was observed in the mock-treated well (up to 48 h). After three times freezing and thawing of the plates, viral titers were determined by real-time quantitative RT-PCR as described previously (Kim et al., 2017). The TCID50 equivalents/mL was calculated by a standard curve generated from the CT values plotted against the corresponding TCID50 titers/mL. The EC50 and EC90 values were calculated by non-linear regression analysis (four-parameter variable slope) using GraphPad Prism software version 6.07 (GraphPad Software, La Jolla, CA). To determine whether 3CLpro or caspases cleave N protein, GC376 (20 μM), Z-VAD-fmk (100 μM) or DMSO (Mock) was added to PEDV 8aa-infected Vere cells at 24 h after virus inoculation. Cells lysates were prepared at 0, 12 and 24 h (24, 36 and 48 post virus inoculation (hpi), respectively) after the addition of the inhibitors for Western blot analysis as described above.
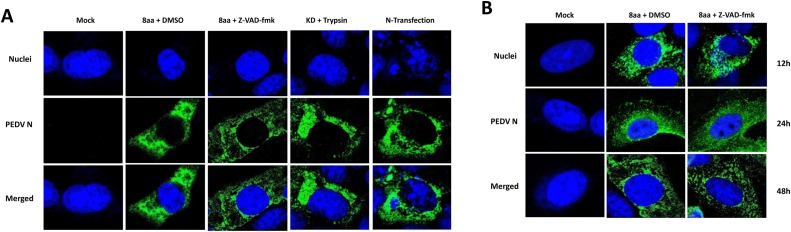
2.9. Confocal laser scanning microscopy for N protein localization
A confocal microscopy study was performed to determine whether N protein cleavage affects the localization of the N protein. Semi-confluent Vero cells on Lab-Tek II CC2 chamber slide (Thermo Fisher Scientific, PA) were inoculated with PEDV 8aa strain at an MOI of 1 in the presence of Z-VAD-fmk (100 μM) or DMSO. Following 1 h incubation at 37 °C, the cells were washed three times with PBS, replenished with fresh media containing the same concentration of Z-VAD-fmk and further incubated for 36 h at 37 °C. In another experiment, PEDV KD strain was used to infect the cells on Lab-Tek II CC2 chamber slide, and the cells were incubated for 12 h in the presence of trypsin (1 μM) at 37 °C. Also, Vero cells were transfected with pIRES-N-nHA and incubated for 24 h. The cells in these experiments were fixed in 4% paraformaldehyde (Sigma-Aldrich, MO) in PBS (pH 7.4) for 15 min at room temperature (RT), permeabilized with 0.1% Triton X-100 in PBS for 10 min at RT, washed three times with PBS and further incubated with 0.5% bovine serum albumin in PBS for 15 min. The cells were then incubated with Mab against PEDV N protein for 1 h at 37 °C. Then, the cells were washed three times with PBS and further incubated with FITC-labeled secondary antibody against mouse IgA, IgG or IgM (KPL, Gaithersburg, MD) for 1 h at 37 °C. The cells were also stained with SYTOX orange (Invitrogen, CA) for 15 min. Coverslips were mounted with ProLong® Gold antifade reagent (Invitrogen, CA), and the cells were observed with a confocal microscope LSM 510 (Zeiss, Oberkochen, Germany) using a 100x oil-immersion objective lens. The images were processed by Image J software 1.52n (http://imagej.nih.gov/ij/). For the evaluation of colocalization between N protein and nucleus, colocalization analysis was performed using JACoP for ImageJ software. Single-channel images were thresholded by Costes’ auto threshold method and the Manders’ split correlation coefficient for colocalization was then determined for each image.
2.10. Statistical analysis
The effects of Z-VAD-fmk in PEDV 8aa replication were compared to the Mock treatment using GraphPad Prism (GraphPad Software, La Jolla, CA, USA). Statistical analysis was performed using the student t-test. P-value of <0.05 was considered as statistically significant. Data were from at least three independent experiments.
3. Results
3.1. The N protein of PEDV 8aa is cleaved during virus replication
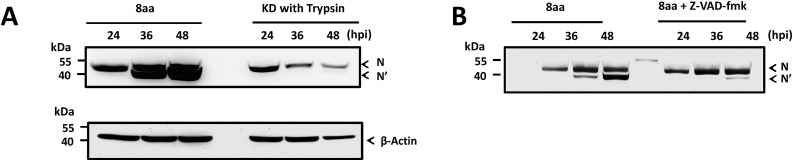
We have previously generated two distinct PEDV strains, 8aa and KD by serially passaging a field-isolated US PEDV strain in different culture conditions (Kim et al., 2017). PEDV 8aa strain shows some distinct characteristics including protease independence, no cell fusion formation, efficient virus replication (viral titers reach over 108 TCID50/mL), and putative N protein cleavage (Kim et al., 2017). In this current study, we confirmed that these bands are both N proteins by analyzing the two protein bands at approximately 49 and 47 kDa on an SDS-PAGE gel using MALDI-TOF mass spectrometry (data not shown). Next, we examined the kinetics of N protein cleavage in the cells (cell lysates). The cleaved N protein appeared at 36 and 48 hpi, but not at 24 hpi (Fig. 1 A, left lanes). The ratios of the intact(N) and cleaved(N′) N protein were approximately 1:1 for both 36 and 48 hpi (Fig. 1A). In contrast, scarcely detectable cleaved N protein from PEDV KD strain grown in the presence of trypsin (1 μg/mL) was observed only at 48 hpi (Fig. 1A, right lanes). Of note, PEDV KD infection produced extensive cell-fusion and cell lysis after 24 hpi, which resulted in reduced levels of N protein in 36 and 48 hpi (Fig. 1A, right lanes).
Fig. 1.
The kinetics of PEDV N protein synthesis and effects of Pan-caspase inhibitor on the N protein cleavage determined by Western Blot analysis (A) The kinetics of N protein. Confluent Vero cells were inoculated with PEDV 8aa (left lanes) or KD (right lanes) at an MOI of 0.1. Cell lysates were prepared at 24, 36 or 48 h post-inoculation (hpi). (B) Effects of Pan-caspase inhibitor on the N protein cleavage. Confluent Vero cells were inoculated with PEDV 8aa at an MOI of 0.1 in the presence of 100 u M Z-VAD-fmk(right lanes) or absence(left lanes). Cell lysates were collected at 24, 36, or 48 h post-inoculation (hpi). Uncleaved or cleaved N protein, designated as N or N′ and as β-actin were indicated by arrowheads.
3.2. N protein cleavage is dependent on the induction of apoptosis
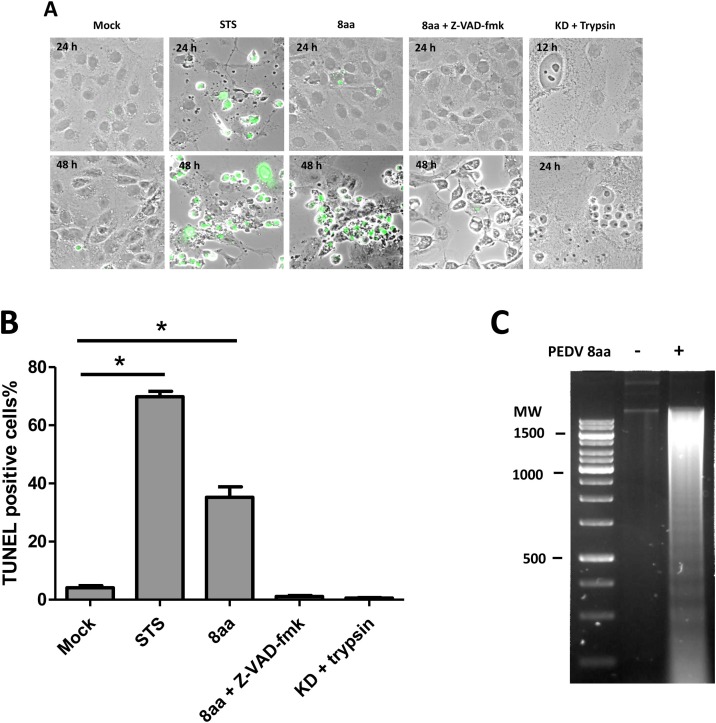
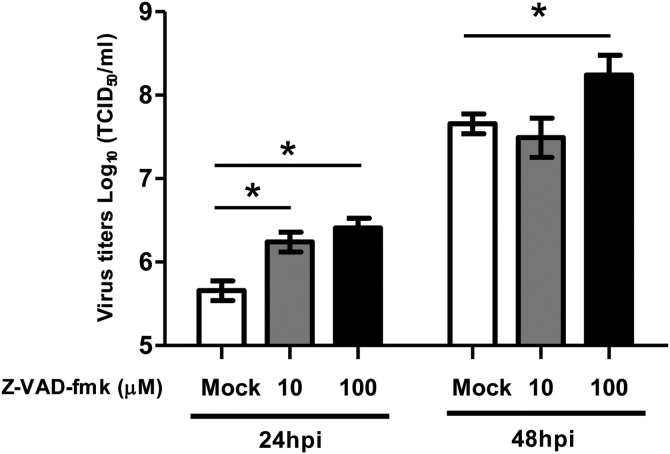
We explored potential protease(s) that can cleave N protein during PEDV 8aa replication using protease inhibitors. As shown in Fig. 1B, the N protein cleavage was abolished by the addition of Z-VAD-fmk (36 and 48 hpi). However, furin or trypsin inhibitors did not block N protein cleavage (Data not shown). Because activation of the caspase family plays central roles in cellular apoptosis, we analyzed the occurrence of apoptosis in PEDV infected Vero cells using TUNEL staining and DNA fragmentation assay. At 48 hpi, 8aa infected cells became TUNEL-positive with a typical cellular feature of apoptosis and the percentage of TUNEL-positive 8aa infected cells was 35%, which was statistically significantly higher than that of mock-treated cells (Fig. 2 A and B). The treatment of Z-VAD-fmk, however, blocked the progression of apoptosis in 8aa infected cells. KD infected cells started to show cell-to-cell fusion from 12 hpi and showed extensive cell-to-cell fusions and syncytium formations at 24 hpi, but no TUNEL positive cell was observed at 12 and 24 hpi. To confirm the progress of apoptosis in 8aa infected cells, the DNA fragmentation assay was conducted. As expected, the low molecular weight DNA from the 8aa infected Vero cells showed a typical feature of apoptosis, fragmentation of DNA (Fig. 2C), whereas the mock-infected cells did not show any sign of DNA fragmentation. Inhibition of caspase by the addition of Z-VAD-fmk led to significantly increased PEDV 8aa replication compared to mock-treated (DMSO) cells (Fig. 3 ).
Fig. 2.
Protease independent PEDV 8aa infection induced apoptosis in Vero cells. (A) TUNEL staining of PEDV infected Vero cells. Confluent Vero cells were infected with 8aa, 8aa with 100 μM Z-VAD-fmk for 48 hpi, or KD with trypsin(1 μg/mL) for 24 hpi. 1 μM of staurosporine (STS) was added as a positive control. The green fluorescence indicated the DNA fragmentation mediated by apoptosis. (B) Percent of TUNEL-positive cells in each treatment. Asterisks indicate the statistical significance compared to the mock treatment. (C) DNA fragmentation assay was performed using Vero cells infected with mock-Medium or PEDV 8aa. Low-molecular-weight DNA was extracted at 48 hpi and loaded on 2% agarose gel. Molecular weight (MW) is a dsDNA marker.
Fig. 3.
Effects of caspase inhibition on PEDV 8aa replication. Confluent Vero cells were inoculated with PEDV 8aa at an MOI of 0.01 for 1 h with Mock (medium) or Z-VAD-fmk(10 or 100 μM). The plates were then washed three times with PBS, replenished with fresh media containing the same concentration of Z-VAD-fmk or Mock (medium) and further incubated at 37 °C. At 24 (open bars) or 48 hpi (closed bars), cells were subjected to three times of freezing and thawing for virus titration using the TCID50 assay. The mean and the standard deviations of the mean was acquired from three independent experiments. Asterisks indicate the statistical significance compared to the mock treatment.
3.3. Caspase 6 or 7 cleave N protein at D424G425
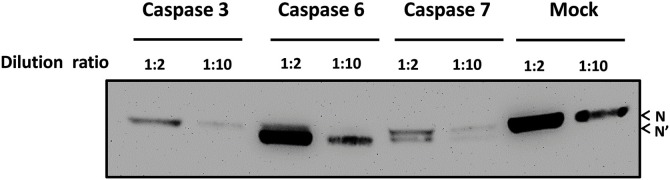
To determine which caspase cleaves N protein, a cell-free cleavage assay was performed with mock treatment (PBS) or the recombinant caspase 3, 6 or 7. The Western blot analysis demonstrated that caspase 6 or 7 cleaved PEDV N protein, whereas caspase 3 did not cleave the N protein (Fig. 4 ). The mutation study on the four mutated N proteins carrying a single mutation of D345G, D372G, D424G or D427G at the putative caspase cleavage site showed that D424G mutation abolished the N protein cleavage by caspase 6 (Fig. 5 ), which revealed N protein cleavage occurs between D424G425 (Fig. 5).
Fig. 4.
Cleavage of the PEDV N protein with recombinant caspase 3, 6 or 7. Expressed N protein was incubated with recombinant caspases with different dilution ratios (1:2 or 1:10) at 37 °C for 2 h. The mixtures were subjected to SDS-PAGE, and Western blot analysis was performed with the anti-HA monoclonal antibody. Uncleaved (N) and cleaved (N′) N protein are indicated by arrowheads.
Fig. 5.
Identification of the N protein cleavage site by caspase 6 with mutagenesis assay and the Western blot analysis. (A) A schematic representation of PEDV N protein with putative C-terminal caspase cleavage sites predicted by the Cascleave. (B) Four mutant recombinant N proteins carrying a single mutation of D345 G, D372 G, D424 G or D427 G were generated by site-directed mutagenesis. (C) Wild-type N protein (WT) or mutant N proteins (D345G, D372G, D424G, or D427G) were incubated for 2 h at 37 °C with a recombinant caspase 6. The mixtures were analyzed by the Western blot with anti-HA antibody. Uncleaved (N) and cleaved (N′) N protein are indicated by arrowheads.
3.4. PEDV 3CLpro did not affect N protein cleavage
First, the inhibitory effects of GC376 against PEDV 8aa were determined with the EC50 and EC90 at 2.75 and 3.84 μM, respectively (Fig. 6 A). To examine the effect of 3CLpro on N protein cleavage, GC376 at 20 μM was added at 24 hpi, and the N cleavage was monitored at 36 or 48 hpi. The addition of GC376 or Mock (DMSO) did not block N protein cleavage at 36 or 48 hpi (Fig. 6B). As a positive control, Z-VAD-fmk at 100 μM prevented the N protein cleavage at 36 or 48 hpi (Fig. 6B).
Fig. 6.
Effects of a 3CLpro inhibitor (GC376) on PEDV replication and N protein cleavage. (A) Inhibition of PEDV replication by GC376. Confluent cells were inoculated with PEDV 8aa at an MOI of 0.01 and incubated with serial dilutions of GC376 for 48 h. Viral titers were measured by real-time qRT-PCR and converted to TCID50 equivalents/mL, and EC50 and EC90 were calculated. (B) The effects of GC376 on the cleavage of PEDV N protein. Cells were inoculated with PEDV 8aa at an MOI of 0.1, and Z-VAD-fmk (100 μM), GC376 (20 μM) or Mock(DMSO) was added at 24 hpi. Cells were further incubated for an additional 12 or 24 h. Cell lysates were then prepared, and Western blot analysis was performed with an anti-PEDV positive serum. Uncleaved (N) and cleaved (N′) N protein are indicated by arrowheads.
3.5. PEDV N protein localizes in the cytoplasm
Because previous studies suggested PEDV N protein was localized in both nucleus and cytoplasm (Shi et al., 2014), and the nuclear localization of SARS-CoV N protein determined N protein cleavage by caspases (Diemer et al., 2008), we analyzed the localization of N protein of PEDV 8aa (and KD) with or without Z-VAD-fmk (100 μM) using the confocal microscopy. With or without Z-VAD-fmk, N protein was exclusively localized in the cytoplasm at 36 hpi of PEDV 8aa (Fig. 7 A). The N protein of PEDV KD (with trypsin) or the expressed N protein were also localized in the cytoplasm at 12 hpi or 36 hpi, respectively (Fig. 7A) (Pearson’s coefficient of <0.1). There is a possibility that N enters the nucleus at earlier or later time of infection. In order to test this possibility, we examined cells infected with 8aa or 8aa and Z-VAD-fmk at 12, 24, or 48 hpi. PEDV N protein was consistently distributed in the cytoplasm and no detectable nuclear translocation of N protein was observed(Fig. 7B) (Pearson’s coefficient of <0.1).
Fig. 7.
Subcellular localization of PEDV N protein in Vero cells. Vero cells were mock-infected or infected at MOI of 1 with PEDV 8aa with DMSO, 8aa with Z-VAD-fmk (100 μM) or KD with trypsin (1 μg/mL). For the expression of N protein, 24 h-old semi-confluent cells were transfected with pIRES-N-nHA, and cells were incubated at 37 °C for (A) 12 h (KD) or 36 h (8aa and N transfection), or (B)12, 24 or 48 h (8aa and 8aa with Z-VAD-fmk). Fixed and permeabilized cells were stained with anti-PEDV N monoclonal antibody (green) and SYTOX orange (Blue) and examined by confocal microscopy.
4. Discussion
Induction of apoptosis by coronavirus infection and caspase cleavage of N protein by caspases have been reported in TEGV and SARS studies (Diemer et al., 2008; Eléouët et al., 2000). TGEV N protein is cleaved by caspase 6 and 7 during replication in HRT18 cells, which resulted in 22 amino acid fragment from the C-terminal of N protein (Eléouët et al., 2000). SARS-CoV N protein is also cleaved by caspase 6 and potentially by caspase 3, and the possible cleavage site is located at residues 400 and 403 on the C-terminal of N protein (Diemer et al., 2008). The biological significance of the virus-induced apoptosis and N protein cleavages is still unclear, but it is suggested to be involved in the pathogenicity (Diemer et al., 2008; Eléouët et al., 2000). In this study, we demonstrated similar findings to TEGV and SARS mediated apoptosis in protease independent PEDV 8aa strain. The cleaved N band (N′) appeared at the late stage (after 36 hpi) of 8aa infection (Fig. 1A) and required caspase activities because the N′ product was abolished by the addition of pan-caspase inhibitor (Z-VAD-fmk) (Fig. 1B). This cleavage was closely associated with the virus-induced apoptosis evident with TUNEL staining and the DNA fragmentation at 48 hpi (Fig. 2). However, the N protein of PEDV KD was barely cleaved during virus replication (Fig. 1A). Because PEDV KD, a trypsin-dependent strain, causes extensive cell fusion formation with lysis at 24 hpi without cellular apoptosis(Fig. 2), there were significant differences in the virus-induced cell death by these two different PEDV strains.
The induction of apoptosis of virus-infected cells raised a discussion of whether apoptosis is favorable to host or virus. In a host defense point of view, apoptosis curtails the use of cellular machinery necessary for viral protein synthesis and facilitates the elimination of viral proteins through caspase-mediated proteolysis, resulting in the reduction of viral replication and spread (Danthi, 2011; Yan et al., 2010). Thus, viruses in diverse families encode anti-apoptotic proteins (Clem et al., 1991) to limit apoptosis. On the other hand, viruses can take advantage of the apoptosis of host cells to facilitate virus replication. Various viruses have utilized caspases to cleave their viral proteins for successful replication, which is aborted when the caspase cleavage sites on the viral proteins are eliminated (Best et al., 2003; Moody and Laimins, 2009). In this study, the replication of 8aa was slightly but significantly increased in the presence of Z-VAD-fmk (Fig. 3), suggesting the apoptosis is not favorable for viruses. Of note, the replication of PEDV KD in the presence of trypsin induced extensive fusions with cell lysis starting 12 hpi, and the addition of Z-VAD-fmk in the medium did not affect virus replication (data not shown).
We demonstrated that caspase 6 or 7, but not caspase 3, is responsible for the N protein cleavage (Fig. 4) and the cleave site located at the C-terminal of the PEDV N protein between D424/G425 (Fig. 5). This cleavage location is similar to that of TGEV N protein (Eléouët et al., 2000). This site is located within domain 3 on the PEDV N protein. Because domain 3 is not involved in N-N interaction and RNA binding, the cleavage may not affect these functions of N protein. However, the cleavage resulted in the reduction of 8aa replication (Fig. 3) probably because domain 3 interacts with M protein to plays an important role in coronavirus replication (Narayanan et al., 2000) and the N cleavage disrupts the N-M interaction. Interestingly, we observed that both forms of N proteins were present in the concentrated (purified) PEDV 8aa virus at the ratio of 1:1 (Kim et al., 2017) suggesting that the cleaved form of N protein could still integrate into virions at the similar rates as the uncleaved form.
It was previously reported in a study that PEDV N protein is processed by viral 3CLpro glutamine at the position 382 (P1) and leucine at the position 381 (P2), and this cleavage was speculated to be associated with cell culture adaptation of PEDV (Jaru-Ampornpan et al., 2017). In this study, we used a specific 3CLpro inhibitor (GC376) to investigate whether the N protein of PEDV 8aa is cleaved by 3CLpro or not. The treatment of GC376 did not abolish N protein cleavage for PEDV 8aa (Fig. 6). 3CLpro-mediated N protein cleavage may dependent on the different PEDV strains.
A nucleus localization of N protein has been reported for several coronaviruses including PEDV, TGEV, Murine Hepatitis Virus(MHV), SARS-CoV, and infectious bronchitis virus (IBV) (Diemer et al., 2008; Hiscox et al., 2000; Shi et al., 2014; Wurm et al., 2001). Shi et al. (2014) reported nuclear localization of PEDV N protein in Vero E6 cells after transfection with N genes. Amino acid 71-90 region sufficiently mediated nuclear localization of the N protein and R87 and R89 were crucial for its function. This 71–90 region is almost conserved N protein of 8aa and KD strains with a single mutation (G84-to-A84). The N protein of 8aa and KD also poses two potential nuclear localization signals (NLS) that are composed of “pat 7” NLS sequences at 261-PKKNKSR-267 and 381-PQRKKEK-387. However, during the examination of Vero cells with 8aa, KD infection or N transfection using confocal microscopy, N proteins were exclusively localized in the cytoplasm (Fig. 7). The cytoplasmic localization of N protein was consistently shown when it was examined at different time points for both PEDV 8aa and KD (Fig. 7). It is possible a minor population of N protein (with or without N protein cleavage) may be localized to the nucleus, and there could be variations among strains. To further elucidate the functional role of N protein and its cleavage, we are currently investigating the cleaved (and uncleaved) N protein in viral assembly and replication.
Author statement
The authors thank David George for technical assistance.
Funding
The research was supported in part by grants from the National Institutes of Health (R01 AI130092 to K.O.C.).
Declaration of competing interest
Authors declare no competing interest.
CRediT authorship contribution statement
Changin Oh: Investigation, Writing - original draft. Yunjeong Kim: Concentualization, Writing - review & editing. Kyeong-Ok Chang: Conceptualization, Writing - review & editing.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198026.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Anastasia N.V., Douglas M., Qiuhong W., Marie R.C., Kurt R., Albert R., James C., Kwonil J. Distinct characteristics and complex evolution of PEDV strains, North America, may 2013–February 2014. Emerg. Infect. Dis. J. 2014;20:1620. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S.M., Shelton J.F., Pompey J.M., Wolfinbarger J.B., Bloom M.E. Caspase cleavage of the nonstructural protein NS1 mediates replication of Aleutian mink disease parvovirus. J. Virol. 2003;77:5305–5312. doi: 10.1128/JVI.77.9.5305-5312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Gao Y., Ren X., Ren Y., Ge X., Li G. Porcine epidemic diarrhea virus infection induces NF-κB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 2015;96:1757–1767. doi: 10.1099/vir.0.000133. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Cavanaugh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014;52:234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cima G. 2014. PED Virus Reinfecting U.S. Herds. pp. 245:166-167. [PubMed] [Google Scholar]
- Clem R.J., Fechheimer M., Miller L.K. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- Danthi P. Enter the kill zone: initiation of death signaling during virus entry. Virology. 2011;411:316–324. doi: 10.1016/j.virol.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemer C., Schneider M., Seebach J., Quaas J., Frösner G., Schätzl H.M., Gilch S. Cell type-specific cleavage of nucleocapsid protein by effector caspases during SARS coronavirus infection. J. Mol. Biol. 2008;376:23–34. doi: 10.1016/j.jmb.2007.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte M., Gelfi J., Lambert P., Rasschaert D., Laude H. Springer; Boston, MA: 1994. Genome Organization of Porcine Epidemic Diarrhoea Virus; pp. 55–60. [DOI] [PubMed] [Google Scholar]
- Eléouët J.F., Slee E.A., Saurini F., Castagné N., Poncet D., Garrido C., Solary E., Martin S.J. The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and -7 during TGEV-induced apoptosis. J. Virol. 2000;74:3975–3983. doi: 10.1128/jvi.74.9.3975-3983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasland B., Bigault L., Bernard C., Quenault H., Toulouse O., Fablet C., Rose N., Touzain F., Blanchard Y. Complete genome sequence of a porcine epidemic diarrhea s gene indel strain isolated in france in december 2014. Genome Announc. 2015;3:e00535–00515. doi: 10.1128/genomeA.00535-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke D., Jenckel M., Petrov A., Ritzmann M., Stadler J., Akimkin V., Blome S., Pohlmann A., Schirrmeier H., Beer M., Höper D. Comparison of porcine epidemic diarrhea viruses from Germany and the United States, 2014. Emerging Infect. Dis. 2015;21:493–496. doi: 10.3201/eid2103.141165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw V.S., Olsen C.W., Dybdahl-Sissoko N., Evans D. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J. Virol. 1994;68:3667–3673. doi: 10.1128/jvi.68.6.3667-3673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2000;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4:e00737–00713. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Kuo L., Koetzner C.A., Ye R., Hsue B., Masters P.S. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 2005;79:13285–13297. doi: 10.1128/JVI.79.21.13285-13297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Ye R., Goebel S.J., Jayaraman P., Masters P.S. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 2010;84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaru-Ampornpan P., Jengarn J., Wanitchang A., Jongkaewwattana A. Porcine epidemic diarrhea virus 3C-like protease-mediated nucleocapsid processing: possible link to viral cell culture adaptability. J. Virol. 2017:91. doi: 10.1128/JVI.01660-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Oh C., Shivanna V., Hesse R.A., Chang K.-O. Trypsin-independent porcine epidemic diarrhea virus US strain with altered virus entry mechanism. BMC Vet. Res. 2017;13 doi: 10.1186/s12917-017-1283-1. 356-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23:137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg. Infect. Dis. 2014;20:1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-N., Chung W.-B., Chang S.-W., Wen C.-C., Liu H., Chien C.-H., Chiou M.-T. US-like strain of porcine epidemic diarrhea virus outbreaks in Taiwan, 2013–2014. J. Vet. Med. Sci. 2014;76:1297–1299. doi: 10.1292/jvms.14-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody C.A., Laimins L.A. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000605. e1000605-e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasma T., Furness M.C., Alves D., Aubry P. Outbreak investigation of porcine epidemic diarrhea in swine in Ontario. Can. Vet. J. 2016;57:84–89. [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent ENDPOINTS12. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Schulz L.L., Tonsor G.T. Assessment of the economic impacts of porcine epidemic diarrhea virus in the United States. J. Anim. Sci. 2015;93:5111–5118. doi: 10.2527/jas.2015-9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Lv M., Chen J., Shi H., Zhang S., Zhang X., Feng L. Molecular characterizations of subcellular localization signals in the nucleocapsid protein of porcine epidemic diarrhea virus. Viruses. 2014:1253–1273. doi: 10.3390/v6031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke H.R., Salvesen G.S. Biochemical characteristics of caspases-3, -6, -7, and -8. J. Biol. Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of <i>Porcine epidemic diarrhea virus</i> in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Investig. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Theuns S., Conceição-Neto N., Christiaens I., Zeller M., Desmarets L.M.B., Roukaerts I.D.M., Acar D.D., Heylen E., Matthijnssens J., Nauwynck H.J. Complete genome sequence of a porcine epidemic diarrhea virus from a novel outbreak in Belgium, january 2015. Genome Announc. 2015;3:e00506–00515. doi: 10.1128/genomeA.00506-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiew K.C., He G., Aravapalli S., Mandadapu S.R., Gunnam M.R., Alliston K.R., Lushington G.H., Kim Y., Chang K.O., Groutas W.C. Design, synthesis, and evaluation of inhibitors of Norwalk virus 3C protease. Bioorg. Med. Chem. Lett. 2011;21:5315–5319. doi: 10.1016/j.bmcl.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Diep N., Norimine J., Sueyoshi M., Lan N.T., Hirai T., Yamaguchi R. US-like isolates of porcine epidemic diarrhea virus from Japanese outbreaks between 2013 and 2014. SpringerPlus. 2015;4 doi: 10.1186/s40064-015-1552-z. 756-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Bednar V., Blount A., Hogue B.G. Identification of functionally important negatively charged residues in the carboxy end of mouse hepatitis coronavirus A59 nucleocapsid protein. J. Virol. 2006;80:4344–4355. doi: 10.1128/JVI.80.9.4344-4355.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm T., Chen H., Hodgson T., Britton P., Brooks G., Hiscox J.A. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y., Liu H.-J., Tong D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Xia D., lv S., Qi Y., Xu H. Functional analysis of the orf390 gene of the White Spot Syndrome Virus. Virus Res. 2010;151:39–44. doi: 10.1016/j.virusres.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Zúñiga S., Sola I., Moreno J.L., Sabella P., Plana-Durán J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Cruz J.L.G., Sola I., Mateos-Gomez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.