Abstract
Certain species of the genus Bifidobacterium represent human symbionts. Many studies have shown that the establishment of symbiosis with such bifidobacterial species confers various beneficial effects on human health. Among the more than ten (sub)species of human gut-associated Bifidobacterium that have significantly varied genetic characteristics at the species level, Bifidobacterium bifidum is unique in that it is found in the intestines of a wide age group, ranging from infants to adults. This species is likely to have adapted to efficiently degrade host-derived carbohydrate chains, such as human milk oligosaccharides (HMOs) and mucin O-glycans, which enabled the longitudinal colonization of intestines. The ability of this species to assimilate various host glycans can be attributed to the possession of an adequate set of extracellular glycoside hydrolases (GHs). Importantly, the polypeptides of those glycosidases frequently contain carbohydrate-binding modules (CBMs) with deduced affinities to the target glycans, which is also a distinct characteristic of this species among members of human gut-associated bifidobacteria. This review firstly describes the prevalence and distribution of B. bifidum in the human gut and then explains the enzymatic machinery that B. bifidum has developed for host glycan degradation by referring to the functions of GHs and CBMs. Finally, we show the data of co-culture experiments using host-derived glycans as carbon sources, which underpin the interesting altruistic behavior of this species as a cross-feeder.
Keywords: Bifidobacterium bifidum, human milk oligosaccharide, mucin O-glycan, glycoside hydrolase, carbohydrate-binding module, cross-feeding
1. Introduction
Bifidobacteria are Gram-positive obligate anaerobes that represent common inhabitants of the gastrointestinal tract of animals [1]. In humans, it is well known that a bifidobacteria-rich microbiota (so-called, bifidus-flora) is formed in the intestinal tracts of breast-fed infants. These “first colonizers” are believed to confer various beneficial effects on human health, including protection against pathogen infection [2,3], nutritional supplementation [4], reduced inflammation [5,6], and the development of the immune system [7]. Notably, a recent research revealed that bifidus-flora formation is associated with the increased fecal concentration of indole lactic acid, a ligand of arylhydrocarbon receptor (AhR) [8]. In the Bifidobacterium genus, four species, i.e., Bifidobacterium bifidum, Bifidobacterium breve, Bifidobacterium longum ssp. infantis (B. infantis), Bifidobacterium longum ssp. longum (B. longum), are frequently detected in the stool of breast-fed infants. Many studies [9,10,11,12,13,14,15,16,17,18,19] have indicated that the formation of a bifidus-flora in the gut of breast-fed infants can be attributed to human milk oligosaccharides (HMOs), which are the third most abundant solid component in breastmilk after lactose and lipids [20,21,22]. Mothers produce the energy-rich HMOs, even though HMOs have no direct nutritional value for infants, as HMOs are resistant to digestive enzymes secreted in the gastrointestinal tract. Recent studies have unequivocally revealed that HMOs are utilized by infant gut-associated bifidobacteria to proliferate in their specific ecosystem [23,24]. Interestingly, the pathways for HMO assimilation differ among the Bifidobacterium species and even among strains of the same species, suggesting strong selective pressure and adaptive evolution under symbioses with individuals with different gut environments [11,13]. After weaning, the bifidobacterial population gradually decreases and its dominance is replaced with other microbes that are adapted to plant-derived glycan utilization, such as Bacteroides and Clostridium, to form an adult-type microbiota. Nonetheless, in many cases, Bifidobacterium continues to be a member of the human gut microbiome (described later).
Among Bifidobacterium species, B. bifidum is quite unique in that this species possesses many extracellular glycosidases specified for degrading host-derived glycans, including HMOs, the sugar chains of high molecular-weight glycoproteins, and glycosphingolipids [11,25,26,27]. B. bifidum liberates mono- and disaccharides from host glycans and releases them into the living environmental niches or culture medium in vitro. By virtue of these characteristics, B. bifidum can support the growth of other bifidobacteria by sharing the glycan degradation products within the community [28]. In this review, when compared with other human gut-associated Bifidobacterium species, we reveal the unique characteristics of B. bifidum that have adaptively evolved for host glycan utilization.
2. Prevalence and Dominance of B. bifidum in the Gut
The abundance of the genus Bifidobacterium in human fecal samples is reported to vary by country [29,30] or dwelling environment (urban industrialized area vs. remote/rural area) [31]. The factors that affect the prevalence and dominance of bifidobacteria include not only diet, birth delivery mode, and living environment but also include host genetic statuses. For example, studies have reported that secretor status or lactose intolerance (i.e., FUT2 [32] or LCT [33]) is associated with the intestinal bifidobacterial population. However, it should be mentioned that the variability of the bifidobacterial abundance across the studies can be caused by varied sample processing and/or primer design in 16S metagenomic analyses [34].
B. bifidum is a common colonizer of the gut of mammals, including humans [35,36,37]. This species is, in general, detected in healthy individuals with a high inter-subject variability. Matsuki et al. [38] reported that B. bifidum was detected in 38% and 22% of adult and infant subjects, respectively. In a report examining its dominance in the intestinal samples of Finnish adults, B. bifidum was found to be the third most abundant, after B. longum and Bifidobacterium adolescentis [39]. Guglielmetti et al. [40] quantified the cell numbers of Bifidobacterium species in fecal samples from healthy male volunteers in Italy and demonstrated that B. bifidum was less represented than B. longum and B. adolescentis by approximately two orders of magnitude. Turroni et al. [35] have examined the diversity of bifidobacterial populations in mucosal and fecal samples of the human intestinal tract by culture-based analysis, and isolated B. bifidum from both mucosal and fecal samples, suggesting that this species has a mucosa-adherent property. This mucosa-adherent property is conferred by the presence of sortase-dependent pili, which may be involved in the attachment to the extracellular matrix components [41], and also by glycan-binding properties of extracellular glycoside hydrolases (described below) [42].
The age-related compositional changes of human gut bifidobacterial species have been investigated [43,44,45]. Kato et al. [44] have examined the prevalence of 11 different Bifidobacterium species/subspecies in the stools of 441 healthy Japanese subjects across a wide age range (0 to 104 years) by quantitative-PCR. With the exception of centenarians, B. bifidum was detected at all ages, but their prevalence was lower (28.3%) than that of the B. longum group (B. longum and B. infantis) (88.1%). A similar pattern was observed for the Bifidobacterium catenulatum group. Nagpal et al. [45] have examined the early-life dynamics of the bifidobacterial population in 76 full-term vaginally-born Japanese infants. They showed that B. bifidum became prevalent (~60%) six-months after birth and became less prevalent (~20%) after 3 years. B. breve was detected in 71.4% of children under 3 years old and its prevalence was consistently high in individuals younger than 10, but the number of B. breve cells decreased with age and was very scarce past the age of 50. The prevalence of B. breve also decreased during the transition from childhood to adulthood.
Collectively, B. breve and B. longum are, in general, more abundantly detected than B. bifidum during the first three years of life. The prevalence of B. breve is, however, largely limited to infants. B. longum is widely distributed across age groups and frequently found in adult fecal samples. B. bifidum is also widely distributed across age groups, but with a lesser prevalence than the B. longum group. B. adolescentis and the B. catenulatum group are also abundant in adult intestinal microbiota. This transition of bifidobacterial species in human intestinal microbiota with age is primarily due to changes in diet and possibly host glycans (described below). Considering the age-related compositional changes and nutritional adaptation, Bifidobacterium species can be classified into at least four groups: the HMO-dedicated group (B. infantis), the dietary fiber-adapted group (B. adolescentis, B. catenulatum group, and other adult-type species except B. longum), the both HMO and dietary fiber-adapted group (B. breve and B. longum), and the both HMO and mucin-adapted group (B. bifidum). Thus, as the sole known mucin-degrader within the genus Bifidobacterium, B. bifidum is uniquely positioned in the context of carbohydrate utilization strategies. This may be the reason it is detected in a wide range of age groups, except for centenarians. It is known that intestinal mucin production becomes low in elderly adults [46]. Therefore, the nutritional switch, as well as host glycan availability, may have an impact on bifidobacterial prevalence and abundance.
3. Structures of Human Milk Oligosaccharides and Mucin O-Glycans
“HMOs” is a collective term for the mixture of more than 100 structurally different free oligosaccharides (degree of polymerization ≥ 3) produced in the mammary gland [20,47]. The synthesis of HMOs begins with the modification or the elongation of lactose (Galβ1-4Glc). Fucosylation occurs at the C2 position of Gal or the C3 position of Glc of lactose by the activity of fucosyltransferases (FUTs) to form 2′-fucosyllactose (2′-FL) and 3-fucosyllactose (3-FL), respectively. Sialylation with N-acetylneuraminic acid occurs at the C3 or C6 position of Gal of lactose to form 3′/6′-sialyllactose (SL). The elongation or branching of lactose is initiated with the addition of N-acetylglucosamine (GlcNAc) onto the C3 position of Gal of lactose to form lacto-N-triose II, then subsequent addition of Gal onto the C3 position or the C4 position of GlcNAc occurs to form the type-1 chain unit (Galβ1-3GlcNAc or lacto-N-biose I, LNB)-containing lacto-N-tetraose (LNT, Galβ1-3GlcNAcβ1-3Galβ1-4Glc), or the type-2 chain LacNAc unit (Galβ1-4GlcNAc)-containing lacto-N-neotetraose (LNnT, Galβ1-4GlcNAcβ1-3Galβ1-4Glc). LNT and LNnT can then be subjected to further modification and/or elongation. GlcNAc branching also occurs at the C6 position of the Gal residue of Lac after the synthesis of lacto-N-triose II, and the branch is further elongated by the addition of Gal in a similar manner. When compared to the milk oligosaccharides of other mammals, HMOs are characterized by the richness of type-1 disaccharide units [20]. It should be noted that the genetic background of mothers significantly influences the structural variety of HMOs. For example, non-secretor FUT2-deficient mothers (Se-negative, 0% to 50%, depending upon ethnic background) do not produce H-antigen (Fucα1-2Gal-OR)-containing oligosaccharides such as 2′-FL or lacto-N-fucopentaose (LNFP) I in their milk [16], and FUT3-deficient mothers with the Lewis negative phenotype produce HMOs with differential amounts of Lewis antigen structures [48].
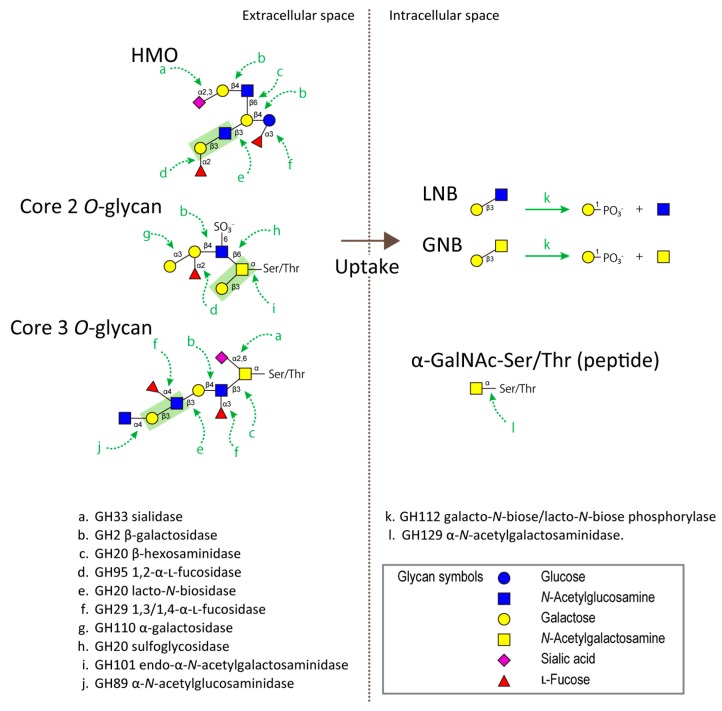
While HMOs are present as a free form in the aqueous phase of milk, mucin O-glycans are covalently attached to the polypeptide backbone through N-acetylgalactosaminyl Ser/Thr residues. O-Glycosylation on mucin glycoproteins densely occurs in regions that are rich in the amino acid residues of Pro/Thr/Ser (mucin domain) [49], and the resulting overall molecular structures of mucin glycoproteins are thought to be shaped like bottle brushes. O-Glycans are synthesized by sequential addition of monosaccharides by the orchestrated action of glycosyltransferases in mucin-producing cells, i.e., goblet cells, and they are classified into several groups based on their “core” types (primarily core 1, core 2, core 3, and core 4, as reviewed by [50,51,52]). Therefore, the structure of mucin O-glycans is highly heterogeneous and it is also dependent on which mucin molecules they are attached to [53,54]; however, they still share a structural similarity in part with HMOs. O-Glycans contain many type-1/2 LacNAc units as terminal and internal structures, and fucosylation/sialylation as terminal modifications (Figure 1). The structural similarities between HMOs and mucin O-glycans, which may reflect their overlapping function as a defense from pathogens by acting as anti-adhesive decoys [55], can also serve as prime targets for decomposition by glycan-degrading enzymes of B. bifidum to assimilate sugars.
Figure 1.
Enzymes involved in the degradation and assimilation of human milk oligosaccharides (HMOs) and mucin O-glycans by B. bifidum. The sugars are depicted according to the nomenclature committee of the Consortium for Functional Glycomics (http://www.functionalglycomics.org/static/index.shtml).
4. Glycoside Hydrolases Involved in the Degradation of HMOs/Mucin O-Glycans
Biochemical studies of glycoside hydrolases (GHs) from B. bifidum have revealed the mechanism of adaptation to HMOs and mucin O-glycans. The reports on a GH95 1,2-α-l-fucosidase (AfcA) from B. bifidum JCM 1254 by Katayama et al. [56] and a galactose operon containing a gene encoding a GH112 galacto-N-biose (GNB)/LNB phosphorylase from B. longum JCM 1217 by Kitaoka et al. [57] were the key findings that revived attention on HMOs by linking them to bifidobacterial biology, long after the first notion, proposed in 1954 by Gauhe et al. [58], that milk oligosaccharides serve as a bifidus factor. Since then, using the strain B. bifidum JCM 1254, extensive studies have been carried out and the gene products relevant to the HMO/mucin utilization pathways have been successfully isolated and characterized [26,27,59,60,61,62,63,64,65,66]. These efforts have illustrated how B. bifidum extracellularly degrades HMOs and mucin O-glycans into smaller sugars by the concerted action of the many cell surface-anchored GHs summarized in Figure 1. Terminal modifications such as fucosylation and sialylation can be removed by either a GH95 1,2-α-l-fucosidase (AfcA) [56,67], a GH29 1,3-1,4-α-l-fucosidase (AfcB) [62], or a GH33 sialidases (SiaBb1 [65,68] and SiaBb2 [42,60]) to expose the internal core structures. The internal type-1/2 LacNAc units are removed by GH20 lacto-N-biosidase (LnbB) to liberate LNB [66] and by a sequential digestion with GH2 β-galactosidase, BbgIII [63], and GH20 β-N-acetylhexosaminidase, BbhI, to liberate Gal and GlcNAc [63], respectively. Furthermore, B. bifidum possesses specific GHs for degrading various glycan epitopes that are frequently found on mucin O-glycans. GH89 α-N-acetylglucosaminidase acts on terminal α-linked GlcNAc linkages attached to gastric mucin O-glycans [59], while GH110 α-galactosidase acts on the blood group antigen B to liberate Gal [64]. Interestingly, B. bifidum does not have a gene encoding GH109 α-N-acetylgalactosaminidase that acts on the blood group antigen A [69]. A recent report [70] has shown that gut microbial enzymes convert A-antigens via a novel mechanism to H-antigens through a galactosamine intermediate involving a deacetylase and a GH36 galactosaminidase. Although B. bifidum has one GH36 gene, it likely encodes for raffinose synthase.
Sialic acid residues are frequently modified with O-acetylation [71]. One of the GH33 sialidases, SiaBb1, contains an esterase-domain in its polypeptide sequence and was shown to remove the acetyl group from 9-O-acetylated N-acetylneuraminic acids [65]. Additionally, sulfated Gal and GlcNAc residues are present on intestinal mucin O-glycans [72]. We have recently identified a GH20 enzyme, BbhII, as a sulfoglycosidase that can remove terminal 6-sulfated GlcNAc residues from mucin [61].
Intracellular GH112 GNB/LNB phosphorylase acts on both GNB and LNB, converting them into galactose-1-phosphate and N-acetylhexosamine (GalNAc and GlcNAc, respectively) [57,73]. GNB is released from mucin core 1 by the action of extracellular GH101 endo-α-N-acetylgalactosaminidase (EngBF) [74], and LNB is formed not only from type-1 HMOs but also from mucin O-glycans by the action of LnbB. GNB and LNB are known to be imported by the GNB/LNB transporter, an ATP-binding cassette-type (ABC) transporter [75,76]. GH129 α-N-acetylgalactosaminidase (NagBb) is able to act on α-N-acetylgalactosaminyl Ser (Tn antigen) to liberate GalNAc and Ser [26,77]. GalNAc-Ser/Thr can be generated from core 3 or core 4 mucin O-glycans after complete trimming by concerted actions of GHs and peptidases. NagBb does not have a signal peptide, indicating that the hydrolysis of GalNAc-Ser/Thr takes place in the cytosol after the uptake of GalNAc-Ser/Thr into the cell by an unknown transporter.
5. Enzymatic Adaptation to HMO- and Mucin O-Glycan Degradation from the View of the “Spectra” of GHs and CBMs
The complete genomic sequence of B. bifidum was determined for the strain JCM 1255 by Hattori’s group [78] and for the strain PRL2010 by Ventura’s group [25], and, currently, a total of eleven sequences are available in public databases (NCBI genome assembly database, as of October 2019). A comparison of the annotated GH genes with those of other human-associated Bifidobacterium highlights its distinct mechanism that is exclusive to the assimilation of HMOs/mucin O-glycans. At the genus level, the total number of GHs ranges from 40 to 60 in the genomes of Bifidobacterium; however, the “GH spectrum”, or the distribution of GH families and the cellular localization of those GHs, is species-dependent and should reflect its carbohydrate preference and distinct assimilation mechanisms. As visualized in Figure 2, B. bifidum [strains PRL2010, LMG 13195 (= JCM 7004), and TMC 3115] has 14 extracellular GHs involved in HMO/mucin O-glycan degradation, as predicted in silico, whereas B. infantis ATCC 15697 (= JCM 1222) and B. breve UCC2003 are equipped with the intracellular counterpart enzymes (e.g., GH33 sialidase and GH95 1,2-α-l-fucosidase). Indeed, B. infantis ATCC 15697 (= JCM 1222) has been shown to assimilate HMOs but not mucin that was supplemented in vitro [25]. Furthermore, it is worth noting that although some GHs, including GH33 sialidases, GH2 β-galactosidase, and GH20 β-N-acetylhexosaminidase, are also able to act on complex N-glycans, B. bifidum does not contain N-glycan-specific enzymes such as GH38 α-mannosidase or GH18/GH85 endo-β-N-acetylglucosaminidase. Among infant-gut associated Bifidobacterium, B. breve and B. infantis possess such N-glycan-specific GHs and are reportedly able to grow on media containing N-glycosylated proteins [79]. B. bifidum, B. breve, and B. infantis are all adapted to HMO utilization. However, in terms of the degradation of glycan chains attached to glycoproteins, B. breve and B. infantis are specific to N-glycans, while B. bifidum prefers O-glycans to perhaps avoid competition with other Bifidobacterium species.
Figure 2.
Distribution of glycoside hydrolases (GHs) and their cellular localization in the selected Bifidobacterium species/strains. The circles represent the occurrence of enzymes (domains) classified in the respective GH families in the CAZy database (http://www.cazy.org). The extracellular and intracellular enzymes are shown by solid and open circles, respectively. Protein localization was predicted by signalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/) and TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The pseudogenes that encode incomplete protein sequences are excluded from the analysis. The GHs that are (possibly) involved in the degradation of HMOs and mucin O-glycans are colored in red.
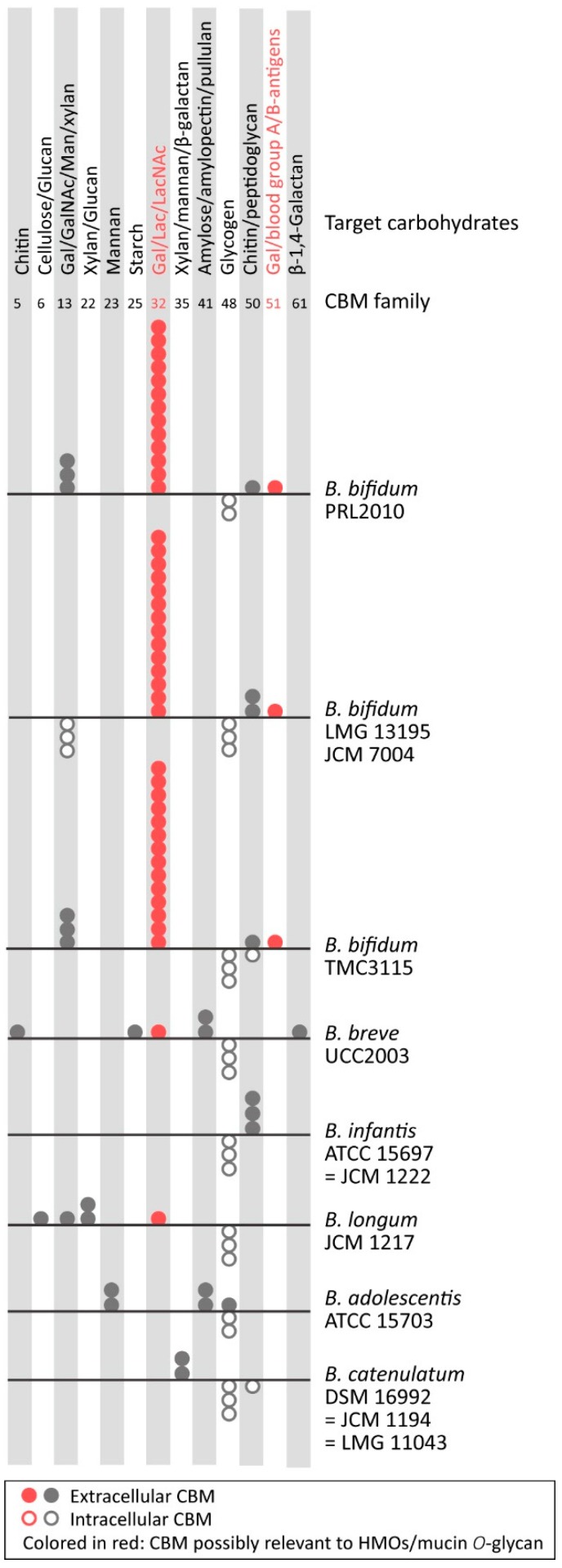
Interestingly, the number of carbohydrate-binding modules (CBMs) in B. bifidum is also higher than that of other species: 20–23 in B. bifidum vs. six in B. infantis ATCC 15697, eight in B. longum JCM 1217, and nine in B. breve UCC2003 (Figure 3). In general, the CBMs are present in tandem with the peptide chains of the GH domains and serve to enhance the catalytic activities of GHs by increasing their affinity toward the polymeric carbohydrates [80]. In particular, B. bifidum has 13 CBM32 domains that are specifically associated with the recognition of Gal/Lac/LacNAc and their derivatives [81], possibly to enhance catalytic activities for mucin O-glycans. The quantity of CBMs is outstanding within the genus Bifidobacterium. This could represent the evolutionary adaptation toward mucins and the basis for carbohydrate-preference of B. bifidum. Interestingly, Nishiyama et al. [42] found that SiaBb2 from B. bifidum can interact with α2,6-linked sialic acid and the blood group A trisaccharide moieties by the GH33 sialidase domain, not the CBM domain, suggesting that there are more unidentified protein sequences involved in the interaction with glycans.
Figure 3.
Distribution of carbohydrate-binding modules (CBMs) and their cellular localization in the selected Bifidobacterium species/strains. The circles represent the occurrence of CBMs (domains) classified in the CAZy database (http://www.cazy.org). The extracellular and intracellular CBMs are shown by solid and open circles, respectively. The localization of proteins including the CBMs was predicted by signalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/) and TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The pseudogenes that encode incomplete protein sequences are excluded from the analysis. The CBMs possibly associated with HMO- and mucin O-glycan degradation are colored in red.
These HMO/mucin-related GHs are distributed separately within the B. bifidum genomes, rather than being clustered [25]. However, HMO/mucin supplementation induces the expression of some of these genes [25,61,82]. Although the mechanism of the transcriptional regulation remains unclear, a conserved 24 bp sequence is found upstream of these genes, which suggests the presence of a common transcriptional regulator(s) with a shared ligand(s), presumably a sugar metabolite(s) [25].
A repertoire of the transporters may also be an important factor that dictates the carbohydrate availability for bacteria. Although the GNB/LNB [75,76], fucosyllactose [12,24,83,84], and LNnT transporters [85] have been identified in Bifidobacterium, our knowledge regarding HMO/mucin-related transporters remains limited. However, Turroni et al. [86] reported that the genome of B. bifidum PRL2010 contains 25 genes predicted to encode components of a transport system dedicated to carbohydrate uptake. These are classified into different groups: eight genes of ABC-type transporters, 12 genes for a component of the phosphoenolpyruvate-phosphotransferase systems, one major intrinsic protein, and four secondary carriers. The number of genes predicted to encode the proteins for carbohydrate uptake in B. bifidum PRL2010 is relatively small when compared with the numbers of such genes in other species of human-associated Bifidobacterium. For example, B. infantis ATCC 15697 has 68 genes predicted to encode proteins for carbohydrate uptake, including 20 of the solute-binding protein family 1 of the ABC transporter [9], while B. bifidum PRL2010 has only 4 of the solute-binding proteins [86], which is a stark contrast with the numbers of extracellular GHs (22 extracellular GHs in B. bifidum PRL2010 vs. 10 in B. infantis ATCC 15697). The differences in the diversity and richness of sugar transporters and extracellular GHs may thus be complementary to carbohydrate acquisition strategies within the bifidobacterial community, which simultaneously allows for the differentiation of accessible carbohydrates amongst the community members. Further studies determining the substrate specificity of transporters will reveal how these transporters serve to define the adaptation strategy of each Bifidobacterium species for diet-derived and/or host-derived carbohydrates.
6. Cross-Feeding of the Oligosaccharide Degradants among Bifidobacterial Community
Regarding the bifidobacterial population in breast-fed infant guts, B. breve and B. longum are generally more abundant than B. infantis and B. bifidum. This observation is not paralleled with their in vitro growth ability in HMO-containing media, i.e., B. infantis and B. bifidum grow avidly with HMOs as a sole carbon source and obtain high cell density, but B. breve and B. longum show very limited growth in single culture experiments [87]. This apparent inconsistency between the abundance in the intestinal tracts and abundance in vitro suggest that there is a more complicated mechanism underlying the bifidus-flora formation in infant guts.
The HMO consumption behavior of B. bifidum was analyzed by in vitro culture experiments followed by a high performance liquid chromatography analysis of the sugars in the corresponding spent media. The results reveal the release of monosaccharides (Fuc, Gal, and Glc) and the disaccharides (Lac and LNB) into the media [28,87]. B. bifidum grew well on HMOs; however, high amounts of Fuc and Gal remained unconsumed, even after prolonged culture. The leftover HMO mono- and disaccharidic breakdown products in the spent media were observed for four different strains of B. bifidum, and thus it appears to be a common characteristic of this species [28]. B. infantis is able to consume almost all HMOs by directly internalizing them with their highly abundant ABC transporters, and degrading them using cytoplasmic GHs [9,24]. Most of the B. breve strains catabolize limited species of HMOs, such as LNT and LNnT through direct uptake and intracellular degradation [85,88]. B. longum also shows limited HMO utilization ability, and, in most cases, it consumes LNT only. However, recent studies revealed that several strains of B. breve and B. longum have the ability to assimilate 2′-FL, 3-FL, lactodifucotetraose (LDFT), and LNFP I using specific ABC transporters and intracellular GHs [12,24,83]. Interestingly, the corresponding transporter (fucosyllactose transporter) was shown to be enriched in breast-fed infant guts [24].
Tannock et al. [89] reported that the proportion of B. bifidum within the total Bifidobacterium population was positively correlated with the elevated abundance of the genus Bifidobacterium within the total microbiota of breast-fed infant guts, suggesting that B. bifidum plays an important role for an efficient bifidus-flora formation. Given the capability of B. bifidum to liberate sugars into the spent media (the surrounding environments), it was suggested that cross-feeding may occur to mediate the increased abundance of total Bifidobacterium. Indeed, it was shown that sialic acid and other sugars liberated by B. bifidum can be utilized by B. breve in in vitro co-cultures and in vivo mouse models [68,90,91], though B. breve does not grow in single culture experiments in media supplemented with mucin as a sole carbon source. Additionally, a correlation with the higher abundance of Bifidobacterium in the total microbiota was observed not only for B. bifidum in breast-fed infants, but also for B. longum, B. adolescentis, and B. catenulatum group in human subjects with a wide age range [44]. Interestingly, some B. longum and B. adolescentis strains were shown to have the capability to serve as cross-feeders of plant-derived polysaccharide degradants [92,93,94]. These observations suggested that intragenus cross-feeding might continue in the bifidobacterial community after weaning. As for extracellular GHs targeting plant-derived polysaccharides, B. longum JCM 1217 possesses eleven extracellular GH43 β-xylosidase/α-l-arabinofuranosidases (Figure 2). B. adolescentis ATCC 15703 and B. catenulatum DSM 16992 (= JCM 1194) have two extracellular GH13 (amylase family) and one extracellular GH121 (β-l-arabinobiosidase), respectively.
A recent study suggested that cross-feeding of the HMO/mucin degradation products is not limited to the bifidobacterial community. A Cutibacterium avidum isolate from infant feces was shown to produce a higher amount of propionate when co-cultured with B. bifidum in yeast extract, casitone and fatty acid (YCFA) medium containing HMOs than when cultured alone in the same medium, suggesting inter-genus utilization of B. bifidum-released sugar metabolites [95].
7. Co-Culture Experiments
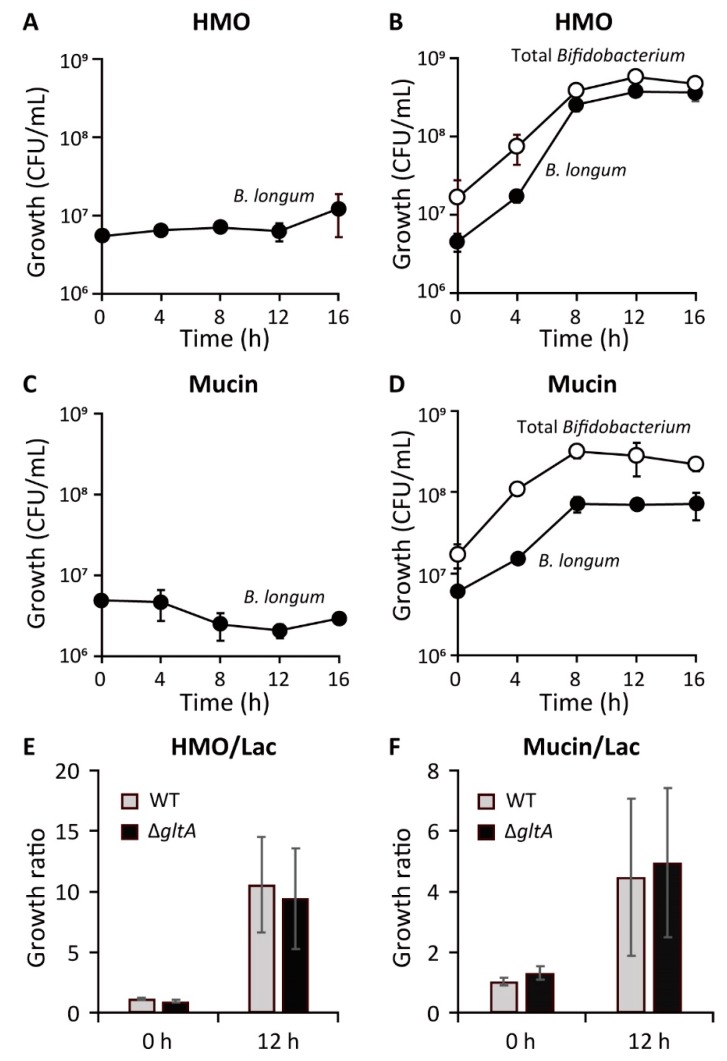
The co-culture experiments in our previous study showed that B. bifidum supported the growth of other bifidobacterial species. B. longum 105-A strain grew in medium containing 1% HMOs as the sole carbon source, but only when it is co-cultured with B. bifidum [28] (Figure 4A,B). The data were reproduced from [28]. The supportive growth effect of B. bifidum for B. longum was also observed in medium containing 1% porcine gastric mucin (PGM) as a sole carbon source (Figure 4C,D). To examine how LNB and GNB disaccharides liberated from HMOs and mucin O-glycans affect the growth of B. longum by cross-feeding from B. bifidum, we disrupted the gltA gene that encodes the solute-binding protein of the GNB/LNB transporter of B. longum [75,76] and made the knockout strain compete against the wild-type strain. Unexpectedly, in the growth competition assays in the presence of B. bifidum, the colony-forming units of ΔgltA strain at 12 h culture were comparable with that of the wild-type in the media containing either HMOs or mucin (Figure 4E,F). These results indicate that the utilization of GNB/LNB was not prioritized by B. longum in the presence of many other sugars, such as Gal, GlcNAc, or lactose released from HMOs/mucin O-glycans under our experimental conditions. It should be noted that the growth ability on LNB was completely abrogated by the disruption of gltA (Supplementary Figure S1), which is well-conserved among the infant gut-associated Bifidobacterium [11,96].
Figure 4.
B. bifidum-mediated cross-feeding supports the growth of B. longum on HMO- and mucin-containing media. (A–D): The growth of B. longum 105-A in a basal medium supplemented with 1% human milk oligosaccharides (HMOs) (A,B) or 1% porcine gastric mucin (C,D) in the absence (A,C) and presence (B,D) of B. bifidum JCM 1254. The wild-type B. longum 105-A strain carrying the chloramphenicol (A,B) or spectinomycin (C–F) resistance gene on a plasmid (pBFS38 or pBFO2, respectively) [97] was used for monitoring the growth. Colony-forming units (CFU) of B. longum 105-A were determined by spreading the serial dilution of the cultures on the agar plates containing the antibiotics (closed circles), while the CFU of total Bifidobacterium was determined using the agar plates without antibiotics (open circles). The data used in (A) was obtained from our previous study [28]. (E,F): The growth competition between wild-type and ΔgltA mutant strains of B. longum 105-A in the presence of B. bifidum JCM 1254. Lactose (Lac), HMOs, or PGM was used as the sole carbon source. The ratio of the growth on HMOs (E) or PGM (F) that was normalized by that on Lac was compared. The ΔgltA mutant was transformed with pBFS38 [97] carrying the chloramphenicol resistance gene, and CFU was determined on agar plates containing the antibiotics.
Further evidence for cross-feeding between B. bifidum and other species was provided by fecal culture experiments [28]. We collected fecal samples from healthy Japanese infants, children, and adults, and then incubated the samples in media supplemented with Glc or HMOs as a sole carbon source, with and without the addition of B. bifidum strains. After 24 h of culture, we quantified the abundance of Bifidobacterium species other than B. bifidum by quantitative-polymerase chain reaction (qPCR). The results indicate that the addition of B. bifidum markedly increased both the abundance and ratio of endogenous Bifidobacterium species other than B. bifidum in the samples. Importantly, this enhancement was observed only for HMO-containing media but not for Glc-containing media. Additionally, in the presence of deoxyfuconojirimycin (DFJ), a potent inhibitor for α-fucosidases (GH29 1,3-1,4-α-l-fucosidase and GH95 1,2-α-l-fucosidase), the bifidus-flora formation-promoting effect exerted by B. bifidum was abrogated. Since most of the abundant HMOs, such as 2′-FL, 3-FL, LDFT, LNFP, and LNDFH are modified with fucose, the removal of the fucose residues should be a critical step for other GHs to further degrade the sugars. Taken together, B. bifidum can enrich the bifidobacterial population and abundance by acting as a cross-feeder of the HMO- and mucin O-glycan degradants produced by its membrane-associated GHs with CBMs.
8. Conclusions
B. bifidum resides in the gastrointestinal tract of humans of a wide age range, yet generally does not dominate the intestinal microbiota, like B. longum and B. breve do during infancy. Instead of being the dominant species in the gut ecosystem, however, it influences the microbiota composition through the altruistic cross-feeding of host glycan degradants. Apart from the microbe to microbe interactions, recent studies have revealed that B. bifidum has immunomodulatory activity by inducing regulatory T-cells by microbe-host interaction with its cell-surface polysaccharides [98]. The production of indole lactic acid, an AhR ligand, by infant gut-associated Bifidobacterium species, including B. bifidum, was also recently reported [8,99]. Further investigation is needed to understand the physiology of this species in the gut ecosystem and its role in host interactions.
9. Materials and Methods
9.1. In Silico Analysis of Bifidobacterial Genes
The protein information of GHs and CBMs in B. bifidum PRL 2010, JCM 7004 (=LMG 13195), TMC 3115, B. breve UCC2003, B. infantis ATCC 15697 (=JCM 1222), and B. longum JCM 1217 were retrieved from the carbohydrate-active enzymes (CAZy) database (http://www.cazy.org) [100]. The pseudogenes that encode incomplete protein sequences (fragment) are excluded from the analysis. Protein localization was predicted by signalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/) and TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
9.2. Disruption of gltA in B. longum 105-A
A markerless gene disruption of gltA (BL105A_1604) was carried out by the methods described previously [24]. The upstream and downstream regions of gltA were amplified by PCR from the genome of B. longum 105-A (JCM 31944) [101] using the primer pairs of Pr-15/16 (5′-cggtacccggggatcatctctactccttcgtagtgaaatc-3′ and 5′-caggcatgcaagctttaacatgcggtgtccccgttg-3′) and Pr-11/12 (5′-tatatatgagtactgatgcgaccacgcccggaatg-3′ and 5′-cgagtcgctcgaattcattaacggttagggttccttccag-3′), respectively. The underlined sequences represent 15 bp-extensions required for In-Fusion cloning (Clontech Laboratories, Mountain View, CA, USA). The amplified fragments were ligated with the replicon of the conditional replication plasmid pBS423 ΔrepA so that the marker gene (spectinomycin resistance) was sandwiched between the upstream and downstream regions of gltA [24,102]. Subsequent integration into the genome and excision of the marker gene from the genome were conducted as described previously [24]. Disruption at the targeted site was confirmed by genomic PCR using the primer pair Pr-36/37 (5′-gtcgccgaagaagttcaccttg-3′ and 5′-ttctggaacctgtcttcttgattgc-3′) and Western Blot analysis using anti-GltA antibodies [87] (Supplementary Figure S1).
9.3. Co-Culture of B. longum with B. bifidum
The wild-type (WT) strain of B. longum 105-A with a plasmid carrying the spectinomycin resistance gene on a plasmid (pBFO2) [97] was cultured in basal medium [87] supplemented with 1% porcine gastric mucin (PGM, Sigma-Aldrich, MO, USA) as a carbon source in the presence and absence of B. bifidum JCM 1254. During incubation, aliquots were collected, serially diluted, and spread on GAM agar plates (Nissui Pharmaceutical, Co., Ltd., Tokyo, Japan) supplemented with and without 30 μg/mL of spectinomycin. The colonies appearing on the antibiotic-containing plates were attributed to B. longum cells, while those formed on antibiotic-free plates were assumed to represent the sum of B. longum and B. bifidum cells. When the ΔgltA derivative of B. longum 105-A was competed against its parental WT strain (with pBFO2 carrying the spectinomycin resistance gene [97]) for the growth on PGM in the presence of B. bifidum, pBFS38 with the chloramphenicol resistance gene [97] was used as the marker of the ΔgltA strain. WT and ΔgltA derivative were also cultured for competition in basal medium supplemented with 1% HMOs as a carbon source. During incubation, aliquots were collected, serially diluted, and spread on GAM agar plates containing the respective antibiotics to determine the CFUs of each strain. Spectinomycin and chloramphenicol were used at the final concentrations of 30 and 2.5 μg/mL, respectively.
Acknowledgments
We thank Atsushi Yokota and Satoru Fukiya (Hokkaido University) for providing Bifidobacterium gene manipulation tools, and Hiroyuki Yachi (Ishikawa Prefectural University) for technical assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/4/481/s1, Figure S1: Disruption of gltA in B. longum 105-A.
Author Contributions
Conceptualization, T.K. (Toshihiko Katoh); methodology, T.K. (Toshihiko Katoh), A.G., M.S., and T.K. (Takane Katayama); investigation, M.N.O., and M.S.; resources, T.K. (Toshihiko Katoh), A.G., M.S.; data curation, T.K. (Toshihiko Katoh), M.N.O.; writing—original draft preparation, T.K. (Toshihiko Katoh); writing—review and editing, T.K. (Takane Katayama); visualization, T.K. (Toshihiko Katoh); supervision, H.A., T.K. (Takane Katayama); project administration, T.K. (Takane Katayama); funding acquisition, T.K. (Toshihiko Katoh), A.G., M.S., and T.K. (Takane Katayama). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by JSPS KAKENHI 19K05789 to Toshihiko Katoh, 19K15732 to A.G., 18K14379 to M.S., and 19K22277 to Takane Katayama. Employment of M.N.O., A.G., and M.S. are supported by JSPS Research Fellowships 19J14598, JSPS Research Fellowships 17J08530, and JSPS Overseas Research Fellowships 201860637, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gavini F., Pourcher A.M., Neut C., Monget D., Romond C., Oger C., Izard D. Phenotypic differentiation of bifidobacteria of human and animal origins. Int. J. Syst. Bacteriol. 1991;41:548–557. doi: 10.1099/00207713-41-4-548. [DOI] [PubMed] [Google Scholar]
- 2.Yoshioka H., Iseki K., Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72:317–321. [PubMed] [Google Scholar]
- 3.Harmsen H.J., Wildeboer-Veloo A.C., Raangs G.C., Wagendorp A.A., Klijn N., Bindels J.G., Welling G.W. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 4.Scholz-Ahrens K.E., Schaafsma G., van den Heuvel E.G., Schrezenmeir J. Effects of prebiotics on mineral metabolism. Am. J. Clin. Nutr. 2001;73:459S–464S. doi: 10.1093/ajcn/73.2.459s. [DOI] [PubMed] [Google Scholar]
- 5.Sheil B., MacSharry J., O’Callaghan L., O’Riordan A., Waters A., Morgan J., Collins J.K., O’Mahony L., Shanahan F. Role of interleukin (IL-10) in probiotic-mediated immune modulation: An assessment in wild-type and IL-10 knock-out mice. Clin. Exp. Immunol. 2006;144:273–280. doi: 10.1111/j.1365-2249.2006.03051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanabe S., Kinuta Y., Saito Y. Bifidobacterium infantis suppresses proinflammatory interleukin-17 production in murine splenocytes and dextran sodium sulfate-induced intestinal inflammation. Int. J. Mol. Med. 2008;22:181–185. doi: 10.3892/ijmm_00000006. [DOI] [PubMed] [Google Scholar]
- 7.Olszak T., An D., Zeissig S., Vera M.P., Richter J., Franke A., Glickman J.N., Siebert R., Baron R.M., Kasper D.L., et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laursen M.F., Sakanaka M., von Burg N., Andersen D., Mörbe U., Rivollier A., Pekmez C.T., Moll J.M., Michaelsen K.F., Mølgaard C., et al. Breastmilk-promoted bifidobacteria produce aromatic lactic acids in the infant gut. bioRxiv. 2020:2020.01.22.914994. [Google Scholar]
- 9.Sela D.A., Chapman J., Adeuya A., Kim J.H., Chen F., Whitehead T.R., Lapidus A., Rokhsar D.S., Lebrilla C.B., German J.B., et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward R.E., Niñonuevo M., Mills D.A., Lebrilla C.B., German J.B. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol. Nutr. Food Res. 2007;51:1398–1405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 11.Sakanaka M., Gotoh A., Yoshida K., Odamaki T., Koguchi H., Xiao J.-Z., Kitaoka M., Katayama T. Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation with Bifidobacteria-Rich. Nutrients. 2019;12:1–21. doi: 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuki T., Yahagi K., Mori H., Matsumoto H., Hara T., Tajima S., Ogawa E., Kodama H., Yamamoto K., Yamada T., et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat. Commun. 2016;7:11939. doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayama T. Host-derived glycans serve as selected nutrients for the gut microbe: Human milk oligosaccharides and bifidobacteria. Biosci. Biotechnol. Biochem. 2016;80:621–632. doi: 10.1080/09168451.2015.1132153. [DOI] [PubMed] [Google Scholar]
- 14.Thomson P., Medina D.A., Garrido D. Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization. Food Microbiol. 2018;75:37–46. doi: 10.1016/j.fm.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Zúñiga M., Monedero V., Yebra M.J. Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium species. Front. Microbiol. 2018;9:1–23. doi: 10.3389/fmicb.2018.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis Z.T., Totten S.M., Smilowitz J.T., Popovic M., Parker E., Lemay D.G., Van Tassell M.L., Miller M.J., Jin Y.S., German J.B., et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:15–17. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borewicz K., Gu F., Saccenti E., Arts I.C.W., Penders J., Thijs C., van Leeuwen S.S., Lindner C., Nauta A., van Leusen E., et al. Correlating Infant Fecal Microbiota Composition and Human Milk Oligosaccharide Consumption by Microbiota of 1-Month-Old Breastfed Infants. Mol. Nutr. Food Res. 2019;63:1–13. doi: 10.1002/mnfr.201801214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis J.C.C., Totten S.M., Huang J.O., Nagshbandi S., Kirmiz N., Garrido D.A., Lewis Z.T., Wu L.D., Smilowitz J.T., German J.B., et al. Identification of Oligosaccharides in Feces of Breast-fed Infants and Their Correlation with the Gut Microbial Community. Mol. Cell. Proteomics. 2016;15:2987–3002. doi: 10.1074/mcp.M116.060665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Leoz M.L.A., Kalanetra K.M., Bokulich N.A., Strum J.S., Underwood M.A., German J.B., Mills D.A., Lebrilla C.B. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: A proof-of-concept study. J. Proteome Res. 2015;14:491–502. doi: 10.1021/pr500759e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urashima T., Asakuma S., Leo F., Fukuda K., Messer M., Oftedal O.T. The Predominance of Type I Oligosaccharides Is a Feature Specific to Human Breast Milk. Adv. Nutr. 2012;3:473S–482S. doi: 10.3945/an.111.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunz C., Rudloff S., Baier W., Klein N., Strobel S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu. Rev. Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 22.McGuire M.K., Meehan C.L., McGuire M.A., Williams J.E., Foster J., Sellen D.W., Kamau-Mbuthia E.W., Kamundia E.W., Mbugua S., Moore S.E., et al. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am. J. Clin. Nutr. 2017;105:1086–1100. doi: 10.3945/ajcn.116.139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawson M.A.E., O’Neill I.J., Kujawska M., Gowrinadh Javvadi S., Wijeyesekera A., Flegg Z., Chalklen L., Hall L.J. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020;14:635–648. doi: 10.1038/s41396-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakanaka M., Hansen M.E., Gotoh A., Katoh T., Yoshida K., Odamaki T., Yachi H., Sugiyama Y., Kurihara S., Hirose J., et al. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci. Adv. 2019;5:eaaw7696. doi: 10.1126/sciadv.aaw7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turroni F., Bottacini F., Foroni E., Mulder I., Kim J.-H., Zomer A., Sanchez B., Bidossi A., Ferrarini A., Giubellini V., et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. USA. 2010;107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiyohara M., Nakatomi T., Kurihara S., Fushinobu S., Suzuki H., Tanaka T., Shoda S.-I., Kitaoka M., Katayama T., Yamamoto K., et al. α-N-Acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J. Biol. Chem. 2012;287:693–700. doi: 10.1074/jbc.M111.277384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gotoh A., Katoh T., Sugiyama Y., Kurihara S., Honda Y., Sakurama H., Kambe T., Ashida H., Kitaoka M., Yamamoto K., et al. Novel substrate specificities of two lacto-N-biosidases towards β-linked galacto-N-biose-containing oligosaccharides of globo H, Gb5, and GA1. Carbohydr. Res. 2015;408:18–24. doi: 10.1016/j.carres.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Gotoh A., Katoh T., Ling Y., Sakanaka M., Yamada C., Asakuma S., Urashima T., Tomabechi Y., Katayama-ikegami A., Kurihara S., et al. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci. Rep. 2018;8:13958. doi: 10.1038/s41598-018-32080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishijima S., Suda W., Oshima K., Kim S.W., Hirose Y., Morita H., Hattori M. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23:125–133. doi: 10.1093/dnares/dsw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam Y.D., Jung M.J., Roh S.W., Kim M.S., Bae J.W. Comparative analysis of korean human gut microbiota by barcoded pyrosequencing. PLoS One. 2011;6:7. doi: 10.1371/journal.pone.0022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta V.K., Paul S., Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 2017;8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wacklin P., Makivuokko H., Alakulppi N., Nikkila J., Tenkanen H., Rabina J., Partanen J., Aranko K., Matto J. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE. 2011;6:e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato K., Ishida S., Tanaka M., Mitsuyama E., Xiao J.-Z., Odamaki T. Association between functional lactase variants and a high abundance of Bifidobacterium in the gut of healthy Japanese people. PLoS ONE. 2018;13:e0206189. doi: 10.1371/journal.pone.0206189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mancabelli L., Milani C., Lugli G.A., Fontana F., Turroni F., van Sinderen D., Ventura M. The Impact of Primer Design on Amplicon-Based Metagenomic Profiling Accuracy: Detailed Insights into Bifidobacterial Community Structure. Microorganisms. 2020;8:131. doi: 10.3390/microorganisms8010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turroni F., Foroni E., Pizzetti P., Giubellini V., Ribbera A., Merusi P., Cagnasso P., Bizzarri B., De’Angelis G.L., Shanahan F., et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 2009;75:1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turroni F., Peano C., Pass D.A., Foroni E., Severgnini M., Claesson M.J., Kerr C., Hourihane J., Murray D., Fuligni F., et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE. 2012;7:20–24. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milani C., Mangifesta M., Mancabelli L., Lugli G.A., James K., Duranti S., Turroni F., Ferrario C., Ossiprandi M.C., van Sinderen D., et al. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 2017;11:2834–2847. doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuki T., Watanabe K., Tanaka R., Fukuda M., Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 1999;65:4506–4512. doi: 10.1128/AEM.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mättö J., Malinen E., Suihko M.L., Alander P., Palva A., Saarela M. Genetic heterogeneity and functional properties of intestinal bifidobacteria. J. Appl. Microbiol. 2004;97:459–470. doi: 10.1111/j.1365-2672.2004.02340.x. [DOI] [PubMed] [Google Scholar]
- 40.Guglielmetti S., Fracassetti D., Taverniti V., Del Bo’ C., Vendrame S., Klimis-Zacas D., Arioli S., Riso P., Porrini M. Differential modulation of human intestinal bifidobacterium populations after consumption of a wild blueberry (vaccinium angustifolium) drink. J. Agric. Food Chem. 2013;61:8134–8140. doi: 10.1021/jf402495k. [DOI] [PubMed] [Google Scholar]
- 41.Turroni F., Serafini F., Foroni E., Duranti S., Motherway M.O.C., Taverniti V., Mangifesta M., Milani C., Viappiani A., Roversi T., et al. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. USA. 2013;110:11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishiyama K., Yamamoto Y., Sugiyama M., Takaki T., Urashima T., Fukiya S., Yokota A., Okada N., Mukai T. Bifidobacterium bifidum Extracellular Sialidase Enhances Adhesion to the Mucosal Surface and Supports Carbohydrate Assimilation. MBio. 2017;8:e00928-17. doi: 10.1128/mBio.00928-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arboleya S., Binetti A., Salazar N., Fernández N., Solís G., Hernández-Barranco A., Margolles A., de los Reyes-Gavilán C.G., Gueimonde M. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 2012;79:763–772. doi: 10.1111/j.1574-6941.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 44.Kato K., Odamaki T., Mitsuyama E., Sugahara H. Age-Related Changes in the Composition of Gut Bifidobacterium Species. Curr. Microbiol. 2017;74:987–995. doi: 10.1007/s00284-017-1272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagpal R., Kurak T., Tsuji H., Takaha T. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: A quantitative assessment. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-10711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elderman M., Sovran B., Hugenholtz F., Graversen K., Huijskes M., Houtsma E., Belzer C., Boekschoten M., de Vos P., Dekker J., et al. The effect of age on the intestinal mucus thickness, microbiota composition and immunity in relation to sex in mice. PLoS ONE. 2017;12:e0184274. doi: 10.1371/journal.pone.0184274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bode L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kunz C., Meyer C., Collado M.C., Geiger L., Garcia-Mantrana I., Bertua-Rios B., Martinez-Costa C., Borsch C., Rudloff S. Influence of Gestational Age, Secretor, and Lewis Blood Group Status on the Oligosaccharide Content of Human Milk. J. Pediatr. Gastroenterol. Nutr. 2017;64:789–798. doi: 10.1097/MPG.0000000000001402. [DOI] [PubMed] [Google Scholar]
- 49.Lang T., Hansson G.C., Samuelsson T. Gel-forming mucins appeared early in metazoan evolution. Proc. Natl. Acad. Sci. USA. 2007;104:16209–16214. doi: 10.1073/pnas.0705984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brockhausen I. Pathways of O-glycan biosynthesis in cancer cells. Biochim. Biophys. Acta-Gen. Subj. 1999;1473:67–95. doi: 10.1016/S0304-4165(99)00170-1. [DOI] [PubMed] [Google Scholar]
- 51.Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: Glycodynamics and functions. EMBO Rep. 2006;7:599–604. doi: 10.1038/sj.embor.7400705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corfield A.P. Mucins: A biologically relevant glycan barrier in mucosal protection. Biochim. Biophys. Acta-Gen. Subj. 2015;1850:236–252. doi: 10.1016/j.bbagen.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Ravn V., Dabelsteen E. Tissue distribution of histo-blood group antigens. Apmis. 2000;108:1–28. doi: 10.1034/j.1600-0463.2000.d01-1.x. [DOI] [PubMed] [Google Scholar]
- 54.Arike L., Hansson G.C. The Densely O-Glycosylated MUC2 Mucin Protects the Intestine and Provides Food for the Commensal Bacteria. J. Mol. Biol. 2016;428:3221–3229. doi: 10.1016/j.jmb.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newburg D.S., Ruiz-Palacios G.M., Morrow A.L. Human Milk Glycans Protect Infants Against Enteric Pathogens. Annu. Rev. Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 56.Katayama T., Sakuma A., Kimura T., Makimura Y., Hiratake J., Sakata K., Yamanoi T., Kumagai H., Yamamoto K. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase Family 95) J. Bacteriol. 2004;186:4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitaoka M., Tian J., Nishimoto M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl. Environ. Microbiol. 2005;71:3158–3162. doi: 10.1128/AEM.71.6.3158-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gauhe A., György P., Hoover J.R.E., Kuhn R., Rose C.S., Ruelius H.W., Zilliken F. Bifidus factor. IV. Preparations obtained from human milk. Arch. Biochem. Biophys. 1954;48:214–224. doi: 10.1016/0003-9861(54)90326-4. [DOI] [PubMed] [Google Scholar]
- 59.Shimada Y., Watanabe Y., Wakinaka T., Funeno Y., Kubota M., Chaiwangsri T., Kurihara S., Yamamoto K., Katayama T., Ashida H. α-N-Acetylglucosaminidase from Bifidobacterium bifidum specifically hydrolyzes α-linked N-acetylglucosamine at nonreducing terminus of O-glycan on gastric mucin. Appl. Microbiol. Biotechnol. 2015;99:3941–3948. doi: 10.1007/s00253-014-6201-x. [DOI] [PubMed] [Google Scholar]
- 60.Kiyohara M., Tanigawa K., Chaiwangsri T., Katayama T., Ashida H., Yamamoto K. An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 2011;21:437–447. doi: 10.1093/glycob/cwq175. [DOI] [PubMed] [Google Scholar]
- 61.Katoh T., Maeshibu T., Kikkawa K., Gotoh A., Tomabechi Y., Nakamura M., Liao W.-H., Yamaguchi M., Ashida H., Yamamoto K., et al. Identification and characterization of a sulfoglycosidase from Bifidobacterium bifidum implicated in mucin glycan utilization. Biosci. Biotechnol. Biochem. 2017;81:2018–2027. doi: 10.1080/09168451.2017.1361810. [DOI] [PubMed] [Google Scholar]
- 62.Ashida H., Miyake A., Kiyohara M., Wada J., Yoshida E., Kumagai H., Katayama T., Yamamoto K. Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 63.Miwa M., Horimoto T., Kiyohara M., Katayama T., Kitaoka M., Ashida H., Yamamoto K. Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology. 2010;20:1402–1409. doi: 10.1093/glycob/cwq101. [DOI] [PubMed] [Google Scholar]
- 64.Wakinaka T., Kiyohara M., Kurihara S., Hirata A., Chaiwangsri T., Ohnuma T., Fukamizo T., Katayama T., Ashida H., Yamamoto K. Bifidobacterial α-galactosidase with unique carbohydrate-binding module specifically acts on blood group B antigen. Glycobiology. 2013;23:232–240. doi: 10.1093/glycob/cws142. [DOI] [PubMed] [Google Scholar]
- 65.Ashida H., Tanigawa K., Kiyohara M., Katoh T., Yamamoto K. Bifunctional properties and characterization of a novel sialidase with esterase activity from Bifidobacterium bifidum. Biosci. Biotechnol. Biochem. 2018;82:2030–2039. doi: 10.1080/09168451.2018.1497944. [DOI] [PubMed] [Google Scholar]
- 66.Wada J., Ando T., Kiyohara M., Ashida H., Kitaoka M., Yamaguchi M., Kumagai H., Katayama T., Yamamoto K. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl. Environ. Microbiol. 2008;74:3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sugiyama Y., Gotoh A., Katoh T., Yoshida E., Honda Y., Kurihara S., Ashida H., Kumagai H., Yamamoto K., Kitaoka M., et al. Introduction of H-antigen structures onto oligosaccharides and sugar chains of glycoproteins using highly efficient 1,2-α-L-fucosynthase. Glycobiology. 2016;26:1235–1247. doi: 10.1093/glycob/cww085. [DOI] [PubMed] [Google Scholar]
- 68.Nishiyama K., Nagai A., Uribayashi K., Yamamoto Y., Mukai T., Okada N. Two extracellular sialidases from Bifidobacterium bifidum promote the degradation of sialyl-oligosaccharides and support the growth of Bifidobacterium breve. Anaerobe. 2018;52:22–28. doi: 10.1016/j.anaerobe.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 69.Liu Q.P., Sulzenbacher G., Yuan H., Bennett E.P., Pietz G., Saunders K., Spence J., Nudelman E., Levery S.B., White T., et al. Bacterial glycosidases for the production of universal red blood cells. Nature. 2007;25:454–464. doi: 10.1038/nbt1298. [DOI] [PubMed] [Google Scholar]
- 70.Rahfeld P., Sim L., Moon H., Constantinescu I., Morgan-Lang C., Hallam S.J., Kizhakkedathu J.N., Withers S.G. An enzymatic pathway in the human gut microbiome that converts A to universal O type blood. Nat. Microbiol. 2019;4:1475–1485. doi: 10.1038/s41564-019-0469-7. [DOI] [PubMed] [Google Scholar]
- 71.Campbell F., Appleton M.A.C., Fuller C.E., Greeff M.P., Hallgrimsson J., Katoh R., Ng O.L.I., Satir A., Williams G.T., Williams E.D. Racial variation in the O-acetylation phenotype of human colonic mucosa. J. Pathol. 1994;174:169–174. doi: 10.1002/path.1711740305. [DOI] [PubMed] [Google Scholar]
- 72.Larsson J.M.H., Karlsson H., Sjövall H., Hansson G.C. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19:756–766. doi: 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- 73.Nishimoto M., Kitaoka M. Identification of the putative proton donor residue of lacto-N-biose phosphorylase (EC 2.4.1.211) Biosci. Biotechnol. Biochem. 2007;71:1587–1591. doi: 10.1271/bbb.70064. [DOI] [PubMed] [Google Scholar]
- 74.Fujita K., Oura F., Nagamine N., Katayama T., Hiratake J., Sakata K., Kumagai H., Yamamoto K. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum. J. Biol. Chem. 2005;280:37415–37422. doi: 10.1074/jbc.M506874200. [DOI] [PubMed] [Google Scholar]
- 75.Suzuki R., Wada J., Katayama T., Fushinobu S., Wakagi T., Shoun H., Sugimoto H., Tanaka A., Kumagai H., Ashida H., et al. Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem. 2008;283:13165–13173. doi: 10.1074/jbc.M709777200. [DOI] [PubMed] [Google Scholar]
- 76.Wada J., Suzuki R., Fushinobu S., Kitaoka M., Wakagi T., Shoun H., Ashida H., Kumagai H., Katayama T., Yamamoto K. Purification, crystallization and preliminary X-ray analysis of the galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum JCM1217. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 2007;63:751–753. doi: 10.1107/S1744309107036263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sato M., Liebschner D., Yamada Y., Matsugaki N., Arakawa T., Wills S.S., Hattie M., Stubbs K.A., Ito T., Senda T., et al. The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors. J. Biol. Chem. 2017;292:12126–12138. doi: 10.1074/jbc.M117.777391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morita H., Toh H., Oshima K., Nakano A., Shindo C., Komiya K., Arakawa K., Suda W., Honda K., Hattori M. Complete genome sequence of Bifidobacterium bifidum JCM 1255(T) isolated from feces of a breast-fed infant. J. Biotechnol. 2015;210:66–67. doi: 10.1016/j.jbiotec.2015.06.413. [DOI] [PubMed] [Google Scholar]
- 79.Garrido D., Nwosu C., Ruiz-Moyano S., Aldredge D., German J.B., Lebrilla C.B., Mills D.A. Endo-β-N-acetylglucosaminidases from Infant Gut-associated Bifidobacteria Release Complex N-glycans from Human Milk Glycoproteins. Mol. Cell. Proteomics. 2012;11:775–785. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boraston A.B., Bolam D.N., Gilbert H.J., Davies G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ficko-Blean E., Boraston A.B. Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr. Opin. Struct. Biol. 2012;22:570–577. doi: 10.1016/j.sbi.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 82.Ruas-Madiedo P., Gueimonde M., Fernandez-Garcia M., de los Reyes-Gavilan C.G., Margolles A. Mucin Degradation by Bifidobacterium Strains Isolated from the Human Intestinal Microbiota. Appl. Environ. Microbiol. 2008;74:1936–1940. doi: 10.1128/AEM.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garrido D., Ruiz-Moyano S., Kirmiz N., Davis J.C., Totten S.M., Lemay D.G., Ugalde J.A., German J.B., Lebrilla C.B., Mills D.A. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci. Rep. 2016;6:35045. doi: 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.James K., Bottacini F., Contreras J.I.S., Vigoureux M., Egan M., Motherway M.O., Holmes E., van Sinderen D. Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Sci. Rep. 2019;9:1–20. doi: 10.1038/s41598-019-51901-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.James K., Motherway M.O.O.O., Bottacini F., van Sinderen D., Engfer M.B., Stahl B., Finke B., Sawatzki G., Daniel H., Kunz C., et al. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci. Rep. 2016;6:38560. doi: 10.1038/srep38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turroni F., Strati F., Foroni E., Serafini F., Duranti S., van Sinderen D., Ventura M. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl. Environ. Microbiol. 2012;78:5002–5012. doi: 10.1128/AEM.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Asakuma S., Hatakeyama E., Urashima T., Yoshida E., Katayama T., Yamamoto K., Kumagai H., Ashida H., Hirose J., Kitaoka M. Physiology of Consumption of Human Milk Oligosaccharides by Infant Gut-associated Bifidobacteria. J. Biol. Chem. 2011;286:34583–34592. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ruiz-Moyano S., Totten S.M., Garrido D.A., Smilowitz J.T., German J.B., Lebrilla C.B., Mills D.A. Variation in Consumption of Human Milk Oligosaccharides by Infant Gut-Associated Strains of Bifidobacterium breve. Appl. Environ. Microbiol. 2013;79:6040–6049. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tannock G.W., Lawley B., Munro K., Gowri Pathmanathan S., Zhou S.J., Makrides M., Gibson R.A., Sullivan T., Prosser C.G., Lowry D., et al. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl. Environ. Microbiol. 2013;79:3040–3048. doi: 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Egan M., Motherway M.O., Kilcoyne M., Kane M., Joshi L., Ventura M., van Sinderen D. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014;14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O’Connell Motherway M., O’Brien F., O’Driscoll T., Casey P.G., Shanahan F., van Sinderen D. Carbohydrate Syntrophy enhances the establishment of Bifidobacterium breve UCC2003 in the neonatal gut. Sci. Rep. 2018;8:10627. doi: 10.1038/s41598-018-29034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rivière A., Gagnon M., Weckx S., Roy D., De Vuyst L. Mutual cross-feeding interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl. Environ. Microbiol. 2015;81:7767–7781. doi: 10.1128/AEM.02089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moens F., Weckx S., De Vuyst L. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int. J. Food Microbiol. 2016;231:76–85. doi: 10.1016/j.ijfoodmicro.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 94.Ozcan E., Sun J., Rowley D.C., Sela D.A. A Human Gut Commensal Ferments Cranberry Carbohydrates to Produce Formate. Appl. Environ. Microbiol. 2017;83:e01097-17. doi: 10.1128/AEM.01097-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rocha Martin V.N., Lacroix C., Killer J., Bunesova V., Voney E., Braegger C., Schwab C. Cutibacterium avidum is phylogenetically diverse with a subpopulation being adapted to the infant gut. Syst. Appl. Microbiol. 2019;42:506–516. doi: 10.1016/j.syapm.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 96.Xiao J.J.-Z.J., Takahashi S., Nishimoto M., Odamaki T., Yaeshima T., Iwatsuki K., Kitaoka M. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl. Environ. Microbiol. 2010;76:54–59. doi: 10.1128/AEM.01683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sakanaka M., Tamai S., Hirayama Y., Onodera A., Koguchi H., Kano Y., Yokota A., Fukiya S. Functional analysis of bifidobacterial promoters in Bifidobacterium longum and Escherichia coli using the alpha-galactosidase gene as a reporter. J. Biosci. Bioeng. 2014;118:489–495. doi: 10.1016/j.jbiosc.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 98.Verma R., Lee C., Jeun E.-J., Yi J., Kim K.S., Ghosh A., Byun S., Lee C.-G., Kang H.-J., Kim G.-C., et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci. Immunol. 2018;3:28. doi: 10.1126/sciimmunol.aat6975. [DOI] [PubMed] [Google Scholar]
- 99.Sakurai T., Odamaki T., Xiao J.-Z. Production of Indole-3-Lactic Acid by Bifidobacterium Strains Isolated fromHuman Infants. Microorganisms. 2019;7:340. doi: 10.3390/microorganisms7090340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matsumura H., Takeuchi A., Kano Y. Construction of Escherichia coli-Bifidobacterium longum shuttle vector transforming B. longum 105-A and 108-A. Biosci. Biotechnol. Biochem. 1997;61:1211–1212. doi: 10.1271/bbb.61.1211. [DOI] [PubMed] [Google Scholar]
- 102.Hirayama Y., Sakanaka M., Fukuma H., Murayama H., Kano Y., Fukiya S., Yokota A. Development of a double-crossover markerless gene deletion system in Bifidobacterium longum: Functional analysis of the alpha-galactosidase gene for raffinose assimilation. Appl. Environ. Microbiol. 2012;78:4984–4994. doi: 10.1128/AEM.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.