Abstract
Background: Systemic Sclerosis (SSc), also known as scleroderma, is an autoimmune rheumatic disease, which is clinically subdivided into two major subgroups; limited (lcSSc) and diffuse cutaneous scleroderma (dcSSc). Even though the SSc etiologies remains unclear, some HLA and non-HLA genetic variants have been associated with the disease.
Aim: This study was designed to evaluate the associations between several HLA-related genetic variants and SSc in the Greek-Cypriot population.
Methods: Forty-one SSc patients and 164 controls were genotyped at 18 selected single nucleotide polymorphisms (SNPs) using restriction fragment length polymorphism analyses, Sanger sequencing, and a multiplex SNaPshot minisequencing assay. Logistic regression analysis under the log-additive model was used to evaluate all possible associations between these SNPs and SSc; nominal statistical significance was assumed at p < 0.05.
Results: Associations of SSc with SNPs rs3117230, rs3128930, and rs3128965 within the HLA-DPB1 and HLA-DPB2 regions were observed in the Greek-Cypriot population at the level of p < 0.05. However, none of these associations survived a Bonferroni correction. The direction of the effect is consistent with the direction reported in previous studies. In addition, allele frequencies of the majority of the selected SNPs in the Greek-Cypriot population are similar to those reported in the European population.
Conclusion: This study initiates the genetic investigation of SSc in the Greek-Cypriot population, a relatively small newly investigated population. Further investigation with a larger sample size and/or additional SSc susceptibility loci may confirm the association of some of these variants with SSc in the Greek-Cypriot population that could potentially be used for predictive testing.
Keywords: systemic sclerosis, susceptibility loci, autoimmunity, population study
Introduction
Systemic sclerosis (SSc) is a rare autoimmune disease characterized by vasculopathies, inflammation, and fibrosis (Bossini-Castillo et al., 2011; Allanore et al., 2015). SSc is divided into two subgroups; limited (lcSSc) and diffused cutaneous SSc (dcSSc), based on the clinical manifestations. It occurs more frequently in women and onset is between the second and fifth decade of life (Alba et al., 2014). Although the etiopathogenesis of SSc remains unclear, it is a multifactorial disease caused by a combination of genetic, epigenetic, and environmental factors (Broen et al., 2014).
Many studies focused on the investigation of genetic factors and mechanisms that may be implicated in the triggering of the disease. Multiple genetic loci from the HLA as well as non-HLA regions have been associated with predisposition to SSc (Chairta et al., 2017). Genome-wide association studies (GWAS) and HLA studies have shown that genetic variants in the HLA-Class II region are more frequently associated with the development of SSc, as compared with variants in the HLA-Class I and III regions (Radstake et al., 2010; Allanore et al., 2011; Chairta et al., 2017). For example, the HLA-DRB1*1104 allele, which is involved in HLA-Class II region was markedly associated with SSc in the Greek population (Vlachoyiannopoulos et al., 2000).
In addition, single nucleotide polymorphisms (SNPs) in genes with different molecular functions like transient receptor potential melastatin channel genes (Oztuzcu et al., 2015), Rho/Rho-kinase (Pehlivan et al., 2016), Th17 pathway genes (Mellal et al., 2018), and the Vitamin D receptor (Li et al., 2019) were reported to be significantly associated with SSc susceptibility in the neighboring Turkish, Algerian, and Han Chinese populations, respectively.
SSc is genetically heterogeneous among populations and its prevalence in Europe ranges from 31 to 277 cases per million individuals (Silman et al., 1988; Allcock et al., 2004; Le Guern et al., 2004; Alamanos et al., 2005; Arias-Nunez et al., 2008; Lo Monaco et al., 2011). The prevalence of SSc in the Greek-Cypriot population has not been estimated and SSc susceptibility loci have not been evaluated.
Through this study, we aimed at the investigation of the Greek-Cypriot population for some already reported genetic variants that have been associated with SSc in other populations. The rationale of this study was to assess whether Greek-Cypriot patients with SSc have similar genetic susceptibility with patients from other populations, and evaluate whether some variants could be used in the future for prediction of the disease. In addition, this study initiated the collection of biomaterials and data for biobanking, as well as the prospective epidemiological investigation of SSc in our population.
Materials and Methods
Study participants
Forty-one patients with SSc were recruited for this study between February 2017 and January 2019, through their annual follow-up appointment at the Rheumatology Department of the Nicosia General Hospital. All patients fulfilled the 2013 American College of Rheumatology and the European League Against Rheumatism classification criteria of SSc (van den Hoogen et al., 2013). Clinical and serological data were recorded for all patients (Table 1). One hundred and sixty four age- and sex-matched Greek-Cypriot healthy controls were recruited. Due to the small size of the population (∼700,000 individuals), the number of recruited patients with SSc, was relatively small. Therefore, we used an increased number of control individuals per case with a ratio of 4:1, as suggested by Hong et al. (Hong and Park, 2012), to increase the statistical power of the study. This study was approved by the Cyprus National Bioethics Committee (EEBK/EΠ/2013/28, May 14, 2015 and EEBK/EΠ/2015/31, February 9, 2016) and conducted in accordance with the 1964 Declaration of Helsinki.
Table 1.
The Main Features of the Patients with Systemic Sclerosis and Healthy Control Participants
| Trait | Patients with SSc | Healthy controls |
|---|---|---|
| Number | 41 | 164 |
| Sex (female: male) | (36:5) | (144:20) |
| Age (years, mean ± SD)a | 60.10 ± 12.97 | 61.70 ± 11.56 |
| Age of onset (years, mean ± SD) | 49.05 ± 13.80 | — |
| Subtype | — | |
| lcSSc, n (%) | 20 (48.78) | — |
| dcSSc, n (%) | 21 (51.22) | — |
| Autoantibodies | — | |
| ANA+, n (%) | 39 (95.12) | — |
| ATA+, n (%) | 14 (34.15)b | — |
| ACA+, n (%) | 21 (51.22)c | — |
| Raynaud's phenomenon (%) | 41 (100) | — |
| Smoking | — | |
| Current smoker n (%) | 6 (14.63) | — |
| Past smoker n (%) | 8 (19.51) | — |
| Never n (%) | 27 (65.85) | — |
p = 0.47 (patients age vs. control age).
13 (92.86%) out of 14 patients with dcSSc.
18 (85.71%) out of 21 patients with lcSSc.
ACA, anticentromere autoantibodies; ANA, antinuclear autoantibodies; ATA, antitopoisomerase autoantibodies; dcSSc, diffused cutaneous scleroderma; lcSSc, limited cutaneous scleroderma; SD, standard deviation.
Genomic DNA extraction
DNA was extracted from whole blood samples using the DNA purification system Gentra Puregene Blood Core Kit C (Qiagen Sciences), according to the manufacturer's protocol.
SNPs selection
Eighteen SNPs previously associated with SSc in other populations, were selected (Appendix Table A1) based on our systematic review (Chairta et al., 2017).
Genotyping
SNP genotyping was performed using either Sanger sequencing, restriction fragment length polymorphism (RFLP) (BsrBI and ApoI restriction enzymes [New England Biolabs]), or a SNaPshot Multiplex Minisequencing Assay (SNaPshot Multiplex Kit [Applied Biosystem, United Kingdom]). PCR and SNaPshot primers (Metabion International, Germany) were designed using the Primer3 (available upon request). Sanger sequencing and SNaPshot multiplex minisequencing samples were analyzed on an ABI 3130xl genetic analyzer (Applied Biosystems).
Statistical analysis
Quality control (QC) check was carried out for samples and SNPs. A chi-square test was used to evaluate the deviation from Hardy–Weinberg equilibrium (HWE) in healthy control samples. SNPs deviating from HWE were excluded from further consideration (p < 0.05 in controls). Minor allele frequency (MAF) for each SNP was calculated (MAF >0.01) in the control samples. The difference in frequency distribution of SNPs between patients and healthy controls was examined using logistic regression analysis (log-additive model), having the common allele as reference.
A p-value of <0.05 was considered as nominally statistically significant, and corrected for multiple testing using Bonferroni correction for the SNPs that passed the HWE. Analyses were carried out with the R software. Odds ratio (OR) and 95% confidence intervals (CI) were calculated. Comparison of age distribution between case and control samples was performed using the independent samples t-test.
Results
Study participants
This study included 41 Greek-Cypriot unrelated patients with SSc (36 female and 5 male) with a mean ± standard deviation (SD) age of 60.10 ± 12.97 years, and 164 age- and sex-matched Greek-Cypriot unrelated healthy controls (144 female and twenty male) with a mean ± SD age of 61.70 ± 11.56. The main clinical and serological features of the patients are described in Table 1.
Association studies and statistical analysis
Eighteen SNPs were selected and investigated using three different methodologies (Fig. 1). Seventeen out of eighteen SNPs passed QC checks. SNP rs2298428 was excluded as it deviated from the HWE (Appendix Table A1). Each SNP was analyzed for association with SSc and the relevant results are shown in Table 2. Associations of SSc with three SNPs (rs3117230, p = 0.004; rs3128930, p = 0.006; rs3128965, p = 0.02) were observed in the Greek-Cypriot population at the p < 0.05 level. However, none of these associations survives Bonferroni correction.
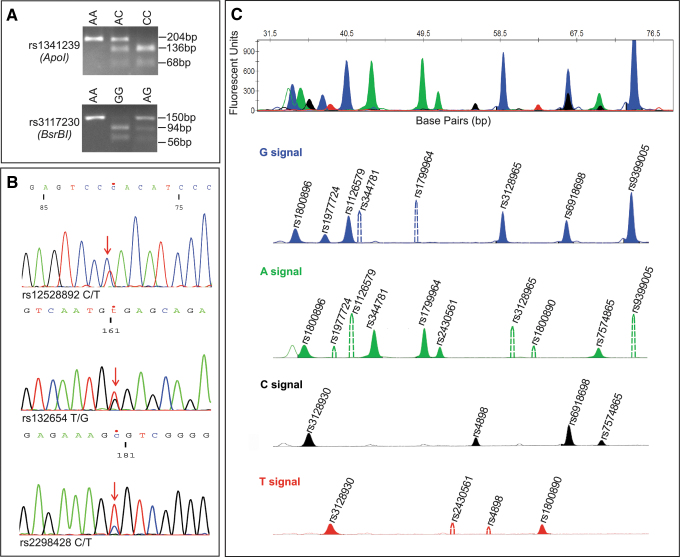
FIG. 1.
Genotyping of eighteen SSc associated SNPs by RFLP, Sanger sequencing and multiplex SNaPshot minisequencing assay. (A) Genotyping of SNPs rs1341239 and rs3117230 using restriction enzymes ApoI and BsrBI, respectively. (B) Sanger sequencing results show a heterozygous genotype of SNPs rs12528892, rs132654, and rs2298428, respectively. (C) The first electropherogram shows the detection of the thirteen SNPs in a random sample. Four separate electropherograms are presented, according to the color of peak (ddNTP extension). Blue, green, black, and red-filled peaks correspond to the fluorescence signal of G, A, C and T alleles of detected SNP, respectively. Colored nonfilled solid peak indicates nonspecific allele; this is not associated with the genotype. Dotted peaks represent the position of the alternative allele in the cases of homozygous SNP results. SNP genotype of the selected sample is the following: rs1800896 (GA), rs3128930 (CT), rs1799724 (GG), rs1126579 (GG), rs344781 (AA), rs1799964 (AA), rs2430561 (AA), rs4898 (CC), rs3128965 (GG), rs1800890 (TT), rs6918698 (GC), rs7574865 (CA), rs9399005 (GG). RFLP, restriction fragment length polymorphism; SNPs, single nucleotide polymorphisms; SSc, systemic sclerosis.
Table 2.
Genotypes/Alleles Frequencies and Association of Selected Single Nucleotide Polymorphisms with Systemic Sclerosis Under Logistic Regression Analysis in the Greek-Cypriot Population (Patients with Systemic Sclerosis Versus Healthy Controls)
| SNP | Genotypes frequency, n (%) | Alleles frequency, n (%) | OR (95% CI)a | pa,b | |||
|---|---|---|---|---|---|---|---|
| rs4898c | T/T | T/C | C/C | T | C | 1.06 (0.65–1.74) | 0.82 |
| Controls (144) | 58 (40.28) | 59 (40.97) | 27 (18.75) | 175 (60.76) | 113 (39.24) | ||
| Cases (36) | 13 (36.11) | 17 (47.22) | 6 (16.67) | 43 (59.72) | 29 (40.28) | ||
| rs344781 | A/A | A/G | G/G | A | G | 0.97 (0.54–1.73) | 0.91 |
| Controls (164) | 93 (56.71) | 61 (37.20) | 10 (6.10) | 247 (75.30) | 81 (24.70) | ||
| Cases (41) | 21 (51.22) | 20 (48.78) | 0 (0.00) | 62 (75.61) | 20 (24.39) | ||
| rs1126579 | C/C | C/T | T/T | C | T | 1.07 (0.66–1.75) | 0.78 |
| Controls (164) | 66 (40.24) | 74 (45.12) | 24 (14.63) | 206 (62.80) | 122 (37.20) | ||
| Cases (41) | 16 (39.02) | 18 (43.90) | 7 (17.07) | 50 (60.98) | 32 (39.02) | ||
| rs1341239 | G/G | G/T | T/T | G | T | 1.01 (0.59–1.71) | 0.98 |
| Controls (164) | 67 (40.85) | 79 (48.17) | 18 (10.98) | 213 (64.94) | 115 (35.06) | ||
| Cases (41) | 15 (36.59) | 23 (56.10) | 3 (7.32) | 53 (64.63) | 29 (35.37) | ||
| rs1799724 | C/C | C/T | T/T | C | T | 1.22 (0.68–2.17) | 0.51 |
| Controls (164) | 110 (67.07) | 48 (29.27) | 6 (3.66) | 268 (81.71) | 60 (18.29) | ||
| Cases (41) | 26 (63.41) | 12 (29.27) | 3 (7.32) | 64 (78.05) | 18 (21.95) | ||
| rs1799964 | T/T | T/C | C/C | T | C | 0.94 (0.50–1.76) | 0.84 |
| Controls (164) | 98 (59.76) | 60 (36.59) | 6 (3.66) | 256 (78.05) | 72 (21.95) | ||
| Cases (41) | 24 (58.54) | 17 (41.46) | 0 (0.00) | 65 (79.27) | 17 (20.73) | ||
| rs1800890 | T/T | T/A | A/A | T | A | 0.82 (0.44–1.49) | 0.51 |
| Controls (164) | 90 (54.88) | 66 (40.24) | 8 (4.88) | 246 (75.00) | 82 (25.00) | ||
| Cases (41) | 24 (58.54) | 16 (39.02) | 1 (2.44) | 64 (78.05) | 18 (21.95) | ||
| rs1800896 | A/A | A/G | G/G | A | G | 1.25 (0.77–2.03) | 0.35 |
| Controls (164) | 62 (37.80) | 79 (48.17) | 23 (14.02) | 203 (61.89) | 125 (38.11) | ||
| Cases (41) | 16 (39.02) | 14 (34.15) | 11 (26.83) | 46 (56.10) | 36 (43.90) | ||
| rs2430561 | T/T | A/T | A/A | T | A | 1.00 (0.59–1.69) | 1 |
| Controls (164) | 37 (22.56) | 92 (56.10) | 35 (21.34) | 166 (50.61) | 162 (49.39) | ||
| Cases (41) | 7 (17.07) | 27 (65.85) | 7 (17.07) | 41 (50.00) | 41 (50.00) | ||
| rs3117230 | A/A | A/G | G/G | A | G | 2.27 (1.30–3.98) | 0.004 |
| Controls (164) | 123 (75.00) | 37 (22.56) | 4 (2.44) | 283 (86.28) | 45 (13.72) | ||
| Cases (41) | 23 (56.10) | 13 (31.71) | 5 (12.20) | 59 (71.95) | 23 (28.05) | ||
| rs3128930 | G/G | G/A | A/A | G | A | 2.08 (1.23–3.53) | 0.006 |
| Controls (164) | 118 (71.95) | 39 (23.78) | 7 (4.27) | 275 (83.84) | 53 (16.16) | ||
| Cases (41) | 21 (51.22) | 15 (36.59) | 5 (12.20) | 57 (69.51) | 25 (30.49) | ||
| rs3128965 | G/G | G/A | A/A | G | A | 1.97 (1.10–3.54) | 0.02 |
| Controls (164) | 123 (75.00) | 37 (22.56) | 4 (2.44) | 283 (86.28) | 45 (13.72) | ||
| Cases (41) | 24 (58.54) | 14 (34.15) | 3 (7.32) | 62 (75.61) | 20 (24.39) | ||
| rs6918698 | G/G | G/C | C/C | G | C | 0.88 (0.55–1.41) | 0.59 |
| Controls (164) | 48 (29.27) | 79 (48.17) | 37 (22.56) | 175 (53.35) | 153 (46.65) | ||
| Cases (41) | 15 (36.59) | 17 (41.46) | 9 (21.95) | 47 (57.32) | 35 (42.68) | ||
| rs7574865 | G/G | G/T | T/T | G | T | 1.35 (0.79–2.33) | 0.27 |
| Controls (164) | 76 (46.34) | 70 (42.68) | 18 (10.98) | 222 (67.68) | 106 (32.32) | ||
| Cases (41) | 20 (48.78) | 20 (48.78) | 1 (2.44) | 60 (73.17) | 22 (26.83) | ||
| rs9399005 | C/C | C/T | T/T | C | T | 0.76 (0.43–1.35) | 0.35 |
| Controls (164) | 87 (53.05) | 64 (39.02) | 13 (7.93) | 238 (72.56) | 90 (27.44) | ||
| Cases (41) | 25 (60.98) | 14 (34.15) | 2 (4.88) | 64 (78.05) | 18 (21.95) | ||
| rs12528892 | C/C | C/T | T/T | C | T | 0.80 (0.09–7.08) | 0.84 |
| Controls (164) | 159 (96.95) | 5 (3.05) | 0 (0.00) | 323 (98.48) | 5 (1.52) | ||
| Cases (41) | 40 (97.56) | 1 (2.44) | 0 (0.00) | 81 (98.78) | 1 (1.22) | ||
| rs131654 | T/T | T/G | G/G | T | G | 1.71 (0.99–2.96) | 0.05 |
| Controls (164) | 93 (56.71) | 63 (38.41) | 8 (4.88) | 249 (75.91) | 79 (24.09) | ||
| Cases (41) | 17 (41.46) | 20 (48.78) | 4 (9.76) | 54 (65.85) | 28 (34.15) | ||
| rs2298428 | C/C | C/T | T/T | C | T | — | — |
| Controls (164) | 104 (63.41) | 32 (19.51) | 28 (17.07) | 240 (73.17) | 88 (26.83) | ||
| Cases (41) | 30 (73.17) | 7 (17.07) | 4 (9.76) | 67 (81.71) | 15 (18.29) | ||
Bold in the last column indicates significant p-values.
OR (CI 95%) and p value were calculated based on log-additive model (alleles).
Nominal significance threshold = 0.05; Bonferroni corrected significance threshold = 0.003.
Calculations for this SNP were performed using only the female patient genotypes since it is located on the X-chromosome.
CI, confidence intervals; OR, odds ratio; SNP, single nucleotide polymorphism.
Discussion
A large number of studies support that HLA/non-HLA genetic variants and environmental factors play a key role in the triggering of SSc (Chairta et al., 2017). Genetic heterogeneity has been observed among populations suggesting that investigation of SSc susceptibility in additional populations might provide a clearer understanding of disease etiology. The aim of this study was to evaluate SNP associations already discovered in other populations, within the Greek-Cypriots.
Forty-one Greek-Cypriot patients with SSc and 164 age- and sex-matched Greek-Cypriot healthy controls were included in this study. More females than males with SSc were recorded (7.2:1) and this difference follows the pattern also observed in other studies (Mayes, 2003; Chifflot et al., 2008; Barnes and Mayes, 2012; Alba et al., 2014; Ngo et al., 2014). The mean age of onset of patients with SSc in our study was 49.05, within the ranges that have been reported worldwide (Alba et al., 2014). Clinical and serological features did not differ from those of patients with SSc described in other studies.
Almost all Greek-Cypriot patients with SSc (95.12%) were positive for antinuclear autoantibodies (ANA). Similarly, Mierau et al. showed that 94.2% of patients with SSc in the German Network for Systemic Scleroderma were also positive for ANA (Mierau et al., 2011). In our study lcSSc and dcSSc subgroups were mainly associated with anticentromere autoantibodies (ACA) and antitopoisomerase autoantibodies (ATA), respectively, in agreement with findings of other studies, where ACA has been strongly related with CREST syndrome (lcSSc) patients, while ATA was found in ∼40% of patients with dcSSc and <10% of patients with lcSSc (Tan et al., 1980; Spencer-Green et al., 1997; Ho and Reveille, 2003; Reveille and Solomon, 2003). Raynaud's phenomenon, which is one of the initial and obvious features in patients with SSc (Sunderkötter and Riemekasten, 2006), was observed in all patients of our study.
Through this study, we performed a replication/evaluation study because the number of recruited Greek-Cypriot patients with SSc was relatively small and thus statistical power for a discovery study could not be reached. A nominally significant association between SSc and three SNPs (rs3117230; rs3128930; rs3128965) has been detected under log-additive model (Table 2). Interestingly, these three SNPs are located on chromosome 6p in the region of HLA-DPB1 and HLA-DPB2. These SNPs were significantly associated with SSc and the ATA+ SSc subgroup in the discovery phase of the Korean population study, but this association did not survive in the replication phase (Zhou et al., 2009). Our results are consistent with the above study and may support the HLA region genetic susceptibility to SSc predisposition (Table 2 and Appendix Table A2).
Ten out of eighteen selected SNPs were previously reported to be significantly associated with SSc under log-additive model. In the current study, five (significantly associated: rs3128965, rs3117230, and rs3128930; nonsignificantly associated: rs1800896 and rs7574865) out of these ten SNPs have effects on the disease in the same direction as reported in previous studies (Appendix Table A2). Lack of association of the rest of the SNPs with SSc in the Greek-Cypriot population could be either attributed to genetic heterogeneity or small power of the study. In addition, the majority of investigated SNPs had similar frequencies with those reported in European populations in published studies/dbSNP (Appendix Tables A1 and A2).
SSc prevalence in Europe ranges from 31 to 277 cases/million individuals (Silman et al., 1988; Allcock et al., 2004; Le Guern et al., 2004; Alamanos et al., 2005; Arias-Nunez et al., 2008; Lo Monaco et al., 2011). To our knowledge, no epidemiological or genetic data on SSc in the Greek-Cypriot population have been previously reported. Based on epidemiological data reported in other populations, a relatively small number of SSc cases (21 to 193) is expected in our population. Thus, the number of patients analyzed through this study is a comparatively good representation of SSc in the Greek-Cypriot population. However, because a relatively small number of study subjects might lead to false results, we increased the number of controls per case (4:1) to limit this chance.
This is the first SSc susceptibility loci study in the Greek-Cypriot population and the majority of the investigated SNPs, do not confirm any statistically significant association with SSc in our population. Further investigation of the Greek-Cypriot population with a larger sample size may increase statistical power and enable identification of SSc susceptibility loci in this newly investigated population.
Acknowledgments
The authors would like to thank (1) all SSc patients and healthy controls for their participation in the study and (2) the nursing staff of Nicosia General Hospital and Cyprus Institute of Neurology and Genetics that contributed to the recruitment of SSc samples.
Appendix
Appendix Table A1.
Details of Single Nucleotide Polymorphisms Selected for This Study
| SNP | Chromosomal position (GRCh38.p12) | Gene: consequence | Allelesa | MAFb | MAF in controls (current study) | HWE in controls (current study) | References |
|---|---|---|---|---|---|---|---|
| rs4898c | chrX:47585586 | TIMP1: Synonymous Variant | T/C | 0.46 | 0.39 | 0.09 | Indelicato et al. (2006), Skarmoutsou et al. (2012) |
| SYN1: Intron Variant | |||||||
| MIR4769: 2KB Upstream Variant | |||||||
| rs344781 | chr19:43670636 | PLAUR: 2KB Upstream Variant | A/G | 0.25 | 0.25 | 1.00 | Manetti et al. (2011) |
| rs1126579 | chr2:218136011 | CXCR2: 3 Prime UTR Variant | C/T | 0.49 | 0.37 | 0.66 | Renzoni et al. (2000), Salim et al. (2012) |
| rs1341239 | chr6:22303975 | PRL: 2KB Upstream Variant | G/T | 0.35 | 0.35 | 0.46 | Fojtíková et al. (2010) |
| rs1799724 | chr6:31574705 | TNF: 2KB Upstream Variant | C/T | 0.09 | 0.18 | 0.79 | Sato et al. (2004), Otieno et al. (2007) |
| LTA: 500B Downstream Variant | |||||||
| rs1799964 | chr6:31574531 | TNF: 2KB Upstream Variant | T/C | 0.21 | 0.22 | 0.39 | Sato et al. (2004), Otieno et al. (2007) |
| LTA: 500B Downstream Variant | |||||||
| LOC100287329: 2KB Upstream Variant | |||||||
| rs1800890 | chr1:206776020 | IL19: Intron Variant | T/A | 0.37 | 0.25 | 0.35 | Hudson et al. (2005), Peng et al. (2012b) |
| rs1800896 | chr1:206773552 | IL19: Intron Variant | A/G | 0.45 | 0.38 | 0.79 | Ates et al. (2008), Salim et al. (2013) |
| IL10: 2KB Upstream Variant | |||||||
| rs2430561 | chr12:68158742 | IFNG: Intron Variant | T/A | 0.46 | 0.49 | 0.12 | Wastowski et al. (2009) |
| rs3117230 | chr6:33107858 | HLA-DPB1: Downstream Variant | A/G | 0.23 | 0.14 | 0.55 | Zhou et al. (2009) |
| rs3128930 | chr6:33107889 | HLA-DPB1: Downstream Variant | G/A | 0.26 | 0.16 | 0.12 | Zhou et al. (2009) |
| rs3128965 | chr6:33088122 | HLA-DPB1: 3 Prime UTR Variant | G/A | 0.19 | 0.14 | 0.55 | Zhou et al. (2009) |
| rs6918698 | chr6:131952117 | CCN2: 2KB Upstream Variant | G/C | 0.49 | 0.47 | 0.68 | Fonseca et al. (2007), Kawaguchi et al. (2009) |
| rs7574865 | chr2:191099907 | STAT4: Intron Variant | G/T | 0.23 | 0.32 | 0.76 | Dieude et al. (2009), Gourh et al. (2009), Rueda et al. (2009), Tsuchiya et al. (2009), Allanore et al. (2011), Liang et al. (2012), Peng et al. (2012a), Yi et al. (2013), Zheng et al. (2013), Zochling et al. (2014), Xu et al. (2016) |
| rs9399005 | chr6:131947824 | CCN2: 500B Downstream Variant | C/T | 0.30 | 0.27 | 0.80 | Granel et al. (2010) |
| rs12528892 | chr6:32725729 | HLA-DQA2:Upstream variant | C/T | 0.07 | 0.02 | 0.84 | Mayes et al. (2014) |
| rs131654 | chr22:21562901 | UBE2L3: Intron Variant | T/G | 0.33 | 0.24 | 0.52 | Hasebe et al. (2012) |
| rs2298428 | chr22:21628603 | YDJC: Missense Variant | C/T | 0.18 | 0.27 | 0.00 | Hasebe et al. (2012) |
Major/Minor allele based on the current and published studies.
MAF of European population submitted in 1000 Genome Project (dbSNP).
Calculations for this SNP in the current study were performed using only the female patient genotypes since it is located on the X-chromosome.
HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Appendix Table A2.
Previously Reported Single Nucleotide Polymorphism Associations Based on Systemic Sclerosis Patients Versus Healthy Controls
| SNP | Alleles | Cases alleles n (%) | Controls alleles n (%) | OR (95% CI) | p | Cases/controls | Population | References |
|---|---|---|---|---|---|---|---|---|
| rs4898a | T | 269 (65.29) | 180 (57.69) | 1 | — | 206/156 | Italian | Skarmoutsou et al. (2012) |
| C | 143 (34.71) | 132 (42.31) | 0.72 (0.53–0.98) | 0.04 | ||||
| rs344781 | G | 210 (27) | 180 (23) | 1.29 (1.03–1.63) | 0.03 | 388/391 | Italian (Caucasian) | Manetti et al. (2011) |
| A | 566 (73) | 602 (77) | — | |||||
| G | 366 (25) | 256 (21) | 1.22 (1.02–1.47) | 0.03 | 732/607 | Italian and French (Caucasian) | ||
| A | 1098 (75) | 959 (79) | — | — | ||||
| rs1126579 | T | 287 (56) | 341 (44) | NA | 0.002 | 256/388 | United Kingdom (Caucasian) | Renzoni et al. (2000) |
| C | 225 (44) | 435 (56) | — | |||||
| rs1800890 | T vs. A | — | — | 0.75 (0.61–0.93) | NA | 382/1125 | Meta-analysis (4 studies) | Peng et al. (2012b) |
| rs1800896 | G | 45 (50) | 78 (26) | 2.85 (1.74–4.63) | <0.000 | 45/150 | Turkish | Ates et al. (2008) |
| A | 45 (50) | 222 (74) | — | — | ||||
| rs3117230 | G | 48 (18) | 102 (9.2) | 2.20 (1.50–3.22) | 3.52E-05 | 133/557 | Discovery, Koreans | Zhou et al. (2009) |
| A | 218 (82) | 1012 (90.8) | — | |||||
| rs3128930 | A | 90 (34) | 167 (15) | 3.0 (2.20–4.10) | 8.16E-13 | 133/557 | Discovery, Koreans | Zhou et al. (2009) |
| G | 176 (66) | 947 (85) | — | — | ||||
| rs3128965 | A | 48 (18) | 104 (9.3) | 2.18 (1.49–3.18) | 4.47E-05 | 133/557 | Discovery, Koreans | Zhou et al. (2009) |
| G | 218 (82) | 1114 (90.7) | — | — | ||||
| rs6918698 | G | 435 (55) | 241 (45) | 1.5 (1.2–1.9) | <0.001 | 395/269 | Asian (Japanese) | Kawaguchi et al. (2009) |
| C | 355 (45) | 297 (55) | ||||||
| rs7574865 | T | 218 (27.1) | 220 (22.9) | 1.26 (1.01–1.56) | 0.039 | 402/481 | Discovery, French | Dieude et al. (2009) |
| G | 586 (72.9) | 742 (77.1) | — | — | ||||
| T | 212 (26.6) | 206 (21.3) | 1.35 (1.07–1.66) | 0.0099 | 399/483 | Replication, French | ||
| G | 586 (73.4) | 760 (78.7) | — | — | ||||
| T | 429 (26.8) | 426 (22.1) | 1.29 (1.11–1.51) | 0.001 | 801/964 | Combination, French | ||
| G | 1173 (73.2) | 1502 (77.9) | — | — | ||||
| T | 458 (26) | 213 (21) | 1.31 (NA) | 0.01 | 880/507 | North American | Gourh et al. (2009) | |
| G | 1302 (74) | 801 (79) | — | — | ||||
| T | 757 (27) | 315 (22.5) | 1.26 (1.1–1.5) | 0.004 | 1402/698 | North American | ||
| G | 2047 (73) | 1082 (77.5) | — | — | ||||
| T | 231 (41) | 401 (34) | 1.35 (1.10–1.66) | 0.023 | 282/590 | Japanese | Tsuchiya et al. (2009) | |
| G | 333 (59) | 779 (66) | — | — | ||||
| T | 307 (27.2) | 778 (21.9) | 1.33 (1.14–1.55) | 2.50E-04 | 564/1776 | Discovery, French | Allanore et al. (2011) | |
| G | 821 (72.8) | 2774 (78.1) | ||||||
| T | 976 (29) | 1727 (22) | 1.40 (1.26–1.56 | 1.9E-10 | 1682/3926 | Replication (French, Italians, German & Eastern European) | ||
| G | 2388 (71) | 6124 (78) | — | — | ||||
| T vs. G | — | — | 0.72 (0.66–0.79) | 0.00 | — | Meta-analysis (8 studies) | Peng et al. (2012a) | |
| T vs. G | — | — | 1.34 (1.25–1.44) | <0.00001 | — | Meta-analysis (11 studies) | Liang et al. (2012) | |
| T allele | — | — | 1.38 (1.27–1.50) | <1.44E-14 | — | Meta-analysis (8 studies) | Zheng et al. (2013) | |
| G | 565 (62.4) | 744 (69.7) | 0.72 (0.48–1.09) | 0.1 | 453/534 | Han Chinese | Yi et al. (2013) | |
| T | 341 (37.6) | 324 (30.3) | 1.58 (1.22–2.05) | 0.00041 | ||||
| T | 278 (28.6) | 2006 (22.5) | 1.35 | 0.00012 | 486/4458 | Discovery, Australian SSc, British controls | Zochling et al. (2014) | |
| G | 694 (71.40) | 6909 (77.5) | — | — | ||||
| T | NA | NA | NA | 5.7 E-5 | 1319/6396 | Combination, Australian SSc, British controls | ||
| G | NA | NA | — | — | ||||
| T vs. G | — | — | 1.37 (1.27–1.48) | <0.00001 | — | Meta-analysis (4 studies) | Xu et al. (2016) |
Calculations for this SNP were performed in females as it is located in X-chromosome.
CI, confidence intervals; OR, odds ratio; SSc, systemic sclerosis.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This study has been financially supported by the Cyprus Institute of Neurology and Genetics and the Cyprus School of Molecular Medicine.
References
- Alamanos Y, Tsifetaki N, Voulgari PV, et al. (2005) Epidemiology of systemic sclerosis in northwest Greece 1981 to 2002. Semin Arthritis Rheum 34:714–720 [DOI] [PubMed] [Google Scholar]
- Alba MA, Velasco C, Simeon CP, et al. (2014) Early- versus late-onset systemic sclerosis: differences in clinical presentation and outcome in 1037 patients. Medicine 93:73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allanore Y, Saad M, Dieude P, et al. (2011) Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allanore Y, Simms R, Distler O, et al. (2015) Systemic sclerosis. Nat Rev Dis Primers 1:15002. [DOI] [PubMed] [Google Scholar]
- Allcock RJ, Forrest I, Corris PA, et al. (2004) A study of the prevalence of systemic sclerosis in northeast England. Rheumatology 43:596–602 [DOI] [PubMed] [Google Scholar]
- Arias-Nunez MC, Llorca J, Vazquez-Rodriguez TR, et al. (2008) Systemic sclerosis in northwestern Spain: a 19-year epidemiologic study. Medicine 87:272–280 [DOI] [PubMed] [Google Scholar]
- Ates O, Musellim B, Ongen G, et al. (2008) Association between ‘interleukin’ 10 gene (IL10) polymorphisms and systemic sclerosis with interstitial lung involvement. Rheumatol Int 28:1123–1126 [DOI] [PubMed] [Google Scholar]
- Barnes J, Mayes MD (2012) Epidemiology of systemic sclerosis: incidence, prevalence, survival, risk factors, malignancy, and environmental triggers. Curr Opin Rheumatol 24:165–170 [DOI] [PubMed] [Google Scholar]
- Bossini-Castillo L, Simeon CP, Beretta L, et al. (2011) Confirmation of association of the macrophage migration inhibitory factor gene with systemic sclerosis in a large European population. Rheumatology 50:1976–1981 [DOI] [PubMed] [Google Scholar]
- Broen JC, Radstake TR, Rossato M (2014) The role of genetics and epigenetics in the pathogenesis of systemic sclerosis. Nat Rev Rheumatol 10:671–681 [DOI] [PubMed] [Google Scholar]
- Chairta P, Nicolaou P, Christodoulou K (2017) Genomic and genetic studies of systemic sclerosis: a systematic review. Hum Immunol 78:153–165 [DOI] [PubMed] [Google Scholar]
- Chifflot H, Fautrel B, Sordet C, et al. (2008) Incidence and prevalence of systemic sclerosis: a systematic literature review. Semin Arthritis Rheum 37:223–235 [DOI] [PubMed] [Google Scholar]
- Dieude P, Guedj M, Wipff J, et al. (2009) STAT4 is a genetic risk factor for systemic sclerosis having additive effects with IRF5 on disease susceptibility and related pulmonary fibrosis. Arthritis Rheum 60:2472–2479 [DOI] [PubMed] [Google Scholar]
- Fojtíková M, Cejkova P, Becvar R, et al. (2010) Polymorphism of the extrapituitary prolactin promoter and systemic sclerosis. Rheumatol Int 30:1691–1693 [DOI] [PubMed] [Google Scholar]
- Fonseca C, Lindahl GE, Ponticos M, et al. (2007) A polymorphism in the CTGF promoter region associated with systemic sclerosis. N Engl J Med 357:1210–1220 [DOI] [PubMed] [Google Scholar]
- Gourh P, Agarwal SK, Divecha D, et al. (2009) Polymorphisms in TBX21 and STAT4 increase the risk of systemic sclerosis: evidence of possible gene–gene interaction and alterations in Th1/Th2 cytokines. Arthritis Rheum 60:3794–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granel B, Argiro L, Hachulla E, et al. (2010) Association between a CTGF gene polymorphism and systemic sclerosis in a French population. J Rheumatol 37:351–358 [DOI] [PubMed] [Google Scholar]
- Hasebe N, Kawasaki A, Ito I, et al. (2012) Association of UBE2L3 polymorphisms with diffuse cutaneous systemic sclerosis in a Japanese population. Ann Rheum Dis 71:1259–1260 [DOI] [PubMed] [Google Scholar]
- Ho KT, Reveille JD (2003) The clinical relevance of autoantibodies in scleroderma. Arthritis Res Ther 5:80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EP, Park JW (2012) Sample size and statistical power calculation in genetic association studies. Genomics Inform 10:117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson LL, Rocca KM, Kuwana M, et al. (2005) Interleukin-10 genotypes are associated with systemic sclerosis and influence disease-associated autoimmune responses. Genes Immun 6:274–278 [DOI] [PubMed] [Google Scholar]
- Indelicato M, Chiarenza V, Libra M, et al. (2006) Analysis of TIMP-1 gene polymorphisms in Italian sclerodermic patients. J Clin Lab Anal 20:173–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Ota Y, Kawamoto M, et al. (2009) Association study of a polymorphism of the CTGF gene and susceptibility to systemic sclerosis in the Japanese population. Ann Rheum Dis 68:1921–1924 [DOI] [PubMed] [Google Scholar]
- Le Guern V, Mahr A, Mouthon L, et al. (2004) Prevalence of systemic sclerosis in a French multi-ethnic county. Rheumatology 43:1129–1137 [DOI] [PubMed] [Google Scholar]
- Li J, Chen SY, Liu HH, et al. (2019) Associations of vitamin D receptor single nucleotide polymorphisms with susceptibility to systemic sclerosis. Arch Med Res 50:368–376 [DOI] [PubMed] [Google Scholar]
- Liang YL, Wu H, Shen X, et al. (2012) Association of STAT4 rs7574865 polymorphism with autoimmune diseases: a meta-analysis. Mol Biol Rep 39:8873–8882 [DOI] [PubMed] [Google Scholar]
- Lo Monaco A, Bruschi M, La Corte R, et al. (2011) Epidemiology of systemic sclerosis in a district of northern Italy. Clin Exp Rheumatol 29:12. [PubMed] [Google Scholar]
- Manetti M, Allanore Y, Revillod L, et al. (2011) A genetic variation located in the promoter region of the UPAR (CD87) gene is associated with the vascular complications of systemic sclerosis. Arthritis Rheum 63:247–256 [DOI] [PubMed] [Google Scholar]
- Mayes MD. (2003) Scleroderma epidemiology. Rheum Dis Clin North Am 29:239–254 [DOI] [PubMed] [Google Scholar]
- Mayes MD, Bossini-Castillo L, Gorlova O, et al. (2014) Immunochip analysis identifies multiple susceptibility loci for systemic sclerosis. Am J Hum Genet 94:47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellal Y, Allam I, Tahiat A, et al. (2018) Th17 pathway genes polymorphisms in Algerian patients with systemic sclerosis. Acta Reumatol Port 43:269–278 [PubMed] [Google Scholar]
- Mierau R, Moinzadeh P, Riemekasten G, et al. (2011) Frequency of disease-associated and other nuclear autoantibodies in patients of the German network for systemic scleroderma: correlation with characteristic clinical features. Arthritis Res Ther 13:R172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo ST, Steyn FJ, McCombe PA (2014) Gender differences in autoimmune disease. Front Neuroendocrinol 35:347–369 [DOI] [PubMed] [Google Scholar]
- Otieno FG, Lopez AM, Jimenez SA, et al. (2007) Allograft inflammatory factor-1 and tumor necrosis factor single nucleotide polymorphisms in systemic sclerosis. Tissue Antigens 69:583–591 [DOI] [PubMed] [Google Scholar]
- Oztuzcu S, Onat AM, Pehlivan Y, et al. (2015) Association of TRPM channel gene polymorphisms with systemic sclerosis. In Vivo 29:763–770 [PubMed] [Google Scholar]
- Pehlivan Y, Yolbas S, Cetin GY, et al. (2016) Investigation of the association between Rho/Rho-kinase gene polymorphisms and systemic sclerosis. Rheumatol Int 36:421–427 [DOI] [PubMed] [Google Scholar]
- Peng WJ, Pan HF, Tao JH, et al. (2012a) A meta-analysis of the association between cytokine gene polymorphisms and systemic sclerosis. Mod Rheumatol 22:695–703 [DOI] [PubMed] [Google Scholar]
- Peng WJ, Wang BX, Pan HF, et al. (2012b) Association of the interleukin-10 1082G/A, 819C/T and 3575T/A gene polymorphisms with systemic sclerosis: a meta-analysis. Mol Biol Rep 39:6851–6855 [DOI] [PubMed] [Google Scholar]
- Radstake TRDJ, Gorlova O, Rueda B, et al. (2010) Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet 42:426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzoni E, Lympany P, Sestini P, et al. (2000) Distribution of novel polymorphisms of the interleukin-8 and CXC receptor 1 and 2 genes in systemic sclerosis and cryptogenic fibrosing alveolitis. Arthritis Rheum 43:1633–1640 [DOI] [PubMed] [Google Scholar]
- Reveille JD, Solomon DH (2003) Evidence-based guidelines for the use of immunologic tests: anticentromere, Scl-70, and nucleolar antibodies. Arthritis Rheum 49:399–412 [DOI] [PubMed] [Google Scholar]
- Rueda B, Broen J, Simeon C, et al. (2009) The STAT4 gene influences the genetic predisposition to systemic sclerosis phenotype. Hum Mol Genet 18:2071–2077 [DOI] [PubMed] [Google Scholar]
- Salim PH, Jobim M, Bredemeier M, et al. (2012) Combined effects of CXCL8 and CXCR2 gene polymorphisms on susceptibility to systemic sclerosis. Cytokine 60:473–477 [DOI] [PubMed] [Google Scholar]
- Salim PH, Jobim M, Bredemeier M, et al. (2013) Interleukin-10 gene promoter and NFKB1 promoter insertion/deletion polymorphisms in systemic sclerosis. Scand J Immunol 77:162–168 [DOI] [PubMed] [Google Scholar]
- Sato H, Lagan AL, Alexopoulou C, et al. (2004) The TNF-863A allele strongly associates with anticentromere antibody positivity in scleroderma. Arthritis Rheum 50:558–564 [DOI] [PubMed] [Google Scholar]
- Silman A, Jannini S, Symmons D, et al. (1988) An epidemiological study of scleroderma in the West Midlands. Br J Rheumatol 27:286–290 [DOI] [PubMed] [Google Scholar]
- Skarmoutsou E, D'Amico F, Marchini M, et al. (2012) Association of TIMP-1 + 372 SNP with digital ulcer manifestation in female systemic sclerosis patients. Hum Immunol 73:950–953 [DOI] [PubMed] [Google Scholar]
- Spencer-Green G, Alter D, Welch HG (1997) Test performance in systemic sclerosis: anti-centromere and anti-Scl-70 antibodies. Am J Med 103:242–248 [DOI] [PubMed] [Google Scholar]
- Sunderkötter C, Riemekasten G (2006) Pathophysiology and clinical consequences of Raynaud's phenomenon related to systemic sclerosis. Rheumatology 45:iii33–iii35 [DOI] [PubMed] [Google Scholar]
- Tan EM, Rodnan GP, Garcia I, et al. (1980) Diversity of antinuclear antibodies in progressive systemic sclerosis. Anti-centromere antibody and its relationship to CREST syndrome. Arthritis Rheum 23:617–625 [DOI] [PubMed] [Google Scholar]
- Tsuchiya N, Kawasaki A, Hasegawa M, et al. (2009) Association of STAT4 polymorphism with systemic sclerosis in a Japanese population. Ann Rheum Dis 68:1375–1376 [DOI] [PubMed] [Google Scholar]
- van den Hoogen F, Khanna D, Fransen J, et al. (2013) 2013 classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann Rheum Dis 72:1747–1755 [DOI] [PubMed] [Google Scholar]
- Vlachoyiannopoulos PG, Dafni UG, Pakas I, et al. (2000) Systemic scleroderma in Greece: low mortality and strong linkage with HLA-DRB1*1104 allele. Ann Rheum Dis 59:359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wastowski IJ, Sampaio-Barros PD, Martelli-Palomino G, et al. (2009) Association of interferon-gamma gene polymorphism (+874 T/A) with systemic sclerosis. Dis Markers 27:93–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang W, Tian Y, et al. (2016) Polymorphisms in STAT4 and IRF5 increase the risk of systemic sclerosis: a meta-analysis. Int J Dermatol 55:408–416 [DOI] [PubMed] [Google Scholar]
- Yi L, Wang JC, Guo XJ, et al. (2013) STAT4 is a genetic risk factor for systemic sclerosis in a Chinese population. Int J Immunopathol Pharmacol 26:473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yin J, Huang R, et al. (2013) Meta-analysis reveals an association of STAT4 polymorphisms with systemic autoimmune disorders and anti-dsDNA antibody. Hum Immunol 74:986–992 [DOI] [PubMed] [Google Scholar]
- Zhou X, Lee JE, Arnett FC, et al. (2009) HLA-DPB1 and DPB2 are genetic loci for systemic sclerosis: a genome-wide association study in Koreans with replication in North Americans. Arthritis Rheum 60:3807–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zochling J, Newell F, Charlesworth JC, et al. (2014) An immunochip-based interrogation of scleroderma susceptibility variants identifies a novel association at DNASE1L3. Arthritis Res Ther 16:438. [DOI] [PMC free article] [PubMed] [Google Scholar]