Abstract
Taste bud cells are specialized epithelial cells that undergo continuous turnover, and thus require active progenitors for their renewal and an intact taste function. Our previous studies suggested that a population of taste bud cells originates from outside of the surrounding tongue epithelium—previously regarded sole source of taste bud progenitors. In this study, we demonstrated that SOX10 (SRY-related HMG-box gene 10)-expressing cells, known to be in the migrating neural crest, were also distributed in taste bud-surrounding tissue compartments under the tongue epithelium, that is, the connective tissue core of taste papillae and von Ebner's glands. By lineage tracing of SOX10-expressing cells using SOX10-Cre, a Cre model driven by the endogenous SOX10 promoter, crossing with a Cre reporter line R26-tdTomato (tdT), we found SOX10-Cre-labeled tdT+ cells within taste buds in all three types of taste papillae (fungiform, circumvallate, and foliate) as well as in the soft palate in postnatal mice. The tdT+ taste bud cells were progressively more abundant along the developmental stages, from virtually zero at birth to over 35% in adults. Most of tdT+ taste bud cells had a low intensity of immunosignals of Keratin 8 (a widely used taste bud cell marker). In circumvallate taste buds, tdT signals were co-localized principally with a type III taste bud cell marker, less so with type I and II cell makers. Together, our data demonstrate a novel progenitor source for taste buds of postnatal mice—SOX10-Cre-labeled cells in the connective tissue core and/or von Ebner's glands.
Keywords: SOX10, taste buds, progenitor, taste papilla, soft palate
Introduction
Taste bud cells are postmitotic, specialized epithelial cells that undergo continuous turnover [1–5]. Thus, progenitors in the immediate surrounding tissue compartments are essential for cell renewal and homeostasis of taste buds. Previous studies have reported that taste bud progenitor cell populations reside in the basal layer of taste bud-surrounding tongue epithelium [3,4,6–10] and express K14, K5, p63, SOX2, and Lgr5 [8,11,12]. Another possible origin of taste bud cells from the underlying connective tissue, largely derived from neural crest, is also indicated [13,14], but remains inconclusive. Using P0-Cre [13,14] and Dermo1-Cre [13] for cell lineage tracing, we observed a large dispersal of labeled cells in taste buds, in concurrence with those in the underlying connective tissue. However, Wnt1-Cre-labeled cells, while extensive in the connective tissue, were rare in taste buds [14,15]. Lineage tracing of stromal cells in the connective tissue using Vimentin-CreER indicated a Vimentin+ cell lineage for taste bud cells [13]. However, the use of a membrane-bound green fluorescent protein reporter made it difficult to draw concrete conclusions because the membranes of the taste buds cells would be difficult to distinguish from the labeled nerve fibers, which are thin and fibrous and protrude into the taste bud. Thus, the fundamental issue of whether taste bud progenitors exist in tissue compartments beyond the tongue epithelium remains unsettled.
Discrepancies among these mouse models, for example, P0-Cre, Dermo1-Cre, and Wnt1-Cre, when tracking cell populations may account for the disparity in the observations [16]. The use of a model system with clear information on the expressing cell populations is needed to address this issue. SOX10 (SRY-related HMG-box gene 10) is expressed specifically in neural crest cells during early embryonic stages [17]. Sustained SOX10 expression in later embryonic stages and in postnatal mice is evident in neural crest lineage cell types such as glial cells [18,19], melanocytes [20], and salivary gland cells [21]. To advance our understanding as to whether taste bud progenitors exist in tissue compartments other than in tongue epithelium, we located the SOX10-expressing (SOX10+) cells in taste bud-surrounding tissue compartments and mapped the lineage of SOX10-Cre+ cells using a SOX10-Cre mouse line in which Cre expression is under the control of the endogenous promoter of SOX10 [22,23].
In this study, we found that SOX10, in addition to the expression in neural crest, was expressed in the taste bud-surrounding tissue compartments under, but not within, the tongue epithelium (ie, connective tissue core of taste papillae and von Ebner's glands in postnatal mice). SOX10-Cre/tdTomato (tdT)-labeled cells were progressively distributed in taste buds after birth and plateaued at 4 weeks, when most taste buds are mature. The labeled cells in taste buds were predominantly neuronal-like type III taste cells and not noticeably immunostained with the pan-taste cell marker Keratin (Krt)8. Our results complement with the findings that taste papilla placodal cells give rise to type I and II, but not type III, taste cells [15,24,25], and thus demonstrate a novel source of stem/progenitor cells for taste buds during their maturation and maintenance—SOX10-Cre+ cells in the connective tissue and/or von Ebner's glands.
Materials and Methods
Animals
Animal use was approved by The University of Georgia Institutional Animal Care and Use Committee and was in accordance with the National Institutes of Health Guidelines for care and use of animals for research.
Mice were maintained and bred in the animal facility of the Animal and Dairy Science department at the University of Georgia. Wild type (C57BL/6J, Stock#000664), SOX10-Cre [B6; CBA-Tg (Sox10-cre) 1Wdr/J, Stock#025807], and R26-tdTomato (hereafter tdT) Cre reporter mice [B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, Stock#007914] were obtained from The Jackson Laboratory. The hemizygous SOX10-Cre mice and homozygous tdT reporter breeders were crossed to generate SOX10-Cre/tdT mice. Both male and female mice were used at the examined stages. Cre-negative littermates served as controls.
PCR genotyping was performed using the following primers: (1) Cre allele forward 5′-ATTGCTGTCACTTGGTCGGC-3′ and reverse 5′-GGAAAATGCTTCTGTCCGTTTGC-3′ to detect the Cre recombinase allele; and (2) msSRYz_SexDet forward 5′-TTGTCTAGAGAGCATGGAGGGCCATGTCAA-3′ and reverse 5′-CCACTCCTCTGTGACACTTTAGCCCTCCGA-3′ to determine the sex of embryos.
Tissue collections
Mouse embryos on embryonic (E) day 8.0–8.5 were collected between 10 am and 4 pm. Noon of the day of vaginal plug detection was designated E0.5. The embryos were also staged by counting somite pairs. Embryos with 7–15 somites were used. Dams were euthanized with CO2 followed by cervical dislocation. The uterus was removed and placed in a Petri dish containing 0.1 M phosphate-buffered saline (PBS) (Cat. No. CP4390-48; Denville Scientific, Inc., Metuchen, NJ). Embryos were dissected from the uterus under a stereomicroscope and processed for in situ hybridization.
Postnatal mice were harvested at various stages corresponding to different phases of taste bud development, that is, newborn when early taste buds emerge; 2 weeks when more taste buds become mature; and 4, 8, and 16 weeks when taste buds are mature and undergo continuous cell renewal for homeostasis. The day on which the pups were born was designated postnatal day 1. Mice were euthanized with CO2 for tongue and soft palate tissue collections.
Immunohistochemistry
Postnatal mice were transcardially perfused using 10 mL warm 0.1 M PBS, followed by 10 mL warm and 20 mL cold 2% paraformaldehyde (PFA) (Cat. No. AAJ19943k2; Fisher Healthcare, Houston, TX) in 0.1 M PBS. Postnatal mouse tongues were dissected from mandible and further fixed in 2% PFA at 4°C for 2 h. All fixed tissues were cryoprotected in 30% sucrose in 0.1 M PBS for at least 48 h at 4°C. Tissues were trimmed and dissected, embedded in O.C.T. (Cat. No. 23-730-571; Fisher Healthcare), and rapidly frozen as such for the following: (1) sagittal sections of the left and right halves of the anterior 2/3 of the oral tongue where fungiform papillae are distributed, sagittal sections of both foliated papillae, left and right halves of soft palate; and (2) sagittal or transverse sections of the single circumvallate papilla.
Frozen sections were cut at 8 μm in thickness and mounted onto charged slides (Fisher Brand™ Superfrost™ Plus Microscope Slides, Cat. No. 12-550-15; Fisher Scientific, Waltham, MA). Nonspecific binding was blocked with 10% normal donkey serum (Cat. No. SLBW2097; Sigma-Aldrich, St. Louis, MO) in 0.1 M PBS containing 0.3% Triton X-100 (Cat. No. X100-100ML; Sigma-Aldrich) for 30 min, followed by overnight incubation with primary antibody diluted with 0.1 M PBS containing 0.3% Triton X-100 and 1% normal donkey serum. Primary antibodies used in this study are listed in Table 1.
Table 1.
Primary Antibodies That Were Used
| Antibodies | Dilution | Source |
|---|---|---|
| Goat anti-E-cadherin | 1:500 | Cat. No. AF748; Fish Scientific (Waltham, MA) |
| Rat anti-Keratin 8 (Krt8) | 1:1,000 | Cat. No. TROMA-1; Developmental Studies Hybridoma Bank (Iowa City, IA) |
| Rabbit anti-NTPDase II | 1:1,000 | Centre de recherché du CHUL Rhumatologie-Immunologie (Québec, Canada) |
| Rabbit anti-PLCβ2 | 1:500 | Cat. No. sc-515912; Santa Cruz Biotechnology (Dallas, TX) |
| Rabbit anti-SNAP25 | 1:5,000 | Cat. No. S9684; Sigma-Aldrich (St. Louis, MO) |
After rinses in 0.1 M PBS (three times, 10 min each), sections were incubated in Alexa Fluor® 488 (for E-cadherin)- and/or Alexa Fluor® 647 (for all the other markers)-labeled secondary antibody (1:500; Invitrogen, Eugene, OR) for 1 h at room temperature. Sections were rinsed and then counterstained with DAPI (200 ng/mL, Cat. No. D1306; Fisher Scientific). After rinses with 0.1 M PBS followed by dipping in Milli-Q water (Direct-Q® 3 UV water purification system; Millipore, MA), sections were air dried and coverslipped with Prolong® diamond antifade mounting medium (Cat. No. P36970; Fisher Scientific).
In situ hybridization
Postnatal mice were transcardially perfused using 10 mL warm 0.1 M PBS solution, followed by 10 mL warm and 20 mL cold 4% PFA in 0.1 M PBS, and 20 mL cold 4% PFA. Mouse embryos and postnatal tongues were fixed in 4% PFA in 0.1 M PBS overnight, followed by cryoprotection in 30% sucrose in 0.1 M PBS for at least 48 h at 4°C. Rapidly frozen tissues were sectioned at 15 μm in thickness. In situ hybridization for SOX10 was performed, as previously described [26], using digoxigenin-labeled riboprobes. DCGS-10 Plasmid carrying SOX10 probe template was a gift from William Pavan (Addgene plasmid# 24752) [17]. Digoxigenin-labeled antisense RNA probe was prepared by linearizing with EcoR1, followed by transcription with T7 RNA polymerase. A sense RNA probe was prepared by linearizing with XhoI, followed by transcription with T3 RNA polymerase, and used as control.
Photomicroscopy and quantification
Immunostained slides were analyzed under a fluorescent light microscope (EVOS FL, Life Technologies, CA) and images were taken using a laser scanning confocal microscope (Zeiss LSM 710; Zeiss, Germany). In situ hybridized slides were examined and imaged using a Zeiss Axio imager (Zeiss).
Quantitative analyses (n = 3 each group) were performed using confocal images and ImageJ software to (1) quantify the number of SOX10-Cre/tdT-labeled and SOX10-Cre/tdT-unlabeled taste bud cells in circumvallate taste buds of SOX10-Cre/tdT mice at 2, 4, 8, and 16 weeks of age; (2) quantify the number of SOX10-Cre/tdT-labeled tdT+ type II (PLCβ2+) and tdT+ type III (SNAP25+) taste bud cells, total PLCβ2+ type II cells, total SNAP25+ type III cells, and total tdT+ taste bud cells in circumvallate taste buds of SOX10-Cre/tdT mice at 8 weeks; and (3) evaluate the intensity of Krt8 immunosignals by measuring Area, Integrated Density, and Mean Gray Value of individual taste bud cells in circumvallate taste buds of 8-week-old wild-type and SOX10-Cre/tdT mice. The circumvallate taste papilla was cut in a sagittal orientation to produce serial sections that were perpendicular to the longitudinal axis of taste bud cells. A representative section, with the most taste buds from each wall of the circumvallate papilla trench, was selected for analyses. Taste bud cells with a DAPI+ nucleus were included to analyze the proportion of SOX10-Cre/tdT in labeling taste bud cell types. Taste bud cells with both strong and weak (but apparently above background) tdT labeling were considered SOX10-Cre-labeled tdT+ cells. Taste bud cells with a clear boundary marked by E-cadherin immunoproducts were included to analyze the proportion of SOX10-Cre/tdT-labeled taste bud cells at different stages and measure the intensity of Krt8 immunosignals in individual taste bud cells.
Statistical data analysis
The percentages of SOX10-Cre/tdT-labeled tdT+ type II (PLCβ2+) and tdT+ type III (SNAP25+) relative to total tdT+ taste bud cells, total PLCβ2+ type II cells, or total SNAP25+ type III cells in the circumvallate taste buds were calculated. Due to the difficulty of identifying individual type I cells with NTPDase II immunosignals, the percentage of tdT+ type I (NTPDase II+) relative to total tdT+ taste bud cells was extrapolated from the quantitative data for type II and III cells. The corrected total cell fluorescence (CTCF) of Krt8 immunosignals in individual taste bud cells was calculated to represent the intensity using the formula [CTCF = Integrated Density − (Area of selected cell × Mean fluorescence of background readings)].
Quantitative data are represented as histograms (mean ± SD, n = 3) using JMP 14 software. Males and females were grouped together because no apparent difference was found between male and female mice regarding proportion and distribution of labeled cells in taste buds, and SOX10 expression in embryos and postnatal mice. Student's t-test was used for comparisons between the two groups. One-way analysis of variance (ANOVA) was used to evaluate statistical differences across the groups followed by post hoc Bonferroni tests. A P-value less than 0.05 was taken as statistical significance.
Results
SOX10-Cre labels taste bud cells in postnatal mice: few in early and abundant in mature taste buds
To map the lineage of SOX10-expressing cells (hereafter SOX10+) in taste buds, SOX10-Cre in which Cre expression is under control of the endogenous SOX10 promoter was used. The distribution of SOX10-Cre/tdT-labeled cells in taste buds was thoroughly analyzed in the soft palate and all three types of lingual taste papillae, that is, fungiform, foliate, and circumvallate, in young adult (8 week) mice when taste buds are mature. In serial sections of the soft palate and tongue tissues, SOX10-Cre/tdT-labeled cells were observed in most of the taste buds located by the marker Krt8 in the soft palate (Fig. 1A), and in all three types of lingual taste papillae, that is, fungiform (Fig. 1B), foliate (Fig. 1C), and circumvallate (Fig. 1D). Concurrently, SOX10-Cre-driven tdT+ cells were extensively distributed in the connective tissue (Fig. 1A–D) and von Ebner's glands (Supplementary Fig. S1). Of note, tdT+ cells were not found in the taste bud-surrounding tongue epithelium, including basal epithelial cells that are known as progenitors of taste buds [3,4,6–10].
FIG. 1.
SOX10-Cre/tdT (tdTomato)-labeled taste bud cells in all examined tissue regions, soft palate (A) and taste papillae, that is, fungiform (B), foliate (C), and circumvallate (D), in young adult (8-week old) SOX10-Cre/tdT mice. Krt8 (green) indicates presence of taste buds. White dashed lines demarcate lingual epithelium from the underlying connective tissue. Green dashed lines in (A, B) bracket a taste bud. Arrowheads point to tdT+ taste bud cells. Scale bars: 50 μm for all images (single-plane laser scanning confocal). Krt8, Keratin 8; SOX10, SRY-related HMG-box gene 10.
To evaluate at what stages SOX10-Cre+ cells contribute to taste buds, the distribution of SOX10-Cre/tdT-labeled cells in palatal and lingual taste buds was analyzed in newborn and in 2- to 16-week-old mice. At birth when taste buds are structurally recognizable, tdT+ cells were sporadically seen in Krt8+ taste buds in the soft palate and fungiform papillae (Fig. 2A, Soft palate and Fungiform); however, no SOX10-Cre/tdT-labeled taste bud cells were observed in the foliate and circumvallate papillae (Fig. 2A). At 2 and 4 weeks, tdT+ cells were found in taste buds more frequently in the soft palate and in all three types of lingual papillae (Fig. 2B, C). At 16 weeks, the distribution of tdT+ cells within taste buds (Fig. 2D) was similar to that at 8 weeks in the foliate and circumvallate papillae (Fig. 1), while a trend of reduced proportion of labeled cells was noticed in the fungiform papillae and soft palate (Fig. 2D). In the mice at all examined stages, tdT+ cells were extensive in the connective tissue of all the tissues (Fig. 2A–D) and in von Ebner's glands, as shown in Supplementary Fig. S1.
FIG. 2.
SOX10-Cre/tdT-labeled cells in taste buds were rare in newborn and abundant in adult mice. (A–D) Single-plane laser scanning confocal images of tissue sections of the soft palate and fungiform, foliate, and circumvallate papillae in newborn (A), 2-week- (B), 4-week- (C), 16-week- (D) old SOX10-Cre/tdT mice. Taste buds were Krt8+ (green). White dashed lines demarcate the lingual epithelium from the underlying connective tissue. Scale bars: 50 μm for all images. (E) Histograms (X ± SD, n = 3) to illustrate the percentages of SOX10-Cre/tdT-labeled tdT+ relative to total taste bud cells in the circumvallate papilla in newborn, 2-, 4-, 8-, and 16-week-old mice. The diamond dots represent the values of individual samples. **P < 0.01 one-way AONVA followed by Bonferroni post hoc test.
To understand the proportion of taste bud cells that are derived from SOX10-Cre+ cells, sagittal sections of the circumvallate papilla were used to visualize the transverse plane of taste bud cells for quantifications in newborn, 2-, 4-, 8-, and 16-week-old SOX10-Cre/tdT mice (Fig. 2E). Taste buds were recognized by Krt8 immunosignals and the boundary of each taste bud cell was marked by E-cadherin immunoproducts. The percentage of SOX10-Cre/tdT-labeled tdT+ versus total taste bud cells progressively increased (one-way ANOVA, P < 0.01) from zero at birth to 13.8% ± 2.1% at 2 weeks and 30.0% ± 2.0% at 4 weeks (Fig. 2E). After 4 weeks, when most taste bud cells are mature [27,28], a slight increase was shown by our quantitative data, up to 35.1% ± 3.4% at 8 weeks and 36.8% ± 5.7% at 16 weeks (Fig. 2E).
SOX10-Cre labels all three types, but predominantly type III differentiated taste bud cells
Taste buds comprise a group of heterogeneous cells, including type I, II, and III differentiated taste bud cells. To better understand which type(s) of taste bud cells are produced by SOX10-Cre+ progenitors, co-localization of tdT with immunosignals of markers for a specific type of differentiated taste bud cells, that is, NTPDase II for type I, PLCβ2 for type II, and SNAP25 for type III, was examined in the circumvallate taste buds of young adult (8 week) SOX10-Cre/tdT mice. In sections of the circumvallate papilla, with orientations showing both longitudinal and transverse planes of the taste bud cells, we observed that SOX10-Cre-driven tdT+ cells were prevalent throughout the taste buds. Once again, tdT+ cells were widespread in the connective tissue (Fig. 3A1–C1) and in von Ebner's glands, as shown in Supplementary Fig. S1. A subpopulation of taste cells, immunoreacted with a type I, II, or III cell marker, appeared as tdT+ (Fig. 3, solid arrowheads) in taste buds. Of interest, tdT+ taste bud cells were more frequently co-labeled with type III taste cell marker SNAP25 (Fig. 3C2) than type II cell marker PLCβ2 (Fig. 3B2).
FIG. 3.
SOX10-Cre/tdT-labeled distinct proportions of types I, II, and III differentiated taste bud cells. (A–C) Representative images (single-plane laser scanning confocal) of transverse (A1, B1, C1) and sagittal (A2, B2, C2) sections of the circumvallate papilla in 8-week-old SOX10-Cre/tdT mice. Specific types of taste bud cells were labeled by NTPDase II for type I (A1–2), PLCβ2 for type II (B1–2), and SNAP25 for type III (C1–2) cells. Solid arrowheads point to the cells co-labeled by tdT and markers for specific types of taste bud cells. Open arrowheads point to tdT+ cells in taste buds that were negative for specific taste cell type markers. Scale bars: 50 μm in (A1, B1, C1) and 20 μm for (A2, B2, C2). (D, E) Histograms (X ± SD, n = 3) to illustrate the percentages of tdT+ specific type (I, II, and III) versus total tdT+ (D) or versus total type II or III (E) taste bud cells in circumvallate taste buds. The diamonds represent data points of individual samples. **P < 0.01 (D) one-way ANOVA followed by Bonferroni post hoc test. **P < 0.01 (E) Student's t-test. ANOVA, analysis of variance.
Quantitative analyses were performed to understand the proportions of SOX10-Cre-driven tdT in labeling each specific type of differentiated taste bud cells (Fig. 3D). Type II and type III taste bud cells were easily distinguishable and counted. In all of the SOX10-Cre/tdT-labeled taste bud cells, tdT labeling was distributed unevenly among the three types (I, II, and III) of differentiated taste bud cells (one-way ANOVA, P < 0.01). Relative to total tdT+ taste bud cells, the quantity of tdT+ type III cells (73.9% ± 3.5%) was significantly higher than tdT+ type II (10.2% ± 1.6%) (P < 0.01) and tdT+ type I (15.9% ± 2.2%, extrapolated) (P < 0.01) cells. No significant difference was found between tdT+ type I and tdT+ type II cells (P = 0.10) (Fig. 3D). Relative to the total number of a specific type of taste bud cells, 79.1% ± 4.4% of type III and 16.5% ± 4.7% of type II taste bud cells were tdT+, and the difference was statistically significant (Student's t-test, P < 0.01) (Fig. 3E).
Taste bud cells vary in the intensity of Krt8 immunosignals with SOX10-Cre/tdT-labeled taste bud cells having a low level of Krt8
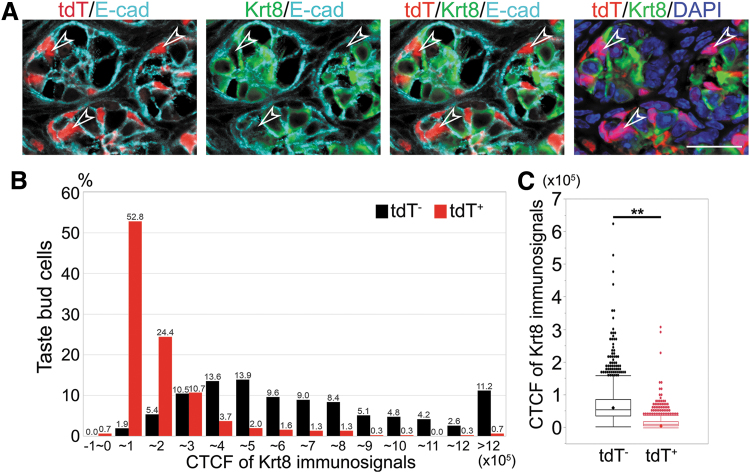
Krt8 is a widely used marker for taste bud cells. We observed that the intensity of Krt8 immunolabeling in individual taste bud cells varies considerably and that SOX10-Cre-/tdT-labeled taste bud cells could not be obviously identified with Krt8 immunostaining. We performed thorough examinations of the co-localization of tdT with Krt8 immunosignals in individual taste bud cells at a perpendicular plane to the elongated taste bud cells of the circumvallate papilla. Within the boundaries of taste bud cells labeled with E-cadherin, tdT+ taste bud cells did not have significant Krt8 immunoproducts (Fig. 4A, open arrowheads) (Fig. 4A, tdT/Krt8/E-cad or tdT/Krt8/DAPI). To illustrate the intensity of Krt8 immunosignals in taste bud cells in a quantitative manner, the CTCF of individual taste bud cells was calculated to represent Krt8 fluorescence intensity. CTCF values in individual taste bud cells were distributed in a broad range (Fig. 4B). The average of CTCF values of tdT+ taste bud cells (15,854 ± 23,950) were significantly lower compared with tdT− taste bud cells (68,425 ± 54,225) (P < 0.01) (Fig. 4C). A small population of tdT+ taste bud cells was completely absent of Krt8 immunoproducts (Fig. 4B). Moreover, 52% of tdT+, in contrast to only 2% of tdT−, taste bud cells had very low CTCF values, from 0 to 10,000 (Fig. 4B).
FIG. 4.
SOX10-Cre/tdT-labeled taste bud cells had a low intensity of Krt8 immunosignals (green). (A) Images of sagittal sections of the circumvallate papilla in an 8-week-old SOX10-Cre/tdT mouse. E-Cadherin (E-cad, cyan), Krt8 (green), and counterstained with DAPI (blue). Open arrowheads point to tdT+ taste bud cells that were not obvious in Krt8 immunolabeling. Scale bars: 20 μm (single-plane laser scanning confocal images). (B, C) Histograms (B) and box plots (C) to show the CTCF distribution of intensity of Krt8 immunosignals in tdT+ and tdT− taste bud cells in SOX10-Cre/tdT mouse circumvallate papilla (n = 3). The boxes in (C) represent the interquartile range, the whiskers represent the range of normal distribution, and dots represent the outliners. The line and diamond within each box represent the median and average value, respectively. **P < 0.01 (C) Student's t-test. CTCF, corrected total cell fluorescence.
To confirm our observation and verify the diversity of Krt8 in labeling taste bud cells, a cocktail of antibodies for type I (NTPDase II), II (PLCβ2), and III (SNAP25) was used to label all differentiated taste bud cells and double labeled with Krt8 (Supplementary Fig. S2A) in young adult (8 week) C57BL/6J wild-type mice. The heterogeneity of the intensity of Krt8 immunosignals was apparent among the taste bud cells labeled with taste cell markers (type I + II+III). Brightly (Supplementary Fig. S2A, solid arrowheads), mildly, and faintly labeled taste bud cells were seen (Supplementary Fig. S2A, open arrowheads). Interestingly, SNAP25+ type III taste bud cells were always faint in Krt8 labeling (Supplementary Fig. S2B).
To rule out the possibility that variability in Krt8 staining was due to differences in tissue fixation procedures, CTCF values in taste bud cells were collected from our routinely fixed and more lightly fixed tissues (Supplementary Fig. S2C, fixation scheme, Routine and Light). Our data showed that CTCF ranges and distributions in individual taste bud cells were similar between lightly and routinely fixed taste bud cell population (Supplementary Fig. S2C, D). No significant difference (P > 0.05) of CTCF was found between routinely and lightly fixed taste bud cell populations (44,341 ± 53,986 vs. 45,646 ± 52,921) (Supplementary Fig. S2D). The range of CTCF values was broad in both groups, with a distribution from −1,971 to 565,427 (routinely fixed) and −3,078 to 532,718 (lightly fixed).
SOX10 expression is detected in multiple tissue compartments, including neural crest, connective tissue core of taste papillae, and von Ebner's glands
To identify tissue compartments with SOX10+ progenitors for taste bud cells, the distribution of SOX10 mRNA was examined using in situ hybridization. As with previous reports [17], SOX10 mRNA expression was detected in cranial neural crest cells, including the cells in the trigeminal neural crest (Fig. 5A, TN), branchial arch 1 (Fig. 5A, BA1), and optic eminence (Fig. 5A, OE) of E8.5 embryos. Signals were not seen in the adjacent sections hybridized with sense RNA probe for SOX10 (Supplementary Fig. S3A).
FIG. 5.
SOX10 mRNA was detected with in situ hybridization using the digoxigenin-labeled antisense RNA probe (blue). SOX10 mRNA was detected in (A) neural crest cells in TN, BA1, and OE in the transverse sections of E8.5 embryo at the hindbrain level. Black dashed lines demarcate the NE from surrounding tissue; (B, C) the CTC of fungiform and circumvallate papillae, acinar of von Ebner's glands (Acinar of EG), and the duct (Duct of EG) opening (arrowhead) to the circumvallate papilla in newborn mice; (D, E) the CTC of fungiform and circumvallate papilla, acinar of von Ebner's glands (Acinar of EG), and the duct (Duct of EG) opening (arrowhead) to the circumvallate papilla in young adult (8-week old) mice. Green dashed lines in (B, D) bracket a taste bud. Black dashed lines mark the borders between the epithelium and underlying tissue. Scale bars: 50 μm for all images. BA1, branchial arch 1; CTC, connective tissue core; NE, neuroepithelium; OE, optic eminence; TN, trigeminal neural crest stream.
In newborn and 8-week-old mouse tongue, SOX10 mRNA was found in the connective tissue core of taste papillae, for example, fungiform (Fig. 5B, D) and circumvallate (Fig. 5C, E), although the staining was less intense in the adult compared to the newborn. SOX10 mRNA was also observed in the ducts and acini (Fig. 5C, E) of von Ebner's glands adjacent to the circumvallate papilla. Also, signals were seen in the opening of the ducts of von Ebner's glands at the bottom trench areas of circumvallate papilla (Fig. 5C, E, arrowheads).
Of note, SOX10 mRNA was not detected in the tongue epithelium—neither in the taste buds nor surrounding tongue epithelium. Furthermore, signals were not observed in lamina propria outside of the taste papillae. This included the filiform papillae and the surrounding tissue of the circumvallate papilla. Again, no signals were detected in the control sections of tongue tissue with sense RNA probe (Supplementary Fig. S3B–E).
Discussion
SOX10-Cre+ cells: newly discovered progenitors for taste buds in postnatal mice
Previous studies have reported that taste bud progenitor cells reside in the basal layer of the surrounding lingual epithelium and express Krt14, K5, p63, SOX2, and Lgr5 [8,11,12]. However, our recently published data revealed that a population of taste bud cells is labeled concurrently with cells under, but not within, the tongue epithelium [13,29]. In this study, using a model of Cre under the control of endogenous SOX10 promoter (SOX10-Cre), we found that in mature taste buds of adult mice, SOX10-Cre/tdT labeled a large population (up to around 35%) of taste bud cells. These taste bud cells had a significantly low level of immunoproducts of Krt8, a widely used pan-taste cell marker. SOX10 mRNA was not detected in taste buds or in the stratified lingual epithelium, suggesting the taste bud cells labeled by SOX10-Cre are derived from cells in tissue other than tongue epithelium. Together, our data indicate that the SOX10-Cre+ cells are a previously unrecognized source of progenitors for taste bud cells.
Lineage tracing of sonic hedgehog (Shh)+ cells in the epithelium of tongue rudiment with Shh-CreER mouse model demonstrated that early taste buds are from Shh+Krt8+ epithelial cells of the tongue primordium, not the surrounding tissue compartments [15,30]. Our data from this study showing that SOX10-Cre/tdT-labeled cells within taste buds are progressively more abundant with age in postnatal mice indicate a dual origin of taste buds during maturation and maintenance after birth, that is, from both surrounding basal cells of stratified tongue epithelium and SOX10-Cre+ cells under the tongue epithelium. Further studies are needed to demonstrate whether SOX10-Cre+ cells temporarily transit to Krt14+ basal epithelial cells, which, if so, will reconcile the seemingly contradictory reports [8].
Tissue locations of SOX10-expressing taste bud progenitors
SOX10 has been detected in multiple tissues and cell types, including migrating neural crest cells [17] and neural crest-derived cell populations from embryo to adult, including glial cells [18,19], melanocytes [20], peripheral ganglia in the gut [31,32], dorsal root ganglia [33–35], and stem cells in cranial bone marrow [35]. SOX10 expression has also been detected in submandibular salivary glands [21]. Moreover, SOX10 expression has been found in non-neural crest derived cell lineages such as glial cells in the central nervous system in postnatal mice [36–39].
To identify the tissue compartments that contain SOX10+ cells, in situ hybridization was performed and robust expression of SOX10 mRNA in migrating neural crest cells was detected in the migrating neural crest cells in E8.5 embryos, which is consistent with the previous reports [17]. Of particular interest, SOX10+ cells were detected in taste bud-surrounding tissue compartments in postnatal mouse tongues—the connective tissue core of taste papillae and von Ebner's glands under the circumvallate papilla, which add two novel reservoirs of SOX10+ cells.
Of note, SOX10 mRNA was not detected in the tongue epithelium, including taste buds and surrounding stratified tongue epithelium, nor the connective tissue outside of the taste papillae. These results lead us to propose three candidates of SOX10+ taste bud progenitors: neural crest- or non-neural crest-derived connective tissue cells in the core of taste papillae, or von Ebner's glands. To further define the specific tissue compartments that host SOX10+ taste bud (TB) progenitors, it will be necessary to mark lineages of each specific cell population.
Krt8 does not label all taste bud cells and SOX10-Cre+ progenitor-derived taste bud cells are not noticeably Krt8+
Taste bud cells are intragemmal, simple epithelial cells that are distinguishable from the surrounding stratified squamous epithelium. Krt8, a type II intermediate filament, is specifically expressed in simple epithelial cells and has been widely used as a pan taste bud cell marker [40]. We have observed that the intensity of Krt8 immunosignals is considerably variable in individual TB cells. Measurements of CTCF in individual TB cells to represent the intensity of Krt8 immunosignals showed a broad range of CTCF value distribution. Of note, the CTCF values in a small population of taste bud cells are below 0, suggesting the absence of Krt8 immunoproducts in these cells. Also, over half the population of taste bud cells had very low CTCF values. Our results indicate that taste bud cells vary remarkably in Krt8 expression, which prompts a reevaluation to address the question whether Krt8 can serve as a pan-taste bud cell marker.
Interestingly, SOX10-Cre/tdT-labeled taste bud cells were faint in Krt8 immunolabeling. The CTCF values in tdT+ taste bud cells were significantly lower than those in tdT− ones. Intermediate filaments, including Krt8, are key elements of cytoskeleton [41–43] and provide mechanical support for the plasma membrane to withstand the stress [44], which enhances cell structural and morphological integrity. In taste bud cells, the intermediate filaments are organized into bundles around the nucleus and in the periphery of the cytoplasm [45]. The difference of Krt8 levels in individual taste bud cells may represent the status/phase, type, or functions of taste cells.
Quantitative analyses demonstrated that SOX10-Cre+ cells contribute mainly to type III (74%), and a small proportion of type I (16%) and type II (10%) taste bud cells, indicating that SOX10-Cre+ cells tend to differentiate into neuronal-like taste bud cells (Type III). In our previous study using P0-Cre/tdT to trace the lineage of neural crest and derived connective tissue cells, 30% of tdT+ taste bud cells were SNAP25+ type III taste bud cells in the circumvallate papilla [13], suggesting that the SOX10-Cre+ cells represent an overlapping, but different progenitor cell population from P0-Cre+ cells. In addition, among type III taste bud cell population, 79% of them are labeled by SOX10-Cre/tdT, indicating that SOX10-Cre+ cells serve as a major progenitor source for type III taste bud cells. This complements with the previous finding that only type I and II taste bud cells are found as offspring of taste papilla placodal cells expressing Shh [15,24,25]. Overall, our data demonstrate that SOX10-Cre+ cells in the connective tissue and/or von Ebner's glands represent another source of progenitors, as well as those in the tongue epithelium, which give rise to a significant population of taste bud cells.
Supplementary Material
Acknowledgments
The authors thank Drs. Shi-You Chen, Luke Mortensen, and Franklin West (The University of Georgia, Athens, GA), for the discussion and feedback Dr. William Pavan for the SOX10 probe template; and Dr. Guiqian Chen and Mr. Brett Marshall for assistance in mouse maintenance.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This study was supported by the National Institutes of Health, grant number R01DC012308 and R21DC018089 to Hong-Xiang Liu
Supplementary Material
References
- 1. Beidler LM and Smallman RL (1965). Renewal of cells within taste buds. J Cell Biol 27:263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Conger AD and Wells MA (1969). Radiation and aging effect on taste structure and function. Radiat Res 37:31–49 [PubMed] [Google Scholar]
- 3. Hamamichi R, Asano-Miyoshi M and Emori Y (2006). Taste bud contains both short-lived and long-lived cell populations. Neuroscience 141:2129–2138 [DOI] [PubMed] [Google Scholar]
- 4. Cohn ZJ, Kim A, Huang L, Brand J and Wang H (2010). Lipopolysaccharide-induced inflammation attenuates taste progenitor cell proliferation and shortens the life span of taste bud cells. BMC Neurosci 11:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perea-Martinez I, Nagai T and Chaudhari N (2013). Functional cell types in taste buds have distinct longevities. PLoS One 8:e53399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen HM and Barlow LA (2010). Differential expression of a BMP4 reporter allele in anterior fungiform versus posterior circumvallate taste buds of mice. BMC Neurosci 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stone LM, Finger TE, Tam P and Tan S-S (1995). Taste receptor cells arise from local epithelium, not neurogenic ectoderm. Proc Natl Acad Sci U S A 92:1916–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okubo T, Clark C and Hogan BL (2009). Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells 27:442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sullivan JM, Borecki AA and Oleskevich S (2010). Stem and progenitor cell compartments within adult mouse taste buds. Eur J Neurosci 31:1549–1560 [DOI] [PubMed] [Google Scholar]
- 10. Miura H and Barlow LA (2010). Taste bud regeneration and the search for taste progenitor cells. Arch Ital Biol 148:107. [PMC free article] [PubMed] [Google Scholar]
- 11. Takeda N, Jain R, Li D, Li L, Lu MM and Epstein JA (2013). Lgr5 identifies progenitor cells capable of taste bud regeneration after injury. PLoS One 8:e66314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yee KK, Li Y, Redding KM, Iwatsuki K, Margolskee RF and Jiang P (2013). Lgr5-EGFP marks taste bud stem/progenitor cells in posterior tongue. Stem Cells 31:992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boggs K, Venkatesan N, Mederacke I, Komatsu Y, Stice S, Schwabe RF, Mistretta CM, Mishina Y and Liu H-X (2016). Contribution of underlying connective tissue cells to taste buds in mouse tongue and soft palate. PLoS One 11:e0146475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu H-X, Komatsu Y, Mishina Y and Mistretta CM (2012). Neural crest contribution to lingual mesenchyme, epithelium and developing taste papillae and taste buds. Dev Biol 368:294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thirumangalathu S, Harlow DE, Driskell AL, Krimm RF and Barlow LA (2009). Fate mapping of mammalian embryonic taste bud progenitors. Development 136:1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen G, Ishan M, Yang J, Kishigami S, Fukuda T, Scott G, Ray MK, Sun C, Chen SY and Komatsu Y (2017). S pecific and spatial labeling of P0-Cre versus Wnt1-Cre in cranial neural crest in early mouse embryos. Genesis 55:e23034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Southard-Smith EM, Kos L and Pavan WJ (1998). Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet 18:60. [DOI] [PubMed] [Google Scholar]
- 18. Bremer M, Fröb F, Kichko T, Reeh P, Tamm ER, Suter U and Wegner M (2011). Sox10 is required for Schwann-cell homeostasis and myelin maintenance in the adult peripheral nerve. Glia 59:1022–1032 [DOI] [PubMed] [Google Scholar]
- 19. Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I and Wegner M (1998). Sox10, a novel transcriptional modulator in glial cells. J Neurosci 18:237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shakhova O, Zingg D, Schaefer SM, Hari L, Civenni G, Blunschi J, Claudinot S, Okoniewski M, Beermann F and Mihic-Probst D (2012). Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat Cell Biol 14:882. [DOI] [PubMed] [Google Scholar]
- 21. Athwal HK, Murphy G III, Tibbs E, Cornett A, Hill E, Yeoh K, Berenstein E, Hoffman MP and Lombaert IM (2019). Sox10 regulates plasticity of epithelial progenitors toward secretory units of exocrine glands. Stem Cell Reports 12:366–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hari L, Miescher I, Shakhova O, Suter U, Chin L, Taketo M, Richardson WD, Kessaris N and Sommer L (2012). Temporal control of neural crest lineage generation by Wnt/β-catenin signaling. Development 139:2107–2117 [DOI] [PubMed] [Google Scholar]
- 23. Matsuoka T, Ahlberg PE, Kessaris N, Iannarelli P, Dennehy U, Richardson WD, McMahon AP and Koentges G (2005). Neural crest origins of the neck and shoulder. Nature 436:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mbiene JP and Roberts JD (2003). Distribution of keratin 8-containing cell clusters in mouse embryonic tongue: evidence for a prepattern for taste bud development. J Comp Neurol 457:111–122 [DOI] [PubMed] [Google Scholar]
- 25. Mistretta CM and Liu H-X (2006). Development of fungiform papillae: patterned lingual gustatory organs. Arch Histol Cytol 69:199–208 [DOI] [PubMed] [Google Scholar]
- 26. Rakowiecki S and Epstein DJ (2013). Divergent roles for Wnt/β-catenin signaling in epithelial maintenance and breakdown during semicircular canal formation. Development 140:1730–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krimm RF and Hill DL (1998). Quantitative relationships between taste bud development and gustatory ganglion cells. Ann N Y Acad Sci 855:70–75 [DOI] [PubMed] [Google Scholar]
- 28. Harada S and Kanemaru N (2005). Developmental changes of the taste sensation depending on the maturation of the taste bud and its distribution in mammals. Chem Senses 30:i56–i57 [DOI] [PubMed] [Google Scholar]
- 29. Iwatsuki K, Liu H-X, Grónder A, Singer MA, Lane TF, Grosschedl R, Mistretta CM and Margolskee RF (2007). Wnt signaling interacts with Shh to regulate taste papilla development. Proc Natl Acad Sci U S A 104:2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kramer N, Chen G, Ishan M, Cui X and Liu H-X (2019). Early taste buds are from Shh+ epithelial cells of tongue primordium in distinction from mature taste bud cells which arise from surrounding tissue compartments. Biochem Biophys Res Commun 515:149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Le Douarin NM and Teillet M-A (1973). The migration of neural crest cells to the wall of the digestive tract in avian embryo. Development 30:31–48 [PubMed] [Google Scholar]
- 32. Burns AJ and Le Douarin NM (1998). The sacral neural crest contributes neurons and glia to the post-umbilical gut: spatiotemporal analysis of the development of the enteric nervous system. Development 125:4335–4347 [DOI] [PubMed] [Google Scholar]
- 33. Hagedorn L, Suter U and Sommer L (1999). P0 and PMP22 mark a multipotent neural crest-derived cell type that displays community effects in response to TGF-beta family factors. Development 126:3781–3794 [DOI] [PubMed] [Google Scholar]
- 34. Bixby S, Kruger GM, Mosher JT, Joseph NM and Morrison SJ (2002). Cell-intrinsic differences between stem cells from different regions of the peripheral nervous system regulate the generation of neural diversity. Neuron 35:643–656 [DOI] [PubMed] [Google Scholar]
- 35. Nagoshi N, Shibata S, Kubota Y, Nakamura M, Nagai Y, Satoh E, Morikawa S, Okada Y, Mabuchi Y and Katoh H (2008). Ontogeny and multipotency of neural crest-derived stem cells in mouse bone marrow, dorsal root ganglia, and whisker pad. Cell Stem Cell 2:392–403 [DOI] [PubMed] [Google Scholar]
- 36. Touraine RL, Attié-Bitach T, Manceau E, Korsch E, Sarda P, Pingault V, Encha-Razavi F, Pelet A, Augé J and Nivelon-Chevallier A (2000). Neurological phenotype in Waardenburg syndrome type 4 correlates with novel SOX10 truncating mutations and expression in developing brain. Am J Hum Genet 66:1496–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng Y-C, Cheung M, Abu-Elmagd MM, Orme A and Scotting PJ (2000). Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Dev Brain Res 121:233–241 [DOI] [PubMed] [Google Scholar]
- 38. Inoue K, Tanabe Y and Lupski JR (1999). Myelin deficiencies in both the central and the peripheral nervous systems associated with a SOX10 mutation. Ann Neurol 46:313–318 [DOI] [PubMed] [Google Scholar]
- 39. Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U and Wegner M (2002). Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Knapp L, Lawton A, Oakley B, Wong L and Zhang C (1995). Keratins as markers of differentiated taste cells of the rat. Differentiation 58:341–349 [DOI] [PubMed] [Google Scholar]
- 41. Herrmann H, Bär H, Kreplak L, Strelkov SV and Aebi U (2007). Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol 8:562. [DOI] [PubMed] [Google Scholar]
- 42. Chang L and Goldman RD (2004). Intermediate filaments mediate cytoskeletal crosstalk. Nat Rev Mol Cell Biol 5:601. [DOI] [PubMed] [Google Scholar]
- 43. Traub P. (2012). Intermediate Filaments: A Review Springer Science & Business Media, Berlin/Heidelberg, Germany
- 44. Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D and Darnell J (2000). Molecular Cell Biology, 4th ed. National Center for Biotechnology Information, Bookshelf, New York [Google Scholar]
- 45. Takeda M, Obara N and Suzuki Y (1988). Intermediate filaments in mouse taste bud cells. Arch Histol Cytol 51:99–108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.