Abstract
Epithelial ovarian cancer (EOC) is the most lethal gynecological malignancy. The insulin-like growth factor (IGF) system plays a key role in regulating growth and invasiveness in several malignancies, including ovarian cancer. IGF1R targeting showed antiproliferative activity of EOC cells. However, clinical studies failed to show significant benefit. EOC cells suppress antitumor immune responses by inducing dendritic cell (DC) dysfunction. The IGF1 axis can regulate DC maturation.
The current study evaluated involvement of the IGF1 axis in DC differentiation in EOC. Studies were conducted on EOC and on a human monocyte cell line. Tissue microarray analysis (TMA) was performed on 36 paraffin blocks from EOC patients. Expression of IGF1R, p53, Ki67, BRCA1, and DC markers was evaluated using immunohistochemistry. Co-culture of EOC cells with DC pretreated with IGF1R inhibitor blocked cancer cell migration. TMA demonstrated higher rate of IGF1R protein expression in patients with advanced (76.9%) as compared to early (40%) EOC. A negative correlation between IGF1R protein expression and the CD1c marker was found. These findings provide evidence that IGF1R axis inhibition could be a therapeutic strategy for ovarian cancer by restoring DC-mediated antitumor immunity.
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecologic malignancy and the fifth most common cause of cancer-related death in women. Worldwide, more than 200,000 women are diagnosed with ovarian cancer and 152,000 die each year [1]. Among most EOC patients, the disease is diagnosed at an advanced stage and the prognosis is poor. Although most patients will respond to primary treatment, 80% will have recurrent disease, ultimately resistant to chemotherapy and targeted biologic therapies [2].
The insulin-like growth factor (IGF) axis, which plays a key role in regulating growth and development, was identified as a potential therapeutic target, at least 10 years ago [[3], [4], [5]], purportedly due to hyperactivation of the IGF signaling pathway, which is implicated in the development, progression, and survival of many types of cancer, including ovarian [[6], [7], [8], [9], [10], [11]]. The IGF system regulates both physiological and pathophysiological processes involved in glucose metabolism and cell proliferation. It is comprised of the transmembrane receptor, insulin-like growth factor receptor type I (IGF1R), the growth factor ligands IGF1 and IGF2, and IGF binding proteins [[12], [13], [14], [15], [16]]. Ligand-receptor interaction transduces downstream signaling via the canonical phosphatidylinositol 3-kinase (PI3K)-AKT and RAS-extracellular signal–regulated kinase (ERK) pathways [12,17,18].
The success of IGF1R targeting in ovarian cancer models, which demonstrated a significant inhibitory effect on ovarian cancer cell proliferation, initiated great hope [[19], [20], [21], [22], [23], [24], [25]]. However, in clinical settings, IGF1R targeted monotherapy failed to demonstrate significant inhibition of various human malignancies. This lack of clinical effect and the need for new strategies have been widely discussed [[26], [27], [28], [29]]. A possible approach proposed by Liefers-Visser et al. is the combination of IGF1R targeting and immunotherapy [27]. As with other types of cancer, immunotherapy has great potential for improving EOC outcomes. Several studies demonstrated a correlation between tumor infiltrating lymphocytes and increased EOC patient survival [[30], [31], [32]]. Combining IGF1R inhibitors with immunotherapy entails a deep understanding of the interplay between the IGF1 axis and the immune environment in EOC.
Immune cells produce a variety of factors that influence the function of ovarian cancer cells [33]. In addition to cytotoxic T cells, which display antitumor features, ovarian tumors contain an abundance of immune cells that create an immunosuppressive tumor microenvironment (TME), including myeloid dendritic cells (DCs) [34]. A recent study demonstrated that ovarian tumors block the immune response and induce DC dysfunction by expressing immunosuppressive factors, including IDO [35], arginase I [36], IL-10 [37], TGF-β [38], and VEGF [39,40]. These factors can impair the differentiation, maturation, and function of the host DC. Dysfunction of DCs ultimately blocks the local activation and expansion of the intratumoral T cells [41]. In addition, human ovarian cancer was shown to upregulate immunosuppressive ligands such as PD-L1 and CD277 on the surface membrane of DCs [42,43], which led to inhibition of TCR-mediated proliferation of human T cells as well as Th1-related cytokine secretion. Several recent studies have shown that a vaccine of autologous dendritic cells enhanced immune response against ovarian cancer by induction of T cells that reduced the tumor mass and decreased the number of regulatory T cells [[44], [45], [46]].
DCs are professional antigen-presenting cells with specialized features, such as pathogen recognition and antigen-capturing and -processing machinery, which stimulate proliferation of naive T cells and initiate an immune response [47,48]. Conventional DCs can be divided into two main subsets: DC1 (CD141+) and DC2 (CD1c+, CD11c+, and CD11b+) [49]. CD11b is predominantly expressed in monocytes and granulocytes. CD11b+ DCs preferentially induce CD4+ T-cell immunity [49,50]. CD1c and CD141 are specific DC markers. Stimulated CD1c+ DCs can secrete interleukin (IL)-12, tumor necrosis factor (TNF)-α, IL-8, and IL-10. CD141+ DCs secrete high levels of type I interferon (IFNa) and IL-12 upon activation [51]. Both CD1c+ and CD141+ DCs can cross-present antigens to CD4+ and CD8+ T cells and suppress tumor growth [52]. However, CD141+ DCs may show more suppressive potential depending on the type of antigens.
DCs regulate humoral and cellular functions of the immune system and therefore were suggested as valuable targets for therapeutic approaches. Data regarding involvement of the IGF1 axis in the (TME) immune cells such as DCs are limited. Our recent study [53] provided a glimpse of the involvement of IGF1R in DCs in EOC cell lines. In the current study, we delve further into the nature of this relationship.
Materials and Methods
Cell Lines and Treatments
Human ES2 and SKOV3 ovarian cancer cells lines were provided by Prof. Ilan Tsarfaty, Department of Clinical Microbiology and Immunology, Sackler Faculty of Medicine, Tel Aviv University. They were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (FBS), 2 mM glutamine, and 100 μg/ml streptomycin in 5% CO2. THP-1, a human monocyte cell line derived from a patient with acute monocytic leukemia, was obtained from Prof. Isaac Witz lab, Faculty of Life Sciences, Tel Aviv University. THP-1 was cultured in RPMI-1640 medium (Biological Industries, Kibbutz Beit Haemek, Israel). Media were supplemented with 10% FBS, 2 mM glutamine, and 100 μg/ml streptomycin in 5% CO2. In addition, 10 mM HEPES, 1 mM sodium pyruvate, 1500 mg/l sodium bicarbonate, and 0.05 mM 2-mercaptoethanol were added. All reagents were purchased from Biological Industries. The NVP-AEW541 selective IGF1R inhibitor was obtained from Novartis Pharma (Basel, Switzerland) and kept as a stock solution (10 mM) in DMSO and stored at −20°C. In some of the experiments, cells were treated with IGF1 (50 ng/ml) (Cytolab Ltd., Rehovot, Israel).
Study Population
The study was approved by the Hillel Yaffe Medical Center, Institutional Helsinki Committee (0019-16HYMC). Formalin-fixed, paraffin-embedded (FFPE) tissue samples from 36 patients with EOC who were operated at Hillel Yaffe Medical Center during 2012-2018 were collected from the pathology department. Clinical and pathological data of EOC patients, including age, weight, surgical-pathological stage, treatment protocol after diagnosis, pathological type, and histological grade were obtained from their medical records.
Inducing Differentiation of Mature DCs
THP-1 cells were harvested by centrifugation, resuspended in complete RPMI at a concentration of 2 × 105 cells/ml, and transferred into six-well culture plates. To induce differentiation, 100 ng IL-4 and 100 ng GM-CSF were added. Cells were cultured for 5 days to acquire the properties of immature DCs. Medium was exchanged every 2 days with fresh cytokine-supplemented medium. Mature DCs were generated from immature DCs by the addition of 200 ng IL-4, 100 ng GM-CSF, 20 ng/ml TNFα, and 200 ng/ml Ionomycin in serum-free culture media, at a concentration of 2 × 105 cells/ml for 2 days, in a humidified incubator at 37°C and 5% CO2 [54].
Co-Culturing of DCs and Ovarian Cancer Cell Lines
THP-1 and differentiated mature DCs were grown on six-well culture plates in RPMI-1640 and treated with 5 μM NVP-AEW541. Following DC differentiation for 7 days, media were removed, and 6*105 ovarian cancer cell lines (ES2 and SKOV3) were seeded along with Dulbecco's modified Eagle's medium 10% FBS media (without IGF1R inhibitor). The co-culture cells (DCs and ES2/SKOV3) were incubated at 37°C in 5% CO2.
In Vitro Scratch Assay
After 24 hours of DC and SKOV3/ ES2 co-culture, a wound scratch assay was performed using a pipette tip. Migration of SKOV3 and ES2 into the scratch area was evaluated. Images were captured at 0, 24, and 48 hours using a Nikon ECLIPSE Ti microscope. Photomicrographs were analyzed with ImageJ software (National Institutes of Health, version 1.48v). Each experiment was repeated three times, and a representative experiment is presented.
Flow Cytometry Assay
For cell surface staining, THP-1 and differentiated DCs were incubated with CD141 (344,110, Biolegend) and CD11b (101,206, Biolegend) antibodies for 20 minutes on ice in the dark. Staining was terminated by adding FACS buffer [phosphate-buffered saline (PBS) with 3% FBS and 2 mM EDTA]. Samples were run on an LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software, version 9.7.6 (TreeStar, Becton Dickinson & Co., Ashland, OR). The expression of stained molecules at the cell surface was determined as percentage.
For intracellular staining, THP-1 cells were washed twice with PBS and fixed by using 4% paraformaldehyde for 15 minutes at room temperature (RT). Cells were then washed with PBS and permeabilized by adding 1% Triton for 10 minutes at RT. Cells were then washed and incubated with primary antibodies total IGF1R (tIGF1R) (sc-81,167, Santa Cruz Biotechnology, Inc.) and phospho IGF1R (pIGF1R) (Y1135/1136, Cell Signaling) for 1 hour at RT. Cells were washed and incubated with secondary antibodies Alexa Flour donkey Anti-mouse (715-545-150, Jackson Immuno Research, West Grove, PA) and CY3 donkey anti-rabbit (7111-165-152, Jackson Immuno Research) antibodies for 20 minutes in the dark on ice. Staining was terminated by adding FACS buffer (PBS with 3% FBS and 2 mM EDTA). Samples were run on an LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software, version 9.7.6 (BD). The expression of stained molecules at the cell surface was determined as a percentage.
Western Immunoblots
Cells were treated with 5 μM of NVP-AEW541, with or without the addition of IGF1. Cells were then harvested, washed twice with PBS, and incubated with lysis buffer for 20 minutes. The suspension was centrifuged at 13,000 rpm for 10 minutes, diluted with sample buffer, and boiled for 5 minutes. Samples were electrophoresed through 8% SDS-PAGE, followed by electrophoretic transfer of the proteins to nitrocellulose membranes. After blocking with 5% milk in 20 mM Tris-HCl (pH 7.5), 135 mM NaCl, and 0.1% Tween 20, the blots were incubated overnight with antibodies against tIGF1R β-subunit (Cell Signaling, Danvers, MA), pIGF1R (Y1135/1136), total Akt (tAKT) (CST-9272S), phospho Akt (pAKT) (Ser473) (Cell Signaling), and tubulin (Sigma-Aldrich). All antibodies were used at a 1:1000 dilution. After blotting, membranes were washed and incubated with a horseradish peroxidase–conjugated secondary antibody. Proteins were detected using the SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL).
Tissue Microarray (TMA) Construction and Immunohistochemistry
The next-generation TMA was created as previously described [55]. FFPE tissue specimens were chosen based on clinical data and retrieved from the pathology archives. All slides stained with hematoxylin and eosin were reviewed, and representative tumor tissue samples were selected for each case. The selected area was marked for TMA construction: 36 EOC samples, 29 cases of high-grade serous histology, and 7 cases of low-grade serous histology. There were 8 cases with stage I, 2 with stage II, 15 with stage III, and 11 with stage IV. The staging met the criteria of the clinical staging of International Federation of Gynecology and Obstetrics (FIGO). Two TMA blocks (block A and block B) were punched from FFPE human serous ovarian carcinoma specimens with a tissue microarray. From each case, two cores of tumor tissue with a diameter size of 2 mm were punched by TMA grand master (3DHISTECH, Budapest, Hungary). FFPE TMA blocks were cut into 4-μm sections on coated slides and deparaffinized. Peroxidase was blocked using 3% H2O2. Antigens were retrieved using 1 M citrate buffer for 20 minutes. The following primary antibodies were used: IGF1R (sc-81167, Santa Cruz Biotechnology, Inc.), p53 (M7001, DAKO), Ki67 (M 7240, DAKO), BRCA1 (ab16780, Abcam), CD141 (ab109189, Abcam), and CD1c (ab156708, Abcam). Slides were then counterstained with hematoxylin and mounted. Each IHC reaction was accompanied by positive control tissue, according to antibody manufacturer's instructions. Digital slides were scanned using Panoramic MIDI (TMA Scanner), and the images were captured using 3DHISTECH software. Results of the 36 EOC samples were quantified by an experienced pathologist at the Pathology Institute of Haemek Medical Center, who was blinded to clinical data regarding the patients and samples. Protein expression was analyzed and correlated to the clinical data of the EOC patients, including previous hormonal therapy, tumor stage and grading, treatment protocol after diagnosis, progression-free survival (PFS), and overall survival (OS).
Scoring
Levels of staining were subjectively graded by the pathologist based on the number of reactive versus total cells. They were categorized as: (0) no staining and few scattered positive cells (<5%), (1) 5%-25% of cells stained, (2) 26%-50% of cells stained, (3) 51%-75% of cells stained, and (4) 76%-100% of cells stained. Staining intensity was categorized as: (0) no staining, (1) weak staining, (2) moderate staining, and (3) strong staining. Immunohistochemical results of IGF1R, p53, BRCA1, CD1c, and CD141 were divided into low– and high–staining intensity groups, where 0 to 1 was classified as a low-staining group and 2 to 3 as a high-staining group. The percentage of Ki67 was measured using the median value. If the cell density was above the median value (16%), the sample was defined as high staining.
Statistical Analysis
Western blot data and open wound area of scratch assay data were collected using ImageJ software. All statistical analyses were performed using Microsoft Excel. Values reported in figures are expressed as the standard error of the mean, unless otherwise indicated. For normally distributed datasets observed between groups, we used two-tailed Student’s t tests. P values ≤ .05 were considered significant.
Wilcoxon paired test was used to test the similarity of the extensity and intensity values of the different proteins between block A and block B. Also, McNemar-Bowker test and marginal homogeneity test were used to evaluate the similarity of the categorical levels of the proteins.
Differences in protein extensity between groups (high-grade serous vs low grade, neoadjuvant vs chemotherapy, and stage 1 + 2 vs 3 + 4) were tested using the Mann-Whitney U test. Categorical protein intensity levels were tested using Fisher exact test. Spearman's correlation was used to test the relation between proteins. OS and PFS were presented with odds ratios and 95% confidence intervals (CI). SPSS version 25 was used for the statistical analysis.
Descriptive statistics in terms of mean, SD, median, percentiles, and ranges were performed for all the study parameters.
Results
Our recent study [53] provided evidence that inhibition of the IGF1R signaling pathway in monocyte cells might be linked to enhanced DC differentiation. Initial experiments aimed to investigate the IGF1R signaling pathway in dendritic cells. For this purpose, the human leukemic monocyte cell line THP-1 was used.
In Vitro Studies
Differentiation of THP-1 to DC
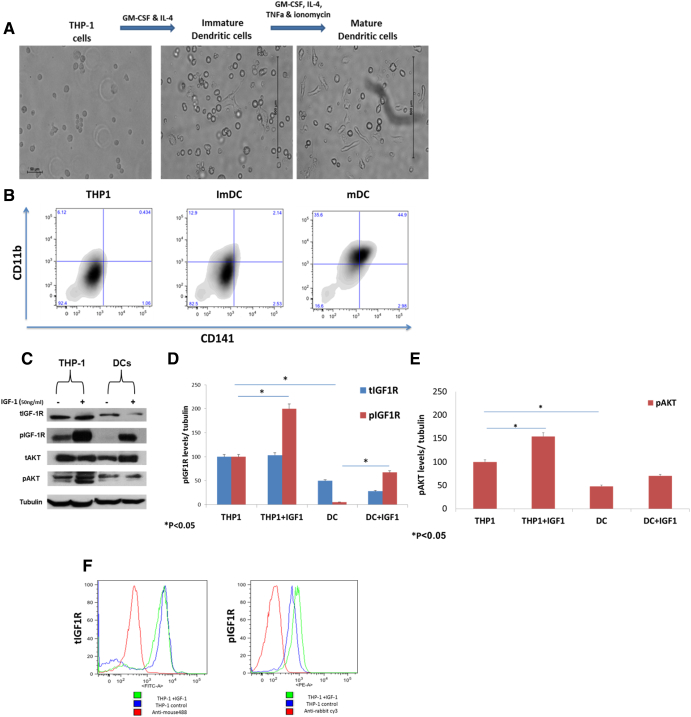
THP-1 cells were differentiated into mature dendritic cells (mDCs) (Figure 1A). To validate cell differentiation, cells were stained with the DC markers CD11b and CD141. Specific antibodies and flow cytometry analysis were applied. The percentage of DC populations is shown in Figure 1B. THP-1 cells treated with IL-4, GM-CSF, TNFα, and Ionomycin expressed significantly higher CD141 and CD11b levels compared to untreated THP-1 cells (Figure 1B).
Figure 1.
Reduced total and phosphorylated IGF1R protein levels in differentiated DCs. (A) Human leukemic THP-1 cells were differentiated to immature DCs by treatment with 100 ng/ml IL-4 and 100 ng/ml GM-CSF for 5 days and then differentiated to DCs by treatment with 200 ng/ml IL-4, 100 ng/ml GM-CSF, 20 ng/ml TNFα, and 200 ng/ml Ionomycin for 2 days. THP1 scale bar = 50 μm, immature DCs scale bar = 500 μm, DCs scale bar = 500 μm. (B) DC marker expression via CD11b and CD141 in THP-1, ImDC, and mDC. (C) THP-1 and THP-1 cells were treated with 200 ng/ml IL-4, 100 ng/ml GM-CSF, 20 ng/ml TNFα, and 200 ng/ml Ionomycin for 72 hours and were incubated with IGF1 for 10 minutes before harvest, after which cell extracts were prepared. Proteins were separated through SDS-PAGE, followed by electrophoretic transfer and incubation with antibodies against tIGF1R, pIGF1R, tAKT, and pAKT. (D) Scanning densitometry analysis of tIGF1R (blue bars) and pIGF1R (red bars) levels in THP-1 and DCs. A value of 100% was given to the expression level of untreated THP-1 cells. Levels of tubulin were measured as a loading control. *P < .05. Bars represent SEM values. (E) Scanning densitometry analysis of pAKT levels in THP-1 and DCs. A value of 100% was given to the expression level of untreated THP-1 cells. Levels of tubulin were measured as a loading control.*P < .05. Bars represent SEM values. The graphs represent average of three independent experiments. (F) Expression of tIGF1R and pIGF1R with (green) or without (blue) IGF1 treatment in THP-1 cells.
IGF1R Protein Expression Levels in Myeloid Differentiated Dendritic Cells
The expression of tIGFR and pIGF1R in THP-1 cells was evaluated by flow cytometry assay (Figure 1E). We found positive expression of tIGFR and pIGF1R in THP-1 cells.
Next, we analyzed the expression of IGF1R in DC compared to THP-1. A representative Western blot analysis (Figure 1C) shows a significant decrease in phosphorylated and total IGF1R levels in differentiated DCs. As shown, pIGF1R levels were reduced by 95% and tIGF1R levels by 50% as compared to THP-1 cells (Figure 1D). In addition, pAKT expression levels were significantly decreased in differentiated DCs as compared to THP-1 cells (Figure 1E). Of interest, undifferentiated THP-1 cells displayed high basal phosphorylated IGF1R levels (Figure 1F).
Effect of IGF1R Inhibition in DCs on Ovarian Carcinoma Cell Migration
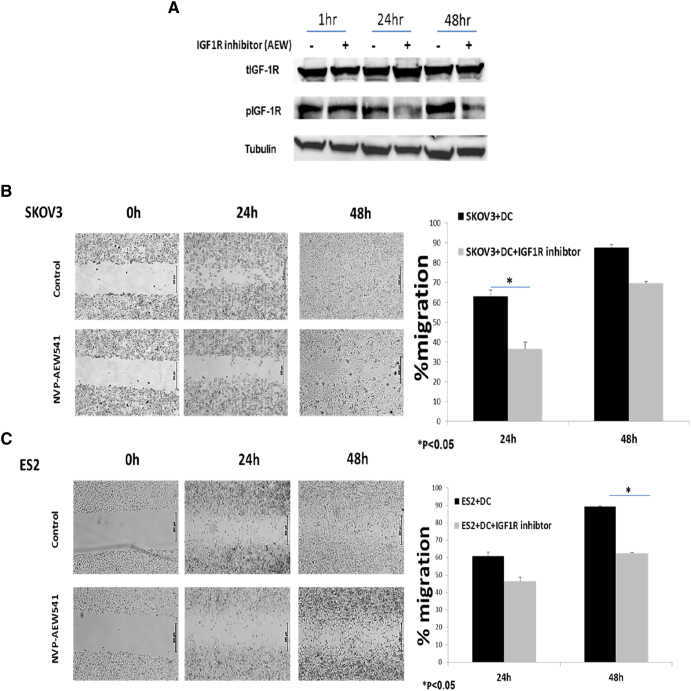
A recent study showed that IGFs treatment resulted in delayed maturation of DCs. Moreover, the suppressive effects of the IGFs on DCs were blocked by IGF1R kinase inhibitor-NVP-AEW541 [56]. We first assessed the effect of IGF1R inhibition on DCs. To confirm IGF1R inhibition, THP-1 cells were treated with NVP-AEW541, and Western blot analysis was applied. As shown in Figure 2A, treatment of THP-1 cells resulted in decreased IGF1R phosphorylation levels after 24 hours. DC differentiation was induced with or without NVP-AEW541 treatment. We then assessed the effect of IGF1R inhibitor-treated DCs on EOC migration. Differentiated DCs pretreated with NVP-AEW541 treatment were co-cultured with SKOV3 (Figure 2B) and ES2 cells (Figure 2C), and a wound scratch assay was performed [57]. A 26.5% decreased migration of SKOV3 cells was observed when co-cultured with NVP-AEW541 pretreated DCs as compared to untreated DCs after 24 hours (Figure 2B). SKOV3 migration was reduced by 18% after 48 hours in pretreated DCs compared to untreated DCs (Figure 2B). Consistently, NVP-AEW541–treated DCs led to 15% reduction in ES2 cell migration as compared to untreated DCs after 24 hours (Figure 2C). Similarly, NVP-AEW541 pretreated DCs led to significantly decreased migration of ES2 cells (26.5%) as compared to untreated DCs after 48 hours (Figure 2C). In addition, IGF1R inhibition in pretreated and untreated DCs led to 39% decreased migration of ES2 cells as compared to 24% in untreated ES2 cells (data not shown).
Figure 2.
NVP-AEW541–treated DCs decrease ovarian cancer cell migration. (A) THP-1 myeloid cells were treated with 5 μM of NVP-AEW541 for 1, 24, and 48 hours. Whole-cell lysates were resolved on SDS-PAGE and immunoblotted with tIGF1R and pIGF1R antibodies. Level of tubulin was used as a loading control. Cell migration was detected by wound scratch assay. (B, left panel) Human leukemic THP-1 cells were differentiated to DCs and treated with 5 μM of NVP-AEW541, after which they were co-cultured with SKOV3; scratch was applied 24 hours after cell merge. The growth of EOC cells into the scratch zone is demonstrated here at time 0, 24,and 48 hours after scratch; scale bar = 500 μm. (B, right panel) Scanning densitometry analysis of % migration in co-culture of SKOV3 with DCs. A value of 100% was given to % migration in untreated DCs at 0 hour. (C, left panel) THP-1 cells were differentiated to DCs and treated with 5 μM of NVP-AEW541, after which they were co-cultured with ES2; scale bar = 500 μm. (C, right panel) Scanning densitometry analysis of % migration in co-culture of ES2 with DCs. A value of 100% was given to the expression level of untreated DCs at 0 hour. The graphs represent average % migration of three independent experiments of ES2 and SKOV3 cells. Images were captured at a magnification of ×4. * P < .05. Bars represent SEM value.
Study of Clinical Tissues
Patients' Baseline Characteristics
To identify the interplay between IGF1R expression and DC markers with clinicopathological data of EOC patients, 36 EOC samples were stained with IGF1R, p53, Ki67, BRCA1, and DC cell-surface molecules (CD141, CD1c).
Table 1 summarizes baseline demographic and clinicopathological characteristics of 36 patients diagnosed with EOC, ages 27 to 87 years (mean 62.8 ± 14 years), with a median age of 61 years. Ten patients had early-stage disease (stage I-II) and 26 (72.3%) had advanced disease (stage III-IV).
Table 1.
Demographic and Pathological Data of 36 Patients with EOC
| Age, mean ± SD [range] | 62.8 ± 14.0; [27-87] |
|---|---|
| Ethnicity mother; n = 22 | |
| Jewish | 17 (77%) |
| Arab | 5 (23%) |
| BMI (n = 33), mean ± SD (range) | 27.8 ± 6.1; [18.3-42.2] |
| Parity | 2.83 ± 1.72; [0-8] |
| Comorbidities | |
| Hypertension | 14 (39%) |
| Diabetes | 7 (19%) |
| Dyslipidemia | 8 (22%) |
| Other | 22 (64%) |
| Primary site | |
| Ovary | 33 (91.6%) |
| Fallopian tube | 1 (2.77%) |
| Primary peritoneal | 3 (8.33%) |
| Unknown | 1 (2.77%) |
| Histology | |
| High-grade serous carcinoma | 29 (81%) |
| Low-grade serous carcinoma | 7 (19%) |
| Stage | |
| 1 | 8 (22%) |
| 2 | 2 (6%) |
| 3 | 15 (42%) |
| 4 | 11 (31%) |
| Other malignancy | |
| Breast | 2 (5.5%) |
| Lymphoma | 2 (5.5%) |
| None | 32 (89%) |
| Neoadjuvant chemotherapy | 17 (47.3%) |
| Primary surgery | 19 (52.7%) |
All patients had histologic subtypes with ovarian serous carcinoma: 7 low grade (19.4%) and 29 high grade (80.5%). Follow-up ranged from 13 to 78 months (median 30). The median PFS was 24 months (range 7-36) and OS was 38 months (range 15-82).
IGF1R, p53, Ki67, BRCA1, CD141, and CD1c Expression in EOC
The immunohistochemical expression of IGF1R, p53, Ki67, BRCA1, and the DC markers CD1c and CD141 is shown in Table 2. Among EOC patients, 66.7% had high IGF1R expression. Furthermore, high staining rates of CD141 and CD1c were found in 36.2% and 44.4% of EOC cases, respectively. High staining of p53, Ki67, and BRCA1 was found in 80.6%, 50%, and 38.8% of EOC cases, respectively.
Table 2.
Protein Expression (Intensity) Rates in EOC Tissue
| Tissue Simples | n=36 | Proportion (%) |
|---|---|---|
| IGF1R | ||
| Low staining | 12 | 33.3 |
| High staining | 24 | 66.7 |
| P53 | ||
| Low staining | 7 | 19.4 |
| High staining | 29 | 80.6 |
| Ki67 | ||
| Low staining | 18 | 50 |
| High staining | 18 | 50 |
| BRCA1 | ||
| Low staining | 22 | 61.2 |
| High staining | 14 | 38.8 |
| CD1c | ||
| Low staining | 20 | 55.55 |
| High staining | 16 | 44.44 |
| CD141 | ||
| Low staining | 23 | 63.8 |
| High staining | 13 | 36.2 |
Association Between Protein Expression in EOC and OS with PFS
No correlation was found between the expression of the stained proteins and PFS or OS.
Correlation Analysis Between Protein Expression and Clinicopathological Characteristics
IGF1R intensity (expression) correlated with FIGO stage (P = .05) and neoadjuvant chemotherapy treatment (P = .014), although not with histological grade (P = .19; Table 3). Moreover, the TMA analysis demonstrated higher rate of p53 (P = .018) and Ki67 (P = .02) mean protein extensity in patients with high-grade as compared to low-grade EOC (Table 4). However, there was no significant correlation between the p53 and Ki67 intensity (protein expression) and FIGO stage or neoadjuvant treatment (Table 3). CD1c expression was 33.8% higher in advanced-stage (53.8%) as compared to early-stage EOC (20%); however, this difference was not statistically significant due to the small sample size.
Table 3.
Relation Between Intensity (Protein Expression) and Clinicopathological Parameters of EOC
| Pathological Data | STAGE |
GRADE |
Neoadjuvant Chemo |
|||
|---|---|---|---|---|---|---|
| I+II | III+IV | Low | High | No | Yes | |
| IGF1R | ||||||
| Low | 6 (60%) | 6 (23.1%) | 4 (57.1%) | 8 (27.6%) | 10 (52.6%) | 2 (11.8%) |
| High | 4 (40%) | 20 (76.9%)* | 3 (42.9%) | 21 (72.4%) | 9 (47.4%) | 15 (88.2%)* |
| p53 | ||||||
| Low | 1 (10%) | 6 (23.1%) | 2 (28.6%) | 5 (17.2%) | 2 (10.5%) | 5 (29.4%) |
| High | 9 (90%) | 20 (76.9%) | 5 (71.4%) | 24 (82.8%) | 17 (85.9%) | 12 (70.6%) |
| Ki67 | ||||||
| Low | 6 (60%) | 12 (46.2%) | 6 (85.7%) | 12 (41.4%) | 7 (36.8%) | 11 (64.7%) |
| High | 4 (40%) | 14 (53.8%) | 1 (14.3%) | 17 (58.6%) | 12 (63.2%) | 6 (35.3%) |
| BRCA1 | ||||||
| Low | 5 (50%) | 17 (65.4%) | 5 (71.4%) | 17 (58.6%) | 11 (57.9%) | 11 (64.7%) |
| High | 5 (50%) | 9 (34.6%) | 2 (28.6%) | 12 (41.4%) | 8 (42.1%) | 6 (53.3%) |
| CD1c | ||||||
| Low | 8 (80%) | 12 (46.2%) | 5 (71.4%) | 15 (51.7%) | 12 (63.2%) | 8 (47.1%) |
| High | 2 (20%) | 14 (53.8%) | 2 (28.6%) | 14 (48.3%) | 7 (36.8) | 9 (52.9%) |
| CD141 | ||||||
| Low | 6 (60%) | 17 (65.4%) | 5 (71.4%) | 18 (62.1%) | 12 (63.2%) | 11 (64.7%) |
| High | 4 (40%) | 9 (34.6%) | 2 (28.6%) | 11 (37.9%) | 7 (36.8%) | 6 (35.3%) |
Table 4.
Correlation Between the Mean Extensity of IGF1R, p53, Ki67, and Histological Grade
| Pathological Data | Low-grade Serous Carcinoma (N = 7) (Mean ± SD) |
High-Grade Serous Carcinoma (N = 29) (Mean ± SD) |
P Value |
|---|---|---|---|
| IGF1R | 80.166 ± 20.5 | 81.44 ± 30.85 | P = .21 |
| P53 | 49 ± 27.38 | 77.87 ± 36.86 | P = .018* |
| Ki67 | 10.26± 9.4167 | 20.19± 27.069 | P = .02* |
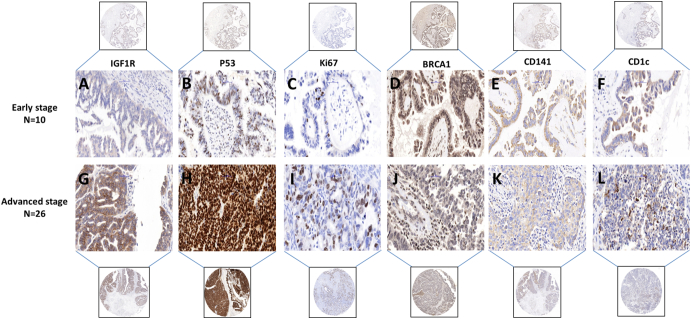
Representative samples with significantly higher IGF1R intensity (37.9%) in advanced-stage EOC (Figure 3G) as compared to early stage (Figure 3A) are shown in Figure 3. Similarly, samples of increased expression in advanced stage EOC are shown in Figure 3, F and L.
Figure 3.
Positive immunohistochemical staining. (A-F) IGFIR, p53, Ki67, BRCA1, CD141, and CD1c in early stage of serous subtypes ovarian carcinoma (×40), respectively. (G-L) IGFIR, p53, Ki67, BRCA1, CD141, and CD1c in advanced stage of serous subtypes ovarian carcinoma (×40), respectively. Scale bar = 50 μm.
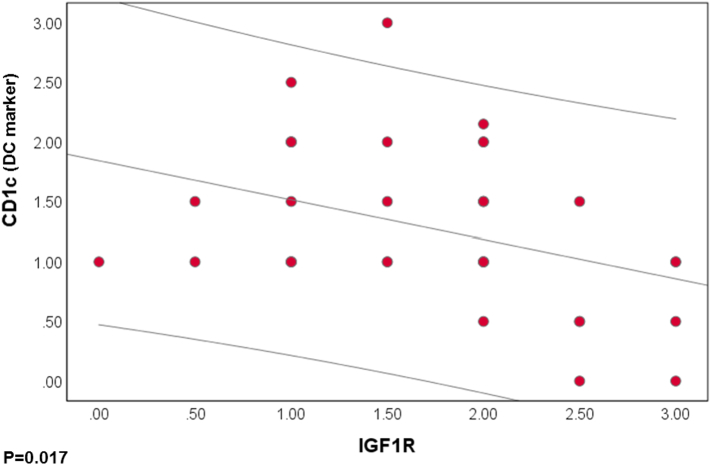
Correlation Between IGF1R and DC Marker Expression Levels
Next, we analyzed the correlation between expression levels of IGF1R and DC markers in EOC cases. A negative correlation between IGF1R and CD1c was found. This was confirmed by a strong Pearson correlation (r = −0.394, P = .017) (Figure 4). IGF1R did not have significant correlations with p53 (r = 0.216, P = .205), Ki67 (0.147, P = .393), BRCA1 (−0.061, P = .725), or CD141 protein expression.
Figure 4.
Correlation coefficient between each individual values of IGF1R expression and CD1c in EOC patients (P˂.05).
Discussion
IGF1 ligand binding stimulates cancer progression through activation of the PI3K/Akt and the Raf-1/MEK/ERK cascades, which induce cell cycle and cellular proliferation. The relation between IGF1R and immune function has remained poorly characterized. Recent findings imply that the IGF1/IGF1R pathway plays diverse roles in regulating immune function. Several immune cells, such as T and B lymphocytes [58], monocyte-macrophage cells, and NK cells [59], display many different surface growth factor receptors, including IGF1R [58]. IGF1 enhances lymphocyte survival [60] and can also block IL-2–dependent lymphocyte growth and function [61]. However, the exact function of IGFs on host immunity and on immune cells such as DC remains unclear. The case is different with DCs, in which activation of the Raf-1/MEK/ERK pathway by IGF1 results in delayed cell maturation [56]. Similarly, Xuan et al. showed that, through PI3K/Akt signaling, IGFs subsequently inhibit the functional maturation of DCs [62]. In turn, IGF1R inhibitor treatment rescued DC maturation [56]. Moreover, IGFs increased IL-10 secretion of DCs, thereby enhancing the immunosuppressive status of the tumor environment.
In the current study, we demonstrated that co-culture of EOC cells with THP1-DCs pretreated with IGF1R inhibitor reduced cancer cell migration (Figure 3). This experiment was performed based on the finding that DCs (derived from human PBMCs) can directly inhibit proliferation of various human tumor lines apart from their ability to execute antitumor effect by stimulation of T lymphocytes [63]. Our recently published study presented similar findings using HL-60-DCs [53]. Hence, the direct effect of DCs on cancer cells is influenced by the IGF1 axis activation status. The TMA led to several assessments. We found high expression of IGF1R in advanced-stage EOC patients and in samples of patients who underwent neoadjuvant chemotherapy (Table 2). On the other hand, OS and PFS were not correlated with IGF1R expression, as we have a small sample size limitation of N = 36. A large body of evidence indicates that IGF1R enhances proliferation, survival, and migration of EOC cells in vitro [24,64,65]. Nevertheless, few studies have analyzed IGF1R expression in serous ovarian cancer using immunohistochemical staining. Studies demonstrated that IGF1R expression was significantly increased in epithelial ovarian cancer tissues [66,67]. Along with our results, Singh et al. reported that IGF1R expression in ovarian cancer patients increased after chemotherapy [68]. These results provide evidence that IGF1R could act as a mediator of chemoresistance [69,70]. King et al. showed IGF1 overexpression in low-grade serous as compared to high-grade serous ovarian carcinomas [20]. Previous studies showed that the pathogenesis of high-grade serous carcinoma is characterized by high levels of p53 mutations [[71], [72], [73], [74]] and that the Ki67 proliferation index is markedly elevated in high-grade serous carcinoma [75]. In line with this, we found significantly higher p53 and Ki67 expression in high-grade serous carcinoma as compared to low-grade serous (Table 4).
Interestingly, we found a negative correlation between IGF1R protein expression levels and the CD1c DC marker in EOC tumor samples (Figure 4). CD1c+ DCs are known to prime cytotoxic T cell response, which plays an important role in the control of tumor growth by activating antitumor immune response [76]. Here, we infer that IGFs may lead to suppression of antitumor immunity by inhibiting DC maturation. Consequently, rescuing the impaired function of the DCs by blocking the IGF signaling pathway constitutes a possible approach to generating a potent antitumor immunity. Here, we present a preliminary effort to explore the involvement of the IGF1R signaling pathway in DC differentiation in the tumor microenvironment. Additional experiments are needed to reveal the nature of this interaction.
Ovarian cancer is an immune responsive disease [77,78]. Expression of Treg cells and immunosuppressive TGFβ isoforms is increased in ovarian tumors as compared to normal ovarian tissue [79,80]. TGFβ reduces secretion of GM-CSF by activated memory CD8+ T cells, which results in delayed maturation of DCs [81]. Preclinical and early clinical data have confirmed the ability of DC vaccines to induce potent immune responses that in some instances can lead to measurable clinical responses [82,83]. A phase I/II study demonstrated that the autologous DC vaccine provides a safe and feasible therapy for advanced ovarian and primary peritoneal cancers in remission. The autologous DC vaccine elicits modest immune response by presenting tumor antigens and improves overall survival in ovarian cancer [46]. These new insights become particularly important in the context of our study results and in discovering a potential new therapy for EOC patients.
Taken together, we suggest that the involvement of the IGF1 axis in DC maturation affects the antitumor immunity in the tumor microenvironment. Consequently, IGF1R blocking may play a key role in reversing the immune escape in patients with advanced serous ovarian carcinoma.
Acknowledgments
Acknowledgements
This work was performed in partial fulfillment of the requirements for a PhD degree by Lina Somri-Gannam at the Gynecology Laboratory, Department of Obstetrics and Gynecology, Hillel Yaffe Medical Center, Israel. The support of the Israel Cancer Research Foundation (grant 2026011 to I.B.) is kindly acknowledged. In addition, the authors wish to thank the support of The Ruth and Bruce Rappaport Faculty of Medicine, Technion, Haifa, Israel. Finally, we wish to thank the director of the histopathology laboratory at Haemek Medical Center, Shulamit Goez, and Natalia Edison for their major help with the TMA analysis.
Conflicts of Interest
The authors declare no conflict of interest.
Funding
This research was funded by the Israel Cancer Research Foundation, Montreal, Canada, grant number 2026011, and The Ruth and Bruce Rappaport Faculty of Medicine, Technion, Institute of Technology, Haifa, Israel, to Dr. Ilan Bruchim.
References
- 1.Ferlay J. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012: Globocan 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Bukowski R.M., Ozols R.F., Markman M. The management of recurrent ovarian cancer. Semin Oncol. 2007;34:S1–S15. doi: 10.1053/j.seminoncol.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Rohlik Q.T., Adams D., Kull F.C., Jacobs S. An antibody to the receptor for insulin-like growth factor I inhibits the growth of MCF-7 cells in tissue culture. Biochem Biophys Res Commun. 1987;149(1):276–281. doi: 10.1016/0006-291X(87)91635-4. [DOI] [PubMed] [Google Scholar]
- 4.Gualberto A., Pollak M. Emerging role of insulin-like growth factor receptor inhibitors in oncology: early clinical trial results and future directions. Oncogene. 2009;28(34):3009–3021. doi: 10.1038/onc.2009.172. [DOI] [PubMed] [Google Scholar]
- 5.Z. Pietrzkowski, G. Mulholland, L. Gomella, B. A. Jameson, and R. Baserga, “Inhibition of growth of prostatic cancer cell lines by peptide analogues of insulin-like growth factor I,” p. 6. [PubMed]
- 6.Resnicoff M., Ambrose D., Coppola D., Rubin R. Insulin-like growth factor-1 and its receptor mediate the autocrine proliferation of human ovarian carcinoma cell lines. Lab Investig. 1993;69:756–760. [PubMed] [Google Scholar]
- 7.Ouban A., Muraca P., Yeatman T., Coppola D. Expression and distribution of insulin-like growth factor-1 receptor in human carcinomas. Hum Pathol. 2003;34(8):803–808. doi: 10.1016/S0046-8177(03)00291-0. [DOI] [PubMed] [Google Scholar]
- 8.Yee D., Morales F.R., Hamilton T.C., Hoff D.D.V. 51(19) 1991. Expression of insulin-like growth factor I, its binding proteins, and its receptor in ovarian cancer; pp. 5107–5112. [PubMed] [Google Scholar]
- 9.Lu L. Promoter-specific transcription of insulin-like growth factor-II in epithelial ovarian cancer. Gynecol Oncol. 2006;103(3):990–995. doi: 10.1016/j.ygyno.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Lu L. The relationship of insulin-like growth factor-II, insulin-like growth factor binding protein-3, and estrogen receptor- expression to disease progression in epithelial ovarian cancer. Clin Cancer Res. 2006;12(4):1208–1214. doi: 10.1158/1078-0432.CCR-05-1801. [DOI] [PubMed] [Google Scholar]
- 11.Brokaw J. IGF-I in epithelial ovarian cancer and its role in disease progression. Growth Factors. 2007;25(5):346–354. doi: 10.1080/08977190701838402. [DOI] [PubMed] [Google Scholar]
- 12.Pollak M.N., Schernhammer E.S., Hankinson S.E. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 13.Hwa V., Oh Y., Rosenfeld R.G. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999;20:761–787. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- 14.Rother K.I., Accili D. Role of insulin receptors and IGF receptors in growth and development. Pediatr Nephrol. 2000;14(7):558–561. doi: 10.1007/s004670000351. [DOI] [PubMed] [Google Scholar]
- 15.Giudice L., Maternal-Fetal C. Conflict — lessons from a transgene. J Clin Invest. 2002;110:307–309. doi: 10.1172/JCI200216389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8(12):915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 17.Girnita L., Worrall C., Takahashi S.-I., Seregard S., Girnita A. Something old, something new and something borrowed: emerging paradigm of insulin-like growth factor type 1 receptor (IGF-1R) signaling regulation. Cell Mol Life Sci. 2014;71(13):2403–2427. doi: 10.1007/s00018-013-1514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crudden C., Ilic M., Suleymanova N., Worrall C., Girnita A., Girnita L. The dichotomy of the Insulin-like growth factor 1 receptor: RTK and GPCR: friend or foe for cancer treatment? Growth Hormon IGF Res. 2015;25(1):2–12. doi: 10.1016/j.ghir.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Beltran P.J. Ganitumab (AMG 479) inhibits IGF-II–dependent ovarian cancer growth and potentiates platinum-based chemotherapy. Clin Cancer Res. 2014;20(11):2947–2958. doi: 10.1158/1078-0432.CCR-13-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King E.R. The insulin-like growth factor 1 pathway is a potential therapeutic target for low-grade serous ovarian carcinoma. Gynecol Oncol. 2011;123(1):13–18. doi: 10.1016/j.ygyno.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotlieb W.H. Insulin-like growth factor receptor I targeting in epithelial ovarian cancer. Gynecol Oncol. 2006;100(2):389–396. doi: 10.1016/j.ygyno.2005.09.048. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y. Inhibition of insulin-like growth factor-I receptor (IGF-IR) signaling and tumor cell growth by a fully human neutralizing anti-IGF-IR antibody. Mol Cancer Ther. 2005;4(8):1214–1221. doi: 10.1158/1535-7163.MCT-05-0048. [DOI] [PubMed] [Google Scholar]
- 23.An Y. Inhibitory effect of small interfering RNA targeting insulin-like growth factor-I receptor in ovarian cancer OVCAR3 cells. Cancer Biother Radiopharm. 2010;25(5):545–552. doi: 10.1089/cbr.2009.0712. [DOI] [PubMed] [Google Scholar]
- 24.J. Tang et al., “Antisense oligonucleotide suppression of human IGF-1R inhibits the growth and survival of in vitro cultured epithelial ovarian cancer cells,” Journal of Ovarian Research, vol. 6, no. 1, p. 71, 2013, 10.1186/1757-2215-6-71. [DOI] [PMC free article] [PubMed]
- 25.Ller M.M., Dietel M., Turzynski A., Wiechen K. 77(4) 1998. Antisense phosphorothioate oligodeoxynucleotide down-regulation of the insulin-like growth factor I receptor in ovarian cancer cells; pp. 567–571. https://doi.org/10.1002/(SICI)1097-0215(19980812)77:4<567::AID-IJC16>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Lodhia K.A., Tienchaiananda P., Haluska P. Understanding the key to targeting the IGF axis in cancer: a biomarker assessment. Front Oncol. 2015;5 doi: 10.3389/fonc.2015.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liefers-Visser J.A.L., Meijering R.A.M., Reyners A.K.L., van der Zee A.G.J., de Jong S. IGF system targeted therapy: therapeutic opportunities for ovarian cancer. Cancer Treat Rev. 2017;60:90–99. doi: 10.1016/j.ctrv.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 28.Baserga R. The decline and fall of the IGF-I receptor. J Cell Physiol. 2013;228(4):675–679. doi: 10.1002/jcp.24217. [DOI] [PubMed] [Google Scholar]
- 29.J. A. M. J. L. Janssen and A. J. Varewijck, “IGF-IR targeted therapy: past, present and future,” Frontiers in Endocrinology, vol. 5, Dec. 2014, 10.3389/fendo.2014.00224. [DOI] [PMC free article] [PubMed]
- 30.Barnett J.C. Ovarian cancer tumor infiltrating T-regulatory (Treg) cells are associated with a metastatic phenotype. Gynecol Oncol. 2010;116(3):556–562. doi: 10.1016/j.ygyno.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 31.Hamanishi J. The comprehensive assessment of local immune status of ovarian cancer by the clustering of multiple immune factors. Clin Immunol. 2011;141(3):338–347. doi: 10.1016/j.clim.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Ventriglia J. Immunotherapy in ovarian, endometrial and cervical cancer: State of the art and future perspectives. Cancer Treat Rev. 2017;59:109–116. doi: 10.1016/j.ctrv.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Zamarron B.F., Chen W. Dual roles of immune cells and their factors in cancer development and progression. Int J Biol Sci. 2011;7(5):651–658. doi: 10.7150/ijbs.7.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wertel I., Nowicka A., Rogala E., Kotarski J. Peritoneal immune system in patients with advance epithelial ovarian cancer. Int Rev Immunol. 2011;30(2–3):87–101. doi: 10.3109/08830185.2011.569902. [DOI] [PubMed] [Google Scholar]
- 35.Belladonna M.L. Cutting edge: autocrine TGF-β sustains default tolerogenesis by IDO-competent dendritic cells. J Immunol. 2008;181(8):5194–5198. doi: 10.4049/jimmunol.181.8.5194. [DOI] [PubMed] [Google Scholar]
- 36.Liu Q., Zhang C., Sun A., Zheng Y., Wang L., Cao X. Tumor-educated CD11b high Ia low regulatory dendritic cells suppress T cell response through arginase I. J Immunol. 2009;182(10):6207–6216. doi: 10.4049/jimmunol.0803926. [DOI] [PubMed] [Google Scholar]
- 37.Bellone G. Cooperative induction of a tolerogenic dendritic cell phenotype by cytokines secreted by pancreatic carcinoma cells. J Immunol. 2006;177(5):3448–3460. doi: 10.4049/jimmunol.177.5.3448. [DOI] [PubMed] [Google Scholar]
- 38.Cubillos-Ruiz J.R. Reprogramming tumor-associated dendritic cells in vivo using miRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res. 2012;72(7):1683–1693. doi: 10.1158/0008-5472.CAN-11-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi A., Kono K., Ichihara F., Sugai H., Fujii H., Matsumoto Y. Vascular endothelial growth factor inhibits maturation of dendritic cells induced by lipopolysaccharide, but not by proinflammatory cytokines. Cancer Immunol Immunother. 2004;53(6):543–550. doi: 10.1007/s00262-003-0466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fricke I. Vascular endothelial growth factor-trap overcomes defects in dendritic cell differentiation but does not improve antigen-specific immune responses. Clin Cancer Res. 2007;13(16):4840–4848. doi: 10.1158/1078-0432.CCR-07-0409. [DOI] [PubMed] [Google Scholar]
- 41.Chae C.-S., Teran-Cabanillas E., Cubillos-Ruiz J.R. Dendritic cell rehab: new strategies to unleash therapeutic immunity in ovarian cancer. Cancer Immunol Immunother. 2017;66(8):969–977. doi: 10.1007/s00262-017-1958-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curiel T.J. Blockade of B7-H1 improves myeloid dendritic cell–mediated antitumor immunity. Nat Med. 2003;9(5):562–567. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- 43.J. R. Cubillos-Ruiz et al., “CD277 is a negative co-stimulatory molecule universally expressed by ovarian cancer microenvironmental cells,” Oncotarget, vol. 1, no. 5, Sep. 2010, 10.18632/oncotarget.165. [DOI] [PMC free article] [PubMed]
- 44.J. L. Tanyi et al., “Personalized cancer vaccine effectively mobilizes antitumor T cell immunity in ovarian cancer,” Sci. Transl. Med., vol. 10, no. 436, p. eaao5931, Apr. 2018, 10.1126/scitranslmed.aao5931. [DOI] [PubMed]
- 45.Bapsy P.P. Open-label, multi-center, non-randomized, single-arm study to evaluate the safety and efficacy of dendritic cell immunotherapy in patients with refractory solid malignancies, on supportive care. Cytotherapy. Feb. 2014;16(2):234–244. doi: 10.1016/j.jcyt.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 46.Chu C.S. Phase I/II randomized trial of dendritic cell vaccination with or without cyclophosphamide for consolidation therapy of advanced ovarian cancer in first or second remission. Cancer Immunol Immunother. May 2012;61(5):629–641. doi: 10.1007/s00262-011-1081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banchereau J. Immunobiology of dendritic cells. Annu Rev Immunol. Apr. 2000;18(1):767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 48.Collin M., McGovern N., Haniffa M. British society for Immunology; Apr: 2013. Human dendritic cell subsets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collin M., Bigley V. Human dendritic cell subsets: an update. Immunology. May 2018;154(1):3–20. doi: 10.1111/imm.12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mildner A., Jung S. Development and function of dendritic cell subsets. Immunity. May 2014;40(5):642–656. doi: 10.1016/j.immuni.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 51.Sabado R.L., Balan S., Bhardwaj N. Dendritic cell-based immunotherapy. Cell Res. Jan. 2017;27(1):74–95. doi: 10.1038/cr.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohn L. Antigen delivery to early endosomes eliminates the superiority of human blood BDCA3 + dendritic cells at cross presentation. J Exp Med. May 2013;210(5):1049–1063. doi: 10.1084/jem.20121251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. A. Yahya, S. M. Sharon, S. Hantisteanu, M. Hallak, and I. Bruchim, “The role of the insulin-like growth factor 1 pathway in immune tumor microenvironment and its clinical ramifications in gynecologic malignancies,” Frontiers in Endocrinology, vol. 9, Jun. 2018, 10.3389/fendo.2018.00297. [DOI] [PMC free article] [PubMed]
- 54.Berges C. A cell line model for the differentiation of human dendritic cells. Biochem Biophys Res Commun. Aug. 2005;333(3):896–907. doi: 10.1016/j.bbrc.2005.05.171. [DOI] [PubMed] [Google Scholar]
- 55.I. Zlobec, G. Suter, A. Perren, and A. Lugli, “A next-generation tissue microarray (ngTMA) protocol for biomarker studies,” Journal of Visualized Experiments, no. 91, Sep. 2014, 10.3791/51893. [DOI] [PMC free article] [PubMed]
- 56.Huang C.-T., Chang M.-C., Chen Y.-L., Chen T.-C., Chen C.-A., Cheng W.-F. Insulin-like growth factors inhibit dendritic cell-mediated anti-tumor immunity through regulating ERK1/2 phosphorylation and p38 dephosphorylation. Cancer Lett. Apr. 2015;359(1):117–126. doi: 10.1016/j.canlet.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 57.Liang C.-C., Park A.Y., Guan J.-L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. Feb. 2007;2(2):329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 58.Smith T.J. Insulin-like growth factor-i regulation of immune function: a potential therapeutic target in autoimmune diseases? Pharmacol Rev. Jun. 2010;62(2):199–236. doi: 10.1124/pr.109.002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Badolato R. Differential expression of surface membrane growth hormone receptor on human peripheral blood lymphocytes detected by dual fluorochrome flow cytometry. The Journal of Clinical Endocrinology & Metabolism. Oct. 1994;79(4):984–990. doi: 10.1210/jcem.79.4.7962309. [DOI] [PubMed] [Google Scholar]
- 60.Walsh P.T., Smith L.M., O'Connor R. Insulin-like growth factor-1 activates Akt and Jun N-terminal kinases (JNKs) in promoting the survival of T lymphocytes. Immunology. Dec. 2002;107(4):461–471. doi: 10.1046/j.1365-2567.2002.01525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hunt P., Eardley D.D. Suppressive effects of insulin and insulin-like growth factor-1 (IGF1) on immune responses. J Immunol. 1986;136(11):3994–3999. [PubMed] [Google Scholar]
- 62.Xuan N.T., Hoang N.H., Nhung V.P., Duong N.T., Ha N.H., Hai N.V. Regulation of dendritic cell function by insulin/IGF-1/PI3K/Akt signaling through klotho expression. Journal of Receptors and Signal Transduction. May 2017;37(3):297–303. doi: 10.1080/10799893.2016.1247862. [DOI] [PubMed] [Google Scholar]
- 63.A. I. Chapoval, K. Tamada, and L. Chen, “In vitro growth inhibition of a broad spectrum of tumor cell lines by activated human dendritic cells,” vol. 95, no. 7, p. 7, 2000. [PubMed]
- 64.Wang X. Crosstalk between TEMs and endothelial cells modulates angiogenesis and metastasis via IGF1-IGF1R signalling in epithelial ovarian cancer. Br J Cancer. Oct. 2017;117(9):1371–1382. doi: 10.1038/bjc.2017.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu L., Wang X., Li X., Wu X., Tang M., Wang X. Oncology Reports; Dec.: 2017. Upregulation of IGF1 by tumor-associated macrophages promotes the proliferation and migration of epithelial ovarian cancer cells. [DOI] [PubMed] [Google Scholar]
- 66.Liu D. The stimulation of IGF-1R expression by Lewis(y) antigen provides a powerful development mechanism of epithelial ovarian carcinoma. Int J Mol Sci. Oct. 2011;12(10):6781–6795. doi: 10.3390/ijms12106781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.An Y., Cai L., Wang Y., Zhu D., Guan Y., Zheng J. Local expression of insulin-like growth factor-I, insulin-like growth factor-I receptor, and estrogen receptor alpha in ovarian cancer. Onkologie. 2009;32(11):638–644. doi: 10.1159/000242253. [DOI] [PubMed] [Google Scholar]
- 68.Singh R.K., Gaikwad S.M., Jinager A., Chaudhury S., Maheshwari A., Ray P. IGF-1R inhibition potentiates cytotoxic effects of chemotherapeutic agents in early stages of chemoresistant ovarian cancer cells. Cancer Lett. Nov. 2014;354(2):254–262. doi: 10.1016/j.canlet.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 69.Singh R.K., Dhadve A., Sakpal A., De A., Ray P. An active IGF-1R-AKT signaling imparts functional heterogeneity in ovarian CSC population. Sci Rep. 2016;6(1):36612. doi: 10.1038/srep36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan J., Yin Z., Tao K., Wang G., Gao J. Function of insulin-like growth factor 1 receptor in cancer resistance to chemotherapy (review) Oncol Lett. 2017 doi: 10.3892/ol.2017.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuo K.-T. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res. May 2009;69(9):4036–4042. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho K.R., Shih I.-M. Ovarian cancer. Annual review of pathology: mechanisms of disease. Feb. 2009;4(1):287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bonome T. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. Nov. 2005;65(22):10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- 74.O'Neill C.J., Deavers M.T., Malpica A., Foster H., McCluggage W.G. An immunohistochemical comparison between low-grade and high-grade ovarian serous carcinomas. Am J Surg Pathol. 2005;29(8):8. doi: 10.1097/01.pas.0000166367.68459.7d. [DOI] [PubMed] [Google Scholar]
- 75.Vang R., Shih I.-M., Kurman R.J. Ovarian low-grade and high-grade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv Anat Pathol. Sep. 2009;16(5):267–282. doi: 10.1097/PAP.0b013e3181b4fffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nizzoli G. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood. 2013;122(6):932–942. doi: 10.1182/blood-2013-04-495424. [DOI] [PubMed] [Google Scholar]
- 77.Zhang L. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. Jan. 2003;348(3):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 78.Sabbatini P., Odunsi K. Immunologic approaches to ovarian cancer treatment. JCO. 2007;25(20):2884–2893. doi: 10.1200/JCO.2007.11.0775. [DOI] [PubMed] [Google Scholar]
- 79.Shah C.A., Allison K.H., Garcia R.L., Gray H.J., Goff B.A., Swisher E.M. Intratumoral T cells, tumor-associated macrophages, and regulatory T cells: association with p53 mutations, circulating tumor DNA and survival in women with ovarian cancer. Gynecol Oncol. May 2008;109(2):215–219. doi: 10.1016/j.ygyno.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 80.Bristow R.E., Baldwin R.L., Yamada S.D., Korc M., Karlan B.Y. Altered expression of transforming growth factor-β ligands and receptors in primary and recurrent ovarian carcinoma. Cancer American Cancer Society. 2000;85(3):658–668. doi: 10.1002/(sici)1097-0142(19990201)85:3<658::aid-cncr16>3.0.co;2-m. https://doi.org/10.1002/(SICI)1097-0142(19990201)85:3<658::AID-CNCR16>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 81.Yamaguchi Y., Tsumura H., Miwa M., Inaba K. Contrasting effects of TGF-beta;1 and TNF-alpha; on the development of dendritic cells from progenitors in mouse bone marrow. Stem Cells. 1997;15(2):144–153. doi: 10.1002/stem.150144. [DOI] [PubMed] [Google Scholar]
- 82.Banchereau J., Palucka A.K. Dendritic cells as therapeutic vaccines against cancer. Nat Rev Immunol. 2005;5(4):296–306. doi: 10.1038/nri1592. [DOI] [PubMed] [Google Scholar]
- 83.Tacken Paul J., de Vries I. Jolanda M., Torensma Ruurd, Carl G. Figdor. Dendritic-cell immunotherapy: from ex vivo loading to in vivo targeting. Nat Rev Immunol. 2007;7:790–802. doi: 10.1038/nri2173. [DOI] [PubMed] [Google Scholar]