Abstract
Adeno-associated virus (AAV) vectors are quickly becoming the vectors of choice for therapeutic gene delivery. To date, hundreds of natural isolates and bioengineered variants have been reported. While factors such as high production titer and low immunoreactivity are important to consider, the ability to deliver the genetic payload (physical transduction) and to drive high transgene expression (functional transduction) remains the most important feature when selecting AAV variants for clinical applications. Reporter expression assays are the most commonly used methods for determining vector fitness. However, such approaches are time consuming and become impractical when evaluating a large number of variants. Limited access to primary human tissues or challenging model systems further complicates vector testing. To address this problem, convenient high-throughput methods based on next-generation sequencing (NGS) are being developed. To this end, we built an AAV Testing Kit that allows inherent flexibility in regard to number and type of AAV variants included, and is compatible with in vitro, ex vivo, and in vivo applications. The Testing Kit presented here consists of a mix of 30 known AAVs where each variant encodes a CMV-eGFP cassette and a unique barcode in the 3′-untranslated region of the eGFP gene, allowing NGS-barcode analysis at both the DNA and RNA/cDNA levels. To validate the AAV Testing Kit, individually packaged barcoded variants were mixed at an equal ratio and used to transduce cells/tissues of interest. DNA and RNA/cDNA were extracted and subsequently analyzed by NGS to determine the physical/functional transduction efficiencies. We were able to assess the transduction efficiencies of immortalized cells, primary cells, and induced pluripotent stem cells in vitro, as well as in vivo transduction in naïve mice and a xenograft liver model. Importantly, while our data validated previously reported transduction characteristics of individual capsids, we also identified novel previously unknown tropisms for some AAV variants.
Keywords: AAV, adeno-associated virus, gene therapy, next-generation sequencing, viral vectors, iPSC
Introduction
In recent years, several gene therapy products based on recombinant adeno-associated viral (AAV) vectors have reached clinical maturity and are available for the general public.1–3 Discovery of novel variants in nature as well as optimization of AAV's efficiency and specificity through directed evolution and rational design steadily increases the number of novel bioengineered AAVs with clinical potential.4–6 While this has opened up new opportunities for preclinical development and basic research in model organisms, to ensure maximal clinical impact, technologies that allow quick and precise comparison of a large number of variants in respect to their performance in specific target cells and tissues are needed. This is especially important when a given tissue is difficult to transduce, or when complex and rate-limiting model systems, such as humanized rodents or novel organoid systems, are used. Multiple groups have undertaken efforts to compare the performance of various AAV variants by testing them individually, usually using expression of fluorescent reporters as a readout of transduction efficiency in cell lines, primary cells, and animal tissues.7–13 While these approaches, at least in principle, can quickly yield accurate results, significant labor intensity limits the number of capsids that can be efficiently and accurately compared at the same time, especially when in vivo animal models are used. This has often resulted in a restricted comparison where a novel AAV variant is only compared with “established” AAVs like AAV1–AAV9 or even fewer variants. Furthermore, the availability of the test model often presents an additional limitation, such as in the case of primary human tissues or organoid cultures, often forcing investigators to select a limited number of “top candidates” for testing, increasing the risk of excluding potentially highly functional variants. Moreover, recent evidence suggests that, depending on the vector dose, conventional reporter screenings in vivo heavily under-represent the true transduction of a given capsid.13
Novel high-throughput approaches based on next-generation sequencing (NGS) coupled with bespoke bioinformatic analysis pipelines have recently been established14,15 and discussed16 as an alternative method for the detection of vector genomes. In this case, identical AAV cassettes containing a unique signature sequence, called a barcode (BC), can be packaged into multiple vector variants allowing for simultaneous NGS-based detection and quantification of vector genomes delivered by individual variants. Strategic placement of the BC sequence in the untranslated region of a reporter gene under the control of a ubiquitous promoter permits analyses at both the DNA and RNA/cDNA levels in different cell types and tissues.14,15
Tracking the BC in DNA recovered from cells of interest gives insight into which AAV capsids facilitate attachment to the cell surface and cell entry (referred to as physical transduction throughout the article), but does not provide information on the vectors' ability to successfully complete the intracellular path that ultimately leads to the generation of dsDNA vector genomes and transgene expression (referred to as functional transduction throughout the article). However, the DNA data can be supplemented by NGS on purified RNA, after cDNA generation, providing insight into the vectors' ability to functionally transduce the cells.14,17 The combination of the NGS DNA and RNA data, which allows simultaneous evaluation of multiple AAV variants for their ability to physically and functionally transduce cells of interest, makes this a very powerful tool that has the potential to revolutionize preclinical and translational studies, enabling time and cost-effective identification of the most suitable vectors with precise tropism for specific models.
In this study, we examined the power of this technology to study the performance of 30 published AAV variants assembled into an AAV Testing Kit. The Kit was evaluated on immortalized cell lines, induced pluripotent stem cells (iPSCs) and primary cells, as well as in vivo using naïve mice and a xenograft liver mouse model. The results demonstrate the power of this approach and validate the NGS-based testing protocol as a powerful tool for screening a large number of AAV variants and selection of the top AAV candidates for individual testing in relevant in vitro and in vivo models. To facilitate adaptation the approach described by the wider research community, the AAV constructs used in the study as well as the prepackaged AAV Testing Kit are available from the Vector and Genome Engineering Facility (Children's Medical Research Institute [CMRI], Australia). Furthermore, the description provided allows individual researchers to generate an identical AAV Testing Kit in their own laboratories or create similar Kits customized for their individual needs. Finally, the study highlights important differences between results obtained at the DNA, RNA/cDNA, and protein levels for individual variants that may have a significant impact on their use, depending on the need to actually express the transgene or simply to deliver the transgene as a template for gene editing.
Materials and Methods
Cell culture conditions and cell origins
AAV production and transduction experiments were performed in a validated human embryonic kidney HEK293T cell line purchased from ATCC and grown in Dulbecco's modified Eagle's medium (DMEM, Cat# 11965; Gibco) supplemented with 10% fetal bovine serum (FBS, Cat# F9423; Sigma-Aldrich), 1 × Pen Strep (Cat# 15070; Gibco), and 25 mM HEPES (Cat# 15630; Gibco). Human hepatocarcinoma 7 (HuH-7) cells were provided by Dr. Jerome Laurence (University of Sydney) and cultured in DMEM supplemented with 10% FBS, 1 × Pen Step, and 1 × non-essential amino acids (Cat# 11140; Gibco). Henrietta Lacks (HeLa) cells used in this study were a generous gift from Dr. Hilda Picket (CMRI). HeLa cells were maintained in DMEM supplemented with 10% FBS and 1 × Pen Strep. An immortalized line of iPSCs (KUCG_2) was provided by Dr. Anai Gonzalez-Cordero (CMRI) and maintained in TeSR-E8 (Cat# 05990; Stemcell) in Geltrex (Cat# A1413202; Gibco)-coated 24-well plates. Primary human fibroblasts, derived from pediatric healthy donor's foreskin, were cultured in DMEM/F12 (1:1) (Cat# 11330; Gibco) supplemented with 10% FBS. Primary human T cells derived from peripheral blood mononuclear cells of adult healthy volunteers using Lymphoprep (Cat# 07811; StemCell Technologies) and isolated by EasySep Release Human CD3-positive selection kit (Cat#17751; StemCell Technologies). T cells were activated using CD3/CD28 T Cell TransAct microbeads (Cat# 130-111-160; Mytenyi Biotec) and expanded using OpTmizer T Cell Expansion SFM (Cat# A1048501; Gibco) supplemented with Immune Cell SR (Cat# A2596102; Gibco), 10 ng/mL hIL-7 (Cat# 130-093-937; Miltenyi Botech), and 5 ng/mL hIL-15 (Cat# 130-093-955; Miltenyi Botech). The primary cells were provided by Dr. Wendy Gold and Dr. Kenneth Hsu (both The Children's Hospital at Westmead).
AAV production
All 30 AAVs in the Kit were produced independently to package a unique single-stranded ITR2-CMVp-eGFP-N6Barcode(BC)-WPRE-ITR2 transgene. For each serotype, 5 × 15 cm dishes of HEK293T cells were transfected using PEI (polyethylenimine, Cat# 115-100; PolyPlus) with a 1:1:2 molar ratio of pRep2CapX:pTransgene:pAd5. Three days post-transfection, the cells and media were harvested, and the AAVs were recovered by iodixanol gradient purification as previously described.18 A final buffer exchange (phosphate-buffered saline [PBS, Cat# 14190; Gibco], 50 mM NaCl [Cat# S5150-1L; Sigma-Aldrich], 0.001%, Pluronic F68 [v/v, Cat# 24040; Gibco]) and concentration step were performed using Amicon Ultra-4 Centrifuge Filter Units with Ultracel-100 kDa membrane (Cat# UFC810024; EMD Millipore). The resulting AAVs were titrated using real-time quantitative polymerase chain reaction (qPCR) master mix (Cat# 172-5125; Bio-Rad) with serial dilutions of a linearized plasmid as a standard curve and eGFP primers (F: 5′-TCAAGATCCGCCACAACATC-3′; R: 5′-TTCTCGTTGGGGTCTTTGCT-3′) as previously described.19
All vector preparations underwent an additional quality control step to confirm the sequence of the unique N6BC. To do so, primers N6BC-Seq_F (5′-TACGGCGTGCAGTGCTTCAGC-3′) and N6BC-Seq_R (5′-GCTGGCAACTAGAAGGCACAG-3′) were used to amplify a 1,198 bp amplicon containing the barcode sequence using Q5 High-Fidelity DNA Polymerase (Cat# M0491L; New England Biolabs) using the following protocol: initial denaturation at 98°C for 10 min, followed by 42 cycles of 98°C for 10 s, 59°C for 10 s, and 72°C for 45 s, and a final cycle of 72°C for 10 min.
The PCR amplicons were purified using the QIAquick Gel Extraction Kit (Cat# 28706; Qiagen) according to the manufacturer's instructions. Ten to 30 ng of the purified PCR products were subsequently submitted for Sanger Sequencing at the Garvan Molecular Genetics Facility (Sydney, Australia) using N6BC-Seq_R (5′-GCTGGCAACTAGAAGGCACAG-3′) and N6BC-Seq _internal (5′-ACGGCAACATCCTGGGGCA-3′) primers. For individual AAV variants and their assigned and verified N6BC sequence, see Supplementary Table S1.
In vitro transductions
The cells were seeded 16 h before transduction. HEK293T, HeLa, and HuH-7 cells were seeded at 100,000 cells per well, while T cells and iPSCs (KUCG_2) were seeded at 400,000 cells per well in 24-well tissue-culture-treated plates (Cat# 353047; Falcon). Fibroblasts were seeded at 225,000 cells per well in 6-well plates (Cat# 353046; Falcon). The adherent cells were incubated with the AAV Testing Kit overnight (16 h, apart from iPSCs, which were incubated with AAVs for 3 h only, due to concerns of vector toxicity20), followed by two washes with PBS. The T cells were harvested after a 16 h transduction and underwent two cycles of PBS washes after spins at 300 g for 10 min.
After PBS washes, fresh medium was provided, and all cells were allowed to grow for an additional 32 h before harvest. Adherent cells were harvested by incubating with Trypsin-EDTA solution (Cat# T3924; Sigma-Aldrich) for 10 min. The cells were then recovered in fresh media, spun at 300 g for 10 min, and the dry pellet was used for DNA and RNA extraction. T cells were processed with the same steps, but harvest (just as the PBS wash) required centrifugation and washes as described above.
Experiments with the single AAV variants for verification of the NGS results were performed following the above-outlined protocol, with the exception that the final for adherent cells harvest was performed using TryPLE Express (Cat# 12604021; Gibco) for HEK293T, HeLa, HuH-7 cells, and fibroblasts, while iPSCs were harvested using StemPro ACCUTASE (Cat# 07922; StemCell). In both cases, the enzymatic process was stopped by diluting the samples 3 × with standard growth media.
Flow cytometry
Harvested cells from the AAV Testing Kit and single transduction experiments were collected into 5 mL fluorescence-activated cell sorting (FACS) tubes (Cat# 352235; Falcon) and pelleted for 10 min at 300 g. The medium was removed, and the pellet was resuspended in FACS buffer (PBS, 2% FBS, and 5 mM EDTA [Cat# 15575-020; Invitrogen]). The flow cytometry analysis was performed using a BD LSRII cell analyzer at the Flow Cytometry Facility, Westmead Institute for Medical Research (Westmead, NSW, Australia). The data were analyzed using FlowJo 7.6.1 (FlowJo, LLC).
Animal work
Mouse studies were supported by the Bio Resources Core Facility at CMRI. All animal care and experimental procedures were approved by the joint CMRI and The Children's Hospital at Westmead Animal Care and Ethics Committee. Fah−/−/Rag2−/−/Il2rg−/− (FRG) mice were housed inside a BSL2 facility in individually ventilated cages with 10% 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclohexanedione (NTBC)-supplemented in drinking water.21 Six- to 8-week-old FRG females were engrafted with primary human hepatocytes (Lonza Group Ltd., Basel, Switzerland) as described previously, followed by on/off NTBC cycles to promote the expansion of human cells.21,22 Levels of human hepatocyte engraftment in the chimeric liver of humanized FRG (hFRG) mice were assessed by measuring the levels of human albumin in peripheral blood using a Human Albumin ELISA Quantitation Kit (Cat# E80-129; Bethyl) as previously described.21 Mice were transduced with 1 × 1011 vgc per mouse of the AAV Testing Kit diluted in 100 μL of sterile saline by intravenous (lateral tail vein) injection.
Mice were euthanized by isofluorane inhalation 1 week after transduction for organ and hepatocyte harvests. For FACS, hepatocytes were harvested by collagenase perfusion as previously described.22 Single-cell suspensions were obtained by cannulation of the inferior vena cava, and pumped with an osmotic minipump (Gilson Minipuls 3) in the following order: 25 mL of Hank's balanced salt solution (HBSS) (Cat# H9394; Sigma), 25 mL of HBSS supplemented with 0.5 mM EDTA, 25 mL HBSS, and 25 mL of HBSS supplemented with 5 mM CaCl2, 0.05% w/v collagenase IV and 0.01% w/v DNase I. After perfusion, the chimeric liver was harvested and placed in a sterile cell culture dish containing DMEM supplemented with 10% FBS.
The cells were collected after opening the liver capsule followed by centrifugation at 50 g for 3 min at 4°C. The cell pellet was resuspended in DMEM and passed through a 100 μm nylon cell strainer. Isotonic Percoll (10% 10 × PBS and 90% Percoll, Cat# 17089102; GE Healthcare) was added to the cell suspension to separate the live and dead cells. Live cells were pelleted at 650 g for 10 min at 4°C, and the pellet was resuspended in FACS buffer. To distinguish between murine and human hepatocytes, cells were labeled with phycoerythrin-conjugated antihuman-HLA-ABC (clone W6/32, Cat# 12-9983-42, 1:20; Invitrogen), biotin-conjugated antimouse-H2Kb (clone AF6-88.5, Cat# 553568, 1:100; BD Pharmigen), and allophycocyanin-conjugated streptavidin (Cat# 17-4317-82, 1:500; eBioscience). Labeled samples were sorted to a minimal 95% purity using a BD AriaIII cell sorter. The purity of the cell sorting was verified using droplet digital (dd)PCR for murine albumin in human cells and human albumin in mouse cells (see Vector DNA Copy Number per Cell section for the exact method). FACS was performed in the Flow Cytometry Facility, Westmead Institute for Medical Research. The data were analyzed using FlowJo 7.6.1 (FlowJo, LLC).
DNA and RNA extraction from cells and tissue samples
DNA and RNA were isolated from the cell pellets from the in vitro experiments using the AllPrep DNA/RNA Mini Kit (Cat# 80204; Qiagen) following the manufacturer's instructions.
DNA from the mouse and human hepatocytes and homogenized mouse tissues was isolated using phenol–chloroform extraction after proteinase K digest. In brief, the cells were resuspended in 200 μL lysis buffer (100 mM Tris-HCl pH 8 [Cat# 15575-020; Invitrogen], 5 mM EDTA [Cat# 15575-020; Invitrogen], 0.2% [w/v] sodium dodecyl sulfate [Cat# 71736-100ML; Sigma-Aldrich], 200 mM NaCl [Cat# S5150-1L; Sigma-Aldrich]), 50 μg/mL Proteinase K (Cat# 19131; Qiagen) and incubated overnight at 55°C with rotation. The next day, 200 μL of phenol:chloroform:isoamyl alcohol (25:24:1, Cat# P3803-100ML; Sigma-Aldrich) was added and mixed well with the sample. The phases were separated by centrifugation at 15,000 g for 5 min; the top aqueous phase containing DNA was transferred to a new tube and mixed with 0.1 × v/v 3 M sodium acetate and 2.5 × v/v 100% ethanol, incubated for 1 h at −80°C, and spun at 15,000 g, 4°C for 20 min. The resulting DNA pellet was washed with 75% (v/v) ethanol and finally resuspended in nuclease-free water.
RNA from the mouse cells and homogenized tissues was extracted using the Direct-zol RNA Microprep Kit (Cat# R2063; Zymogen) following the manufacturer's instructions.
Vector DNA copy number per cell
The vector copy numbers were measured using ddPCR (Bio-Rad, Berkeley) using QX200 ddPCR EvaGreen Supermix (Cat# 1864034; Bio-Rad) with eGFP primers (see AAV Production section above). Six nanograms of genomic DNA was used as a starting concentration, and was diluted 1:100 and 1:10,000 for vector genome quantification. Vector genomes were normalized to either human or mouse albumin using hAlb_F: 5′-TGCTGTCATCTCTTGTGGGCTG-3′, hAlb_R: 5′-AACTCATGGGAGCTGCTGGTTC-3′ for cells and tissues from human origin as well as mAlb_F: 5′-AACTGCTACTCCCCTCCTAC-3′, mAlb_R: 5′-TTTACCCCAGTGCAGGAAAG-3′ for murine tissues. Six nanograms of genomic DNA was used to establish the albumin copy number in the samples.
Reverse transcription of extracted RNA
Seven hundred fifty nanograms of total RNA was incubated with 2 units of TURBO DNase (Cat# AM2238; Invitrogen) for 3 h followed by 10 min DNase heat inactivation at 75°C. The DNase-treated RNA was then used for cDNA synthesis using SuperScript IV First-Strand Synthesis System (Cat# 18091050; Invitrogen) following the manufacturer's instructions using 2 μM of a WPRE-binding primer (5′-GGATTTATACAAGGAGGAGAAAATGAAAG-3′) to specifically synthesize AAV encoded transgene cDNA.
Next-generation sequencing
Fifty nanograms of extracted total genomic and vector DNA as well as 5 μL final cDNA product were used as templates for amplification of the barcode region in each AAV transgene. To do so, a 150 bp amplicon containing the N6BC in the middle was generated using Q5 High-Fidelity DNA Polymerase (Cat# M0491L; New England Biolabs) and N6BC-NGS_F (5′-GCTGGAGTTCGTGACCGCCG-3′) and N6BC-NGS_R (5′-CAACATAGTTAAGAATACCAGTCAATCTTTCAC-3′) primers, using the following PCR program: denaturation step at 98°C for 2 min (for purified gDNA and cDNA) or 10 min (for packaged AAVs), followed by 42 amplification cycles: 98°C for 10 s, 62°C for 10 s, 72°C for 10 s, and a final extension at 72°C for 10 min.
The amplicons were separated on a 2% agarose (Cat# BIO-41025; Bioline) gel in 1 × Tris-acetate-EDTA (Cat#24710-030; Invitrogen), and the DNA was extracted using Zymoclean Gel DNA Recovery Kit (Cat# D4001; Zymogen) following the manufacturer's instructions. For mouse organs except the liver, the forward primer had four random nucleotides at the 5′-end, which served as additional barcodes, allowing for the pooling of the PCR products and sending them as one sample, which was later deconvoluted using the “Separate Barcodes” function integrated in Geneious (v11.1.4). One microgram of amplicon DNA per sample was sent to GENEWIZ Genomics (Suzhou, China) for 150 bp paired-end amplicon sequencing service on an Illumina HiSeq/MiSeq instrument. Paired reads were merged using BBMerge Paired Read Merger Version 37.64 and subsequently filtered using the integrated Geneious Workflow (v11.1.4; Geneious).23 A custom-made N6BC counter was written in Python, and used to quantify and map each barcode to the corresponding AAV variant. For the overall sum of read counts of each sample, see Supplementary Table S2.
Normalization of NGS reads
NGS data obtained from DNA and RNA/cDNA from all cells and tissues were normalized to the distribution of AAVs in the final AAV Testing Kit mix. Read counts for each sample and each variant were multiplied by the variant-specific “normalization co-efficient,” which was calculated as follows:

Results and Discussion
Preparation of the AAV Testing Kit mix
The AAV Testing Kit contains 30 previously published AAV variants (Table 1), including naturally occurring serotypes and isolates as well as bioengineered variants developed through DNA shuffling, peptide display, domain swapping, and in silico-guided ancestral reconstruction. All capsid variants were used to package the ITR2-CMVp-eGFP-N6BC-WPRE-ITR2 transgene, where N6BC represents a unique barcode (Fig. 1A and Supplementary Table 1).
Table 1.
Capsid variants used in the study
| No. | Variant | Capsid Origin | Ref. |
|---|---|---|---|
| 1 | AAV1 | Natural serotype | 24 |
| 2 | AAV2 | Natural serotype | 25 |
| 3 | AAV3b | Natural serotype | 25,27 |
| 4 | AAV4 | Natural serotype | 28 |
| 5 | AAV5 | Natural serotype | 29 |
| 6 | AAV6 | Natural serotype | 27 |
| 7 | AAV7 | Natural serotype | 31 |
| 8 | AAV8 | Natural serotype | 31 |
| 9 | AAV9 | Natural serotype | 33 |
| 10 | AAV10 | Natural serotype | 34 |
| 11 | AAV11 | Natural serotype | 34 |
| 12 | AAV12 | Natural serotype | 36 |
| 13 | AAV13 | Natural serotype | 38 |
| 14 | AAVrh10 | Natural isolate | 40 |
| 15 | AAVDJ | DNA family shuffling | 8 |
| 16 | AAVDJ/8 | DNA family shuffling | 8 |
| 17 | AAVLK01 | DNA family shuffling | 26 |
| 18 | AAVLK02 | DNA family shuffling | 26 |
| 19 | AAVLK03 | DNA family shuffling | 26 |
| 20 | AAVLK19 | DNA family shuffling | 26 |
| 21 | AAVNP22 | DNA family shuffling | 30 |
| 22 | AAVNP40 | DNA family shuffling | 32 |
| 23 | AAVNP59 | DNA family shuffling | 32 |
| 24 | AAVNP66 | DNA family shuffling | 30 |
| 25 | AAVNP94 | DNA family shuffling | 30 |
| 26 | AAV7m8 | Peptide display | 35 |
| 27 | AAVAnc80 | Reconstructed capsid | 37 |
| 28 | AAVPHP.eB | Peptide display | 39 |
| 29 | AAV2i8 | Domain swapping | 41 |
| 30 | AAV2retro | Peptide display | 42 |
AAV, adeno-associated virus.
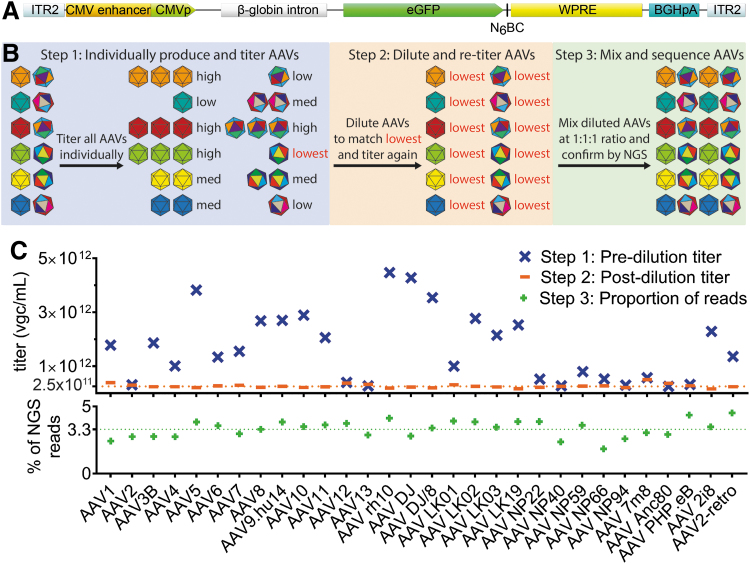
Figure 1.
Experimental setup and preparation of the AAV Testing Kit mix. (A) The ITR2-CMVp-eGFP-N6BC-WPRE-ITR2 transgene packaged in the individual AAV vectors used in the study. (B) Individual steps involved in the preparation of the AAV Testing Kit. (C) Upper panel: titration of individually produced AAVs (Step 1) and diluted AAV variants (Step 2). The dilution benchmark of 2.5 × 1011 vgc/mL is indicated by the orange dotted line. Lower panel: Results of the NGS analysis of the AAV distribution in the AAV Testing Kit used in this study. The expected % contribution of individual variants (100% ÷ 30 = ∼3.33%) is indicated by the green dotted line. AAV, adeno-associated virus; N6BC, unique 6 nucleotide barcode; NGS, next-generation sequencing.
The qPCR quantification of packaged AAV variants revealed an ∼20-fold difference in vector titer between the variants with the lowest titer at 2.5 × 1011 vg/mL for AAV Anc80 and the highest titer at 4.5 × 1012 vgc/mL for AAV rh.10 (Fig. 1C, upper panel—”pre-dilution titer”; Supplementary Fig. S1A; mean: 1.7 × 1012 vgc/mL, median: 1.6 × 1012 vgc/mL, SD: ±1.3 × 1012 vgc/mL). To ensure the generation of a mixed library containing all 30 variants at an equimolar ratio, the initial titer data were used to dilute all AAVs to the concentration of the variant with the lowest titer (2.5 × 1011 vgc/mL). After qPCR-based confirmation of the titers (Fig. 1C, upper panel—”post-dilution titer”; Supplementary Fig. 1A; mean: 2.6 × 1011 vgc/mL, median: 2.5 × 1011 vgc/mL, SD: ±0.7 × 1011 vgc/mL), the diluted variants were mixed at an equimolar ratio. As a final quality control step, the mixed AAV Testing Kit was subjected to NGS-based N6BC analysis.
The analysis of NGS data (Fig. 1C) revealed narrow distribution of the AAV variants in the AAV Testing Kit mix, with AAV NP66 as the least represented variant within the mix, at 1.9% of NGS reads corresponding to its assigned N6BC and AAV2-retro as the most over-represented variant that accounted for 4.5% of total NGS reads (Fig. 1C, lower panel; Supplementary Fig. S1A; mean: 3.33%, median: 3.47%, SD: ±0.006%). The small variations in AAV distribution are most likely the consequence of small additive errors in the titration and mixing of the individual vectors that formed the final AAV Testing Kit. The observed differences will not affect the experimental readout as they will be corrected through normalization of the DNA and RNA NGS data by multiplying the raw read numbers for each variant by its corresponding normalization coefficient (see Material and Methods section). Encouraged by these results, we proceeded to the second phase of the project—in vitro and ex vivo transduction studies.
Transduction of immortalized cell lines, primary cells, and iPSCs
Immortalized cell lines (HEK293T, HeLa, and HuH-7), primary human cells (T cells and fibroblasts) as well as iPSCs (line KUCG_2) were transduced with the AAV Testing Kit at multiplicities of transduction (MOTs) of 100,000 (high dose) and 10,000 (low dose) vgc/cell, which corresponded to doses of 3,333 and 333 vgc/cell for each of the AAV variants. In addition, in light of published reports suggesting potential AAV vector toxicity,20 iPSCs were transduced at a low total MOT of 1,000 vgc/cell. The cell lines used were selected based on their frequent use for studying cell biology, cell–vector interactions and use in producing viral vectors, while the primary cells and iPSCs were selected due to their increasing applications in regenerative medicine. In fact, AAV vectors have been shown recently to facilitate high efficiency in on-target genome editing in T cells (AAV6)43 and iPSCs (AAV DJ),44 establishing them as vectors of choice for a wide range of preclinical and clinical applications targeting those cells.
Because the NGS analysis at the DNA level cannot distinguish between vectors inside the cells and those still present outside the cells, all cells underwent extensive washing after transduction to remove any free AAV particles. After the removal of free AAVs, cells were cultured for 32 h (see Materials and Methods section) to allow the vectors that entered the cells to complete the intracellular journey to the nucleus and to initiate transgene expression. Furthermore, to remove any AAVs that could be attached to the cell surface, cells were trypsinized at harvest, a critical step introduced to reduce background signal from vector particles exposed on the cell surface that have not fully entered. The harvested cells were analyzed by flow cytometry for GFP to determine the overall efficiency of transduction. HEK293T, HeLa, and HuH-7 were >95% positive for GFP expression at both vector doses, while the fibroblasts were 65% GFP positive at the high dose and 40% GFP positive at the low dose. The T cells were 39% GFP positive at the high dose and 18% GFP positive at the low dose. No GFP signal was detected in the iPSCs (KUCG_2) at any of the three MOTs used. Based on the FACS results, all the cells transduced at the low dose were selected for NGS analysis, except for primary T cells where the high dose was selected for further analysis to more closely match the transduction rate of the primary fibroblasts.
After genomic DNA and RNA extraction and subsequent cDNA synthesis (Fig. 2A), PCR was used to amplify the 150 bp region of the AAV genome that contained the signature N6BC. All samples were submitted for NGS analysis to establish the contribution of the individual AAV variants to the overall physical and functional transduction of the cells, using the NGS results from the pretransduction mix for input normalization. Furthermore, the genomic DNA was analyzed by ddPCR to measure the average AAV vector copy number inside the cells, using human albumin as an endogenous control.
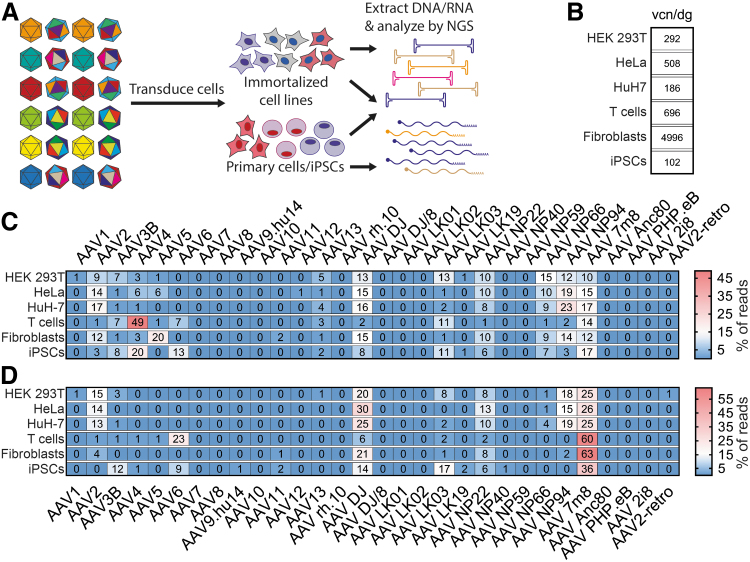
Figure 2.
Experimental setup and results of in vitro and ex vivo transductions with the AAV Testing Kit. (A) Schematic representation of the experimental procedure. (B) The average total vector copy number in cells transduced with the AAV Testing Kit. The values are given as vcn/dg. (C) Results of NGS-based quantification of vector-encoded DNA. Values are given as percentage of total reads for each cell type. (D) Results of NGS-based analysis of vector function based on quantification of vector genome-transcribed mRNA. Values are given as percentage of total reads for each cell type. The values presented in panels (C, D) were rounded to the nearest whole number. vcn/dg, vector copy number per diploid genome.
The analysis of vector genome copy number revealed that AAVs were very efficient at entering (physical transduction) each of the cell types (Fig. 2B). The iPSCs had the lowest average vector copy number of 102 per diploid genome (vcn/dg), while primary human fibroblasts were the most transducible with an average of 4,996 vcn/dg. It is important to keep in mind that the immortalized cells are known to be polyploid and have frequent genomic duplications, translocations, and other chromosomal aberrations.45 While it is thus impossible to accurately estimate the average vector copy number per cell using endogenous genes as reference, it would be reasonable to anticipate that the average number of vector genomes per cell was underestimated in our analysis.
Analysis of the genomic DNA-NGS results revealed that for each cell type, many capsids contributed to the observed average vector copy number (Fig. 2C). Considering only the AAVs that contributed to >1% of the total reads, the HEK293T cells were transduced by the highest number of individual AAV variants (n = 13), and we observed the lowest variability of the transduction efficiencies among the AAVs that entered the cells (Supplementary Fig. S1), with AAV NP66 detected as the most abundant variant accounting for 15% of input-normalized total reads, which is equivalent to 44.6 AAV NP66 copies per diploid genome.
The most abundant AAV in HeLa and HuH-7 cells was AAV NP94 with 19% (98.4 vcn/dg) and 23% (43.1 vcn/dg) of DNA-NGS read contributions, respectively (Fig. 2C). Entry into primary human T cells was dominated by AAV4, which accounted for 49% (338.8 vcn/dg) of DNA NGS reads (Fig. 2C). Primary human fibroblasts and iPSCs had a broader distribution among DNA-contributing AAV variants compared with T cells, which showed a strong peak for AAV4 (Supplementary Fig. S1B), with AAV5 (20% of DNA reads, 998.5 vcn/dg) and AAV4 (20% of DNA reads, 20.7 vcn/dg) as the most abundant variants for those cells, respectively.
Interestingly, the DNA data were not predictive of the results obtained for RNA/cDNA, highlighting the fact that cell entry (physical transduction) may not be the best predictor of vector function (transgene expression, functional transduction) (compare Fig. 2C, D). For example, AAV4 was the most abundant variant in T cells at the DNA level, but its relative expression at the RNA level in those cells was very low, implying that this serotype may be inefficient at one or multiple steps the vector must complete between cell entry and triggering transgene expression. While it has been previously shown that barriers such as inefficient nuclear translocation,46 nuclear uncoating,47,48 and second-strand synthesis/transcriptional regulation by the capsid49 can affect function of AAV vectors, it was interesting and unexpected that a vector capable of such efficient cell entry was so inefficient at transgene expression. These results position AAV4 as a powerful genetic tool to study AAV biology and the interactions between AAV and human T cells.
Based on published data and our own experience (data not shown), we expected AAV DJ, a chimeric AAV2/AAV8/AAV9 capsid, to perform with high efficiency in the immortalized cell lines tested.8 Indeed, AAV DJ was the top variant in HeLa cells (30% of total RNA reads), and also performed very well in HEK293T (20% of RNA reads) and HuH-7 cells (25% of RNA reads). In fact in the HEK293T and HuH-7 cells, AAV DJ was surpassed only by AAV 7m8 (25% of RNA reads for both), an AAV2 with several point mutations as well as a peptide insertion after amino acid position N587, which was originally selected using directed evolution in murine photoreceptors (Fig. 2D).35 Strikingly, AAV 7m8 was also the highest expressing variant in primary human T cells (60% of RNA reads), fibroblasts (63% of RNA reads), and iPSCs (36% of RNA reads) (Fig. 2D) positioning it as the most efficient variant for in vitro and ex vivo transductions in our study.
Based on published results, AAV6 was expected to functionally transduce primary T cells with superior efficiency.43,50 However in our NGS screen, it only contributed 23% of the total RNA reads, substantially less than the 60% observed for AAV 7m8 (Fig. 2D). Compared with other cells used in this study, significantly less is known about the efficiency of AAV vectors for targeting primary human fibroblasts and iPSCs. AAV2 has been shown to transduce two different lines of human foreskin fibroblasts and the human fibroblast cell line HT1180 very efficiently.8,11,51 While we were able to detect AAV2 in primary fibroblasts at the RNA level (4% of total reads), it was outperformed by AAV 7m8 and AAV DJ (21% of total RNA reads). Our RNA data indicated that human iPSCs were most efficiently transduced by AAV 7m8, AAV LK03, AAV DJ, AAV3B, and AAV6 (in descending order). These findings are in agreement with a previously published report that human iPSCs were most efficiently transduced by AAV 7m8, AAV3B, and AAV6 (AAV LK03 and AAV DJ were not included in that study).52 In summary, the high performance of AAV 7m8, both in vitro and ex vivo, positions this bioengineered variant as a strong candidate for nucleic acid delivery into iPSCs, T cells, and fibroblasts. These newly identified properties of AAV 7m8 with respect to T cells and fibroblasts may prove invaluable for applications such as gene editing, making this AAV variant a potential candidate for preclinical and clinical applications in cell therapy and regenerative medicine.43,44
Next, we wanted to investigate whether the RNA NGS results were predictive of the transgene expression at the protein level. To do so, we tested the five most efficient candidates for each cell type using a single-stranded AAV-CMV-GFP reporter construct (Fig. 3A). Based on the expected differences in AAV permissiveness between the individual cell types, the cells were transduced at a range of doses followed by GFP expression quantification using flow cytometry (Fig. 3B and Supplementary Fig. S2). Analysis of the selected groups revealed that in the cell lines, the differences observed at the RNA level from the NGS data did not correlate perfectly with the observed differences at the GFP level, especially for the top performers. For example, while in HEK293T cells based on the RNA-NGS results AAV 7m8 > AAV DJ > AAV NP59 > AAV2 >> AAV LK03, at the GFP level the order changed to AAV2 > AAV 7m8 > AAV NP94 > AAV DJ >> AAV LK03 (Fig. 3B). Similar trends were observed for HeLa and HuH-7 cells (Fig. 3B).
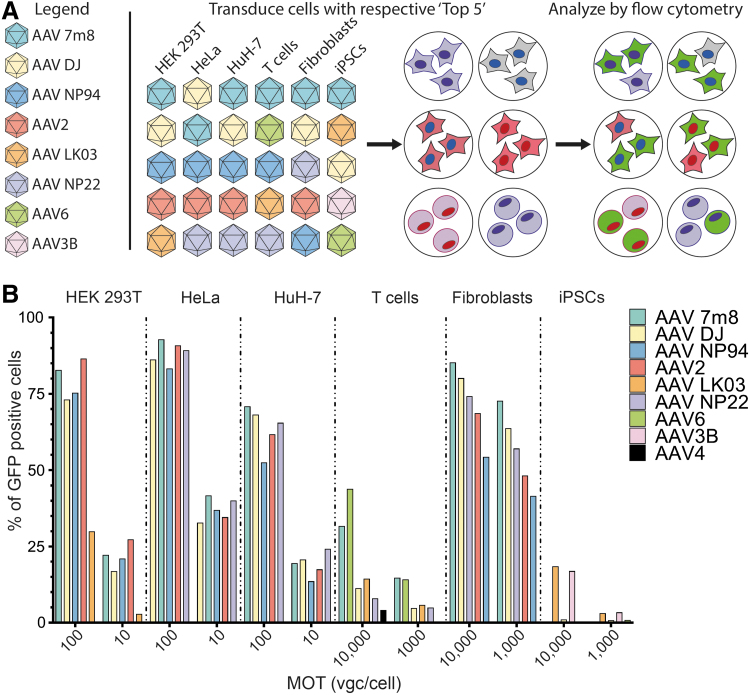
Figure 3.
Analysis of selected AAVs based on transgene expression at the protein level. (A) Schematic representation of the experimental design. The top performing variants (n = 5) for each cell type were used individually to transduce cells, and the vector performance was established by quantifying GFP expression. Individual variants used are color coded for clarity. (B) Results of FACS analysis for each cell type transduced with its respective “Top 5” AAV variants. FACS, fluorescence-activated cell sorting.
There could be a number of possible explanations for the observed differences between NGS RNA reads and GFP results. First, the different half-lives of the two molecules could explain why the RNA readout does not correspond to protein levels measured at the same timepoint after transduction. Second, cotransduction with a pool of AAVs may affect the transduction efficiency of individual variants, highlighting the need to individually evaluate the efficiency of the top candidates identified through the NGS screen. Moreover, the exact vector dose might have also played a role. In the NGS screen, the cell lines analyzed were transduced with 333 vgc per variant per cell. For example, in agreement with NGS data, AAV 7m8 was the most efficient variant in the HuH-7 cells transduced at MOT 100 vgc/cell. However, at MOT 10 vgc/cell, AAV 7m8 (19% GFP-positive cells) was less efficient than AAV NP22 (24% GFP-positive cells) and AAV DJ (21% GFP-positive cells) (Fig. 3B), indicating dose-dependent variations at the GFP expression level.
Interestingly, while the RNA NGS data predicted AAV 7m8 and AAV6 as top performers in T cells (60% and 23% of total NGS RNA reads, respectively), at the GFP level AAV6 was more efficient than AAV 7m8 at MOT 10,000 vgc/cell, and both vectors performed similarly at the lower dose of 1,000 vgc/cell. Moreover, the relative efficiencies of the top performers in human fibroblasts at the RNA level were consistent with the GFP data, where the functional order of variants remained the same (AAV 7m8 > AAV DJ > AAV NP22 > AAV2 > AAV NP94) (compare Figs. 2D and 3B). However, it is important to point out that the NGS analysis was not able to perfectly predict the fold differences in performance between individual variants. AAV 7m8, for example, was only slightly better at transducing fibroblasts based on GFP expression (73% vs. 64% at MOT 1,000, Fig. 3B), while the relative difference in RNA signal was more substantial (63% vs. 21%, Fig. 2D).
These observed differences between NGS and GFP readouts could be the result of a relatively long half-life of GFP protein than mRNA, and could indicate that RNA results may be a more sensitive method to assess differences between AAV variants. Furthermore, NGS RNA/cDNA analysis requires an amplification step that may artificially exacerbate subtle differences between variants. Studies performed by Marsic et al. suggest that the PCR bias should not have affected the NGS readout; however, the study analyzed vectors at the DNA level and did not include barcoded RNA sequences, thereby omitting the potential effect of signal amplification resulting from mRNA transcription.15
The iPSC line KUGC_2 showed similar dose-dependent toxicity when transduced with the AAV Testing Kit and individual top variants selected based on the NGS RNA results (Fig. 2D) where AAV 7m8 > AAV LK03 > AAV DJ > AAV 3B > AAV6. No viable cells could be recovered after transduction with AAV 7m8 at MOT of 10,000 and 1,000 vgc/cell, which made it impossible to validate the RNA-NGS results. It may be possible that AAV 7m8 transduced the iPSCs with very high efficiency, leading to cell death as suggested by Brown et al.20 In fact, from the five variants tested in individual transductions, only AAV LK03 and AAV3B did not cause significant toxicity and allowed us to obtain reliable GFP results (Fig. 3B). As AAV LK03 is largely homologous to AAV3B, our data are in agreement with published work, where AAV3B has previously been shown to transduce iPSCs more efficiently than other AAVs.52
It is possible that the observed toxicity was enhanced by the single-stranded AAV genome used in our experiments, culture conditions, transduction protocol, and/or vector preparation, all of which have been suggested as potential sources of AAV-induced toxicity in iPSCs.20 Interestingly, AAV DJ and AAV6, which accounted for 14% and 6% of total NGS RNA reads, respectively, and caused only moderate toxicity in individual transductions, did not produce detectable GFP signal (Fig. 3B). While the specific explanation for the lack of GFP expression is currently unknown and is being investigated, our current hypothesis is that in the context of transgenes delivered by AAV6 and AAV DJ the expression in iPSC cells may be inhibited at the translation step, and not at the ssDNA to dsDNA conversion or DNA to mRNA transcription.
Of note, AAV4, which in T cells contributed to 49% of the DNA NGS reads (Fig. 2C) but only ∼1% of the RNA reads (Fig. 2D), led to detectable GFP signal only in 4% of T cells (Fig. 3B). This highlights an additional feature of the DNA/RNA-NGS readout—the power to identify capsids that interact with the target cells but are not able to drive functional transduction, a feature that may be of interest for specific applications (see Conclusions section).
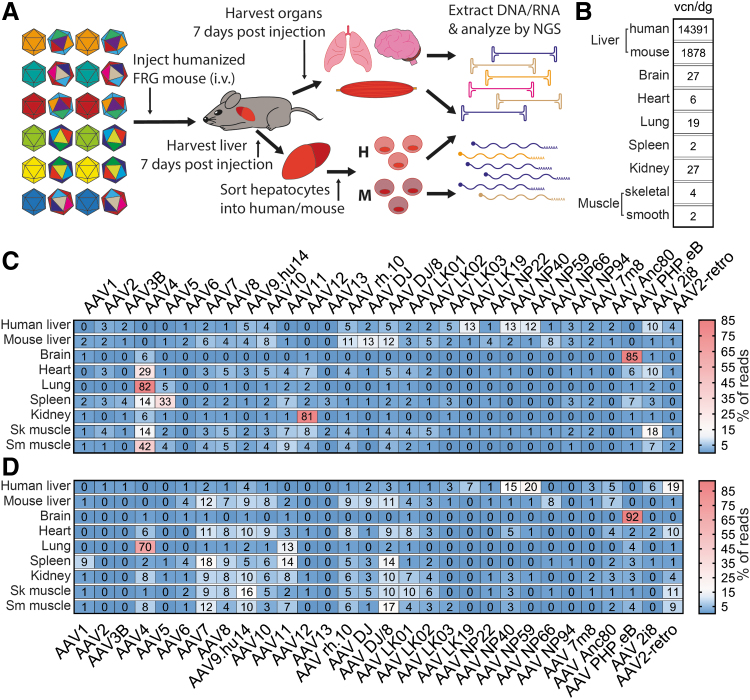
In vivo screening of the AAV Testing Kit
We next set out to test our AAV Testing Kit in vivo. To do so we intravenously injected 1 × 1011 vgc of the AAV Testing Kit into a naïve male Fah−/−/Rag2−/−/Il2rg−/− (FRG) mouse.21 This total vector dose corresponded to an individual dose of 3.3 × 109 vgc per variant per mouse. The mouse was euthanized 7 days postinjection, and the indicated organs and tissues (Fig. 4) were harvested for NGS analysis. The liver was perfused (see Materials and Methods section) to harvest hepatocytes and exclude extracellular matrix. After DNA and RNA extraction, RNA was used for cDNA synthesis and DNA was used for average vector copy determination. DNA and cDNA were used for NGS analysis following the same general protocol as in the context of cell lines and primary cells (Fig. 4A).
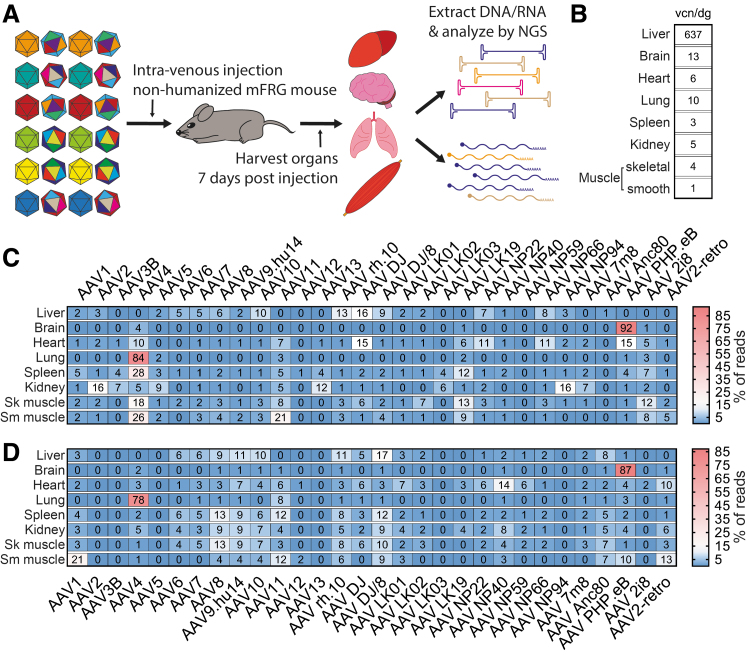
Figure 4.
Experimental setup and in vivo results from naive FRG mouse. (A) Schematic representation of the experimental setup. (B) Results of AAV vector copy quantification in selected organs. The values are given as vcn/dg. (C) Results of NGS-based quantification of vector-encoded DNA in selected tissues. Values are given as percentage of total reads for each organ. (D) Results of NGS analysis of vector function based on quantification of vector-encoded transgene mRNA. Values are given as percentage of total reads for each cell type. The values presented in panels (C, D) were rounded to the nearest whole number. Sk, skeletal; Sm, smooth.
As anticipated based on the known natural tropism of AAV vectors for the liver,10,22 the analysis of vector copy numbers per diploid genome (normalized to the mouse albumin gene) revealed higher average AAV vector copy numbers in mouse hepatocytes compared with the other organs tested (Fig. 4B).
The most abundant AAV in the liver based on DNA-NGS analysis was AAV DJ (16% of DNA reads, 102.8 vcn/dg), a bioengineered variant known for its murine hepatic tropism.8 The hepatocytes were also efficiently transduced by other well-characterized variants with broad tropism, such as AAV6, AAV7, AAV8, AAV10, AAV rh.10, and AAV DJ/8 (Fig. 4C).8,10,53 The brain yielded almost opposite results to those of the liver, due to the limited number of AAVs able to cross the blood–brain barrier. The AAV9-based bioengineered AAV PHP.eB was the most abundant variant in the brain (92% of DNA reads, 11.6 vcn/dg), which is in accordance with previous reports.39 The cardiac muscle was efficiently targeted by AAV DJ and AAV PHP.eB as well as few other variants, but interestingly AAV9, which was shown to efficiently transduce cardiomyocytes after systemic administration,7 performed quite weakly in our experiment at the DNA level.
The lungs showed a very restricted penetrance with AAV4, accounting for 84% of DNA reads (8.4 vcn/dg) in agreement with published data.7 AAV4 was also the most successful in the spleen (28% of DNA reads, 0.7 vcn/dg), skeletal, and smooth muscles (18% of DNA reads, 0.8 vcn/dg and 26% of DNA reads, 0.2 vcn/dg, respectively). Finally, the kidney was most efficiently transduced by AAV2 (16% of DNA reads, 0.8 vcn/dg). As previously reported, with the exception of the brain and the lung, most murine organs were physically transduced by a variety of AAV variants, especially AAV7, AAV8, AAV9, AAV DJ, AAV DJ/8, and AAV rh.10, although individual reports differed in regard to the exact performance of individual variants depending on the detection method and the dose.7,8,53
Similarly to what was observed in vitro, the AAVs most effective at physical transduction and DNA reads did not necessarily perform with the same efficiency at the RNA (expression) level (Fig. 4D). The highest relative functional transduction of the murine liver was achieved with AAV DJ/8 (17% of RNA reads), while AAV8, which is well known for its superior transduction of murine hepatocytes,10 was the fifth top AAV variant. The heart was most efficiently transduced by AAV NP40 (14% of RNA reads), which was unexpected given the fact that AAV NP40 was selected for transduction of human hepatocytes in vivo.32 The spleen, kidney, and skeletal muscle were most efficiently transduced by AAV8 (13% of RNA reads), AAV9 (9% of RNA reads), and AAV8 (13% of RNA reads), respectively (Fig. 4D). These organs also showed broad penetrance, and we did not detect a single variant that would show significantly higher tropism compared with other AAVs tested. Considering overall low average vcn/dg in those tissues, it appears that while several AAV can enter and express the transgene in those organs, the overall accessibility to AAV vectors was low under our experimental setup (Fig. 4B). The transgene expression in the smooth muscle was mostly driven by AAV1 (21% of RNA reads), but this tissue had also the lowest average vcn/dg of all the targets tested, indicating that it is quite resistant to AAV transduction (Fig. 4B, D). Overall, in organs other than the liver, brain, and lungs, we observed that AAV4 and AAV5 were able to physically transduce (DNA reads) the tissues with reasonably high efficiency, but in most cases the AAV variants failed to express their transgene (RNA reads), a similar trend to the one observed in ex vivo primary cells and iPSCs (compare Figs. 2C, D and 4C, D).
Finally, the brain and the lungs were the only organs for which the same AAV was responsible for the superior physical (DNA reads) and functional (RNA reads) transduction: AAV PHP.eB in case of the brain (87% of RNA reads) and AAV4 in case of the lungs (78% of RNA reads). While it is known that the brain is protected by the blood–brain barrier,54 the data suggest that a barrier may also protect the lungs from transduction after systemic delivery. To this end, AAV4 is the only AAV variant included in our screen that is known to enter cells independently of the recently described AAV receptor (AAVR).55,56 Studies show that AAVR is only weakly expressed on airway epithelial cells, and the expression is limited to the basolateral side of the epithelium.57 This specific and restricted AAVR expression pattern could thus impose a strong barrier for most AAVs, which rely on this receptor for cellular entry. It would be interesting, and remains to be tested, if similar results would be observed after intranasal AAV application to the lungs.
In vivo testing in a xenograft model of the human liver
In a final experiment, we set out to test the applicability of our AAV Testing Kit in a biologically predictive xenograft model of the human liver—the hFRG mouse.21 The hFRG mouse was injected with the same total vector dose as the naïve mouse discussed above. However, in the case of the hFRG mouse we were able to sort human and mouse GFP-positive hepatocytes after liver perfusion (Fig. 5A). Before the sort, the human hepatocytes were 90% GFP positive, whereas the murine hepatocytes were 70% positive (data not shown). This extra step, which allowed us to achieve >95% purity for each hepatocyte population, partly accounts for the high vcn observed for mouse (1,878 vcn/dg) and human cells (14,391 vcn/dg) (Fig. 5B). Apart from the kidney (27 vcn/dg in hFRG vs. 5 vcn/dg in the naïve FRG), most other organs have very similar average vector copy numbers in both humanized and naïve mice (compare Figs. 4B and 5B).
Figure 5.
Experimental setup and results of in vivo transductions in xenograft model of the human liver. (A) Schematic representation of the AAV Testing Kit study performed in hFRG animal. (B) Results of AAV vector copy number quantification in selected organs. The values are given as vcn/dg. The quantity of endogenous genome was obtained by measuring the copy number of human genomic albumin locus in genomic DNA purified from human hepatocytes, while murine albumin genomic locus was targeted in mouse hepatocytes and other organs. (C) Results of NGS-based quantification of vector-encoded DNA in selected tissues. Values are given as percentage of total reads for each organ. (D) Results of NGS analysis of vector function based on quantification of vector-encoded transgene mRNA. Values are given as percentage of total reads for each cell type. The values presented in panels (C, D) were rounded to the nearest whole number. i.v., intravenous.
The most efficient physical transduction of human hepatocytes was achieved with AAV NP40 (13% of DNA reads, 1917.1 vcn/dg), which is consistent with the fact that as mentioned, this capsid was selected by Directed Evolution on primary human hepatocytes in hFRG xenograft animals.32 As seen also in the naïve FRG mouse, AAV DJ was most abundant in murine hepatocytes (13% of DNA reads, 237.6 vcn/dg) (Fig. 5C). Other AAV variants that remained the same between naïve FRG and hFRG were PHP.eB for the brain (85% of DNA reads, 23.3 vcn/dg), as well as AAV4 in the lungs (82%, 16 vcn/dg) and smooth muscle (42% of DNA reads, 1 vcn/dg) (Fig. 5C). In contrast to naïve FRG, in the hFRG the heart was most efficiently transduced by AAV4 (29% of DNA reads, 1.8 vcn/dg), the spleen by AAV5 (33% of DNA reads, 0.8 vcn/dg), the kidney by AAV12 (81% of DNA reads, 21.4 vcn/dg), and the skeletal muscle was most efficiently transduced by AAV 2i8 (18% of DNA reads, 0.7 vcn/dg), in agreement with previously published data41 (Fig. 5C).
There are a number of possible reasons for these observed differences. First of all, the human hepatocytes can efficiently take up a range of AAV variants as evident by very high average vector copy number (Fig. 5B), which would affect the overall availability of vectors to transduce other targets, most notably, the availability of free AAV NP40 to transduce the heart. Also, while the results presented for the naïve FRG mouse (Fig. 4) were obtained using a male mouse, only the female FRG mice undergo efficient repopulation with human hepatocytes. Thus, the hFRG mouse used in the study was a female, and it has been previously reported that the transduction pattern in mice is sexually dimorphic.58 Finally, the presence of human hepatocytes affects the composition of proteins and other macro/micromolecules the vectors interact with in the blood, which could affect their function and thus tropism.59,60
At the level of functional transduction (RNA/cDNA) of the organs and tissues analyzed, we observed a very similar trend to what was observed for the naïve FRG mouse. Specifically, the highest vector contribution at the DNA level did not correspond to the highest expression in most cases, except for the brain (AAV PHP.eB; 92% of RNA reads) and lungs (AAV4; 70% of RNA reads) (Fig. 5D). Human hepatocytes were most efficiently functionally transduced by AAV NP59, a bioengineered variant designed to target human hepatocytes in hFRGs (Fig. 5D).32 The mouse hepatocytes in the hFRG were most efficiently functionally transduced by AAV7 (12% of RNA reads), which differs from what was observed in the naïve mouse. However, AAV DJ/8, which was the most efficient at functional transduction in naïve FRG, was the second most efficient variant in mouse cells in hFRG (Fig. 5D), supporting the known fact that many AAVs can efficiently enter murine hepatocytes.10 In contrast to data obtained in the naïve FRG moue, the heart was no longer most efficiently transduced by AAV NP40, most likely because AAV NP40 was now very successful at targeting its designated target, the human liver at the DNA and RNA levels (Fig. 5D). Instead, the heart was now functionally transduced by several natural serotypes, with AAV7 contributing most of the RNA reads (11%) (Fig. 5D).
AAV7 was also the highest expressing variant in the spleen in hFRG (18% of RNA reads), instead of AAV8 observed in naïve FRG, but AAV11 and AAVDJ/8 were the second best variants in both animals. The kidney was functionally most efficiently transduced by AAV9 (10% of RNA reads) and not by AAV12, which contributed the vast majority of vector copies in the DNA reads, indicating that AAV12 is not efficient at completing the intracellular journey, which culminates in RNA expression. AAV9 functionally transduced the skeletal muscle of the hFRG mouse most efficiently (17% of RNA reads), consistent with data from the naïve animal. Finally, the smooth muscle was not highly transduced by AAV1 as seen in the non-hFRG mouse, but rather by AAV DJ/8 (17% of RNA reads). Similar to data from the previously discussed in vivo study, the smooth muscle tissue in hFRG had a very low overall vector copy number. The low average vector copy number in this organ and several other organs is most likely due to the low dose of individual vector (3.3 × 109 vgc/variant) in the injection mix and may contribute to experimental variations.
Conclusions
The studies presented herein validate the NGS-based method of vector-encoded barcod analysis at the DNA and RNA/cDNA levels as a powerful and efficient approach to evaluate the performance of a large cohort of AAV vectors in vitro, ex vivo, and in vivo. The described method allows for robust and cost-efficient high-throughput analysis of vector performance at the level of physical and functional transduction, further increasing the impact of this method. In addition, recently published data show that, at least for a single capsid, the DNA/RNA BC-NGS approach is highly scalable between a large range of vector doses in vivo.17 Using this approach, as presented herein, we were able to identify AAV 7m8 as a powerful capsid for ex vivo applications in primary human fibroblasts and in vitro application, as well as verify known highly efficient capsids for human hepatocytes (AAV NP59, NP40), the mouse brain (AAV PHP.eB), and lungs (AAV4).7,32,39 The results obtained were overall well aligned with published reports, providing an important level of validation that increases the impact of the data presented.
An interesting trend that was observed in primary human cells, iPSCs, and several mouse organs was that a few AAV variants, such as AAV4 (multiple cell types and tissues, except the lungs), AAV5, and AAV12 (in a few targets, but especially in the case of kidney in the hFRG mouse), were very efficient at cell entry, but were unable to express the transgene at the RNA level. While we do not understand the mechanism behind these observations, it is possible that these naturally occurring serotypes evolved to enter cells independently but require factors from helper viruses such as human adenovirus 5 to be able to express their transgene. This possibility and other intracellular trafficking hurdles have been previously discussed elsewhere.61
Despite the fact that the NGS RNA data did not always correlate perfectly with the GFP results for individual AAV transductions, the AAVs identified as top performers in the in vitro and ex vivo NGS screen generally were very efficient at the GFP level, and none of the identified variants was shown not to functionally transduce target cells per GFP expression readout. This fact adds a future directive toward the interpretation of NGS screen results, as the experimental settings and the intended application may influence the choice of the AAV variant. For example, functional transduction data may not be the best at predicting which AAV to use to deliver repair template for gene editing, especially if homology-directed repair (HDR) is considered where single-stranded DNA templates have been shown to yield increased rates of HDR.62,63 Thus, while vector screens can also be performed in a 96-well format using highly parallel individual transductions and subsequent detection of the reporter transgene expression, NGS screens can be a powerful method to predict which subset of AAVs should be considered for in-depth validation for such specific applications.
Importantly, our data revealed that the bioengineered capsids outperform the natural serotypes at the level of RNA expression for targeting primary human cells, iPSCs, and human hepatocytes in vivo, supporting further efforts to develop bioengineered variants to serve as second-generation gene therapy vectors with the potential to replace the natural variants currently in clinical applications.
Supplementary Material
Acknowledgments
We thank CMRI Vector and Genome Engineering Facility for help in vector preparation and the Stem Cell & Organoid Facility for maintenance of the iPSC line.
Authors' Contributions
A.W., M.C.-C., A.R., I.E.A., and L.L. designed the experiments. A.W., M.C.-C., A.R., G.B., R.N., E.Z., M.D., B.H.N., M.K., R.F.A., P.K., K.H., W.G., A.G.-C., and L.L. generated reagents, protocols, performed experiments, and analyzed data. A.W. and L.L. wrote the article and generated the figures. All authors reviewed, edited, and commented on the article.
Author Disclosure
L.L. and A.J.T. have commercial affiliations. L.L. and I.A.E. have consulted on technologies discussed in this article. L.L. and I.A.E. have stock and/or equity in companies with technologies broadly related to this article. L.L. is a coinventor of, and receives licensing royalties from, four AAV variants used in the study. All other authors declare that no competing financial interests exist.
Funding Information
This work was supported by project grants from the Australian National Health and Medical Research Council (NHMRC) to Leszek Lisowski and Ian E. Alexander (APP1108311, APP1156431 and APP1161583) and Paediatrio Paediatric Precision Medicine Program to Leszek Lisowski and Anai Gonzalez-Cordero (PPM1 K5116/RD274). Leszek Lisowski was also supported by research grants from the Department of Science and Higher Education of Ministry of National Defense, Republic of Poland (“Kościuszko” k/10/8047/DNiSW/T–WIHE/3) and from the National Science Centre, Republic of Poland (OPUS 13) (UMO-2017/25/B/NZ1/02790). The work of Ian E. Alexander was also supported by an Australian Research Council (ARC) Discovery Project (DP150101253). Adrian J. Thrasher was supported by the Wellcome Trust (104807/Z/14/Z). The studies on human cells performed by Giorgia Santilli and Adrian J. Thrasher were also supported by the NIHR Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London.
Supplementary Material
References
- 1. Bennett J. Taking stock of retinal gene therapy: looking back and moving forward. Mol Ther 2017;25:1076–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. George LA, Sullivan SK, Giermasz A, et al. Hemophilia B gene therapy with a high-specific-activity factor IX variant. N Engl J Med 2017;377:2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mendell JR, Al-Zaidy S, Shell R, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med 2017;377:1713–1722 [DOI] [PubMed] [Google Scholar]
- 4. Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet 2014;15:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 2019;18:358–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Büning H, Srivastava A. Capsid modifications for targeting and improving the efficacy of AAV vectors. Mol Ther Methods Clin Dev 2019;12:248–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zincarelli C, Soltys S, Rengo G, et al. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 2008;16:1073–1080 [DOI] [PubMed] [Google Scholar]
- 8. Grimm D, Lee JS, Wang L, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol 2008;82:5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Limberis MP, Vandenberghe LH, Zhang L, et al. Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol Ther 2009;17:294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang L, Wang H, Bell P, et al. Systematic evaluation of AAV Vectors For Liver Directed Gene Transfer In Murine Models. Mol Ther 2010;18:118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ellis BL, Hirsch ML, Barker JC, et al. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1–9) and one engineered adeno-associated virus serotype. Virol J 2013;10:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song L, Kauss MA, Kopin E, et al. Optimizing the transduction efficiency of capsid-modified AAV6 serotype vectors in primary human hematopoietic stem cells in vitro and in a xenograft mouse model in vivo. Cytotherapy 2013;15:986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lang JF, Toulmin SA, Brida KL, et al. Standard screening methods underreport AAV-mediated transduction and gene editing. Nat Commun 2019;10:3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adachi K, Enoki T, Kawano Y, et al. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat Commun 2014;5:3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marsic D, Méndez-Gómez HR, Zolotukhin S. High-accuracy biodistribution analysis of adeno-associated virus variants by double barcode sequencing. Mol Ther Methods Clin Dev 2015;2:15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weinmann J, Grimm D. Next-generation AAV vectors for clinical use: an ever-accelerating race. Virus Genes 2017;53:707–713 [DOI] [PubMed] [Google Scholar]
- 17. Xu M, Li J, Xie J, et al. High-throughput quantification of in vivo adeno-associated virus transduction with barcoded non-coding RNAs. Hum Gene Ther 2019;30:946–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strobel B, Miller FD, Rist W, et al. Comparative analysis of cesium chloride- and iodixanol-based purification of recombinant adeno-associated viral vectors for preclinical applications. Hum Gene Ther Method 2015;26:147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cabanes-Creus M, Ginn SL, Amaya AK, et al. Codon-optimization of wild-type adeno-associated virus capsid sequences enhances DNA family shuffling while conserving functionality. Mol Ther Methods Clin Dev 2019;12:71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brown N, Song L, Kollu NR, et al. Adeno-associated virus vectors and stem cells: friends or foes? Hum Gene Ther 2017;28:450–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Azuma H, Paulk N, Ranade A, et al. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat Biotechnol 2007;25:903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Logan GJ, Dane AP, Hallwirth CV, et al. Identification of liver-specific enhancer–promoter activity in the 3′ untranslated region of the wild-type AAV2 genome. Nat Genet 2017;49:1267. [DOI] [PubMed] [Google Scholar]
- 23. Bushnell B, Rood J, Singer E. BBMerge—accurate paired shotgun read merging via overlap. PLoS One 2017;12:e0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Atchison RW, Casto BC, Hammon WM. Adenovirus-associated defective virus particles. Science 1965;149:754. [DOI] [PubMed] [Google Scholar]
- 25. Hoggan MD, Blacklow NR, Rowe WP. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc Natl Acad Sci U S A 1966;55:1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lisowski L, Dane AP, Chu K, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 2014;506:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rutledge EA, Halbert CL, Russell DW. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J Virol 1998;72:309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parks WP, Melnick JL, Rongey R, et al. Physical assay and growth cycle studies of a defective adeno-satellite virus. J Virol 1967;1:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bantel-Schaal U, Zur Hausen H. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology 1984;134:52–63 [DOI] [PubMed] [Google Scholar]
- 30. Paulk NK, Pekrun K, Charville GW, et al. Bioengineered viral platform for intramuscular passive vaccine delivery to human skeletal muscle. Mol Ther Methods Clin Dev 2018;10:144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao G-P, Alvira MR, Wang L, et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A 2002;99:11854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paulk NK, Pekrun K, Zhu E, et al. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol Ther 2018;26:289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao G, Vandenberghe LH, Alvira MR, et al. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol 2004;78:6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mori S, Wang L, Takeuchi T, et al. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology 2004;330:375–383 [DOI] [PubMed] [Google Scholar]
- 35. Dalkara D, Byrne LC, Klimczak RR, et al. In vivo–directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci Transl Med 2013;5:189ra176. [DOI] [PubMed] [Google Scholar]
- 36. Schmidt M, Voutetakis A, Afione S, et al. Adeno-associated virus type 12 (AAV12): a novel AAV serotype with sialic acid- and heparan sulfate proteoglycan-independent transduction activity. J Virol 2008;82:1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Landegger LD, Pan B, Askew C, et al. A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat Biotechnol 2017;35:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmidt M, Govindasamy L, Afione S, et al. Molecular characterization of the heparin-dependent transduction domain on the capsid of a novel adeno-associated virus isolate, AAV(VR-942). J Virol 2008;82:8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan KY, Jang MJ, Yoo BB, et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci 2017;20:1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao G, Alvira MR, Somanathan S, et al. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci U S A 2003;100:6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Asokan A, Conway JC, Phillips JL, et al. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol 2010;28:79–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tervo D, Hwang B-Y, Viswanathan S, et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 2016;92:372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eyquem J, Mansilla-Soto J, Giavridis T, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 2017;543:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sebastiano V, Zhen HH, Haddad B, et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med 2014;6:264ra163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Landry JJM, Pyl PT, Rausch T, et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 2013;3:1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiao P-J, Samulski RJ. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J Virol 2012;86:10462–10473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomas CE, Storm TA, Huang Z, et al. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J Virol 2004;78:3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rossi A, Dupaty L, Aillot L, et al. Vector uncoating limits adeno-associated viral vector-mediated transduction of human dendritic cells and vector immunogenicity. Sci Rep 2019;9:3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salganik M, Aydemir F, Nam H-J, et al. Adeno-associated virus capsid proteins may play a role in transcription and second-strand synthesis of recombinant genomes. J Virol 2014;88:1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang J, DeClercq JJ, Hayward SB, et al. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res 2015;44:e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weltner J, Anisimov A, Alitalo K, et al. Induced pluripotent stem cell clones reprogrammed via recombinant adeno-associated virus-mediated transduction contain integrated vector sequences. J Virol 2012;86:4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duong TT, Lim J, Vasireddy V, et al. Comparative AAV-eGFP transgene expression using vector serotypes 1–9, 7m8, and 8b in human pluripotent stem cells, RPEs, and human and rat cortical neurons. Stem Cells Int 2019;2019:7281912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu C, Busuttil RW, Lipshutz GS. RH10 provides superior transgene expression in mice when compared with natural AAV serotypes for neonatal gene therapy. J Gene Med 2010;12:766–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lykken EA, Shyng C, Edwards RJ, et al. Recent progress and considerations for AAV gene therapies targeting the central nervous system. J Neurodev Disord 2018;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pillay S, Meyer NL, Puschnik AS, et al. An essential receptor for adeno-associated virus infection. Nature 2016;530:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dudek AM, Pillay S, Puschnik AS, et al. An alternate route for adeno-associated virus (AAV) entry independent of AAV receptor. J Virol 2018;92:e02213–e02217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hamilton BA, Li X, Pezzulo AA, et al. Polarized AAVR expression determines infectivity by AAV gene therapy vectors. Gene Ther 2019;26:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dane A, cunningham S, Graf N, et al. Sexually dimorphic patterns of episomal rAAV genome persistence in the adult mouse liver and correlation with hepatocellular proliferation. Mol Ther 2009;17:1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang M, Sun J, Crosby A, et al. Direct interaction of human serum proteins with AAV virions to enhance AAV transduction: immediate impact on clinical applications. Gene Ther 2017;24:49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Denard J, Beley C, Kotin R, et al. Human galectin 3 binding protein interacts with recombinant adeno-associated virus type 6. J Virol 2012;86:6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nonnenmacher M, Weber T. Intracellular transport of recombinant adeno-associated virus vectors. Gene Ther 2012;19:649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miura H, Gurumurthy CB, Sato T, et al. CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Sci Rep 2015;5:12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoshimi K, Kunihiro Y, Kaneko T, et al. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun 2016;7:10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.