Abstract
The formation of micronuclei (MN) is prevalent in human cancer cells and its role in activating the senescence-associated secretory phenotype (SASP) machinery has been identified recently. However, the role of MN in regulation of SASP signaling still needs to define in practical cancers. Here, we reported that in colorectal cancer cells the expression of NAT10 (N-acetyltransferase 10) could mediate MN formation through DNA replication and NAT10-positive MN could activate SASP by binding to cGAS. The chemical inhibition of NAT10 by Remodelin or genomic depletion could markedly reduce MN formation, SASP activation, and senescence in colorectal cancer cells. Cell stress such as oxidative or hypoxia could upregulate NAT10 and its associated MN formation senescence and expression of SASP factors. Statistical analysis of clinical specimens revealed correlations between NAT10 expression, MN formation, SASP signaling, and the clinicopathological features of colorectal cancer. Our data suggest that NAT10 increasing MN formation and SASP pathway activation, promoting colorectal cancer progression.
Introduction
Senescent cells secrete several proinflammatory factors, such as cytokines, growth factors, proteases, and chemokines, which are collectively termed the SASP [1,2]. SASP-activated senescent cells have tumor suppressive functions, preventing cancer cell growth, but can also induce cancer cell genomic instability and remodel the tumor microenvironment in either an autocrine or paracrine manner [3]. The SASP is activated by the cGAS-cGAMP-STING pathway, in which cytosolic DNA was recognized and combined by cGAS, catalyzing GTP and ATP to form 2′,3′-cGAMP, which then activates STING, enabling the downstream activation of nuclear factor kappa B and CCAAT enhancer binding protein beta, thereby inducing the production of proinflammatory factors such as type I interferon [[4], [5], [6]].
DNA-triggered cGAS activation is a crucial initial step in the pathway, which is believed to occur in the cytoplasm, as STING is a transmembrane protein that is usually anchored in the endoplasmic reticulum network. Therefore, free cytosolic DNA is considered the main initiator of this pathway, and micronuclei (MN) are believed to be its major source. MN, which contain DNA, are encapsulated by nuclear membranes, and may or may not be contiguous with the main nucleus, are prevalent in human cancer cells [7]. MN formation is a pivotal sign of DNA damage and genetic instability [8,9]. Several possible fates have been postulated for MN, including extrusion, reincorporation, degradation, and persistence, but two additional fates, chromothripsis and SASP activation, have been increasingly discussed [10]. However, the exact mechanism by which MN mediate cGAS-STING activation remains unclear.
NAT10 is a nucleolar protein that contains an acetyltransferase domain and a tRNA binding domain. NAT10 has histone acetylation activity and participates in the regulation of human telomerase reverse transcriptase. It is also involved in the DNA damage response and regulates cytokinesis [11,12]. NAT10 is highly expressed in various human cancers, and interestingly, its translocation from the nucleus to the cytoplasm or membrane promotes invasion and metastasis in CRC cells [13]. More recently, the chemical inhibition of NAT10 was reported to ameliorate nuclear lobulation, MN formation, and senescence in Hutchinson-Gilford progeria syndrome cells [14]. In this study, we reveal that NAT10 is involved in MN formation and activates SASP activity in CRC, expanding our understanding of the role of NAT10 in CRC carcinogenesis and progression.
Materials and Methods
Plasmid Construction and Reagents
cGAS (NM_138441) tagged with a C-terminal 3 × FLAG tag was purchased from YouBio Biotechnology (Changsha, HN, China). GFP-RPA43 (#17659) was purchased from Addgene (Cambridge, MA, UK). GFP-NAT10 (Full length), Flag-NAT10 (Full length) and a rabbit polyclonal antibody against human NAT10 have been previously described [13]. Transient transfection was carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's recommendations. Nuclear Fast Red Staining Solution (0.1%; G1320) and DAPI (C0060) were purchased from Solarbio (Beijing, China). Remodelin (S7641) and CX-5461 (S2684) were purchased from Selleck (Houston, TX, USA). Actinomycin D (15021) was purchased from Cell Signaling Technology (Danvers, MA, USA). Nocodazole (M1404) and cobalt chloride (CoCl2, C8661) were purchased from Sigma Aldrich (St Louis, MO, USA). Hydrogen peroxide (H2O2, KHJ001) was purchased from Rockland (Gilbertsville, PA, USA). Exonuclease III (EN0191) was purchased from Fermentas (Burlington, Ontario, Canada). BrdU (5-bromo-2′-deoxyuridine) (ab142567) was purchased from Abcam (Cambridge, MA, UK). The used primary antibodies were listed in Supplementary Table 1.
Cell Culture and Treatment
Colorectal cancer cells (LoVo, HCT116) were purchased from the National Infrastructure of Cell Line Resource. Cells were maintained in Dulbecco's modified Eagle's medium with high glucose (Gibco, Life Technologies) supplemented with 10% foetal bovine serum. Cells were incubated in a humidified atmosphere with 5% CO2 at 37 °C. For cell treatments, 20 μM Remodelin, 0.4 mM H2O2, or 200 μM CoCl2 were added. For long-term treatment (3 weeks), HCT116 cells were cultured with 0.2 mM H2O2.
Cell co-culture experiments were performed using 0.4-μm inserts (BD Biosciences). Control and NAT10 shRNA-transfected LoVo cells (1 × 105) were suspended in 0.2 mL complete medium and loaded into the upper chambers, while LoVo cells (1 × 106) were suspended in 0.2 mL complete medium and loaded into the lower chambers. To examine paracrine effects, cells in the upper and lower chambers were cocultured for 3 days, and cells in the lower chamber were analyzed by western blotting and SA-β-gal staining.
Lentivirus-Mediated shRNA
Lentivirus-mediated NAT10 (shNAT10) and control (NC) shRNA were purchased from Shanghai GenePharma. For cell infection, viral supernatants were transduced into LoVo cells and supplemented with 50 μg/mL polybrene (Sigma, St. Louis, MO, USA) for 3 days after transfection. Cells were then selected with puromycin (2 μg/mL) for 3 d and maintained in 0.2 μg/mL puromycin. The expression of NAT10 was verified by immunofluorescence and western blotting.
Western Blotting
Total cell lysates were obtained by incubating the cells in 0.5% NP40 Lysis Buffer for 30 min on ice. After centrifugation at 10,000×g for 10 min at 4 °C, the supernatant was collected and stored at −20 °C for subsequent analysis. Western blotting was performed as described [15], using polyvinylidene difluoride membranes (Millipore), nonfat dry milk (BD Biosciences), bovine serum albumin (Sigma), and peroxidase-conjugated goat anti-rabbit and goat anti-mouse IgG (Origene). Band intensities from three independent experiments were quantified by optical density using Lab-Works 4.6 software (Bio-Rad). β-Actin was used as an internal control. The used primary antibodies were listed in Supplementary Table 1.
CRISPR-Mediated Deletion of NAT10
Deletion of NAT10 was mediated by LentiCRISPR v2 (Addgene, Cambridge, MA, UK), containing expression cassettes for Streptococcus pyogenes CRISPR-Cas9, and a chimeric guide RNA (5′-GTGAGTTCATGGTCCGTAGG-3′) selected using the Guide Design Resources at http://crispr.mit.edu to target exon 5 of NAT10. Transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific, MA, USA), according to the manufacturer's instructions. Generation of the NAT10+/− cell line was performed as previously described [16]. NAT10 deletion was detected by western blotting, immunofluorescence, and DNA sequencing using primers flanking exon 5 (forward: 5′-GTCCTTTGGGTTGCTATTTG-3′, reverse: 5′-GCTCTTAGCCCA GAGGCTGT-3′). Clones containing the desired mutation were used in downstream studies.
Immunofluorescence
Immunofluorescence was performed as previously described [17], using IgG, Alexa Fluor 488, or tetramethyl rhodamine isothiocyanate (TRITC)-conjugated secondary antibodies (Origene). Nuclei were counterstained with DAPI (Origene) for 10 min. For RPA foci immunodetection, cells were pre-extracted twice with pre-extraction buffer (20 mM HEPES, pH 7.9, 50 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 0.5% Triton X-100) for 10 min on ice before fixation [18]. Images were acquired on a fluorescence microscope (Model CX51; Olympus, Tokyo, Japan) and Photoshop version 7.0 (Adobe Systems Inc.) was used to analyze the results.
Cell Cycle Assay
1 × 106 of cells were fixed with cold 70% ethanol and incubated overnight at 4 °C. After washed with PBS for two times (800 rpm, 5 min), cells were stained with propidium iodide (10 μg/mL) contained with RNase A (100 μg/mL) in PBS for 30 min at 37 °C. Cells were applied to flowcytometry using the Cell Quest software package (Becton–Dickinson FACScalibur, USA).
BrdU Incorporation Assay
BrdU incorporation assays were performed as described [19]. Briefly, BrdU (10 μM) was added to the culture medium for 30 min before analysis. For immunofluorescence, cells were fixed with 4% formaldehyde, permeabilised with 0.1% Triton X-100, and denatured with 20 mM HCl in 150 mM NaCl and 3 mM KCl for 20 min at 25 °C. Cells were then incubated with a primary antibody mixture composed of primary antibodies (targeting BrdU and either RPA2, NAT10, or Lamin B1) and exonuclease III (0.4 U/μL) in exonuclease III buffer (66 mM Tris–HCl, pH 8.0 and 0.66 mM MgCl2) for 30 min at 37 °C, then with secondary antibodies together with DAPI (10 μM) in 25 mM Tris–HCl, pH 7.5 and 150 mM NaCl for 30 min at 37 °C. For flow cytometry, 1 × 106 of cells were fixed with cold 70% ethanol and incubated overnight at 4 °C. Fixed cells were denatured with 20 mM HCl in 150 mM NaCl and 3 mM KCl for 20 min at 25 °C and neutralized with exonuclease III buffer for 10 min at 25 °C. Cells were then incubated with a primary antibody mixture composed of exonuclease III (0.4 U/μL), RNase A (100 μg/mL) and primary antibody in exonuclease III buffer for 30 min at 37 °C, then with a secondary antibody (IgG/Alexa Fluor/488) mixture containing propidium iodide (10 μg/mL) in 25 mM Tris–HCl, pH 7.5 and 150 mM NaCl for 30 min at 37 °C. Cells were applied to flowcytometry using the Cell Quest software package (Becton–Dickinson FACScalibur, USA). The used primary antibodies were listed in Supplementary Table 1.
SA-β-Gal Staining
A SA-β-gal staining kit (Genmed Scientifics, USA) was used according to the manufacturer's instructions. Cells (500) in random fields were counted and the percentage of SA-β-gal-positive cells was calculated. The experiments were repeated three times.
Patients and Tissue Specimens
We collected 140 surgically resected cases of CRC, obtained from 2012–2013, from the archives of the Department of Pathology of the First Hospital Peking University. None of the patients received preoperative chemotherapy. Sections (4-μm thick) that had been fixed in 4% buffered formalin and embedded in paraffin) were stained with hematoxylin and eosin for histological diagnosis, typing, and grading. Each patient's age, sex, depth of invasion, lymphatic invasion, distant organ metastases, presence of poorly differentiated clusters (PDCs), TNM stage, and modified Dukes stage were obtained by reviewing their medical charts and pathologic records. The cohort contained 83 men and 53 women, with a median age of 64.5 years (range: 30–91 years). Patients with stage III or higher CRC received adjuvant chemotherapy. The OS of patients with stages I–III CRC were evaluated. Recurrence occurred in 30.8% (37/120) of patients, and 27.5% (33/120) died of recurrence during the follow-up period. This study was approved by the Peking University Institutional Review Board and ethics committee prior to its onset. All human tissues were collected in accordance with protocols approved by the Ethics Committee of the Peking University Health Science Center, and informed consent was obtained from all patients.
Tissue Microarray and Immunohistochemistry
Tissue microarrays containing seven cases each were prepared and immunohistochemistry was performed as previously described [20]. Lamin B1 was used to mark the nuclear membranes and MN. Neutrophil nuclei were used as internal staining references. MN was counted under 100× magnification. The formation of MN was graded into three categories, respectively. MN: weak: 0–10% positive cells, moderate: 10–20% positive cells, and high: ≥ 20% positive cells.
NAT10 staining displayed nucleolar, nuclear, cytoplasmic, membranous, and combined patterns and was analyzed individually. cGAS and STING were considered positive in the cytoplasm, while only nuclear staining of IRF3 or HIF1A was recorded as positive. Immunostaining results were categorized into four groups: negative (−), 0–10% positive cells; weakly positive (+), 10–25% positive cells; moderately positive (++), 25–50% positive cells; and highly positive (+++), ≥50% positive cells. Stained slides were reviewed separately by three independent reviewers who were blinded to the clinical data.
Statistical Analysis
All statistical analyses were performed using SPSS (Version 17.0, Chicago), Microsoft Excel 2007, and GraphPad Prism software. For data in immunohistochemistry analysis, Relationships between tumor markers and other parameters were studied using the chi-square test, Fisher exact test or the independent t test when appropriate. OS curves were plotted using the Kaplan–Meier method and compared with log-rank tests. For data in all cellular experiments, student t test was used for the comparison of two groups. Statistical comparisons between multiple groups were analyzed using the one-way ANOVA. Experiments were repeated three times independently. Data were expressed as means ± SEM. All statistical tests were two-sided, and Significance was taken as, *P < .05, **P < .01, and ***P < .001.
Results
NAT10 is Involved in MN Formation in CRC Cells
NAT10 was generally stained in cellular nucleolus and nucleoplasm as previously described [21], and besides, almost all MN were positive for NAT10 in LoVo or HCT116 cells (Figure 1A), of which their spontaneous MN were observed in 8–13% of cells. Like normal nuclei, NAT10-positive MN generally contained DNA and LaminB1 or A/C, the nuclear lamina proteins, as well as other nuclear envelope components, including LBR (LaminB receptor), nuclear pore complex components (MAB414, nucleoporin 153), nuclear basket protein TPR (translocated promoter region), and integral membrane components (Sun2 and nesprin2) (Supplementary Figure S1A, Supplementary Table 2). Interestingly, NAT10 staining revealed nucleolus-like regions of varying size in some MN (Figure 1A), which was co-stained with B23 (or NPM1, nucleophosmin 1) (Supplementary Figure S1B). In addition, the transfection of LoVo cells with GFP-RPA43, a subunit of DNA-directed RNA polymerase I, revealed a co-localization with NAT10 in nucleoli and MN (Supplementary Figure S1C). While treatment with either AMD (actinomycin D) or CX-5461 (RNA polymerase I inhibitor) induced dispersal of these nucleolar MN structures, as in the host nucleoli (Supplementary Figure S1B), implying the active transcription of rDNA in the MN.
Figure 1.
NAT10 localized in MN and involved in MN formation in colorectal cancers cells. (A) NAT10 localized in MN of colorectal cancers cells. LoVo or HCT116 were double staining of NAT10 (green) and LaminB1 (red). (B) Ectopic expression of NAT10 promoted MN formation. LoVo cells were transfected with GFP-NAT10 or GFP-C3 plasmid for 24 h, respectively, and were immunostaining of LaminB1. MN in transfected cells were counted and calculated as percentage of total transfected cells. (C) Inhibition of NAT10 decreased MN formation. LoVo (or HCT116) cells were treated with DMSO or Remodelin (20 μM) for 24 h and double staining was performed with NAT10 (green) and LaminB1 (red). MN in each of groups of cells were counted and calculated as percentage of total cells, respectively. The immunofluorescent images from LoVo cells. (D) Down-regulation of NAT10 reduced MN formation. Control shRNAs (NC) and shNAT10 cells (LoVo and HCT116) were subjected to double staining with NAT10 (green) and LaminB1 (red). MN in each of groups of cells were counted and calculated as percentage of total cells, respectively. (E) Genomic depletion of NAT10 reduced MN formation. The expression of NAT10 in LoVo and corresponding CRISPR-Cas9 edited cells (NAT10+/−) were measured by Western blotting, and the formation of MN in these cells were analyzed by immunofluorescent staining of NAT10 (green) and LaminB1 (red). MN in cells was counted and calculated as percentage of total cells, respectively. Scale bars represent 40 μm. The results are the representative of repeated experiments.
To further explore the functional involvement of NAT10 in MN formation, cells were subjected to conditions with over-or down regulation of NAT10 and the consequent MN was analyzed, respectively. GFP-NAT10 was transfected into LoVo cells. Ectopic expression of GFP-NAT10 increased MN formation in LoVo and HCT116 cells (Figure 1B). Consistently, LoVo or HCT116 treated with the NAT10 inhibitor Remodelin (20 μM), displayed markedly reduced MN formation (Figure 1C). In addition, stable cell lines expressing shNAT10 displayed decreased MN formation (Figure 1D). To confirm this, CRISPR-based gene editing was used to knockout NAT10 expression in LoVo cells (NAT10+/− cells). NAT10 knockout was verified by immunofluorescence (Figure 1E), Western blotting (Figure 1E), and sequencing of NAT10 exon 5 (Supplementary Figure S1D). Compared with control cells (LoVo), NAT10+/− cells displayed reduced MN formation (Figure 1E).
Since interference with mitosis can induce MN formation, cells with or without Remodelin treatment were treated with Nocodazole (200 ng/mL) and MN formation was analyzed. Nocodazole-induced MN were significantly reduced in Remodelin-treated cells (Supplementary Figure S1E). Similarly, NAT10+/− cells also revealed decreased Nocodazole-induced MN formation (Supplementary Figure S1F).
Taken together, these results indicate that in CRC cells, NAT10 should be involved in the formation of MN that may contain nucleolar activity.
NAT10 Mediates MN Formation Through Regulation of DNA Replication
MN formation usually results from errors in DNA replication or mitosis. To further investigate the mechanism by which NAT10 mediates MN formation, the distribution of cell cycle of cells (LoVo and HCT116) treated with Remodelin were evaluated by flow cytometry. Both cell lines displayed decreased S phase cells after 24 h Remodelin treatment compared with control cells (Supplementary Figure S2A). Similarly, a marked reduction in S phase cells was observed in NAT10+/− cells compared with LoVo cells (Figure 2A).
Figure 2.
Down-regulation of NAT10 reduced DNA replication. (A) Genomic depletion of NAT10 induced cell cycle arrest. LoVo and NAT10+/− cells were subjected to cell cycle analysis and was analyzed by propidium iodide (PI) staining and flow cytometry. (B) Down-regulation of NAT10 reduced BrdU incorporation of cells. LoVo and NAT10+/− cells were treated with 10 μM BrdU for 30 min and were subjected to flow cytometry after staining of BrdU and PI, or to (C) Double staining of LaminB1 (green) and BrdU (red), respectively. Positive staining of BrdU was counted and calculated as percentage of total cells. At the same time, LoVo cells under the treatment of DMSO and Remodelin (20 μM) for 24 h were further chased with 10 μM BrdU for 30 min, and after double staining of LaminB1 and BrdU positive staining of BrdU was counted and calculated as percentage of total cells. (D) Genomic depletion of NAT10 reduced DNA damage response of cells. LoVo and NAT10+/− cells were doubly stained by NAT10 with either γH2AX or p-ATR. The staining intensity of γH2AX, p-ATR were measured, respectively. Scale bars represent 40 μm (C, D). (E) Genomic depletion of NAT10 altered histone modification of cells. The levels of histone core, acetylated lysine and methylated lysine in LoVo and NAT10+/− cells were measured by Western blotting. The results are the representative of repeated experiments.
To confirm that the reduced MN formation upon down regulation of NAT10 is due to changes in DNA replication, BrdU incorporation assays were performed in NAT10 inhibited cells. NAT10+/− cells, the reduction of S phase population in cell cycle was consistent with their decrease of DNA replication by approximately 50% (Figure 2B). With compared to control cells, BrdU positivity was significantly decreased in NAT10 inhibited cells by either gene editing (NAT10+/−) (Figure 2C, p = .0138) or Remodelin treatment (Figure 2C, p = .027). At the same time, NAT10 inhibited cells by Remodelin treatment or gene editing revealed decreased levels of γH2AX and decreased phosphorylation of ATR and ATM (Figure 2D, Supplementary Figure S2B, S2C) compared with control cells. The results suggest that down regulation of NAT10 reduces both DNA replication and associated damage responses.
Since NAT10 can acetylate histones, which posttranslational modification plays a critical role in DNA replication, total levels of acetylated histones were measured in status of downregulation of NAT10. Not surprisingly, acetylated histones were decreased when NAT10 was inhibited by Remodelin treatment or gene editing (Figure 2E, Supplementary Figure S2D), while displayed increased in histone methylation (Figure 2E, Supplementary Figure S2D). These results implied that the balance between histone acetylation and methylation could be involved in NAT10 regulated DNA replication.
NAT10-Related MN Activates SASP Signaling in CRC Cells
MN and cytosol free DNA fragments are believed to be able to trigger SASP signaling by activating the cGAS-STING pathway, in which upon binding free DNA cGAS synthesizes cGAMP, which activates STING [[22], [23], [24]]. Thus, the effects of NAT10-associated MN on SASP activation in CRC cells were investigated. Actually, cGAS was detected to co-localize with 20–30% of NAT10-positive MN in either LoVo or HCT116 cells (Figure 3A). In further, co-transfection of GFP-NAT10 and FLAG-cGAS also revealed their co-localization in MN (Figure 3B). More importantly, cGAS positive MN was usually also positive for γH2AX (Supplementary Figure S3A) or/and 53BP1 (Figure 3C), while inhibition or down regulation of NAT10 could result in reduction of γH2AX-labeled MN (Supplementary Figure S3A). There were consistent correlations between MN positive for cGAS, γH2AX and 53BP1 and the frequency of Senescence-associated β-gal (SA-β-gal) stained cells, at approximately 5% (Figure 3D). As expected, NAT10 inhibition through either Remodelin treatment or gene editing markedly reduced MN formation, cGAS- and γH2AX-positive MN, in parallel with decreased SA-β-gal-positive cells. At the same time, the expressions of key SASP factors, including STING and IRF3 or their activation of phosphorylation (p-STING and p-IRF3), were markedly decreased when NAT10 was inhibited either by Remodelin treatment (Supplementary Figure S3B) or gene (Figure 3E).
Figure 3.
NAT10 related MN activated SASP signaling. (A) The co-localization of cGAS with NAT10 in some MN of cells. LoVo cells were doubly stained with NAT10 (green) and cGAS (red). (B) The co-localization of ectopic cGAS and NAT10 in MN of cells. LoVo cells were co-transfected with GFP-NAT10 and Flag-cGAS for 24 h and immunofluorescence staining was performed with Flag antibody. (C) The co-localization of cGAS with 53BP1 in some MN of cells. Double staining was performed with 53BP1 (green) and cGAS (red) antibodies in LoVo cells. (D) Down-regulation of NAT10 reduced senescence of cells. LoVo and NAT10+/− cells were subjected to SA-β-gal staining and positive cells were counted and calculated as percentage of total cells, respectively. (E) Down-regulation of NAT10 reduced the levels of SASP associated factors. The level of NAT10, STING, p-STING, IRF3, p-IRF3 and γH2AX in LoVo and NAT10+/− cells were respectively measured by Western blotting. (F, G, H) Down-regulation of NAT10 reduced the paracrine effects of SASP. A co-culture assay as in (F) schematic representation was used. LoVo cells were cultured in a six-well plate and co-culture cells (NC and shNAT10) were seeded on the inserts. (G) LoVo cells were co-cultured with NC or shNAT10 cells for 3 days, and the levels of NAT10, cGAS and STING were measured by Western blotting, or subjected to SA-β-gal staining and positive cells were counted and calculated as percentage of total cells, respectively (H). The results are the representative of repeated experiments.
On another hand, LoVo or HCT116 cells treated with either AMD or CX-5461 presented a reduction of SA-β-gal stained cells (Supplementary Figure S3C), indicating that the transcription of rDNA should be involved in interaction between MN and cGAS-STING.
To further prove the influence of NAT10 on the paracrine effects of SASP signaling, an indirect co-culture assay was performed, in which colorectal cancer cells (LoVo or HCT116) were separately co-cultured with control LoVo cells, a stable LoVo cell line expressing shNAT10, or medium alone for 3 days (Figure 3F). cGAS and STING levels were decreased in LoVo cells co-cultured with shNAT10 cells compared with those co-cultured with control cells or medium alone (Figure 3G). SA-β-gal staining was also reduced in colorectal cancer cells (LoVo or HCT116) co-cultured with NAT10 inhibited (shNAT10 or NAT10+/−) cells compared to those co-cultured with either medium only or control cells (Figure 3H, Supplementary Figure S3D).
These results indicate that NAT10-related MN could activate cGAS-STING, cellular senescence and SASP signaling in CRC cells.
Cell Stress Stimulates the Effects of NAT10 on MN Formation and SASP Signaling in CRC Cells
Our previous research demonstrated that NAT10 expression could be upregulated in oxidative stress or DNA damage [11]. To further explore the role of NAT10 in MN formation and SASP signaling under cell stress, cells were treated with H2O2 or CoCl2. Long-term culture (3 weeks) of HCT116 cells with H2O2 (0.2 mM) increased the levels of NAT10, cGAS, STING, and p-IRF3 (Figure 4A), and increased SA-β-gal staining (Figure 4B), indicating increased SASP activation. MN formation was also elevated in H2O2-treated cells, but not in NAT10 knockdown cells (Figure 4C). Furthermore, when hypoxia was induced by CoCl2 (200 μM) treatment of LoVo cells for 36 h, NAT10, cGAS, STING and SA-β-gal staining increased with increased expression of HIF1A (Figure 4, D and E). Similarly, increased MN formation was also apparent in CoCl2-treated cells, but not with NAT10 knockdown cells (Figure 4F). Both γH2AX and RPA2 foci were dramatically increased in H2O2 (0.4 mM)-treated LoVo cells, but not in NAT10 inhibited cells either by Remodelin treatment or gene editing (Figure 4, G and H). Moreover, H2O2-treated LoVo cells displayed increased phosphorylation of the upstream kinases of γH2AX and p-RPA2, ATM serine/threonine kinase and ATR serine/threonine kinase, suggesting increased DNA damage and replication stress, while NAT10+/− cells did not (Figure 4I).
Figure 4.
Cell stress stimulated the effects of NAT10 on MN formation and SASP signaling. (A) The oxidative stress stimulated the level of NAT10 and SASP associated activities. HCT116 cells, under treatment of H2O2 (0.2 mM) for 3 weeks, were subjected to measuring the level of NAT10, cGAS, STING and p-IRF3 were analyzed by Western blotting, and SA-β-gal staining (B), and SA-β-gal-positive cells were counted and calculated as percentage of total cells, respectively (B). (C) Down-regulation of NAT10 reduced the effects of oxidative stress on MN formation. NC and shNAT10 cells were treated with H2O2 (0.4 mM) for 24 h, respectively, and NAT10 (green) and LaminB1 (red) were doubly stained. The formed MN in each group of cells were counted and calculated as percentage of total cells, respectively. (D) Hypoxia stimulated the level of NAT10 and SASP associated activities. LoVo cells were treated with CoCl2 (200 μM) for 36 h, were subjected to measuring the level of NAT10, HIF1A, cGAS and STING, and SA-β-gal staining (E), and SA-β-gal-positive cells were counted and calculated as percentage of total cells, respectively. (F) Down-regulation of NAT10 reduced hypoxia-induced MN formation. NC and shNAT10 were treated with CoCl2 (200 μM) for 36 h, respectively, and NAT10 (green) and LaminB1 (red) were doubly stained. The formed MN in each group of cells were counted and calculated as percentage of total cells, respectively. (G) Down-regulation of NAT10 reduced oxidative stress induced DNA damage response. LoVo and NAT10+/− cells were treated with DMEM or H2O2 (0.4 mM) for 24 h, and were double staining of LaminB1 (green) and γH2AX (red). (H) Inhibition of NAT10 reduced oxidative induced replication stress. LoVo cells were treated with H2O2 (0.4 mM) with or without Remodelin (20 μM) for 24 h, and were double staining of NAT10 (green) and RPA2 (red). Positive RPA2 cells were counted and calculated as percentage of total cells, respectively. (I) Genomic depletion of NAT10 reduced oxidative-induced replication stress. LoVo and NAT10+/− cells were treated with DMEM or H2O2 (0.4 mM) for 24 h, and the levels of NAT10, p-ATM, p-ATR, p-RPA2, RPA2, γH2AX were analyzed by Western blotting, respectively. The results are the representative of repeated experiments.
These results demonstrated that cell stress responses were dependent on NAT10, which up-expression could stimulate MN formation and activate SASP signaling by accelerating DNA replication and related damage.
Correlations Between NAT10 Expression and MN Formation, SASP Signaling, and Clinicopathological Features of CRC
To examine the correlations between NAT10 expression, MN formation, and SASP signaling activation in CRC, the expression of NAT10, HIF1A, cGAS, STING, and IRF3 and the frequencies of MN were analyzed in 140 surgically resected CRC samples. The results are summarized in Table 1, Table 2.
Table 1.
The mutual correlations between NAT10, MN, cGAS, STING, IRF3 and HIF1A.
| MN | cGAS | STING | IRF3 | HIF1A | |
|---|---|---|---|---|---|
|
P valuea (Spearman r) (1) (2) | |||||
| NAT10 | 0.096 (0.095) | 0.068 (0.184) | 0.040* (−0.181) | 0.066 (0.185) | 0.021* (0.203) |
| MN | 0.402 (0.074) | 0.518 (0.048) | 0.055 (0.168) | 0.065 (0.161) | |
| cGAS | 0.028* (0.194) | 0.924 (−0.008) | 0.737 (0.030) | ||
| STING | 0.405 (−0.073) | 0.870 (−0.014) | |||
| IRF3 | 0.006* (0.195) | ||||
Note:
(1) null probability of each correlation, *P < .05;
(2) spearman rank coefficient for each correlation, positive correlation (Spearman r > 0), negative correlation (Spearman r < 0)
Table 2.
The correlations between NAT10, MN, cGAS, STING, IRF3, HIF1A and clinical features.
| NAT10 | MN | cGAS | STING | IRF3 | HIF1A | |
|---|---|---|---|---|---|---|
| Histological grading | 0.038* | 0.889 | 0.457 | 0.209 | 0.201 | 0.293 |
| PDC | 0.003 | 0.001+ | 0.985 | 0.287 | 0.318 | 0.221 |
| Invasion depth | 0.156 | 0.083 | 0.486 | 0.227 | 0.103 | 0.616 |
| Lymph node metastasis | 0.343 | < 0.001* | 0.942 | 0.832 | 0.152 | 0.814 |
| Distant metastasis | 0.046* | 0.559 | 0.240 | 0.834 | 0.043* | 0.933 |
| TNM stage | 0.260 | 0.049* | 0.671 | 0.928 | 0.102 | 0.811 |
| Duke stage | 0.326 | 0.083 | 0.560 | 0.857 | 0.183 | 0.962 |
Note: *P < .05; For correlations between tumor marker and PDC stage, significance was taken as +P ≤ .001
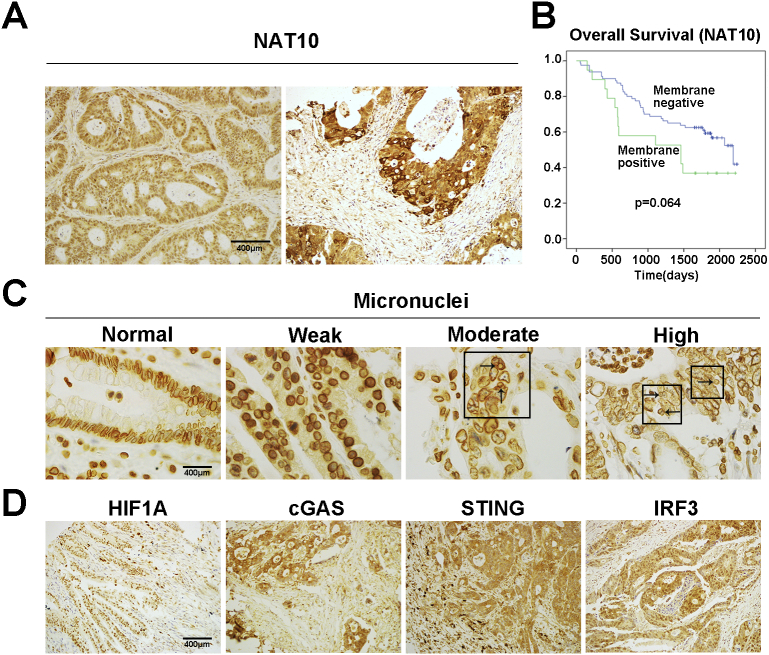
The staining of NAT10 presented in the nucleus, cytoplasm, membrane or combined, respectively (Figure 5A, Supplementary Figure S4A). The expression of NAT10 levels were significantly correlated with histological grading (P = .038) and distant metastasis (P = .046) (Table 2). And as previously reported [13], there was a slight but insignificant decrease in OS in patients with CRC that were positive for NAT10 membrane staining (Figure 5B, p = .064). In addition, NAT10 positivity correlated with strong HIF1A staining (P = .021) and correlated with STING expression (P = .040) (Table 1), while NAT10 was somewhat related to levels of cGAS (P = .068) and IRF3 (P = .066) (Table 1).
Figure 5.
The correlations between expression of NAT10 and MN formation, SASP signaling in clinical colorectal cancers. (A) Representative images of immunostaining of NAT10 in CRC tissues (nuclear and membrane positive). (B) Overall survival (OS) of patients with CRC exhibiting different levels of membranous NAT10 expression as determined by Kaplan–Meier analysis. Data are from 140 CRC cases for which survival information was available. P = .064, log-rank test. (C) Representative immune-staining of MN. The arrows indicate MNs. (D) Representative immune-staining of HIF1A, cGAS, STING and IRF3. Scale bars represent 400 μm.
MN formation in cancer tissues was assayed by LaminB1 staining using both immunofluorescence and immunohistochemistry, both of which proved satisfactory for analysis. For convenience, LaminB1 immunohistochemical staining was used and neutrophils were used as internal references since their lobulated nuclei displayed uniform and consistently strong LaminB1 positivity (Figure 5C). The frequency of MN formation in colorectal cancer tissues was classified (Supplementary Figure S4B), and apparently, MN formation was significantly with the presence of poorly differentiated clusters (PDCs) (P = .001), with node metastasis (P < .001), and TNM stage (P = .049) (Tables 2). There was a positive correlation between the frequency of MN formation and NAT10 expression despite just reach a marginal level in statistics (P = .096). Similarly, MN formation was somewhat related to high levels of IRF3 (P = .055) and HIF1A (P = .065) (Table 1).
There was also a positive correlation between the levels of cGAS and STING (P = .028), consisting with the role of cGAS in promoting STING activation. And high IRF3 levels were correlated with increased distant metastasis (P = .043) (Table 2).
These results suggest that there could be some correlations between NAT10 expression, MN formation, cGAS-STING pathway activation, and the clinicopathological characteristics of CRC.
In summary, the described study demonstrated that NAT10 could promote MN formation by enhancing DNA replication, and NAT10 related MN could consequently activate cGAS-STING-SASP signaling, and while above interactions could be strengthened in cell stress. There were some positive correlations between above events and the clinicopathological features of CRC. The possible interaction and connection were illustrated in Figure 6.
Figure 6.
Schematic diagram illustrating the involvement of NAT10 in MN formation, and activation of cGAS-STING and SASP activity described in present investigation. ① The expression of NAT10 could augment DNA replication or associated damage, at the same time nucleolar NAT10 also regulates rDNA transcription, to collectively induce formation of MN, which ② usually contains NAT10, DNA and rDNA, where DNA replication or damage and rDNA transcription could make ③ MN recruit cytosolic cGAS through an undefined way, activating its catalyzing to form 2′,3′-cGAMP, which then triggered polymerization of STING at ER. ④ the activated STING induces phosphorylation of IRF3 by recruiting TBK1(TANK binding kinase 1), and ⑤ the phosphorylated IRF3 makes nuclear translocation and mediates the activation of downstream nuclear factor kappa B to stimulate the production of proinflammatory factors such as type I interferon. SASP activity could exert both ⑥ the autocrine effects on tumor cell itself, and ⑦ on tumor microenvironment (TME) in paracrine. On another hand, ⑧ cell stresses could causes oxidative or DNA damage response possibly via cytoplasmic mechanism such as mitochondria ⑨ to enhance the expression of NAT10 and HIF-1A in nucleus ①.
Discussion
MN formation is prevalent in a variety of cancer cells, and their existence alongside the primary nucleus indicates genetic instability [25]. MN formation can occur in several ways. Although they can form during interphase through nuclear blebbing [26], in most cases, MN result from either aberrant mitosis or DNA damage. Aneugens (compounds that interfere with mitotic chromosome segregation) induce the formation of MN that contain complete chromosomes, while clastogens (genotoxic agents) result in MN that contain acentric chromosome fragments resulting from DNA breaks [9]. Recently, it was reported that DNA replication stress leads to genomic mutations and chromosome missegregation [27,28]. Even mild replication stress in cancer cells can induces numerical chromosomal instability through the formation of transiently multipolar spindles caused by premature centriole disengagement [29]. Replication stress and MN formation are closely related to G2/M checkpoint adaptation in cancer cells, in which cells exit anaphase prematurely, producing lagging or broken chromosomes that become incorporated in MN. Because of this link to structural chromosomal aberrations, replication stress is a hallmark of cancerous and precancerous lesions [29], and our results suggest that it leads to MN formation. In addition, our observations revealed that a portion of the spontaneously formed MN in CRC cells contained nucleolar components of varying size, implying that they could be derived from randomly broken chromosomes caused by DNA damage. This suggests that NAT10 inhibition slows DNA replication, consequently relieving replication stress and associated genetic damage, and reducing MN formation in CRC cells. Similarly, Nocodazole-induced MN formation was also reduced by NAT10 inhibition, via decreased DNA replication and consequently, decreased mitosis. The definitive mechanisms controlling MN formation remain unclear, and our study provides insightful clues regarding their formation in CRC cells.
Chemical inhibition of NAT10 reduces MN formation and nuclear lobulation in Hutchinson-Gilford progeria syndrome cells, possibly by redistributing the microtubules, and NAT10 may be involved in microtubule acetylation [14]. Contrary to mitosis, in which microtubules play a major role, it is not clear how microtubules impact DNA replication. Recently, a study revealed that replication stress triggers increased microtubule plus-end growth rates during mitosis, an abnormality previously identified to cause chromosome missegregation in cancer cells [25]. This missegregation is suppressed after restoration of proper microtubule growth rates and upon rescue of replication stress [25]. NAT10-inhibited cells displayed reduced histone acetylation and increased histone methylation, indicating that its acetylation activity may also be involved.
MN formation is prevalent in precancerous or cancerous cells, but its biological effects have only recently been established. First, MNs are a source of chromothripsis, in which a severely damaged chromosome undergoes complicated rearrangements, promoting genetic instability and cancer progression [27]. Second, MN activate cGAS-STING to activate SASP, promoting the production of pro-inflammatory cytokines that regulate tumor progression. cGAS is the cytosolic sensor that recognizes exposed DNA and synthesizes cGAMP, which activates STING [22,28]. However, how MN mediate cGAS-STING activation in details remains unclear. An early report demonstrated that >60% of MN experience nuclear envelope disruptions due to defective nuclear lamina, and that exposure of MN DNA recruits cGAS [28]. However, more recent studies have demonstrated that DNA damage occurring in MN is crucial to activate the cGAS-STING pathway, in which DNA damage could cause nuclear intact of MN although the detailed mechanism is still clear [30]. Our results demonstrate that MN in CRC cells generally contain all nuclear envelope components, such as nuclear lamina, pore complexes, and integral membrane proteins, suggesting that they are intact like the host nuclei. Only a subset of MNs were positive for cGAS, γH2AX, and 53BP1, all of which were consistent with the frequency of SA-β-gal-positive cells, confirming that MN harboring DNA damage have activated cGAS-STING pathways. Moreover, our investigation revealed that some MN contain rDNA elements that are actively transcribed by RNA polymerase I. Previous studies have indicated that MN can undergo DNA replication and subsequent nuclear envelope breakdown as cells enter mitosis [28]. Therefore, the replication and transcription occurring in MN may be closely related to the DNA damage observed in them, as a lack of the complete complement of necessary enzymes results in incomplete transcription or replication [28]. This could also explain how NAT10 inhibition suppresses MN formation and replication and transcription activities, consequently diminishing cGAS-STING activity.
SASP signaling is a double-edged sword in tumorigenesis. On one hand, cellular senescence is considered a barrier against transformation and overcoming it is crucial for cancer development. Moreover, SASP activation by a STING agonist can suppress cancer growth. On the other hand, there is extensive evidence that SASP promotes the production of pro-inflammatory factors that remodel the tumor microenvironment, promoting tumor progression. One study suggested that cGAS-STING-mediated SASP is a source of genetic instability that promotes cancer metastasis. Our in vitro experiments demonstrate that NAT10 inhibition decreased SASP signaling and cellular senescence in CRC cells. Furthermore, analysis of clinical CRC specimens revealed positive correlations between NAT10 and MN formation as well as SASP signaling, indicating that SASP signaling is oncogenic in advanced CRC, and is regulated by NAT10 and associated MN formation. This study also revealed that cell stress, including oxidative stress and hypoxia, regulates SASP signaling through the overexpression of NAT10 and HIF1A and increased MN formation. Oxidative stress could also disturb replication and promote MN formation.
Taken together, this study demonstrates that NAT10 promotes MN formation by enhancing DNA replication, and that NAT10 related MN activates cGAS-STING-SASP signaling, above of which could be strengthened by cell stress, and correlate with the clinicopathological features of CRC. The investigation expands our understanding of the role NAT10 plays in CRC development and progression.
Authors' Contributions
The investigation was proposed by Bo Zhang. The experiments were mainly performed by Yanan Cao. Mengfei Yao and Yaqian Wu contributed to tissue microarray preparation and some Western analysis. Ningning Ma took part in evaluation of the immunohistochemical staining. Some of experiments and data analysis were instructed by Haijing Liu. The manuscript was written by Yanan Cao and revised by Bo Zhang. All authors reviewed, edited, and approved the final manuscript.
Acknowledgments
Acknowledgements
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited. This project was supported by the National Natural Science Foundation of China (No. 81872018, No. 81372292), and Key Project from the Chinese Ministry of Science and Technology (No. 2017YFC0110200).
Declarations of Competing Interest
No potential conflicts of interest were disclosed.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100783.
Appendix A. Supplementary data
Supplementary figures
Supplementary tables
References
- 1.Coppe J.P., Patil C.K., Rodier F., Sun Y., Munoz D.P., Goldstein J., Nelson P.S., Desprez P.Y., Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loaiza N., Demaria M. Cellular senescence and tumor promotion: is aging the key? Biochim. Biophys. Acta. 2016;1865:155–167. doi: 10.1016/j.bbcan.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q., Sun L., Chen Z.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 5.Pépin G., Gantier M.P. cGAS-STING Activation in the Tumor Microenvironment and Its Role in Cancer Immunity. In: Xu D., editor. Regulation of Inflammatory Signaling in Health and Disease. Springer Singapore; Singapore: 2017. pp. 175–194. [Google Scholar]
- 6.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kisurina-Evgenieva O.P., Sutiagina O.I., Onishchenko G.E. Biogenesis of Micronuclei. Biochemistry. Biokhimiia. 2016;81:453–464. doi: 10.1134/S0006297916050035. [DOI] [PubMed] [Google Scholar]
- 8.Lewis C.W., Golsteyn R.M. Cancer cells that survive checkpoint adaptation contain micronuclei that harbor damaged DNA. Cell cycle (Georgetown, Tex.) 2016;15:3131–3145. doi: 10.1080/15384101.2016.1231287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalsbeek D., Golsteyn R.M. 2017. G2/M-Phase Checkpoint Adaptation and Micronuclei Formation as Mechanisms That Contribute to Genomic Instability in Human Cells, International journal of molecular sciences, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hintzsche H., Hemmann U., Poth A., Utesch D., Lott J., Stopper H., G.f.U.-M. Working Group In vitro micronucleus test, fate of micronuclei and micronucleated cells. Mutation research. 2017;771:85–98. doi: 10.1016/j.mrrev.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Liu H., Ling Y., Gong Y., Sun Y., Hou L., Zhang B. DNA damage induces N-acetyltransferase NAT10 gene expression through transcriptional activation. Mol. Cell. Biochem. 2007;300:249–258. doi: 10.1007/s11010-006-9390-5. [DOI] [PubMed] [Google Scholar]
- 12.Shen Q., Zheng X., McNutt M.A., Guang L., Sun Y., Wang J., Gong Y., Hou L., Zhang B. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp. Cell Res. 2009;315:1653–1667. doi: 10.1016/j.yexcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H., Hou W., Wang H.L., Liu H.J., Jia X.Y., Zheng X.Z., Zou Y.X., Li X., Hou L., McNutt M.A., Zhang B. GSK-3beta-regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clin. Cancer Res. 2014;20:4717–4729. doi: 10.1158/1078-0432.CCR-13-3477. [DOI] [PubMed] [Google Scholar]
- 14.Larrieu D., Britton S., Demir M., Rodriguez R., Jackson S.P. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527–532. doi: 10.1126/science.1252651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurien B.T., Scofield R.H. Western blotting: an introduction. In: Kurien B.T., Scofield R.H., editors. Western Blotting: Methods and Protocols. Springer New York; New York, NY: 2015. pp. 17–30. [Google Scholar]
- 16.Xiang X., Li C., Chen X., Dou H., Li Y., Zhang X., Luo Y. CRISPR/Cas9-mediated gene tagging: a step-by-step protocol. In: Luo Y., editor. CRISPR Gene Editing: Methods and Protocols. Springer New York; New York, NY: 2019. pp. 255–269. [Google Scholar]
- 17.Odell I.D., Cook D. Immunofluorescence techniques. The Journal of investigative dermatology. 2013;133 doi: 10.1038/jid.2012.455. [DOI] [PubMed] [Google Scholar]
- 18.Ismail I.H., Gagne J.P., Genois M.M., Strickfaden H., McDonald D., Xu Z., Poirier G.G., Masson J.Y., Hendzel M.J. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat. Cell Biol. 2015;17:1446–1457. doi: 10.1038/ncb3259. [DOI] [PubMed] [Google Scholar]
- 19.Ligasova A., Konecny P., Frydrych I., Koberna K. Cell cycle profiling by image and flow cytometry: the optimised protocol for the detection of replicational activity using 5-Bromo-2′-deoxyuridine, low concentration of hydrochloric acid and exonuclease III. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin K.M., Lu M., Liu F.F., Gu J., Du X.J., Xing B.C. N-WASP is highly expressed in hepatocellular carcinoma and associated with poor prognosis. Surgery. 2013;153:518–525. doi: 10.1016/j.surg.2012.08.067. [DOI] [PubMed] [Google Scholar]
- 21.Lv J., Liu H., Wang Q., Tang Z., Hou L., Zhang B. Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem. Biophys. Res. Commun. 2003;311:506–513. doi: 10.1016/j.bbrc.2003.09.235. [DOI] [PubMed] [Google Scholar]
- 22.de Oliveira Mann C.C., Kranzusch P.J. cGAS conducts micronuclei DNA surveillance. Trends Cell Biol. 2017;27:697–698. doi: 10.1016/j.tcb.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Li T., Chen Z.J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018;215:1287–1299. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding S.M., Benci J.L., Irianto J., Discher D.E., Minn A.J., Greenberg R.A. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466–470. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohly N., Kistner M., Bastians H. Mild replication stress causes aneuploidy by deregulating microtubule dynamics in mitosis. Cell cycle (Georgetown, Tex.) 2019;18:2770–2783. doi: 10.1080/15384101.2019.1658477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu N., Itoh N., Utiyama H., Wahl G.M. Selective entrapment of extrachromosomally amplified DNA by nuclear budding and micronucleation during S phase. J. Cell Biol. 1998;140:1307–1320. doi: 10.1083/jcb.140.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatch E.M., Fischer A.H., Deerinck T.J., Hetzer M.W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilhelm T., Olziersky A.M., Harry D., De Sousa F., Vassal H., Eskat A., Meraldi P. Mild replication stress causes chromosome mis-segregation via premature centriole disengagement. Nat. Commun. 2019;10:3585. doi: 10.1038/s41467-019-11584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackenzie K.J., Carroll P., Martin C.A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., Osborn R.T., Wheeler A.P., Nowotny M., Gilbert N., Chandra T., Reijns M.A.M., Jackson A.P. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary tables