Abstract
This study aims at evaluating the performances of the multiplex PCR AllplexTM Gastrointestinal Panel-Parasite Assay (GIPPA), which detects G. duodenalis, Cryptosporidium spp., E. histolytica, D. fragilis, B. hominis, and C. cayetanensis, by comparison to microscopy. A retrospective evaluation was conducted on a series of positive clinical samples (n = 99) stored at −80 °C or at +4 °C. A five-month prospective study was then conducted on all samples sent to our lab for parasite detection (n = 586). In the retrospective cohort, sensitivity was 81% for both G. duodenalis (26/32) and D. fragilis (21/26) and 100% for Cryptosporidium spp. (26/26, including 6 different species), B. hominis (26/26), and C. cayetanensis (4/4). During the prospective study, 95 samples were positive by microscopy and 207 by multiplex PCR assay. The molecular assay showed a significantly higher sensitivity of PCR, especially for G. duodenalis (100% vs. 60.7%, p < 0.01), D. fragilis (97.2% vs. 14.1%, p < 0.001), and B. hominis (99.4% vs. 44.2%, p < 0.001) but also for E. histolytica (100% vs. 50.0%). The sensitivity of the AllplexTM GIPPA on the first stool sample was equivalent to the sensitivity of microscopy on multiple stool samples but inferior to multiplex PCR on multiple stool samples. Taken together, the AllplexTM GIPPA is suitable for the routine detection of protozoa in fecal samples.
Keywords: Giardia duodenalis, Entamoeba histolytica, Cryptosporidium spp., Dientamoeba fragilis, Cyclospora cayetanensis, Blastocystis hominis
1. Introduction
Infectious diarrheas are among the most life-threatening and invalidating infectious diseases in the world, particularly in children under five years. In 2015, they caused 1.3 million deaths in the world [1]. Diagnosis is sometimes difficult because of the great diversity of pathogens potentially responsible for these intestinal symptoms. For these reasons, and because they are globally less frequent than viral and bacterial infections, parasitic diseases that are due to soil-transmitted helminths and protozoa are often neglected. Yet protozoa represent a major cause of infection, (i) in terms of mortality, with amebiasis and cryptosporidiosis being responsible for respectively 11,000 and 42,000 deaths yearly [2,3], and (ii) in terms of frequency, with pathologies such as giardiasis and dientamoebiasis [4,5]. These protozoa are also very prevalent in high income countries. Microscopic examination of stools remains the reference method for the diagnosis of most intestinal protozoa. This technique however requires three successive samples for the same patient and trained operators and several concentration techniques for optimal results. This approach is time-consuming and yields limited sensitivity. There is, therefore, a need for new methods for the diagnosis of enteric protozoa. Molecular biology—particularly multiplex PCR—seems to offer performances similar to microscopy [6,7] but is limited by the number of parasite species detected.
The recently marketed assay AllplexTM Gastrointestinal Panel-Parasite Assay (GIPPA) (Seegene, Seoul, Korea) is able to detect most protozoa pathogens, i.e., Giardia duodenalis, Cryptosporidium spp., Entamoeba histolytica, Dientamoeba fragilis, Blastocystis hominis, and Cyclospora cayetanensis. In this study, the performances of this assay are evaluated on both retrospective and prospective cohorts.
2. Materials and Methods
2.1. Clinical Samples
First, 89 clinical samples positive for G. duodenalis, Cryptosporidium spp., E. histolytica/dispar/moshkovskii, D. fragilis and/or B. hominis by routine microscopic examination were retrospectively analyzed. For each stool analyzed over a three-year period (2015–2018), an aliquot was stored at −80 °C until DNA extraction. This allowed the selection of positive samples for the PCR evaluation. The routine procedure consists of the wet mount examination of fresh stool and various in-house concentration methods based on clinical data (Bailenger’s, Thebault’s, and/or merthiolate-iodine-formalin biphasic methods). Cryptosporidium spp. and Cyclospora cayetanensis detection relied on Henriksen’s modified Ziehl–Neelsen staining. Finally, four positive samples for C. cayetanensis (collected between 2005 and 2009, stored at +4 °C), and 10 Cryptosporidium-positive stools provided by the French National Reference Centre for Cryptosporidiosis and identified at species-level were also included. The final retrospective panel contained 103 positive samples including 33 G. duodenalis, 15 E. histolytica/dispar/moshkovskii, 27 Cryptosporidium sp., 26 D. fragilis, 27 B. hominis and 4 C. cayetanensis, possibly associated with other protozoa and helminths.
Secondly, during a five-month period (September 2019–February 2020), all stool samples routinely analyzed in our laboratory were prospectively included for analysis with the AllplexTM assay. The prospective panel consisted of 588 stools from 350 patients.
2.2. Multiplex PCR Testing
DNA extraction was performed using the automated device MICROLAB® STARlet (Hamilton Company, Reno, NV, USA) with the STARMag 96 Universal Cartridge kit, following the manufacturer’s instructions. Briefly, a small amount (140–180 mg) of stool was suspended in a Cary-Blair Medium (FecalSwabTM, Copan Diagnostics Inc, Murrieta, CA, USA), vigorously mixed, and, after a 10 min incubation at room temperature, was centrifuged 10 min at 2000 g before processing. Extraction was then performed on 50 µL of supernatant and eluted in 100 µL. For amplification with the AllplexTM assay, an internal control DNA (provided) was added to the medium before extraction. The reaction mix and DNA extract were displayed in 96-wells plates by the MICROLAB® STARlet. All PCR runs included both positive and negative controls. Amplification was realized on a CFX96 (Bio-Rad, Marnes-la-Coquette, France) and managed with CFX Manager IVD 1.6 software. Results were analyzed with Seegene Viewer® software. Positive stools for E. histolytica detection were confirmed with the G-DiaParaTM assay (Diagenode Diagnostics, Liège, Belgium) following the manufacturer’s instructions.
2.3. DNA Preservation in Cary-Blair Suspension
We evaluated whether the FecalSwabTM stool suspensions could be reliably analysed after different conditions of storage. The aim was to assess the possibility of analysing grouped samples. Hence, the differences in signal intensities (expressed in CT values) before and after storage were computed for each stool suspension. Different storage conditions were tested (room temperature and +4 °C) between 0 and 7 days. The samples included in this study were positive for B. hominis (n = 15), D. fragilis (n = 9), G. duodenalis (n = 6), Cryptosporidium sp. (n = 2), and E. histolytica (n = 1).
2.4. Statistical Analysis
Differences in sensitivities were analyzed with McNemar’s Test. To determine the sensitivity of the assay, true positive and false negative results were determined according to microscopy or to microscopy and PCR assay when specified. The impact of the storage conditions were analyzed through the calculation of differences in cycle threshold (CT) values before storage (CT(D0)) and after storage (CT(DX)). This was done for various storage times and temperatures (storage either at 4 °C or at room temperature). For each condition, the median was compared to a hypothetical value of 0 using the Wilcoxon signed-rank test to assess the preservation of the DNA.
3. Results
3.1. Retrospective Cohort
Four samples presented invalid results (no amplification of an internal control DNA) and were excluded from the subsequent calculations (1 G. duodenalis, 1 E. histolytica/dispar/moshkovskii, 1 B. hominis and 1 Cryptosporidium spp.). The final composition of the cohort is available in Table 1.
Table 1.
Number of samples included in the retrospective and prospective cohorts and parasites detected by microscopy.
| Sampling Numbers | ||
|---|---|---|
| Retrospective Cohort | Prospective Cohort | |
| Included samples: | 99 | 586 |
| Total number | 103 | 588 |
| Invalid results (excluded for analysis) | 4 | 2 |
| Parasites detected by microscopy: | 99 | 95 |
| Giardia duodenalis | 32 | 17 |
| Cryptosporidium spp. 1 | 26 | 2 |
| C. parvum | 13 | Nd 2 |
| C. hominis | 5 | Nd 2 |
| C. felis | 4 | Nd 2 |
| C. canis | 1 | Nd 2 |
| C. cuniculus | 1 | Nd 2 |
| C. meleagridis | 1 | Nd 2 |
| Dientamoeba fragilis | 26 | 10 |
| Blastocystis hominis | 26 | 72 |
| Cyclospora cayetanensis | 4 | 0 |
| Entamoeba histolytica/dispar/moshkovskii | 14 | 4 |
| E. histolytica (identified by PCR) | 2 | 3 |
1 Molecular identification; 2 The 2 Cryptosporidium spp. observed in the prospective cohort were not identified at species level.
Sensitivity for the G. duodenalis detection was 81% (26/32) (Table 2); the six false negative results were observed for low parasitic loads. The detection of D. fragilis had a 81% sensitivity (21/26), with false negative results also related to low parasitic loads. The sensitivity reached 100% for C. cayetanensis (4/4), Cryptosporidium spp. (26/26), and B. hominis (26/26) positive samples. All Cryptosporidium species tested (C. parvum (n = 13), C. hominis (n = 5), C. felis (n = 4), C. canis (n = 1), C. cuniculus (n = 1), C. meleagridis (n = 1)) were detected. As the identification at the species level is not possible through microscopy for E. histolytica/dispar/moshkovskii, no sensitivity value could be calculated. However, two positive results were obtained and verified with another molecular assay. The 13 other samples that contained E. histolytica/dispar/moshkovskii were negative by PCR.
Table 2.
Performances of the AllplexTM PCR assay compared to microscopy on the retrospective cohort (n = 99).
| Sensitivity% (n/N) | |
|---|---|
| Giardia duodenalis | 81% (26/32) |
| Cryptosporidium spp. | 100% (26/26) |
| Dientamoeba fragilis | 81% (21/26) |
| Blastocystis hominis | 100% (26/26) |
| Cyclospora cayetanensis | 100% (4/4) |
| Entamoeba histolytica/dispar/moshkovskii | nd 1 |
1 As microscopy cannot allow the species identification, sensitivity and specificity could not be determined for E. histolytica detection. However, two positive results were confirmed by other molecular assay.
3.2. Prospective Cohort
Ninety-five out of 588 samples were positive by microscopy, consisting of 17 G. duodenalis, 4 E. histolytica/dispar/moshkovskii, 2 Cryptosporidium sp., 10 D. fragilis, and 72 B. hominis. Among them, 10 samples were positive for multiple targets: 6 for G. duodenalis and B. hominis, 2 for D. fragilis and B. hominis, 1 for E. histolytica and G. duodenalis, and 1 for E. histolytica and B. hominis. With the AllplexTM assay, 207 samples were positive for at least one target: 28 for G. duodenalis, 6 for E. histolytica, 2 for Cryptosporidium sp., 69 for D. fragilis and 162 for B. hominis. Two samples yielded invalid results (absence of amplification of the internal control) and were excluded from the calculations; both were negative by microscopy and no signal was observed in PCR runs. The sensitivity was 100% (17/17) for G. duodenalis, 100% (2/2) for Cryptosporidium spp., 98.6% (71/72) for B. hominis and 80.0% (8/10) for D. fragilis, taking microscopy as the gold standard. During the inclusion period, no C. cayetanensis was diagnosed in the laboratory. One sample positive for hematophagous E. histolytica and 2 samples positive for E. histolytica/dispar/moshkovskii cysts by microscopy were also positive for E. histolytica with the AllplexTM assay and confirmed with another PCR assay.
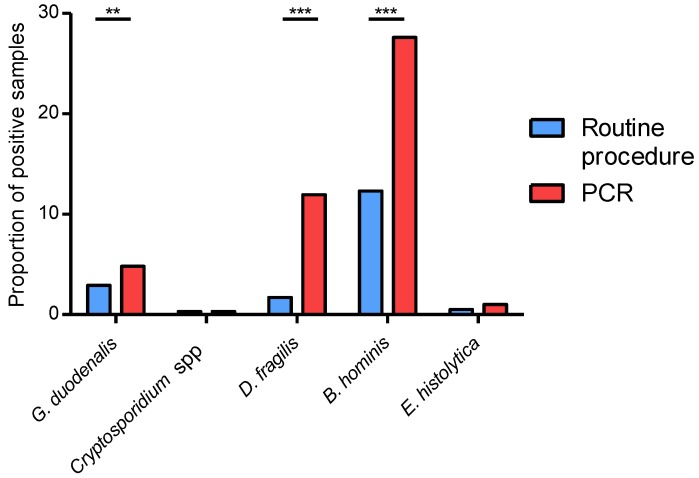
Among the samples deemed negative by microscopy, several were positive with the AllplexTM assay greatly increasing the proportion of positive samples compared to routine procedure (Figure 1). This increase was significant for G. duodenalis (4.8% vs. 2.9%, p < 0.01), D. fragilis (11.9% vs. 1.7%, p < 0.001), and B. hominis (27.6% vs. 12.2%, p < 0.001). The difference was not statistically significant for E. histolytica detection (1.0% vs. 0.5%), due to the low number of positive samples, but it should be noticed that PCR assay detected two times as many positive samples than microscopy. No additional Cryptosporidium sp. was detected with the AllplexTM assay. Finally, the sensitivity was also calculated by combining results of both microscopy and the AllplexTM assay to define positive samples (Table 3). Of note, PCR had a higher sensitivity than microscopy for most pathogens, especially D. fragilis (97.2% vs. 13.8%, p < 0.001) and B. hominis (99.4% vs. 44.2%, p < 0.001), but also G. duodenalis (100% vs. 60.7%, p < 0.01) and E. histolytica (100% vs. 50.0%).
Figure 1.
Proportion of positive samples on the prospective cohort (n = 586), using routine microscopy and multiplex PCR. **: p < 0.01; ***: p < 0.001.
Table 3.
Overall sensitivity of the AllplexTM PCR assay and the routine procedure on the prospective cohort (n = 586).
| Sensitivity | |||
|---|---|---|---|
| By Routine Procedure % (n/N) |
By PCR % (n/N) |
Statistical Significance 1 | |
| Giardia duodenalis | 60.7% (17/28) | 100% (28/28) | ** |
| Cryptosporidium spp. | 100% (2/2) | 100% (2/2) | ns |
| Dientamoeba fragilis | 13.8% (10/72) | 97.2% (70/72) | *** |
| Blastocystis hominis | 44.2% (72/163) | 99.4% (162/163) | *** |
| Cyclospora cayetanensis | na 2 | na 2 | na 2 |
| Entamoeba histolytica | 50% (3/6) | 100% (6/6) | ns |
1 **: p < 0.01; ***: p < 0.001; ns: not significant; 2 no C. cayetanensis was diagnosed during the study.
3.3. Does the AllplexTM GIPPA Need Repeated Samples?
In order to evaluate if clinical laboratories could override the dogma of the multiple stool sampling, the sensitivity of the PCR assay on the first sample was calculated and compared to the sensitivity of the routine procedure on at least three consecutive samples. Only 74 patients had three or more repeated stool samples, and among them 32 had at least one stool positive for B. hominis, 13 for D. fragilis, 4 for G. duodenalis, and none for E. histolytica or Cryptosporidium sp. The sensitivities of the microscopy on consecutive samples were 78% (25/32), 15% (2/13), and 75% (3/4) for B. hominis, D. fragilis, and G. duodenalis, respectively (Table 4). Sensitivities were either equal or higher with the AllplexTM assay on the first stool, reaching 94% (30/32; p < 0.05), 92% (12/13; p < 0.01), and 75% (3/4; not significant) for B. hominis, D. fragilis and G. duodenalis respectively.
Table 4.
Sensitivity of the AllplexTM PCR assay on the first patient sample, compared to the routine procedure on multiple consecutive samples (n = 74).
| Sensitivity | |||
|---|---|---|---|
| By Routine Procedure on Multiple Consecutive Samples % (n/N) |
By PCR on First Sample % (n/N) |
Statistical Significance 1 | |
| Giardia duodenalis | 75% (3/4) | 75% (3/4) | ns |
| Dientamoeba fragilis | 15% (2/13) | 92% (12/13) | ** |
| Blastocystis hominis | 78% (25/32) | 94% (30/32) | * |
1 *: p < 0.05; **: p < 0.01; ns: not significant.
3.4. DNA Preservation in Cary-Blair Medium
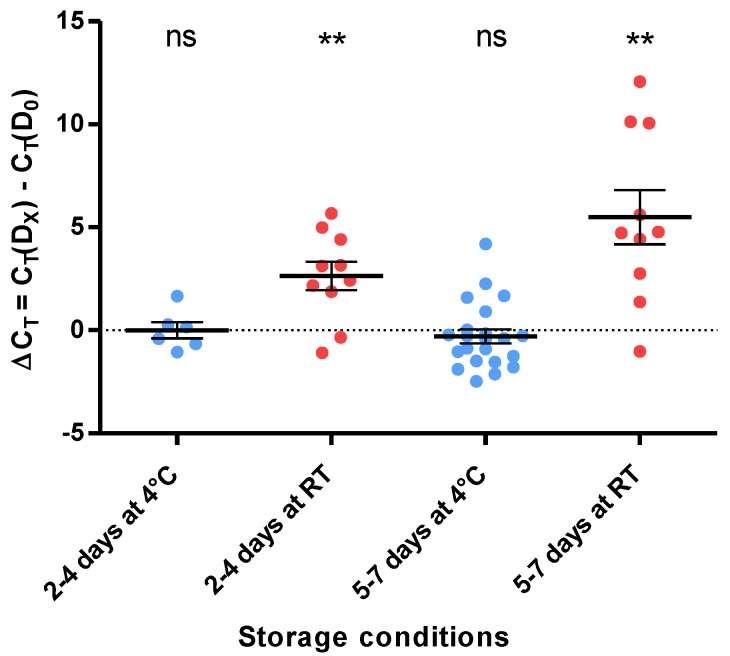
To assess whether the analysis of grouped samples is possible, the impact of different storage conditions of the FecalSwabTM stool suspension on the signal intensity was evaluated. As detailed above, the differences in signal values between the first analysis and after the storage period were calculated (ΔCT = CT(DX) − CT(D0)) for different times (2–4 days vs. 5–7 days) and different temperatures (4 °C vs. room temperature). After a storage at 4 °C, the medians of ΔCT were −0.125 and −0.405 for the “2–4 Days” and “5–7 Days” groups, respectively, and were not significantly different from 0 (Figure 2). After storage at room temperature, the medians of ΔCT were 2.780 and 4.750 for the “2–4 Days” and “5–7 Days” groups, respectively, and were in both cases significantly different from 0 by the Wilcoxon signed-rank test (p < 0.01).
Figure 2.
Impact of the storage conditions on the detection signal. Representation of the signal variation in the function of the FecalSwabTM storage conditions. **: p < 0.01; ns: not significant; RT: room temperature.
4. Discussion
In the retrospective study, while the sensitivity was excellent for Cryptosporidium spp., C. cayetanensis, and B. hominis, some false negative results were observed for G. duodenalis and D. fragilis. All Cryptosporidium species tested were positive, which is an important point, as many species can infect humans [8]. During the prospective study, sensitivity was excellent for all species, and only rare false negative results were observed for B. hominis and D. fragilis. No false negative results were observed for G. duodenalis in the prospective study. Performances of the AllplexTM GIPPA assay performed on the first stool were equivalent to that of microscopy on multiple consecutive stools, but repeating PCR on consecutive samples yielded even higher sensitivities. Finally, this study assessed that PCR performances were not affected by the storage of the FecalSwabTM stool suspension at 4 °C until 7 days.
The multiplex PCR assay AllplexTM GIPPA showed excellent performances for protozoa detection, even higher than that of other marketed PCR assays such as the BD MaxTM Enteric Parasite Panel, G-DiaParaTM, or ParaGENIE G-Amoeba assays [9,10,11,12] and with additional targets. Indeed, the sensitivity of these assays ranged from 41% to 96% for G. duodenalis [9,10,11], depending on the extraction method and the assay, and the ParaGENIE G-Amoeba assay had 67% sensitivity for E. histolytica detection [12]. Moreover, the BD MaxTM and G-DiaParaTM assays detected only C. parvum/hominis, and none of these detected C. cayetanensis, D. fragilis, or B. hominis. Few studies evaluated the performances of the AllplexTM GIPPA. Among them, one included a unique stool positive for parasites (Cryptosporidium spp.) which was detected by the assay [13]. Another study, with a more consistent cohort, observed performances slightly lower to ours, with 92%, 100%, and 78% sensitivity for G. duodenalis, E. histolytica \, and Cryptosporidium spp., respectively [14]. However, this study was conducted retrospectively, on DNA which had not been extracted with the recommended Hamilton device (MICROLAB® STARlet or NIMBUS).
Regarding the G. duodenalis detection, sensitivity was only 81% during the retrospective study, but it should be noticed that the analysis was not performed on fresh stools but on frozen samples without preservative, and this could possibly explain some false negative results [15]. Moreover, the undetected samples contained very low parasite loads and underwent long-time freezing, which could have led to DNA degradation. Interestingly, during the prospective study, PCR showed higher sensitivity than microscopy, which supports this hypothesis. For D. fragilis and B. hominis few false negative results occurred in retrospectively and prospectively analyzed samples. This appears as surprising, as during the prospective study, the AllplexTM GIPPA assay was significantly more sensitive than the microscopy for these parasites. As an explanation, it could be hypothesized that (i) DNA could have been degraded in retrospectively analyzed samples, as per the G. duodenalis example, and (ii) some positive results by microscopy could be a “false positive”. It should indeed be remembered that the B. hominis and D. fragilis morphological diagnosis is challenging, even for experienced operators, especially in France, where permanent staining (as Wheatley’s trichrome or iron hematoxylin) are not current practice. In the case at hand, almost all but one of the samples that were positive only by microscopy for B. hominis and D. fragilis contained numerous leukocytes, arthroconidias, or other protozoa which could have led to misdiagnosis.
The numerous samples that were negative by microscopy and positive by PCR raised the question of the specificity of the assay. For E. histolytica detection, specificity has been ensured by other molecular assays and clinical data: the positive samples were collected from patients with amoebic abscess or with E. histolytica observed in another stool sample. A great number of samples were positive for B. hominis only by PCR. These were not verified by other molecular assays but are most likely a true positive of the PCR. Indeed, most of them (71%, 65/91) were confirmed on another stool sample from the same patient, 34% by microscopy (31/91) and 37% by PCR only (34/91). Besides, the relative sensitivity of microscopy was 44%, which is in line with the 47% sensitivity observed in previous works [16]. Likewise, 84% (52/62) of the samples positive by PCR only for Dientamoeba fragilis were confirmed on another sample from the patient (16% by microscopy (10/62) and 68% by PCR only (42/62)). In this study, microscopy showed dramatically poor results for D. fragilis detection (14% sensitivity). This is in line with data from the literature and explains why PCR is now considered as the reference method for D. fragilis diagnosis. Additionally, the detection of D. fragilis trophozoites relied only on the direct wet mount of non-fixed stool samples, which is known to be of poor value [4] and it has to be underlined that there is no cyst for this protozoa. Finally, for G. duodenalis detection, 82% (9/11) of the samples positive only by PCR were confirmed on another stool sample. Moreover, the two remaining samples were clinically evocative of giardiasis.
The major limit of this study was the poor number of C. cayetanensis tested, in line with the low prevalence of this intestinal parasite in France. However, as the oocyst wall of C. cayetanensis is similar to that of Cryptosporidium spp. [17], the DNA extraction performance should be hypothetically equivalent for both parasites. Further studies are necessary to confirm this. Another limit is that all human-infecting Cryptosporidium species could not be tested. Nevertheless, we assessed the most encountered species in human pathology. In France, the six species tested were shown to be involved in 98% of human cases: 54% were due to C. parvum, 37% to C. hominis, 5% to C. felis, 1% to C. meleagridis, and 1% to C. canis [18].
5. Conclusions
To our knowledge, this study is the first evaluation of the AllplexTM GIPPA for protozoa detection, performed on both an exhaustive retrospective cohort and an important prospective cohort. First, this assay combines several advantages, i.e., the ease of use (almost fully automated process), a high number of protozoa detected, and excellent sensitivity results. The use of such a technique could improve routine diagnosis of protozoan infections by clinical laboratories, while being far easier to implement, compared to microscopy. However, a limitation of the technique is that helminths and few other protozoa are not targeted. Finally, we assessed that stool suspension in the Cary-Blair medium was stable until 7 days when stored at +4 °C, allowing the analysis of grouped samples, which is more convenient for most clinical laboratories.
Acknowledgments
The authors gratefully thank Loïc Favennec, Romy Razakandrainibe, and the French National Reference Centre for Cryptosporidiosis (University Hospital of Rouen, France) for the species identification of Cryptosporidium-positive stools and the Seegene Company for providing the reagents for the retrospective evaluation.
Author Contributions
Conceptualization, F.R.-G.; methodology, F.R.-G.; formal analysis, B.A.; investigation, B.A.; resources, J.-P.G.; writing—original draft preparation, B.A.; writing—review and editing, F.R.-G. and J.-P.G.; visualization, B.A.; supervision, F.R.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Troeger C., Forouzanfar M., Rao P.C., Khalil I., Brown A., Reiner R.C., Fullman N., Thompson R.L., Abajobir A., Ahmed M., et al. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017;17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abubakar I.I., Tillmann T., Banerjee A. GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan U., Hijjawi N., Xiao L. Foodborne cryptosporidiosis. Int. J. Parasitol. 2018;48:1–12. doi: 10.1016/j.ijpara.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Stark D., Barratt J., Chan D., Ellis J.T. Dientamoeba fragilis, the neglected trichomonad of the human bowel. Clin. Microbiol. Rev. 2016;29:553–580. doi: 10.1128/CMR.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams D.A., Thomas K.R., Jajosky R.A., Foster L., Baroi G., Sharp P., Onweh D.H., Schley A.W., Anderson W.J., Nationally Notifiable Infectious Conditions Group Summary of notifiable infectious diseases and conditions—United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 2017;64:1–143. doi: 10.15585/mmwr.mm6453a1. [DOI] [PubMed] [Google Scholar]
- 6.Verweij J.J. Application of PCR-based methods for diagnosis of intestinal parasitic infections in the clinical laboratory. Parasitology. 2014;141:1863–1872. doi: 10.1017/S0031182014000419. [DOI] [PubMed] [Google Scholar]
- 7.Madison-Antenucci S., Relich R.F., Doyle L., Espina N., Fuller D., Karchmer T., Lainesse A., Mortensen J.E., Pancholi P., Veros W., et al. Multicenter evaluation of BD Max Enteric Parasite real-time PCR assay for detection of Giardia duodenalis, Cryptosporidium hominis, Cryptosporidium parvum, and Entamoeba histolytica. J. Clin. Microbiol. 2016;54:2681–2688. doi: 10.1128/JCM.00765-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zahedi A., Paparini A., Jian F., Robertson I., Ryan U. Public health significance of zoonotic Cryptosporidium species in wildlife: Critical insights into better drinking water management. Int. J. Parasitol. Parasites Wildl. 2015;5:88–109. doi: 10.1016/j.ijppaw.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mölling P., Nilsson P., Ennefors T., Ögren J., Florén K., Thulin Hedberg S., Sundqvist M. Evaluation of the BD Max Enteric Parasite panel for clinical diagnostics. J. Clin. Microbiol. 2016;54:443–444. doi: 10.1128/JCM.02100-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laude A., Valot S., Desoubeaux G., Argy N., Nourrisson C., Pomares C., Machouart M., Le Govic Y., Dalle F., Botterel F., et al. Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review. Clin. Microbiol. Infect. 2016;22:e1–e190. doi: 10.1016/j.cmi.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Autier B., Belaz S., Razakandrainibe R., Gangneux J.-P., Robert-Gangneux F. Comparison of three commercial multiplex PCR assays for the diagnosis of intestinal protozoa. Parasite. 2018;25:48. doi: 10.1051/parasite/2018049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morio F., Valot S., Laude A., Desoubeaux G., Argy N., Nourrisson C., Pomares C., Machouart M., Le Govic Y., Dalle F., et al. Evaluation of a new multiplex PCR assay (ParaGENIE G-Amoeba Real-Time PCR kit) targeting Giardia intestinalis, Entamoeba histolytica and Entamoeba dispar/moshkovskii from stool specimens: Evidence for the limited performances of microscopy-based approach for amoeba species identification. Clin. Microbiol. Infect. 2018;24:1205–1209. doi: 10.1016/j.cmi.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Yoo J., Park J., Lee H.K., Yu J.K., Lee G.D., Park K.G., Oak H.C., Park Y.J. Comparative evaluation of Seegene Allplex Gastrointestinal, Luminex xTAG Gastrointestinal Pathogen Panel, and BD MAX Enteric assays for detection of gastrointestinal pathogens in clinical stool specimens. Arch. Pathol. Lab. Med. 2019;143:999–1005. doi: 10.5858/arpa.2018-0002-OA. [DOI] [PubMed] [Google Scholar]
- 14.Paulos P., Saugar J.M., de Lucio A., Fuentes I., Mateo M., Carmena D. Comparative performance evaluation of four commercial multiplex real-time PCR assays for the detection of the diarrhoea-causing protozoa Cryptosporidium hominis/parvum, Giardia duodenalis and Entamoeba histolytica. PLoS ONE. 2019;14:e0215068. doi: 10.1371/journal.pone.0215068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuk S., Yazar S., Cetinkaya U. Stool sample storage conditions for the preservation of Giardia intestinalis DNA. Mem. Inst. Oswaldo Cruz. 2012;107:965–968. doi: 10.1590/S0074-02762012000800001. [DOI] [PubMed] [Google Scholar]
- 16.Roberts T., Barratt J., Harkness J., Ellis J., Stark D. Comparison of microscopy, culture, and conventional polymerase chain reaction for detection of Blastocystis sp. in clinical stool samples. Am. J. Trop. Med. Hyg. 2011;84:308–312. doi: 10.4269/ajtmh.2011.10-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belli S.I., Smith N.C., Ferguson D.J.P. The coccidian oocyst: A tough nut to crack! Trends Parasitol. 2006;22:416–423. doi: 10.1016/j.pt.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 18.The ANOFEL Cryptosporidium National Network Laboratory-based surveillance for Cryptosporidium in France, 2006–2009. Euro Surveill. 2010;15:19642. [PubMed] [Google Scholar]