Abstract
Injuries from exposure to explosions rose dramatically during the Iraq and Afghanistan wars, which motivated investigations of blast-related neurotrauma and operational breaching. In this study, military “breachers” were exposed to controlled, low-level blast during a 10-day explosive breaching course. Using an omics approach, we assessed epigenetic, transcriptional, and inflammatory profile changes in blood from operational breaching trainees, with varying levels of lifetime blast exposure, along with daily self-reported symptoms (with tinnitus, headaches, and sleep disturbances as the most frequently reported). Although acute exposure to blast did not confer epigenetic changes, specifically in DNA methylation, differentially methylated regions (DMRs) with coordinated gene expression changes associated with lifetime cumulative blast exposures were identified. The accumulative effect of blast showed increased methylation of PAX8 antisense transcript with coordinated repression of gene expression, which has been associated with sleep disturbance. DNA methylation analyses conducted in conjunction with reported symptoms of tinnitus in the low versus high blast incidents groups identified DMRS in KCNE1 and CYP2E1 genes. KCNE1 and CYP2E1 showed the expected inverse correlation between DNA methylation and gene expression, which have been previously implicated in noise-related hearing loss. Although no significant transcriptional changes were observed in samples obtained at the onset of the training course relative to chronic cumulative blast, we identified a large number of transcriptional perturbations acutely pre- versus post-blast exposure. Acutely, 67 robustly differentially expressed genes (fold change ≥1.5), including UFC1 and YOD1 ubiquitin-related proteins, were identified. Inflammatory analyses of cytokines and chemokines revealed dysregulation of MCP-1, GCSF, HGF, MCSF, and RANTES acutely after blast exposure. These data show the importance of an omics approach, revealing that transcriptional and inflammatory biomarkers capture acute low-level blast overpressure exposure, whereas DNA methylation marks encapsulate chronic long-term symptoms.
Keywords: blast overpressure, epigenetics, sleep, tinnitus, traumatic brain injury
Introduction
Injuries from exposure to explosive blasts rose dramatically during Operation Iraqi Freedom and Operation Enduring Freedom (OIF, OEF) attributable to increased use of improvised explosive devices in military settings and has also increased in civilian populations through acts of terrorism,1–3 thus motivating investigations of blast-related neurotrauma. Congressional acts, including the John S. McCain National Defense Authorization Act for Fiscal Year 2019, have called for “review of guidance on blast exposure during training” (Section 253), which emphasize the importance of understanding and mitigating the effects of blast after successive days of training and over the course of a military career.4 Despite the increase in occurrence and public awareness, our understanding of the effects of blast and the mechanisms behind subsequent injury are limited.5,6 Blast overpressure (BOP) events are capable of injuring the brain by means of high-energy pressure waves that are rapidly emitted from the explosive, which propagate from the object and turn into shock waves upon interacting with a medium—in this case, being the military warfighter.7
The clinical and pathological effects of blast exposure varies depending on the magnitude of the detonation, proximity to the blast, and the use of protective gear7–9 and includes, but is not limited to, neuronal swelling, subdural hematomas, myelin deformation, inflammation, loss of consciousness, temporary disorientation, sleep disturbances, memory deficits, and tinnitus.10–18 A better understanding of the systemic biomarkers underlying blast exposure responses is critical to not only the identification of blast-related injury, but also to the development of effective diagnostics and potential treatments.
In this effort, we have undertaken prospective human studies involving “breachers,” military and law enforcement personnel who are exposed to repeated blast as part of their occupational duty. Breachers are typically in close proximity to controlled, repeated, low-level blast during operations and training and have reported a range of physical, emotional, and cognitive symptoms, including headache, sleep disturbance, anxiety, and impaired cognitive performance.19 It has been previously shown that BOP exposure is capable of inducing changes in gene expression in military personnel20 and in animal models of blast injury.21
We took an omics approach, performing large-scale epigenetics, as well as transcriptional, and inflammatory profiling to identify blood-based biomarkers associated with acute and chronic blast exposure. We investigated DNA methylation, a highly stable epigenetic marker associated with gene repression as well as gene expression patterns, in addition to transcriptome analysis by RNA sequencing (RNA-seq) to identify potential epigenetic and coordinated gene expression abnormalities. Further, we assessed inflammatory changes after blast exposure, in addition to self-reported symptoms. These data allow us to investigate regulatory, transcriptional, and inflammatory biosignatures of blast exposure.
Methods
Samples and subjects
This study was approved by the Institutional Review Board (NMRC#2011.0002; WRAIR#1796). The present study obtained samples from 34 healthy, male participants at U.S. Army explosive entry training sites (special operations and combat engineer courses). Biological specimens for epigenetics and transcriptional studies were collected at the start (baseline) and at the end of the training course (pre- vs. post-blast exposure), and serum samples for inflammatory analyses were collected daily. Demographic information, including sex, age, lifetime operational exposure to blast, as well as self-reported traumatic brain injury (TBI) history, were recorded at the start of the training. During the course of the training, self-report symptom assessments were completed daily.
DNA methylation sample processing and quality control
Blood was collected using ethylenediaminetetraacetic acid tubes and processed to separate peripheral blood mononuclear cells (PBMCs). PBMCs were purified by Ficoll gradient, washed with phosphate-buffered saline (PBS), and stored at −80°C. DNA was extracted with the QIAamp DNA Micro Kit (QIAGEN, Hilden, Germany). Genomic DNA was bisulfite converted (Zymo Research, Irvine, CA), and CpG methylation was determined using Illumina Infinium HumanMethylationBeadChip microarrays (HM450; Illumina, Inc., San Diego, CA), as described previously.22 Data and quality-control (QC) analyses were performed using R Language 3.03 (R Foundation for Statistical Computing, Vienna, Austria),23 an environment for statistical computing, and Bioconductor 2.13.24 Raw data files (.idat) were processed by the minfi package.25 All samples displayed a mean probe-wise detection call for the 485512 array probes <0.0005 (Supplementary Fig. S1). Sex QC analysis also confirmed methylation-based sex prediction with those reported (Supplementary Fig. S2). For QC sample tracking of pre- versus post-breacher training, we used the 65 single-nucleotide polymorphism (SNP) probes included in the HumanMethylation 450k BeadChip, confirming that subjects for which multiple samples were available grouped together (Supplementary Fig. S3).
DNA methylation data analysis
For all DNA methylation analyses, we used the matrix of M-values (logit transformation of beta-values), which correspond to methylation levels. Surrogate variable analysis (SVA) was performed to add surrogate variables and rule out potential batch effects. A linear model was used for the binary variable of interest, while including age and history of TBI as covariates in the model. Performing the comparative analysis in limma26 implemented in R (R Foundation for Statistical Computing), we obtained t-statistics and associated p values for each CpG site. The point-wise p values were then used for the identification of differentially methylated regions (DMRs) using the combined p-value tool.27
Total RNA sample/library preparation and sequencing
Blood was collected using Paxgene RNA tubes (PreAnalytiX; QIAGEN/BD, Hombrechtikon, Switzerland), according to the manufacturer's instructions, and stored at −80°C. RNA was extracted with the Paxgene Blood RNA Kit (PreAnalytiX; QIAGEN/BD). Globin messenger RNA (mRNA) was removed with the Globin Clear Human Globin mRNA Removal kit (Ambion, Inc., Austin, TX). All RNA samples had RNA integrity numbers ≥6.0. Total RNA-seq libraries were prepared using the Illumina Stranded Total RNA Library Prep Kit with Ribo-Zero Gold (Illumina, Inc.), in accord with the manufacturer's instructions. Briefly, 290–500 ng of total RNA was used for ribosomal depletion and fragmented by divalent cations under elevated temperatures.
Fragmented RNA underwent first strand synthesis using reverse-transcriptase and random primers followed by second strand synthesis to generate complementary DNA (cDNA). The cDNA fragments underwent end repair, adenylation, and ligation of Illumina sequencing adapters. The cDNA library was enriched using 11 cycles of polymerase chain reaction and purified. Final libraries were evaluated using PicoGreen (Life Technologies, Carlsbad, CA) and Fragment Analyzer (Advanced Analytical, Agilent Technologies, Santa Clara, CA) and were sequenced on an Illumina HiSeq2500 sequencer (v4 chemistry) using 2 × 125-base-pair read lengths.
RNA-sequencing data preprocessing and bioinformatics analysis
Reads were aligned to the human reference hg19 using STAR aligner (v2.4.0c).28 Quantification of genes annotated in Gencode v18 was performed using featureCounts (v1.4.3). QC metrics were collected with Picard (v1.83) and RSeQC 29 (http://broadinstitute.github.io/picard/). Normalization of feature counts was done. Further, for gene expression analysis, we used the voom function in limma to get logCPM (log counts per minute) matrix, where the design matrix consists of intercept, age, history of TBI, and variable of interest. We used SVA to rule out potential batch effects. Analyses were performed in limma, and point-wise as well as multiple testing adjusted p values are reported.
Gene Ontology and gene set enrichment analyses
We performed Gene Ontology (GO) analysis using the goseq R package, with gene length bias considered. Gene set enrichment analysis (GSEA) version 3.030 was run on our ranked list of 8157 genes, which is filtered from the logCPM matrix obtained in pre-post analysis by criteria that average logCPM is ≥4 in either of the comparison groups considered here, and ordered by t-statistics. GSEA pre-ranked was run with 1000 permutations using gene sets from the Molecular Signatures Database (MsigDB)31 as follows: 1) GO gene sets (C5) including biological processes (BP), cellular components (CC), and molecular function (MF) and 2) hallmark gene set (H).
Cytokine sample processing by Luminex
For assays of inflammatory cytokines, we used the Luminex 63-Plex assay (eBiosciences/Affymetrix, Inc., Santa Clara, CA), which has the ability to multi-plex, measuring levels of 63 inflammatory cytokines simultaneously. Assays were performed by the Human Immune Monitoring Center at Stanford University (Stanford, CA), and kits were used according to the manufacturer's recommendations, with modifications, as described here briefly. Beads were added to a 96-well plate and washed in a BioTek Elx405 washer (BioTek Instruments, Inc., Winooski, VT). Serum samples were added to the plate containing the mixed antibody-linked beads and incubated at room temperature for 1 h followed by overnight incubation at 4°C with shaking. Cold and room temperature incubation steps were performed on an orbital shaker at 500–600 rpm.
After the overnight incubation, plates were washed in a BioTek Elx405 washer, and then biotinylated detection antibody was added for 75 min at room temperature with shaking. Plates were washed as above and streptavidin-phycoerythrin was added. After incubation for 30 min at room temperature, a wash was performed as above and reading buffer was added to the wells. Each sample was measured in duplicate. Plates were read using a Luminex 200 instrument (Millipore Sigma, Burlington, MA) with a lower bound of 50 beads per sample per cytokine. Custom assay control beads by Radix Biosolutions (Georgetown, TX) were added to all wells.
Cytokine data analysis
Cytokine data were available for a subset of subjects, for 5 days: day 4, day 7, day 8, day 9, and day 10. Two levels were taken for each subject for each day, and all values were analyzed. All cytokine levels were log-transformed and then graphed. Given that outliers were present in many of the cytokines, values were capped (winsorized) from both below and above, at the lower quartile minus 1.5 times the interquartile range (IQR), and the upper quartile plus 1.5 times the IQR, respectively. Separate mixed-effect models were fit for each log-transformed and capped cytokine measure, with day as fixed, categorical predictor, and subject-specific random intercepts. p values for each level of the fixed effect were recorded and corrected for multiple testing (for K = 63 cytokines) using the Bonferroni method. To calculate standardized effect sizes for the contrast of each experimental day compared to the baseline, the model coefficients were divided by the standard deviation (SD) of the baseline measure, with the scaleless effect sizes then represented by heatmap plots.
Results
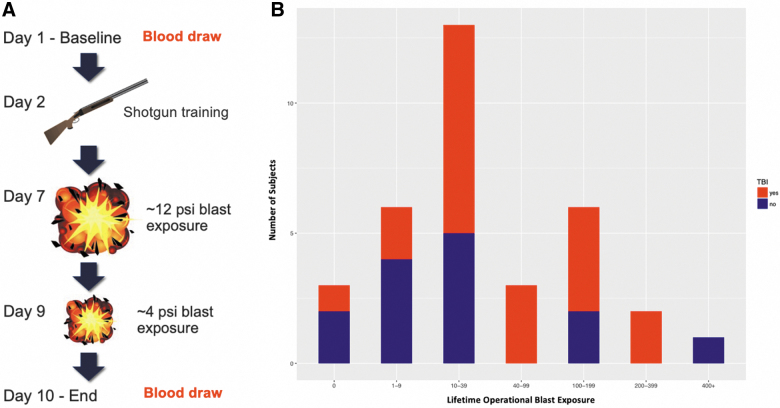
The present study collected data from 34 participants during three separate 2-week data collection cycles at U.S. Army explosive entry training sites (special operations and combat engineer courses). In these advanced training courses, both trainees and instructors have a career history of repeated exposure to low-level blasts. Blood samples were obtained pre- and post-training for epigenetics, transcriptional, and protein assays, subsequently referred to as pre- versus post-blast exposure. All participants were male, with an average age of 30.79 years (SD, 4.57). Self-report history of injury and blast exposure was recorded at baseline, and daily self-report symptom assessment during the training course was also recorded (Supplementary Fig. S4). The chronology of exposures during the 2-week breacher training and participants' reported history of lifetime exposure to blast and TBI history is provided in Figure 1, demonstrating participant exposures ranging from tens to hundreds over a military career. A total of 60% of these 34 breachers self-reported at least one lifetime mild TBI event; however, there was no correlation between history of TBI and the number of lifetime blast exposures (p > 0.8).
FIG. 1.
(A) Protocol for operational breaching and blast exposure throughout the 10-day training course, showing pre- and post-blast exposure blood draws on days 1 and 10, respectively. (B) Distribution of number of lifetime operational blast exposures with history of self-reported TBI (red) and no history of TBI (blue). psi, pounds per square inch; TBI, traumatic brain injury. Color image is available online.
DNA methylation and transcriptional changes associated with chronic cumulative career blast exposures
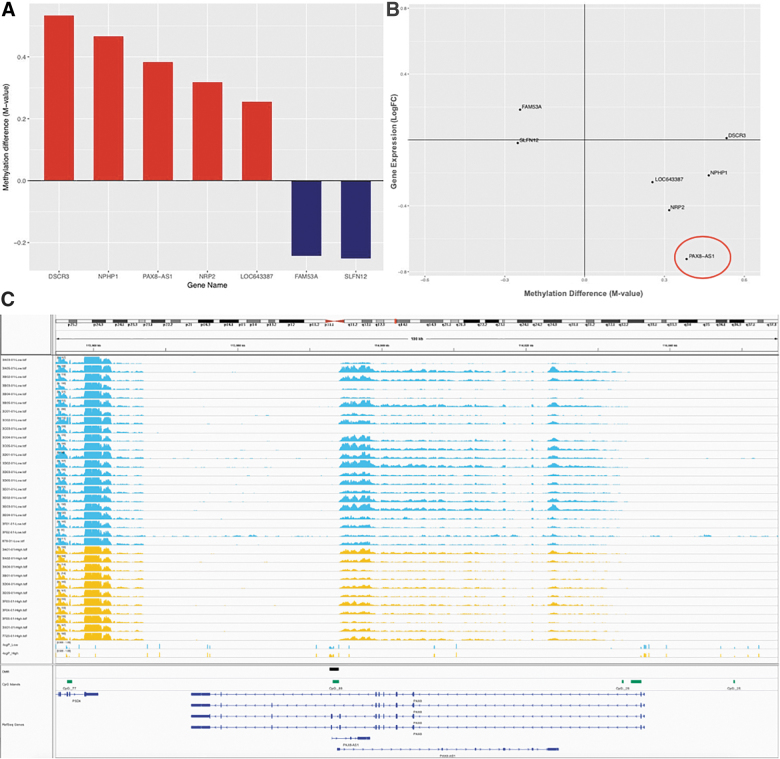
For baseline low versus high career blast exposure DNA methylation analyses, we used 34 subjects for which baseline data were available to examine whether the number of cumulative blast exposure events during a career in military service was associated with changes in transcriptional regulation. We empirically defined the low-exposure group as those with <40 reported blast exposures and the high group reporting greater numbers. Whole-genome transcriptional profiling by RNA-seq did not show significant gene expression changes between low versus high blast lifetime exposed groups after multiple testing correction (data not shown). However, DNA methylation analyses of these samples by Illumina 450K methylation microarrays identified significant methylation differences. We found 10 significantly DMRs and genes associated with cumulative blast (Fig. 2A; Supplementary Table S1).
FIG. 2.
Differentially methylated regions (DMRs) associated with cumulative blast exposure. (A) DMRs with corresponding observed methylation data, showing gain (red) and loss (blue) of DNA methylation in groups with high relative to low lifetime exposure to blast events. (B) DNA methylation and gene expression changes, with the majority of genes showing the expected anticorrelation between DNA methylation and gene expression. (C) Genome browser representation of PAX8 antisense transcript expression relative to accumulative lifetime blast exposure in low (blue) versus high (yellow) exposed breacher groups. After an anticorrelated pattern of gain of DNA methylation and repression of gene expression, we see that the low accumulative blast-exposed group shows a loss of DNA methylation DMR track in blue, whereas the high accumulative blast-exposed group shows a gain of methylation indicated in red in the DMR track with coordinated repression of gene expression, as shown per subject. It should be noted that the DMR seems to be specifically regulating the PAX8 antisense transcript given that the upstream gene, PSD4, does not seem to be impacted by the differential methylation at the DMR. AS1, antisense RNA 1; DSCR3, Down syndrome critical region gene 3; FAM53A, family with sequence similarity 53 member A; LOC643387, TAR DNA-binding protein pseudogene; LogFC, log fold-change; NPHP1, nephrocystin 1; NRP2, neuropilin 2; PAX8, paired box gene 8; PSD4, pleckstrin and Sec7 domain-containing 4. Color image is available online.
The majority of DMRs exhibited gain of methylation associated with cumulative blast exposure, with corresponding gene expression changes (Fig. 2B). The paired box gene 8 (PAX8) gene is an antisense transcript within the PAX8 gene, wherein the DMR is localized in promoter of this antisense transcript (Fig. 2C). Interestingly, gain of DNA methylation in those subjects with high cumulative blast exposure corresponds to loss of gene expression in the PAX8 antisense transcript (NR_047570; Fig. 2B,C). PAX8 transcription factor is responsible for control of expression of thyroid-specific genes involved in thyroid function, and, more recently, genome-wide association studies have implicated variants associated with PAX8 and sleep duration.32–34
DNA methylation and transcriptional changes associated with acute blast exposure
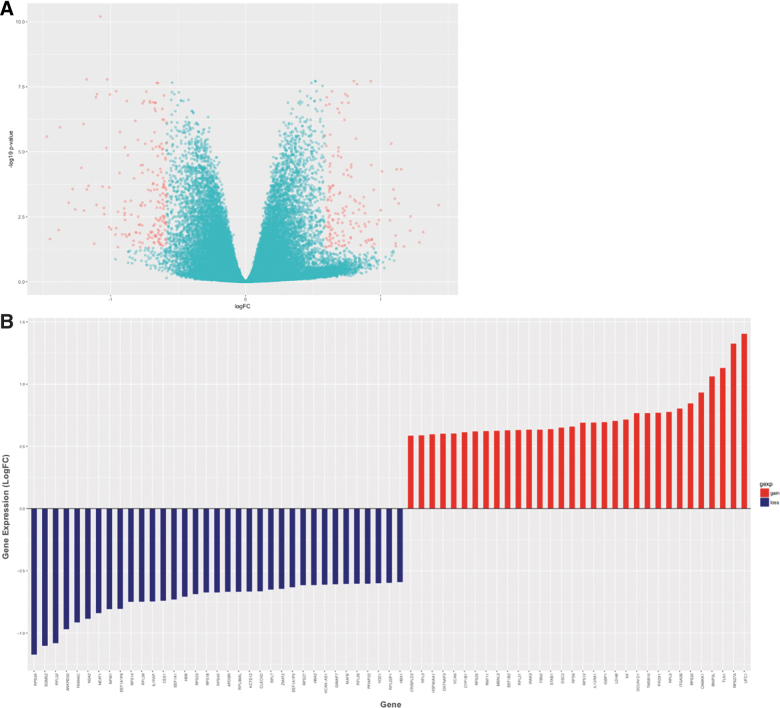
Thirty-one of the subjects had available samples for both the baseline time point and at day 10 (completion of breacher training). Comparing DNA methylation patterns pre- vs. post-blast exposure, no differences were identified. Interestingly, however, gene expression analyses of RNA-seq data revealed 6362 genes with statistically significant differential expression after multiple testing corrections (Fig. 3A; Supplementary Table S2). To investigate robust gene expression changes associated with acute blast exposure, we focused on those genes with moderate-to-large fold change (FC) in expression (≥1.5 FC) and at least moderate expression levels in pre- or post-assessments (logCPM ≥4). Using these stringent criteria, we identified 67 genes that show robust FC pre- vs. post-blast exposure (Fig. 3B). An overwhelming number of these genes are involved in ribosomal function, which were dysregulated after blast. Transcripts of ribosomal proteins constituted 30% of these genes, with additional related transcripts of chaperone proteins involved in protein biogenesis and degradation (i.e., heatshock protein HSP90, translation elongation factors EEF1B2, EEF1A1P5, and EEF1A1, ribosome biogenesis homolog [NSA2], and ubiquitins UFC1 and YOD1). Some of these loci and genes in related pathways have been previously implicated in responses to cellular stress, neurodegeneration, and TBI.35–38 Of note, ubiquitin proteins have been implicated in TBI and neurodegenerative disease, and the ubiquitin C-terminal hydrolase-L1 (UCH-L1) has been U.S. Food and Drug Administration approved as a robust blood-based biomarker for acute mild brain injury.39
FIG. 3.
Gene expression changes track molecular response to acute exposure to blast. (A) Volcano plot highlighting genes that passed multiple testing correction (6362 genes), that also showed greater than ±1.5 fold change in expression (336 genes, red). The y-axis shows the unadjusted p value. (B) Showing genes (67) with robust expression changes pre- versus post-blast training with fold change ≥|1.5| and excluding rarely expressed genes (logCPM <4). logCPM, log counts per million; LogFC, log fold-change. Color image is available online.
Blast associated physiological and psychological symptoms linked to DNA methylation and transcriptional changes
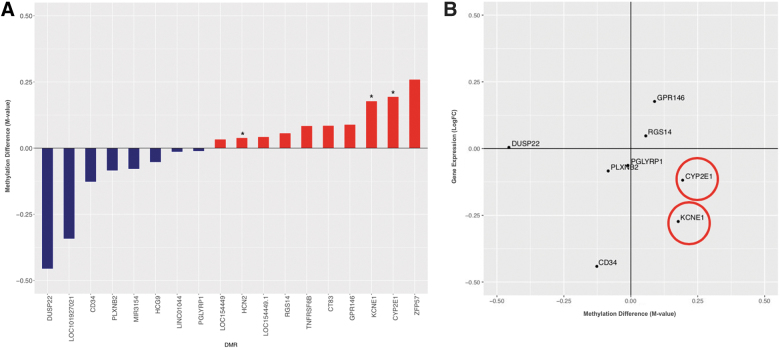
Given that daily symptom reports were ascertained from all participants, we utilized this information to track DNA methylation and gene expression changes associated with symptom reporting. Given the data sparsity, with missing symptom reporting by some participants, we performed initial symptom filtering taking a heuristic approach. Specifically, symptoms were analyzed in DNA methylation analyses if endorsed by ≥10 subjects for each comparison, as well as having a difference in average symptom score in high relative to low cumulative blast-exposed groups (with difference >0.25, defined a priori). For the baseline analysis of high versus low cumulative blast-exposed groups (day 1 symptom report), this filtering produced one symptom, tinnitus (ringing in the ear), with 11 participants reporting (Supplementary Fig. S4A). Tinnitus is a commonly reported symptom by military and veteran subjects with repeated exposure to blast.19 Although we found no significant genome-wide transcriptional changes after multiple testing correction, we report point-wise transcriptional changes for the most robustly differentially expressed loci (with FC ≥±1.5; Supplementary Table S3). We did identify genes with differential DNA methylation changes that track with reported symptoms of tinnitus in the high versus low career breaching groups (Supplementary Fig. 4A). Of these DMRs and associated loci potassium voltage-gated channel, Isk-related family, member 1 (KCNE1), cytochrome P450 family 2, subfamily E, member 1 (CYP2E1), dual specificity phosphatase 22 (DUSP22), and hyperpolarization-activated cyclic nucleotide-gated potassium channel 2 (HCN2) genes have been previously implicated in auditory functioning.40–42 To determine whether these DMRs confer transcriptional regulatory changes, we examined the corresponding gene expression levels in the low versus high breaching groups in these loci. We found that the genes KCNE1 and CYP2E1 showed the expected anticorrelated pattern of gain in DNA methylation and loss of gene expression (Fig. 4B). KCNE1 is associated with noise-related hearing loss through human genetic studies, and animal studies have also implicated CYP2E1 as associated with nitrile exposure and noise-related hearing loss in rodents.43–45
FIG. 4.
Symptom-associated DNA methylation analyses. (A) Differentially methylated genes associated with ringing in the ear/tinnitus in high relative to low blast-exposed groups, denoting genes implicated in auditory functioning by an asterisk (*). (B) Of note, KCNE1 and CYP2E1 genes follow the expected anticorrelated pattern of increase in DNA methylation with decrease in gene expression. CYP2E1, cytochrome P450 family 2 subfamily E member 1; DUSP12, dual specificity phosphatase 12; GPR148, G-protein-coupled receptor 148; KCNE1, potassium voltage-gated channel subfamily E member 1; LogFC, log fold-change; PGLYRP1, peptidoglycan recognition protein 1; PLXNB2, plexin B2; RGS14, regulator of G-protein signaling 14. Color image is available online.
For the pre-post blast exposure analysis, we accounted for the observation that the breachers had a higher exposure to blast on day 7 (average exposure of ∼12 pounds per square inch [psi]; Fig. 1A), well beyond the 4-psi exposure level used as a safety threshold on most training ranges.46 Typical exposures in this type of training are <1 psi.47 For consideration of the large-magnitude blast exposure we recorded, we took the average symptom levels reported in days 1–6 and compared with the average from days 7 to 10, inclusive of the seventh day high-magnitude blast event. Similarly, for pre-post breaching course analyses, we analyzed symptoms that ≥10 subjects had when responding pre- vs. post-blast, where “pre-” corresponds to symptoms reported at least once during training days 2–6 and “post-” days 7–10, with higher (again >0.25) average symptom scores in “post-” vs. “pre-” days. We chose this separation interval, because the breaching cohort was exposed to a higher level of blast (maximum, ∼12 psi) during day 7 of training. Using this a priori symptom criterion, we found that headache was the most highly reported symptom pre-post blast exposure, endorsed by 18 subjects (Supplementary Fig. S4B).
In line with the pre-post DNA methylation results on the total subjects, the symptom analysis also did not show statistically significant genome-wide DMRs that tracked with reported symptoms of headache post-blast exposure, after multiple testing correction. Also, no significant transcriptional changes were detected with multiple testing correction, yet point-wise data for the most significant gene expression changes with ≥±1.5 FC are reported in supplementary data (Supplementary Table S4).
Inflammatory markers of acute blast exposure
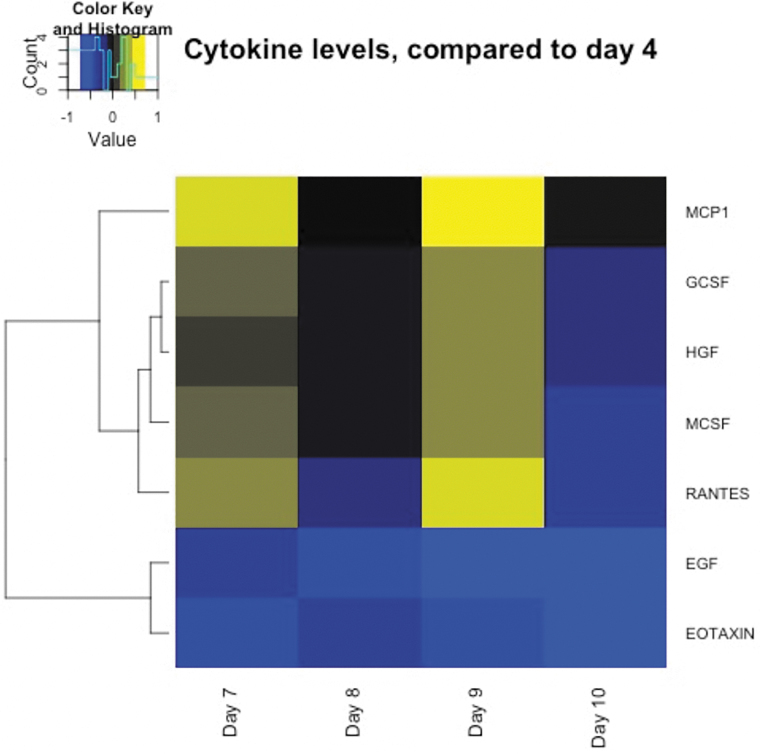
Assaying cytokine levels across the training course allows us to determine how repeated exposure to blast and breaching environment induces perturbations in levels of cytokine proteins. Data were available for 32 subjects. Cytokine measures on day 4 were used as a reference, because by this time the participants had acclimated to the training environment and had no exposure to high explosive blast or physical exertions on that day. Comparatively, days 7–10 were considered post-blast exposure days respectively. We contrasted cytokine levels for each of the post-blast days (7–10) to those from the reference (day 4). Comparing cytokine levels pre-post exposure, we found significant elevation in 5 of 63 cytokines (monocyte chemoattractant protein 1 [MCP1], granulocyte colony-stimulating factor [GCSF], hepatocyte growth factor [HGF], macrophage colony-stimulating factor [MCSF], and regulated upon activation normal T-cell expressed and secreted [RANTES]) associated with acute blast exposure (Fig. 5; p ≤ 0.05 corrected for multiple testing). These data show that changes in cytokines track acute exposure to blast, as indicated by the observed effect sizes depicted in Figure 5 (with effect sizes for all cytokines shown in Supplementary Fig. S5; Supplementary Table S5).
FIG. 5.
Heatmap showing significant changes in cytokine level tracking with blast exposure (days 7 and 9) using day 4 (no exposure to blast activity) as the reference (Bonferroni corrected). EGF, epidermal growth factor; GCSF, granulocyte colony-stimulating factor; HGF, hepatocyte growth factor; MCP1, monocyte chemoattractant protein 1; MCSF, macrophage colony-stimulating factor; RANTES, regulated upon activation normal T-cell expressed and secreted. Color image is available online.
Discussion
In the present study, we have taken a multi-modal omics approach to elucidate changes in inflammatory, epigenetic, and transcriptional profiles of military breachers following a 10-day breacher operations training course. We obtained DNA methylation, RNA-seq, and chemokine/cytokine data for military subjects before and after blast exposure in order to identify changes in DNA methylation, transcriptional, and inflammatory processes, which could serve as potential acute and chronic biomarkers of blast exposure in neurotrauma patients. Cumulative blast exposure history was used in conjunction with epigenetic and transcriptional data in order to determine the extent to which low or high instances of exposure to repeated blast impacts physiological responsivity. Further, self-reported symptom information was used in order to identify novel gene associations with blast-related symptomology.
A major limitation of the study is that DNA methylation and gene expression changes detected may not be reflective of acute changes associated with blast exposure, attributable to the blood collection end point on day 10. Although this likely has a bigger effect in regard to gene expression rather than DNA methylation, future studies would require specimen collection directly after blast exposure training sessions in order to more accurately assess transient changes in methylation and transcriptional processes. An additional limitation of the study is the lack of longitudinal data, for both genetic and symptom-based studies, on the breacher participants. Increased collection points several weeks or months after blast exposure training would allow for a better understanding of the dynamics and long-term effects of blast exposure on the epigenetic, transcriptional, and symptom-based profiles in blast-exposed individuals. Further, we acknowledge that our study lacks representation of both sexes, for the present cohort involved only male participants; thus, findings may not be generalizable to females.
The primary findings of our study suggest that DNA methylation and gene expression are modulated by blast exposure in military personnel after blast exposure training programs. Although we were unable to detect acute changes in DNA methylation pre- vs. post-blast exposure, we show that high cumulative blast exposure history alters DNA methylation patterns relative to subjects with lower blast exposure throughout their military careers. Most notably, these DNA methylation changes associated with cumulative exposures generally conferred functional changes in gene expression. Specifically, increase in DNA methylation in the promoter region of the PAX8 antisense transcript in breachers with high cumulative blast exposure history relative to those with low lifetime blast exposure (Fig. 2A) conferred a decrease in PAX8 gene expression (Fig. 2B). This is potentially important mechanistically, because the PAX8 transcription factor is involved in control of the expression of thyroid-specific genes and thyroid development,33,48 and it has recently been associated with alterations in sleep duration.32,34
The association between PAX8 and sleep duration is particularly relevant, for sleep dysregulation is reported in our breacher symptom data (Supplementary Fig. S4) and in both active duty service members and U.S. Veterans.49,50 Reciprocally, sleep dysregulation has been shown to impact both metabolic and endocrine function,51 possibly linking PAX8 as a modulator between sleep dysregulation and endocrine processes.
This finding associating high lifetime cumulative blast exposure to changes in PAX8 antisense transcript methylation (and its respective association with changes in sleep duration) is important because sleep disturbances are reported in military warfighters in both active duty and post-deployment settings.49,50,52 Sleep disturbances lead to immediate impairments in cognition and alertness, potentially resulting in accidents or death,53 and are also associated with mental health outcomes, including post-traumatic stress disorder and depression, in military populations.49,52 While we hypothesize that the changes in DNA methylation of PAX8 represent more stable marks as chronic biosignatures of lifetime blast exposure history, it is also possible, however, that abnormal sleep patterns inherent in military operations54 contribute to changes in DNA methylation reported here. Although detailed information regarding previous deployment or special forces history for the study participants is not available, we recognize that as part of military service, the participants in the present study may have chronic sleep disturbances.50,54
Such chronic sleep problems may have long-term effects and associated aberrant DNA methylation changes related to circadian regulation. For example, previous studies have shown that poor sleep quality and duration can induce DNA methylation changes in core circadian rhythm genes among shift workers and nurses.55,56 The findings in the present study related to increased methylation of PAX8 are in line with these findings, where cumulative blast may also be compounded by chronic sleep problems in the military environment.
Further, acute transcriptional alteration post-blast exposure in the KCTD12 gene was previously shown to be involved in circadian regulation57 and was also detected in this study. Potassium channel tetramerization domain-containing protein (KCTD) 12, an auxiliary subunit of the gamma-aminobutyric acid (GABA)B receptor gene, has been shown to both increase desensitization and slow onset of GABAB G-protein-coupled receptor responses.58,59 Previous animal studies by Cathomas and colleagues generated Kctd12 null mutant (Kctd12–/–) and heterozygous (Kctd12+/–) mice in order to determine whether behavioral and neurobiological observations could be made that are in line with the endophenotypes observed in psychiatric populations with KCTD12 perturbations.57 Notably, Kctd–/– and Kctd+/– mice show increased electrical excitability of CA1 pyramidal neurons,57 further implicating KCTD12 in mediating synaptic transmission and neuropsychiatric endophenotypes.
Phenotypically, mice are typically inactive during the light phase of their circadian rhythm cycle60; however, Kctd12+/– mice exhibit increased activity during the inactive (light) phase of the circadian cycle,57 suggesting that downregulation of KCTD12 modulates circadian rhythm cycles. Interestingly, we also found KCTD12 as significantly downregulated pre-post blast exposure. Our RNA-seq findings in conjunction with our DNA methylation findings further associate the effects of BOP exposure with biological/molecular perturbations potentially resulting in sleep dysregulation in our military warfighters.
Further, RNA-seq data revealed 67 genes (32 upregulated, 35 downregulated) that had robust changes in gene expression (fold expression changes ≥1.5) after exposure to blast (Fig. 3B). We identified a subset of these 67 significantly differentially expressed genes that are involved in cell-cell adhesion, fibrosis, and accumulation of extracellular matrix (ECM) proteins. Specifically, VCAN encodes the proteoglycan, versican, which is one of the primary proteins that makes up the ECM.61 FBN2 regulates processes related to elastic fiber assembly, and both TLN1 and DSC2 are implicated in cell-cell adhesion.62–64 It may be posited that increased expression of these genes is a protective mechanism after blast exposure in order to allow cells to potentially better resist the harmful effects of mechanical stress.
We speculate that the notable increases in expression in that cysteine-rich secretory protein LCCL domain-containing 2 (CRISPLD2) and stabilin-1 (STAB-1), genes involved in angiogenesis and lung morphogenesis, are likely modulated in response to blast exposure rather than related to respiratory processes. Decrease in expression of HBA1, HBA2, and HBB (hemoglobin subunit alpha 1, hemoglobin subunit alpha 2, and hemoglobin subunit beta, respectively; Fig. 3B) may suggest that the increase in expression of angiogenesis-related genes could be associated to the body's injury response rather than physiological oxygen needs. These gene expression findings were corroborated by GSEA, where positive enrichment in gene sets related to vasculature development, blood vessel morphogenesis, wound healing, and cortical cytoskeleton organization were also observed (Supplementary Table S6).
Further, we observed CRISPLD2 has been shown to be immune responsive, in that patients with asthma, chronic inflammation in the lungs, exhibit significant expression levels of CRISPLD2 after glucocorticoid treatment.65 Glucocorticoids (GCs) are anti-inflammatory, highlighting that anti-inflammatory actions in the lung by GCs occurred in conjunction with increases in CRISPLD2 gene expression. Himes and colleagues investigated this further and found CRISPLD2 to be an immune-responsive gene that increased in response to proinflammatory cytokine interleukin (IL)-1ß.65 We identified an increase in CRISPLD2 expression in our pre- vs. post-blast exposure gene expression analyses (Fig. 3B), highlighting the presence of an inflammatory response after exposure to BOP waves in our study. We further observe inflammation-related gene expression in the downregulation of KCTD12 (Fig. 3B), which was shown to be under expressed in a genome-wide expression microarray study by Miller and colleagues investigating chronic stress within caregivers of brain cancer patients versus controls,66 thus indicating a repressive effect of chronic stress on KCTD12 expression.
Additionally, other inflammatory pathways enriched in our gene expression data set include the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)-mediated TNF-α signaling gene hallmark set, with the highest normalized enrichment score (Supplementary Table S7). The NF-κB pathway responds to elevation of proinflammatory cytokine TNF-α, and further NF-κB activation is known to occur in both acute inflammatory responses and in chronic inflammatory diseases.67–69 Acute changes in circulating inflammatory cytokines after blast exposure were also observed in our assessment of serum cytokine levels across multiple time points of the breacher operations training course.
We observed an increase in MCP-1, GCSF, HGF, MCSF (CSF-1), and RANTES on days of blast exposure (Fig. 5; days 7 and 9). Notably, MCP-1 (CCL-2) has been shown to be involved in neuron-immune cell interactions and, more specifically, is responsible for macrophage movement into peripheral nerves, microglia activation, and recently has been implicated as modulated by the transcription factor, activating transcription factor 3, after TBI.70–73 Generally, these cytokines play a role in the recruitment of white blood cells to injured areas,74 which supports previous observations on inflammatory response after blast exposure, even in short (day to day) time frames.75–77
Moreover, blast-associated physiological and psychological symptom analyses in participants with low versus high cumulative blast exposure revealed that tinnitus was the primary symptom present using daily symptom reports throughout the breaching operations course. This finding is not surprising because not only is tinnitus a commonly reported symptom by military and veteran populations, but it is, in fact, the most prevalent service-connected disability of all Veterans Benefit Administration compensation recipients, according to the 2018 Veterans Affairs (VA) Annual Benefits Report.78 DNA methylation and transcriptional profiles were compared in the low versus high cumulative blast groups, and although we identified no significant genome-wide transcriptional changes after multiple testing correction, we did identify genes with differential DNA methylation changes that track with the reported symptom of tinnitus in the low versus high career blast groups (Fig. 4A).
Two of the genes, KCNE1 and CYP2E1, showed the expected inversely correlated pattern of increase in DNA methylation and decrease in gene expression. KCNE1 is a potassium channel that has been previously implicated in noise-induced hearing loss (NIHL) and tinnitus through genetic studies43,44 and has been shown to play a critical role in K+ recycling in the endolymph of the inner ear.79 CYP2E1 metabolizes acrylonitriles, which have been demonstrated to promote NIHL through oxidative stress mechanisms.45
We also observed changes in hearing-related genes in our pre-post blast exposure gene expression analysis, in that KCTD12 which was robustly downregulated has been associated with tinnitus.80 This is of particular interest to the VA community because tinnitus is a commonly reported symptom by military and veteran personnel with repeated exposure to blast. Our knowledge of the neurobiological mechanisms behind auditory and vestibular injuries in our military warfighters is limited, but particularly relevant given that tinnitus is present in 11% of OIF and OEF veterans and has been demonstrated in active duty medical records to be the primary clinical risk for personnel in military occupational specialties associated with blast exposure.81 Our results suggest that these hearing-related genes may play a direct role in auditory processing and the perception of sound.
Overall, we demonstrate that blast exposure, both acutely and in accumulation, is capable of altering gene expression and DNA methylation patterns, respectively. Further epigenetic, transcriptional, and inflammatory studies are required to deduce the molecular biosignatures of blast and the associated symptoms.
Conclusion
Given that DNA methylation represents highly stable, long-lasting marks, these are likely representative of cumulative blast exposures, whereas changes in gene transcription are facile and represent acute or proximal exposure to blast events. Hence, it is not surprising that we did not detect significant gene expression changes at baseline associated with lifetime blast exposure. We did, however, detect changes in gene expression directly after blast exposure, representing a rapid molecular response to BOP. The present study suggests that physiological responsivity to different environmental factors, in this case BOP exposure, may be captured by differing biomarkers—with DNA methylation encapsulating the chronic cumulative exposures and inflammatory and RNA transcription the acute response. This systems-based approach allows for context-dependent investigations, allowing for discovery of molecular perturbations and symptomatology, both proximally to exposure to BOP and distally across the life span.
Supplementary Material
Acknowledgments
The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as representing the true views, position, or policy of the U.S. government, the Department of the Army, Department of the Navy, the Defense and Veterans Brain Injury Center, the Department of Defense, or the Department of Veterans Affairs. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25. The study protocol was approved by the Walter Reed Army Institute of Research (WRAIR) and the Naval Medical Research Center (NMRC) Institutional Review Boards in compliance with all applicable federal regulations governing the protection of human subjects. Some of the authors are military service members or federal/contracted employees of the United States government. This work was prepared as part of their official duties. Title 17 U.S.C. 105 provides that copyright protection under this title is not available for any work of the United States government. We thank the instructors and students from the Urban Mobility Breaching Course at site 3 for volunteering to participate in this study; the WRAIR field research team for collecting biological samples and performance data as well as formatting the data for analytical use; and Dr. Angela Boutté for her critical review and editorial comments of the manuscript. We thank the Stanford Human Immune Monitoring Center (HIMC) for running the Luminex assay. The authors thank the New York Genome Center for performing the DNA methylation and RNA sequencing assays.
Funding Information
This work was supported by the U.S. Army Medical Research and Development Command/USAMRDC and the Veterans Affairs Office of Research and Development. This research was supported, in part, by an appointment to the Research Participation Program at the Walter Reed Army Institute of Research (WRAIR) administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and Medical Research and Development Command/USAMRDC. The WRAIR and NMRC support further derives from Joint Program Committee Five intramural awards, The Development of Blast Exposure Standards, Evaluation of the Effects of High Level Overpressure (8+ psi) on Cognitive Performance, Brain Blood Biomarkers and Symptom Reporting, and Environmental Sensors in Training (ESiT). Dr. Haghighi is the recipient of a Research Career Scientist Award (#1IK6CX002074) from the United States Department of Veterans Affairs. Dr. Haghighi's research is supported by Veterans Affairs Merit Grants RX001705, CX001395, BX003794, and CX001728.
Author Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- 1. Tsao J.W., Alphonso A.L., Griffin S.C., Yurkiewicz I.R., and Ling G.S.F. (2013). Neurology and the military: five new things. Neurol. Clin. Pract. 3, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mathews Z.R., and Koyfman A. (2015). Blast Injuries. J. Emerg. Med. 49, 573–587 [DOI] [PubMed] [Google Scholar]
- 3. Edwards D.S., McMenemy L., Stapley S.A., Patel H.D.L., and Clasper J.C. (2016). 40 years of terrorist bombings—a meta-analysis of the casualty and injury profile. Injury 47, 646–652 [DOI] [PubMed] [Google Scholar]
- 4. Public Law No: 115-232: John S. McCain National Defense Authorization Act for Fiscal Year 2019. U.S. Government Publishing Office: Washington, DC
- 5. Kuriakose M., Rama Rao K.V., Younger D., and Chandra N. (2018). Temporal and spatial effects of blast overpressure on blood-brain barrier permeability in traumatic brain injury. Sci. Rep. 8, 8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schouten J.W. (2007). Neuroprotection in traumatic brain injury: a complex struggle against the biology of nature. Curr. Opin. Crit. Care 13, 134–142 [DOI] [PubMed] [Google Scholar]
- 7. Ling G., Bandak F., Armonda R., Grant G., and Ecklund J. (2009). Explosive blast neurotrauma. J. Neurotrauma 26, 815–825 [DOI] [PubMed] [Google Scholar]
- 8. Ling G.S., and Ecklund J.M. (2011). Traumatic brain injury in modern war. Curr. Opin. Anaesthesiol. 24, 124–130 [DOI] [PubMed] [Google Scholar]
- 9. Shanker T. (2007). Iraqi bombers thwart efforts to shield G.I.s, in: The New York Times
- 10. Murthy J.M., Chopra J.S., and Gulati D.R. (1979). Subdural hematoma in an adult following a blast injury. Case report. J. Neurosurg. 50, 260–261 [DOI] [PubMed] [Google Scholar]
- 11. Cernak I., Wang Z., Jiang J., Bian X., and Savic J. (2001). Cognitive deficits following blast injury-induced neurotrauma: possible involvement of nitric oxide. Brain Inj. 15, 593–612 [DOI] [PubMed] [Google Scholar]
- 12. Mayorga M.A. (1997). The pathology of primary blast overpressure injury. Toxicology 121, 17–28 [DOI] [PubMed] [Google Scholar]
- 13. Okie S. (2005). Traumatic brain injury in the war zone. N. Engl. J. Med. 352, 2043–2047 [DOI] [PubMed] [Google Scholar]
- 14. Valiyaveettil M., Alamneh Y., Miller S.A., Hammamieh R., Wang Y., Arun P., Wei Y., Oguntayo S., and Nambiar M.P. (2012). Preliminary studies on differential expression of auditory functional genes in the brain after repeated blast exposures. J. Rehabil. Res. Dev. 49, 1153–1162 [DOI] [PubMed] [Google Scholar]
- 15. Valiyaveettil M., Alamneh Y.A., Miller S.A., Hammamieh R., Arun P., Wang Y., Wei Y., Oguntayo S., Long J.B., and Nambiar M.P. (2013). Modulation of cholinergic pathways and inflammatory mediators in blast-induced traumatic brain injury. Chem. Biol. Interact. 203, 371–375 [DOI] [PubMed] [Google Scholar]
- 16. Ruff R.L., Ruff S.S., and Wang X.F. (2008). Headaches among Operation Iraqi Freedom/Operation Enduring Freedom veterans with mild traumatic brain injury associated with exposures to explosions. J. Rehabil. Res. Dev. 45, 941–952 [DOI] [PubMed] [Google Scholar]
- 17. Huber B.R., Meabon J.S., Martin T.J., Mourad P.D., Bennett R., Kraemer B.C., Cernak I., Petrie E.C., Emery M.J., Swenson E.R., Mayer C., Mehic E., Peskind E.R., and Cook D.G. (2013). Blast exposure causes early and persistent aberrant phospho- and cleaved-tau expression in a murine model of mild blast-induced traumatic brain injury. J. Alzheimers Dis. 37, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ling G.S.F., and Marshall S.A. (2008). Management of traumatic brain injury in the intensive care unit. Neurol. Clin. 26, 409–426 [DOI] [PubMed] [Google Scholar]
- 19. Carr W., Polejaeva E., Grome A., Crandall B., LaValle C., Eonta S.E., and Young L.A. (2015). Relation of repeated low-level blast exposure with symptomology similar to concussion. J. Head Trauma Rehabil. 30, 47–55 [DOI] [PubMed] [Google Scholar]
- 20. Gill J., Cashion A., Osier N., Arcurio L., Motamedi V., Dell K.C., Carr W., Kim H.-S., Yun S., Walker P., Ahlers S., LoPresti M., and Yarnell A. (2017). Moderate blast exposure alters gene expression and levels of amyloid precursor protein. Neurol. Genet. 3, e186–e186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pun P., Kan E.M., Salim A., Li Z.H., Ng K.C., Moochhala S., Ling E.-A., Tan M.H., and Lu J. (2011). Low level primary blast injury in rodent brain. Front. Neurol. 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bibikova M., Lin Z., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Goldmann T., Seifart C., Jiang W., Barker D.L., Chee M.S., Floros J., and Fan J.B. (2006). High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 16, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. R Development Core Team. (2014). R: a language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria [Google Scholar]
- 24. Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y., and Zhang J. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., and Irizarry R.A. (2014). Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pedersen B.S., Schwartz D.A., Yang I.V., and Kechris K.J. (2012). Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics 28, 2986–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang L., Wang S. and Li W. (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 [DOI] [PubMed] [Google Scholar]
- 30. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., and Tamayo P. (2015). The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 1, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gottlieb D.J., Hek K., Chen T.H., Watson N.F., Eiriksdottir G., Byrne E.M., Cornelis M., Warby S.C., Bandinelli S., Cherkas L., Evans D.S., Grabe H.J., Lahti J., Li M., Lehtimäki T., Lumley T., Marciante K.D., Pérusse L., Psaty B.M., Robbins J., Tranah G.J., Vink J.M., Wilk J.B., Stafford J.M., Bellis C., Biffar R., Bouchard C., Cade B., Curhan G.C., Eriksson J.G., Ewert R., Ferrucci L., Fülöp T., Gehrman P.R., Goodloe R., Harris T.B., Heath A.C., Hernandez D., Hofman A., Hottenga J.J., Hunter D.J., Jensen M.K., Johnson A.D., Kähönen M., Kao L., Kraft P., Larkin E.K., Lauderdale D.S., Luik A.I., Medici M., Montgomery G.W., Palotie A., Patel S.R., Pistis G., Porcu E., Quaye L., Raitakari O., Redline S., Rimm E.B., Rotter J.I., Smith A.V., Spector T.D., Teumer A., Uitterlinden A.G., Vohl M.C., Widen E., Willemsen G., Young T., Zhang X., Liu Y., Blangero J., Boomsma D.I., Gudnason V., Hu F., Mangino M., Martin N.G., O'Connor G.T., Stone K.L., Tanaka T., Viikari J., Gharib S.A., Punjabi N.M., Räikkönen K., Völzke H., Mignot E., and Tiemeier H. (2015). Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol. Psychiatry 20, 1232–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mansouri A., Chowdhury K., and Gruss P. (1998). Follicular cells of the thyroid gland require Pax8 gene function. Nat. Genet. 19, 87–90 [DOI] [PubMed] [Google Scholar]
- 34. Jones S.E., Tyrrell J., Wood A.R., Beaumont R.N., Ruth K.S., Tuke M.A., Yaghootkar H., Hu Y., Teder-Laving M., Hayward C., Roenneberg T., Wilson J.F., Del Greco F., Hicks A.A., Shin C., Yun C.-H., Lee S.K., Metspalu A., Byrne E.M., Gehrman P.R., Tiemeier H., Allebrandt K.V., Freathy R.M., Murray A., Hinds D.A., Frayling T.M., and Weedon M.N. (2016). Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 12, e1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Beckelman B.C., Zhou X., Keene C.D., and Ma T. (2016). Impaired eukaryotic elongation Factor 1A expression in Alzheimer's disease. Neurodegener. Dis. 16, 39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beckelman B.C., Day S., Zhou X., Donohue M., Gouras G.K., Klann E., Keene C.D., and Ma T. (2016). Dysregulation of elongation factor 1A expression is correlated with synaptic plasticity impairments in Alzheimer's disease. J. Alzheimers Dis. 54, 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Doig J., Griffiths L.A., Peberdy D., Dharmasaroja P., Vera M., Davies F.J.C., Newbery H.J., Brownstein D., and Abbott C.M. (2013). In vivo characterization of the role of tissue-specific translation elongation factor 1A2 in protein synthesis reveals insights into muscle atrophy. FEBS J. 280, 6528–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Newbery H.J., Gillingwater T.H., Dharmasaroja P., Peters J., Wharton S.B., Thomson D., Ribchester R.R., and Abbott C.M. (2005). Progressive loss of motor neuron function in wasted mice: effects of a spontaneous null mutation in the gene for the eEF1A2 translation factor. J. Neuropathol. Exp. Neurol. 64, 295–303 [DOI] [PubMed] [Google Scholar]
- 39. Welch R.D., Ayaz S.I., Lewis L.M., Unden J., Chen J.Y., Mika V.H., Saville B., Tyndall J.A., Nash M., Buki A., Barzo P., Hack D., Tortella F.C., Schmid K., Hayes R.L., Vossough A., Sweriduk S.T., and Bazarian J.J. (2016). Ability of serum glial fibrillary acidic protein, ubiquitin C-terminal hydrolase-L1, and S100B to differentiate normal and abnormal head computed tomography findings in patients with suspected mild or moderate traumatic brain injury. J. Neurotrauma 33, 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen H., Liu W., Geng Q., Li H., Lu M., Liang P., Zhang B., Yamoah E.N., and Lv P. (2018). Age-dependent up-regulation of HCN channels in spiral ganglion neurons coincide with hearing loss in mice. Front. Aging Neurosci. 10, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim Y.H., and Holt J.R. (2013). Functional contributions of HCN channels in the primary auditory neurons of the mouse inner ear. J. Gen. Physiol. 142, 207–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramakrishnan N.A., Drescher M.J., Khan K.M., Hatfield J.S., and Drescher D.G. (2012). HCN1 and HCN2 proteins are expressed in cochlear hair cells: HCN1 can form a ternary complex with protocadherin 15 CD3 and F-actin-binding filamin A or can interact with HCN2. J. Biol. Chem. 287, 37628–37646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van Laer L., Carlsson P.-I., Ottschytsch N., Bondeson M.-L., Konings A., Vandevelde A., Dieltjens N., Fransen E., Snyders D., Borg E., Raes A., and Van Camp G. (2006). The contribution of genes involved in potassium-recycling in the inner ear to noise-induced hearing loss. Hum. Mutat. 27, 786–795 [DOI] [PubMed] [Google Scholar]
- 44. Pawełczyk M., Rajkowska E., Kotyło P., Dudarewicz A., Van Camp G., and Śliwińska-Kowalska M. (2012). Analysis of inner ear potassium recycling genes as potential factors associated with tinnitus. Int. J. Occup. Med. Environ. Health 25, 356–364 [DOI] [PubMed] [Google Scholar]
- 45. Fechter L.D., Gearhart C., and Shirwany N.A. (2004). Acrylonitrile potentiates noise-induced hearing loss in rat. J. Assoc. Res. Otolaryngol. 5, 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carr W., Dell K.C., Yanagi M.A., Hassan D.M., and LoPresti M.L. (2017). Perspectives on repeated low-level blast and the measurement of neurotrauma in humans as an occupational exposure risk. Shock Waves 27, 829–836 [Google Scholar]
- 47. Carr W., Stone J.R., Walilko T., Young L.A., Snook T.L., Paggi M.E., Tsao J.W., Jankosky C.J., Parish R.V., and Ahlers S.T. (2016). Repeated low-level blast exposure: a descriptive human subjects study. Mil. Med. 181, 5 Suppl., 28–39 [DOI] [PubMed] [Google Scholar]
- 48. Ruiz-Llorente S., Carrillo Santa de Pau E., Sastre-Perona A., Montero-Conde C., Gómez-López G., Fagin J.A., Valencia A., Pisano D.G., and Santisteban P. (2012). Genome-wide analysis of Pax8 binding provides new insights into thyroid functions. BMC Genomics 13, 147–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seelig A.D., Jacobson I.G., Smith B., Hooper T.I., Boyko E.J., Gackstetter G.D., Gehrman P., Macera C.A., and Smith T.C.; Millennium Cohort Study Team. (2010). Sleep patterns before, during, and after deployment to Iraq and Afghanistan. Sleep 33, 1615–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luxton D.D., Greenburg D., Ryan J., Niven A., Wheeler G., and Mysliwiec V. (2011). Prevalence and impact of short sleep duration in redeployed OIF soldiers. Sleep 34, 1189–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spiegel K., Leproult R., and Van Cauter E. (1999). Impact of sleep debt on metabolic and endocrine function. Lancet 354, 1435–1439 [DOI] [PubMed] [Google Scholar]
- 52. Mysliwiec V., McGraw L., Pierce R., Smith P., Trapp B., and Roth B.J. (2013). Sleep disorders and associated medical comorbidities in active duty military personnel. Sleep 36, 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Drake C., Roehrs T., Breslau N., Johnson E., Jefferson C., Scofield H., and Roth T. (2010). The 10-year risk of verified motor vehicle crashes in relation to physiologic sleepiness. Sleep 33, 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peterson A.L., Goodie J.L., Satterfield W.A., and Brim W.L. (2008). Sleep disturbance during military deployment. Mil. Med. 173, 230–235 [DOI] [PubMed] [Google Scholar]
- 55. Cedernaes J., Osler M.E., Voisin S., Broman J.-E., Vogel H., Dickson S.L., Zierath J.R., Schiöth H.B., and Benedict C. (2015). Acute sleep loss induces tissue-specific epigenetic and transcriptional alterations to circadian clock genes in men. J. Clin. Endocrinol. Metab. 100, E1255–E1261 [DOI] [PubMed] [Google Scholar]
- 56. Bukowska-Damska A., Reszka E., Kaluzny P., Wieczorek E., Przybek M., Zienolddiny S., and Peplonska B. (2017). Sleep quality and methylation status of core circadian rhythm genes among nurses and midwives. Chronobiol. Int. 34, 1211–1223 [DOI] [PubMed] [Google Scholar]
- 57. Cathomas F., Stegen M., Sigrist H., Schmid L., Seifritz E., Gassmann M., Bettler B., and Pryce C.R. (2015). Altered emotionality and neuronal excitability in mice lacking KCTD12, an auxiliary subunit of GABAB receptors associated with mood disorders. Transl. Psychiatry 5, e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schwenk J., Metz M., Zolles G., Turecek R., Fritzius T., Bildl W., Tarusawa E., Kulik A., Unger A., Ivankova K., Seddik R., Tiao J.Y., Rajalu M., Trojanova J., Rohde V., Gassmann M., Schulte U., Fakler B., and Bettler B. (2010). Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature 465, 231–235 [DOI] [PubMed] [Google Scholar]
- 59. Seddik R., Jungblut S.P., Silander O.K., Rajalu M., Fritzius T., Besseyrias V., Jacquier V., Fakler B., Gassmann M., and Bettler B. (2012). Opposite effects of KCTD subunit domains on GABA(B) receptor-mediated desensitization. J. Biol. Chem. 287, 39869–39877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peirson S.N., Brown L.A., Pothecary C.A., Benson L.A., and Fisk A.S. (2018). Light and the laboratory mouse. J. Neurosci. Methods 300, 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu Y.J., Pierre D.P.L.A., Wu J., Yee A.J. and Yang B.B. (2005). The interaction of versican with its binding partners. Cell Res. 15, 483–494 [DOI] [PubMed] [Google Scholar]
- 62. Frederic M.Y., Monino C., Marschall C., Hamroun D., Faivre L., Jondeau G., Klein H.G., Neumann L., Gautier E., Binquet C., Maslen C., Godfrey M., Gupta P., Milewicz D., Boileau C., Claustres M., Beroud C., and Collod-Beroud G. (2009). The FBN2 gene: new mutations, locus-specific database (Universal Mutation Database FBN2), and genotype-phenotype correlations. Hum. Mutat. 30, 181–190 [DOI] [PubMed] [Google Scholar]
- 63. Nieswandt B., Moser M., Pleines I., Varga-Szabo D., Monkley S., Critchley D., and Fässler R. (2007). Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J. Exp. Med. 204, 3113–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dubash A.D., and Green K.J. (2011). Desmosomes. Curr. Biol. 21, R529–R531 [DOI] [PubMed] [Google Scholar]
- 65. Himes B.E., Jiang X., Wagner P., Hu R., Wang Q., Klanderman B., Whitaker R.M., Duan Q., Lasky-Su J., Nikolos C., Jester W., Johnson M., Panettieri R.A. Jr., Tantisira K.G., Weiss S.T., and Lu Q. (2014). RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One 9, e99625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miller G.E., Chen E., Sze J., Marin T., Arevalo J.M., Doll R., Ma R., and Cole S.W. (2008). A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biol. Psychiatry 64, 266–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lawrence T., Gilroy D.W., Colville-Nash P.R., and Willoughby D.A. (2001). Possible new role for NF-κB in the resolution of inflammation. Nat. Med. 7, 1291. [DOI] [PubMed] [Google Scholar]
- 68. Greten F.R., Arkan M.C., Bollrath J., Hsu L.-C., Goode J., Miething C., Göktuna S.I., Neuenhahn M., Fierer J., Paxian S., Van Rooijen N., Xu Y., O'Cain T., Jaffee B.B., Busch D.H., Duyster J., Schmid R.M., Eckmann L., and Karin M. (2007). NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell 130, 918–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lawrence T. (2009). The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1, a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kohl B., Wessig C., Groh J., Greeske J., Heinl K., Fischer S., and Martini R. (2010). Attenuation of MCP-1/CCL2 expression ameliorates neuropathy in a mouse model for Charcot–Marie–Tooth 1X. Hum. Mol. Genet. 19, 3530–3543 [DOI] [PubMed] [Google Scholar]
- 71. Kwon M.J., Shin H.Y., Cui Y., Kim H., Thi A.H.L., Choi J.Y., Kim E.Y., Hwang D.H., and Kim B.G. (2015). CCL2 mediates neuron-macrophage interactions to drive proregenerative macrophage activation following preconditioning injury. J. Neurosci. 35, 15934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Muessel M.J., Klein R.M., Wilson A.M., and Berman N.E.J. (2002). Ablation of the chemokine monocyte chemoattractant protein-1 delays retrograde neuronal degeneration, attenuates microglial activation, and alters expression of cell death molecules. Mol. Brain Res. 103, 12–27 [DOI] [PubMed] [Google Scholar]
- 73. Förstner P., Rehman R., Anastasiadou S., Haffner-Luntzer M., Sinske D., Ignatius A., Roselli F., and Knöll B. (2018). Neuroinflammation after traumatic brain injury is enhanced in activating transcription factor 3 mutant mice. J. Neurotrauma 35, 2317–2329 [DOI] [PubMed] [Google Scholar]
- 74. Orman M.A., Ierapetritou M.G., Berthiaume F., and Androulakis I.P. (2011). The dynamics of the early inflammatory response in double-hit burn and sepsis animal models. Cytokine 56, 494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shetty A.K., Mishra V., Kodali M., and Hattiangady B. (2014). Blood brain barrier dysfunction and delayed neurological deficits in mild traumatic brain injury induced by blast shock waves. Front. Cell. Neurosci. 8, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Elder G.A., Gama Sosa M.A., De Gasperi R., Stone J.R., Dickstein D.L., Haghighi F., Hof P.R., and Ahlers S.T. (2015). Vascular and inflammatory factors in the pathophysiology of blast-induced brain injury. Front. Neurol. 6, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kamnaksh A., Ahmed F., Kovesdi E., Barry E.S., Grunberg N.E., Long J.B., and Agoston D.V. (2014). Molecular mechanisms of increased cerebral vulnerability after repeated mild blast-induced traumatic brain injury. Translational Proteomics 3, 22–37 [Google Scholar]
- 78. U.S. Department of Veterans Affairs. (2018). Annual Benefits Report: Fiscal Year 2018. U.S. Department of Veterans Affairs: Washington, D.C., pps. 1–232 [Google Scholar]
- 79. Wangemann P. (2002). K+ cycling and the endocochlear potential. Hearing Res. 165, 1–9 [DOI] [PubMed] [Google Scholar]
- 80. Sand P.G., Langguth B., Itzhacki J., Bauer A., Geis S., Cardenas-Conejo Z.E., Pimentel V., and Kleinjung T. (2012). Resequencing of the auxiliary GABA(B) receptor subunit gene KCTD12 in chronic tinnitus. Front. Syst. Neurosci. 6, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Carr W., Weber N.S., Belding J., Kelley A., da Silva U.O., Cowan D.N., Toolin C., Egnoto M., and Thomsen C.J. (2018). Epidemiologic studies of occupational blast exposure. Presented at the 25th Annual Meeting of Military Aspects of Blast and Shock, The Hague, Netherlands [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.