Abstract
This report describes and characterizes a novel entomobirnavirus, designated Port Bolivar virus (PTBV), that was isolated from a pool of Aedes sollicitans mosquitoes collected in a saltwater marsh in East Texas, USA. Full genome sequencing and phylogenetic analyses indicate that PTBV is distinct but genetically related to Drosophila X virus and mosquito X virus, which are assigned to species in the genus Entomobirnavirus, family Birnaviridae. PTBV produced cytopathic effect (CPE) in cultures of mosquito (C6/36) cells, but not in Vero cell cultures. Ultrastructural studies of PTBV in infected C6/36 cells demonstrated unenveloped virus particles about 55 nm in diameter.
Keywords: Birnaviridae, Entomobirnavirus, Port Bolivar virus, mosquito viruses
1. Introduction
The family Birnaviridae consists of viruses with two segmented dsRNA genomes, that form icosahedral, non-enveloped single-shelled particles with a diameter of approximately 65 nm [1]. The family currently includes seven genera; members of three genera (Aquabirnavirus, Avibunyavirus and Blosnavirus) infect vertebrates, whereas members of four genera (Entomobiravirus, Dronavirus, Ronavirus and Telnavirus) infect invertebrates.
In the genus Entomobirnavirus, only two viruses have been classified taxonomically to date: Drosophila X virus (DXV; species Drosophila X virus) was detected in laboratory-reared fruit flies (Drosophila melanogaster) in France [2] and mosquito X virus (MoXV; species Mosquito X virus) was detected by metagenomic sequencing in mosquitoes (Anopheles sinensis) collected in China [3]. Four other viruses cluster phylogenetically with the entomobirnaviruses but have not yet been classified: Culex Y virus (CuYV) was isolated from mosquitoes (Culex pipiens complex) collected in Germany [4]; Eridge virus (ERV) was detected by metagenomic sequencing of fruit flies (Drosophila immigrans) collected in the United Kingdom [5]; culicine-associated Z virus (CaZV) was detected by metagenomic sequencing in mosquitoes (Ochlerotatus caspius and Oc. detritus) collected in France [6]; and Espirito Santo virus (ESV) was discovered in mosquito cell cultures (Aedes albopictus) during studies of dengue type 2 virus in Brazil [7]. In addition, Thirlmere virus, which was isolated in 1980 from a water sample collected in the UK [8], is related antigenically to DXV but no nucleotide sequence data have yet been reported. In this report, we describe and characterize a novel seventh entomobirnavirus, designated Port Bolivar virus (PTBV), which was isolated from Aedes sollicitans mosquitoes collected in 2013 near Port Bolivar, Texas, USA.
2. Materials and Methods
2.1. Study Area and Mosquito Collection
PTBV was isolated from mosquitoes collected in a salt marsh on the Bolivar Peninsula of Galveston County, Texas, USA. Galveston County is located in the southeastern part of Texas, along the Gulf of Mexico. The county has a total area of 2264 km2 of which 979 km2 is land and 1282 km2 (57%) is water. Fresh and saltwater lakes, marshes and rivers are scattered throughout the county, providing abundant breeding sites for mosquitoes. For this reason and because of the subtropical climate, mosquitoes are a significant pest problem, so the county has an active mosquito surveillance and control program. As part of its surveillance program, the Galveston County Mosquito Control Division (GCMCD) collects adult mosquitoes with CO2-baited CDC light traps placed at designated sites throughout the county to monitor species composition and abundance during most of the year.
2.2. Culture Methods
Light traps were collected early each morning by GCMCD personnel and returned to their field laboratory where the mosquitoes were sorted on a chill table and separated into pools by species, sex and trap locality. Representative mosquito pools collected during July 2013 were frozen and transported on dry ice to the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), Department of Pathology, University of Texas Medical Branch (UTMB) in Galveston for virus isolation.
After thawing, each mosquito pool was homogenized using a TissueLyser (Qiagen, Hilde, Germany) in tubes with 1.5–2.0 mL of phosphate-buffered saline, pH 7.4, containing 10% fetal bovine serum, 1% penicillin–streptomycin–amphotericin stock (Sigma, St Louis, MO, USA) and several 3 mm stainless steel balls. After centrifugation, 150 μL of the supernatant was inoculated into separate 12.5 cm2 flask cultures of Vero E6 and Aedes albopictus C6/36 cells, originally obtained from the American Type Culture Collection, Manassas, VA. After adsorption for 2 h at 28 °C (C6/36) or 1 h at 37 °C (Vero), 5.0 mL of maintenance medium was added to each flask, and they were held in incubators at 28 and 37 °C, respectively. Cell cultures were examined regularly for evidence of viral cytopathic effect (CPE). The supernatant from a pool, designated GMC-202, produced CPE in the C6/36 cell culture beginning on day 6 post-inoculation; however, it did not produce visible CPE in the Vero cell culture.
2.3. Immunofluorescent Studies
A second flask of C6/36 cells was inoculated with culture fluid from the initial passage of GMC-202. Seven days after inoculation the infected cells were scraped from the surface of the culture flask and spotted onto Cell-Line 12-well glass slides (Thermo Fisher Scientific, Waltham, MA, USA) for examination by indirect fluorescent antibody tests (IFAT) [9], using hyperimmune mouse ascitic fluids prepared against eastern equine encephalitis (EEEV), Western equine encephalitis (WEEV), Saint Louis encephalitis (SLEV), West Nile (WNV), San Angelo (SAV) and Flanders (FLAV) viruses provided by WRCEVA.
2.4. Transmission Electron Microscopy (TEM)
For ultrastructural analysis, C6/36 cells infected with the second passage of GMC-202 were fixed for 1 h in a mixture of 2.5% formaldehyde prepared from paraformaldehyde powder and 0.1% glutaraldehyde in 0.05 M cacodylate buffer (pH 7.3), to which 0.01% picric acid and 0.03% CaCl2 were added. The monolayer was washed in 0.1 M cacodylate buffer, and cells were scraped off and processed further as a pellet. The pellet was post-fixed in 1% OsO4 in 0.1 M cacodylate buffer (pH 7.3) for 1 h, washed with distilled water and stained in block with 2% aqueous uranyl acetate for 20 min at 60 °C. The pellet was dehydrated in ethanol, processed through propylene oxide and embedded in Poly/Bed 812 (Polysciences, Warrington, PA, USA), as described previously [10]. Ultrathin sections were cut on a Leica EM UC7 microtome (Leica Microsystems, Buffalo Grove, II, USA), stained with lead citrate and examined in a Phillips 201 transmission electron microscope (FEI Phillips, Hillsboro, OR, USA) at 60 kV.
2.5. RNA Extraction and Next-Generation Sequencing
Fluid supernatants from cultures of infected C6/36 cells were used for RNA extraction and sequencing. Supernatants were harvested on day 6 post-infection and clarified by low-speed centrifugation (2000× g, 10 min at 4 °C). One milliliter of clarified supernatant was treated with a cocktail of DNases (14 U Turbo DNase (Ambion, Austin, TX, USA), 20 U Benzonase (EMD Millipore, Billerica, MA, USA) and 20 U RNase One (Promega, Madison, WI, USA) for 1 h at 37°C. Viral RNA was then extracted using Trizol and resuspended in 50 μL RNase/DNase and protease-free water (Ambion, Austin, TX, USA). Viral RNA (~0.9 µg) was fragmented by incubation at 94°C for 8 min in 19.5 µL of fragmentation buffer (Illumina 15016648). Sequencing libraries were prepared from the sample RNAs using an Illumina TruSeq RNA v2 kit following the manufacturer’s protocol. The samples were sequenced on a HiSeq 1000 using the High-Output 2 × 50 paired-end protocol. Reads in fastq format were quality-filtered, and any adapter sequences were removed, using Trimmomatic (v0.17) [11] software. The de novo assembly program ABySS (v1.3.7) [12] was used to assemble the reads into contigs, using several different sets of reads, and kmer values from 20 to 40. Blastn and blastx searches with contigs over 400 bases were used to identify the viral segments. All other contigs were from host RNA. Contigs covering nearly the full length of the viral segments were obtained from 1 million read pairs and kmer values of 25 and 38 for the two segments. Reads were mapped back to the viral contigs using bowtie2 (v2.1.0) [13] and visualized with the Integrated Genomics Viewer (v2.3.26) [14] to verify that the assembled contigs were correct. There were 7.2 million paired reads after filtering and ~1.5% (109,537) mapped to the viral contigs. Raw sequencing data are available upon request to the corresponding author.
2.6. Phylogenetic Analysis
An alignment of complete birnavirus VP1 protein (RdRp) amino acid sequences was created using ClustalW in MEGA version 7.0, and phylogenetically informative sites were selected using Gblocks. The resulting alignment comprising 274 amino acids was used to infer phylogenetic relationships in MEGA 7.0 using the Maximum Likelihood method based on the Whelan and Goldman + Frequency model of amino acid substitution [15]. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model [16], and then selecting the topology with superior log likelihood value. The phylogenetic robustness of each node was determined using 1000 bootstrap replicates. Trees were annotated using Figtree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree).
3. Results
3.1. Virus Isolation
Mosquito pool GMC-202 consisted of 40 non-blooded, female Aedes sollicitans collected in a light trap placed in a saltwater marsh (Figure 1) near the town of Port Bolivar, Galveston County, Texas (Segment A) on the evening of 25 July 2013. The mosquito pool homogenate produced moderate cytopathic effect (CPE) in the C6/36 cell culture beginning 6 days post-inoculation. No CPE was observed in Vero cells inoculated with GMC-202 after 14 days.
Figure 1.
A saltwater marsh near Port Bolivar, Texas, where the infected Aedes sollicitans mosquitoes were collected. (Courtesy of Galveston County Mosquito Control).
3.2. Immunofluorescent Studies
IFATs performed on the GMC-202-infected C6/36 cells were negative with mouse hyperimmune ascitic fluids (MIAFs), prepared against EEEV, WEEV, SLEV, WNV, SAV and FLAV and used at a 1:20 dilution (data not shown). These six agents are the most common mosquito-borne arboviruses recovered in East Texas.
3.3. Transmission Electron Microscopy (TEM)
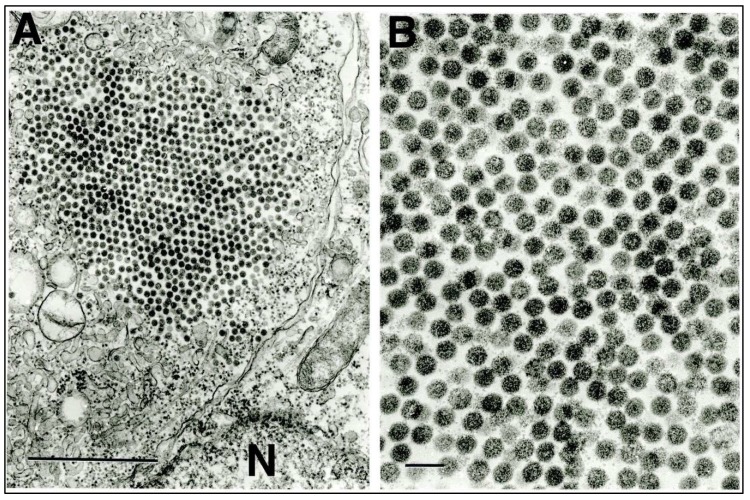
In ultrathin sections of C6/36 cells infected with GMC-202, individual unenveloped virus particles about 55 nm in diameter were observed either scattered free in the cytosol or as paracrystalline agglomerates (Figure 2A,B).
Figure 2.
Ultrastructure of entomobirnavirus GMC-202 in C6/36 mosquito cells. (A) Paracrystalline agglomeration of virus particles in the cell cytosol. N = nucleus of adjacent cell. Bar = 1 μ. (B) Higher-power image of the agglomeration showing the details of virion ultrastructure. Bar = 100 nm.
3.4. Genomic Characterization
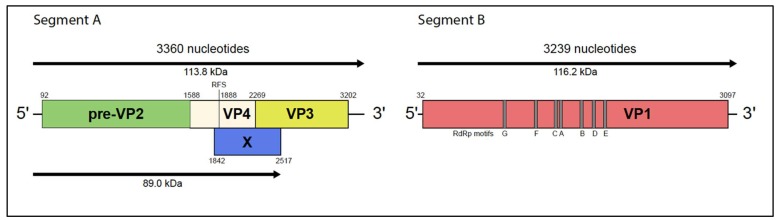
The PTBV genome comprises two segments of double-stranded RNA (Figure 3). As in other entomobirnaviruses, segment A (3360 nt) contains a long open reading frame (ORF) encoding a 113.8 kDa polyprotein (pre-VP2-VP4-VP3) that is co-translationally processed by autocatalysis to generate three polypeptides: pre-VP2 (54.6 kDa), which is further processed to generate major capsid protein VP2; core protein VP3 (35.1 kDa); and serine/lysine protease VP4 (24.2 kDa) [17]. An alternative long ORF (X) in segment A commences 46 nt downstream of a putative –1 ribosomal frame-shift site (UUUUUUAA) that is predicted to generate an 89.0 kDa protein (pre-VP2-VP4N-X) [2]. PTBV RNA segment B (3239 nt) contains a single long ORF encoding VP1, the 116.2 kDa RNA-dependent RNA polymerase (RdRp). PTBV VP1 includes all seven recognized sequence motifs (A–G) in the ‘palm’ and ‘fingers’ domains that form the distinctive topology of the birnavirus RdRp [18]. The PTBV RdRp also shares with other birnaviruses the unusual active site motif (AND) rather than the G/SDD motif that is characteristic of most (+) ssRNA viruses [18,19].
Figure 3.
Genome organization of Port Bolivar virus.
The PTBV genome architecture is similar to that of DXV and other entomobirnaviruses [1]. In particular, segment A encodes a polyprotein (pre-VP2-VP4-VP3) in one long ORF and an alternative ORF (X) that appears likely to be expressed by –1 ribosomal frameshift at a “slippery” site (UUUUUUAA), which is conserved amongst entomobirnaviruses and has been recognized as a particularly shift-prone sequence [2,4]. Although the DXV polyprotein (pre-VP2-VP4-VP3) has been shown to be processed autocatalytically by the VP4 serine-lysine protease [17], the pre-VP2-VP4N-X protein generated by –1 frameshift would lack two critical elements of the VP4 protease active site [20]. As such, it is unlikely to be processed autocatalytically. However, as shown for several birnaviruses, maturation of VP2 involves further processing of the C-terminal region of pre-VP2, most likely by cellular proteases [21,22,23]. Therefore, subsequent processing of PTBV pre-VP2-VP4N-X to generate VP2 and VP4N-X cannot be excluded.
3.5. Amino Acid Sequence Identities
Pairwise amino acid sequence identities (p-distances) were estimated in MEGA 7 from a ClustalW alignment of available birnavirus VP1 (RdRp) proteins (Table 1) and birnavirus polyproteins (Table 2). In VP1, PTBV showed highest sequence identity with entomobirnaviruses and is most closely related to CaZV (87.8% identity) and most distantly related to ERV (69.4% identity). Amongst entomobirnavirus pre-VP2-VP4-VP3 polyproteins, PTBV is most closely related to CaZV (91.7% identity) and most distantly related to DXV (69.6% identity).
Table 1.
Percentage amino acid sequence identities (p-distance) of a ClustalW alignment of birnavirus VP1 (RdRp) proteins.
| Genus | Virus | PTBV | CaZV | CuYV | MXV | ESV | DXV | ERV | IPNV | TABV | VTABV | IBDV | BSHV | LCBV | RBV | DBV | TV-1 | JmBLV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Entomobirnavirus | PTBV | |||||||||||||||||
| CaZV | 87.8 | |||||||||||||||||
| CuYV | 85.8 | 92.2 | ||||||||||||||||
| MXV * | 85.9 | 92.5 | 98.5 | |||||||||||||||
| ESV | 86.1 | 92.1 | 98.8 | 98.1 | ||||||||||||||
| DXV * | 72.3 | 72.5 | 72.5 | 72.8 | 72.5 | |||||||||||||
| ERV | 69.4 | 71.3 | 71.6 | 71.7 | 71.6 | 84.7 | ||||||||||||
| Aquabirnavirus | IPNV * | 28.4 | 28.4 | 28.3 | 28.1 | 28.1 | 29.2 | 28.7 | ||||||||||
| TABV | 28.6 | 28.0 | 28.4 | 28.3 | 28.3 | 28.1 | 28.0 | 90.8 | ||||||||||
| VTABV | 29.0 | 29.0 | 28.7 | 28.6 | 28.6 | 29.4 | 29.1 | 94.5 | 90.4 | |||||||||
| Avibirnavirus | IBDV * | 29.1 | 28.8 | 28.1 | 28.1 | 27.9 | 28.1 | 27.7 | 48.0 | 47.4 | 47.4 | |||||||
| Blosnavirus | BSHV * | 29.6 | 29.8 | 29.9 | 30.1 | 29.8 | 29.5 | 29.8 | 49.2 | 48.2 | 49.0 | 52.3 | ||||||
| LCBV * | 28.6 | 29.0 | 29.5 | 29.6 | 29.1 | 28.6 | 28.4 | 50.4 | 48.9 | 49.7 | 51.0 | 62.2 | ||||||
| Ronavirus | RBV * | 26.1 | 25.1 | 24.7 | 25.0 | 24.9 | 25.0 | 24.9 | 30.9 | 31.3 | 31.0 | 30.2 | 31.7 | 30.7 | ||||
| Dronavirus | DBV * | 24.5 | 23.4 | 23.9 | 24.2 | 23.9 | 23.4 | 22.4 | 29.5 | 29.2 | 29.8 | 27.7 | 29.6 | 29.6 | 24.6 | |||
| Telnavirus | TV-1 * | 24.5 | 24.2 | 23.9 | 23.5 | 23.6 | 25.0 | 25.1 | 30.3 | 30.5 | 30.1 | 31.1 | 31.8 | 33.3 | 28.7 | 25.0 | ||
| unassigned | JmBLV | 28.1 | 27.3 | 27.3 | 27.3 | 27.3 | 27.3 | 28.0 | 31.7 | 31.0 | 31.1 | 30.2 | 30.6 | 30.7 | 26.9 | 23.2 | 26.4 |
* Only these viruses are currently formally classified to species.
Table 2.
Percentage amino acid sequence identities (p-distance) of a ClustalW alignment of birnavirus VP2-VP4-VP3 polyproteins.
| Genus | Virus | PTBV | CaZV | CuYV | MXV | ESV | DXV | ERV | IPNV | TABV | VTABV | IBDV | BSHV | LCBV | RBV | DBV | TV-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Entomobirnavirus | PTBV | ||||||||||||||||
| CaZV | 91.7 | ||||||||||||||||
| CuYV | 85.9 | 89.2 | |||||||||||||||
| MXV * | 86.2 | 89.7 | 98.4 | ||||||||||||||
| ESV | 85.1 | 88.1 | 98.4 | 97.3 | |||||||||||||
| DXV * | 69.6 | 70.5 | 69.3 | 68.8 | 68.9 | ||||||||||||
| ERV | 71.3 | 72.7 | 70.7 | 70.4 | 70.4 | 86.1 | |||||||||||
| Aquabirnavirus | IPNV * | 25.2 | 25.2 | 25.0 | 24.9 | 24.8 | 24.8 | 25.6 | |||||||||
| TABV | 25.1 | 24.5 | 24.4 | 24.4 | 24.2 | 24.3 | 25.1 | 84.7 | |||||||||
| VTABV | 25.3 | 25.0 | 24.5 | 24.4 | 24.4 | 24.3 | 25.3 | 88.8 | 86.8 | ||||||||
| Avibirnavirus | IBDV * | 25.1 | 24.8 | 25.2 | 25.2 | 25.0 | 25.6 | 27.0 | 35.5 | 35.5 | 36.1 | ### | |||||
| Blosnavirus | BSHV * | 26.6 | 26.6 | 26.4 | 26.3 | 26.4 | 25.6 | 27.7 | 35.3 | 36.2 | 35.8 | 39.8 | |||||
| LCBV * | 24.5 | 25.0 | 25.0 | 24.9 | 24.9 | 23.8 | 25.0 | 34.5 | 34.8 | 33.8 | 39.6 | 51.5 | |||||
| Ronavirus | RBV * | 24.9 | 24.7 | 25.0 | 24.5 | 24.7 | 25.7 | 26.5 | 27.5 | 26.6 | 27.0 | 27.7 | 28.2 | 26.3 | |||
| Dronavirus | DBV * | 24.9 | 24.5 | 24.0 | 24.0 | 24.0 | 24.3 | 24.0 | 27.5 | 27.7 | 28.7 | 29.2 | 27.4 | 27.7 | 25.7 | ||
| Telnavirus | TV-1 * | 21.5 | 21.6 | 20.9 | 21.1 | 20.5 | 22.6 | 22.6 | 23.8 | 24.3 | 22.9 | 26.0 | 26.7 | 26.2 | 25.7 | 23.1 |
* Only these viruses are currently formally classified to species.
3.6. Phylogenetic Analysis
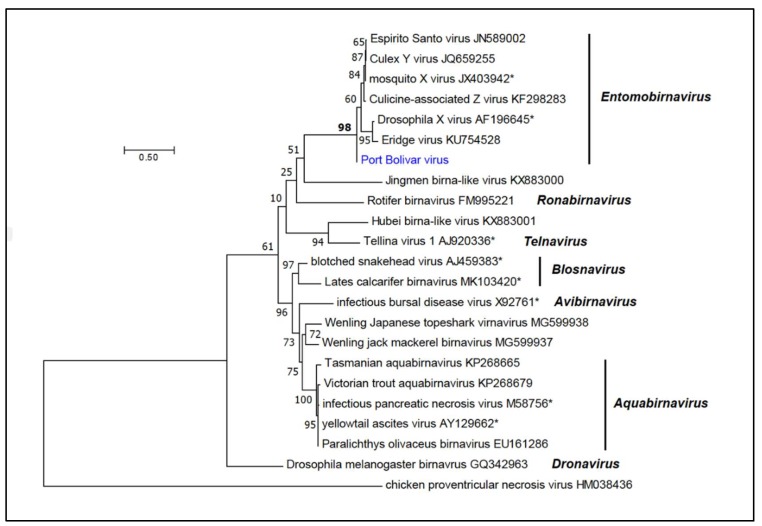
Phylogenetic analysis conducted using a ClustalW alignment of VP1 (RdRp) amino acid sequences of PTBV and 22 birnaviruses indicated that it clustered with strong bootstrap support (BSP = 98%) with the entomobirnaviruses (Figure 4).
Figure 4.
Phylogenetic relationships of Port Bolivar virus. An unrooted phylogenetic tree was inferred by using the Maximum Likelihood method from a ClustalW alignment of complete VP1 protein (RdRp) amino acid sequences of Port Bolivar virus (PTBV) and 22 birnaviruses. Phylogenetically informative sites were selected from the alignment using Gblocks, resulting in 274 positions in the final dataset. The tree with the highest log likelihood (−6338.33) is shown. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. Bootstrap values (100 iterations) are shown for each node. All seven established genera are shown in bold italics; those viruses which have currently been assigned to species are also shown.
4. Discussion
Along with CuYY, PTBV is the second entomobirnavirus to be isolated from free-living mosquitoes (Table 3) [4]. Two others, CaZV and MXV, were detected by next-generation sequencing (without isolation) of field-collected mosquitoes [3,6]. The remaining four known entomobirnaviruses (DXV, ERV, ESV and Thirlmere virus) were isolated from or were detected by next-generation sequencing of insect cell lines, or in a water sample passed in insect cell cultures [2,5,7]. For this reason, there is some uncertainty as to their origin. For example, ESV was discovered during purification of a strain of dengue virus type 2 (DENV-2) that was originally isolated from a Brazilian dengue patient and had been passaged three times in C6/36 cell cultures [7]. DXV, the prototype of the genus Entomobirnavirus, was discovered under similar circumstances. DXV was isolated as a contaminant during infection studies with sigma virus (genus Sigmavirus: Rhabdoviridae) [2].
Table 3.
Names, original source, locality, and accession numbers of currently recognized entomobirnaviruses *.
| Virus Name (Abbreviation) | Source | Locality | Accession Numbers | Reference |
|---|---|---|---|---|
| Drosophila X virus (DXV) | Drosophila melanogaster cell culture | France | U60650; AF196645 | [2] |
| Eridge virus (ERV) | D. melanogaster cell culture | UK | KU754527; KU754528 | [5] |
| Culicine-associated Z virus (CaZV) | Ochlerotatus caspius and Oc. detritus | France | KF298271; KF298272 | [6] |
| Espirito Santo virus (ESV) | Aedes albopictus cell culture | Brazil | JN589003; NJ589002 | [7] |
| Mosquito X virus (MXV) | Anopheles sinensis | China | JX403941; JX403942 | [3] |
| Culex Y virus (CuYV) | Culex pipiens complex | Germany | JQ659254; JQ659255 | [4] |
| Thirlmere virus | Water | UK | Not available | [8] |
| Port Bolivar virus (PTBV) | Aedes sollcitans | USA |
MT263973 MT263974 |
Present paper |
* Entomobirnaviruses reported as of February 2020.
Currently, only two species (Drosophila X virus and Mosquito X virus) have been assigned in the genus Entomobirnavirus by the International Committee on Taxonomy of Viruses (ICTV), and no formal species demarcation criteria appear to have been published. Phylogenetically, PTBV clearly falls within this genus and, based upon amino acid sequence divergence in VP1 RdRp and the VP2-VP4-VP3 polyprotein, it is clearly distinct from both DXV (27.7% and 30.4% divergence, respectively) and MXV (14.1% and 13.8% divergence, respectively) (Figure 4). Although PTBV is most closely related to CaZV, these viruses have been detected in culicine mosquitoes of different species from geographically distant locations (Aedes sollicitans in Texas and Ochlerotatus spp. in France, respectively) and are sufficiently divergent in amino acid sequence (12.2% divergence in VP1; 8.3% divergence in VP2-VP4-VP3) to be assigned to distinct new entomobirnavirus species.
To date, the strongest association of entomobirnaviruses has been with mosquitoes; but the effects of this group of viruses on their mosquito hosts are still unknown. With the current interest in mosquito microbiomes, this should be an area for future study. Also, given their wide geographic distribution and the diversity of mosquito hosts, it seems likely that many other entomobirnaviruses exist in nature in mosquitoes and possibly other insects and arthropods.
Author Contributions
Conceptualization, R.B.T. and N.V.; Data curation, R.B.T., S.G.W., P.J.W., and N.V.; Formal analysis, R.B.T., V.L.P., S.W., P.J.W. and N.V.; Funding acquisition, R.B.T.; Investigation, R.B.T., H.G., B.G.B., V.L.P., A.W., S.G.W., T.G.W., and N.V.; Methodology, R.B.T., H.G., B.G.B., A.W., S.G.W., and N.V.; Project administration, R.B.T., H.G. and N.V.; Resources, R.B.T., H.G. and N.V.; Supervision, R.B.T. and N.V.; Writing—review and editing, R.B.T., V.L.P., S.G.W., P.J.W. and N.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grant R24AI120942 and contracts N01-AI30027, HHS-N2722010000401 and HHSN2720004/D04 from the U.S. National Institutes of Health.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Delmas: B., Attoui H., Ghosh S., Malik Y.S., Mundt E., Vakharia V.N. ICTV Report Comm., ICTV virus taxonomy profile: Birnaviridae. J. Gen. Virol. 2019;100:5–6. doi: 10.1099/jgv.0.001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teninges D., Ohanessian A., Richard-Molard C., Contamine D. Isolation and Biological Properties of Drosophila X Virus. J. Gen. Virol. 1979;42:241–254. doi: 10.1099/0022-1317-42-2-241. [DOI] [Google Scholar]
- 3.Huang Y., Mi Z., Zhuang L., Ma M., An X., Liu W., Cao W., Tong Y. Presence of entomobirnaviruses in Chinese mosquitoes in the absence of Dengue virus co-infection. J. Gen. Virol. 2013;94:663–667. doi: 10.1099/vir.0.048231-0. [DOI] [PubMed] [Google Scholar]
- 4.Marklewitz M., Gloza-Rausch F., Kurth A., Kummerer B.M., Drosten C., Junglen S. First isolation of an Entomobirnavirus from free-living insects. J. Gen. Virol. 2012;93:2431–2435. doi: 10.1099/vir.0.045435-0. [DOI] [PubMed] [Google Scholar]
- 5.Webster C.L., Longdon B., Lewis S.H., Obbard D.J. Twenty-Five New Viruses Associated with the Drosophilidae (Diptera) Evol. Bioinform. Online. 2016;12:13–25. doi: 10.4137/EBO.S39454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook S., Chung B.Y., Bass D., Moureau G., Tang S., McAlister E., Culverwell C.L., Glucksman E., Wang H., Brown T.D., et al. Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in dipteran hosts. PLoS ONE. 2013;8:e80720. doi: 10.1371/journal.pone.0080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vancini R., Paredes A., Ribeiro M., Blackburn K., Ferreira D., Kononchik J.P., Jr., Hernandez R., Brown D. Espirito Santo virus: A new birnavirus that replicates in insect cells. J. Virol. 2012;86:2390–2399. doi: 10.1128/JVI.06614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly D.C., Ayres M.D., Howard S.C., Lescott T., Arnold M.K., Seeley N.D., Primrose S.B. Isolation of a bisegmented double-stranded RNA virus from Thirlmere reservoir. J. Gen. Virol. 1982;62:313–322. doi: 10.1099/0022-1317-62-2-313. [DOI] [PubMed] [Google Scholar]
- 9.Tesh R.B. A method for the isolation and identification of dengue viruses, using mosquito cell cultures. Am. J. Trop. Med. Hyg. 1979;28:1053–1059. doi: 10.4269/ajtmh.1979.28.1053. [DOI] [PubMed] [Google Scholar]
- 10.Popov V.L., Tesh R.B., Weaver S.C., Vasilakis N. Electron Microscopy in Discovery of Novel and Emerging Viruses from the Collection of the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) Viruses. 2019;11:5. doi: 10.3390/v11050477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohse M., Bolger A.M., Nagel A., Fernie A.R., Lunn J.E., Stitt M., Usadel B. RobiNA: A user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J., Birol I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotech. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 16.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 17.Chung H.K., Kordyban S., Cameron L., Dobos P. Sequence analysis of the bicistronic Drosophila X virus genome segment A and its encoded polypeptides. Virology. 1996;225:359–368. doi: 10.1006/viro.1996.0610. [DOI] [PubMed] [Google Scholar]
- 18.Pan J., Vakharia V.N., Tao Y.J. The structure of a birnavirus polymerase reveals a distinct active site topology. Proc. Natl. Acad. Sci. USA. 2007;104:7385–7390. doi: 10.1073/pnas.0611599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shwed P.S., Dobos P., Cameron L.A., Vakharia V.N., Duncan R. Birnavirus VP1 proteins form a distinct subgroup of RNA-dependent RNA polymerases lacking a GDD motif. Virology. 2002;296:241–250. doi: 10.1006/viro.2001.1334. [DOI] [PubMed] [Google Scholar]
- 20.Petit S., Lejal N., Huet J.C., Delmas B. Active residues and viral substrate cleavage sites of the protease of the birnavirus infectious pancreatic necrosis virus. J. Virol. 2000;74:2057–2066. doi: 10.1128/JVI.74.5.2057-2066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Da Costa B., Chevalier C., Henry C., Huet J.C., Petit S., Lepault J., Boot H., Delmas B. The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 2002;76:2393–2402. doi: 10.1128/jvi.76.5.2393-2402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Da Costa B., Soignier S., Chevalier C., Henry C., Thory C., Huet J.C., Delmas B. Blotched snakehead virus is a new aquatic birnavirus that is slightly more related to avibirnavirus than to aquabirnavirus. J. Virol. 2003;77:719–725. doi: 10.1128/JVI.77.1.719-725.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galloux M., Chevalier C., Henry C., Huet J.C., Costa B.D., Delmas B. Peptides resulting from the pVP2 C-terminal processing are present in infectious pancreatic necrosis virus particles. J. Gen. Virol. 2004;85:2231–2236. doi: 10.1099/vir.0.80012-0. [DOI] [PubMed] [Google Scholar]